Abstract
A clinical isolate of Streptococcus pneumoniae (SP#5) that showed decreased susceptibility to evernimicin (MIC, 1.5 μg/ml) was investigated. A 4,255-bp EcoRI fragment cloned from SP#5 was identified by its ability to transform evernimicin-susceptible S. pneumoniae R6 (MIC, 0.03 μg/ml) such that the evernimicin MIC was 1.5 μg/ml. Nucleotide sequence analysis of this fragment revealed that it contained portions of the S10-spc ribosomal protein operons. The nucleotide sequences of resistant and susceptible isolates were compared, and a point mutation (thymine to guanine) that causes an Ile52-Ser substitution in ribosomal protein L16 was identified. The role of this mutation in decreasing susceptibility to evernimicin was confirmed by direct transformation of the altered L16 gene. The presence of the L16 mutation in the resistant strain suggests that evernimicin is an inhibitor of protein synthesis. This was confirmed by inhibition studies using radiolabeled substrates, which showed that the addition of evernimicin at sub-MIC levels resulted in a rapid decrease in the incorporation of radiolabeled isoleucine in a susceptible isolate (SP#3) but was much less effective against SP#5. The incorporation of isoleucine showed a linear response to the dose level of evernimicin. The incorporation of other classes of labeled substrates was unaffected or much delayed, indicating that these were secondary effects.
Everninomicins are a class of oligosaccharide antibiotics isolated from Micromonospora carbonaceae (31). One such compound, evernimicin (SCH 27899) (10, 11, 12) is currently undergoing evaluation as a therapeutic agent. It has been shown to have potent activity against many gram-positive bacteria, including emerging problem organisms such as vancomycin-resistant enterococci, methicillin-resistant staphylococci, and penicillin-resistant pneumococci (16). In fact, there were no staphylococcal, enterococcal, and pneumococcal isolates that displayed resistance to evernimicin in either the investigation by Jones and Barrett (16) or a more-recent worldwide survey of clinical isolates, including isolates known to be resistant to other antibiotics (R. S. Hare, F. J. Sabatelli, and the Ziracin Susceptibility Testing Group, Abstr. 38th Intersci. Conf. Antimicrob. Agents Chemother., abstr. E-119, p. 204, 1998). The paucity of isolates showing resistance to evernimicin is presumably a result of no prior clinical exposure to a drug similar to the family of everninomicins. The lack of cross-resistance to evernimicin, however, would suggest that the mechanism of action is novel and that prior selection leading to resistance to other antimicrobials will not impact the efficacy of evernimicin.
Previous studies with another oligosaccharide antibiotic, avilamycin (33), showed protein synthesis inhibition as the mechanism of action, apparently by interacting with the 30S ribosomal subunit. Nevertheless, avilamycin lacks the nitro-sugar moiety that distinguishes the everninomicin class of antibiotics, and the mechanism of action of everninomicins, including evernimicin, is unknown. In fact, the primarily gram-positive activity and the inconsistent response as a bactericidal agent made it difficult to predict the target site of action for evernimicin. We report on the analysis of Streptococcus pneumoniae mutants that have reduced susceptibility to evernimicin and the in vivo effect of these mutations on macromolecular syntheses in the presence of the drug. The mechanism of action of evernimicin and the identity of a putative drug interaction site in the ribosome are implicated.
(Portions of this work were previously presented at the 38th Interscience Conference on Antimicrobial Agents and Chemotherapy, San Diego, Calif., 1998.)
MATERIALS AND METHODS
Bacterial strains.
Clinical isolates of S. pneumoniae SP#3 and SP#5 are clonally related isolates as determined by serotype, pulsed-field gel electrophoresis, and arbitrarily primed diagnostic PCR fingerprinting (data not shown). SP#3 and SP#5 were derived from a single patient enrolled in a clinical trial conducted in Johannesburg, South Africa. The MIC of evernimicin for strain SP#3 was 0.023 μg/ml, while SP#5 showed reduced susceptibility to evernimicin (MIC, 1.5 μg/ml). Laboratory strains S. pneumoniae R6 and ATCC 49619 were used in transformation experiments and as evernimicin-susceptible controls.
DNA extraction.
Whole chromosomal DNA from S. pneumoniae strains was prepared by detergent lysis followed by phenol-chloroform extraction as described previously (3). Extracted DNA was treated with RNase and then further purified by precipitation with 0.6 volume of 20% polyethylene glycol (PEG) 6000–2.5 M NaCl.
Transformation.
S. pneumoniae R6 was grown in C medium supplemented with yeast extract (C+y) (30). Five milliliters of overnight culture was inoculated into 100 ml of C+y medium and grown at 37°C. Between optical densities at 650 nm (OD650) of 0.01 to 0.5, aliquots of cells were collected, and the efficiencies of cells transforming to streptomycin resistance in the presence of DNA from a streptomycin-resistant pneumococcus were determined. Cells from the aliquot which produced the highest transformation efficiency were stored at −70°C in 15% glycerol for further transformation experiments. S. pneumoniae ATCC 49619 cells for transformation were grown to an OD650 of 0.2 in brain heart infusion (BHI) broth (Difco, Detroit, Mich.) supplemented with 5% horse serum. For S. pneumoniae ATCC 49619, competence was induced by the addition of 1 μg of competence-stimulating peptide/ml (14). Transformations were performed by incubating the thawed cells (1 ml) with 1 μg of donor DNA/ml at 30°C for 30 min. The cells were allowed to express resistance for 60 min at 37°C before being plated out on selection media (Mueller Hinton agar supplemented with 5% horse blood and evernimicin). For routine transformations, a drug concentration of 0.25 μg/ml was used to isolate strains with reduced susceptibility to evernimicin.
