Abstract
Background
CRISPR-Cas systems have expanded the possibilities for gene editing in bacteria and eukaryotes. There are many excellent tools for designing CRISPR-Cas guide RNAs (gRNAs) for model organisms with standard Cas enzymes. GuideMaker is intended as a fast and easy-to-use design tool for challenging projects with (i) non-standard Cas enzymes, (ii) non-model organisms, or (iii) projects that need to design a panel of gRNA for genome-wide screens.
Findings
GuideMaker can rapidly design gRNAs for gene targets across the genome using a degenerate protospacer-adjacent motif (PAM) and a genome. The tool applies hierarchical navigable small world graphs to speed up the comparison of guide RNAs and optionally provides on-target and off-target scoring. This allows the user to design effective gRNAs targeting all genes in a typical bacterial genome in ∼1–2 minutes.
Conclusions
GuideMaker enables the rapid design of genome-wide gRNA for any CRISPR-Cas enzyme in non-model organisms. While GuideMaker is designed with prokaryotic genomes in mind, it can efficiently process eukaryotic genomes as well. GuideMaker is available as command-line software, a stand-alone web application, and a tool in the CyCverse Discovery Environment. All versions are available under a Creative Commons CC0 1.0 Universal Public Domain Dedication.
Keywords: PAM, CRISPR-Cas, gRNA, Perturb-seq, Hierarchical Navigable Small World graph
Introduction
CRISPR-Cas technology enables rapid and efficient genome editing in both prokaryotic and eukaryotic cells [1, 2]. CRISPR-based systems are set apart from other genome-editing tools by the ease with which they can be programmed to target specific sequences. Almost any DNA sequence in the cell can be targeted if it possesses a compatible protospacer-adjacent motif (PAM). The PAM is a sequence that flanks the DNA target site, known as the protospacer, and must be present for target recognition [3]. The target-specifying guide RNA (gRNA) can be supplied as RNA, or encoded in DNA, depending on the organism under investigation. Although CRISPR-Cas is often used to edit single genes in eukaryotes, it is increasingly used for other purposes in prokaryotic and eukaryotic organisms [4].
The Streptococcus pyogenes Cas9 (SpCas9) was the first Cas described [5], and it is still the most widely used enzyme in CRISPR gene editing. Other Cas enzymes described early in the CRISPR revolution, such as the Staphylococcus aureus Cas9 (SaCas9) and the Acidaminococcus Cas12a, are also commonly used [6,7]. Accordingly, the parameters for these enzymes are often included in computational tools to identify CRISPR target sites [8–11]. Cas9 enzymes from other organisms and other Cas-associated proteins that can cleave double-stranded DNA, single-stranded DNA, and single-stranded RNA and insert transposon elements have also been described and have their place in molecular toolkits [12–18]. Each of these enzymes generally has specific requirements, such as PAM sequence constraints, PAM orientation, and protospacer length. Many of these CRISPR-Cas systems have been repurposed to enable molecular genetics techniques such as gene deletions, gene insertions, transcriptional depletion and activation, and translational repression [12,19–22]. Some of these techniques can be scaled to the genome level with chip-synthesized oligonucleotides and pooled approaches to screening [23]. In pooled screens, high-throughput DNA sequencing is used to identify how the pool has changed over time to elucidate genes that affect cells' fitness in specific conditions. Given the diversity of the CRISPR systems and their uses, identifying appropriate target sites is not trivial, especially for the number of targets needed for genome-scale experiments.
Here we introduce GuideMaker, a computational tool to identify target sites and design gRNA sequences that is not limited to any specific CRISPR system or organism. GuideMaker is most useful for a few kinds of CRISPR experiments. The first use case is designing pools of gRNAs for genome-wide screening experiments such as Perturb-seq and CRISPR pool [23,24]. GuideMaker is optimized for making the all-versus-all comparisons necessary to design a genome-wide screen and return candidate gRNAs for every gene locus. The tool allows the user to filter targets on the basis of their proximity to features of interest, such as the start codon for any coding sequence. The second major use case is for researchers working with non-model organisms. Online gRNA design tools often have a limited number of preselected genomes available for analysis because most methods require PAM site positions to be precomputed. GuideMaker rapidly computes all guide positions on demand from user-provided GenBank files or a set of GFF/GTF files and fasta files from any organism. The third use case is experiments with Cas enzymes other than the canonical versions of Cas9 and Cas12a (Cpf1), which have atypical PAM and target site requirements. GuideMaker allows the user to specify a custom PAM with variable length, including degenerate nucleotides, and allows the PAM to be on either the 3′ or 5′ side of the protospacer. These features allow GuideMaker to support any current or future CRISPR-Cas system. Because the determination of which CRISPR-Cas system functions best in any given organism is not predictable, this tool is highly relevant to researchers developing CRISPR tools in new species. For SpCas9 GuideMaker also implements on-target and off-target scoring from Doench et al. [8–11]. Because there are limited experimental data on most Cas/organism combinations, GuideMaker cannot calculate target scoring for other Cas enzymes but instead uses design heuristics that prioritize uniqueness in the seed region of the guide.
