Abstract
Background:
Hepatocellular carcinoma (HCC) is one of the important causes of mortality due to malignancy. Toll-like receptors (TLRs) are very important in liver pathophysiology in terms of their roles in the innate immune system, such as the regulation of inflammation, wound healing, stimulation of adaptive immune responses, promotion of epithelial regeneration, and carcinogenesis. In this study, we planned to examine the role of TLR1 (rs4833095, rs5743551) and nucleotide-binding oligomerization domain (NOD2) (rs2066844, rs2066845, rs2066847) polymorphisms in the development of HCC and their effects on the clinical presentation of HCC patients.
Methods:
Our study was designed prospectively. Cirrhotic and HCC patients who were followed up in our clinic between January 2015 and September 2018 were included in the study. Sex, age, cirrhosis etiology, Child–Pugh class, and MELD scores were recorded. TLR1 and NOD2 polymorphisms were studied by the PCR method.
Results:
HCC developed in 88 (31.4%) of the 280 patients who were followed up, either during the recruitment phase of our study or during the follow-up. The mean follow-up time of our patient group was 17.04 ± 11.72 months, and the mean follow-up time of HCC patients was 12.09 ± 10.26 months. TLR1 (rs5743551) polymorphism was associated with HCC development (P = .003). TLR1 (rs5743551) and NOD2 (rs2066844) polymorphisms were associated with the development of spontaneous bacterial peritonitis (SBP) in the HCC patient group (P = .013 and P = .021, respectively).
Conclusion:
We think that increased bacterial translocation in cirrhotic patients may contribute to HCC development by causing chronic inflammation, especially in patients with TLR 1 (rs5743551) polymorphism.
Keywords: Toll-like receptors, nucleotide-binding oligomerization domain, hepatocellular carcinoma, spontaneous bacterial peritonitis
Introduction
Although hepatocellular carcinoma (HCC) is the fifth most frequently diagnosed malignant tumor, it is the third leading cause of mortality.1 The frequency of HCC varies according to the geographic region, race, or ethnic group. However, the incidence is higher in patients with hepatitis B virus, hepatitis C virus, chronic alcohol use, and non-alcoholic fatty liver disease.2 Although there are positive developments in the clinical diagnosis and treatment of HCC today, it is difficult to detect early and is characterized by a long incubation period, rapid development, high morbidity, and mortality.2,3 In clinical practice, very few biochemical, inflammatory, and genetic markers have a place in the diagnosis of HCC.4,5 For this reason, it is important to identify new diagnostic markers for diagnosis and follow-up.
Pattern recognition receptors (PRR) are divided into 2 groups, the toll-like receptors (TLRs) and nucleotide-binding oligomerization domain (NOD)-like receptors. The NOD-like receptors are members of the PRR family and play an important role in fighting bacterial pathogens. NOD2 plays an important role in activating host defense by recognizing bacterial components.6 There is evidence that NOD2 gene variants contribute to bacterial translocation by various mechanisms.7,8 In addition to an impaired monocyte/macrophage response in patients with NOD2 variants, there is some evidence that they may impair neutrophil activation and migration toward the inflammatory stimulus.9 TLRs have multiple antigen responses, and their specific types are found in high concentrations in a large number of cells in the liver.10,11 TLR1 is a member of the TLR family, forms a heterodimer with TLR2, and is a receptor that facilitates natural immune activation by interacting with bacterial cell wall components such as peptidoglycan, lipoteichoic acid, and lipopeptides.12 Human TLRs consist of a leucine-rich extracellular domain and a Toll/IL-1 receptor area in the intracellular domain. They are responsible for the recognition of bacterial, viral, and fungal structures.13 TLRs stimulate the activation of various signaling pathways, especially transcription factors and IL-6.14 These factors stimulate the induction of inflammatory genes and the release of the tumor necrosis factor.
Hepatocarcinogenesis is a process with serious changes in the genome.15 TLRs can provide the signal that acts as a molecular link between inflammation and tumor cells.16 TLRs are very important in liver pathophysiology in terms of their roles in the innate immune system, such as inflammation regulation, wound healing, stimulation of adaptive immune responses, promotion of epithelial regeneration, and carcinogenesis.17 There is a relationship between chronic inflammation and cancer.18 Continuous exposure of the liver to intestinal-derived bacterial products, viral infection, alcohol, or other products through the portal vein can cause chronic liver inflammation, thus increasing the risk of HCC.5,19 There are studies reporting that the activation of TLRs plays a role in the progression of the inflammation–fibrosis–HCC axis.20,21 Little is known about PRR functions in HCC patients today. In this study, we aimed to examine the role of TLR1 (rs4833095, rs5743551) and NOD2 (rs2066844, rs2066845, rs2066847) polymorphisms in the development of HCC and their effects on the clinical presentation of HCC patients.
