Abstract
Twenty P. aeruginosa isolates were collected from six cystic fibrosis (CF) patients, aged 27 to 33, in 1994 (9 isolates) and 1997 (11 isolates) at the CF Center, Copenhagen, Denmark, and were typed by pulse-field gel electrophoresis (PFGE) or ribotyping. Five of the patients had isolates with the same PFGE or ribotyping patterns in 1997 as in 1994, and ciprofloxacin had a two- to fourfold higher MIC for the isolates collected in 1997 than those from 1994. Genomic DNA was amplified for gyrA, parC, mexR, and nfxB by PCR and sequenced. Eleven isolates had mutations in gyrA, seven isolates had mutations at codon 83 (Thr to Ile), and four isolates had mutations at codon 87 (Asp to Asn or Tyr). Sixteen isolates had mutations in nfxB at codon 82 (Arg to Leu). Increased amounts of OprN were found in six isolates and OprJ in eight isolates as determined by immunoblotting. No isolates had mutations in parC or mexR. Six isolates had mutations in efflux pumps without gyrA mutations. The average number of mutations was higher in isolates from 1997 than in those from 1994. The results also suggested that efflux resistance mechanisms are more common in isolates from CF patients than in strains from urine and wounds from non-CF patients, in which mutations in gyrA and parC dominate (S. Jalal and B. Wretlind, Microb. Drug Resist. 4:257–261, 1998).
Fluoroquinolones (FQ) are the only available antibiotics for oral treatment of Pseudomonas aeruginosa infections in most countries. However, P. aeruginosa easily becomes resistant to these drugs, which severely limits their usefulness. The main mechanisms of resistance are mutations in the target genes, those encoding DNA gyrase (gyrA) and topoisomerase IV (parC) (3, 9), and mutations in regulatory genes for drug efflux pumps. Three different multidrug efflux-pump systems have been identified, MexAB-OprM, MexCD-OprJ, and MexEF-OprN, which are regulated by mexR (nalB), nfxB, and mexT (nfxC), respectively (4, 8, 17).
In this study, 20 quinolone-resistant P. aeruginosa isolates from cystic fibrosis (CF) lungs have been analyzed for gyrA, parC, nfxB, and mexR genes and the expression of membrane proteins mexT (nfxC)-related OprN and nfxB-related OprJ associated with FQ resistance. Our aim was to determine molecular mechanisms for quinolone resistance of repeated isolates of P. aeruginosa from CF patients.
MATERIALS AND METHODS
Bacterial strains.
Twenty P. aeruginosa isolates were collected from six CF patients (ages, 27 to 33) with chronic P. aeruginosa infections (duration, 18 to 26 years) in 1994 (9 isolates) and 1997 (11 isolates) at the CF Center, Copenhagen, Denmark, and were typed by pulsed-field gel electrophoresis (PFGE) (Table 1). However, four isolates (B1, B2, B3, and D1) could not be typed by PFGE, probably because of high DNase activities, and were subjected to ribotyping using Riboprinter (Qualicon, Wilmington, Del.). All patients had been exposed to repeated treatment periods with ciprofloxacin since 1986, and all of them had received at least 15 treatment courses (14 days/course) by 1994. The isolates were considered to be resistant to ciprofloxacin (MIC, ≥2 mg/liter) by the disk diffusion method (Rosco, Taastrup, Denmark).
TABLE 1.