Methods
Main features, input parameters, and workflow
GuideMaker is designed to be easy to use as either a web application or a command-line utility. The key features of GuideMaker are as follows:
All the potential guides in a genome can be quickly designed in 1 run.
It can design gRNAs for any PAM sequence from any Cas system.
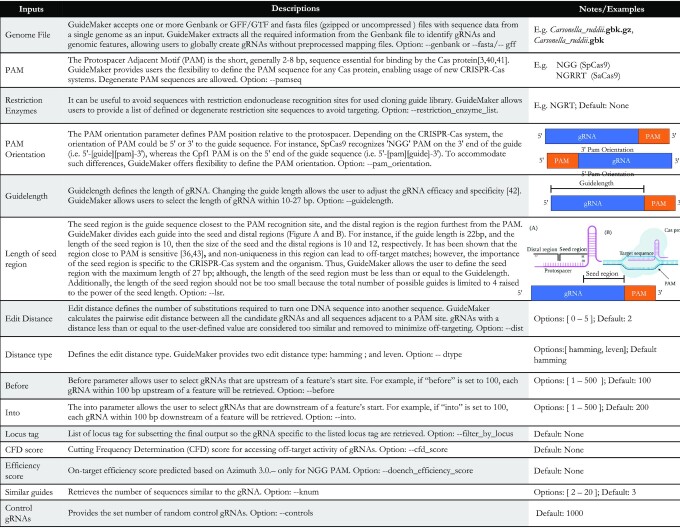
Search is customizable through user-defined guide parameters (as highlighted in Fig. 1). These features are specific to organisms, CRISPR-Cas systems, and experiments. Tuning these parameters can improve the sensitivity and specificity of gRNA.
Users can exclude specific restriction sites from guides to preserve those sites for downstream experiments.
It creates control sequences based on the input genome. In CRISPR experiments it is often desirable to create negative control sequences to evaluate off-target binding. GuideMaker provides the user with realistic control gRNAs that are highly divergent from sequences adjacent to PAM sites.
It provides an option to select a subset of results by locus tags of interest.
It provides off-target Cutting Frequency Determination (CFD) scores for gRNAs [8].
It provides on-target efficacy score for canonical “NGG” PAM. These efficiency scores are based on Azimuth algorithm [8].
It provides tabular result files, which can be used for the design and ordering of gRNA pools.
It provides an interactive visualization and exploratory tool to evaluate the guides.
The software can be run as a web application [25], a CyVerse application, or a command-line application [26]. Server code is included for running local instances of the web application as well.
Figure 1:
Input parameters for GuideMaker
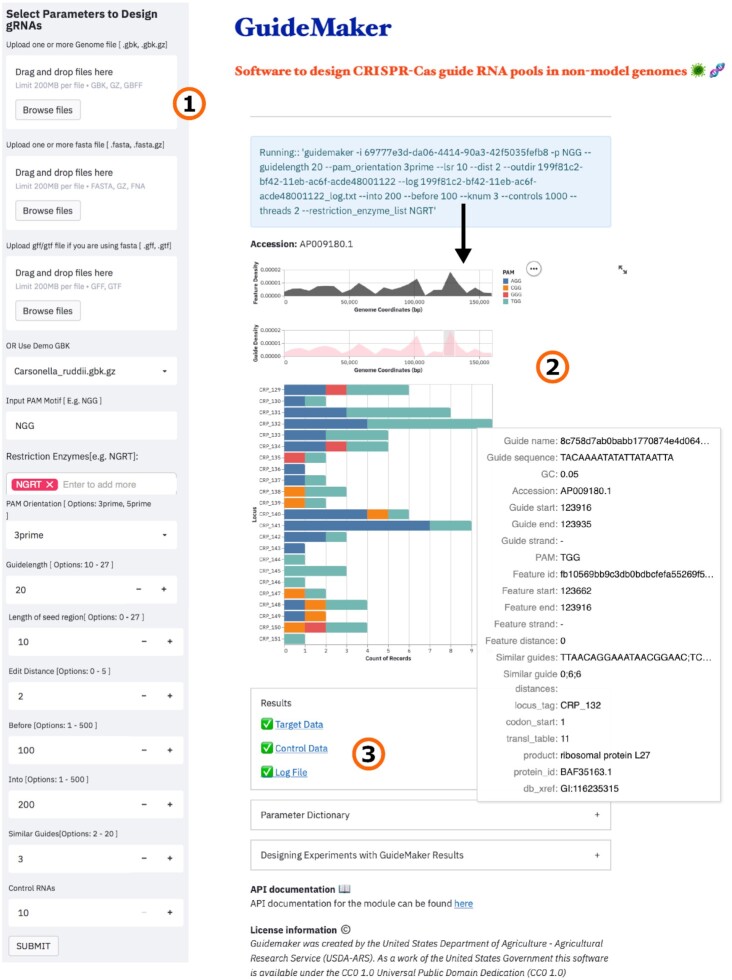
A typical workflow of GuideMaker involves 3 major steps (Fig. 2). In the first step, the user uploads the input genome in 1 or more GenBank or GFF/GTF and fasta files (gzipped or uncompressed) and defines the PAM and gRNA parameters (as highlighted in Fig. 1). GuideMaker identifies and filters target sites, then returns summary data to the graphical environment (Fig. 2). Users can inspect the interactive plots to learn more about the identified gRNAs and sort them by genome coordinates or locus tag. In the final step, GuideMaker provides the results as downloadable files under the Results section. These files are used for synthesizing the guides. The command-line version of GuideMaker has input parameters similar to those of the web application, with the flexibility to generate plots, configure the underlying hyperparameters for the hierarchical navigable small world (HNSW) graph, filter the results by specific locus tag, select Hamming or Levenshtein as the edit distance, predict on-target scores for “NGG” PAM or off-target CFD scores, or to run the web application locally. To make the application easier to install we distribute the application as a Bioconda environment [27], Docker container [28], Python package on GitHub [26], through the CyVerse discovery environment [29], or as an online web application [25]. Detailed information on accessing the software through various methods is available on the project home page [30].
