Abstract
Snake venoms represent a danger to human health, but also a gold mine of bioactive proteins that can be harnessed for drug discovery purposes. The evolution of snakes and their venom has been studied for decades, particularly via traditional morphological and basic genetic methods alongside venom proteomics. However, while the field of genomics has matured rapidly over the past 2 decades, owing to the development of next-generation sequencing technologies, snake genomics remains in its infancy. Here, we provide an overview of the state of the art in snake genomics and discuss its potential implications for studying venom evolution and toxinology. On the basis of current knowledge, gene duplication and positive selection are key mechanisms in the neofunctionalization of snake venom proteins. This makes snake venoms important evolutionary drivers that explain the remarkable venom diversification and adaptive variation observed in these reptiles. Gene duplication and neofunctionalization have also generated a large number of repeat sequences in snake genomes that pose a significant challenge to DNA sequencing, resulting in the need for substantial computational resources and longer sequencing read length for high-quality genome assembly. Fortunately, owing to constantly improving sequencing technologies and computational tools, we are now able to explore the molecular mechanisms of snake venom evolution in unprecedented detail. Such novel insights have the potential to affect the design and development of antivenoms and possibly other drugs, as well as provide new fundamental knowledge on snake biology and evolution.
Keywords: snake genomics, DNA sequencing, venom, venom evolution, snakes, snake toxins
Background
Snakes (Squamata: Serpentes) represent a monophyletic lineage, comprising ∼3,600 extant species found in all continents, except Antarctica [1,2]. From an evolutionary perspective, these reptiles stand out for their characteristic lack of limbs, elongated body shape, and exclusively carnivorous diet. Even before the advent of genetic approaches, conventional anatomical and morphology-based phylogenetic evidence unambiguously suggested that snakes are nested within lizards, with the Anguimorpha lineage (e.g., monitor lizards, glass lizards, beaded lizards) as their closest relatives [3–5]. Together with amphisbaenians, snakes and all other lizards thus form the largest branch of terrestrial vertebrates, the squamate reptiles [3–5]. Snakes have many specialized adaptations compared to other reptile lineages. For example, the evolution of infrared sensing pits in pit vipers (Viperidae: Crotalinae), boas (Boidae), and pythons (Pythonidae), and of a venom apparatus in several snake families (Fig. 1), provides these animals with exceptional predatory capabilities despite the loss of limbs and the degradation of visual and auditory perception in many (but not all) species [6–8]. Moreover, severe jaw modifications and low metabolic rates enable snakes to swallow and digest large prey whole, further consolidating their position as formidable predators [9,10]. Thus, snakes are important model organisms for evolutionary studies and have yielded insights into limb development [11–13], sex chromosome evolution [14], and venom evolution [15].
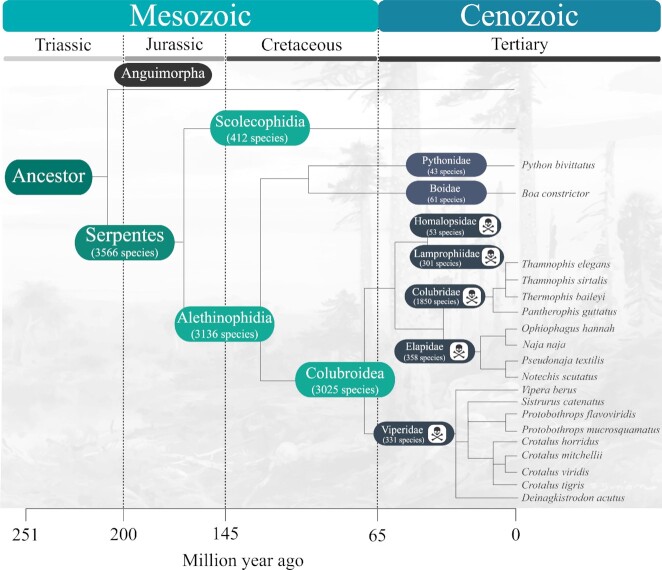
Figure 1:
Schematic diagram of snake evolution based on data from Reptile-database.org [16]. Snakes (Serpentes) are divided into 3 main infraorders, Scolecophidia, Henophidia, and Alethinophidia, which together encompass ∼24 families (7 shown here). Families comprising venomous species are indicated with a skull and crossbones symbol. Colubridae constitutes the largest family of snakes, encompassing 52% of the ∼3,566 snake species currently described. The total number of currently described venomous snake species is 2,901, predominantly falling within the families Homalopsidae, Lamprophiidae, Colubridae, Elapidae, and Viperidae. Only snake species that have undergone whole-genome sequencing and assembly are listed in this figure.
The development of next-generation sequencing (NGS) technologies in recent decades has allowed researchers to generate large genomic datasets and rendered the assembly and characterization of complete genomes a routine task. Despite the availability of NGS since the early 2000s, the use of these technologies to assemble and study complete snake genomes has been limited, especially when compared to the amount of research that has been conducted in the fields of snake venom proteomics and transcriptomics [17]. It was not until 2013 that the first snake genomes based on high-throughput sequencing data were published for the Burmese python (Python bivittatus), the red-tailed boa (Boa constrictor constrictor), and the king cobra (Ophiophagus hannah) [9, 18,19]. Fortunately, snake genome research has eventually gained more attention, with 18 new genomes being released since 2013 and several more currently in progress [15,20–31]. These increased sequencing efforts have already revealed intriguing insights into the regulation and expression of venom-related genes. As an example, a large number of dormant toxin-encoding genes with unknown bioactivity were identified in the Okinawan habu (Protobothrops flavoviridis) [15]. Such discoveries could be of high scientific value and may improve our basic understanding of the interplay between protein function and evolution. Furthermore, because toxins from several animal lineages are known to possess different types of bioactivity, some of them could find utility in a variety of applications, from the development of novel therapeutics [32] to biopesticides [33] and molecular research tools [34]. With only 21 snake genomes publicly available to date, there is great unexplored scientific potential in sequencing and analysing more snake genomes [17,35].
From a broader perspective, having access to a complete or nearly complete assembled genome provides an excellent basis for addressing a wide range of biological research questions. For example, genomic data can be used to predict protein-coding exons [36] (including exons in genes that recently underwent pseudogenization), non-expressed genes, translated proteins, and microRNA genes [37]. Genomic data may also allow for the identification of toxin orthologs using comparative studies and homology searches [38]. Knowledge of homology is crucial for the reliability of functional annotation of genomes and can provide fundamental information on evolution and speciation processes [39,40]. Therefore, complete genomes are crucial to the field of proteomics as well, because the absence of reliable genome-derived protein libraries forces researchers to rely on homologous proteins from other organisms as a benchmark against which to compare newly characterized protein sequences. This results in severely limited accuracy in identifying potentially homologous proteins, which consequently leads to overlooking and/or misrepresenting evolutionary patterns. This is especially relevant considering the likely widespread occurrence of alternative splicing in snake genomes, which gives rise to multiple messenger RNA products that in turn result in various isoforms of a particular toxin [41–43]. Extensive post-genomic and post-translational modifications are also at play, leading to often remarkable discrepancies between genome, transcriptome, and proteome in terms of expression and sequence identity [44–46]. Along this line, comparative analysis of whole snake genomes could likely provide invaluable insight on the evolution and structure of the gene regulatory network responsible for the expression of venom genes in these animals (and arguably venomous amniotes in general) [47].
Several approaches are available to obtain reliable genomic data. Among them is reduced-representation sequencing, in which only a part of the genome is sequenced [48]. For instance, capture sequencing techniques allow for specific areas of interest (e.g., the exon part of the genome) to be targeted and sequenced at a lower cost compared to whole-genome sequencing (WGS) [49]. Although less suitable for studying venom genes, restriction site associated–DNA sequencing (RAD-seq) uses restriction enzymes to obtain genome-wide sequencing data, which have recently been used to study population demographic trends that underly venom variation [50]. Nevertheless, reliable detection of homologous genes across species and/or lineages can be hindered by the acquisition, loss, or pseudogenization of genes [40]. One way to overcome this challenge is to use WGS, which represents a more comprehensive resource for the detection of homologous genes because it provides the entire genotype of the target organism(s) [40]. WGS can also provide information on genomic variability of a species, and potentially discover and quantify the extent of selective (e.g., positive/purifying selection and hitchhiking effects) and neutral forces (e.g., genetic drift) driving venom evolution [51].
This review aims to provide a comprehensive summary of the current knowledge on snake genomics, with a particular focus on the current use and future potential of high-throughput DNA sequencing technologies in the field of snake toxinology. Moreover, we discuss how these technologies can be used to expand our current knowledge on snake venom evolution and toxin diversification.
Current Status of Snake Venom Research
Overview of snake toxin families
Studies have estimated that between 19,000 and 25,000 toxins are found in venoms from the Elapidae and Viperidae snake families, but only a few thousands have been characterized [52]. Nonetheless, this body of knowledge has proven sufficient for the systematic classification of snake venom toxins into 63 families, most of which are, however, only found in a small percentage of snake species and/or in negligible amounts within venom mixtures [53]. The 4 families generally considered to be of highest relevance both from a clinical (human envenoming cases) and an ecological perspective (e.g., prey incapacitation) are the 3-finger toxins (3FTxs), phospholipases A2 (PLA2s), snake venom metalloproteinases (SVMPs), and snake venom serine proteinases (SVSPs). Other widespread snake venom protein families include cysteine-rich secretory proteins (CRISPs), L-amino acid oxidases (LAAOs), and C-type lectin-like proteins (CTLPs) [53]. An overview of the main snake venom toxin families is provided in Table 1.
