Abstract
Purpose
The detection of clinically significant prostate cancer (PCa) is shown to greatly benefit from MRI–ultrasound fusion biopsy, which involves overlaying pre-biopsy MRI volumes (or targets) with real-time ultrasound images. In previous literature, machine learning models trained on either MRI or ultrasound data have been proposed to improve biopsy guidance and PCa detection. However, quantitative fusion of information from MRI and ultrasound has not been explored in depth in a large study. This paper investigates information fusion approaches between MRI and ultrasound to improve targeting of PCa foci in biopsies.
Methods
We build models of fully convolutional networks (FCN) using data from a newly proposed ultrasound modality, temporal enhanced ultrasound (TeUS), and apparent diffusion coefficient (ADC) from 107 patients with 145 biopsy cores. The architecture of our models is based on U-Net and U-Net with attention gates. Models are built using joint training through intermediate and late fusion of the data. We also build models with data from each modality, separately, to use as baseline. The performance is evaluated based on the area under the curve (AUC) for predicting clinically significant PCa.
Results
Using our proposed deep learning framework and intermediate fusion, integration of TeUS and ADC outperforms the individual modalities for cancer detection. We achieve an AUC of 0.76 for detection of all PCa foci, and 0.89 for PCa with larger foci. Results indicate a shared representation between multiple modalities outperforms the average unimodal predictions.
Conclusion
We demonstrate the significant potential of multimodality integration of information from MRI and TeUS to improve PCa detection, which is essential for accurate targeting of cancer foci during biopsy. By using FCNs as the architecture of choice, we are able to predict the presence of clinically significant PCa in entire imaging planes immediately, without the need for region-based analysis. This reduces the overall computational time and enables future intra-operative deployment of this technology.
Keywords: Information fusion, Multimodality training, Deep learning, Prostate cancer detection, Image-guided biopsy, Temporal enhanced ultrasound, Magnetic resonance imaging
Introduction
Prostate Cancer (PCa) is among the most commonly diagnosed cancers in men. Observation of an elevated prostate-specific antigen (PSA) is usually the first step of the diagnostic pathway for PCa. Recent studies [1] have suggested that acquiring multiparametric magnetic resonance imaging (mp-MRI) for diagnostic purposes would reduce unnecessary biopsies and associated potential harmful side effects for men with elevated PSA. Despite the diagnostic power of mp-MRI, the gold standard for PCa diagnosis has remained as transrectal ultrasound-guided biopsy (TRUS biopsy) in most centers due to the availability and lower costs of ultrasound. In contrast to mp-MRI, ultrasound lacks the sensitivity to be used as a primary diagnostic modality. The fusion of the pre-biopsy mp-MRI of the prostate with TRUS-guided biopsy promises to increase the yield of aggressive cancer [23,28,29]. In the literature and in clinical practice, MR-TRUS fusion biopsy refers to the alignment of pre-biopsy mp-MRI volumes with TRUS (acquired at the beginning of biopsy) and targeting of the lesions identified on mp-MRI. mp-MRI is projected and fused with TRUS images either through commercially available systems [28,29] or using cognitive co-location of targets in the TRUS imaging space [23]. Although registration techniques do not lead to an ideal true fusion of imaging spaces, providing the spatial information of the mp-MRI lesion delineations during real-time TRUS-biopsy has been shown to improve the detection of high-risk prostate cancer [23,28,29]. A significant missing piece of the puzzle and a natural question that follows is: In addition to the spatial information of the MR-identified lesions on TRUS, can a machine learn from the explicit integration of information in MRI and TRUS to improve detection of PCa, and reduce unnecessary biopsies?
In the past few years, advancements in Deep Learning (DL) have dominated the field of computer-assisted PCa detection using mp-MRI or ultrasound information, as individual modalities. Most of the research have utilized Convolutional Neural Networks (CNN) and various optimization strategies; achieving state-of-the-art performance in PCa detection. Several groups have taken advantage of the multisequence nature of the mp-MRI data by stacking each modality as input channels similar to RGB images [3,14], and integrating their information early in the training. Mehrtash et al. [17] used 3D CNNs on images of apparent diffusion coefficient (ADC), high b-value and KTrans for PCa diagnosis. Kiraly et al. [12] used fully convolutional networks (FCN) for localization and classification of prostate lesions and achieved area under the curve (AUC) of 0.83 by training on 202 patients. In a recent study, Schelb et al. [25] proposed a U-Net architecture on biparametric prostate MRI (T2-Weighted and ADC), and achieved performance similar to that of Prostate Imaging–Reporting and Data System (PI-RADS), the clinical standard of mp-MRI scoring [31]. Several ultrasound-based methods have been proposed for the detection of PCa including the analysis of spectral features in a single frame of ultrasound [6], analysis of tissue elasticity in the presence of external excitation [4], and more recently, temporal enhanced ultrasound (TeUS) for analysis of radio frequency (RF) time series [11,18]. Although analysis of a single frame of ultrasound has been studied in the past [6,7], a consistent yet differentiable tissue property has not been derived, as a result of heterogeneity of PCa and its variability across patients. TeUS has emerged as a promising imaging modality for tissue characterization, within a deep learning framework. It involves acquisition and analysis of a sequence of ultrasound data from a stationary tissue location without explicit excitation. Previous in vivo [9,10] and ex vivo [19] studies have shown promising results for differentiating PCa from healthy tissue. Azizi et al. [2] used a deep belief network (DBM) and achieved AUCs of as high as 0.84. Sedghi et al. [27] used an unsupervised learning approach to capture representations of TeUS signals associated with variations of PCa. No studies to date have attempted to quantitatively fuse the information from the two modalities.