Figure 2:
A typical workflow of GuideMaker: (1) A user uploads the input genome as GenBank file(s) or a Fasta and GTF/GFF file, then defines the PAM sequence along with all the associated parameters and submits them to run the program. (2) GuideMaker processes the input files and generates the interactive plots. Users can use these interactive plots to explore the results and sort them by locus tag and genome coordinates. (3) GuideMaker provides all the results and log files as downloads under the “Results” section.
Search method
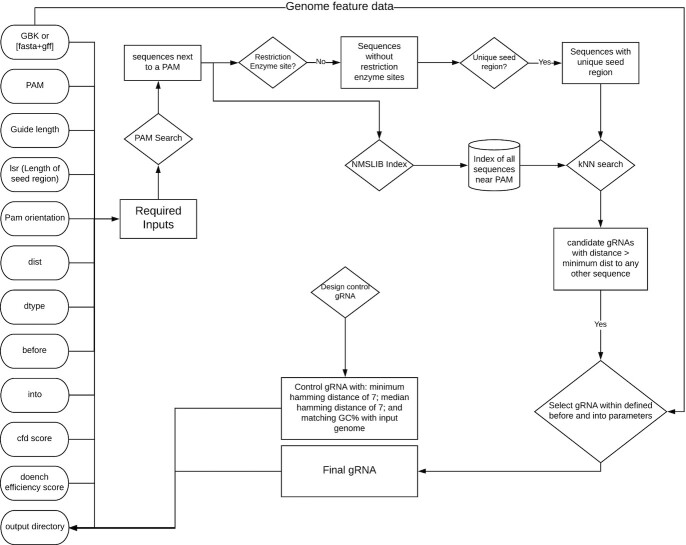
GuideMaker initially scans the genome, recording all candidate guide sequences adjacent to the specified PAM sequence on both DNA strands (Fig. 3). Candidate guides are then optionally checked for the restriction sites. Next, the candidate guides are searched for a unique “seed region” closest to the PAM site and candidate gRNAs that are not unique in their seed region are removed. Then, approximate nearest neighbor search is used to remove candidate guides too similar to PAM adjacent sequences in the genome, based on Hamming distance by default (the number of substitutions required to turn 1 DNA sequence into another equal-length sequence). Users can also select Levenshtein distance in the command-line version. The approximate nearest neighbor search is performed using the HNSW graph method in the Non-Metric Space Library (NMSLIB) [31,32]. An index of all the initial candidate guides is created using the selected edit distance. Each guide with a unique seed region is compared to all candidate guides, and any guides with edit distances below the user-set threshold are removed. This differs from the standard procedure of indexing the genome and mapping each candidate guide against the whole genome then parsing each result. HNSW has a search complexity of 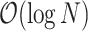 and index complexity of
and index complexity of 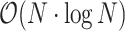 [31]. Finally, user-defined criteria are applied to specify the proximity and orientation of guides relative to genomic features like genes. A list of guides is then returned to the user with relevant information about the guide and its target genomic features.
[31]. Finally, user-defined criteria are applied to specify the proximity and orientation of guides relative to genomic features like genes. A list of guides is then returned to the user with relevant information about the guide and its target genomic features.
Figure 3:
Entity relationship diagram showing the operation of the GuideMaker core program.