Table 1:
Number of toxin-encoding genes for 22 toxin families in selected venomous and non-venomous reptile species
| Venom protein family | Non-venomous | Venomous | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Venom family abbreviation | Anolis carolinensis* | Boa constrictor | Python bivittatus | Thamnophis sirtalis | Ophiophagus hannah | Naja naja | Deinagkistrodon acutus | Protobothrops flavoviridis | Crotalus viridis | Crotalus tigris | Bothrops jararaca | |
| 5′-nucleotidases | 5Nase | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 5 | 1 | ||
| Acetylcholinesterase | ACeH | 22 | 11 | 12 | 16 | 2 | 14 | 7 | ||||
| Kunitz-type peptide | 86 | 39 | 49 | 53 | 3 | 70 | 2 | 2 | ||||
| Bradykinin-potentiating peptides and C-type natriuretic peptides | BNP | 1 | 3 | 1 | 6 | 3 | 2 | 1 | 1 | 1 | 1 | |
| Cysteine-rich secretory proteins | CRISPs | 2 | 1 | 1 | 2 | 3 | 7 | 2 | 2 | 4 | 2 | 1 |
| C-type lectins and C-type lectin-like proteins | CTLPs | 5 | 7 | 6 | 13 | 2 | 22 | 10 | 6 | 5 | 6 | |
| Disintegrins | Dis | 3 | 2 | |||||||||
| Factor V | 5 | 5 | 6 | 5 | 5 | 3 | ||||||
| Factor X | 9 | 11 | 11 | 11 | 11 | |||||||
| Hyaluronidases | HYAL | 5 | 6 | 6 | 1 | 6 | 3 | 6 | 1 | 4 | 1 | 1 |
| L-amino acid oxidases | LAAO | 4 | 5 | 6 | 2 | 3 | 3 | 4 | 1 | 4 | 2 | 2 |
| Nerve growth factors or neurotrophins | NGF | 5 | 5 | 5 | 5 | 3 | 4 | 1 | 2 | 1 | 1 | |
| Phosphodiesterases | PDE | 6 | 6 | 5 | 5 | 1 | 5 | 1 | 1 | |||
| Phospholipases A2 | PLA2 | 1 | 1 | 1 | 1 | 4 | 8 | 1 | 9 | 5 | 3 | 1 |
| Phospholipases B | PLB | 1 | 1 | 1 | 4 | 1 | 1 | 1 | 1 | 1 | ||
| Snake venom metalloproteinases | SVMP (PI) | 2 | 1 | |||||||||
| SVMP (PII) | 1 | 4 | 3 | 3 | 7 | |||||||
| SVMP (PIII) | 1 | 1 | 2 | 7 | 4 | 8 | 5 | 6 | 11 | 2 | 20 | |
| Snake venom serine proteinases | SVSP | 4 | 6 | 7 | 1 | 8 | 8 | 22 | 11 | 9 | 15 | 12 |
| Three-finger toxins | 3FTx | 5 | 19 | 4 | 2 | 3 | ||||||
| Vascular endothelial growth factors | VEGF | 4 | 7 | 7 | 5 | 6 | 6 | 1 | 3 | 1 | 1 | |
| Venom ficolins | Veficolins | 11 | 9 | 9 | 11 | 10 | 4 | 1 | ||||
| Vespryns/ohanin-like proteins | 90 | 40 | 52 | 39 | 1 | 42 | 1 | 1 | ||||
| Waprin | 5 | 3 | 3 | 4 | 3 | 1 | 1 | |||||
In venomous snake genomes, the numbers refer to the venom gland genome only. Non-venomous species lack venom glands, and the indicated numbers refer to homologous proteins expressed in other organs.
The green anole (Anolis carolinensis) was selected as outgroup taxon because it is a non-venomous, non-snake squamate with a complete genome sequence available.
3FTxs belong to a superfamily of non-enzymatic proteins and are a major component in the venoms of most elapids, while they generally feature less prominently in viperid and colubrid venoms. These toxins have 3 β-stranded loops extending from a central core, contain 4 or 5 conserved disulfide bonds, and cause a wide range of pharmacological effects [54–56]. A prominent group of 3FTxs, α-neurotoxins, interfere with neuromuscular signal transmission of cholinergic neurons by binding to nicotinic acetylcholine receptors, causing flaccid paralysis [53, 55]. Other 3FTxs are toxic to cardiomyocytes and can lead to increased heart rate and ultimately cardiac arrest, while yet others function as calcium channel blockers or platelet aggregation inhibitors [54].
PLA2s are found in the venoms of vipers, elapids, and certain rear-fanged species [57–60] and exert a wide variety of cytotoxic, myotoxic, cardiotoxic, and neurotoxic effects [57,58,60]. Of particular interest is a catalytically inactive, myotoxic category of PLA2s stemming from a single substitution of a highly conserved amino acid residue (Asp49 to Lys49/Asn49) [57]. Both non-catalytic and enzymatic PLA2s are able to form heterodimeric complexes with other PLA2s or other toxins in certain venoms, whereby their toxicity is greatly potentiated [58]. Most snake genomes contain multiple PLA2 genes, which likely originated from repeated gene duplication events [60,61]. These paralogs have diverse pharmacological activities, which were likely acquired through neofunctionalization (i.e., recruitment of a paralog to the venom gland following gene duplication and its subsequent evolution into a toxin-coding gene) [62,63]. Pseudogenization and deletion of PLA2 genes are also frequent in snakes, making this toxin family one of the most dynamic in terms of evolutionary history [28,39,64]. The annotation of more snake genomes, and the likely consequent discovery of more PLA2 genes, might provide an improved understanding of the evolution and the mechanisms of action of these proteins (including how the phenomenon of toxin synergism has evolved) and potentially assist in the characterization of similar evolutionary processes for other enzymes.
Another major category of enzymes found in snake venoms are SVMPs [65,66]. These proteinases are enzymes that cleave peptide bonds in other proteins, which may result in the degradation or activation of the target [66]. Zinc-dependent SVMPs are often the major venom component in vipers [67], and these toxins hydrolyze extracellular matrix components, leading to rupture of capillaries and local and systemic bleeding [59]. Other clinical manifestations induced by SVMPs include edema, inflammation, myonecrosis, and reduced muscle regeneration [67]. Additionally, these enzymatic toxins can have anticoagulant, clotting factor–activating, or platelet-aggregating effects [68,69]. SVMPs are divided into 3 distinct classes depending on the domains present in the mature enzymes: P-I (metalloproteinase [M] domain only), P-II (M domain and disintegrin-like domain), and P-III (M domain, disintegrin-like domain, and cysteine-rich domain) [65]. Elucidation of snake genomes could help shed light on how these enzymes evolved from the ancestral P-III type via loss of domains [70–72] and postgenomic modifications, acquiring different functions and specificities in the process [65]. A better understanding of SVMP evolution via snake genomics could also provide insight into the evolutionary process that led to the diversification of SVMPs as a whole from the ancestral A disintegrin and metalloprotease (ADAM) family of metalloproteinases, which play significant roles in all stages of development and survival of higher-order organisms [73].
Finally, SVSPs are typically present in the venoms of vipers [74] but can also be found in elapid venoms [75]. SVSPs contain 2 6-stranded β-barrels and consist of ∼245 amino acid residues. SVSPs also have a unique extended C-terminus that forms a disulfide bridge, which contributes to structural stability [76]. These toxins can induce blood coagulation through fibrin formation, Factor V activation, prothrombin activation, actin dissolvement, or platelet aggregation; conversely, they can also act as anticoagulants via fibrinolysis, fibrinolytic enzyme activation, or protein C activation [59, 77–79]. This toxin family has received increased attention with recent genome studies on P. flavoviridis and B. jararaca, where the evolutionary pathway as well as the molecular regulation of SVSP expression was systematically investigated [15, 29].
In summary, snake toxin families are numerous and their pharmacological actions are complex [80]. Knowledge on the toxicity and structure of different snake toxin families is essential to further our understanding of snake venom evolution, as well as to understand venoms as drug targets for antivenom development. Much knowledge has been gained from venom proteomics and transcriptomics, and new genomics technologies now allow for the investigation of the evolutionary relationships between toxins in different families in unprecedented detail.
State of the art in snake genomics
With the rapid development of high-throughput sequencing technology, large-scale genomic projects have generated rich sequence information data of billions of base pairs and have paved the way for a new era in the field of phylogenetics, whereby the evolutionary history of organisms can be reconstructed from genomic data. The supermatrix method is the most well-known approach for analysing concatenation of multiple gene sequences, and using genomic data sets with improved resolution can potentially mitigate phylogenetic problems previously caused by sampling errors [81]. However, because only 21 (∼0.6%) of the ∼3,600 existing snake species have undergone WGS so far [9,15, 17,18,20–28,30,82–85], snake genomics will likely develop significantly in the coming years. A complete list of currently available snake genomes is provided in Table 2.