Characteristics of gyrA and nfxB gene products and expression of OprN and OprJ in quinolone-resistant P. aeruginosa isolated from six CF patients in 1994 and 1997a
| Patient | Characteristics of 1994 isolate(s)
|
Characteristics of 1997 isolate(s)
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Isolate | Alginate | MIC (μg/ml) of:
|
Amino acid change
|
Isolate | Alginate | MIC (μg/ml) of:
|
Amino acid change
|
|||||||||
| Nor | Cip | GyrA | NfxB | OprN | OprJ | Nor | Cip | GyrA | NfxB | OprN | OprJ | |||||
| CF166 | A2 | NM | 16 | 4 | T83I | R82L | ||||||||||
| A1 | NM | 2 | 1 | R82L | A3 | M | 2 | 1 | R82L | |||||||
| CF222 | B1 | NM | 8 | 2 | T83I | R82L | B3 | NM | 16 | 8 | T83I | R82L | wk | wk | ||
| B2 | M | 2 | 2 | R82L | wk | wk | B4 | M | 2 | 0.5 | wk | |||||
| CF86 | C1 | NM | 4 | 2 | D87N | R82L | C3 | NM | 16 | 8 | T83I | R82L | wk | + | ||
| C2 | M | 4 | 2 | D87N | R82L | C4 | M | 8 | 2 | D87N | R82L | wk | ||||
| CF59 | D1 | NM | 8 | 2 | T83I | R82L | + | D2 | NM | 8 | 4 | D87Y | R82L | |||
| D3 | M | 2 | 0.5 | R82L | wk | |||||||||||
| CF21 | E1 | NM | 2 | 0.5 | E3 | NM | 16 | 4 | T83I | R82L | wk | |||||
| E2 | M | 2 | 1 | + | E4 | M | 2 | 1 | + | |||||||
| CF89 | F | NM | 2 | 0.5 | R82L | G | NM | 8 | 2 | T83I | R82L | + | ||||
| Control | PAO1 | NM | 0.25 | 0.12 | PAO1 | NM | 0.25 | 0.12 | ||||||||
Nor, norfloxacin; Cip, ciprofloxacin; +, strongly positive; wk, weak; NM, nonmucoid; M, mucoid; T, threonine; I, isoleucine; D, aspartate; N, asparagine; R, arginine; L, leucine; Y, tyrosine. Patient CF89 yielded different strains in 1994 and 1997, but the other five patients had the same strains, as determined by PFGE (patients CF86, CF59, and CF21) or ribotyping (patients CF222 and CF59). Strain PAO1 is quinolone sensitive.
Antibiotic sensitivity testing.
Each MIC was determined by an agar dilution method using Isosensitest agar medium (Oxoid, Basingstoke, United Kingdom). The size of the inoculum of bacterial cells was about 100,000 CFU/spot. The MIC was defined as the lowest concentration inhibiting visible cell growth, as evaluated after an 18-h incubation at 37°C.
PCR.
Chromosomal DNA from the isolates of P. aeruginosa was prepared with guanidium thiocyanate as described by Pitcher et al. (13) and subjected to PCR. A master mix was prepared on ice containing 1× UlTma buffer (Gene Amp; Perkin-Elmer Cetus), 200 μM concentrations of each deoxynucleoside triphosphate, 0.6 pmol of each primer (Scandinavian Gene Synthesis, Köping, Sweden) per μl, 1.5 mM MgCl2, 1.5 U of UlTma DNA polymerase per 50 μl, an enzyme with proofreading activity (Gene Amp; Perkin-Elmer Cetus), and ∼5.0 ng of genomic DNA from P. aeruginosa per μl. The master mix was aliquoted in volumes of 50 μl. The quinolone-resistance-determining region of gyrA and parC, a 385-bp region (441 bp) of mexR, and a 924-bp region including the whole nfxB gene (561 bp) were amplified by a single-step PCR. The same temperature profile was applied for all genes except for the annealing temperatures. The annealing temperatures were 59°C for gyrA, 60°C for parC and mexR, and 64°C for nfxB gene amplification. The mix was denatured at 97°C for 10 min and cycled at 94°C for 90 s following the respective annealing temperature for 30 s and then at 72°C for 2 min. The cycles were repeated for 30 cycles, with a final 7-min extension at 72°C. The DNA amplification was done in a DNA thermal cycler, GeneAmp PCR System 2400 (Perkin-Elmer). Five microliters of PCR product was analyzed by electrophoresis on 1% (wt/vol) agarose gel, stained with ethidium bromide, and visualized under UV light. The corresponding primer pairs were gyrA-3 and -4, parC-X and -Y, mexR-U and -L, and nfxB-1 and -2 (7), which revealed PCR fragments of 360, 186, 385, and 924 bp, respectively.
Nucleotide sequencing.
Primers and free nucleotides from the PCR products were removed by the QIAquick-spin PCR purification kit (Qiagen Inc., Chatsworth, Calif.). The purified PCR products were directly processed for DNA sequencing by Big-Dye terminator using capillary electrophoresis technology in the ABI 310 Genetic Analyzer (Perkin-Elmer). Both strands of each PCR-amplified fragment were sequenced twice, but no errors in PCR amplification or sequencing were detected. Nucleotide and deduced amino acid sequences were analyzed by Macintosh DNA programs FACTURA and EditView; ClustalW Interactive Multiple Sequence alignment at the European Bioinformatics Institute (http://www2.ebi.ac.uk), Cambridge, United Kingdom; and ExPASy Molecular Biology Server at Swiss Institute of Bioinformatics, Geneva, Switzerland (http://www.expasy.ch). The sequences of the quinolone-resistance-determining regions of gyrA and parC, the amplified region of mexR, and the whole nfxB gene were compared with those from the corresponding quinolone-susceptible strain and the sequences present in the GenBank databases. The results are presented in Table 1.