The core of GuideMaker's search method is the HNSW method in NMSLIB [32]. The method builds a multilayer graph index of the input data and has several parameters that can be optimized for index building and search to trade off speed and accuracy. Graph construction is the most time-consuming step in our tests, and thus grid optimization was run to minimize run time while keeping recall >99% relative to the ground truth exact nearest-neighbor search. The grid optimization parameters (M, efc, ef, and post) used in the HNSW graph for approximate nearest neighbor search have been optimized for bacterial genomes. A Jupyter notebook [33] script for re-optimization and visualization of these hyperparameters is included in the test directory of the command-line version of the software, and optimized parameters can be passed to GuideMaker with the “–config” flag.
Target specificity
Estimating the on-target and off-target performance of a guide requires experimental data; while these are not available for most Cas systems, they are available for SpCas9. Guidemaker re-implements Two gRNA scoring methods from [8] to provide on-target and off-target scoring for the common SpCas9 enzyme with 25 nt guides. The on-target scoring method is the Doench Rule Set 2 method, specifically the “Azimuth Version 3 no position” model. The model applies boosted regression trees to nucleotide features. The featurization script was rewritten and parallelized for increased speed and updated to Python 3. The original Python Pickle model data object was converted to Open Neural Network Exchange (ONNX) format [34], and parameters were moved to a JSON file for better reproducibility and security. GuideMaker uses the ONNX Runtime [35] rather than Scikit-Learn [36] to make predictions from the model. For off-target scoring GuideMaker calculates Cutting Frequency Distribution (CFD) scores using the scoring matrix from [8], converted to JSON format for better reproducibility and security.
Computational performance
Genomes of different sizes, GC content, and chromosome numbers were used to test the speed and scalability of GuideMaker (Supplementary Table S1). For benchmarking the performance, the same parameters were used unless a specific parameter was being tested: a PAM motif of “NGG,” 3′ PAM orientation, target length of 20, lsr (length of seed region) of 11, before and after parameters of 500, knum of 10, controls of 10, dist of 3, and threads of 16. We profiled the performance of GuideMaker with different threads (1, 2, 4, 8, and 16) in processors with and without the Advanced Vector Extensions (AVX2) processor instruction set. The human genome was run with separate parameters described in Supplementary Table S2. All tests were run on a single compute node with 2 × 24 core Intel Xeon® Platinum 8260 CPU at 2.40 GHz with Cascade Lake microarchitecture. Three bacterial genomes, 1 fungal genome, 2 plant genomes, and 1 human genome were used in performance benchmarking: Escherichia coli K12 (NC_000913), Pseudomonas aeruginosa PAO1 (NC_002516), Burkholderia thailandensis E264 (NC_007651), Aspergillus fumigatus (NC_007194), Arabidopsis thaliana (NC_003070), Phaseolus vulgaris (NC_023759), and Homo sapiens (GRCh38.p13). For the gene- or locus-specific comparisons, only the guides within the locus coordinates (i.e., zero feature distance) were considered.
Comparison to existing design method
We compared the results of GuideMaker with the results of the online and command-line versions of CHOPCHOP (CHOPCHOP, RRID:SCR_015723) [37]. GuideMaker and CHOPCHOP parameters were set to approximate the same search. The length of the target sequence was set to 20, and zero mismatches were allowed in the seed region (11 nt) of the target. The E. coli (strain K-12/MG1655) genome was used with the online version of CHOPCHOP. Targets were searched in 40-kb increments to account for CHOPCHOP's online size limitations. Target sequences were searched across multiple 40-kb segments of E. coli genome (NC_000913.3:2001–42000, NC_000913.3:80001–120000, NC_000913.3:160001–200000, NC_000913.3:240001–280000, and NC_000913.3:320001–360000). We also searched for target sequences and genes/locus_tags within 40 kb of NC_000913.3:2001–42000 to compare identifications at the locus level. The ratio between the tools was calculated by dividing the number of gRNA identified with GuideMaker by the number of guides identified by CHOPCHOP to represent the proportion of guides identified by both GuideMaker and CHOPCHOP.
The command-line version of CHOPCHOP was used to compare the memory usage and computation time of CHOPCHOP and GuideMaker over an entire genome. The E. coli K-12 genome was chosen for comparison because the precomputed 2-bit genome files and Bowtie indexes were provided with CHOPCHOP v 3. The matching GenBank file was downloaded for GuideMaker, and both programs were run 5 times on the same machine using different numbers of processor cores (1, 2, 4, 8, 16).
Results
The time for GuideMaker to complete a typical run identifying all SpCas9 gRNAs (PAM "NGG") in a bacterial genome using 8 compute cores was 75 seconds for E. coli and 130 seconds for P. aeruginosa (Fig. 4). For SaCas9 and Streptococcus thermophilus Cas9 (StCas9), which have a longer PAM sequence (“NGRRT” and “NNAGAAW,” respectively, with 3′ PAM orientation) and thereby fewer potential targets, the same genomes ran in 19 or 5 seconds (Supplementary Fig. S1). The fungus A. fumigatus (28 Mb) and the plants A. thaliana (114 Mb) and P. vulgaris (537 Mb) have larger genomes but were still processed quickly. A. fumigatus processed in 23–304 seconds, while A. thaliana processed in 250–921 and P. vulgaris processed in 333–4,162 seconds depending on the number of cores, AVX2 instructions, and PAM sequence (Supplementary Fig. S2). Guidemaker designed guides for the entire human genome in 2–22 hours depending on the PAM used (Supplementary Table S2).
