Abstract
Genes encoding the proteins required for clavulanic acid biosynthesis and for cephamycin biosynthesis are grouped into a “supercluster” in Streptomyces clavuligerus. Nine open reading frames (ORFs) associated with clavulanic acid biosynthesis were located in a 15-kb segment of the supercluster, including six ORFs encoding known biosynthetic enzymes or regulatory proteins, two ORFs that have been reported previously but whose involvement in clavulanic acid biosynthesis is unclear, and one ORF not previously reported. Evidence for the involvement of these ORFs in clavulanic acid production was obtained by generating mutants and showing that all were defective for clavulanic acid production when grown on starch asparagine medium. However, when five of the nine mutants, including mutants defective in known clavulanic acid biosynthetic enzymes, were grown in a soy-based medium, clavulanic acid-producing ability was restored. This ability to produce clavulanic acid when seemingly essential biosynthetic enzymes have been mutated suggests that paralogous genes encoding functionally equivalent proteins exist for each of the five genes but that these paralogues are expressed only in the soy-based medium. The five genes that have paralogues encode proteins involved in the early steps of the pathway common to the biosynthesis of both clavulanic acid and the other clavam metabolites produced by this organism. No evidence was seen for paralogues of the four remaining genes involved in late, clavulanic acid-specific steps in the pathway.
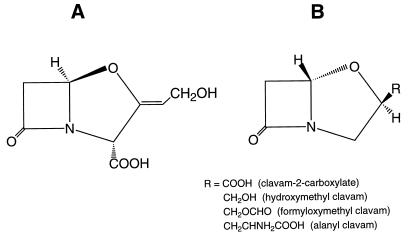
Streptomyces clavuligerus produces an array of β-lactam compounds including cephamycin C, clavulanic acid, and at least four other clavam metabolites. Clavulanic acid has considerable chemotherapeutic and economic value because of its β-lactamase-inhibitory activity. In contrast, the other clavam metabolites are ineffective as β-lactamase inhibitors. These clavam metabolites have the same nuclear structure, a fused bicyclic β-lactam–oxazolidine ring system, as does clavulanic acid. However, the stereochemistry of the ring system is 5R in the clavams rather than 5S as in clavulanic acid, and the clavam metabolites also have different side chain substituents at C-2 (Fig. 1).
FIG. 1.
Structures of the clavam metabolites produced by Streptomyces clavuligerus. (A) Clavulanic acid; (B) other clavam metabolites.
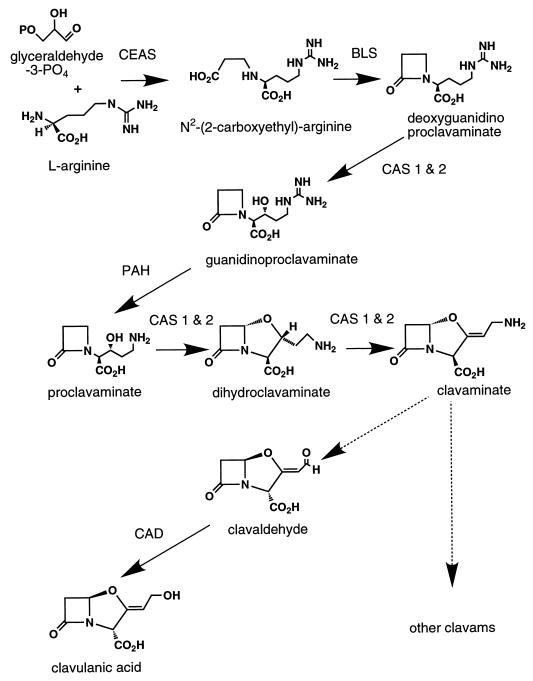
The biosynthetic pathway leading to clavulanic acid has been partially elucidated (Fig. 2), and evidence has been obtained to indicate that both clavulanic acid and the antipodal clavam metabolites share a common pathway, at least to the level of proclavaminate or clavaminate (5, 9). The structural similarity between clavulanic acid and the other clavam metabolites, together with the evidence for a shared pathway, has led to the assumption that these compounds comprise a family of biosynthetically related metabolites.
FIG. 2.
Biosynthetic pathway leading to clavulanic acid and the other clavams. CEAS, N-carboxyethylarginine synthase; BLS, β-lactam synthetase; PAH, proclavaminate amidinohydrolase; CAS, clavaminate synthase; CAD, clavaldehyde dehydrogenase.
The genes encoding enzymes involved in clavulanic acid biosynthesis are known to be clustered in S. clavuligerus and located adjacent to the genes involved in cephamycin C biosynthesis (1, 7, 23). In early studies, we localized the gene that encodes proclavaminate amidinohydrolase (pah) and showed that clavulanic acid production on starch asparagine (SA) medium was lost when pah was disrupted (1). Involvement in clavulanic acid biosynthesis was further substantiated when the gene encoding one of the isozymes of clavaminate synthase (cas2 [11]) was found to be located immediately downstream of pah. In subsequent studies, additional genes involved in clavulanic acid biosynthesis have been located through sequence analysis of DNA on either side of pah and cas2. The gene orf2 (S. E. Jensen, K. A. Aidoo, and A. S. Paradkar, U. S. patent application 08/134,018) has very recently been shown to encode a carboxyethylarginine synthase (10) responsible for condensing 3-phosphoglyceraldehyde with arginine as the first dedicated step in the clavulanic acid biosynthetic pathway. The gene bls (7) encodes a β-lactam synthetase which forms the monocyclic β-lactam ring at an early stage in the pathway (2, 12), while the gene cad (18) encodes clavaldehyde dehydrogenase, which oxidizes clavaldehyde to clavulanic acid as the final step in the pathway (14). Late pathway steps involved in the inversion of stereochemistry of the bicyclic ring nucleus and modification of the C-2 side chain have yet to be elucidated, and candidate genes have not been identified. Similarly, the biosynthetic enzymes and corresponding genes responsible for the formation of the clavam metabolites are unknown.