Table 2:
Whole-genome sequencing studies on snakes, published or in progress
| Assembly | Annotation | Venomous | INSDC ID | Source | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Scientific classification | Sequencing� | Scaffold | |||||||||||
| Superfamily | Family | Genus | Species | Sequencing platform | DoC | GC% | N50 Size (kb) | Contig N50 Size (kb) | Genome Size (Gb) | Protein-encoding genes identified | |||
| Colubroidea | Viperidae | Bothrops | jararaca | Illumina; PacBio; BAC-SeqSc | 150 IL20 PB | 163.5 | 2.1 | Yes | PRJNA691605 | [29] | |||
| Crotalus | viridis | Illumina; PacBio | 100 | 36.6 | 139 | 15.74 | 1.3 | Yes | PDHV00000000.1 | [20,21] | |||
| tigris | Illumina; PacBio | 190 IL33 PB | 39.9/39.8 | 207,720 | 2,110 | 1.6 | 18,240 | Yes | VORL00000000 | [28] | |||
| pyrrhus | Illumina | 40 | 38.5 | 5.1 | 4.1 | 1.1 | Yes | JPMF00000000.1 | [26] | ||||
| horridus | Illumina | 135 | 34.3 | 23.8 | 5.8 | 1.5 | Yes | LVCR00000000.1 | [82] | ||||
| Protobothrops | flavoviridis | Illumina | 96 | 38.2 | 467 | 3.8 | 1.4 | 20,540 | Yes | BFFQ00000000.1 | [15] | ||
| mucrosquamatus | Illumina | 86 | 40.6 | 424 | 22 | 1.6 | 20,122 | Yes | BCNE00000000.2 | [24] | |||
| Sistrurus | catenatus | Illumina, PacBio | 1,045,000 | 1.6 | Yes | PRJNA750087 | [31] | ||||||
| Vipera | berus | Illumina | 121 | 41.3 | 126.6 | 11.7 | 1.5 | Yes | JTGP00000000.1 | [25] | |||
| Deinagkistrodon | acutus | Illumina | ♂ 114 ♀ 238 | 2,120 | 22.42 | 1.4 | 21,194 | Yes | DQ343647.1 | [86] | |||
| Colubridae | Pantherophis | guttatus | Illumina | 13 | 38.3 | 4.3 | 2.39 | 1.4 | No | JTLQ00000000.1 | [22] | ||
| Thermophis | baileyi | Illumina | 185 | 43.6 | 2,414 | 16.8 | 1.8 | 20,995 | No | QLTV00000000 | [23] | ||
| Thamnophis | sirtalis | Illumina | 72 | 41.8 | 516 | 10.45 | 1.4 | Yes | LFLD00000000.1 | [87] | |||
| elegans | Illumina; PacBio | 62 | 41.1 | 100,851 | 4,620 | 1.6 | 18,900 | Yes | PRJNA561996 | ||||
| Elapidae | Ophiophagus | hannah | Illumina | 28 | 40.6 | 226 | 3.98 | 1.6 | Yes | AZIM00000000.1 | [18] | ||
| Pseudonaja | textilis | 73 | 40.1 | 14,685 | 50.44 | 1.6 | 19,358 | Yes | ULFR00000000.1 | [84] | |||
| Notechis | scutatus | Illumina; PacBio | 71 | 40.2 | 5,997 | 31.76 | 1.6 | 19,770 | Yes | PRJEB27871 | [28] | ||
| Naja | naja | PacBio; Nanopore; Illumina | 250 | 40.4 | 223,350 | 303.98 | 1.79 | 23,248 | Yes | SOZL00000000.1 | [27] | ||
| Hydrophis | curtus | Illumina NovaSeq | 120 | 37.2 | 1,346 | 183 | 1.62 | 21,863 | Yes | PRJNA597425 | [30] | ||
| Pythonoidea | Pythonidae | Python | bivittatus | Illumina; Roche 454 | 20 | 39.7 | 214 | 10.66 | 1.4 | 19,793 | No | AEQU00000000.2 | [9] |
| Booidea | Boidae | Boa | constrictor | Illumina; Roche 454; PacBio | 125 | 1.6 | No | [19] | |||||
PB stands for PacBio and IL for Illumina.
GC% refers to the percentage of the guanine (G) and cytosine (C) bases in a genome, scaffold N50 is a measure of the assembly quality (see below), DoC is a measure of the depth of coverage (see below), and INSDC ID is the NCBI gene bank accession number of the respective genome.
Available snake genomes differ notably in their assembly and annotation qualities, which makes evaluation of genome quality an important factor in determining the suitability of a genome for addressing a given set of questions. For instance, while estimation of nucleotide composition and genomic repeat content can be achieved from a relatively fragmented genome assembly, high-quality genome assemblies are required for analyses of multi-gene families and regulatory elements [88]. The reason for this is that the majority of the known venom gene families form tandem-arrayed “gene islands” (significantly enriched in microchromosomes, see, e.g., [13]), which generally represent a challenge for performing a continuous assembly. To achieve the best quality of assembly of venom genes, the use of long-read technology (e.g., PacBio HiFi or MinIon) is therefore essential (Fig 2). Genome assembly quality is assessed using statistics that measure fragmentation of the genome assembly, such as total assembly length, total contig number, contig N50, and scaffold N50. The total length of the assembly represents the total length of all the contigs that are part of the de novo assembled genome. A high total assembly length usually indicates a high-quality genome assembly. The contig N50 expresses the contiguity of the assembled genome. For instance, a contig N50 of 10 kb implies that 50% of the entire genome assembly is contained in contigs that are longer than 10 kb. Thus, a high contig N50 value represents a high-quality assembly without too many gaps. Currently, the contig N50 values of most published snake genomes are <25 kb; exceptions include 7 species with better assembly quality, namely, T. elegans (western terrestrial garter snake; 4,620 kb) [89], C. tigris (tiger rattlesnake; 2,110 kb) [32], N. naja (Indian cobra; 304 kb) [31], H. curtus (Shaw's sea snake; 183 kb) [30], B. jararaca (Brazilian lancehead; 163.5 kb) [29], P. textilis (eastern brown snake; 51 kb) [84], and N. scutatus (tiger snake; 32 kb) [83].
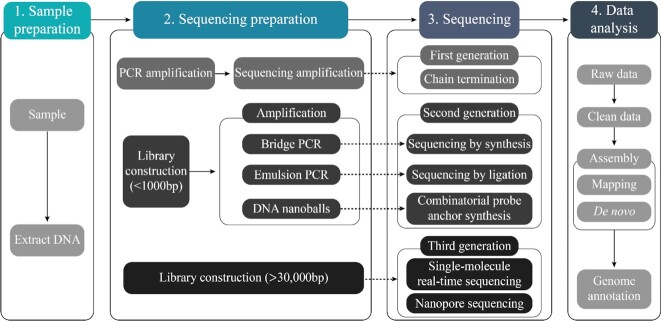
Figure 2:
Schematic representation of the next-generation sequencing pipeline for genomic assembly. (1) Multiple companies have marketed sequencing platforms for genomic and transcriptomic studies, the most commonly used being Illumina (left), PacBio (middle), and Nanopore (right). (2) The 3 platforms differ in read length and accuracy of their generated sequences. Whilst Illumina sequencing generally yields short reads with low error rates, Nanopore sequences are substantially longer (≤2 Mb), yet subject to frequent sequencing errors. Last, PacBio generates sequences with lengths and error rates in between the 2 other platforms. (3) After sequencing, reads are computationally processed and assembled into contigs, which in turn (4) serve as the building blocks for scaffolds.The scaffolds are then aligned and annotated to produce the complete target genome.
Another important parameter is the contig L50, which represents the minimum number of contigs required to cover 50% of the total assembly length. N50 and L50 values can be computed both at the contig and scaffold level. The most complete published snake genomes to date are those of N. naja and C. tigris, which were assembled by combining data obtained from long-read platforms (Pacific Biosciences [PacBio] and Nanopore) and short-read platforms (Illumina), as well as Chicago, Hi-C, and optical mapping in the case of N. naja [31, Fig. 2]. The resulting assemblies have a scaffold N50 reaching a staggering 207.72 Mb (C. tigris) and 223.35 Mb (N. naja) in length, which is ∼2.5 times greater than the previously assembled human reference genome (87 Mb) [31, 32, 90].
In addition to measures of genome contiguity — such as N50 scores — evaluating the representation of genes in a genome assembly via tools such as BUSCO provides great insight into genome assembly and annotation completeness [91]. A recent study using 611 published eukaryotic genomes showed that assemblies with high contig and scaffold N50 scores were shown to have high BUSCO values as well. However, the study revealed that assemblies with poor N50 scores may also (albeit rarely) show high BUSCO scores [92]. One example of this scenario in snakes is the case of the P. flavoviridis genome assembly, where contig N50 was 3.8 kb but percentages of complete and partial coverages for a set of 233 core vertebrate genes were 92.7% and 97.0%, respectively [18].
Furthermore, much can be learned about the quality of a genome from its reported depth of coverage (DoC). A DoC of 10× implies that each position in the genome has been read on average 10 times from independent sequencing reads. High DoC values imply that each position (i.e., each nucleotide) can be determined with greater confidence. Consequently, the 21 snake genomes published to date or in progress can be categorized into 2 groups: (1) a high DoC group (>50×) comprising B. jararaca (Brazilian lancehead) [29], C. viridis (prairie rattlesnake) [20,21], C. horridus (timber rattlesnake) [82], P. flavoviridis [15], P. mucrosquamatus (brown-spotted pit viper) [24], Vipera berus (European adder) [25], T. baileyi (Tibetan hot-spring snake) [23], T. sirtalis (common garter snake) [85], T. elegans (western terrestrial garter snake), P. textilis [84], C. tigris (tiger rattlesnake) [28], N. scutatus (tiger snake) [93], D. acutus (5-paced viper) [86], B. constrictor (red-tailed boa) [19], H. curtus (Shaw's sea snake) [30], and N. naja [27]; and (2) a low DoC group (13–40×), which includes S. catenatus (eastern massasauga rattlesnake), C. pyrrhus (southwestern speckled rattlesnake) [26], P. guttatus (corn snake) [22], O. hannah [18], and P. bivittatus [9]. Unsurprisingly, the earliest published snake genomes are characterized by lower DoCs, whereas the more recently sequenced genomes benefitted from technological advancement and thus generally obtained better coverages. The best example of this is the N. naja genome, which reached a DoC of 250× [27]—by far the highest DoC reported for a snake genome to date (Table 2). This high DoC enabled the discovery of 43 new toxin-encoding genes, some of which are likely to be unique to N. naja [27].
Genome size (the total amount of DNA contained within 1 copy of a single complete genome [94]), number of genes, and guanine-cytosine content [95] vary from species to species and therefore may help elucidate phylogenetic relationships and molecular events (e.g., gene/genome duplication, pseudogenization, gene loss) in the evolution of species. Genome size can vary greatly and is typically correlated with organism size and complexity, as well as with genome repeat content [94]. The reported genome sizes of snakes range from 1.3 to 1.8 Gb, except for C. pyrrhus (1.1 Gb) and B. jararaca (2.1 Gb) (Table 2). This is consistent with previous findings that squamate reptiles and birds generally have smaller genomes than mammals (1–3 Gb for squamates vs 1–2 Gb for birds vs 2–6 Gb for mammals) (Table 3) [22].