Figure 4:
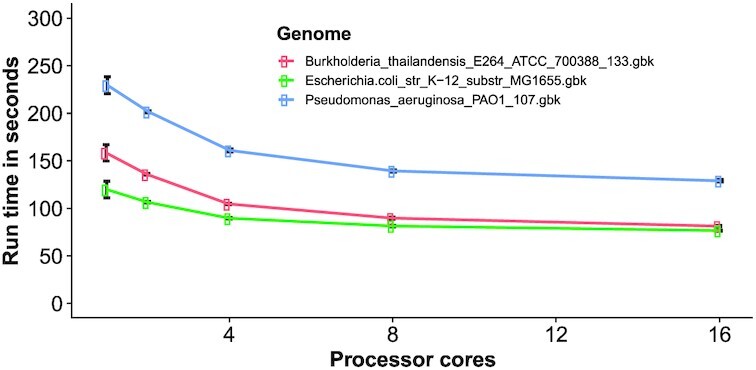
Performance of GuideMaker for SpCas9. Evaluating the performance of GuideMaker across 3 bacterial genomes using the “NGG” PAM motif with a target length of 20, unique zone of 11, 3′ PAM orientation, before and into parameters of 500, knum of 10, controls of 10, and dist of 3. The mean of 10 runs was used for the evaluation, where dot and error bar represent the mean and standard error, respectively.
GuideMaker can take advantage of AVX2 on newer x86 processors, which improves the search speed because HNSW search is accelerated with AVX2 (Supplementary Fig. S3). The acceleration was larger when fewer processors were available (Supplementary Fig. S3). The HNSW algorithms are parallelized, and indexing-and-search takes most of the compute time in GuideMaker so the software scales well when additional cores are added up to 8 cores (Supplementary Fig. S3). In practice it scaled up sublinearly with genome size, globally estimating Cas9 guides for E. coli MG1655 (4.6 Mb) in 75 seconds and P. vulgaris (537 Mb) in 1,549 seconds, both on 8 cores (memory usage: 1.9 GB for E. coli and 46.9 GB for P. vulgaris, Supplementary Fig. S4).
The results of GuideMaker were compared with those of the popular guide design software CHOPCHOP version 3 [37]. When GuideMaker's filtering settings were set to match CHOPCHOP, the results were very similar and 99.9% of the targets identified by GuideMaker fell within 2 nt of target coordinates returned by CHOPCHOP. When GuideMaker's unique seed region criterion was not applied at the locus level, the mean number of guides identified by the 2 approaches was similar per locus (mean GuideMaker = 116.8, mean CHOPCHOP = 113.6, P = 0.86, Supplementary Table 3). Although the number of guides identified per gene locus differed, none of the genes were missed by either tool. GuideMaker's default requirement of a unique seed region is more stringent than CHOPCHOP, and with it enabled, GuideMaker returned (count = 1,787) 38.4% of the targets compared to CHOPCHOP (count = 4,651) over a 2–42 kb test region in E. coli strain K12 substrain MG1655. At the sequence level, 96.8% of the identified gRNA (1,729/1,787) from both tools had identical sequences. The ratio of gRNA found by both the tools across the multiple 40-kb regions was 39.2% (sd = 1.9%, Supplementary Table S4) when using GuideMaker's more stringent default settings. This ratio was calculated by dividing the number of gRNA from GuideMaker by the number from CHOPCHOP for each 40-kb region. The effect of the stringent filtering heuristic used by GuideMaker was investigated computationally by applying on-target and off-target scoring to the guides designed by GuideMaker with and without the filtering heuristic (Supplementary Fig. S5). As expected, the filtering heuristic did not affect on-target scoring but did reduce the off-target CFD scores, suggesting that GuideMaker heuristics could decrease off-target binding. This result remains to be validated experimentally. The speed and memory usage of the command-line versions of CHOPCHOP and Guidemaker were also compared. When using 8 cores to process the E. coli strain K-12 substrain MG165 genome, Guidemaker was 65 times faster and used 2.7 times less memory than CHOPCHOP (Supplementary Fig. S6).