An intriguing aspect of this biosynthetic gene cluster is the observation that cas2 is only one of a pair of paralogous biosynthetic genes that encode very similar enzymes (11). The location of cas1, the second of this pair of genes, is unknown, but it is more than 25 kb away from cas2. The rationale for having two separate genes encoding isozymes with very similar substrate specificities and kinetic properties is unclear. However, the two genes are known to be regulated differently, since cas1 is expressed in a soy-based (Soy) medium but not in SA medium, whereas cas2 is expressed in both media (17). Hypotheses put forward to explain the existence of two genes have included the suggestion that two genes may be required to ensure sufficient supplies of this potentially rate-limiting enzyme, since clavaminate synthase is responsible for three separate steps in the pathway. Alternatively, it has been suggested that one form of Cas may be associated with the biosynthesis of clavulanic acid while the second form is involved in the biosynthesis of other clavam metabolites. In this regard, Mosher et al. (13) have recently shown that the DNA sequence flanking cas1 contains a number of open reading frames (ORFs) and that disruption of these genes interferes specifically with the production of the clavam metabolites, while clavulanic acid production is unaffected.
In the present study we show that, like cas, several more of the genes involved in early steps of clavulanic acid biosynthesis have paralogues located elsewhere in the genome and that these paralogues are associated with the biosynthesis of the clavam metabolites rather than with that of clavulanic acid.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
S. clavuligerus NRRL 3585 was obtained from the Northern Regional Research Laboratories, Peoria, Ill., and maintained either on MYM agar (21) or on ISP medium no. 3 (Difco, Detroit, Mich.). Escherichia coli MV1193 (25), used as the host strain for all of the cloning and subcloning experiments, was grown in Luria broth (LB [19]) or on LB agar plates containing ampicillin (50 μg/ml) or tetracycline (10 μg/ml). The indicator organism E. coli ESS was obtained from A. L. Demain, Massachusetts Institute of Technology, Cambridge), Staphylococcus aureus N2 was obtained from the Department of Biological Sciences, University of Alberta, and Bacillus sp. strain ATCC 27860 was obtained from the American Type Culture Collection, Manassas, Va.
Plasmid vectors pUC118 and pUC119 were obtained from J. Vieira (22), and pSL1180 was obtained from Amersham Pharmacia Biotech (Baie d'Urfe, Quebec, Canada). Plasmid pFDNEO-S contains a bifunctional E. coli-Streptomyces aminoglycoside resistance gene (3). Plasmid pCAT is a pUC119 derivative that contains a chloramphenicol resistance gene inserted into the ampicillin resistance marker (4). The Streptomyces vector pJOE829 was generously provided by J. Altenbucher (1); pIJ702 was obtained from the American Type Culture Collection, and pIJ486 was obtained from D. Hopwood, John Innes Institute.
Spores of S. clavuligerus were inoculated into seed medium consisting of Trypticase soy broth containing 1% starch and were incubated for 48 h at 28°C on a rotary shaker (250 rpm). Seed cultures were harvested by centrifugation at 10,000 × g for 10 min, and the mycelia were washed with sterile distilled water and then used to inoculate a defined SA medium (17) or a complex Soy medium (17) to 2% (vol/vol). The production cultures were incubated under the same conditions as those used for seed cultures.
DNA isolation, manipulation, and Southern analyses.
Plasmid and genomic DNA preparations from Streptomyces spp. were isolated as described by Hopwood et al. (8). Protoplasts of S. clavuligerus were prepared, transformed, and regenerated as described previously (17). Plasmid DNA isolation from E. coli cultures, restriction enzyme digestion and analysis, and ligation and transformation of E. coli were all performed using standard techniques (19).
DNA sequencing and analysis.
Sequence information was obtained for approximately 15 kb of S. clavuligerus DNA extending downstream from pcbC and ending at a BglII site. Ordered sets of deletions were generated (19) using fragments of the DNA insert from the cosmid clone K6L2 (1) and subcloned into the E. coli plasmids pUC118 and pUC119. The deletion-generated fragments were sequenced in both orientations by the dideoxynucleotide chain termination method of Sanger et al. (20), using Sequenase (version 2.0) DNA polymerase and commercially available universal primers. Areas of compression in the sequence band pattern were relieved by carrying out reactions using 7-deaza-dGTP in place of dGTP. The nucleotide sequence data were analyzed for the presence of restriction sites, ORFs, and codon usage by the PC-gene programme (Intelligenetics Corp.). Similarity searches were accomplished using the online BLAST program at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/) and the online FASTA program available through European Bioinformatics Institute (http://www2.ebi.ac.uk/fasta3).
Insertional inactivation of ORFs in the 15 kb of DNA sequence.
In order to determine the roles of the various ORFs contained within the 15 kb of DNA, mutants were constructed by a gene replacement procedure based on that described by Paradkar and Jensen (17).