MICs.
MICs of evernimicin were determined by Etest (AB Biodisk, Solna, Sweden) on Mueller Hinton agar supplemented with 5% sheep blood according to the manufacturer's recommendations. Plates were incubated at 37°C for 24 h under 5% CO2.
Cloning of DNA conferring evernimicin resistance.
Whole chromosomal DNA from an R6 derivative (ZR1) which was transformed with chromosomal DNA from SP#5 to increase its resistance to evernimicin (MIC, 1.5 μg/ml) was restricted with EcoRI and electrophoresed on a 0.8% agarose gel at 2 V/cm for 16 h. Sections of agarose containing EcoRI restriction fragments of different sizes were extracted from the gel, and the DNA was recovered using the Geneclean kit (Bio 101, La Jolla, Calif.). Restriction fragments ranging from 4.17 to 4.5 kb) were shown to transform R6 at the highest frequency (4.7 × 10−6) and were retained for further cloning experiments. The evernimicin MICs for R6 transformed with fractionated DNA were identical to that of the original clinical isolate. Ligation reactions were set up with 1 μg of dephosphorylated vector DNA (LAMBDA ZAP II; Stratagene, La Jolla, Calif.) which had been predigested with EcoRI and 100 ng of target DNA (4.17- to 4.5-kb fraction of EcoRI-restricted DNA). The DNA was ligated with the Fast-Link DNA ligation kit (Epicentre Technologies, Madison, Wis.) according to the manufacturer's recommendations. The ligated DNA was ethanol precipitated and resuspended in 5 μl of 10 mM Tris-Cl, pH 8.0. The ligated DNA was packaged with the Gigapack II Gold Packaging Extract (Stratagene) as recommended by the manufacturer. The packaged phage library was diluted in 1 ml of SM buffer, plated out with E. coli XL1-Blue, and titered as recommended by the manufacturer.
PCR amplification of cloned fragments.
Inserts from the lambda clones were amplified with the Expand Long Template PCR system (Boehringer GmbH, Mannheim, Germany) in 50-μl reaction mixtures containing 3 U of DNA polymerase mix, 1× polymerase buffer 1, 350 μM deoxynucleoside triphosphates (dNTPs), 300 nM each primer (M13 forward and reverse) and 2 μl of phage suspension in SM buffer. The reactions were performed with an Omnigene (Hybaid, Middlesex, United Kingdom) as follows: 93°C for 60 s, followed by 25 cycles of 92°C for 2 s, 55°C for 30 s, and 68°C for 195 s; for the last 15 cycles, the 68°C extension time was extended by 12 s for each cycle.
Screening the library by transformation.
The phage library was plated out at approximately 10 to 50 PFU/cm2. Cores containing approximately 10 to 20 plaques were extracted from the top agar with the rear end of a sterile Pasteur pipette (internal diameter, 6 mm) and resuspended in 1 ml of SM buffer. Two microliters from each of these phage pools was amplified by PCR, and 10 μl of the PCR product was used in the transformation experiments. The phage pools from which the PCR products were shown to transform R6 to reduced susceptibility to evernimicin (MIC, 1.5 μg/ml) (transformation frequency, 3 × 10−5 to 6 × 10−5) were selected for further evaluation. Approximately one out of six phage pools from each library produced transformation-positive PCR products. Transformation-positive phage pools were plated out at low density, and individual plaques were cored from the top agar with the narrow end of a Pasteur pipette and vortex mixed in 1 ml of SM buffer to form a stock. Two microliters from each stock containing individual plaques was again amplified by PCR, and the PCR product (10 μl) was used to determine whether the cloned fragment was capable of transforming R6 to reduced susceptibility to evernimicin (MIC, 1.5 μg/ml). Two transformation-positive clones were selected for sequencing and analysis.
Sequencing and analysis.
DNA templates were prepared by the PCR described above. Primers for amplifying the corresponding sequence from the evernimicin-susceptible S. pneumoniae R6 and the original clinical isolate SP#5 were designed from the 5′ and 3′ ends of the cloned fragment. The PCR products were purified through a spin column (catalog no. UFC3LTK00; Millipore, Bedford, Mass.) and resuspended at a DNA concentration of 0.5 μg/μl for sequencing. Nucleotide sequencing was performed by the chain termination method with the Sequenase version 2.0 DNA Sequencing Kit (U.S. Biochemicals, Cleveland, Ohio) and [S35]dATP label (Amersham, Little Chalfont, Buckinghamshire, United Kingdom) according to the manufacturer's recommendations. Sequencing of two transformation-positive clones and the evernimicin-susceptible and -resistant control strains was performed by primer walking in both directions with both the dGTP and the dITP labeling mix. The putative open reading frames were identified with the DNAstar software. The putative polypeptides were identified with the BLAST program (National Center for Biotechnology Information) by comparison with the SWISSPROT amino acid sequence database.
Site-directed mutagenesis.
Mutations to the gene for ribosomal protein L16 were introduced by a PCR-based “megaprimer” method (23).
In vivo macromolecular labeling assay.