Discussion
Designing gRNAs is a 2-step process where GuideMaker first identifies potential guides adjacent to PAM sequences and then filters the potential guides on the basis of multiple criteria. The most important criterion is that each guide has a minimum edit distance from any other sequence adjacent to a PAM site in the genome; this decreases the likelihood of off-target binding. The second way GuideMaker reduces off-target binding is by requiring that a set number of bases near the PAM site be unique from any other candidate guide. The 8 bases nearest the PAM are the most important for target specificity, and any mismatch is sufficient to prevent binding [38,39]. The length of the unique region should be set with consideration for the size of the genome because requiring short unique regions will limit the number of total guides that can be found. For example, requiring that every gRNA be unique in the first 3 nt would only allow for 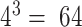 possible guides to be designed. For normal –lsr values of 9–12 this is only limiting for human-sized genomes and can be disabled by setting –lsr to 0. All guides designed by GuideMaker are perfect matches to a single site in the genome. Additional specificity is obtained by requiring all similar PAM-adjacent sequences to be unique in the critical seed region and have a total number of mismatches that exceed the user-defined threshold. This double criterion is expected to increase specificity.
possible guides to be designed. For normal –lsr values of 9–12 this is only limiting for human-sized genomes and can be disabled by setting –lsr to 0. All guides designed by GuideMaker are perfect matches to a single site in the genome. Additional specificity is obtained by requiring all similar PAM-adjacent sequences to be unique in the critical seed region and have a total number of mismatches that exceed the user-defined threshold. This double criterion is expected to increase specificity.
The primary goal of the current version of our software is to support the design of gRNAs for non-standard Cas enzymes or non-model organisms at the genome scale. Guide RNAs do not perform equally; thus empirical experiments will be needed to fully validate the functionality and efficacy of gRNA predictions. Given the similarity in targets identified by GuideMaker and CHOPCHOP, we anticipate that performance is similar to the current state of the art but applicable to more design use cases. When a unique seed region and edit distance–based filters were applied, GuideMaker created guides more conservatively, generating only ∼40% of the guides created by CHOPCHOP. While CHOPCHOP has an option to specify the maximum number of mismatches in the first 9 nt or the whole guide, it does not allow the application of both criteria. While there are differences in the number and position of guides generated by GuideMaker, with GuideMaker being more conservative by default, both programs create enough guides to target nearly all gene loci in the genome of E. coli. The current version of GuideMaker provides options to predict off-target CFD scores and on-target scores for the canonical NGG PAM. Both scoring approaches are based on the publicly available models trained on empirical data with SpCas9. If experimentally validated data become available from genome-wide screens with different Cas enzymes, future versions of GuideMaker could potentially incorporate new scoring models to help rank candidate guides.
GuideMaker is a fast and flexible tool for designing guide RNA across the entire genome in non-model organisms or with non-canonical Cas enzymes. It takes advantage of fast HNSW search to quickly index and search new genomes. Several parameters can be tuned to ensure compatibility with the specific application of the user. For example, GuideMaker checks the designed gRNA for a given restriction enzyme site to prevent incompatibility with the cloning strategy. Second, the maximum distance from a target sequence from the start of an annotated feature can be chosen to disrupt promoters or the beginning of the coding sequence because these sites are preferred for CRISPR interference experiments. GuideMaker also creates off-target control RNA sequences for use as negative controls in high-throughput experiments. Finally, the program plots the results for visual exploration of the targets and exports the data as .csv files. The software is available as a command-line application or a web application and is integrated into the CyVerse Discovery Environment to provide users with a range of use options. Guidemaker is a fast, flexible design tool for the creation of challenging gRNA pools.
Availability and Requirements
Project name: GuideMaker
Project home page: https://guidemaker.org
Operating systems: Linux or macOS
Programming language: Python ≥3.6
Other requirements:
License: CC0 1.0 Public Domain Dedication
biotoolsID: guidemaker
Data Availability
The source code and command-line executables for GuideMaker are available and can be installed directly from GitHub [26], Bioconda [27], or as a Docker container [28]. Data and code to reproduce the analysis in the article are available in Zenodo [40]. As a work of the United States Department of Agriculture, GuideMaker is released to the public domain under a Creative Commons (CC0) public domain attribution. The program is also available as a web application through the CyVerse discovery environment [29], and as a stand-alone web application [25].
Additional Files
Supplementary Figure S1: Performance of GuideMaker for SaCas9 and StCas9 in selected bacteria
Supplementary Figure S2: Performance of GuideMaker for SpCas9, SaCas9, and StCas9 in selected eukaryotes
Supplementary Figure S3: Performance of GuideMaker with AVX2 settings
Supplementary Figure S4: Memory usage of GuideMaker for SpCas9, SaCas9, and StCas9
Supplementary Figure S5: Comparison of efficiency and CFD scores with or without GuideMaker-based filters
Supplementary Figure S6: Performance and memory usage comparisons between CHOPCHOP (CLI version) and GuideMaker
Supplementary Table S1: Organism features
Supplementary Table S2: Comparison of processing times and the number of gRNAs with different PAMs in Homo sapiens (GRCh38.p13)
Supplementary Table S3: Comparison of the mean number of gRNAs predicted by GuideMaker and CHOPCHOP
Supplementary Table S4: Comparison of consensus ratio between GuideMaker and CHOPCHOP
Supplementary Table S5: Comparison of processing times and guide similarity for Levenshtein and Hamming distances with different PAMs in Escherichia coli and Phaseolus vulgaris.