Materials and Methods
Patient Population
Our study was designed prospectively. There were 305 cirrhotic patients included in the study who were followed up at Necmettin Erbakan University, Meram Faculty gastroeterology clinic Medical Faculty gastroenterology clinic, between January 1, 2015 and September 1, 2018. Informed written consent was obtained from all patients. The study included cirrhotic patients, cirrhotic patients undergoing paracentesis for diagnostic purposes, and patients with HCC. Patients with any malignancy other than primary liver tumors, who used immunosuppressive therapy, who were not followed up regularly, or who did not consent to participate in the study were excluded from the study. Secondary peritonitis was ruled out with clinical and radiological findings, and patients with secondary peritonitis were excluded from the study.
The diagnosis of cirrhosis and HCC was based on the histological criteria obtained from a liver biopsy, and/or on the evaluation of clinical, biochemical, and imaging data. According to the 2012 American Association for the Study of Liver Diseases Practice Guidelines,22 patients admitted to the hospital with ascites and patients who developed signs, symptoms, or laboratory signs suggestive of infection (e.g., abdominal pain/discomfort, fever, encephalopathy, kidney insufficiency, acidosis, or leukocytosis) were diagnosed with spontaneous bacterial peritonitis (SBP) based on the number of polymorph nuclear leukocyte (PNL) in ascites fluid in diagnostic paracentesis being ≥250 cells/mm3. In patients with a red blood cell count of >10 000 cells/mm3 in ascites fluid, a PNL was obtained per 250 red blood cells.22 Age, sex, cirrhosis etiology, Child–Pugh class,23 and Model for End-Stage Liver Disease (MELD) scores24 were recorded as soon as patients were included in the study.
The patients were followed up regularly, from the moment of recruitment until the moment of death.
Sample and Data Collection
Ascites were sampled from each patient under sterile conditions. For each patient, 2.7 mL of ascites was placed in a tube with ethylenediamine tetra acetic acid (EDTA), and the number of ascites cells was determined by an automatic blood cell counting machine. Patients with leukocyte count ≥ 250/mL in the ascites sample were confirmed by cell count under a light microscope. For TLR 1 and NOD2 gene analysis, 9 mL of venous blood was taken into the tube with EDTA and stored at −80°C until the day of analysis.
DNA Isolation and Genotyping
Genomic DNA was isolated from the blood samples using the Genomic DNA Isolation Kit From Blood (RTA, Turkey), according to the manufacturer’s protocol. All extracted DNA samples were immobilized to 10 ng/µL for 2 SNPs in the TLR1 gene (rs5743551 and rs4833095) and 3 SNPs in the NOD2 gene (rs2066847, rs2066844, and rs2066845). Polymorphisms in the TLR1 and NOD2 genes were determined by real-time polymerase chain reaction (RT-PCR) using TaqMan SNP Genotyping Assay (Assay ID: C_1180670_30 for rs5743551, Assay ID: C_44103606_10 for rs4833095, Assay ID: C_11717468_20 for rs2066844, and Assay ID: C_11717466_20 for rs2066845). The RT-PCR system was used according to the manufacturer’s instructions (LightCycler 96). RT-PCR was conducted in a total volume of 50 µL, using the following reagents: 25 µL Thermo Fisher TaqMan assay master mix, 2.5 μL TaqMan assay primer probe, 17.5 μL PCR-grade water, and 5 μL genomic DNA. Probes specific for rs2066847 were obtained from Macrogen. A total volume of 20 μL, consisting of 1 μL forward primer, 1 μL reverse primer, 1 μL FAM probe, 1 μL VIC probe, 10 μL TaqMan master mix, 1 μL PCR-grade water, and 5 μL genomic DNA was used for RT-PCR. Briefly, the PCR conditions were as follows: denaturation at 95°C for 10 minutes, amplification at 95°C for 15 seconds, and at 60°C for 1 minute. Melting temperature (Tm) was 60°C for optimum results. All results were analyzed using the LightCycler 96 software endpoint genotyping.
Statistics
The results of our study were analyzed with the SPSS 19.0 program. Data that received continuous values were given as mean (±standard deviation), and categorical data as frequency and percentage (n, %). The data were tested for compliance with the normal distribution using the Kolmogorov–Smirnov test, histogram, and standard deviation. Non-parametric data of the groups were compared using the Mann–Whitney U-test, and parametric data were compared used using the independent t-test. The chi-square test was used to test categorical data. The effect of risk factors on survival was examined with the log-rank (Mantel–Cox) test. Survival rates were calculated by the Kaplan–Meier method. A value of P < .05 was considered statistically significant.