GenBank accession numbers.
GenBank accession numbers for the nucleotide sequences of gyrA, parC, mexR, and nfxB genes are L29417, D89652, U23763, and X65646, respectively.
Isolation of cell envelope, SDS-PAGE, and immunoblot assay of OprN and OprJ.
Isolation of cell envelope proteins, fractionation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), electrophoretic transfer to nitrocellulose paper, and binding of the primary antibody (anti-OprN antibody TN005 [8] and anti-OprJ antibody HJ001 [5]) were done as described by Poole et al. (14).
PFGE.
Genome fingerprinting was done by PFGE as previously described (12). In brief, each of the 20 P. aeruginosa isolates was grown in trypsin broth until it reached the exponential phase. These bacterial suspensions were used in preparation of agar blocks (type VII; Sigma), which were incubated for 15 h at 56°C in buffer containing proteinase K (Boehringer Mannheim, Mannheim, Germany), lauroyl sarcosine, and EDTA. Two halves of agarose blocks were incubated for 15 h at 37°C in buffer containing 10 U of the restriction end nucleases XbaI and DraI (Boehringer Mannheim), dithiothreitol, and bovine serum albumin. The restriction fragments were separated in 1% agarose (SeaKem GTG; FMC, Rockland, Maine) by field inversion gel electrophoresis (Bio-Rad). The field strength was 5.6 V/cm in recirculating Tris-borate-EDTA buffer, pH 8.2. The forward-to-reverse ratio was 3:1, and the pulses were increased linearly from 3 to 15 s. The gel was stained with ethidium bromide and photographed with a Polaroid camera. The relatedness of the P. aeruginosa isolates was determined by analyzing the band pattern of the restriction digests by side-to-side visual comparison of the bands produced on the gel. Four P. aeruginosa isolates showed only a smear of DNA on the agarose gel and were typed by ribotyping.
Ribotyping.
Ribotyping (riboprinting) was performed with the Riboprinter, as recommended by the manufacturer (Qualicon). In brief, single colonies from a 24-h culture at 37°C on a 5% blood agar plate were suspended in a sample buffer. Mucoid isolates were grown in brain heart infusion broth for 6 h, and cells were harvested by centrifugation and were suspended in sample buffer. Samples were heated until 80°C for 15 min. After addition of lytic enzymes, samples were transferred to the Riboprinter. Further analysis, including EcoRI restriction of DNA, was carried out automatically. The Riboprint profiles were aligned according to the position of the molecular-size standard and compared with patterns obtained previously, including a database consisting of 750 validated riboprints supplied from the manufacturer. The riboprint profiles were analyzed visually (15).
Isolates were considered to be the same strain if their PFGE or ribotyping patterns were identical, closely related if the patterns differed by two or three bands, possibly related if the patterns differed by four to six bands, and distinct if the patterns differed by seven or more bands.
RESULTS AND DISCUSSION
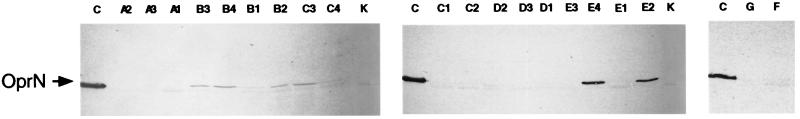
In this study, we analyzed quinolone resistance genes in 20 P. aeruginosa isolates collected in 1994 and 1997 from six CF patients with long-time exposure to ciprofloxacin treatment. Five patients had the same type of strain in their lungs in 1994 and 1997, as judged by PFGE (patients CF86, CF59, and CF21) or ribotyping (patients CF222 and CF59). It has been shown previously that CF patients harbor the same strain over several years (12). However, PFGE analysis showed that patient CF89 had different strains in 1994 and 1997. For each patient, one or two isolates from 1994 and 1997 were compared for susceptibility to FQs, for production of OprN (Fig. 1) and OprJ, and for DNA sequences of the gyrA, parC, mexR, and nfxB genes (Table 1).
FIG. 1.
SDS-PAGE of cell envelope proteins prepared from P. aeruginosa CF isolates. C, control strain PAO4222nfxC; K, control strain PAO1. CF isolate designations are marked on the top of each lane. Proteins were electroblotted to nitrocellulose membranes and probed with anti-OprN antibody (TN005). Samples (20 μg of total protein) were solubilized in sample buffer at 95°C for 5 min prior to electrophoresis.