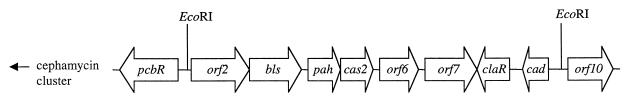
Disruption of bls, cas2, orf6, and claR was accomplished using an approximately 12-kb EcoRI fragment subcloned from the cosmid K6L2 into pCAT to give pCATL2. The location of the 12-kb EcoRI fragment within the 15-kb sequenced region is shown in Fig. 3. The 12-kb fragment contains four NcoI sites located within the bls, cas2, orf6, and claR genes. Insertion of an apramycin resistance gene (apr) cassette, modified by adding NcoI restriction sites to both ends, into each of the four NcoI sites individually resulted in a series of four plasmids with disruptions in either bls, cas2, orf6, or claR. From each of these plasmids, a smaller fragment carrying only the disrupted gene and some flanking sequence was subcloned and finally inserted into the Streptomyces vector pIJ486 for transformation into S. clavuligerus. In the case of bls, the disrupted gene was subcloned as a 4-kb EcoRI-KpnI fragment. In the case of orf6, an 8-kb BglII fragment carrying the disrupted gene was subcloned. Details for the creation of cas2 and claR disruption mutants have already been published elsewhere (16, 17).
FIG. 3.
Arrangement of ORFs in the clavulanic acid region of the supercluster.
Disruption of orf2 was accomplished by subcloning a 2.1-kb EcoRI-BglII fragment carrying the orf2 gene from cosmid K6L2 into pUC119. The apr cassette was modified by attachment of NotI-NcoI linker oligonucleotides to both ends, and the modified apr cassette was then inserted into the NotI site within orf2. Subsequently, the EcoRI-BglII fragment carrying the disrupted orf2 was inserted into pIJ486 for transformation into S. clavuligerus.
Disruption of orf7 was accomplished using the plasmid vector pSL1180 which had been modified to remove the NruI site from the multiple cloning site by digestion with enzymes flanking NruI and religation of the resulting plasmid. A 1.9-kb BglII-NcoI fragment of S. clavuligerus DNA encompassing the orf7 gene was then cloned into the modified pSL1180 plasmid vector. A neomycin resistance gene cassette was removed from pFDNEO-S, made blunt by treatment with the Klenow fragment of DNA polymerase, and introduced into the NruI site of the cloned orf7 gene. The Streptomyces plasmid pJOE829 was fused to the vector carrying the disrupted orf7 gene to create a shuttle vector for transformation into S. clavuligerus.
Disruption of cad was achieved using an apr cassette that was first cloned as an EcoRI/PstI fragment into the E. coli vector pBluescript and reisolated as an EcoRV/SmaI fragment. The cad gene was subcloned into pUC119 as a 2.4-kb BglII fragment encompassing both cad and orf10, and it was then disrupted by insertion of the apr cassette into a unique MscI site. Subsequently, the BglII fragment carrying the disrupted cad was transferred into pIJ486.
Disruption of orf10 was achieved using the same 2.4-kb BglII fragment of DNA cloned into pUC119 that was described above for the disruption of cad. A blunt-ended apr cassette (isolated as a SmaI/EcoRV fragment) was used to disrupt the orf10 gene by insertion into the BclI site after treatment with the Klenow fragment of DNA polymerase to blunt the overhanging ends. The disruption plasmid construct was converted to a shuttle vector by fusion with pIJ486 via the HindIII site.
Once the disruption gene constructs were in Streptomyces-compatible vectors, they were introduced into S. clavuligerus by protoplast transformation. Transformants were identified by growth on selective media corresponding to the resistance gene used to create the disruption. Transformants were then allowed to sporulate on nonselective media, and from the resulting spores, clones which retained the antibiotic resistance marker associated with the gene disruption, but which lost the antibiotic resistance marker associated with the plasmid vector, were identified by replica plating. The authenticity of the mutations in these clones was confirmed by Southern analysis of genomic DNA.
Disruption of pcbR and pah have already been described in previous studies (1, 15).
HPLC analysis of clavulanic acid and other clavam metabolites.
High-pressure liquid chromatography (HPLC) analyses were performed as described previously (13). In brief, samples of culture supernatants were derivatized with an imidazole reagent that reacts specifically with β-lactam-containing compounds. Derivatized samples were analyzed by HPLC under conditions which allow detection of clavulanic acid and the other clavam metabolites; they were compared to authentic standards of clavulanic acid, clavaminic acid, and clavaminic acid-2-carboxylate (all generously provided by SmithKline Beecham Pharmaceuticals). Although no standard was available for 2-hydroxymethylclavam, the relevant peak could also be identified based on its chromatographic properties relative to the known standards. Alanylclavam was not eluted under these chromatographic conditions and could be detected only by bioassay.
Bioassays of β-lactam antibiotics and of alanylclavam.
Bioassays were conducted by the agar plate disk diffusion method with appropriate indicator organisms. Total antibiotic was bioassayed using E. coli ESS as the indicator organism, clavulanic acid was bioassayed using S. aureus N2 as the indicator organism, and alanylclavam was bioassayed using Bacillus sp. strain ATCC 27860 as the indicator organism (17).
Nucleotide sequence accession number.
The sequence data obtained for approximately 15 kb of S. clavuligerus DNA have been deposited in GenBank under accession no. AF205427.
RESULTS
Nucleotide sequence of the clavulanic acid gene cluster.