Fusion of information has been studied in computer vision and medical imaging literature in the context of early, intermediate, and late fusion. The most common approach involves concatenating information from multiple modalities (early fusion) prior to learning. There are challenges with early fusion of information. As mentioned before, image registration often does not result in perfect alignment between modalities; thus, finding the corresponding information between different modalities is challenging. In addition, without any post-processing or latent space extraction, the relationships between modalities are highly complex, and hard for machines to learn in the case of early fusion. In late fusion, different models are independently learned from different modalities and the outputs of models are combined (either averaging, or weighted sum) to make a final decision. A shortcoming of this approach is that it is possible to miss the low-level interactions between different data types [20]. As a result, intermediate fusion (fusion of information at some point between data inputs and model prediction) has been pursued as an appropriate strategy in the literature [8,13,20,32]. In segmentation literature, Havaei et al. [8] proposed heteromodal network architecture for multimodal brain MRI segmentation. In their study, they learned an embedding of each modality in a shared common space as well as an average value for this space, and performed segmentation using the fused information. In another study, Valindria et al. [32] used different encoder–decoder architectures for segmentation of MRI and CT of liver. They evaluated different strategies for information fusion and demonstrated that learning from multimodal data increased accuracy compared to single modality. In computer vision, Kuga et al. [13] proposed a multimodal, multitask encoder–decoder network with shared latent and skip connection for simultaneous learning from RGB and depth images. They also demonstrated improved performance using all modalities.
To the best of our knowledge, integration of imaging information for improving TRUS-guided biopsies have not been studied yet. The motivation for integration of mp-MRI and ultrasound is the complementary nature of information in each modality and their individual limitations. We demonstrate the improved performance of detecting PCa by utilizing both sources of information to provide cancer likelihood maps that can augment biopsies. Although rule-based addition of MR-identified labels for lesions with TeUS has shown promise [10], the explicit incorporation of the information content of each modality has not been investigated yet. In this work, for the first time, we propose an application of multimodality FCN for information fusion between MRI and TeUS. Our work lays the foundation for effective utilization of all available imaging data for improving targeting of biopsies, and reducing the need for unnecessary biopsies.
Materials and method
Data
The data used in this study consist of mp-MRI and TeUS from patients who underwent prostate MR-TRUS fusion biopsy. All patients provided informed consent to participate in the institutional review board (IRB)-approved study. Patients cohort include those with at least one suspicious lesion visualized on mp-MRI acquired on a 3-T MR scanner (Achieva-TX, Philips Medical Systems, Best, NL) with an endorectal coil (BPX-30, Medrad, Indianola, PA). T2-weighted MRI (T2), diffusion-weighted MRI (DWI) and dynamic contrast-enhanced MRI (DCE) were acquired for each patient. Two independent radiologists with 6 and 13 years of experience evaluated the mp-MRI through a criteria based on the number of positive parameters on T2, ADC, and DCE. More details about the mp-MRI scoring approach used for this data is available in [33]. Subsequently, subjects underwent MR-TRUS fusion-guided biopsy using commercial UroNav system (Invivo, Philips Healthcare, Gainesville, FL). More specifically, MR-identified suspicious lesions were imported into the UroNav and were displayed on triplanar images as biopsy targets. Patients had 12-core sextant biopsy, and two (axial and sagittal) targeted MR-TRUS fusion biopsy per identified MRI lesion. Prior to needle firing for the targeted MR-identified lesion, the ultrasound transducer was held steady freehand for 5 s to capture 100 frames of beam-formed radio frequency (RF) data. More specifically, an endocavity curvilinear probe (Philips C9–5ec) with frequency of 6.6 MHz was used. Finally, tissue samples were examined under microscope, and histopathology information was provided, which included the length and type of the tissue.
Our dataset consists of 107 patients with a total of 145 biopsy cores. Of the three mp-MRI sequences acquired from patients, we only had access to T2-weighted and ADC images (biparametric MRI). All patients had TeUS from their TRUS-imaging plane. In our dataset, 51 cores have cancer and 94 are benign. The average Tumor in Core Length (TIL) in cores with cancer is 6.2 mm. We use stratified sampling on both TIL and the ratio of cancerous to benign cores to split the data into 65% training and validation and 35% testing. As a result, training and validation set consists of 59 benign cores and 31 cancerous cores with average TIL of 6.5 mm for cancerous cores, and test set consists of 35 benign and 20 cancerous cores with average TIL of 6.3 mm. Throughout the study, we use the biopsy-proven MR-identified targets as ground truth for our labels. An example of our TeUS and mp-MRI data is shown in Fig. 1.
Fig. 1.
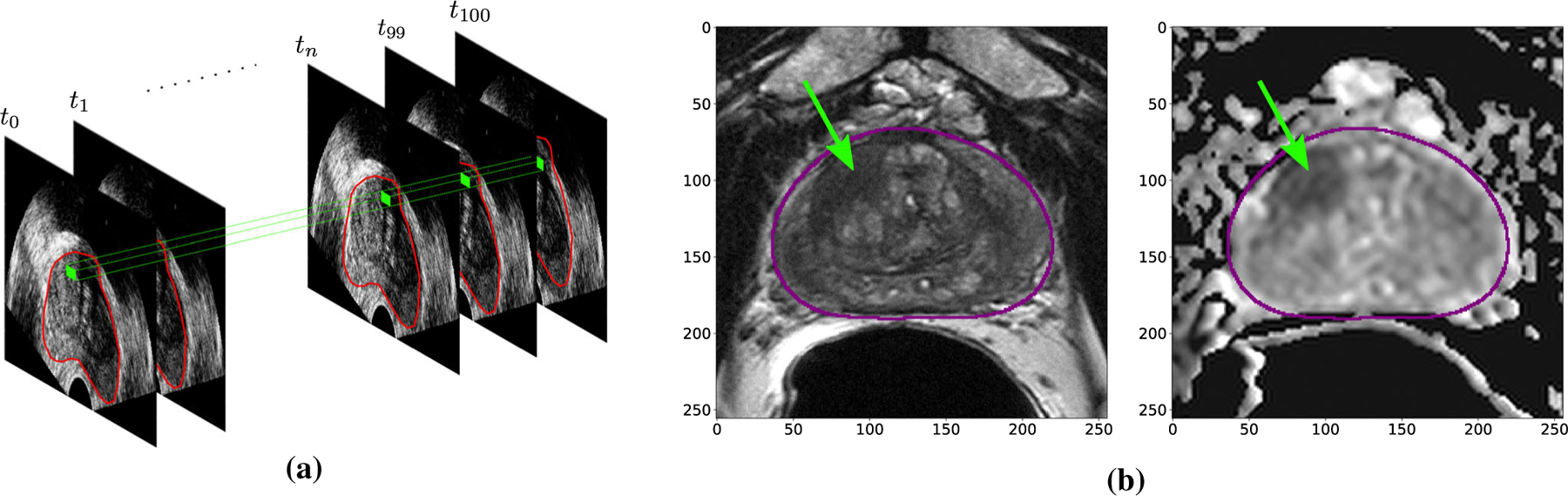
Examples of temporal enhanced ultrasound (TeUS) (a) and Bi-parametric MRI (b) in our dataset. Green dots and arrows correspond to biopsy target
Preprocessing
MRI
We start preprocessing of mp-MRI images by correcting the signal inhomogeneity as a result of endorectal coil using N4 algorithm in 3D Slicer [5]. We then resample all MRI images to 0.25×0.25×3 mm voxel spacing. To segment the prostate gland, we use a publicly available deep learningbased segmentation tool, DeepInfer [16]. Next, exploratory data analysis is utilized to find the largest bounding box (size 256×256 pixels) containing the prostate gland in our dataset. The reported location of the biopsy is utilized to select the 256 × 256 image patch from the axial planes of MRI. To overcome overfitting to the data, we augment the images 10 times via intensity augmentation with additive Gaussian noise of σ = 10. In addition, images are flipped left-to-right to increase the size of the dataset. Overall, the training data size is increased by a factor of 20. Finally, the intensity values are normalized to 0–1 range. As previous studies [17] demonstrated superior performance of deep models trained on ADC images compared to T2-weighted images, we utilize the preprocessed ADC images as our main MRI data.