Results
Patients’ Demographic and Clinical Data
A total of 305 patients were included in the study. Four of the patients were excluded from the study because secondary malignancy developed in 6 patients and we were not technically able to study gene polymorphism, and 15 patients did not follow-up or their follow-up data were missing. A total of 280 patients were evaluated within the scope of the study. In 88 (31.4%) of 280 patients who were followed up during our study, HCC developed at the time of recruitment or follow-up. Our patient group was followed up for 36 months. The mean follow-up time of our patient group was 17.04 ± 11.72 months and the mean follow-up time of HCC patients was 12.09 ± 10.26 months. The mean age of all patients was 60.7 ± 12.6, and 168 (60%) of the patients were male. The etiologies of the cases investigated were as follows: viral hepatitis was found in 163 (58.2%), autoimmune hepatitis in 18 (6.4%), alcohol in 12 (4.3%), non-alcoholic steatohepatitis in 10 (3.6%), the etiology was cryptogenic in 61 (21.8%), and 16 (5.7%) cases had other causes. The Child–Pugh A/B/C classification at the time of admission was 99/134/47 (35.3%/47.8%/16.9%), respectively, and the MELD score was 10.8 ± 6.5 (6-41) (Table 1).
Table 1.
Clinical and Demographic Characteristics and Laboratory Data of the Groups
| HCC (+) (n = 88) | HCC (−) (n = 192) | All Patients (n = 280) | P | |
|---|---|---|---|---|
| Age(years) | 62.3 ± 10.0 | 59.9 ± 13.5 | 60.7 ± 12.5 | .142 |
| Sex | .007 | |||
| Male, n (%) | 63 (71.6) | 105 (54.7) | 159 (60.2) | |
| Female, n (%) | 25 (28.4) | 87 (45.3) | 105 (39.8) | |
| MELD Score | 10.7 ± 6.6 | 10.9 ± 6.4 | 10.8 ± 6.5 | .714 |
| Child–Pugh Class | .564 | |||
| A, n (%) | 34 (38.6) | 65 (33.8) | 99 (35.3) | |
| B, n (%) | 42 (47.7) | 92 (47.9) | 134 (47.8) | |
| C, n (%) | 12 (13.6) | 35 (18.3) | 47 (16.9) | |
| Etiology | <.001 | |||
| Chronic hepatitisB, n (%) | 57 (64.8) | 68 (35.4) | 125 (44.6) | |
| Chronic hepatitisC, n (%) | 15 (17.0) | 23 (12.0) | 38 (13.6) | |
| Cryptogenic, n (%) | 9 (10.2) | 52 (27.1) | 61 (21.8) | |
| Autoimmune, n (%) | 0 (0) | 18 (9.4) | 18 (6.4) | |
| Alcohol, n (%) | 2 (2.3) | 10 (5.2) | 12 (4.3) | |
| Other, n (%) | 5 (5.7) | 21 (10.9) | 26 (9.3) | |
| Creatinine (mg/dL) | 0.83 ± 0.25 | 0.85 ± 0.37 | 0.85 ± 0.34 | .666 |
| Albumin (g/dL) | 3.30 ± 0.67 | 3.10 ± 0.59 | 3.17 ± 0.62 | .045 |
| Total bilirubin (mg/dL) | 2.37 ± 3.28 | 3.76 ± 5.89 | 3.32 ± 5.23 | .029 |
| Leukocyte (1000/mm3) | 7.03 ± 4.36 | 6.25 ± 3.10 | 6.50 ± 3.50 | .128 |
| Platelet (1000/mm3) | 126.0 ± 64.30 | 124.00 ± 66.60 | 124.6 ± 65.80 | .832 |
| INR | 1.40 ± 0.30 | 1.50 ± 0.41 | 1.47 ± 0.40 | .075 |
| AFP (U/mL) | 892.8 ± 3869.8 | 7.31 ± 20.44 | 278.61 ± 2170.0 | .006 |
HCC, hepatocellular cancer; AFP, alpha-fetoprotein; MELD, model for end-stage liver disease; INR, international normalized ratio.
Relationship of TLR 1 and NOD 2 Polymorphism with HCC and SBP
TLR 1 (rs4833095) polymorphism was not associated with HCC development in our patient group (P = .081), while TLR 1 (rs5743551) polymorphism was associated with HCC development (P = .003). NOD2 polymorphisms (rs2066844, rs2066845, rs2066847) were not associated with HCC development. Laboratory, clinical, and demographic characteristics of all patients with and without HCC are given in Table 2. Sixty-one patients with HCC developed ascites. During the follow-up of the patient group with HCC, 26 patients were found to have developed SBP. TLR1 (rs5743551) and NOD2 (rs2066844) polymorphisms in the HCC patient group were associated with SBP development (P = .013 and P = .021, respectively) (Table 3).
