Abstract
Individual tumors comprise genetically and epigenetically heterogeneous subclones, each of which is presumably associated with a distinct function, such as self-renewal or drug sensitivity. The dissection of such intratumoral heterogeneity is crucial to understand how tumors evolve during disease progression and under the selection of therapeutic intervention. As a paradigm of cancer intratumoral heterogeneity and clonal evolution, acute myeloid leukemia (AML) has been shown to possess complex clonal architecture based on karyotype studies, as well as deep sequencing of mixed cellular populations using next-generation sequencing (NGS) technologies. The recent development of single-cell sequencing (SCS) methods provides a powerful tool to allow analysis of genomes, transcriptomes, proteomes, and epigenomes at an individual cell level. The technologies applied in AML have broadened our understanding of AML heterogeneity and provided new insights for the development of novel therapeutic strategies. In this review, we summarize the progress in the research of AML heterogeneity using SCS technology and discuss the limitations and future direction regarding how SCS can contribute to AML prognosis and treatment.
Keywords: Acute myeloid leukemia, Heterogeneity, Single-cell sequencing
1. INTRODUCTION
Peter Nowell first proposed that the formation of tumors is an evolutionary process1 and that genetic mutations are traditionally recognized as molecular determinants of this process. Although unique combinations of mutations are commonly used to define different subclones, it is now widely accepted that genetic and epigenetic alterations collectively contribute to functional heterogeneity, such as self-renewal and drug resistance within individual tumors.2 Intratumoral heterogeneity is a key determinant in disease progression and treatment response; therefore, a comprehensive analysis of tumor heterogeneity has important clinical implications and may provide valuable insights for the development of anticancer therapies.3
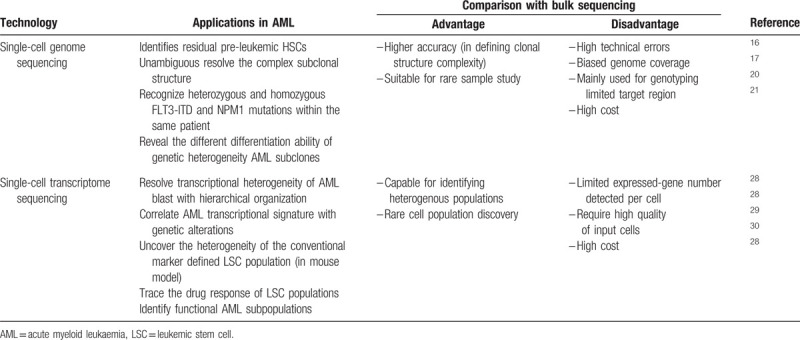
Acute myeloid leukemia (AML) is one of the best studied cancer type owing to the easy accessibility of samples and well-defined cellular hierarchy in the normal hematopoietic system.4 This unique feature with the relatively low recurrent somatic mutation rate of the disease5 make AML an excellent model to explore intratumoral heterogeneity and clinical significance.6 Laboratory-based technologies such as fluorescence in situ hybridization and flow cytometry as well as high-throughput next-generation sequencing (NGS) have revealed that AML comprises heterogeneous subpopulations characterized by distinct genetic and epigenetic alterations.7,8 However, the multiple layers of heterogeneity have remained ambiguous until the introduction of single-cell sequencing (SCS). SCS has empowered the cancer field to achieve an unprecedented understanding of the molecular basis of intratumoral heterogeneity. In this review, we summarize how SCS technology contributes to our understanding of AML heterogeneity, with a particular focus on insights that are only revealed from single-cell analyses (Table 1).
Table 1.
Applications of Single-cell Sequencing technologies in AML studies.