TeUS
For every biopsy core, TeUS data are available from the entire imaging frame. Due to consistency in imaging acquisition, the prostate gland and biopsied areas are always in the first half of the full-image data; therefore, we crop and analyze only the 2560 samples along the axial direction in TeUS. As previous studies in TeUS [2,9] have shown promising results with learning from frequency domain signals, we compute the frequency components of each RF time series. More specifically, we first remove the DC component from the signal, followed by fast Fourier transform (FFT) with 128 samples to compute the frequency response. Similar to previous studies [2,18], we average FFT signals (every 50 samples with step size of 10) along the axial direction of the RF for smoothing.
Unsupervised dimensionality reduction of TeUS
As a result of the preprocessing step, the dimensionality of TeUS data reduces to 256 × 256 × 128 pixels, representing number of samples, number of RF lines and number of frequency features, respectively. Since the dimensionality of data in TeUS still imposes complexity in training and information fusion, we use 1D convolutional astronomers (AE) to compress the information in the feature space. AEs are trained in a way that they learn to nonlinearly reduce the dimensionality of the input, while preserving the reconstructability of the latent space to the original input. We use TeUS signals from all the pixels of training and validation set for training the AE. The input to the AE has a shape of 128 × 1, which is the same for the output. Our AE architecture is composed of an encoder with 6 blocks of 1D convolutions with kernel size of 3, ReLU activation function, and max-pooling. After the final block, we flatten the filters to feed into a dense neural network with 8 nodes. Correspondingly, we utilize a symmetric architecture for decoder with 6 blocks of 1D convolutions with upsampling modules (instead of max-pooling) to reconstruct the input again. The AE is optimized with mini-batch stochastic gradient descent with Adam update rule with initial learning rate of 10−4. After training, we use the latent features with the most variability in training and validation to convert the feature space to the final size of 256 × 256 × 1, similar to ADC images.
Label generation
Since the groundtruth in our dataset comes from biopsy, we do not have access to the whole imaging plane labels. To overcome this issue for training deep models, we consider a Region of Interest (ROI) in the shape of a disk with a radius of 3.5 mm centered at biopsy location to assign biopsy-proven labels to the region. For the tumor cores in our training set, we increased the disk radius proportional to the TIL to artificially increase our labeled data. We mask this ROI with the extracted prostate boundary to avoid out of gland regions for training. The generated labels are one-hot vector for each pixel representing its class. More specifically, the label for pixels representing biopsy-proven cancer is z = [0, 1 ], and for biopsy-proven normal tissue is z = [1, 0 ]. As a result of label generation for both TeUS and MRI, our outputs have the same size as inputs with two channels (normal, cancer).
Model architecture and training
Prior works in TeUS have used deep models for learning the characteristics of cancer from each ROI. As a result of this strategy, to generate class-specific labels of the whole imaging plane in TRUS-biopsy, every ROI needs to be fed into the model for prediction which is time-consuming. More recently, semantic segmentation by FCNs and their variants (U-Nets) has emerged as the de facto standard for image segmentation in medical imaging [15,24]. FCNs are composed of contraction (encoder) and expansion (decoder) paths. During encoding, the model extracts and propagates spatially invariant features by convolutional layers and downsampling. As a result, it understands the what but at the cost of losing the where. Correspondingly, the decoder uses upsampling to expand the shrunk feature maps to their original resolution to recover lost spatial location. Training FCNs requires dense labeling of each pixel, which is not available for the biopsy data in our case. Here, we first introduce our model architectures, and then we propose to use a sparse loss function evaluated only on the biopsy-proven locations for training.
Model architecture
We employ a two-dimensional U-Net architecture for assigning class-specific probabilities to each pixel in our data. Skip connections from intermediate-low level features of encoder to decoder are shown to be beneficial for semantic segmentation since upsampling is a sparse operation. Additionally, we perform experiments with attention-based U-Nets [22,26]. Oktay et al. proposed Attention U-Nets by incorporating attention gates into a standard U-Net architecture and performed pancreas segmentation [22]. An attention mechanism is often utilized to allow the decoder to automatically identify and focus on relevant information from the lower-level encoder feature maps. As stated in the preprocessing section, our input size and output size for both modalities are 256 × 256 × 1 and 256 ×256 × 2, respectively.
Unimodal architecture
For training on each modality alone, we use five blocks in the encoding path of our U-Net. Each block includes 2D convolutions with kernel size of 3 × 3, followed by Batch Normalization (BN) and ReLU activation. Before moving to the next block, the features are down-sampled by 2 × 2 max-pooling to extract spatially invariant features. The number of convolutional filters for our encoder stage is 14, 28, 56, 112, and at the last scale 224. Correspondingly at the decoding stage, we replace max-pooling in each block with upsampling layers to expand the features. Intermediate features from the encoder are concatenated with the upsampled features, and mixed with 2D convolutions after. Finally, we use a SoftMax layer at the end to generate class probability distribution for Normal and Cancer for each pixel. Using our unimodal structures trained on TeUS and mp-MRI separately, we experiment late fusion strategy for integrating the information in both modalities. More specifically, we average the output of each modality for the defined ROI around the target location.