The location of the clavulanic acid biosynthetic genes adjacent to the cephamycin gene cluster in S. clavuligerus was first reported by Ward and Hodgson (23). In parallel to these studies, we mapped pah to a region about 5.7 kb downstream of pcbC, the gene encoding isopenicillin N synthase (1). In view of the tight linkage of pah to the cephamycin cluster, we undertook a sequence analysis of the DNA flanking pah to look for additional genes encoding clavulanic acid biosynthetic enzymes. Ordered sets of deletion subclones were generated which extended from the 3′ end of pcbC downstream for a distance of about 15 kb, ending at a BglII site. The deletion-generated subclones were sequenced on both strands, and additional sequence information was obtained to cross all subclone junctions and ensure that no small fragments were inadvertently missed. This sequence information was originally documented as part of a patent application (Jensen et al., U.S. patent application 08/134,018) and has now been deposited in GenBank under accession no. AF205427.
The nucleotide sequence data were analyzed for the presence of ORFs. Ten ORFs were identified, originally named orf1 through orf10, but those for which functions are known have since been given appropriate gene designations. A diagrammatic representation of the region showing the relationship of the ORFs to each other and to the cephamycin cluster is shown in Fig. 3. Sequence information for a number of these genes has since been published in other studies. pcbR (15) encodes a penicillin binding protein responsible for the self-resistance of S. clavuligerus to its own β-lactam metabolites. As such, it is part of the cephamycin gene cluster and will not be considered further. bls (2, 7, 12), pah (1, 6, 7, 24), cas2 (7, 11), and cad (18) are all structural genes which encode known enzymes of the clavulanic acid biosynthetic pathway. The gene sequence for orf2 has not been published previously, except in the patent literature (Jensen et al., U.S. patent application 08/134,018), but the protein encoded by orf2 has recently been shown to exhibit carboxyethylarginine synthase activity, corresponding to the first step in the clavulanic acid biosynthetic pathway (10). claR (16, 18) encodes a transcriptional activator that specifically controls the production of enzymes responsible for the late steps of clavulanic acid biosynthesis. The complete sequence of orf6 and a partial sequence of orf7 have also been reported previously (referred to as orf4 and orf5 by Hodgson et al. [7]), but no gene designations have been assigned because their involvement in clavulanic acid biosynthesis is unclear. The putative proteins encoded by these genes show the greatest similarity to ornithine acetyltransferases (53.4% identity over 562 amino acids [aa] to the ornithine acetyltransferase from Rhodococcus fasciens, SWISS-PROT no. P96426) and oligopeptide binding proteins (36.8% identity over 562 aa to the putative oligopeptide-binding protein from Streptomyces coelicolor, SWISS-PROT no. 086572), respectively. Finally, orf10 represents new sequence information that extends our knowledge of the components of the clavulanic acid gene cluster. The putative Orf10 protein shows greatest similarity to cytochrome P450s (46.8% identity over 404 aa to the cytochrome P450 from Streptomyces griseolus, SWISS-PROT no. P18327). The characteristics of orf2 and orf10, genes which have not previously been described, and of orf7, for which additional sequence information now allows a complete description, are presented in Table 1.
TABLE 1.
Newly sequenced ORFs located in the 15-kb region downstream from pcbC
| ORF | Size (bp) | G+C content (%) | Molecular size of encoded protein (Da) | Similarity to known proteinsa |
|---|---|---|---|---|
| orf2 | 1,722 (573 aa) | 69 | 60,869 | N-Carboxyethylarginine synthasea |
| orf7b | 1,668 (555 aa) | 73 | 61,217 | Oligopeptide-binding proteins |
| orf10 | 1,227 (408 aa) | 72 | 45,009 | Cytochrome P450s |
Similarities were detected by database searching using the BLASTp program. orf2 has recently been shown to encode a carboxyethylarginine synthase.
A partial sequence for orf7 has been published previously (7).
The sequence information reported here was obtained independently from that reported by other groups, and while the agreement is generally good, some differences were noted between our data for claR and cad and those obtained by Perez-Redondo et al. (18). Careful examination of our data for the regions that showed discrepancies did not reveal any ambiguities, and so these may represent sequencing errors in the previously published data, or actual differences in DNA sequence between the strains used in the various studies.
Mutation of ORFs involved in clavulanic acid biosynthesis.