The median MIC of norfloxacin was 2 to 4 mg/liter for the nine isolates from 1994 and 4 to 8 mg/liter for the 11 isolates from 1997. Mutations were found in all but one isolate, in nfxB (n = 16) and gyrA (n = 11), but in neither parC nor mexR. The mean number of mutations was 1.2 in nfxB and gyrA and 1.4 in parC and mexR. Moreover, four of nine isolates of the former pair and 10 of 12 isolates of the latter pair also produced a detectable level of OprN or OprJ proteins related to the genes mexT (nfxC) and nfxB, respectively.
Mutations in the target enzymes, DNA gyrase (gyrA) and topoisomerase IV (parC), and reduced intracellular accumulation of the drug either by decreased uptake and/or by increased efflux have been reported to be the main mechanisms of resistance to FQs in several bacterial species (1, 7, 10). A high level of resistance is commonly associated with gyrA and parC or two or more point mutations in the same gene or mutations involving more than one gene.
All six patients had been exposed to at least 15 treatment courses (14 days/course) of ciprofloxacin by 1994 and additional courses during the following years. Isolates collected in 1997 had two- to fourfold higher MICs of quinolones than those collected in 1994 and had a higher number of mutations. However, none of the isolates had extreme levels of ciprofloxacin resistance in spite of repeated exposure to the drug.
The isolates from 1997 from patients CF166 and CF21 harbored one (gyrA, Thr 83 to Ile) and two (gyrA, Thr 83 to Ile, and nfxB, Arg 82 to Leu) more mutations, respectively, than the isolates from 1994, which had the same DNA fingerprinting patterns but lower FQ MICs. All isolates from patients CF86 and CF222 exhibited detectable levels of OprJ and/or OprN in 1997, with a similar pattern of mutations in gyrA and nfxB as in 1994. Patient CF59 had one isolate from 1997 with a different mutation in gyrA (Asp 87 to Tyr) compared to the isolate from 1994 (Thr 83 to Ile). This discrepancy may be due to selection of different clones with the same fingerprinting pattern from a mixed culture in the highly viscous CF sputum. One isolate from patient CF59 with the Arg 82 to Leu nfxB mutation expressed OprJ in 1994, but not in 1997, possibly because different clones were isolated or because the immunoblot method applied may not be 100% sensitive.
Results of the present study showed that 11 of 20 isolates had mutations in gyrA, and 19 isolates had mutations affecting efflux systems. Isolates with MICs of norfloxacin of ≥8 mg/liter had two or more mutations. We did not find any mutations in one low-level resistant isolate (E1), and we found no mutations in parC or mexR in any of the isolates. Our previous study showed that most clinical P. aeruginosa strains with high-level FQ MICs (MIC, ≥32 mg/liter) had additional parC and/or mexR mutations (7).
Sixteen isolates had a mutation in nfxB in position 82 (Arg to Leu), but eight of them did not express OprJ. The role of this mutation in quinolone resistance is not clear; however, it is likely that the mutation is advantageous to the bacterium, since it was found in unrelated strains in all patients and other mutations in nfxB confer resistance to FQs (4, 7). None of the isolates in this study had a very high level of FQ resistance compared to strains from urine or wounds (7). This is probably due to lower concentrations of antibiotic in the viscous secretions of CF lungs (2) and explains why we found comparatively few mutations in the target genes for FQ. Mutations in parC are usually associated with higher levels of FQ resistance than in the CF isolates. Other antibiotics, such as beta-lactams, are able to cause mutations in efflux pumps, which may influence the pattern of mutations in the different regulatory genes (11, 18). It has also been shown that the proportion of mutations in gyrA, nfxB, and mexR in resistant strains is dependent on the conditions during the selection procedure (6, 16).
In conclusion, alterations in two efflux systems, MexCD-OprJ and MexEF-OprN, were the predominant mechanisms of FQ resistance in P. aeruginosa strains from the lungs of CF patients. This is in contrast to strains isolated from urine and wounds, where mutations in gyrA and parC were the most common finding (7). Five of the six patients had isolates with the same PFGE or ribotyping pattern in 1997 as in 1994, but the number of mutations and MIC levels were higher in 1997, probably due to treatment with ciprofloxacin and other antibiotics during these three years.
ACKNOWLEDGMENT
This work was supported by a grant from the Scandinavian Society for Antimicrobial Chemotherapy.