Multimodal architecture
Considering the significant differences in the tissue appearance in MRI and TeUS, instead of fusing the information as channels in the input, we used a dual-stream U-Net structure to implement an intermediate fusion of the information between modalities. Inspired by [13], our multimodal architecture for the integration of TeUS and MRI employs a two-stream of 2D U-Nets, which share the final encoding layer as depicted in Fig. 2. More specifically, each modality is fed into a separate encoder network. After four scales of separate convolutional and maxpooling layers (similar to unimodal architecture explained earlier), both streams share weights at the last scale of encoder. This can be interpreted as a shared (intermediate) representation between MRI and TeUS (shared what). Next, this shared representation is fed into separate decoders to recover spatial localization of where for each modality separately. We used separate decoders in particular since the field of view in each modality is different; using the same decoder will result in an undesirable mixture of spatial information between modalities at output.
Fig. 2.
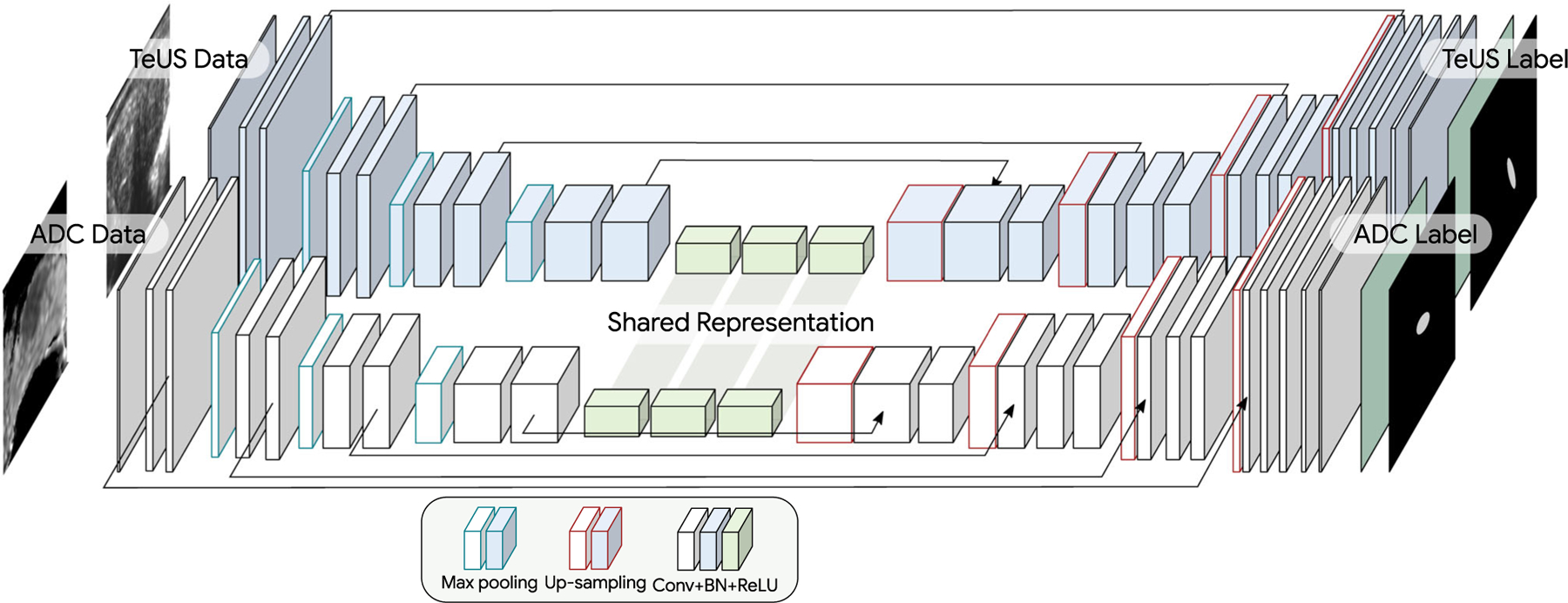
Our multimodal U-Net architecture consists of a modality-specific encoder and a modality-specific decoder with shared latent representation
Training
As explained, training FCNs requires pixel-level class labels, which is missing in the biopsy data. Calculating the loss for labeled data only is referred to as partial cross-entropy, in the segmentation literature [30]. Tang et al. [30] showed that using partial cross-entropy, more than 80% of the performance can be achieved, compared to using the full labeled data for training. Here, we explain our strategy for training FCN models on biopsy data. We consider a supervised training problem for segmentation of the cancer with data: D = {(I i , Si , zi ), ...}, where I is a collection of voxel intensities, Si denotes the sparse locations in the image (biopsy targets) for which we have label z j ∈ 0, 1 representing the histopathology result (healthy or cancer).
Let p(z j = 1 I,θ) be the probability of cancer tissue at location j for image I parameterized by θ . A standard classifier can be trained by maximum likelihood (ML) parameter estimation, which is also known as cross-entropy (CE) in machine learning:
| (1) |
Here, the location of the biopsy targets j in the dataset is considered as known values, and includes the biopsy target and an approximated vicinity of it (see “Preprocessig” section). In summary, we utilize a partial CE loss and optimize the weights of the network based on the loss calculated only at the locations that we have label for. The model is trained with mini-batch stochastic gradient descent with Adam update rule. The initial learning rate is set to 10−3 and exponentially decayed to reach its 0.75 value every 10 epochs. Networks are trained in an end-to-end manner. For multimodality training, final loss is the summation of each modality loss function.
Final label for a biopsy location is calculated by averaging the probability of cancer in the defined ROI around the biopsy target. We perform quantitative analysis of the performance of models by calculating AUC for each experiment.