Bacterial strains SP#3 and SP#5 were grown from glycerol stocks overnight at 37°C on TSA II agar plus 5% sheep blood (BBL, Cockeysville, Md.) in an atmosphere containing 5% CO2. Cells were scraped from the plates and inoculated into Todd-Hewitt broth containing 0.2% yeast extract (wt/vol) and 20 mM N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid (TES) (pH 7.5). The OD540 of the inoculum was adjusted to 0.1, and the cells were allowed to grow at 37°C with shaking at 250 rpm for 2 h. The OD540 of the culture was checked, and the cells were diluted into fresh media containing [14C]isoleucine to give a final specific activity of 0.7 μCi/ml and an OD540 of ∼0.1. Seven separate tubes of inoculum were prepared for each strain for the addition of various concentrations of evernimicin. Triplicate samples of 200 μl were taken from each tube after 30, 60 and 90 min following inoculation and were mixed into 600 μl of ice-cold 7% trichloroacetic acid (TCA) in 96-well deep-well plates. Immediately after the 90-min sample was taken, various concentrations of evernimicin or carrier for the drug was added to individual tubes. Samples were taken as described above at 5, 10, 15, 20, 25, 45, 65, 85 and 105 min after the addition of drug or carrier. The growth of the strains was checked as indicated by the OD540 every 30 min following inoculation. After the last time point, the samples were applied to 96-well glass fiber (type B; 1 μm) filtration plates (Millipore, Ann Arbor, Mich.) and were washed three times with 200 μl of ice-cold 5% TCA and once with 200 μl of 100% ethanol. Plates were briefly air dried, and then 100 μl of aqueous scintillation fluid was added. Plates were counted on a TopCount (Packard Instruments, Meriden, Conn.). Labeling with other radioactive substrates was performed with only SP#3 using either carrier or evernimicin at 0.025 or 0.00625 μg/ml. Experiments were performed as described above using either 0.7 μCi of [14C]thymidine/ml, 0.7 μCi of [3H]UTP/ml, 3.3 μCi of [3H]GlcNAc/ml, or 3.3 μCi of [14C]acetate/ml. All radiolabeled substrates were purchased from DuPont, NEN (Wilmington, Del.) except for the [3H]GlcNAc, which was purchased from Amersham. The growth measurements were made with a 20D+ reader (Spectronic Instruments, Inc., Rochester, N.Y.) set at 540 nm to read the cultures in 150- by 25-mm glass culture tubes.
Nucleotide sequence accession numbers.
The nucleotide sequences of the clones reported here have been assigned the following GenBank accession numbers: for the R6 wild-type sequence, AF126059; for the ZR1 transformation into R6, AF126060; and for the SP#5 original mutant sequence, AF126061.
RESULTS
Transformation.
Nonencapsulated S. pneumoniae R6 and the ATCC 49619 strain could be transformed with whole chromosomal DNA (1 μg/ml) from the clinical isolate SP#5, which was shown to have reduced susceptibility to evernimicin (MIC, 1.5 μg/ml). The transformation frequency per milliliter of cells (total count, 6 × 107 CFU/ml) selected at different drug concentrations is shown in Table 1. The transformation frequencies were approximately fivefold lower in ATCC 49619 than in R6, and the number of transformants which could be isolated decreased with increasing evernimicin concentrations in the selection media. The MICs for transformants selected at different drug concentrations were shown to be identical to that of the original clinical isolate. The transformation frequency from the same batch of competent S. pneumoniae R6 to reduced evernimicin susceptibility was similar to that for streptomycin resistance (data not shown). Furthermore, PCR products from the lambda clones which harbored the genes for evernimicin resistance transformed S. pneumoniae R6 so that the evernimicin MIC was the same (1.5 μg/ml) as chromosomal DNA of SP#5 at a frequency of 3 × 10−5 to 6 × 10−5. The spontaneous mutation rate of R6 and the transformation rate for the SP#3 chromosomal DNA negative control were <10−7.
TABLE 1.
Transformation of S. pneumoniae strains with whole chromosomal DNA from resistant clinical isolate SP#5
| Concn of evernimicin (μg/ml) | Transformation frequency
|
|
|---|---|---|
| R6 | ATCC 69419 | |
| 0.06 | 3.5 × 10−5 | 1.0 × 10−5 |
| 0.12 | 3.3 × 10−5 | 4.6 × 10−6 |
| 0.25 | 2.4 × 10−5 | 3.6 × 10−6 |
| 0.5 | 1.2 × 10−5 | 2.2 × 10−6 |
| 1.0 | <2 × 10−7 | <2 × 10−7 |
| 2.0 | <2 × 10−7 | <2 × 10−7 |
Nucleotide sequence and analysis.