Table 3:
Selected genomic features compared across several vertebrate lineages [21]
| Transposable elements content (%) | ||||
|---|---|---|---|---|
| Tetrapod taxon | Genome size (Gb) | GC content (%) | Range | Mean |
| Mammals | 2.2–6.0 | ∼40.9 | 33.4–56.4 | 44.5 |
| Birds | 1.2–2.1 | ∼40.2 | 4.6–10.1 | 7.8 |
| Colubroidea | 1.5–3.0 | 39.3–47.8 | 33.0–56.3 | 46.2 |
| Non-colubroid snakes | 1.7–2.1 | 38.8–43.4 | 28.7–48.7 | 38.7 |
| Scincoidea (skinks) | 1.3–2.6 | 43.2–46.1 | 34.3–44.0 | 37.6 |
Somewhat counterintuitively, genome size is not necessarily correlated with the number of genes in the genome. For example, although the Homo sapiens genome (2.90 Gb) is ∼2 times larger than the T. sirtalis genome (1.42 Gb), the number of genes is similar between the two (20,186 genes for T. sirtalis compared to 21,407 genes for H. sapiens) [87]. This implies higher average gene density (genes/Mb) in T. sirtalis than in H. sapiens, which is likely rooted in the larger percentage of repeat elements in the human genome compared to that of T. sirtalis (∼70% and 37.12%, respectively) [87, 96, 97]. Thus, a considerably larger portion of the H. sapiens genome is not composed of protein-coding regions compared to the genome of T. sirtalis, which may compensate for the difference between their genome sizes. Furthermore, even though the average gene length of T. sirtalis (13,384 bp) is significantly smaller than that of H. sapiens (23,247 bp), exon length is comparable between the two (280.12 vs 249.22 bp, respectively) [87].
Unlike their genome sizes and gene lengths, the genomic GC contents for mammals, birds, and squamates are similar (∼40%) (Table 3), and the GC contents of reported snake genomes range from 34.3% to 43.6% (Table 3). Interspecies variation in GC content is thought to be caused by selective variation, mutation bias, and biased DNA repair-related recombination [95]. High GC content might also be an indication of biased sequencing results [98]. It is advisable to obtain information regarding both genome size and GC content prior to de novo assembly of a genome because these key genomic features can guide the choice of the most appropriate assembly strategy.
Understanding Snake Venom Evolution Through Snake Genomes
Genetic research on snake toxins
Phylogenetics is the cornerstone of our understanding of evolutionary relationships at all taxonomic levels and provides a historical basis for testing and inferring ecological and evolutionary processes [99–102]. In the past few decades, snake venom and its evolutionary origins have received considerable attention [46,103–105]. Although there is uncertainty and controversy about the origin of the venom system in squamate reptiles [29,103,106–108] a prevalent hypothesis is that the core snake venom system evolved in the common ancestor of snakes and lizards [103].
Venom is a polygenic trait that has evolved many times in the tree of life, and it serves a role in both prey capture and defence against predators [104,109]. Unlike many polygenic traits [110,111], venom has a relatively direct pathway from transcription of toxin genes to translation into toxin proteins, which are then stored in the venom gland [46,112]. Thus, by combining venom-gland transcriptomics and venom proteomics, we can accurately map the progression from genotype to phenotype in this adaptive trait [104]. Because transcriptome data will vary depending on the geographical origin of the population, the age and sex of the snake, and time since the last expulsion of venom [113,114] that the snake was subjected to at the time of collection and/or sampling, as well as on the characteristics of the underlying genotype, transcriptomes represent a sample of the spatiotemporally expressed genome and can be used as an entry into genome divergence analysis. Genome divergence analysis takes advantage of whole-genome and/or transcriptome data to reconstruct phylogenies that chart the relationships among snakes, thus representing a precious resource for studies of snake venom evolution.
Structural characteristics of the toxin genes in snake genomes
More than 10,000 species of squamate reptiles have evolved over the past 200 million years, making this clade a major component of the vertebrate lineage [115]. The number of protein-coding genes is remarkably constant across vertebrates (including snakes), but vertebrate genomes differ considerably in size, structure, and composition [21]. An important genomic feature in this regard are transposable elements (TEs), which are self-replicating DNA sequences with the ability to insert themselves in new positions in the genome, thereby altering genome structure and gene regulation [116,117]. Having a high abundance of TEs could lead to a high degree of evolvability in structural features of the genome, where pseudogenization and gene duplication may occur more frequently, thus creating opportunities for neofunctionalization. As such, it is perhaps hardly surprising that TEs are consistently involved in the evolution of snake venoms [17,18].
Preliminary research indicates that one of the main differences across snake genomes is the abundance and diversity of TEs, which ranges between 33.0–56.3% in Colubroidea and 28.7–8.7% in non-colubroid snakes [20,27,86, 87]. For comparison, other reptiles, such as members of the order Scincoidea, have a lower variation in their number of TEs (34.3–44.0%) (Table 3) [21,27,86]. Both abundance and diversity of TEs in snake genomes are exemplified by the genomes of D. acutus and B. jararaca. The former is made up of 13.84% long interspersed elements (LINEs, e.g., CR1, L1, and L2), 7.96% DNA transposons (e.g., hAT and TcMar elements), and 2.59% retrotransposons (e.g., Gypsy and DIRS elements) [86], whereas the latter comprises 14.6% LINEs with L2/CR1/Rex as the most abundant (8.8% of whole genome). The observed differences in the repeat content cannot be attributed only to varying sequencing technologies, as shown by the comparison of genome assembly qualities between snakes. For instance, while B. constrictor has a higher scaffold N50 (4,505.2 kb) and less total gap length (55,688.38 kb) compared to D. acutus (N50 2,122.2 kb; gap length 82,553.36 kb), the latter shows a higher total TE content (47.47 vs 39.59%) [118]. The genomes of D. acutus and O. hannah have a fairly low divergence level (<10%) of CR1 and hAT elements from the inferred ancestral consensus sequences, while snakes belonging to more basal-branching clades (e.g., B. constrictor and P. bivittatus) have >20% divergence level [86]. Conversely, CR1 and hAT content is >3 times higher in D. acutus and O. hannah than in B. constrictor and P. bivittatus, but the latter 2 species have undergone independent expansion of L2 repeat contents [86]. Another study that highlights genomic differences in TE content in snakes showed that repeat element abundances in the genomes of D. acutus, T. sirtalis, and O. hannah (all part of the Colubroidea clade) were characterized by a higher CR1-like and DNA transposon content compared to the genome of P. bivittatus [87]. Overall, repeat elements in the genomes of venomous snakes are generally more active, diverse, and dynamic compared to those of non-venomous species, indicating that different types of TEs may have played multiple important roles in functional regulation of snake genes throughout evolution.
Another TE category that has attracted research attention is microsatellites (short repeated DNA sequences). Microsatellites are so ubiquitous in certain snake species that a snake genome holds the record for containing the highest microsatellite content in any known eukaryote [21]. Bolstering this claim, a study of 11 viper species found an unprecedented average microsatellite content of 16,214 bp/Mb [21]. In comparison, the mean microsatellite density of 4 non-venomous snakes was ∼55% of that amount, i.e., 8,953 bp/Mb [21]. The same study found that the mean genome density of simple sequence repeat (SSR) loci (448–896 loci/Mb) was roughly twice as large in venomous snake microsatellites as in non-venomous snake homologs [21]. The study further found that the AATAG loci (which tend to be immediately adjacent to CR1-L3 LINEs in colubroid genomes) in venomous colubroids were increased 75-fold compared to other squamate reptiles and 71-fold compared to non-colubroid snakes [21]. Based on the significant expression of SSRs and LINE-SSR hybrid element content in venomous snakes compared to non-venomous snakes, the study also concluded that SSRs and LINE-SSR hybrid elements may have played key roles in the evolution of snake venoms [21]. The dynamics and extent of the influence of SSRs and LINE-SSR on venom evolution therefore represent an intriguing venue for further research.
However, microsatellite content alone cannot explain the course of venom evolution. Indeed, another important factor is the chromosomal location of venom genes. What is known about snake chromosomes is largely based on cytogenetic experimental studies, which have revealed that the majority of snakes have 18 chromosomes (8 macrochromosomes and 10 microchromosomes) [20]. It has been observed that a high proportion of venom genes are located on microchromosomes [15,21], revealing a consistent pattern of homologous chromosomal location for multiple venom gene families arranged in tandem-array gene clusters. For example, 37% of all venom genes in the C. viridis genome and ∼57% (27/47 genes) of all annotated venom-related genes in the P. flavoviridis genome are located on microchromosomes (Fig. 3) [21,22]. This is the case for C. tigris as well, with all genes belonging to the major toxin family in the venom of this species (PLA2s) located on microchromosome 7 [33]. Phylogenetic analysis of the 3 most abundant and well-characterized toxin families in C. viridis venom (SVMPs, SVSPs, and PLA2s, all located on microchromosomes) revealed that each toxin gene family represents a distinct set of duplicated genes derived from a single ancestral homolog that produced a monophyletic cluster of venomous paraphyletic lineages [21]. Notably, microchromosomes have higher GC content and faster recombination rates than macrochromosomes [21], as is evident in the C. viridis genome [20]. Therefore, it appears that microchromosomes are generally enriched with venom genes, which together with their high recombination rate could explain the huge radiation and rapid evolution of venom-related genes [15].