Results and discussion
Our qualitative results are shown in Fig. 3 depicting the cancer likelihood maps for a benign (two right columns of Fig. 3) and cancer target (two left columns of Fig. 3) based on U-Net predictions (top row) and Attention U-Net predictions (bottom row). The range of the colormap is from blue to red, representing the likelihood of benign and cancer, respectively. We observe that for the biopsy-proven cancer core, the multimodality trained network with intermediate fusion of information in TeUS and MRI has successfully identified the location of the cancerous region in each space (red means higher cancer likelihood). Moreover, as shown for the biopsy-proven benign core, the target location in MRI has been predicted as benign, and on TeUS, we can observe the majority benign near the target. It should be noted that although the clinician performing the biopsy was aiming for the axial plane in ultrasound in order to match pre-biopsy mp-MRI, there is no guarantee that the two imaging planes depicted in Fig. 3 are exactly aligned. The images of the two modalities have the best alignment at the biopsy target locations.
Fig. 3.
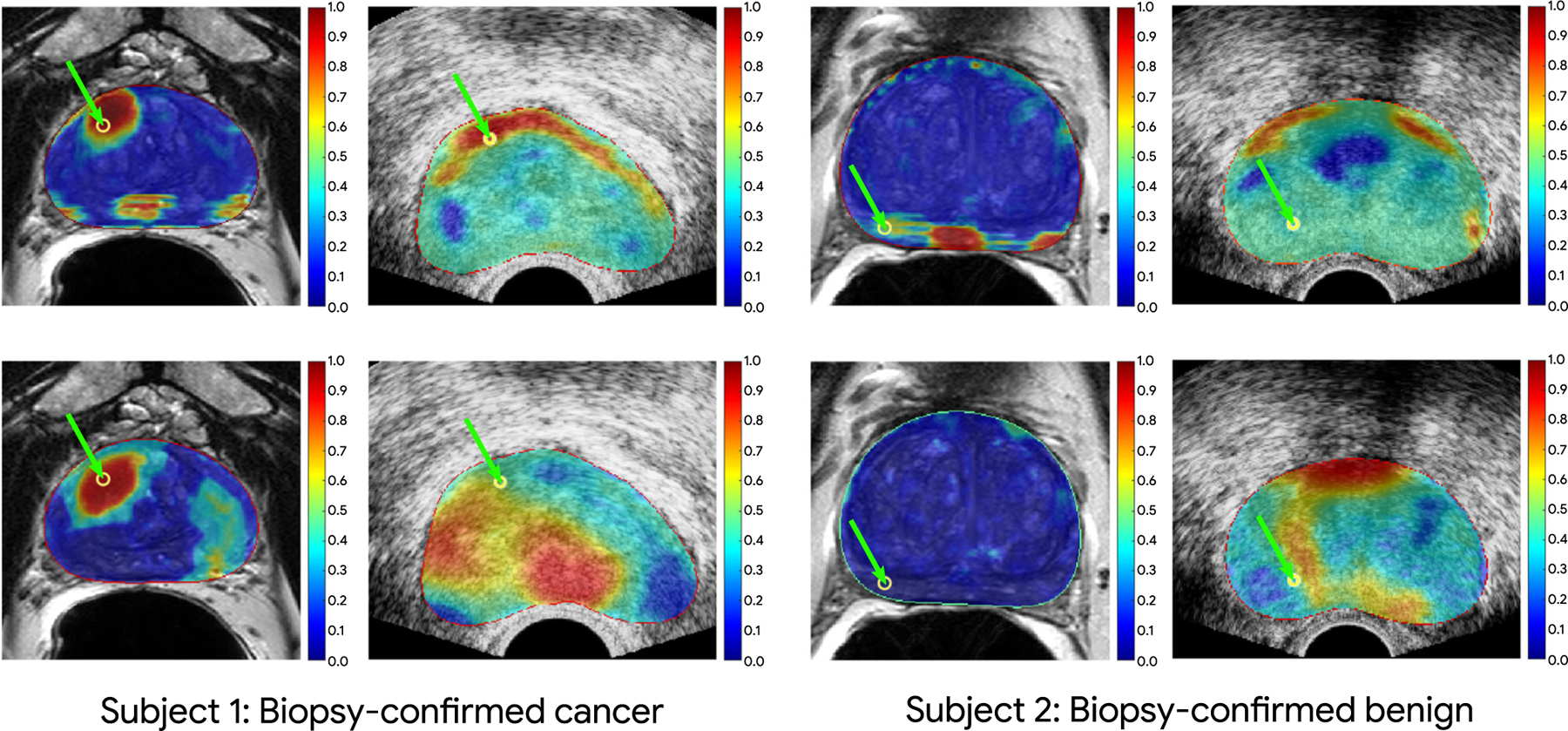
Color maps of cancer prediction from our intermediate fusion models overlaid on ultrasound and MRI for a biopsy-confirmed cancerous case (on the left) and a biopsy-confirmed normal case (on the right). The first and second rows for each case depict the prediction of the U-Net and Attention U-Net models, respectively. The locations of green arrows for each image show the corresponding fusion biopsy target between MRI and TeUS. The image registration accuracy should be the highest at the biopsy target
Results in Table 1 provide an overview of different experiments performed with U-Net and Attention U-Net architectures for uni- and multimodality training. The experiments on the multimodal training with both TeUS and MRI suggested improvement over training with each modality alone for the U-Net and Attention U-Net. As seen in Table 1, the AUC of the late fusion model with U-Net is closer to the AUC of the ADC model since the final results are averages of the unimodality probabilities in this approach. Moreover, the increased performance of Attention U-Net model on unimodality training led to an increased late fusion performance compared to U-Net experiment. We also performed a grid search over the possible weighting combinations of the MRI and TeUS outputs in our validation set; however, no significant improvements were found based on weighted averaging.
Table 1.