2. USING SINGLE-CELL GENOME SEQUENCING TO UNAMBIGUOUSLY DEFINE THE GENETIC HETEROGENEITY OF AML
AML is thought to originate from a subset of hematopoietic stem/progenitor cells (HSCs/HPCs).9,10 By gradually accumulating mutations and numerous additional genetic and epigenetic abnormalities, the found cells take a complex trajectory path to finally evolve into functionally diversified subpopulations.11–13 Obtaining genomic information at the single-cell level is the ultimate solution to dissect clonal diversity. To achieve this goal, current technology requires whole-genome amplification (WGA) of DNA extracted from an individual cell to generate enough DNA for sequencing. WGA methods continue to face several technical limitations, such as a high allele dropout rate, high false-positive rate, and chimera formation, which have been well described in other reviews.14,15 Currently, most single-cell genomic studies of AML have relied on genotyping of somatic mutations pre-detected by genome sequencing of a mixed cell population, with the genotyping libraries constructed from single-cell WGA or colony-forming units derived from a single cell.16–19
Single-cell genotyping of limited somatic mutations (usually <10) in a small number of cells (several to several hundreds) allows the determination of a relatively clearer clonal structure of de novo AML compared with NGS of bulk samples. AML was originally viewed as evolving from a linear accumulation of multiple driver mutations. However, the estimates of subclone size in deep bulk sequencing indicate that AML is likely evolved with a complex branching architecture.6 Despite this improved understanding, it remains difficult to determine the exact evolutionary trajectory and clonal structure based on the variant allele frequency (VAF), particularly when the VAFs of two subclonal mutations are similar. Single-cell DNA sequencing can resolve the combined pattern of multiple mutations in an individual cell and subsequently provide evidence for either linear or branched structures in individual AMLs. Remarkably, single-cell genotyping of pre-detected mutations identifies residual preleukemic HSCs that exist in bulk AML cells and constitute a cellular reservoir with linearly accumulated founder mutations. These pre-leukemic cells are predisposed to acquire additional mutations and evolve into a distinct clonal hierarchy observed in the full-blown disease.16 Single-cell gene capture of 1953 somatic single nucleotide variants (SNVs) from patients with AML unambiguously demonstrates the origin of two separate subclones by detecting mutually exclusive SNVs, which represent an evolutionary branch point from the founding clone.17 Interestingly, most VAF in bulk sequencing is calculated under the assumption that all mutations occur heterozygously in single cells, and a study using single-cell genome analysis has revealed the existence of both heterozygous and homozygous FLT3-ITD and NPM1 mutations in an individual patient.20 These data suggest that bulk sequencing overestimates the number of leukemic cells but underestimates the clonal diversity of the blast. In an intriguing study, single-cell genotyping was performed on flow-sorted AML blasts and mature myelomonocytes. These two cell populations contain the same founding clone but distinct subclones, indicating that genetic heterogeneity impacts the differentiation ability of AML subclones.21 A recent study adopted droplet microfluidics for single-cell DNA genotyping, which enabled a comprehensive analysis of thousands of AML cells in parallel.22 This single-cell DNA sequencing has potential in the development of high-throughput genome-wide detection of genetic alterations in the future.
These single-cell genomic studies16–20 strongly support the clonal architecture inferred from the analysis of bulk samples. Importantly, analyses of AML at single-cell resolution unambiguously resolve the subclonal structure that cannot be appropriately assigned through mutation frequencies or unknown mutation status (heterozygous or homozygous). Thus, these studies reveal the previously unappreciated complexity of the clonal structure of AML.
3. USING SINGLE-CELL TRANSCRIPTOME SEQUENCING TO REVEAL THE FUNCTIONAL HETEROGENEITY OF AML
The transcriptome is the dynamic output of all genetic information, which closely reflects the functional status of cells. Compared with genomic analyses, single-cell transcriptome sequencing (scRNA-seq) is a more developed technology that allows for a high-throughput (several hundreds to ten thousands cells) detection of tens of thousands of transcripts. Considerable technical progress has been made over the past years, including the introduction of the Moloney murine leukemia virus, which has template-switching activity to ensure amplification of full-length mRNA transcripts,23 and the development of unique molecules index for reduced amplification bias.24 A commercial provider (10× Genomics®, 7068 Koll Center Parkway, Suite 401, Pleasanton, California 94566) ensures the widespread use of high-throughput SCS technology. The scRNA-seq of a normal hematopoietic cell population confirms the structured hierarchy of a normal blood development of stem cell derived-myeloid and -lymphoid lineages,25,26 and more importantly, leads to the discovery of a novel dendritic cell (DC) subset.27 Transcriptomic profiling of AML has significantly improved our understanding of tumor hierarchical structure and led to the identification of novel cell types, leukemic stem cells (LSCs), and drug-resistant subpopulations.28–30