Kornel Labun -- 7/16/2021 Reviewed
Kornel Labun -- 11/4/2021 Reviewed
Wen Wang, Ph.D -- 7/21/2021 Reviewed
Wen Wang, Ph.D -- 11/3/2021 Reviewed
Mudra Hegde -- 7/29/2021 Reviewed
Abbreviations
API: Application Programming Interface; AVX2: Advanced Vector Extensions 2; bp: base pairs; Cas: CRISPR-associated protein; Cas12a: CRISPR-associated protein 12a (previously known as Cpf1); CFD: Cutting Frequency Determination; Cpf1: see Cas12a; CPU: central processing unit; CRISPR: clustered regularly interspaced short palindromic repeats; GFF: General Feature Format; gRNA: guide RNA; GTF: General Transfer Format; HNSW: hierarchical navigable small world; JSON: JavaScript Object Notation; kb: kilobase pairs; Mb: megabase pairs; NMSLIB: non-metric space library; nt: nucleotides; ONNX: Open Neural Network Exchange; PAM: protospacer-adjacent motif; SaCas9: Staphylococcus aureus CRISPR-associated protein 9; SpCas9: Streptococcus pyogenes CRISPR-associated protein 9.
Competing Interests
The authors declare that they have no competing interests.
Funding
The research was supported by the United States Department of Agriculture (USDA), Agricultural Research Service (ARS) project No. 6066–21310-005-D, and ARS cooperative agreement 6066–21310-005–28-S to the University of Florida. This research used resources provided by the SCINet scientific computing initiative of the USDA-ARS, ARS project No. 0500–00093-001–00-D.
Authors' Contributions
All authors conceived and designed the study. R.P. and A.R.R. developed and optimized the software and performed the experiments. All authors tested the software, wrote and revised the manuscript, and read and approved the final manuscript.
Contributor Information
Ravin Poudel, Genomics and Bioinformatics Research Unit, USDA Agricultural Research Service, Gainesville, FL 32608, USA; Department of Microbiology and Cell Science, Institute of Food and Agricultural Sciences, University of Florida, Gainesville, FL 32601, USA.
Lidimarie Trujillo Rodriguez, Department of Microbiology and Cell Science, Institute of Food and Agricultural Sciences, University of Florida, Gainesville, FL 32601, USA.
Christopher R Reisch, Department of Microbiology and Cell Science, Institute of Food and Agricultural Sciences, University of Florida, Gainesville, FL 32601, USA.
Adam R Rivers, Genomics and Bioinformatics Research Unit, USDA Agricultural Research Service, Gainesville, FL 32608, USA.
References
- 1. Jiang W, Bikard D, Cox D, et al. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31(3):233–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ran FA, Hsu PD, Wright J, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8(11):2281–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mojica FJM, Díez-Villaseñor C, García-Martínez J, et al. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155(3):733–40. [DOI] [PubMed] [Google Scholar]
- 4. Pickar-Oliver A, Gersbach CA.. The next generation of CRISPR–Cas technologies and applications. Nat Rev Mol Cell Biol. 2019;20(8):490–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Deltcheva E, Chylinski K, Sharma CM, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471(7340):602–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ran FA, Cong L, Yan WX, et al. In vivo genome editing using Staphylococcus aureusCas9. Nature. 2015;520(7546):186–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zetsche B, Gootenberg JS, Abudayyeh OO, et al. Cpf1 Is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163(3):759–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Doench JG, Fusi N, Sullender M, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34(2):184–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hiranniramol K, Chen Y, Liu W, et al. Generalizable sgRNA design for improved CRISPR/Cas9 editing efficiency. Bioinformatics. 2020;36(9):2684–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xu H, Xiao T, Chen CH, et al. Sequence determinants of improved CRISPR sgRNA design. Genome Res. 2015;25(8):1147–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Perez AR, Pritykin Y, Vidigal JA, et al. GuideScan software for improved single and paired CRISPR guide RNA design. Nat Biotechnol. 2017;35(4):347–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Anzalone AV, Koblan LW, Liu DR.. Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat Biotechnol. 2020;38(7):824–44. [DOI] [PubMed] [Google Scholar]
- 13. Hale CR, Zhao P, Olson S, et al. RNA-Guided RNA Cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139(5):945–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Abudayyeh OO, Gootenberg JS, Konermann S, et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353(6299):doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ma E, Harrington LB, O'Connell MR, et al. Single-stranded DNA cleavage by divergent CRISPR-Cas9 enzymes. Mol Cell. 2015;60(3):398–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Klompe SE, Vo PLH, Halpin-Healy TS, et al. Transposon-encoded CRISPR–Cas systems direct RNA-guided DNA integration. Nature. 2019;571(7764):219–25. [DOI] [PubMed] [Google Scholar]