Table 2.
Hepatocellular Carcinoma Development: The Relationship Between TLR1 and NOD2 Polymorphisms
| HCC (+) (n = 88) | HCC (−) (n = 192) | All Patients (n = 280) | P | |
|---|---|---|---|---|
| TLR1 | ||||
| rs4833095 | .081 | |||
| Wild type, n (%) | 17 (19.3) | 56 (29.2) | 73 (26.1) | |
| Variant, n (%) | 71 (80.7) | 136 (70.8) | 207 (73.9) | |
| rs5743551 | .003 | |||
| Wild type, n (%) | 36 (40.9) | 115 (59.9) | 137 (51.9) | |
| Variant, n (%) | 52 (59.1) | 77 (40.1) | 127 (48.1) | |
| NOD2 | ||||
| rs2066844 | .156 | |||
| Wild type, n (%) | 73 (83.0) | 171 (89.1) | 244 (87.1) | |
| Variant, n (%) | 21 (17.0) | 15 (10.9) | 36 (12.9) | |
| rs2066845 | .251 | |||
| Wild type, n (%) | 84 (95.5) | 188 (97.9) | 272 (97.1) | |
| Variant, n (%) | 4 (4.5) | 4 (2.1) | 8 (2.9) | |
| rs2066847 | .340 | |||
| Wild type, n (%) | 76 (86.4) | 157 (81.8) | 233 (83.2) | |
| Variant, n (%) | 12 (13.6) | 35 (18.2) | 47 (16.8) |
HCC, hepatocellular cancer; TLR, toll-like receptors; NOD2, nucleotide-binding oligomerization domain 2.
Table 3.
Relationship Between TLR1 and NOD2 Polymorphisms and the Development of Spontaneous Bacterial Peritonitis in Patients with Hepatocellular Carcinoma
| Peritonitis (+) (n = 26) | Peritonitis (−) (n = 35) | All Patients (n = 61) | P | |
|---|---|---|---|---|
| TLR1 | ||||
| rs4833095 | .070 | |||
| Wild type, n (%) | 2 (7.7) | 9 (25.7) | 11 (18.0) | |
| Variant, n (%) | 24 (92.3) | 26 (74.3) | 50 (82.0) | |
| rs5743551 | .013 | |||
| Wild type, n (%) | 4 (15.4) | 16 (45.7) | 20 (32.8) | |
| Variant, n (%) | 22 (84.6) | 19 (54.3) | 41 (67.2) | |
| NOD2 | ||||
| rs2066844 | .021 | |||
| Wild type, n (%) | 19 (73.1) | 33 (94.3) | 52 (85.2) | |
| Variant, n (%) | 7 (26.9) | 2 (5.7) | 9 (14.8) | |
| rs2066845 | .388 | |||
| Wild type, n (%) | 24 (92.3) | 34 (97.1) | 58 (95.1) | |
| Variant, n (%) | 2 (7.7) | 1 (2.9) | 3 (4.9) | |
| rs2066847 | .224 | |||
| Wild type, n (%) | 20 (76.9) | 31 (88.6) | 51 (83.6) | |
| Variant, n (%) | 6 (23.1) | 4 (11.4) | 10 (16.4) |
HCC, hepatocellular carcinoma; TLR, toll-like receptors; NOD2, nucleotide-binding oligomerization domain 2.
During the whole follow-up, the survival rates of the patient group with HCC and the patient group without HCC were compared, and the survival of the group without HCC was significantly higher (Log-Rank (Mantel–Cox), chi-square: 39.053, P < .001) (Figure 1). When survival was evaluated in the 1-year follow-up of patients who developed peritonitis in the HCC patient group and those without peritonitis, survival was low in the patient group who developed peritonitis (Log-Rank (Mantel–Cox), chi-square: 4.954, P = .026) (Figure 2).
Figure 1.