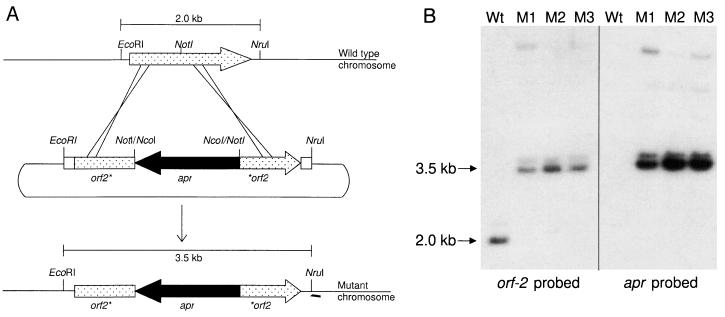
The involvement of each of the ORFs contained in the 15-kb region in clavulanic acid production was investigated by preparing gene disruption mutants and examining the effect on production. In each case, a cloned copy of the gene of interest was disrupted by insertion of an antibiotic resistance marker. The disrupted gene was then crossed into the chromosome by homologous recombination replacing the wild-type copy of the gene (17). Mutants were isolated based on their antibiotic resistance phenotype and then confirmed by Southern analysis. A diagrammatic representation of the gene disruption strategy is shown in Fig. 4A, using orf2 as an example. Southern analysis of the resulting mutants is shown in Fig. 4B. Genomic DNA isolated both from the wild type and from orf2 mutants was digested with EcoRI/NruI, separated by agarose gel electrophoresis, blotted onto nylon membranes, and then probed with a radiolabeled orf2 DNA fragment. Whereas wild-type DNA showed hybridization to a 2.0-kb EcoRI/NruI fragment, the orf2 mutant showed hybridization to a 3.5-kb fragment. The increased size of the hybridizing fragment corresponded to the 1.5-kb size of the apr cassette used to disrupt the gene. When the same blot was hybridized with a probe derived from the apr cassette, only the 3.5-kb band in the orf2 mutants was labeled. The orf2 mutant is designated orf2::apr to indicate that apr was used to create the disruption. In other cases, a thiostrepton resistance gene (tsr) or a neomycin resistance gene (neo) was used for the disruption, and those mutants are designated accordingly. In this way, orf2::apr, bls::apr, pah::tsr, cas2::apr, orf6::apr, orf7::neo, claR::apr, cad::apr, and orf10::apr mutants were created as described in Materials and Methods and confirmed by Southern analysis.
FIG. 4.
Disruption of the orf2 gene. (A) The stippled arrow represents orf2, the solid arrow represents the disrupting apramycin resistance cassette, and the open boxes represent DNA flanking orf2. The solid lines represent chromosomal or plasmid DNA. (B) Southern analysis of genomic DNA from S. clavuligerus orf2::apr mutants M1, M2, and M3 and from the wild-type strain using an orf2-specific probe and an apr-specific probe.
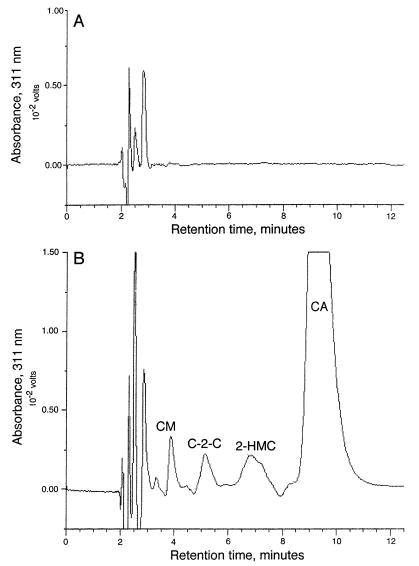
Each mutant was then assayed for production of clavulanic acid both by HPLC and by bioassay. All nine of the mutant strains failed to produce clavulanic acid when grown on SA medium, while the wild-type strain showed good clavulanic acid production. Examples of the HPLC profiles obtained for a representative bls::apr mutant and for the wild-type strain are shown in Fig. 5. The wild-type strain (Fig. 5B) showed a large peak due to clavulanic acid and small amounts of several of the other clavam metabolites. In contrast, the bls::apr mutant (Fig. 5A) showed no evidence of clavulanic acid or clavam production. These results agree with the findings of Bachmann et al. (2), who also prepared a bls disruption mutant and found that clavulanic acid production on SA medium was abolished.
FIG. 5.
HPLC analysis of culture filtrates from wild-type and bls::apr strains of S. clavuligerus grown on SA medium. Samples of culture filtrate were analyzed by HPLC either after derivatization with imidazole reagent or without derivatization. The data presented are difference chromatograms in which underivatized traces have been subtracted from the derivatized traces. (A) bls::apr mutant; (B) wild type. CA, clavulanic acid; 2-HMC, 2-hydroxymethyl clavam; C-2-C, clavam 2-carboxylate; CM, clavaminic acid.
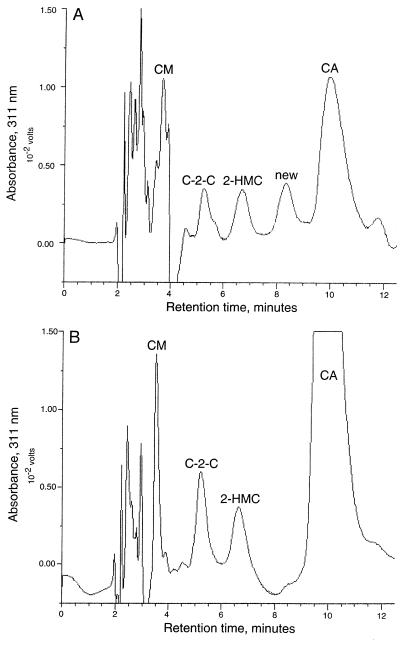
SA medium was used to cultivate the wild-type and mutant strains because it gives good clavulanic acid production. However, we knew from past studies that production of the other clavams is generally poor in SA medium. In order to assess the effect of these mutations on the production of the other clavam metabolites, the wild-type and mutant strains were cultivated on Soy medium, where production of the other clavam metabolites is much more pronounced. Culture filtrates were assayed for clavulanic acid by both bioassay and HPLC, for the other clavam metabolites by HPLC, and for alanylclavam by a bioassay procedure based on its methionine-antagonistic properties. HPLC profiles obtained for the bls::apr mutant and the wild-type strain grown on Soy medium are shown in Fig. 6. The wild-type culture (Fig. 6B) again produced very large amounts of clavulanic acid and lesser amounts of several of the other clavams. However, the bls::apr mutant (Fig. 6A) now regained some ability to produce clavulanic acid and the other clavams, as determined by both HPLC and bioassay. The bls::apr mutants also produced a new clavam peak that was not seen, or was seen only in small amounts, in the wild type.