Quantitative results of the area under the curve (AUC) for different experiments for all the test set (All), and for filtered test set including benign subjects + cancerous subjects with Tumor in Core Length (TIL) greater than 2 (TI L > 2) and 4 (TI L > 4), respectively
| Experiments | Area under the curve (AUC) |
||
|---|---|---|---|
| All | TIL > 2 mm | TIL > 4 mm | |
| U-Net | |||
| Unimodal | |||
| ADC | 0.69 | 0.78 | 0.88 |
| TeUS | 0.66 | 0.73 | 0.80 |
| Multimodal | |||
| Late fusion | 0.70 | 0.80 | 0.89 |
| Intermediate fusion | 0.76 | 0.82 | 0.88 |
| Attn U-Net | |||
| Unimodal | |||
| ADC | 0.70 | 0.79 | 0.89 |
| TeUS | 0.70 | 0.75 | 0.77 |
| Multimodal | |||
| Late fusion | 0.73 | 0.79 | 0.89 |
| Intermediate fusion | 0.75 | 0.83 | 0.89 |
We performed statistical analysis with bootstrapping (n = 1000) and calculated the 95% confidence intervals (CI) for the AUCs in intermediate fusion experiments for both U-Net and Attention U-Net architectures. The results are shown in Fig. 4. As seen, due to the limited number of samples in the training set, the width of CIs are large. While AUC values increased for all TIL, improvements were more pronounced for smaller TIL. Our hypothesis is that with a larger dataset for training, the differences between the performance of unimodal and multimodal techniques will be further increased.
Fig. 4.
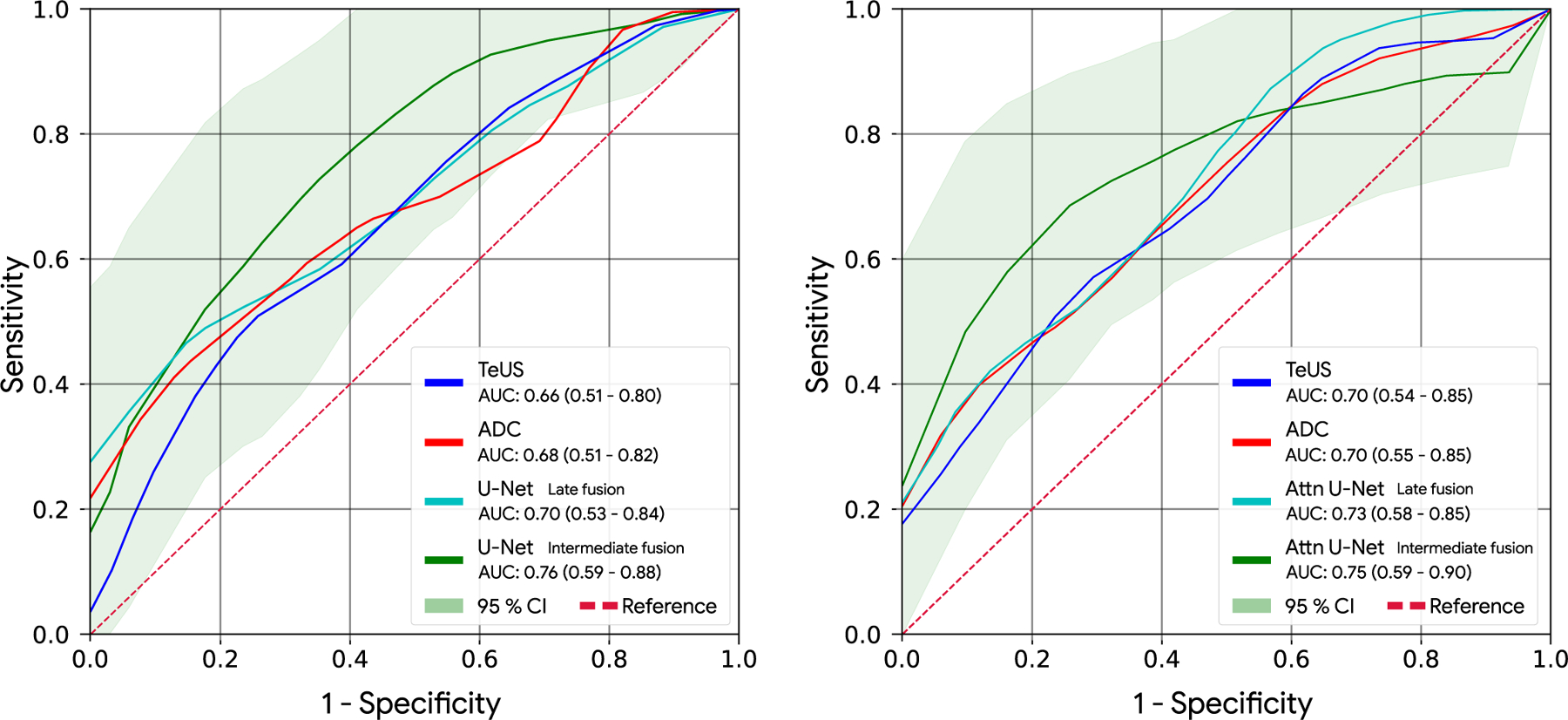
AUC of unimodal TeUS, and ADC models along with AUC of multimodal fusion experiments for U-Net (left) and Attention U-Net (right). The 95% confidence interval region is depicted for the intermediate fusion experiments. The results represent the significant improvement of multimodal training
Further research needs to be performed to investigate the full potential of multimodal training to improve targeting of biopsy in PCa. For instance, in practice, many patients may have missing data from one modality. Several groups [20,21] have proposed different strategies for training that can deal with missing data from one of the modalities at test-time. Given that TRUS-guided biopsy is the current standard-of-care, these strategies could have significant potential for centers that do not include pre-biopsy mp-MRI in their care plan.
Finally, as mentioned previously, our deep FCN model structure in both unimodal and multimodal training enables fast and computationally efficient prediction at test time. As an example, the computation time for our most complex model (multimodal intermediate fusion) is 44 ms on NVIDIA GTX 1080 Ti GPU.
Conclusion and future directions
In this paper, we demonstrated that we can improve targeting of PCa biopsies through generation of cancer likelihood maps using information fusion between MRI and TeUS. We utilized different FCN architectures to train models for PCa detection using sparse biopsy data. We investigated strategies for information fusion between TeUS and MRI, and showed superior performance of using the two-stream U-Net architecture with shared representation. Our approach is computationally efficient and can predict cancer likelihoods for each pixel of ultrasound in near real time. Future directions of this study include investigation of the transfer-learned networks from publicly available mp-MRI data, and different strategies in training for handling missing modalities. Additionally, alternative late fusion mechanisms, e.g., nonlinear modeling of outputs, will be investigated in the future.
Acknowledgements
We would like to thank Nvidia Corp. for the donated GPUs.