Unique leukemia-enriched plasma membrane proteins (e.g., CD25 and IL1RAP) enabled the prospective isolation of two AML blast populations from individual patients, and the separate subclones were shown to be genetically and epigenetically distinct and to have different functions in their drug sensitivity, growth, and engraftment behavior.31 These data demonstrate the existence of functional diverse subpopulations within individual AMLs. scRNA-seq data were used to profile 16 patients with AML and 5 healthy donors and to classify AML cells into 6 subpopulations along the HSC to myeloid axis of the normal cell hierarchy. Each population was characterized by a unique expression signature, including HSCs, progenitors, granulocyte-macrophage progenitors (GMPs), promonocytes, monocytes, and conventional DCs. Such signatures exhibit a close correspondence to the genetics of the AMLs. For example, TCGA tumors with high GMP-like and promonocyte-like scores derived from scRNA-seq perfectly overlap with PML-RARA fusions. Different AML cell signatures are further correlated with clinical outcomes such the outcomes are better in patients with a higher GMP-like score than in those with a higher HSC/Progenitor-like score.28 The molecular classification of AMLs has also been explored by mass cytometry and chromatin accessibility at the single-cell level. By comparing surface and intracellular signaling proteins simultaneously in healthy and leukemic cells by single-cell mass cytometry, Levine et al32 have reported that the tightly coupled surface and signaling phenotypes in healthy cells were perturbed in AML cells. Compared with cell surface markers, the signatures of intracellular signaling proteins are more reliable to predict clinical outcomes in patients.32 The analysis of single-cell chromatin accessibility also reveals the regulatory heterogeneity of AML cells resembling a normal blood cell hierarchy, with a similar pattern distributed along the HSC to myeloid axis.33 These studies indicate an intrinsic relationship among the transcriptomes, proteomes, and epigenomes in AML cells.
AML LSCs have been recognized as the apex of the AML cellular hierarchy within the heterogeneous population, with properties of self-renewal, cell cycle quiescence, and chemoresistance.34 It is believed that a cure for AML depends on eradicating the LSCs. However, no definitive phenotypic markers are known to purify LSCs. Intriguingly, the LSC-enriched population can be recognized by characteristic transcriptional signatures, such as the “LSC score.”35 The use of the LSC score in a single-cell classification can help identify and trace the LSC population during treatment and provide opportunities to improve identification of the LSC population. Indeed, the LSC expression signature has been applied in single-cell analysis of the MLL-AF9 mouse model and has uncovered the heterogeneity of the conventional marker-defined LSC population (Lin-Il7r-Kit+Sca1-CD34+CD16/CD32+) that contains two independent self-renewing lineages with different clonal activities.29 In contrast, LSCs have metabolic features that require active oxidative phosphorylation status.36 scRNA-seq has been used to trace the response of the LSC population during venetoclax with azacitidine treatment, which can disrupt the energy metabolism in AML patients.30 Interestingly, SCTS also leads to the identification of a differentiated monocyte-like AML cell population, which suppresses T-cell activation in vitro and might have immunomodulatory functions that contribute to the pathogenesis of the disease.28
4. CONCLUSION: LIMITATIONS AND PROSPECTIVE
Multiple SCS technologies have been applied in cancer research and have achieved remarkable progress. However, significant technical improvements are needed to achieve a comprehensive view of tumor heterogeneity. First, SCS technology is still limited by amplification bias and extensive technical errors, particularly in single-cell genome sequencing.37 Second, the need for simultaneous acquisition of multiomics data of a single cell has not been fulfilled. For example, the co-existence of leukemic, preleukemic, and normal blood cells in AML samples requires the integration of mutational spectrum and transcriptional states to optimally classify the functional heterogeneous subpopulations. To date, most attempts at systematically detecting both genetic and RNA expression information from the same single cell are either expensive or have low sensitivity,38–40 thus restricting the widespread use of integrative analysis. However, some limited success has been achieved with a relatively low throughput in the studies of hematopoietic malignancies. These include single-cell transcriptome analyses in combination with successful detection of the BCR–ABL breakpoint in chronic myeloid leukemia41 and a novel method that allows mutational analysis (up to 12 different mutations) and parallel RNA sequencing from the same single cell in myeloproliferative neoplasm.42 Furthermore, efforts have been made to combine transcriptome detection with surface-marker protein analysis43 or assay for transposase-accessible chromatin using sequencing44 within a single cell. These studies hold promise for the future development of multiomics integrated at the single-cell level.