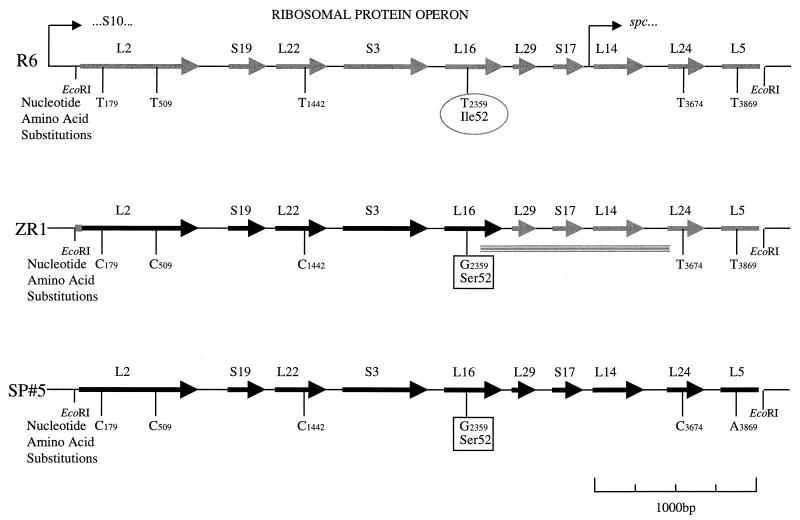
An analysis of the open reading frames of the nucleotide sequence from the cloned fragment showed high levels of amino acid identity (70 to 90%) with ribosomal protein genes from other bacteria and showed that they were arranged in a manner identical to that of the S10-spc ribosomal protein operons of Escherichia coli (18) and Bacillus subtilis (15, 24) from ribosomal protein L2 to L5 (Fig. 1). The nucleotide sequence of the cloned fragment from the evernimicin-resistant transformant differed from that of the susceptible R6 at 4 nucleotides (Table 2). Only one of these nucleotide substitutions (thymine 2359-guanine) resulted in an amino acid substitution (Ile52-Ser) which was present in ribosomal protein L16. The identical point mutations occurred in the clinical isolate SP#5, including an additional two nucleotide substitutions which occurred 3′ of the mutation resulting in the Ile52-Ser substitution in ribosomal protein L16, and may indicate one of the crossover points of homologous recombination which occurred in the R6 transformant (Fig. 1). The cloning experiment was repeated, and an identical nucleotide sequence was obtained from the cloned fragment.
FIG. 1.
Gene organization of the 4,255-bp EcoRI fragment of the S10/spc ribosomal protein operon showing the nucleotide and amino acid sequence differences between the evernimicin-susceptible S. pneumoniae R6, the evernimicin-resistant transformant (ZR1), and the original clinical isolate (SP#5). The hatched lines indicate the putative points of crossing over during homologous recombination.
TABLE 2.
Nucleotide substitutions in evernimicin-susceptible and -resistant strains of S. pneumoniae
| Position | Nucleotide in:
|
|||
|---|---|---|---|---|
| R6 | ZR1, clone 1 | ZR1, clone 2 | Clinical isolate SP#5 | |
| 179 | T | C | C | C |
| 509 | T | C | C | C |
| 1442 | T | C | C | C |
| 2395 | T | G | G | G |
| 3674 | T | T | T | C |
| 3869 | T | T | T | A |
Role of rplP in resistance.
To confirm the role of the Ile52-Ser mutation in evernimicin resistance, a 1,546-bp (positions 1625 to 3171 in the sequence submitted to GenBank [accession no. AF126061]) PCR product from SP#5 containing rplP and part of the 5′ and 3′ flanking structures and a 613-bp (positions 2096 to 2709) pGEM3Zf+ clone encoding the Ile52-Ser mutant rplP were shown to transform R6 to evernimicin resistance (MIC, 1.5 μg/ml) at rates of 5 × 10−5 and 1.6 × 10−4, respectively. No transformants were generated when whole chromosomal DNA from evernimicin-susceptible R6 was used as a control; however, the 1,546-bp rplP PCR products from R6 were able to transform R6 competent cells at a rate of 10−7. Two of these R6 transformants were selected, and the nucleotide sequences of rplP from these strains revealed two novel alterations to ribosomal protein L16: a thymine 2395-to-adenine change which resulted in an Ile52-Asn substitution (ZR4) and a thymine 2391-to-adenine change which resulted in an Arg51-Cys (ZR5) substitution. The evernimicin MICs for these two strains were 1.5 and 0.75 μg/ml, respectively. These fortuitous mutations are thought to have been introduced during the PCR amplification by Taq polymerase, which has been shown to have an error rate of 10−3 to 10−4.
To further clarify the role of ribosomal protein L16 in resistance, mutants were constructed by mutagenic PCR of rplP followed by subsequent cloning into pGEM3Zf+ and transformation back into R6. An rplP clone with the mutation of Ile52 to Thr (ZR3), an amino acid similar to Ser, resulted in the selection of evernimicin-resistant R6 transformants for which the MICs were 0.38 μg/ml, fourfold lower than that for the Ile52-Ser mutant. When Ile52 was replaced with Arg (ZR6), the consensus amino acid found in rplP at a position homologous to that of Ile52 in gram-negative bacteria, no resistant transformants were selected at any evernimicin concentration above the MIC for susceptible S. pneumoniae. The characteristics of the resistant rplP mutants are shown in Table 3.
TABLE 3.
MICs of S. pneumoniae with different amino acid substitutions at position 52 on ribosomal protein L16
| Strain | Recipient strain | Source of transforming DNA | RPL16a substitution | Evernimicin MIC (μg/ml) |
|---|---|---|---|---|
| R6 | Wild type | 0.03 | ||
| ATCC 49619 | Wild type | 0.03 | ||
| SP#5 | Ile52 to Ser | 1.5 | ||
| ZR1 | R6 | SP#5 chromosomal DNA | Ile52 to Ser | 1.5 |
| ZR2 | ATCC 49619 | SP#5 chromosomal DNA | Ile52 to Ser | 1.5 |
| ZR3 | R6 | Site-directed mutagenic PCR product | Ile52 to Thr | 0.38 |
| ZR4 | R6 | Random-mutagenic PCR product from R6 | Ile52 to Asn | 1.5 |
| ZR5 | R6 | Random-mutagenic PCR product from R6 | Arg51 to Cys | 0.75 |
| ZR6 | R6 | Site-directed mutagenic PCR product | Ile52 to Arg | NTSb |
RP, ribosomal protein.