FIG. 6.
HPLC analysis of culture filtrates from wild-type and bls::apr strains of S. clavuligerus grown on Soy medium. Samples of culture filtrate (diluted fivefold with water in the case of the wild-type culture, undiluted in the case of the bls::apr mutant) were analyzed by HPLC either after derivatization with imidazole reagent or without derivatization. The data presented are difference chromatograms in which underivatized traces have been subtracted from the derivatized traces. (A) bls::apr mutant; (B) wild type. For abbreviations, see the legend to Fig. 5.
Data for the entire set of mutants grown on both SA and Soy media are summarized and expressed as approximate percentages of the amount of metabolite seen in the wild-type strain grown on Soy medium (Table 2). Since all of the clavam metabolites other than clavulanic acid tended to behave as a group, they are summarized together. When grown on Soy medium, the orf2::apr, bls::apr, pah::tsr, cas2::apr, and orf6::apr mutants all regained the ability to produce clavulanic acid and the other clavams, at least to some degree. While the production of clavulanic acid and clavam metabolites by the cas2::apr mutant was understandable because of the presence of cas1, the observation of a similar phenomenon for the orf2::apr, bls::apr, pah::tsr, and orf6::apr mutants was unexpected. In the case of the orf2::apr mutant, the degree of restoration of clavulanic acid and clavam production was minimal, about 2 to 5% of wild-type levels. However, with other genes, the degree of restoration was substantial. bls::apr mutants produced both clavulanic acid and the other clavams at about 10 to 15% of wild-type levels (see Fig. 6). pah::tsr and cas2::apr mutants both produced large amounts of clavulanic acid, about 60% of wild-type levels, and correspondingly high levels of the other clavam metabolites. The orf6::apr mutant also showed good levels of clavulanic acid production, about 40% of that in the wild type, but all of the other clavams were absent except for trace amounts of alanylclavam (detected in the highly sensitive bioassay).
TABLE 2.
Production of clavulanic acid and the other clavam metabolites by mutants with defects in the ORFs located in the region downstream from pcbC
| Mutant | Amt of metabolite produceda on the indicated medium (% of wild type on Soy)
|
|||
|---|---|---|---|---|
| Clavulanic acid
|
Other clavams
|
|||
| SA | Soy | SA | Soy | |
| Wild type | 20 | 100 | 5–10 | 100 |
| orf2::apr | 0 | 2–5 | 0 | 2–5 |
| bls::apr | 0 | 10 | 0 | 15 |
| pah::tsr | 0 | 60 | NDb | 100 |
| cs2::apr | 0 | 60 | 0 | 60 |
| orf6::apr | 0 | 40 | 0 | 2–5c |
| orf7::neo | 0 | 0 | 0 | 40 |
| claR::apr | 0 | 0 | 0 | 80 |
| cad::apr | 0 | 0 | 0 | 80 |
| orf10::apr | 0 | 0 | 0 | 20 |
Metabolite production was estimated by bioassay (for clavulanic acid and alanylclavam) and by HPLC analysis (for clavulanic acid and the other clavams). For each metabolite, the amount of production seen in the wild type when grown on Soy medium was assigned a value of 100%.
ND, Not done.
None of the other clavams were seen by HPLC, but some alanylclavam was detected by bioassay.
In contrast to this observed restoration of clavulanic acid and clavam metabolite production when mutants defective in orf2, bls, pah, cas2, or orf6 were grown in Soy medium, mutants defective in orf7, claR, cad, or orf10 produced no clavulanic acid when grown in either SA or Soy medium. However, production of the other clavam metabolites persisted in these mutants, in amounts varying from 20 to 80% of wild-type levels. Production of cephamycin C was not appreciably affected in any of the mutants on either SA medium or Soy medium.
DISCUSSION
A 15-kb stretch of chromosomal DNA located downstream from pcbC in the cephamycin gene cluster of S. clavuligerus was subjected to sequence analysis. Ten ORFs were identified, one associated with self-resistance to penicillins and nine associated with clavulanic acid biosynthesis. orf2, bls, pah, cas2, cad, and claR are structural or regulatory genes already known to be involved in clavulanic acid biosynthesis. orf6 and part of orf7 have been sequenced previously, but it is unclear how they are involved in clavulanic acid biosynthesis (7). Finally, orf10, which resembles a cytochrome P450, is a new ORF that has not been described previously.
Evidence for the importance of this region in clavulanic acid production was obtained by creating mutants with disruptions of each of the ORFs and analyzing their phenotypes. Individual disruption of each of the ORFs resulted in a blockage of clavulanic acid production when the mutants were grown on SA medium, indicating that all were involved in the production of clavulanic acid. However, when the same mutants were grown on Soy medium, clavulanic acid production was restored at least partially for mutants defective in orf2, bls, pah, cas2, and orf6.