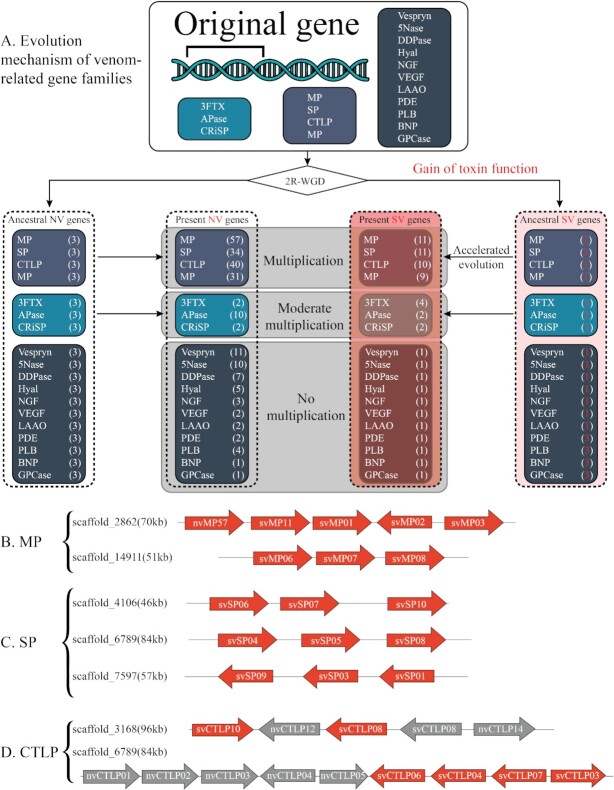
Figure 3:
Venom-related gene families in the P. flavoviridis genome. (A) Deduced evolutionary history of venom-related gene families through 2 rounds of whole-genome duplication (2R-WGD). An original set of 18 genes (shown in the top box) became 72 (4 copies each). Then, a single copy of each family was likely co-opted to develop toxic functions, resulting in 1 snake venom (SV) copy (shown in a pale red box in the right column) and 3 non-venom (NV) paralogs (shown in the see-through box to the left). (B) Tandem duplications of SVMP genes. (C) Tandem duplications of SVSP genes. (D) Tandem duplications of CTLP genes. Based on Fig. 2 and Fig. S8 from [15].
Nonetheless, it should be noted that a substantial percentage of toxin-coding genes are found on macrochromosomes as well. This is evident in N. naja, where as many as 16 toxin gene families are located on macrochromosomes [32]. WGS of other venomous snake species will be essential to investigate how and to what extent chromosomal location of genes influences venom evolution. Interestingly, the chromosome structure of C. viridis is comparable to that of N. naja. In fact, chromosome 4 of N. naja shares syntenic regions with C. viridis chromosomes 3 and 5, and chromosomes 5 and 6 of N. naja are syntenic with chromosome 5 of C. viridis [27]. This might indicate the occurrence of fusion and fission events, respectively [27]. The N. naja genome has also been compared to that of O. hannah (another elapid, and thus more closely related to N. naja than C. viridis), where 139 venom gland toxin genes from the N. naja genome were cross-referenced with genes in the O. hannah genome to find orthologs [27]. The results showed that 96 of the N. naja genes had counterparts in the O. hannah genome, while 43 did not [27]. Although some of these 43 genes may be unique to N. naja, others may simply not have been annotated in the O. hannah genome, possibly owing to the high fragmentation of its assembly (which relied on short reads) [14].
In the future, widespread access to different types of sequencing platforms providing researchers with both short and long reads, complementary tools for genome analysis (Hi-C and CHiCAGO), and higher quality sequence data will likely enable researchers to study snake genomes in greater detail. In turn, this will help elucidate differences and similarities between snake genomes and allow for more fine-grained studies of the structural characteristics of snake venom genes.
Molecular origin and regulation of snake venom genes
Snake venoms and their evolutionary origins have received substantial attention over the past decades, with >15,000 studies published on this topic [15]. Snake venoms have the dual functions of defense against predators and subduction of prey, with predation typically being the primary function [104]. This locks snakes and their prey in an evolutionary arms race, where the prey evolves biological strategies that make it resistant to toxins, and snakes are constantly pressured to optimize and adjust the composition of toxins in their venoms [104]. Indeed, dietary habits have often been indicated as a key driver of adaptive venom evolution in snakes, featuring among the main reasons behind inter- and intraspecific variation in venom composition [119].
Reports on trophic adaptations of snakes are plentiful. As an example, a study showing that venom variation in the Malayan pit viper (Calloselasma rhodostoma) throughout its range is significantly associated with the types of prey locally available [120]. This is also the case for the Mangrove catsnake (Boiga dendrophila), which was found to possess a 3FTx specific for birds and lizards (the bulk of this snake's diet) but scarcely effective on mammals [121]. However, recent research reported that venom composition in the Mojave rattlesnake (Crotalus scutulatus) was associated with environmental factors (e.g., temperature, seasonality) rather than with diet [122]. This suggests that a more complex scenario of factors could be affecting venom diversity than prey-related drivers alone, as confirmed by the dynamics behind venom variation in the Northern Pacific rattlesnake (Crotalus oreganus). In fact, the dichotomy in venom composition observed in this species is consistently influenced not only by coevolution with its prey but also by genetic distance and elevation-based habitat gradients, in a pattern described as “phenotype matching” of venom characteristics to multiple variables in the snake's native ecosystem [123,124]. The genetic basis underlying such complex adaptive processes could likely provide intriguing insight into the influence of natural selection and phylogenetic relatedness on the evolution of a highly dynamic trait such as snake venom. To this end, WGS of snakes will likely be key to conclusively determining the structural and evolutionary features of toxin genes and gene clusters. Analysing such patterns in a comparative framework would then enable researchers to identify similarities and differences in adaptive drivers of venom evolution at all levels of snake taxonomy and phylogeny.
In recent years, venom evolution has been further explored through genome studies on several species of venomous snakes [15,21,27,86]. One of these studies revealed that the venom gene repertoire of D. acutus has a very different composition from those of O. hannah and the non-venomous A. carolinensis (outgroup), B. constrictor, and P. bivittatus. These differences are exemplified both by the absence of characteristic venom genes from the D. acutus genome relative to the O. hannah genome and by the increased gene copy number of other venom gene families, including SVMPs, CTLPs, and SVSPs (Table 1) [86]. Expression of most toxin-encoding genes shared by D. acutus and O. hannah (especially older genes derived from the last common ancestor of these species) is limited to venom glands or accessory glands [86]. Similarly, newer viper-specific toxin genes are expressed in the venom and accessory glands of D. acutus, while equally recent elapid-specific toxin genes are expressed in the venom and accessory glands of O. hannah [86]. Interestingly, genes closely related to the elapid-specific toxin genes expressed in the venom glands of O. hannah are expressed in the liver of D. acutus, and genes related to viper-specific toxin genes expressed in the venom glands of D. acutus are expressed in pooled organs from O. hannah [86].
These special expression patterns suggest that these venom genes may originate from metabolic proteins that have undergone subfunctionalization (i.e., paralogs retaining only part of the functional features of the original gene following duplication) or neofunctionalization, as well as that changes in tissue-specific expression have occurred [17, 86]. This is in accordance with previous protein-based findings [125,126]. Similarly, analysis of the O. hannah genome demonstrated that the regulatory components of the venomous secretion system may have evolved from the pancreas [18]. Several mechanisms likely contribute to the enhanced expression of toxin-coding genes in the venom gland. At the chromosome level, methylation and chromatin accessibility were recently shown to play a prominent role in gene regulation in C. tigris. In fact, methylation appears to be significantly more prevalent in non-toxin and unexpressed toxin genes compared to expressed toxin counterparts in the venom gland and pancreas of this species [33]. Furthermore, chromatin accessibility and methylation levels are positively related with the high expression of toxin genes compared to non-expressed counterparts and non-toxin genes in C. tigris, further supporting a joint role for these 2 factors in toxin gene expression [28]. Another important factor in regulation and expression of toxin genes is the gene regulatory network associated with them (recently termed “metavenom network”), which comprises ∼3,000 genes that do not code for toxins but actively influence their expression and postgenomic modifications (e.g., protein folding) in the venom gland as housekeeping genes [48]. Interestingly, this network presents highly conserved elements common to even distantly related lineages such as snakes and venomous mammals; on the other hand, snakes (specifically P. flavoviridis and P. mucrosquamatus) also displayed several unique regulatory genes that were likely co-opted together with neofunctionalized toxin genes absent in other lineages [48].
Gene duplication is thought to be one of the main mechanisms behind venom diversification [127]. The current consensus is that 2 rounds of whole-genome duplication (2R-WGD) occurred during the evolution of vertebrates [15,128]. A study of the P. flavoviridis genome identified 18 families of venom-related genes, including both toxin and non-toxin gene copies. These include metalloproteinases (SVMPs), serine proteases (SVSPs), CTLPs, PLA2s, 3FTxs, aminopeptidases (APaseNs), CRISPs, vespryns/SPla and ryanodine receptor domain proteins (Vespryns), 5′-nucleotidases (5Nases), dipeptidyl peptidases (DDPases), hyaluronidases (Hyals), nerve growth factors or neurotrophins (NGF), vascular endothelial growth factors (VEGFs), LAAOs, phosphodiesterases (PDEs), phospholipases B (PLBs), bradykinin-potentiating peptides and C-type natriuretic peptides (BNPs), and glutaminyl peptide cyclotransferases (GPCases). [15]. The study suggested that 2R-WGD resulted in the creation of 4 paralogs from each of the 18 genes and that during the later evolution of venomous snakes, 1 of these 4 gene copies underwent neo- or subfunctionalization and evolved toxic properties, while the remaining 3 copies did not [15]. Both the toxin and non-toxin encoding genes subsequently underwent multiplication to different extents (Fig. 4) [15], as is demonstrated by the multiple gene duplication events detected in the SVMP, SVSP, CTLP, PLA2, 3FTx, and CRISP gene families in P. flavoviridis and N. naja [15,27]. However, this phenomenon was investigated to the greatest detail in rattlesnakes (Crotalus spp.), with comparative genomics between species revealing multiple duplication events in neurotoxic PLA2 genes as well as all SVMP classes. Chromosome mapping of the complete genomes of C. viridis and C. tigris provided further support for the occurrence of this phenomenon, highlighting similar duplication events for both gene families as well as SVSP genes (all of which are arranged in tandem-array single clusters) [18, 26].