NTS, no transformants were selected on media containing evernimicin concentrations of ≥0.06 μg/ml.
In vivo macromolecular labeling.
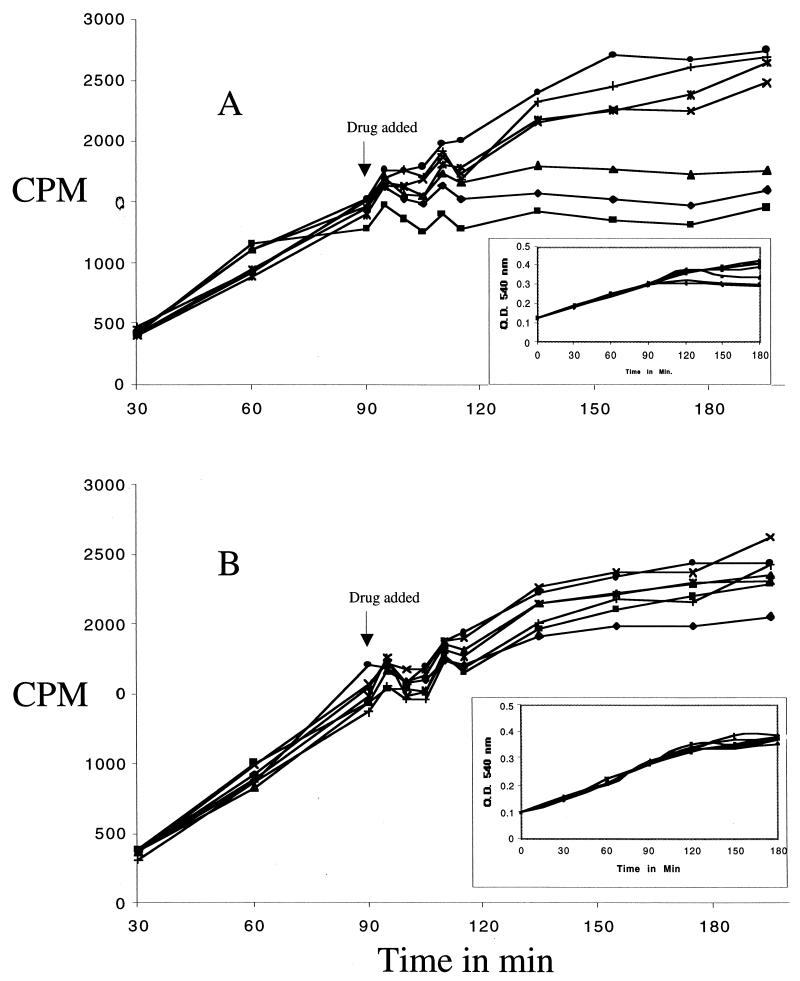
Evernimicin treatment of the susceptible pneumococcal isolate SP#3 led to a rapid drop in the incorporation of isoleucine into TCA-precipitable material (Fig. 2A). This rapid and dramatic effect on incorporation was seen only for isoleucine and not for UTP, thymidine, acetate, or GlcNAc (data not shown). The growth rate of SP#3 was also rapidly decreased with 0.4 and 0.1 μg of evernimicin/ml, while a slight reduction was seen with 0.025 μg/ml. The less-susceptible isolate, SP#5, displayed only a slight decrease in isoleucine incorporation even at the highest levels of added evernimicin (Fig. 2B), and the growth of SP#5 was not affected by the levels of evernimicin used in the experiment (0.4 μg/ml).
FIG. 2.
In vivo labeling with [14C]isoleucine in SP#3 (A) and SP#5 (B). The results shown are from a single experiment that is representative of at least two additional experiments. Each point is the average value of triplicate samples that were taken at every time point. The peculiar sinusoidal response seen following the addition of drug or carrier alone occurred consistently in the labeling of S. pneumoniae and may be a response to the high pH or hydrophobic nature of the carrier agent. Symbols correspond to the evernimicin levels used in the experiment, which were added at the 90-min time point as follows: ⧫, 0.4 μg/ml; ■, 0.1 μg/ml; ▴, 0.025 μg/ml; ✖, 0.00625 μg/ml; ∗, 0.00156 μg/ml; ●, 0.0004 μg/ml; ✚, drug carrier, used at the same level used for 0.4 μg of evernimicin/ml. The growth curves shown in the insets are single OD540 readings.