This pattern of failure to produce on SA medium, but ability to produce on Soy medium, is the same phenotype that was seen previously for cas2 mutants, and it suggests that, as for cas2, paralogues exist for each of these genes. These paralogues are regulated differently from the genes in the clavulanic acid cluster, such that they are not expressed in SA medium but are expressed in Soy medium (17). The existence of a paralogue for cas2 is well established, since the cas1 gene has been cloned and sequenced, and Cas1 has been purified and characterized. A compelling case can also be made for the existence of paralogues for orf2, bls, and pah, because these genes are known to encode essential enzymes in the clavulanic acid biosynthetic pathway. The fact that orf2, bls, and pah disruption mutants, in which these genes have been destroyed by insertion of antibiotic resistance markers, can still form clavulanic acid and other clavams implies that they must have alternative genes that produce equivalent enzymes. The only other simple explanation that comes to mind is that the complex Soy medium already contains intermediates of the clavulanic acid pathway, thereby obviating the need for all of these biosynthetic enzymes. However, the complex chemical structures of these intermediates (Fig. 3) make such an explanation extremely unlikely. The enzymatic steps catalyzed by Orf2, Bls, and Pah are also sufficiently unusual that it is very unlikely that they could be carried out by other enzymes of broad specificity. The case for the existence of a paralogue of orf6 is less compelling because the role of this gene in the biosynthesis of clavulanic acid and the clavams is unknown. However, the striking similarity in phenotype of all five of these mutant classes (blocked on SA medium but producing on Soy medium) argues for a similar mechanism at work in all cases. Of the group of mutants showing evidence for the presence of paralogues, orf2 is the most ambiguous because production of clavulanic acid and clavams was very poor, even on Soy medium. However, since the orf2 gene product is required for clavulanic acid production, the production even of small amounts of clavulanic acid and clavams in the knockout mutant is possible only if a paralogue exists.
The locations of the paralogues relative to the genes in the cephamycin-clavulanic acid supercluster is unknown, just as the location of cas1 relative to cas2 is unknown. Inspection of the DNA sequence in the region flanking cas1 shows no evidence of candidate paralogues, but only a limited amount of sequence information is available at present (13). However, the same studies have shown that the genes surrounding cas1 are involved in clavam biosynthesis, since disruption of the genes flanking cas1 specifically affects clavam production.
While paralogues apparently exist for orf2, bls, pah, cas2, and orf6, individual disruption of orf7, claR, cad, and orf10 results in mutants in which the defect in clavulanic acid production is not growth medium dependent. These mutants are defective in clavulanic acid production on both SA and Soy media. In a recent study, claR was shown to regulate the transcription of orf7, cad, and orf10, as well as regulating its own transcription (16). In contrast, orf2, bls, pah, cas2, and orf6 were transcribed independently from claR. Since mutants defective in claR were unable to produce clavulanic acid but did accumulate clavaminic acid, this suggests that orf2, bls, pah, cas2, and orf6 are involved in early steps of the clavulanic acid biosynthetic pathway, before the level of clavaminic acid. Certainly, this is known to be the case for orf2, bls, pah, and cas2, but it can only be inferred for orf6, since its role in clavulanic acid biosynthesis is not known. All of the early steps in clavulanic acid biosynthesis leading to clavaminic acid are now understood, and yet the role of orf6 remains unclear. The similarity of the Orf6 protein to acetyltransferase type enzymes suggests intriguingly that an as-yet-unrecognized acetylation step may be involved in the early stages of the pathway.
Based on these results, it appears that the genes encoding early enzymes of the biosynthetic pathway, the part of the pathway that is common to both clavulanic acid and the clavam metabolites, all have paralogues. In contrast, the genes encoding biosynthetic enzymes specific for the steps that convert clavaminic acid into clavulanic acid are unique. The simplest explanation for these data would be that S. clavuligerus contains two biosynthetic gene clusters, one for the production of clavulanic acid and a second for the production of the other clavam metabolites. Inasmuch as these two groups of structurally related compounds share a common biosynthetic pathway to the level of clavaminic acid, mutations in the early steps of one pathway can be compensated by the corresponding paralogue from the other pathway. Mutations in late steps of the pathway specifically knock out production of the metabolite involved, while having little effect on production of the product(s) of the other pathway.
ACKNOWLEDGMENTS
We thank A. Wong and C. Anders for excellent technical assistance.
This work was supported by the Natural Sciences and Engineering Research Council of Canada.