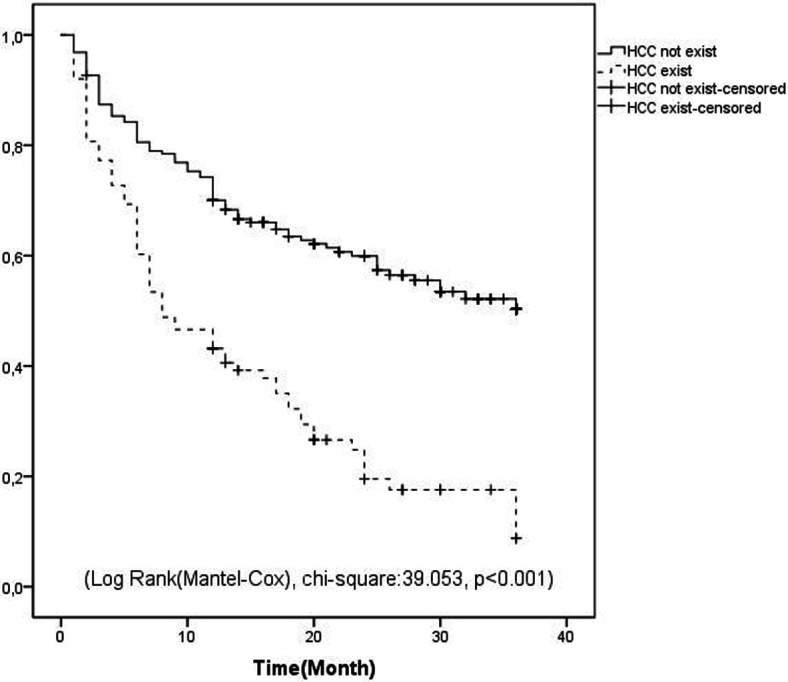
Kaplan–Meier curve indicating the survival of patients with and without hepatocellular carcinoma.
Figure 2.
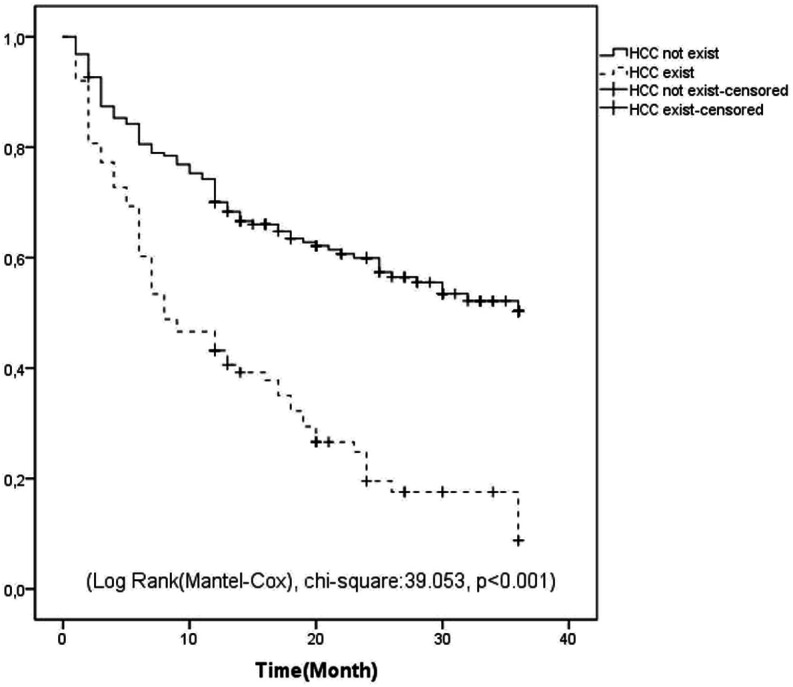
Relationship of spontaneous development of bacterial peritonitis with mortality, in patients with hepatocellular carcinoma.