DISCUSSION
By utilizing in vivo labeling of macromolecules, Black et al. (T. A. Black, W. Zhao, K. J. Shaw, and R. S. Hare, Abstr. 38th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C-106, p. 99, 1998) had shown that evernimicin specifically inhibited protein synthesis in Staphylococcus aureus. The identification in our study of a ribosomal protein mutation in S. pneumoniae that causes reduced susceptibility to the drug lends credence to the assertion that evernimicin is a specific inhibitor of protein synthesis. The macromolecular synthesis experiments with S. pneumoniae confirm that treatment of the susceptible SP#3 strain with evernimicin specifically inhibits the incorporation of amino acid precursors and does not affect the incorporation of RNA, DNA, murein, or lipid precursors. Furthermore, amino acid incorporation in the less-susceptible strain, SP#5, is not affected by the lower concentrations of evernimicin. The apparent ribosome target site of evernimicin is unique in that L16 has not previously been implicated in resistance to other known inhibitors of protein synthesis. However, L16 has been implicated in the binding and mechanism of action for macrolides and streptogramin B (7, 20). Mutations in L4 and L22 confer resistance to erythromycin (5, 32); S12 mutations affect streptomycin resistance (9); L6 mutations confer gentamicin resistance (6); S5 mutations confer spectinomycin resistance (6); and L11 mutations confer thiostrepton resistance (22). Furthermore, the Ile52-Ser mutant form of L16 did not confer co-resistance to any other inhibitors of protein synthesis such as the macrolides, streptogramins, lincosamides, aminoglycosides, tetracyclines, chloramphenicol, and oxazolidinones (data not shown). In addition to this, no cross-resistance to evernimicin has been found in clinical isolates of staphylococci, streptococci, and enterococci shown to be resistant to erythromycin or streptogramins (Hare et al., 38th ICAAC).
The exact role of L16 in ribosome function is not clearly defined. Studies with E. coli ribosomes using a hot tritium bombardment technique have shown that L16 occurs on the surface of the 50S subunit (2). Further studies using immunoelectron microscopy have mapped L16 to a position beside the central protuberance of the 50S particle on the side away from the L7/L12 stalk at the interface between the large and small subunits (21). Cross-linking between L16 and S19, a tRNA binding protein of the small subunit, confirms that L16 lies at the interface of the two subunits (17). Functional analysis of L16 has shown that ribosomes stripped of L16 lose the functions associated with the peptidyl transferase center (4). Activity can be restored by the addition of L16 to deficient ribosomes (13). Furthermore, it has not been possible to isolate mutants that are deficient in L16. The contribution of L16 to the peptidyl transferase catalytic center has been poorly defined. Under special conditions, ribosomes deficient in L16 are capable of, but not optimal for, peptidyl transferase activity (28), and ribosomes stabilized by the additional loss of L11 are capable of release factor 2-dependent peptidyl-tRNA hydrolysis (27). These conditions presumably have a profound effect on the orientation of the bound substrates, resulting in a thermodynamically and kinetically favorable situation in which appropriately positioned functional groups can react without the intervention of L16. In addition, modifications to the histidine residue of L16 have resulted in an unstable assembly of proteins within the ribosome and a reduced rate of activity at the peptidyl transferase center (26). L16 is a late assembly protein of the large subunit (8) and induces a major conformational change with a high activation energy (123 kJ/mol) when reconstituted into cores (29). This suggests that L16 does not play an essential catalytic role in the peptidyl transferase center, but rather a role in the correct conformation of the ribosome where the constituents of the reaction are optimally arranged and placed in contact with non-L16 catalytic sites elsewhere in the ribosome.
A further clarification of the role of L16 in protein synthesis comes in the form of UV cross-linking experiments on E. coli ribosomes with poly(U) and Phe-tRNAphe which have shown that L16 forms covalent links with tRNA bound in the A site of the 70S ribosome (1). Further experiments by Maimets et al. (19) have shown that despite the ability of L16-stripped subunits to perform peptidyl transferase activity with puromycin (a tRNA analogue) as an acceptor substrate, the L16-deficient particles were unable to utilize an oligonucleotide, CACCA-Phe, as a substrate. This suggests that fixation by L16 of the 3′ end of the tRNA in the A site is required for protein synthesis. This proposed interaction is supported by the ability of L16 on its own to bind to and protect the 3′ end of tRNA from RNase digestion. L16 on its own has been shown to bind nonspecifically to a variety of tRNAs, including fMet-tRNA and AcPhe-tRNA. The ability of L16 to bind tRNA has also been demonstrated for other basic ribosomal proteins which are not usually implicated in the formation of the A and P sites (25). This does not, however, preclude L16 from a direct role in tRNA binding within the A site of the ribosome.
From these data it is difficult to predict the exact mode of action of evernimicin in inhibiting protein synthesis. Based on the known function of L16, we suggest that evernimicin may inhibit protein synthesis either by altering the conformation of the A site and preventing the correct positioning of the bound tRNA or by competing with the tRNA molecule for a position within the A site, thus preventing the catalytic activities of the peptidyl transferase center. This is supported by data that show that stoichiometric amounts of avilamycin, a close analogue of evernimicin, are able to inhibit Phe-tRNA binding to 70S ribosomes in the presence of poly(U) by 50%, and it was suggested that the drug acts by preventing the attachment of tRNA to the ribosome (33).
The tolerance of L16 to a variety of mutations at and around position 52 may indicate that evernimicin does not bind specifically to L16. Rather, resistance may occur as a result of a conformational change in the protein that reduces the binding affinity of the drug to the assembled ribosome or increases the ability of the ribosome to function in the presence of the drug. Also, since some inhibitors of protein synthesis are known to interact directly with the rRNA, it is possible that evernimicin interacts with rRNA in a manner that is potentiated by either a direct or an allosteric interaction with L16.
An alignment of the amino acid sequence of L16 from an assortment of gram-positive and gram-negative bacteria is shown in Fig. 3. The amino acid at position 52 differs between these two groups from a consensus Ile52 in gram-positive organisms to Arg52 in gram-negative organisms. It could conceivably be the reason for the lack of activity of evernimicin in gram-negative bacteria. However, the inability to transform R6 to evernimicin resistance with an Ile52-Arg substitution in a tightly controlled experiment suggests that resistance to evernimicin in gram-negative organisms does not occur as a result of this substitution, but rather as a result of other host-specific differences such as drug permeability.