- 17. Strecker J, Ladha A, Gardner Z, et al. RNA-guided DNA insertion with CRISPR-associated transposases. Science. 2019;365(6448):48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jinek M, Chylinski K, Fonfara I, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hsu PD, Lander ES, Zhang F.. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157(6):1262–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Qi LS, Larson MH, Gilbert LA, et al. Repurposing CRISPR as an RNA-γuided platform for sequence-specific control of gene expression. Cell. 2013;152(5):1173–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cox DBT, Gootenberg JS, Abudayyeh OO, et al. RNA editing with CRISPR-Cas13. Science. 2017;358(6366):1019–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yan WX, Chong S, Zhang H, et al. Cas13d Is a compact RNA-targeting type VI CRISPR effector positively modulated by a WYL-domain-containing accessory protein. Mol Cell. 2018;70(2):327–39.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dixit A, Parnas O, Li B, et al. Perturb-Seq: dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell. 2016;167(7):1853–66.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Peters JM, Colavin A, Shi H, et al. A comprehensive, CRISPR-based functional analysis of essential genes in bacteria. Cell. 2016;165(6):1493–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Poudel R, Rodriguez LT, Reisch CR, et al. The GuideMaker web application. 2022. https://guidemaker.app.scinet.usda.gov. Accessed 6 January 2022. [Google Scholar]
- 26. Poudel R, Rodriguez LT, Reisch CRet al. , GuideMaker (Version 0.3.4). Zenodo2021. 10.5281/zenodo.5655842. [DOI] [Google Scholar]
- 27. Poudel R, Rodriguez LT, Reisch CR, et al. The GuideMaker Bioconda installation (version 0.3.4). 2022. https://anaconda.org/bioconda/guidemaker.Accessed 6 January 2022. [Google Scholar]
- 28. Poudel R, Rodriguez LT, Reisch CR, et al. The GuideMaker Docker container. 2022. https://github.com/USDA-ARS-GBRU/GuideMaker/releases.Accessed 6 January 2022. [Google Scholar]
- 29. Merchant N, Lyons E, Goff S, et al. The iPlant collaborative: cyberinfrastructure for enabling data to discovery for the life sciences. PLoS Biol. 2016;14(1):e1002342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Poudel R, Rodriguez LT, Reisch CR, et al. The GuideMaker project homepage. 2022. https://guidemaker.org. Accessed 6 January 2022. [Google Scholar]
- 31. Malkov YA, Yashunin DA. Efficient and robust approximate nearest neighbor search using Hierarchical Navigable Small World graphs. arXiv2016:1603.09320. [DOI] [PubMed] [Google Scholar]
- 32. Naidan B, Boytsov L, Malkov Y, et al. Non-metric space library manual. arXiv2015:1508.05470. [Google Scholar]
- 33. Kluyver T, Ragan-Kelley B, Pérez F, et al. Jupyter Notebooks - a publishing format for reproducible computational workflows. In: Loizides F, Schmidt B, eds. Positioning and Power in Academic Publishing: Players, Agents and Agendas. Amsterdam, Netherlands: IOS Press; 2016:87–90. [Google Scholar]
- 34. Microsoft . ONNX (Version 1.10.0). 2021. https://github.com/onnx/onnx/releases/tag/v1.10.0.Accessed 6 January 2022. [Google Scholar]
- 35. Microsoft . ONNX Runtime (Version 1.8.1). 2021. https://github.com/microsoft/onnxruntime/releases/tag/v1.8.1. Accessed 6 January 2022. [Google Scholar]
- 36. Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: machine learning in Python. J Mach Learn Res. 2011;12:2825–30. [Google Scholar]
- 37. Labun K, Montague TG, Krause M, et al. CHOPCHOP v3: Expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 2019;47(W1):W171–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Semenova E, Jore MM, Datsenko KA, et al. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci U S A. 2011;108(25):10098–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hsu PD, Scott DA, Weinstein JA, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31(9):827–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Poudel Ravin. USDA-ARS-GBRU/GuideMaker_paper: v0.4.0. Zenodo. 2022; 10.5281/zenodo.5825817. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Kornel Labun -- 7/16/2021 Reviewed
Kornel Labun -- 11/4/2021 Reviewed
Wen Wang, Ph.D -- 7/21/2021 Reviewed
Wen Wang, Ph.D -- 11/3/2021 Reviewed
Mudra Hegde -- 7/29/2021 Reviewed
Data Availability Statement
The source code and command-line executables for GuideMaker are available and can be installed directly from GitHub [26], Bioconda [27], or as a Docker container [28]. Data and code to reproduce the analysis in the article are available in Zenodo [40]. As a work of the United States Department of Agriculture, GuideMaker is released to the public domain under a Creative Commons (CC0) public domain attribution. The program is also available as a web application through the CyVerse discovery environment [29], and as a stand-alone web application [25].