This work is funded in part by the Natural Sciences and Engineering Research Council of Canada (NSERC) and in part by the Canadian Institutes of Health Research (CIHR).
Footnotes
Compliance with ethical standards
Conflict of interest A. Sedghi, A. Mehrtash, A. Jamzad, A. Amalou, W. Wells III, T. Kapur, J.T. Kwak, B. Turkbey, P. Choyke, P. Pinto, B. Wood, S. Xu, P. Abolmaesumi, and P. Mousavi confirm that there are no known conflicts of interest with this publication.
Ethical approval All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Informed consent Informed consent was obtained from all individual participants included in the study.
References
- 1.Ahmed HU, Bosaily AES, Brown LC, Gabe R, Kaplan R, Parmar MK, Collaco-Moraes Y, Ward K, Hindley RG, Freeman A, Kirkham AP, Oldroyd R, Parker C, Emberton M (2017) Diagnostic accuracy of multi-parametric mri and trus biopsy in prostate cancer PROMIS: a paired validating confirmatory study. Lancet 389(10071):815–822 [DOI] [PubMed] [Google Scholar]
- 2.Azizi S, Bayat S, Yan P, Tahmasebi AM, Nir G, Kwak JT, Xu S, Wilson S, Iczkowski KA, Lucia MS, Goldenberg L, Salcudean SE, Pinto PA, Wood BJ, Abolmaesumi P, Mousavi P (2017) Detection and grading of prostate cancer using temporal enhanced ultrasound: combining deep neural networks and tissue mimicking simulations. Int J Comput Assisted Radiol Surg 12:1293–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen Q, Xu X, Hu S, Li X, Zou Q, Li Y (2017) A transfer learning approach for classification of clinical significant prostate cancers from mpMRI scans. In: Medical imaging 2017: computer-aided diagnosis, vol 10134. International Society for Optics and Photonics, p 101344F [Google Scholar]
- 4.Correas JM, Tissier AM, Khairoune A, Khoury G, Eiss D, Hélénon O (2013) Ultrasound elastography of the prostate: state of the art. Diagn Interv Imaging 94(5):551–560 [DOI] [PubMed] [Google Scholar]
- 5.Fedorov A, Beichel RR, Kalpathy-Cramer J, Finet J, Fillion-Robin JC, Pujol S, Bauer C, Jennings D, Fennessy F, Sonka M, Buatti JM, Aylward SR, Miller JV, Pieper S, Kikinis R (2012) 3D slicer as an image computing platform for the quantitative imaging network. Magn Reson Imaging 30(9):1323–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feleppa E, Porter C, Ketterling J, Dasgupta S, Ramachandran S, Sparks D (2007) Recent advances in ultrasonic tissue-type imaging of the prostate. In: André MP et al. (eds) Acoustical imaging, vol 28. Springer, Berlin, pp 331–339 [Google Scholar]
- 7.Feleppa EJ, Ketterling JA, Kalisz A, Urban S, Porter CR, Gillespie JW, Schiff PB, Ennis RD, Wuu CS, Fair WR (2001) Advanced ultrasonic tissue-typing and imaging based on radio-frequency spectrum analysis and neural-network classification for guidance of therapy and biopsy procedures. Int Cong Ser 1230:346–351 [Google Scholar]
- 8.Havaei M, Guizard N, Chapados N, Bengio Y (2016) Hemis: Hetero-modal image segmentation In: International conference on medical image computing and computer-assisted intervention. Springer, pp 469–477 [Google Scholar]
- 9.Imani F, Abolmaesumi P, Gibson E, Khojaste A, Gaed M, Moussa M, Gomez JA, Romagnoli C, Leveridge MJ, Chang SD, Siemens R, Fenster A, Ward AD, Mousavi P (2015) Computer-aided prostate cancer detection using ultrasound RF time series: in vivo feasibility study. IEEE Trans Med Imaging 34:2248–2257 [DOI] [PubMed] [Google Scholar]
- 10.Imani F, Ghavidel S, Abolmaesumi P, Khallaghi S, Gibson E, Khojaste A, Gaed M, Moussa M, Gomez JA, Romagnoli C, Cool DW, Bastian-Jordan M, Kassam Z, Siemens DR, Leveridge MJ, Chang SD, Fenster A, Ward AD, Mousavi P (2016) Fusion of multi-parametric MRI and temporal ultrasound for characterization of prostate cancer: in vivo feasibility study. In: Medical imaging 2016: computer-aided diagnosis, vol 9785. International Society for Optics and Photonics, p 97851K [Google Scholar]
- 11.Imani F, Ramezani M, Nouranian S, Gibson E, Khojaste A, Gaed M, Moussa M, Gomez JA, Romagnoli C, Leveridge MJ, Chang SD, Fenster A, Siemens R, Ward AD, Mousavi P, Abolmaesumi P (2015) Ultrasound-based characterization of prostate cancer using joint independent component analysis. IEEE Trans Biomed Eng 62:1796–1804 [DOI] [PubMed] [Google Scholar]
- 12.Kiraly AP, Nader CA, Tuysuzoglu A, Grimm R, Kiefer B, El-Zehiry N, Kamen A (2017) Deep convolutional encoder-decoders for prostate cancer detection and classification In: International conference on medical image computing and computer-assisted intervention. Springer, pp 489–497 [Google Scholar]
- 13.Kuga R, Kanezaki A, Samejima M, Sugano Y, Matsushita Y (2017) Multi-task learning using multi-modal encoder-decoder networks with shared skip connections In: Proceedings of the IEEE international conference on computer vision, pp 403–411 [Google Scholar]
- 14.Liu S, Zheng H, Feng Y, Li W (2017) Prostate cancer diagnosis using deep learning with 3D multiparametric mri. In: Medical imaging 2017: computer-aided diagnosis, vol 10134. International Society for Optics and Photonics, p 1013428 [Google Scholar]
- 15.Long J, Shelhamer E, Darrell T (2015) Fully convolutional networks for semantic segmentation In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 3431–3440 [DOI] [PubMed] [Google Scholar]