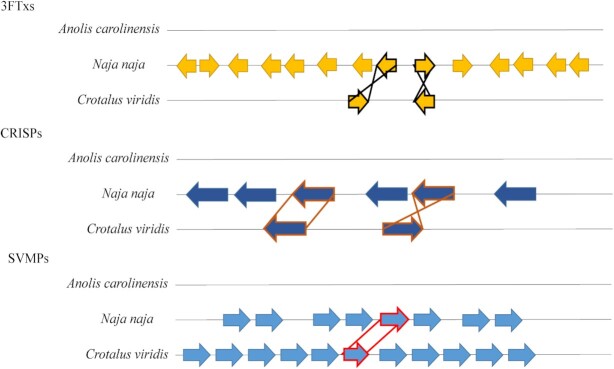
Figure 4:
Syntenic comparison of toxin gene clusters. Comparison showing the 3FTx, CRISP, and SVMP genes in N. naja and C. viridis genomes. Orthologous gene pairs are indicated by the line linked across the corresponding genomic regions. Based on Fig. 4 and Extended Fig. 4 from [27].
Molecular phylogenetic analysis of P. flavoviridis shows that all toxin genes of a given gene family in this species are homologous to the same toxin gene families found in vipers and elapids, such as P. mucrosquamatus (brown-spotted pit viper) and O. hannah [15]. The notion that snake toxin genes massively expanded through gene duplication events and underwent sub- and/or neofunctionalization is also supported by other studies [18,27,108]. For example, the N. naja genome assembly contributes to our understanding of the origin of multiple unlinked venom gene clusters and provides new and conclusive evidence that each toxin family stems from a unique set of tandem duplicate genes [27].
While duplication either before or after gene recruitment to the venom gland is an established driving force of venom evolution in snakes, loss of genetic material has been no less pivotal in facilitating diversification of toxin families in certain venomous snake clades. For instance, the interplay between gene duplication and deletion (of entire genes as well as intragenic regions) is remarkable in rattlesnakes (Crotalus spp.). These pit vipers present signs of multiple independent losses of ancestral genes coding for SVMPs and neurotoxic PLA2s—both of which had previously experienced a rampant expansion via repeated duplication episodes—across their phylogenetic tree [39, 71]. Intriguingly, different genes underwent deletion among and even within species, such as observed in the western diamondback rattlesnake (Crotalus atrox), the Mojave rattlesnake (C. scutulatus), and the Southern Pacific rattlesnake (Crotalus helleri) [39,64,71]. This resulted in great haplotype disparity and differential expression of toxin-encoding genes not only between species but across conspecific individuals as well. WGS of C. tigris further corroborated this pattern, as this species is known for its remarkably simple venom composition largely based on neurotoxic PLA2 isoforms [129]. However, the C. tigris genome revealed a deletion of 3 PLA2 genes on microchromosome 7 and of 10 SVMP genes on microchromosome 1 compared to homologous regions in C. viridis, indicating that even such a simple venom phenotype is the result of extensive genomic modifications over evolutionary time [28]. This pattern is not limited to rattlesnakes. For instance, the B. jararaca genome also displays a great expansion of SVMP genes via duplication upon recruitment in the venom gland, followed by 2 deletions in the exon 14 region of PII-SVMP genes causing loss of the Cys-rich domain found in PIII-SVMPs [29]. This observation sheds further light on the genomic processes responsible for evolution and differentiation via domain loss in SVMPs, which has occurred in other viper lineages as well [46].
Adaptive and neutral evolution in snake venoms
Determining and unraveling the driving factors behind the dynamic evolutionary processes in snake venom gene families has garnered the interest of scientists for decades—a quest that could only benefit from increasing efforts in WGS of venomous snakes. Positive selection appears to be the force behind the evolution of genes involved in predator-prey arms races [130], and it seems to be pervasive across most toxin-related gene families in snakes. Positive selection leaves a well-defined pattern in the genome, with the accumulation of non-synonymous, amino acid–replacing nucleotide substitutions (denoted by Ka), over synonymous substitutions (Ks) in the gene [131]. In P. flavoviridis, the Ka/Ks ratios of the 4 main toxin gene families were consistently > 1 and/or higher than those reported for non-venom genes (SVMPs: 1.047 ± 0.438, SVSPs: 1.253 ± 0.090, CTLPs: 0.871 ± 0.071, PLA2s: 1.093 ± 0.062) [15], suggesting positive selection behind the accelerated evolution of the major toxin gene families in this species. Interestingly, P. flavoviridis also exhibited Ka/Ks > 1, in the 3FTx and CRISP gene families, which therefore also displayed a tendency towards accelerated evolution despite being present in far fewer copies [15]. Similarly, a high Ka/Ks ratio (2.034 ± 0.818) was observed for the 3FTx gene family in the N. naja genome, again pointing towards rapid differentiation and functional diversification for these genes [27]. Conversely, when Ka/Ks < 1 it is indicative of either neutral selection (random substitutions that confer neither evolutionary advantages nor disadvantages) or purifying selection (i.e., removal of mutations that usually tend to be deleterious as they appear in conserved areas of the gene). In the P. flavoviridis genome study, all non-dominant toxin gene families had a Ka/Ks < 1 (mean = 0.512 [SD 0.018]), indicating a more neutral nucleotide substitution and the maintenance of similarity between gene copies [15]. On the other hand, when examining sequence divergence using venom gland transcriptomes in sidewinder rattlesnakes (Crotalus cerastes), data showed evidence of selection being stabilized, which supports that the maintainance of a generalist phenotype is favoured [132]. It must, however, be noted that despite various methods available for studying selection (see [133]), relatively few have been applied for the investigation of selection in venom and only in a small number of species [15,27, 132, 134]. Therefore, additional studies are required before general conclusions can be drawn.
New -omics tools and methods are rapidly advancing our knowledge of the mechanisms behind venom evolution [135]. In particular, WGS has introduced advantages to snake venom research, as WGS data can be used to identify structural variants, including inversions (Fig. 3A-B), insertions (Fig. 3C), deletions, tandem repeats (Fig. 3A-C), TEs, and other repeat content [21, 136]. An increasing number of studies report venom variation at different levels, such as ontogenetic, within-species, and between-species [46,131,137–140]. Once the reference genome of a species is available, population genomics can contribute to the identification of such intra- or interspecific variation. This further enhances the study of venom regulation, helping us to understand the evolution of complex regulatory networks [28]. Although it is generally acknowledged that positive selection seems to be the main driver behind venom evolution, genomic tools allow zooming in on specific venom-related genes to infer the role of neutral evolutionary processes, i.e., genetic drift or random changes in allele frequencies [141]. Genetic drift contributes to the accumulation of random neutral variation, which serves as the basis for natural selection to act upon in response to new evolutionary pressures [142]. Although most research to date has focused on the adaptive processes explaining venom evolution, recent studies have started assessing the role of such neutral forces in shaping venom characteristics. For example, genetic drift was identified as a prominent factor behind sequence divergence in venom genes in P. mucrosquamatus, where dominant toxin-encoding genes displayed relaxed selective constraints for deleterious mutations despite statistically significant rates of positive selection [24]. Furthermore, it has been shown that variation in expression of the myotoxin, crotamine, in the eastern diamondback rattlesnake (Crotalus adamanteus) and the South American rattlesnake (Crotalus durissus) is significantly more correlated with differences in number of duplication-derived gene copies between populations than with adaptive divergence in the sequences themselves [134,143].
The strength at which genetic drift acts on the genome is inversely proportional to effective population size (Ne, namely, the number of reproductive individuals that actually produce offspring) [142]. Ne greatly contributes to sequence variation, as the fate of a favourable mutation spreading is controlled by Ne and the strength of selection [144,145]. A prime example of this pattern in snake venom evolution is presented by the eastern massasauga rattlesnake (Sistrurus catenatus), a threatened species whose range consists of several scattered populations largely isolated from each other. Although the influence of genetic drift on venom evolution in this species is currently weak, it is likely to increase dramatically over time once the impact of drift is augmented due to the low Ne found in most populations [146]. Thus, complete genomes obtained through WGS together with complementary DNA libraries can expand our knowledge of the effects of selection on venom genes, with great potential to either corroborate or challenge the current positive selection–centered view of snake venom evolution.
Conclusions and Perspectives
WGS is a revolutionary advance in genetic research that has only recently been applied to the fields of herpetology and toxinology. Nonetheless, sequencing of complete snake genomes has already shed light on the evolutionary history of toxin-encoding genes as well as their expression patterns in the venom gland. In the future, WGS may be harnessed to obtain a better understanding of the molecular mechanisms involved in snake evolution [6,103], find new bioactive molecules with potential clinical applications, and provide valuable information for antivenom development [35]. Because only 21 complete snake genomes are currently available, there is ample opportunity for genomic research on the remaining thousands of snake species, including medically relevant venomous representatives. With the increasing power of sequencing technologies, the field of snake genomics is indeed likely to expand significantly in the years to come, with multiple complete genomes already in the process of being sequenced or published. However, this will not come without challenges because the interplay of dietary and environmental factors that has fueled venom diversification via gene duplication, recruitment, and neofunctionalization events makes it difficult to assemble whole venomous snake genomes. Another factor adding to the complexity of de novo genome assembly is the high content of repeat sequences in snake genomes. Some of these challenges might be adequately addressed by using third-generation sequencing technology. As the costs and error rates of this and other approaches decrease, they are certain to be used more widely in snake genome research. In turn, the assembly of more venomous snake genomes will allow us to explore adaptation and venom evolution at all phylogenetic levels, bringing a new perspective to the study of snake genomes and venoms.
Data Availability
Not applicable.
Abbreviations
3FTx: 3-finger toxin; bp: base pairs; BUSCO: Benchmarking Universal Single-Copy Orthologues; CRISP: cysteine-rich secretory protein; CTLP: C-type lectin-like protein; DoC: depth of coverage; Gb: gigabase pairs; kb: kilobase pairs; LAAO: L-amino acid oxidase; LINE: long interspersed element; NCBI: National Center for Biotechnology Information; NGS: next-generation sequencing; PLA2: phospholipase A2; SSR: simple sequence repeat; SVMP: snake venom metalloproteinase; SVSP: snake venom serine proteinase; TE: transposable element; WGS: whole-genome sequencing.