FIG. 3.
The amino acid sequence of the S. pneumoniae L16 protein from residues 43 to 60 aligned with the same region of L16 from gram-positive bacteria (S. aureus, Enterococcus faecalis, and B. subtilis) and gram-negative bacteria (E. coli, Haemophilus influenzae, and Neisseria gonorrhoeae.
ACKNOWLEDGMENTS
We thank Richard Goering for performing the pulsed-field gel analysis, Lesley McGee for arbitrarily primed PCR analysis, and Don Low and Avril Wasas for performing the serotyping on the clinical isolates. Cara Mendick performed the cross-resistance analysis on the mutant and transformed strains.
REFERENCES
- 1.Abdurashidova G G, Turchinsky M F, Aslanov K A, Budowsky E I. Polynucleotide-protein interactions in the translation system. Identification of proteins interacting with tRNA in the A- and P- sites of E. coli ribosomes. Nucleic Acids Res. 1979;6:3891–3909. doi: 10.1093/nar/6.12.3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agafonov D E, Kolb V A, Spirin A S. Proteins on ribosome surface: measurements of protein exposure by hot tritium bombardment technique. Proc Natl Acad Sci USA. 1997;94:12892–12897. doi: 10.1073/pnas.94.24.12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons, Inc.; 1995. [Google Scholar]
- 4.Bernabeu C, Vázquez D, Ballesta J P G. The involvement of protein L16 on ribosomal peptidyl transferase activity. Eur J Biochem. 1977;79:469–472. doi: 10.1111/j.1432-1033.1977.tb11829.x. [DOI] [PubMed] [Google Scholar]
- 5.Chittum H S, Champney W S. Ribosomal protein gene sequence changes in erythromycin-resistant mutants of Escherichia coli. J Bacteriol. 1994;176:6192–6198. doi: 10.1128/jb.176.20.6192-6198.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davies C, Bussiere D E, Golden B L, Porter S J, Ramakrishnan V, White S W. Ribosomal proteins S5 and L6: high resolution crystal structures and roles in protein synthesis and antibiotic resistance. J Mol Biol. 1998;279:873–888. doi: 10.1006/jmbi.1998.1780. [DOI] [PubMed] [Google Scholar]
- 7.de Béthune M P, Nierhaus K H. Characterisation of the binding of virginiamycin S to Escherichia coli ribosomes. Eur J Biochem. 1978;86:187–191. doi: 10.1111/j.1432-1033.1978.tb12298.x. [DOI] [PubMed] [Google Scholar]
- 8.Franceschi F J, Nierhaus K H. Ribosomal proteins L15 and L16 are mere late assembly proteins of the large ribosomal subunit. Analysis of an Escherichia coli mutant lacking L15. J Biol Chem. 1990;265:16676–16682. [PubMed] [Google Scholar]
- 9.Funatsu G, Wittmann H G. Ribosomal proteins. Location of amino acid replacements in protein S12 isolated from Escherichia coli mutants resistant to streptomycin. J Mol Biol. 1972;68:547–550. doi: 10.1016/0022-2836(72)90108-8. [DOI] [PubMed] [Google Scholar]
- 10.Ganguly A K, Girijavallabhan V M, Miller G H, Sarre O Z. Chemical modification of everninomicin. J Antibiot. 1982;35:561–570. doi: 10.7164/antibiotics.35.561. [DOI] [PubMed] [Google Scholar]
- 11.Ganguly A K, Pramanik B, Chan T-M, Sarre O Z, Liu Y-T, Morton J, Girijavallabhan V M. The structure of the new oligosaccharide antibiotics, 13-384 components 1 and 5. Heterocycles. 1989;28:83–88. [Google Scholar]
- 12.Ganguly A K, McCormick J L, Chan T-M, Saksena A K, Das P R. Determination of the absolute stereochemistry at the C16 orthoester of everninomicin antibiotics: a novel acid-catalyzed isomerization of orthoesters. Tetrahedron Lett. 1997;38:7989–7992. [Google Scholar]
- 13.Hampl H, Schulze H, Nierhaus K H. Ribosomal components from Escherichia coli 50S subunits involved in the reconstitution of the peptidyltransferase activity. J Biol Chem. 1981;256:12810–12815. [PubMed] [Google Scholar]
- 14.Havarstein L S, Coomaraswamy G, Morrison D A. An unmodified heptadecapeptide induces competence for genetic transformation in Streptococcus pneumoniae. Proc Nat Acad Sci USA. 1995;92:11140–11144. doi: 10.1073/pnas.92.24.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henkin T M, Moon S H, Mattheakis L C, Nomura M. Cloning and analysis of the spc ribosomal protein operon of Bacillus subtilis: comparison with the spc operon of Escherichia coli. Nucleic Acids Res. 1989;17:7469–7486. doi: 10.1093/nar/17.18.7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones R N, Barrett M S. Antimicrobial activity of everninomicin (evernimicin), an oligosaccharide antimicrobial with a potent Gram-positive spectrum. Clin Microbiol Infect. 1995;1:35–43. doi: 10.1111/j.1469-0691.1995.tb00022.x. [DOI] [PubMed] [Google Scholar]