Dissecting AML heterogeneity using SCS has provided novel insights into the molecular mechanisms underlying drug resistance and disease progression. Single-cell targeted mutation analysis in AML treated with the FLT3 inhibitor quizartinib has revealed highly complex drug-resistance clones at relapse, demonstrating marked clonal complexity associated with therapeutic resistance.45 By comparing clonal structures identified by single-cell DNA sequencing at diagnosis and relapse, multiple bypass pathways have been identified in patients with AML having IDH2 mutations that can potentially be targeted for treatment.46
While these studies serve as excellent examples of how new knowledge from SCS can aid in the development of new drugs and treatment options, dissecting intratumoral heterogeneity holds a relatively greater potential for clinical application in the era of precision medicine. For example, new biomarkers identified from previous unknown drug-resistant subpopulations could be used to better stratify patients with AML. Moreover, SCS in combination with a more sensitive targeted mutation detection method can be used to monitor functionally diverse subclones with different drug-sensitivities and stem cell potential during treatment and disease progression. This information will be invaluable in improving prognostic prediction and detecting minimal residual disease. With rapid development of these advanced technologies, it is conceivable that SCS will have a tremendous impact on the clinical practice of AML.
REFERENCES
- [1].Nowell PC. The clonal evolution of tumor cell populations. Science (New York, NY) 1976;194(4260):23–28. [DOI] [PubMed] [Google Scholar]
- [2].Hinohara K, Polyak K. Intratumoral heterogeneity: more than just mutations. Trends Cell Biol 2019;29(7):569–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].McGranahan N, Swanton C. Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell 2015;27(1):15–26. [DOI] [PubMed] [Google Scholar]
- [4].Wilson NK, Göttgens B. Single-cell sequencing in normal and malignant hematopoiesis. HemaSphere 2018;2(2):e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ley TJ, Miller C, Ding L, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 2013;368(22):2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Grove CS, Vassiliou GS. Acute myeloid leukaemia: a paradigm for the clonal evolution of cancer? Dis Model Mech 2014;7(8):941–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Li S, Mason CE, Melnick A. Genetic and epigenetic heterogeneity in acute myeloid leukemia. Curr Opin Genet Dev 2016;36:100–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Cai SF, Levine RL. Genetic and epigenetic determinants of AML pathogenesis. Semin Hematol 2019;56(2):84–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Dick JE. Stem cell concepts renew cancer research. Blood 2008;112(13):4793–4807. [DOI] [PubMed] [Google Scholar]
- [10].Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature 2001;414(6859):105–111. [DOI] [PubMed] [Google Scholar]
- [11].Shlush LI, Zandi S, Mitchell A, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature 2014;506(7488):328–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Duque-Afonso J, Cleary ML. The AML salad bowl. Cancer Cell 2014;25(3):265–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Welch JS, Ley TJ, Link DC, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell 2012;150(2):264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zong C, Lu S, Chapman AR, Xie XS. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science (New York, NY) 2012;338(6114):1622–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ellsworth DL, Blackburn HL, Shriver CD, Rabizadeh S, Soon-Shiong P, Ellsworth RE. Single-cell sequencing and tumorigenesis: improved understanding of tumor evolution and metastasis. Clin Transl Med 2017;6(1):15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Jan M, Snyder TM, Corces-Zimmerman MR, et al. Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia. Sci Transl Med 2012;4(149):149ra118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hughes AE, Magrini V, Demeter R, et al. Clonal architecture of secondary acute myeloid leukemia defined by single-cell sequencing. PLoS Genet 2014;10(7):e1004462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Shouval R, Shlush LI, Yehudai-Resheff S, et al. Single cell analysis exposes intratumor heterogeneity and suggests that FLT3-ITD is a late event in leukemogenesis. Exp Hematol 2014;42(6):457–463. [DOI] [PubMed] [Google Scholar]