Competing Interests
W.R., W.Z., and S.L. are employees at the BGI. The authors declare that they have no other competing interests.
Funding
This research was supported by the Beijing Genomics Institute and the Technical University of Denmark. M.E.A. is funded by the Independent Research Fund Denmark (7027–00147B). C.K. is funded by Innovation Fund Denmark (9065–00007B). T.P.J. is funded under Marie Sklodowska-Curie grant agreement No. 713683 (COFUNDfellowsDTU).
Authors’ Contributions
A.H.L. and S.L. conceived the project. A.H.L., W.R., K.K., T.P.J., C.K., C.T.W., W.Z., S.G., L.S., M.M.D., B.J.M., E.R.T., and M.E.A. structured the draft and provided final editing. A.H.L., K.K., T.P.J., and W.R. coordinated and drafted the manuscript and implemented comments provided by all authors. All authors contributed critically to the scientific content. All authors read and approved the final manuscript.
Supplementary Material
Blair Perry -- 1/18/2022 Reviewed
Matthew Holding -- 1/18/2022 Reviewed
Contributor Information
Wei-qiao Rao, Department of Biotechnology and Biomedicine, Technical University of Denmark, Søltofts Plads 224, 2800 Kongens Lyngby, Denmark; Department of Mass Spectrometry, Beijing Genomics Institute-Research, 518083, Shenzhen, China.
Konstantinos Kalogeropoulos, Department of Biotechnology and Biomedicine, Technical University of Denmark, Søltofts Plads 224, 2800 Kongens Lyngby, Denmark.
Morten E Allentoft, Trace and Environmental DNA (TrEnD) Laboratory, School of Molecular and Life Sciences, Curtin University, Kent Street, 6102, Bentley Perth, Australia; Globe Institute, University of Copenhagen, Øster Voldgade 5, 1350, Copenhagen, Denmark.
Shyam Gopalakrishnan, Globe Institute, University of Copenhagen, Øster Voldgade 5, 1350, Copenhagen, Denmark.
Wei-ning Zhao, Department of Mass Spectrometry, Beijing Genomics Institute-Research, 518083, Shenzhen, China.
Christopher T Workman, Department of Biotechnology and Biomedicine, Technical University of Denmark, Søltofts Plads 224, 2800 Kongens Lyngby, Denmark.
Cecilie Knudsen, Department of Biotechnology and Biomedicine, Technical University of Denmark, Søltofts Plads 224, 2800 Kongens Lyngby, Denmark.
Belén Jiménez-Mena, DTU Aqua, Technical University of Denmark, Vejlsøvej 39, 8600, Silkeborg, Denmark.
Lorenzo Seneci, Department of Biotechnology and Biomedicine, Technical University of Denmark, Søltofts Plads 224, 2800 Kongens Lyngby, Denmark.
Mahsa Mousavi-Derazmahalleh, Trace and Environmental DNA (TrEnD) Laboratory, School of Molecular and Life Sciences, Curtin University, Kent Street, 6102, Bentley Perth, Australia.
Timothy P Jenkins, Department of Biotechnology and Biomedicine, Technical University of Denmark, Søltofts Plads 224, 2800 Kongens Lyngby, Denmark.
Esperanza Rivera-de-Torre, Department of Biotechnology and Biomedicine, Technical University of Denmark, Søltofts Plads 224, 2800 Kongens Lyngby, Denmark.
Si-qi Liu, Department of Mass Spectrometry, Beijing Genomics Institute-Research, 518083, Shenzhen, China.
Andreas H Laustsen, Department of Biotechnology and Biomedicine, Technical University of Denmark, Søltofts Plads 224, 2800 Kongens Lyngby, Denmark.
References
- 1. Burbrink FT, Pyron RA.. The taming of the skew: estimating proper confidence intervals for divergence dates. Syst Biol. 2008;57(2):317–28. [DOI] [PubMed] [Google Scholar]
- 2. Wallach V, Williams KL, Boundy J.. Snakes of the World: A Catalogue of Living and Extinct Species. 1st ed. Boca Raton: CRC Press; 2014. [Google Scholar]
- 3. Da Silva FO, Fabre A-C, Savriama Y, et al. The ecological origins of snakes as revealed by skull evolution. Nat Commun. 2018;9(1):doi: 10.1038/s41467-017-02788-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pyron RA, Burbrink FT, Wiens JJ.. A phylogeny and revised classification of Squamata, including 4161 species of lizards and snakes. BMC Evol Biol. 2013;13(1):93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wiens JJ, Hutter CR, Mulcahy DG, et al. Resolving the phylogeny of lizards and snakes (Squamata) with extensive sampling of genes and species. Biol Lett. 2012;8(6):1043–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vonk FJ, Admiraal JF, Jackson K, et al. Evolutionary origin and development of snake fangs. Nature. 2008;454(7204):630–3. [DOI] [PubMed] [Google Scholar]
- 7. Saviola AJ, Chiszar D, Busch C, et al. Molecular basis for prey relocation in viperid snakes. BMC Biol. 2013;11(1):doi: 10.1186/1741-7007-11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gracheva EO, Ingolia NT, Kelly YM, et al. Molecular basis of infrared detection by snakes. Nature. 2010;464(7291):1006–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Castoe TA, de Koning APJ, Hall KT, et al. The Burmese python genome reveals the molecular basis for extreme adaptation in snakes. Proc Natl Acad Sci U S A. 2013;110(51):20645–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Greene HW, Fogden M, Fogden P.. Snakes: The Evolution of Mystery in Nature. 1st ed. Oxford: University of California Press; 1997. [Google Scholar]
- 11. Cohn MJ, Tickle C.. Developmental basis of limblessness and axial patterning in snakes. Nature. 1999;399(6735):474–9. [DOI] [PubMed] [Google Scholar]
- 12. Di-Poï N, Montoya-Burgos JI, Miller H, et al. Changes in Hox genes’ structure and function during the evolution of the squamate body plan. Nature. 2010;464(7285):99–103. [DOI] [PubMed] [Google Scholar]
- 13. Guerreiro I, Nunes A, Woltering JM, et al. Role of a polymorphism in a Hox/Pax-responsive enhancer in the evolution of the vertebrate spine. Proc Natl Acad Sci U S A. 2013;110(26):10682–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vicoso B, Emerson JJ, Zektser Y, et al. Comparative sex chromosome genomics in snakes: differentiation, evolutionary strata, and lack of global dosage compensation. PLoS Biol. 2013;11(8):e1001643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shibata H, Chijiwa T, Oda-Ueda N, et al. The habu genome reveals accelerated evolution of venom protein genes. Sci Rep. 2018;8(1):doi: 10.1038/s41598-018-28749-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Uetz P, Freed P, Hošek J. The Reptile Database. http://www.reptile-database.org. Accessed 8 June 2020. [Google Scholar]
- 17. Kerkkamp H, Kini R, Pospelov A, et al. Snake genome sequencing: results and future prospects. Toxins. 2016;8(12):360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vonk FJ, Casewell NR, Henkel CV, et al. The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system. Proc Natl Acad Sci U S A. 2013;110(51):20651–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bradnam KR, Fass JN, Alexandrov A, et al. Assemblathon 2: evaluating de novo methods of genome assembly in three vertebrate species. Gigascience. 2013;2(1):doi: 10.1186/2047-217X-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schield DR, Card DC, Hales NR, et al. The origins and evolution of chromosomes, dosage compensation, and mechanisms underlying venom regulation in snakes. Genome Res. 2019;29(4):590–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pasquesi GIM, Adams RH, Card DC, et al. Squamate reptiles challenge paradigms of genomic repeat element evolution set by birds and mammals. Nat Commun. 2018;9(1):doi: 10.1038/s41467-018-05279-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ullate-Agote A, Milinkovitch MC, Tzika AC.. The genome sequence of the corn snake (Pantherophis guttatus), a valuable resource for EvoDevo studies in squamates. Int J Dev Biol. 2014;58(10-11-12):881–8. [DOI] [PubMed] [Google Scholar]
- 23. Li J-T, Gao Y-D, Xie L, et al. Comparative genomic investigation of high-elevation adaptation in ectothermic snakes. Proc Natl Acad Sci U S A. 2018;115(33):8406–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Aird SD, Arora J, Barua A, et al. Population genomic analysis of a pitviper reveals microevolutionary forces underlying venom chemistry. Genome Biol Evol. 2017;9(10):2640–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.: Vipera berus berus isolate:VBER.BE-female (ID 170536) - BioProject - NCBI. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA170536. Accessed 27 November 2019. [Google Scholar]
- 26. Gilbert C, Meik JM, Dashevsky D, et al. Endogenous hepadnaviruses, bornaviruses and circoviruses in snakes. Proc Biol Sci. 2014;281(1791):20141122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Suryamohan K, Krishnankutty SP, Guillory J, et al. The Indian cobra reference genome and transcriptome enables comprehensive identification of venom toxins. Nat Genet. 2020;52(1):106–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Margres MJ, Rautsaw RM, Strickland JL, et al. The Tiger Rattlesnake genome reveals a complex genotype underlying a simple venom phenotype. Proc Natl Acad Sci U S A. 2021;118(4):e2014634118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Almeida DD, Viala VL, Nachtigall PG, et al. Tracking the recruitment and evolution of snake toxins using the evolutionary context provided by the Bothrops jararaca genome. Proc Natl Acad Sci U S A. 2021;118(20):e2015159118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Peng C, Ren J-L, Deng C, et al. The genome of Shaw's sea snake (Hydrophis curtus) reveals secondary adaptation to its marine environment. Mol Biol Evol. 2020;37(6):1744–60. [DOI] [PubMed] [Google Scholar]
- 31. Ochoa A, Gibbs HL.. Genomic signatures of inbreeding and mutation load in a threatened rattlesnake. Mol Ecol. 2021;30(21):5454–69. [DOI] [PubMed] [Google Scholar]