- 17.Lambert J M, Traut R R. The subunit interface of the Escherichia coli ribosome. Identification of proteins at the interface between the 30S and 50S subunits by crosslinking with 2-iminothiolane. J Mol Biol. 1981;149:451–476. doi: 10.1016/0022-2836(81)90481-2. [DOI] [PubMed] [Google Scholar]
- 18.Lindahl L, Sor F, Archer R H, Nomura M, Zengel J M. Transcriptional organization of the S10, spc and α operons of Escherichia coli. Biochem Biophys Acta. 1990;1050:337–342. doi: 10.1016/0167-4781(90)90191-4. [DOI] [PubMed] [Google Scholar]
- 19.Maimets T, Remme J, Villems R. Ribosomal protein L16 binds to the 3′-end of transfer RNA. FEBS Lett. 1984;166:53–56. doi: 10.1016/0014-5793(84)80043-5. [DOI] [PubMed] [Google Scholar]
- 20.Moureau P, di Giambattista M, Cocito C. The lasting ribosome alteration produced by virginiamycin M disappears upon removal of certain ribosomal proteins. Biochim Biophys Acta. 1983;739:164–172. doi: 10.1016/0167-4781(83)90026-x. [DOI] [PubMed] [Google Scholar]
- 21.Nag B, Glitz D G, Tewari D S, Traut R R. Probing the functional role and localization of the Escherichia coli ribosomal protein L16 with a monoclonal antibody. J Biol Chem. 1991;266:11116–11121. [PubMed] [Google Scholar]
- 22.Porse B T, Leviev I, Mankin A S, Garrett R A. The antibiotic thiostrepton inhibits functional transition within protein L11 at the ribosomal GTPase centre. J Mol Biol. 1998;276:391–404. doi: 10.1006/jmbi.1997.1541. [DOI] [PubMed] [Google Scholar]
- 23.Smith A M, Klugman K P. “Megaprimer” method of PCR-based mutagenesis: the concentration of megaprimer is a critical factor. BioTechniques. 1997;22:438–442. doi: 10.2144/97223bm13. [DOI] [PubMed] [Google Scholar]
- 24.Suh J W, Boylan S A, Oh S H, Price C W. Genetic and transcriptional organisation of the Bacillus subtilis spc-α region. Gene. 1996;169:17–23. doi: 10.1016/0378-1119(95)00757-1. [DOI] [PubMed] [Google Scholar]
- 25.Sumpter V G, Tate W P, Nierhaus K H. The complex between ribosomal proteins and aminoacyl-tRNA: the interactions and hydrolytic activities are not confined to the proteins L2 and L16 of Escherichia coli ribosomes. Biochim Biophys Acta. 1990;1048:265–269. doi: 10.1016/0167-4781(90)90065-a. [DOI] [PubMed] [Google Scholar]
- 26.Sumpter V G, Tate W P, Nowotny P, Nierhaus K H. Modification of histidine residues on proteins from the 50S subunit of the Escherichia coli ribosome. Effects on subunit assembly and peptidyl transferase centre activity. Eur J Biochem. 1991;196:255–260. doi: 10.1111/j.1432-1033.1991.tb15812.x. [DOI] [PubMed] [Google Scholar]
- 27.Tate W P, Schulze H, Nierhaus K H. The importance of the Escherichia coli ribosomal protein L16 for the reconstitution of the peptidyl-tRNA hydrolysis activity of peptide chain termination. J Biol Chem. 1983;258:12810–12815. [PubMed] [Google Scholar]
- 28.Tate W P, Sumpter V G, Trotman C N, Herold M, Nierhaus K H. The peptidyltransferase centre of the Escherichia coli ribosome. The histidine of protein L16 affects the reconstitution and control of the active centre but is not essential for release-factor-mediated peptidyl-tRNA hydrolysis and peptide bond formation. Eur J Biochem. 1987;165:403–408. doi: 10.1111/j.1432-1033.1987.tb11453.x. [DOI] [PubMed] [Google Scholar]
- 29.Teraoka H, Nierhaus K H. Protein L16 induces a conformational change when incorporated into a L16-deficient core derived from Escherichia coli ribosomes. FEBS Lett. 1978;88:223–226. doi: 10.1016/0014-5793(78)80179-3. [DOI] [PubMed] [Google Scholar]
- 30.Tomasz A, Hotchkiss R D. Regulation of the transformability of pneumococcal cultures by macromolecular cell products. Proc Natl Acad Sci USA. 1964;61:480–487. doi: 10.1073/pnas.51.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weinstein M J, Luedemann G M, Oden E M, Wagman G H. Everninomicin, a new antibiotic complex from Micromonospora carbonacea. Antimicrob Agents Chemother. 1965;9:24–32. [PubMed] [Google Scholar]
- 32.Wittmann H G, Stöffler G, Apirion D, Rosen L, Tanaka K, Tamaki M, Takata R, Dekio S, Otaka E. Biochemical and genetic studies on two different types of erythromycin resistant mutants of Escherichia coli with altered ribosomal proteins. Mol Gen Genet. 1973;127:175–189. doi: 10.1007/BF00333665. [DOI] [PubMed] [Google Scholar]
- 33.Wolf H. Avilamycin, an inhibitor of the 30S ribosomal subunit function. FEBS Lett. 1973;36:181–186. doi: 10.1016/0014-5793(73)80364-3. [DOI] [PubMed] [Google Scholar]