- 16.Mehrtash A, Pesteie M, Hetherington J, Behringer PA, Kapur T, Wells III WM, Rohling R, Fedorov A, Abolmaesumi P (2017) Deepinfer: Open-source deep learning deployment toolkit for image-guided therapy. In: Proceedings of SPIE–the international society for optical engineering, vol 10135. NIH Public Access; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mehrtash A, Sedghi A, Ghafoorian M, Taghipour M, Tempany CM, Wells WM, Kapur T, Mousavi P, Abolmaesumi P, Fedorov A (2017) Classification of clinical significance of MRI prostate findings using 3D convolutional neural networks. In: SPIE medical imaging International Society for Optics and Photonics, pp 101342A–101342A–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moradi M, Abolmaesumi P, Siemens DR, Sauerbrei EE, Boag AH, Mousavi P (2009) Augmenting detection of prostate cancer in transrectal ultrasound images using SVM and RF time series. IEEE Trans Biomed Eng 56(9):2214–2224 [DOI] [PubMed] [Google Scholar]
- 19.Nahlawi L, Imani F, Gaed M, Gomez JA, Moussa M, Gibson E, Fenster A, Ward AD, Abolmaesumi P, Mousavi P, Shatkay H (2015) Using hidden markov models to capture temporal aspects of ultrasound data in prostate cancer In: 2015 IEEE international conference on bioinformatics and biomedicine (BIBM) pp 446–449 [Google Scholar]
- 20.Ngiam J, Khosla A, Kim M, Nam J, Lee H, Ng AY (2011) Multimodal deep learning In: Proceedings of the 28th international conference on machine learning (ICML-11), pp 689–696 [Google Scholar]
- 21.Oktay O, Ferrante E, Kamnitsas K, Heinrich M, Bai W, Caballero J, Cook SA, de Marvao A, Dawes T, O’Regan DP, Kainz B, Glocker B, Rueckert D (2018) Anatomically constrained neural networks (ACNNs): application to cardiac image enhancement and segmentation. IEEE Trans Med Imaging 37(2):384–395. 10.1109/TMI.2017.2743464 [DOI] [PubMed] [Google Scholar]
- 22.Oktay O, Schlemper J, Folgoc LL, Lee M, Heinrich M, Misawa K, Mori K, McDonagh S, Hammerla NY, Kainz B, others Glocker B, Rueckert D (2018) Attention u-net: Learning where to look for the pancreas. arXiv preprint arXiv:1804.03999 [Google Scholar]
- 23.Puech P, Rouvière O, Renard-Penna R, Villers A, Devos P, Colombel M, Bitker MO, Leroy X, Mege-Lechevallier F, Compérat E, Ouzzane A, Lemaitre L (2013) Prostate cancer diagnosis: multiparametric mr-targeted biopsy with cognitive and transrectal us-mr fusion guidance versus systematic biopsy-prospective multicenter study. Radiology 268(2):461–9 [DOI] [PubMed] [Google Scholar]
- 24.Ronneberger O, Fischer P, Brox T (2015) U-net: convolutional networks for biomedical image segmentation In: International conference on medical image computing and computer-assisted intervention. Springer, pp 234–241 [Google Scholar]
- 25.Schelb P, Kohl S, Radtke JP, Wiesenfarth M, Kickingereder P, Bickelhaupt S, Kuder TA, Stenzinger A, Hohenfellner M, Schlemmer HP, Maier-Hein KH, Bonekamp D (2019) Classification of cancer at prostate MRI: deep learning versus clinical PI-RADS assessment. Radiology 293(3):607–617 [DOI] [PubMed] [Google Scholar]
- 26.Schlemper J, Oktay O, Schaap M, Heinrich M, Kainz B, Glocker B, Rueckert D (2019) Attention gated networks: learning to leverage salient regions in medical images. Med Image Anal 53:197–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sedghi A, Pesteie M, Javadi G, Azizi S, Yan P, Kwak JT, Xu S, Turkbey B, Choyke P, Pinto P, Wood B, Rohling R, Abolmaesumi P, Mousavi P (2019) Deep neural maps for unsupervised visualization of high-grade cancer in prostate biopsies. Int J Comput Assisted Radiol Surg 14(6):1009–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siddiqui MM, Rais-Bahrami S, Turkbey B, George AK, Roth-wax JT, Shakir NA, Okoro C, Raskolnikov D, Parnes HL, Linehan WM, Merino MJG, Simon RM, Choyke PL, Wood BJ, Pinto PA (2015) Comparison of mr/ultrasound fusion-guided biopsy with ultrasound-guided biopsy for the diagnosis of prostate cancer. JAMA 313(4):390–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sonn GA, Chang E, Natarajan S, Margolis DJ, Macairan M, Lieu P, Huang J, Dorey FJ, Reiter RE, Marks LS (2014) Value of targeted prostate biopsy using magnetic resonance-ultrasound fusion in men with prior negative biopsy and elevated prostate-specific antigen. Eur Urol 65(4):809–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang M, Djelouah A, Perazzi F, Boykov Y, Schroers C (2018) Normalized cut loss for weakly-supervised cnn segmentation In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 1818–1827 [Google Scholar]
- 31.Turkbey B, Rosenkrantz AB, Haider MA, Padhani AR, Villeirs G, Macura KJ, Tempany CM, Choyke PL, Cornud F, Margolis DJ, Thoeny HC, Verma S, Barentsz J, Weinreb JC (2019) Prostate imaging reporting and data system version 2.1: 2019 update of prostate imaging reporting and data system version 2. Eur Urol 76(3):340–351. 10.1016/j.eururo.2019.02.033 [DOI] [PubMed] [Google Scholar]
- 32.Valindria VV, Pawlowski N, Rajchl M, Lavdas I, Aboagye EO, Rockall AG, Rueckert D, Glocker B (2018) Multi-modal learning from unpaired images: application to multi-organ segmentation in ct and MRI In: 2018 IEEE winter conference on applications of computer vision (WACV), pp 547–556. IEEE [Google Scholar]
- 33.Yerram NK, Volkin D, Turkbey B, Nix J, Hoang AN, Vourganti S, Gupta GN, Linehan WM, Choyke PL, Wood BJ, Pinto P (2012) Low suspicion lesions on multiparametric magnetic resonance imaging predict for the absence of high-risk prostate cancer. BJU Int 110(11b):E783–E788 [DOI] [PMC free article] [PubMed] [Google Scholar]