Discussion
Carcinogenesis is associated with inflammation in the innate and adaptive immune systems, which are components of the immune system. PRRs are an important component of the innate immune system.1 TLRs are a member of PRR family and activate common signal pathways through TLR-specific adapter proteins. To date, 10 TLRs have been identified in humans. These TLRs have been shown in a variety of innate immune cells, including macrophages, neutrophils, and dendritic cells, as well as non-immune cells, including epithelial and endothelial cells.25 TLRs cause tumorigenesis with different mechanisms. It should be considered that chronic inflammatory components, either directly or as a result of TLR release in the cytokine, play an important role in tumorigenesis.26 Tumor cells can escape from the immune system as a result of suppression of the NF-kB signal pathway, a component of TLRs.12 Immune system cells such as macrophages, T lymphocytes, and granulocytes play an important role in angiogenesis, especially by synthesizing cytokines such as IL-1, IL-6, and vascular endothelial growth factor. These cytokines affect all endothelial cell functions, including activation in the tumor microenvironment.8 TLR4, by binding to microbial lipopolysaccharides, regulates the inflammatory response of the liver, causing hepatic fibrosis and hepatocarcinogenesis.27-29 Studies have shown that the relationship between intestinal microbiota and TLR4 can cause HCC development and mediate HCC progression. Moreover, the relationship between intestinal microbiota and TLR4 has been shown to cause the expression of the hepatomitogen epiregulin and the reduction of apoptosis. On the other hand, gut sterilization or TLR4 inactivation has been shown to significantly reduce HCC development.20 Thus, it seems clear that TLRs play a role in the development of inflammatory hepatic tumor. In addition to their known role in chronic inflammation, TLR2 signals are known to mediate tumor cells’ escape from immunity and tumor progression.30 In our study, the development of HCC was related to TLR 1 (rs5743551) polymorphism. Indeed, there are studies in the literature where TLR variants are associated with the risk of various cancers.31 TLR 4 and TLR 2-deficient mice have marked reductions in the incidence, size, and number of chemical-induced liver cancer, indicating a strong contribution of the TLR signal to hepatocarcinogenesis.32 In their study, Zehran et al. found that the increase of TLR 2 and TLR 4 expression in peripheral monocytes was related with the development of HCC, disease progression, and poor prognosis. In the same study, increased TLR 2 expression was found to be associated with poor response to treatment.5 In another study, TLR 2 and TLR 4 over-expression was shown in HCC cells.33 It is thought that bacterial pathogen-associated molecular patterns may contribute to the development of HCC, due to the increased intestinal mucosal permeability in cirrhotic patients and increased translocation of intestinal bacteria.27 In studies conducted, the increased bacterial translocation in cirrhotic patients has been shown to trigger chronic inflammation.7,21 Considering the relationship between chronic inflammation and cancer development, we attach importance to TLR1 polymorphism and HCC development in our study. In our study group, the more frequent presence of SBP in patients with TLR1 (rs5743551) and NOD2 (rs2066844) polymorphisms supports bacterial translocation and chronic inflammation.
Genetic variations play an important role in the course of infection in patients34 and functional polymorphisms in PRR alter the detection and clearance of bacterial pathogens, affecting their natural defense mechanisms.10,11 The effects of PRR gene polymorphisms on the development of SBP and mortality have been evaluated previously; there are studies in the literature showing the relationship of NOD2 polymorphisms with SBP.35 Appenrodt et al.36 showed that NOD2-risk alleles were related to the development of SBP in cirrhotic patients and increased mortality by a factor of 4. The TLR1 receptor, a component of the natural immune system, is known to facilitate natural immune activation.12
The weakness of our study is particularly that the number of participants is low, and it is not supported by TLR1 and NOD2 gene expressions in HCC tissue.
In our study, a relationship was found between TLR1 (rs5743551) polymorphism and HCC development. Interestingly, we showed that the development of SBP significantly increased in HCC patients with TLR1 (rs5743551) and NOD2 (rs2066844) polymorphism. When these data are evaluated together with the data in the literature, we think that increased bacterial translocation in cirrhotic patients may contribute to the development of HCC by causing chronic inflammation, especially in patients with TLR 1 (rs5743551) polymorphism. We think that this data should be supported by studies conducted in larger patient groups.
Funding Statement
The authors declare that this study has received no financial support.
Footnotes
Ethics Committee Approval: Ethics committee approval was received for this study from the Ethics Committee of Necmettin Erbakan University Meram Medical Faculty, who approved this study protocol (2015/1459).
Informed Consent: Informed consent was obtained from the patients who participated in this study.
Peer review: Externally peer-reviewed.
Author Contributions: Concept – R.D., M.A.; Design – R.D., H.A.; Supervision – R.D., M.A.; Fudıng – R.D.; Materials – R.D., M.B.; Data Collection and/or Processing – A.K., M.K, Y.K, M.B.; Analysis and/or Interpretation – R.D., M.A., M.B., A.K., M.K., Y.K., M.B., H.A.; Literature Search – A.K., M.K., M.B.; Writing – R.D., M.B., H.A.; Critical Reviews – M.A., M.B., H.A.
Acknowledgments: We appreciate all the participants who supported this study.
Conflict of Interest: The authors have no conflict of interest to declare.