REFERENCES
- 1.Aidoo K A, Wong A, Alexander D C, Rittamer R A, Jensen S E. Cloning, sequencing and disruption of a gene involved in clavulanic acid biosynthesis. Gene. 1994;147:41–46. doi: 10.1016/0378-1119(94)90036-1. [DOI] [PubMed] [Google Scholar]
- 2.Bachmann B O, Li R, Townsend C A. β-Lactam synthetase: a new biosynthetic enzyme. Proc Natl Acad Sci USA. 1998;95:9082–9086. doi: 10.1073/pnas.95.16.9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Denis F, Brzezinski R. An improved aminoglycoside resistance gene cassette for use in Gram-negative bacteria and Streptomyces. FEMS Microbiol Lett. 1991;81:261–264. doi: 10.1016/0378-1097(91)90224-x. [DOI] [PubMed] [Google Scholar]
- 4.Doran J L, Leskiw B K, Petrich A K, Westlake D W S, Jensen S E. Production of Streptomyces clavuligerus isopenicillin N synthase in Escherichia coli using two-cistron expression systems. J Ind Microbiol. 1990;5:197–206. doi: 10.1007/BF01569677. [DOI] [PubMed] [Google Scholar]
- 5.Egan L A, Busby R W, Iwata-Reuyl D, Townsend C A. Probable role of clavaminic acid as the terminal intermediate in the common pathway to clavulanic acid and the antipodal clavam metabolites. J Am Chem Soc. 1997;119:2348–2355. [Google Scholar]
- 6.Elson S W, Baggaley K H, Davison M, Fulston M, Nicholson N H, Risbridger G D, Tyler J W. The identification of three new biosynthetic intermediates and one further biosynthetic enzyme in the clavulanic acid pathway. J Chem Soc Chem Commun. 1993;1993:1212–1214. [Google Scholar]
- 7.Hodgson J E, Fosberry A P, Rawlinson N S, Ross H N M, Neal R J, Arnell J C, Earl A J, Lawlor E J. Clavulanic acid biosynthesis in Streptomyces clavuligerus: gene cloning and characterization. Gene. 1995;166:49–55. doi: 10.1016/0378-1119(95)00560-9. [DOI] [PubMed] [Google Scholar]
- 8.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic manipulation of Streptomyces: a laboratory manual. Norwich, United Kingdom: The John Innes Foundation; 1985. [Google Scholar]
- 9.Janc J W, Egan L A, Townsend C A. Purification and characterization of clavaminate synthase from Streptomyces antibioticus: a multifunctional enzyme of clavam biosynthesis. J Biol Chem. 1995;270:5399–5404. doi: 10.1074/jbc.270.10.5399. [DOI] [PubMed] [Google Scholar]
- 10.Khaleeli N, Li R, Townsend C A. Origin of the β-lactam carbons in clavulanic acid from an unusual thiamine pyrophosphate-mediated reaction. J Am Chem Soc. 1999;121:9223–9224. [Google Scholar]
- 11.Marsh E N, Chang M D T, Townsend C A. Two isozymes of clavaminate synthase central to clavulanic acid formation: cloning and sequencing of both genes from Streptomyces clavuligerus. Biochemistry. 1992;31:12648–12657. doi: 10.1021/bi00165a015. [DOI] [PubMed] [Google Scholar]
- 12.McNaughton H J, Thirkettle J E, Zhang Z, Schofield C J, Jensen S E, Barton B, Greaves P. Beta-lactam synthetase: implications for beta-lactamase evolution. J Chem Soc Chem Commun. 1998;1998:2325–2326. [Google Scholar]
- 13.Mosher R H, Paradkar A S, Anders C L, Barton B, Jensen S E. Genes specific for the biosynthesis of clavam metabolites antipodal to clavulanic acid are clustered with the gene for clavaminate synthase 1 in Streptomyces clavuligerus. Antimicrob Agents Chemother. 1999;43:1215–1224. doi: 10.1128/aac.43.5.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nicholson N H, Baggaley K H, Cassels R, Davison M, Elson S W, Fulston M, Tyler J W, Woroniecki S R. Evidence that the immediate biosynthetic precursor of clavulanic acid is its N-aldehyde analogue. J Chem Soc Chem Commun. 1994;1994:1281–1282. [Google Scholar]
- 15.Paradkar A S, Aidoo K A, Jensen S E. Molecular analysis of a β-lactam resistance gene from the cephamycin gene cluster of Streptomyces clavuligerus. J Bacteriol. 1996;178:6266–6274. doi: 10.1128/jb.178.21.6266-6274.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paradkar A S, Aidoo K A, Jensen S E. A pathway-specific transcriptional activator regulates late steps of clavulanic acid biosynthesis in Streptomyces clavuligerus. Mol Microbiol. 1998;27:831–843. doi: 10.1046/j.1365-2958.1998.00731.x. [DOI] [PubMed] [Google Scholar]
- 17.Paradkar A S, Jensen S E. Functional analysis of the gene encoding the clavaminate synthase 2 isoenzyme involved in clavulanic acid biosynthesis in Streptomyces clavuligerus. J Bacteriol. 1995;177:1307–1314. doi: 10.1128/jb.177.5.1307-1314.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perez-Redondo R, Rodriguez-Garcia A, Martin J F, Liras P. The claR gene of Streptomyces clavuligerus, encoding a LysR-type regulatory protein controlling clavulanic acid biosynthesis, is linked to the clavulanate-9-aldehyde reductase (car) gene. Gene. 1998;211:311–321. doi: 10.1016/s0378-1119(98)00106-1. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 20.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stuttard C. Temperate phages of Streptomyces venezuelae: lysogeny and host specificity shown by phages SV1 and SV2. J Gen Microbiol. 1982;128:115–121. [Google Scholar]
- 22.Vieira J, Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- 23.Ward J M, Hodgson J E. The biosynthetic genes for clavulanic acid and cephamycin production occur as a ‘super-cluster’ in three Streptomyces. FEMS Microbiol Lett. 1993;110:239–242. doi: 10.1111/j.1574-6968.1993.tb06326.x. [DOI] [PubMed] [Google Scholar]
- 24.Wu T-K, Busby R W, Houston T A, McIlwaine D B, Egan L A, Townsend C A. Identification, cloning, sequencing, and overexpression of the gene encoding proclavaminate amidinohydrolase and characterization of protein function in clavulanic acid biosynthesis. J Bacteriol. 1995;177:3714–3720. doi: 10.1128/jb.177.13.3714-3720.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zoller M J, Smith M. Oligonucleotide-directed mutagenesis: a simple method using two oligonucleotide primers and a single-stranded DNA template. Methods Enzymol. 1987;154:329–350. doi: 10.1016/0076-6879(87)54083-6. [DOI] [PubMed] [Google Scholar]