REFERENCES
- 1.Cambau E, Perani E, Dib C, Petinon C, Trias J, Jarlier V. Role of mutations in DNA gyrase genes in ciprofloxacin resistance of Pseudomonas aeruginosa susceptible or resistant to imipenem. Antimicrob Agents Chemother. 1995;39:2248–2252. doi: 10.1128/aac.39.10.2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Costerton J W, Lam J, Lam K, Chan R. The role of the microcolony mode of growth in the pathogenesis of Pseudomonas aeruginosa infections. Rev Infect Dis. 1983;5(Suppl. 5):867–873. doi: 10.1093/clinids/5.supplement_5.s867. [DOI] [PubMed] [Google Scholar]
- 3.Drlica K, Zhao X. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol Mol Biol Rev. 1997;61:377–392. doi: 10.1128/mmbr.61.3.377-392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gotoh N, Tsujimoto H, Tsuda M, Okamoto K, Nomura A, Wada T, Nakahashi M, Nishino T. Characterization of the MexC-MexD-OprJ multidrug efflux system in delta-MexA-MexB-OprM mutants of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1998;42:1938–1943. doi: 10.1128/aac.42.8.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hosaka M, Gotoh N, Nishino T. Purification of a 54-kilodalton protein (OprJ) produced in NfxB mutants of Pseudomonas aeruginosa and production of a monoclonal antibody specific to OprJ. Antimicrob Agents Chemother. 1995;39:1731–1735. doi: 10.1128/aac.39.8.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jakics E B, Iyobe S, Hirai K, Fukuda H, Hashimoto H. Occurrence of the nfxB type mutation in clinical isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1992;36:2562–2565. doi: 10.1128/aac.36.11.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jalal S, Wretlind B. Mechanisms of quinolone resistance in clinical strains of Pseudomonas aeruginosa. Microb Drug Resist. 1998;4:257–261. doi: 10.1089/mdr.1998.4.257. [DOI] [PubMed] [Google Scholar]
- 8.Köhler T, Michea-Hamzehpour M, Henze U, Gotoh N, Curty L K, Pechere J C. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol Microbiol. 1997;23:345–354. doi: 10.1046/j.1365-2958.1997.2281594.x. [DOI] [PubMed] [Google Scholar]
- 9.Kureishi A, Diver J M, Beckthold B, Schollaardt T, Bryan L E. Cloning and nucleotide sequence of Pseudomonas aeruginosa DNA gyrase gyrA gene from strain PAO1 and quinolone-resistant clinical isolates. Antimicrob Agents Chemother. 1994;38:1944–1952. doi: 10.1128/aac.38.9.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nikaido H. Multidrug efflux pumps of gram-negative bacteria. J Bacteriol. 1996;178:5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nikaido H. Antibiotic resistance caused by gram-negative multidrug efflux pumps. Clin Infect Dis. 1998;27(Suppl.):S32–S41. doi: 10.1086/514920. [DOI] [PubMed] [Google Scholar]
- 12.Ojeniyi B, Petersen U S, Høiby N. Comparison of genome fingerprinting with conventional typing methods used on Pseudomonas aeruginosa isolates from cystic fibrosis patients. APMIS. 1993;101:168–175. [PubMed] [Google Scholar]
- 13.Pitcher D, Saunders N, Owen R. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol. 1989;8:151–156. [Google Scholar]
- 14.Poole K, Gotoh N, Tsujimoto H, Zhao Q, Wada A, Yamasaki, Neshat S, Yamagishi J, Li X Z, Nishino T. Overexpression of the mexC-mexD-oprJ efflux operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa. Mol Microbiol. 1996;21:713–724. doi: 10.1046/j.1365-2958.1996.281397.x. [DOI] [PubMed] [Google Scholar]
- 15.Wiedmann M, Bruce J L, Keating C, Johnson A E, McDonough P L, Batt C A. Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect Immun. 1997;65:2707–2716. doi: 10.1128/iai.65.7.2707-2716.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoshida H, Nakamura M, Bogaki M, Nakamura S. Proportion of DNA gyrase mutants among quinolone-resistant strains of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1990;34:1273–1275. doi: 10.1128/aac.34.6.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao Q, Li X Z, Srikumar R, Poole K. Contribution of outer membrane efflux protein OprM to antibiotic resistance in Pseudomonas aeruginosa independent of MexAB. Antimicrob Agents Chemother. 1998;42:1682–1688. doi: 10.1128/aac.42.7.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ziha-Zarifi I, Llanes C, Köhler T, Pechère J C, Plesiat P. In vivo emergence of multidrug-resistant mutants of Pseudomonas aeruginosa overexpressing the active efflux system MexA-MexB-OprM. Antimicrob Agents Chemother. 1999;43:287–291. doi: 10.1128/aac.43.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]