- [19].Stirewalt DL, Pogosova-Agadjanyan EL, Tsuchiya K, Joaquin J, Meshinchi S. Copy-neutral loss of heterozygosity is prevalent and a late event in the pathogenesis of FLT3/ITD AML. Blood Cancer J 2014;4:e208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Paguirigan AL, Smith J, Meshinchi S, Carroll M, Maley C, Radich JP. Single-cell genotyping demonstrates complex clonal diversity in acute myeloid leukemia. Sci Transl Med 2015;7(281):281re2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Klco JM, Spencer DH, Miller CA, et al. Functional heterogeneity of genetically defined subclones in acute myeloid leukemia. Cancer Cell 2014;25(3):379–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lan F, Demaree B, Ahmed N, Abate AR. Single-cell genome sequencing at ultra-high-throughput with microfluidic droplet barcoding. Nat Biotechnol 2017;35(7):640–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ramsköld D, Luo S, Wang YC, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol 2012;30(8):777–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Islam S, Zeisel A, Joost S, et al. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat Methods 2014;11(2):163–166. [DOI] [PubMed] [Google Scholar]
- [25].Pellin D, Loperfido M, Baricordi C, et al. A comprehensive single cell transcriptional landscape of human hematopoietic progenitors. Nat Commun 2019;10(1):2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Oetjen KA, Lindblad KE, Goswami M, et al. Human bone marrow assessment by single-cell RNA sequencing, mass cytometry, and flow cytometry. JCI Insight 2018;3(23). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Villani AC, Satija R, Reynolds G, et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 2017;356(6335). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].van Galen P, Hovestadt V, Wadsworth Ii MH, et al. Single-cell RNA-Seq reveals AML hierarchies relevant to disease progression and immunity. Cell 2019;176(6):1265–1281.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Guo G, Luc S, Marco E, et al. Mapping cellular hierarchy by single-cell analysis of the cell surface repertoire. Cell Stem Cell 2013;13(4):492–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Pollyea DA, Stevens BM, Jones CL, et al. Venetoclax with azacitidine disrupts energy metabolism and targets leukemia stem cells in patients with acute myeloid leukemia. Nat Med 2018;24(12):1859–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].de Boer B, Prick J, Pruis MG, et al. Prospective isolation and characterization of genetically and functionally distinct AML subclones. Cancer Cell 2018;34(4):674–689.e8. [DOI] [PubMed] [Google Scholar]
- [32].Levine JH, Simonds EF, Bendall SC, et al. Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell 2015;162(1):184–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Corces MR, Buenrostro JD, Wu B, et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet 2016;48(10):1193–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Thomas D, Majeti R. Biology and relevance of human acute myeloid leukemia stem cells. Blood 2017;129(12):1577–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Ng SW, Mitchell A, Kennedy JA, et al. A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature 2016;540(7633):433–437. [DOI] [PubMed] [Google Scholar]
- [36].Farge T, Saland E, de Toni F, et al. Chemotherapy-resistant human acute myeloid leukemia cells are not enriched for leukemic stem cells but require oxidative metabolism. Cancer Discov 2017;7(7):716–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wang Y, Navin NE. Advances and applications of single-cell sequencing technologies. Mol Cell 2015;58(4):598–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Macaulay IC, Haerty W, Kumar P, et al. G&T-seq: parallel sequencing of single-cell genomes and transcriptomes. Nat Methods 2015;12(6):519–522. [DOI] [PubMed] [Google Scholar]
- [39].Dey SS, Kester L, Spanjaard B, Bienko M, van Oudenaarden A. Integrated genome and transcriptome sequencing of the same cell. Nat Biotechnol 2015;33(3):285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Han KY, Kim KT, Joung JG, et al. SIDR: simultaneous isolation and parallel sequencing of genomic DNA and total RNA from single cells. Genome Res 2018;28(1):75–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Giustacchini A, Thongjuea S, Barkas N, et al. Single-cell transcriptomics uncovers distinct molecular signatures of stem cells in chronic myeloid leukemia. Nat Med 2017;23(6):692–702. [DOI] [PubMed] [Google Scholar]
- [42].Rodriguez-Meira A, Buck G, Clark SA, et al. Unravelling intratumoral heterogeneity through high-sensitivity single-cell mutational analysis and parallel RNA sequencing. Mol Cell 2019;73(6):1292–1305.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Stoeckius M, Hafemeister C, Stephenson W, et al. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods 2017;14(9):865–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Jia G, Preussner J, Chen X, et al. Single cell RNA-seq and ATAC-seq analysis of cardiac progenitor cell transition states and lineage settlement. Nat Commun 2018;9(1):4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Smith CC, Paguirigan A, Jeschke GR, et al. Heterogeneous resistance to quizartinib in acute myeloid leukemia revealed by single-cell analysis. Blood 2017;130(1):48–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Quek L, David MD, Kennedy A, et al. Clonal heterogeneity of acute myeloid leukemia treated with the IDH2 inhibitor enasidenib. Nat Med 2018;24(8):1167–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]