References
- 1. . Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108.. 10.3322/caac.21262) [DOI] [PubMed] [Google Scholar]
- 2. . Altekruse SF, McGlynn KA, Reichman ME. Hepatocellular carcinoma incidence, mortality, and survival trends in the United States from 1975 to 2005. J Clin Oncol. 2009;27(9):1485-1491. 10.1200/JCO.2008.20.7753) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. . Njei B, Rotman Y, Ditah I, Lim JK. Emerging trends in hepatocellular carcinoma incidence and mortality. Hepatology. 2015;61(1):191–199.. 10.1002/hep.27388) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. . Mehmet A, Dertli R. Serum soluble TWEAK concentration is decreased in patients with hepatocellular cancer and low blood levels of sTWEAK are associated with poor survival. Int J Hematol Oncol;28(4):118–126.. [Google Scholar]
- 5. . Zahran AM, Zahran ZAM, El-Badawy O, et al. Prognostic impact of toll-like receptors 2 and 4 expression on monocytes in Egyptian patients with hepatocellular carcinoma. Immunol Res. 2019;67(2-3):157–165.. 10.1007/s12026-019-09075-x) [DOI] [PubMed] [Google Scholar]
- 6. . Ogura Y, Bonen DK, Inohara N, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature. 2001;411(6837):603–606.. 10.1038/35079114) [DOI] [PubMed] [Google Scholar]
- 7. . Harputluoglu MM, Dertli R, Otlu B, et al. Nucleotide-binding oligomerization domain-containing protein 2 variants in patients with spontaneous bacterial peritonitis. Dig Dis Sci. 2016;61(6):1545–1552.. 10.1007/s10620-015-4024-y) [DOI] [PubMed] [Google Scholar]
- 8. . Fritz JH, Ferrero RL, Philpott DJ, Girardin SE. Nod-like proteins in immunity, inflammation and disease. Nat Immunol. 2006;7(12):1250–1257.. 10.1038/ni1412) [DOI] [PubMed] [Google Scholar]
- 9. . Ekman AK, Cardell LO. The expression and function of Nod‐like receptors in neutrophils. Immunology. 2010;130(1):55–63.. 10.1111/j.1365-2567.2009.03212.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. . Henao-Mejia J, Elinav E, Thaiss CA, Licona-Limon P, Flavell RA. Role of the intestinal microbiome in liver disease. J Autoimmun. 2013;46:66–73.. 10.1016/j.jaut.2013.07.001) [DOI] [PubMed] [Google Scholar]
- 11. . Zhu Q, Zou L, Jagavelu K, et al. Intestinal decontamination inhibits TLR4 dependent fibronectin-mediated cross-talk between stellate cells and endothelial cells in liver fibrosis in mice. J Hepatol. 2012;56(4):893–899.. 10.1016/j.jhep.2011.11.013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. . Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on toll-like receptors. Nat Immunol. 2010;11(5):373-384. 10.1038/ni.1863) [DOI] [PubMed] [Google Scholar]
- 13. . Moynagh PN. TLR signalling and activation of IRFs: revisiting old friends from the NF-κB pathway. Trends Immunol. 2005;26(9):469–476.. 10.1016/j.it.2005.06.009) [DOI] [PubMed] [Google Scholar]
- 14. . O’Neill LA, Bryant CE, Doyle SL. Therapeutic targeting of toll-like receptors for infectious and inflammatory diseases and cancer. Pharmacol Rev. 2009;61(2):177–197.. 10.1124/pr.109.001073) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. . Huang YH, Wu JC, Chen CH, et al. Comparison of recurrence after hepatic resection in patients with hepatitis B vs. hepatitis C‐related small hepatocellular carcinoma in hepatitis B virus endemic area. Liver Int. 2005;25(2):236–241.. 10.1111/j.1478-3231.2005.01081.x) [DOI] [PubMed] [Google Scholar]
- 16. . Hornung V, Rothenfusser S, Britsch S, et al. Quantitative expression of toll-like receptor 1-10 mRNA in cellular subsets of human peripheral blood mononuclear cells and sensitivity to CpG oligodeoxynucleotides. J Immunol. 2002;168(9):4531–4537.. 10.4049/jimmunol.168.9.4531) [DOI] [PubMed] [Google Scholar]
- 17. . Kawai T, Adachi S, Ogawa T, Takeda K, Akira S. TLR signaling. Cell Death Differ. 2006;13(5):816–825.. 10.1038/sj.cdd.4401850) [DOI] [PubMed] [Google Scholar]
- 18. . Heidland A, Klassen A, Rutkowski P, Bahner U, et al. The contribution of Rudolf Virchow to the concept of inflammation: what is still of importance? J Nephrol. 2006;19(suppl 10):S102–S109.. [PubMed] [Google Scholar]
- 19. . Hetta HF, Mekky MA, Khalil NK, et al. Association of colonic regulatory T cells with hepatitis C virus pathogenesis and liver pathology. J Gastroenterol Hepatol. 2015;30(10):1543–1551.. 10.1111/jgh.12936) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. . Roh YS, Seki E. Toll‐like receptors in alcoholic liver disease, non‐alcoholic steatohepatitis and carcinogenesis. J Gastroenterol Hepatol. 2013;28(suppl 1):38–42.. 10.1111/jgh.12019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. . Lopes JAG, Borges-Canha M, Pimentel-Nunes P. Innate immunity and hepatocarcinoma: can toll-like receptors open the door to oncogenesis? World J Hepatol. 2016;8(3):162-182. 10.4254/wjh.v8.i3.162) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. . Runyon BA.American Association for the Study of Liver Diseases (AASLD). Introduction to the revised American Association for the Study of Liver Diseases Practice Guideline management of adult patients with ascites due to cirrhosis 2012. Hepatology. 2013;57(4):1651–1653.. 10.1002/hep.26359) [DOI] [PubMed] [Google Scholar]
- 23. . Pugh RN, Murray‐Lyon IM, Dawson JL, Pietroni MC, Williams R. Transection of the oesophagus for bleeding oesophageal varices. Br J Surg. 1973;60(8):646–649.. 10.1002/bjs.1800600817) [DOI] [PubMed] [Google Scholar]
- 24. . Botta F, Giannini E, Romagnoli P, et al. MELD scoring system is useful for predicting prognosis in patients with liver cirrhosis and is correlated with residual liver function: a European study. Gut. 2003;52(1):134–139.. 10.1136/gut.52.1.134) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. . Goutagny N, Estornes Y, Hasan U, Lebecque S, Caux C. Targeting pattern recognition receptors in cancer immunotherapy. Target Oncol. 2012;7(1):29–54.. 10.1007/s11523-012-0213-1) [DOI] [PubMed] [Google Scholar]
- 26. . Pradere JP, Dapito DH, Schwabe RF. The Yin and Yang of toll-like receptors in cancer. Oncogene. 2014;33(27):3485–3495.. 10.1038/onc.2013.302) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. . Yu LX, Yan HX, Liu Q, et al. Endotoxin accumulation prevents carcinogen‐induced apoptosis and promotes liver tumorigenesis in rodents. Hepatology. 2010;52(4):1322–1333.. 10.1002/hep.23845) [DOI] [PubMed] [Google Scholar]
- 28. . Singh A, Koduru B, Carlisle C, et al. NADPH oxidase 4 modulates hepatic responses to lipopolysaccharide mediated by Toll-like receptor-4. Sci Rep. 2017;7(1):14346. 10.1038/s41598-017-14574-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. . Dapito DH, Mencin A, Gwak GY, et al. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell. 2012;21(4):504–516.. 10.1016/j.ccr.2012.02.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. . Shi W, Su L, Li Q, et al. Suppression of toll-like receptor 2 expression inhibits the bioactivity of human hepatocellular carcinoma. Tumour Biol. 2014;35(10):9627–9637.. 10.1007/s13277-014-2268-3) [DOI] [PubMed] [Google Scholar]
- 31. . Stevens VL, Hsing AW, Talbot JT, et al. Genetic variation in the toll‐like receptor gene cluster (TLR10‐TLR1‐TLR6) and prostate cancer risk. Int J Cancer. 2008;123(11):2644–2650.. 10.1002/ijc.23826) [DOI] [PubMed] [Google Scholar]
- 32. . Seki E, Brenner DA. Toll-like receptors and adaptor molecules in liver disease: update. Hepatology. 2008;48(1):322–335.. 10.1002/hep.22306) [DOI] [PubMed] [Google Scholar]
- 33. . Zhe Y, Li Y, Liu D, et al. Extracellular HSP70-peptide complexes promote the proliferation of hepatocellular carcinoma cells via TLR2/4/JNK1/2MAPK pathway. Tumour Biol. 2016;37(10):13951–13959.. 10.1007/s13277-016-5189-5) [DOI] [PubMed] [Google Scholar]
- 34. . Chapman SJ, Hill AV. Human genetic susceptibility to infectious disease. Nat Rev Genet. 2012;13(3):175-188. 10.1038/nrg3114) [DOI] [PubMed] [Google Scholar]
- 35. . Dinya T, Tornai T, Vitalis Z, et al. Functional polymorphisms of innate immunity receptors are not risk factors for the non‐SBP type bacterial infections in cirrhosis. Liver Int. 2018;38(7):1242–1252.. 10.1111/liv.13664) [DOI] [PubMed] [Google Scholar]
- 36. . Appenrodt B, Grünhage F, Gentemann MG, et al. Nucleotide‐binding oligomerization domain containing 2 (NOD2) variants are genetic risk factors for death and spontaneous bacterial peritonitis in liver cirrhosis. Hepatology. 2010;51(4):1327–1333.. 10.1002/hep.23440) [DOI] [PubMed] [Google Scholar]