- 32. Vonk FJ, Jackson K, Doley R, et al. Snake venom: from fieldwork to the clinic: recent insights into snake biology, together with new technology allowing high-throughput screening of venom, bring new hope for drug discovery. Bioessays. 2011;33(4):269–79. [DOI] [PubMed] [Google Scholar]
- 33. Windley MJ, Herzig V, Dziemborowicz SA, et al. Spider-venom peptides as bioinsecticides. Toxins. 2012;4(3):191–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hucho F. Toxins as tools in neurochemistry. Angew Chem Int Ed Engl. 1995;34(1):39–50. [Google Scholar]
- 35. Laustsen AH. Guiding recombinant antivenom development by omics technologies. New Biotechnol. 2018;45:19–27. [DOI] [PubMed] [Google Scholar]
- 36. Majoros WH, Pertea M, Salzberg SL.. TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics. 2004;20(16):2878–9. [DOI] [PubMed] [Google Scholar]
- 37. Liu X, Zheng Q, Vrettos N, et al. A microRNA precursor surveillance system in quality control of microRNA synthesis. Mol Cell. 2014;55(6):868–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Collins JE, White S, Searle SMJ, et al. Incorporating RNA-seq data into the Zebrafish Ensembl Gene Build. Genome Res. 2012;22(10):2067–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dowell NL, Giorgianni MW, Kassner VA, et al. The deep origin and recent loss of venom toxin genes in rattlesnakes. Curr Biol. 2016;26(18):2434–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tekaia F. Inferring orthologs: open questions and perspectives. Genomics Insights. 2016;9:GEI.S37925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Viala VL, Hildebrand D, Trusch M, et al. Venomics of the Australian eastern brown snake (Pseudonaja textilis): detection of new venom proteins and splicing variants. Toxicon. 2015;107:252–65. [DOI] [PubMed] [Google Scholar]
- 42. Ogawa T, Oda-Ueda N, Hisata K, et al. Alternative mRNA splicing in three venom families underlying a possible production of divergent venom proteins of the habu snake, Protobothrops flavoviridis. Toxins. 2019;11(10):581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Siigur E, Aaspõllu A, Siigur J.. Sequence diversity ofVipera lebetina snake venom gland serine proteinase homologs – result of alternative-splicing or genome alteration. Gene. 2001;263(1-2):199–203. [DOI] [PubMed] [Google Scholar]
- 44. Sunagar K, Khochare S, Senji Laxme RR, et al. A wolf in another wolf's clothing: post-genomic regulation dictates venom profiles of medically-important cryptic kraits in India. Toxins. 2021;13(1):69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rokyta DR, Margres MJ, Calvin K.. Post-transcriptional mechanisms contribute little to phenotypic variation in snake venoms. G3 (Bethesda). 2015;5(11):2375–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Casewell NR, Wagstaff SC, Wuster W, et al. Medically important differences in snake venom composition are dictated by distinct postgenomic mechanisms. Proc Natl Acad Sci U S A. 2014;111(25):9205–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Barua A, Mikheyev AS.. An ancient, conserved gene regulatory network led to the rise of oral venom systems. Proc Natl Acad Sci U S A. 2021;118(14):e2021311118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Elshire RJ, Glaubitz JC, Sun Q, et al. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One. 2011;6(5):e19379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jones MR, Good JM. Targeted capture in evolutionary and ecological genomics. Mol Ecol. 2016;25(1):185–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Holding ML, Sovic MG, Colston TJ, et al. The scales of coevolution: comparative phylogeography and genetic demography of a locally adapted venomous predator and its prey. Biol J Linn Soc. 2021;132(2):297–317. [Google Scholar]
- 51. Ellegren H. Genome sequencing and population genomics in non-model organisms. Trends Ecol Evol. 2014;29(1):51–63. [DOI] [PubMed] [Google Scholar]
- 52. Laustsen A, Engmark M, Milbo C, et al. From fangs to pharmacology: the future of snakebite envenoming therapy. Curr Pharm Des. 2016;22(34):5270–93. [DOI] [PubMed] [Google Scholar]
- 53. Tasoulis T, Isbister GK.. A review and database of snake venom proteomes. Toxins. 2017;9(9):290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kini RM, Doley R.. Structure, function and evolution of three-finger toxins: mini proteins with multiple targets. Toxicon. 2010;56(6):855–67. [DOI] [PubMed] [Google Scholar]
- 55. Fry BG, Wüster W, Kini RM, et al. Molecular evolution and phylogeny of elapid snake venom three-finger toxins. J Mol Evol. 2003;57(1):110–29. [DOI] [PubMed] [Google Scholar]
- 56. Nirthanan S. Snake three-finger α-neurotoxins and nicotinic acetylcholine receptors: molecules, mechanisms and medicine. Biochem Pharmacol. 2020;181:114168. [DOI] [PubMed] [Google Scholar]
- 57. Gasanov SE, Dagda RK, Rael ED.. Snake venom cytotoxins, phospholipase A2s, and Zn2+-dependent metalloproteinases: mechanisms of action and pharmacological relevance. J Clin Toxicol. 2014;4(1):1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lomonte B, Gutiérrez JM.. Phospholipases A2 from viperidae snake venoms: how do they induce skeletal muscle damage?. Acta Chim Slov. 2011;58:647–59. [PubMed] [Google Scholar]
- 59. Gutiérrez JM, Calvete JJ, Habib AG, et al. Snakebite envenoming. Nat Rev Dis Primers. 2017;3:17063. [DOI] [PubMed] [Google Scholar]
- 60.. In: Tsai I-H., Gopalakrishnakone P, Calvete JJ. Snake venom phospholipase A2: evolution and diversity, eds. Venom Genomics and Proteomics. Dordrecht: Springer; 2016:291–306. [Google Scholar]
- 61. Kordiš D. Evolution of phospholipase A2 toxins in venomous animals. Acta Chim Slov. 2011;58:638–46. [PubMed] [Google Scholar]
- 62. Manjunatha Kini R. Excitement ahead: structure, function and mechanism of snake venom phospholipase A2 enzymes. Toxicon. 2003;42(8):827–40. [DOI] [PubMed] [Google Scholar]
- 63. Ohno M, Chijiwa T, Oda-Ueda N, et al. Molecular evolution of myotoxic phospholipases A2 from snake venom. Toxicon. 2003;42(8):841–54. [DOI] [PubMed] [Google Scholar]
- 64. Dowell NL, Giorgianni MW, Griffin S, et al. Extremely divergent haplotypes in two toxin gene complexes encode alternative venom types within rattlesnake species. Curr Biol. 2018;28(7):1016–26.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Markland FS, Swenson S.. Snake venom metalloproteinases. Toxicon. 2013;62:3–18. [DOI] [PubMed] [Google Scholar]
- 66. Gutiérrez JM, Rucavado A.. Snake venom metalloproteinases: their role in the pathogenesis of local tissue damage. Biochimie. 2000;82(9-10):841–50. [DOI] [PubMed] [Google Scholar]
- 67. Gutiérrez JM, Escalante T, Rucavado A, et al. A comprehensive view of the structural and functional alterations of extracellular matrix by snake venom metalloproteinases (SVMPs): novel perspectives on the pathophysiology of envenoming. Toxins. 2016;8(10):304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sanchez EF, Flores-Ortiz RJ, Alvarenga VG, et al. Direct fibrinolytic snake venom metalloproteinases affecting hemostasis: structural, biochemical features and therapeutic potential. Toxins. 2017;9(12):392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kini R, Koh C.. Metalloproteases affecting blood coagulation, fibrinolysis and platelet aggregation from snake venoms: definition and nomenclature of interaction sites. Toxins. 2016;8(10):284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Casewell NR, Wagstaff SC, Harrison RA, et al. Domain loss facilitates accelerated evolution and neofunctionalization of duplicate snake venom metalloproteinase toxin genes. Mol Biol Evol. 2011;28(9):2637–49., [DOI] [PubMed] [Google Scholar]
- 71. Giorgianni MW, Dowell NL, Griffin S, et al. The origin and diversification of a novel protein family in venomous snakes. Proc Natl Acad Sci U S A. 2020;117(20):10911–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Brust A, Sunagar K, Undheim EAB, et al. Differential evolution and neofunctionalization of snake venom metalloprotease domains. Mol Cell Proteomics. 2013;12(3):651–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Moura-da-Silva A, Almeida M, Portes-Junior J, et al. Processing of snake venom metalloproteinases: generation of toxin diversity and enzyme inactivation. Toxins. 2016;8(6):183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Serrano SMT. The long road of research on snake venom serine proteinases. Toxicon. 2013;62:19–26. [DOI] [PubMed] [Google Scholar]
- 75. Kunalan S, Othman I, Syed Hassan S, et al. Toxins. 2018;10(11):434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kang TS, Georgieva D, Genov N, et al. Enzymatic toxins from snake venom: structural characterization and mechanism of catalysis. FEBS J. 2011;278(23):4544–76. [DOI] [PubMed] [Google Scholar]
- 77. White J. Snake venoms and coagulopathy. Toxicon. 2005;45(8):951–67. [DOI] [PubMed] [Google Scholar]
- 78. Kini RM. The intriguing world of prothrombin activators from snake venom. Toxicon. 2005;45(8):1133–45. [DOI] [PubMed] [Google Scholar]
- 79. Markland FS. Snake venoms and the hemostatic system. Toxicon. 1998;36(12):1749–800. [DOI] [PubMed] [Google Scholar]
- 80. Meenakshisundaram R, Sweni S, Thirumalaikolundusubramanian P.. Hypothesis of snake and insect venoms against human immunodeficiency virus: a review. AIDS Res Ther. 2009;6(1):doi: 10.1186/1742-6405-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Delsuc F, Brinkmann H, Philippe H.. Phylogenomics and the reconstruction of the tree of life. Nat Rev Genet. 2005;6(5):361–75. [DOI] [PubMed] [Google Scholar]
- 82. Hall JB, Cobb VA, Cahoon AB. Mitochondrial DNA. 2013;24(2):94–6. [DOI] [PubMed] [Google Scholar]
- 83. St Pierre L, Masci PP, Filippovich I, et al. Comparative analysis of prothrombin activators from the venom of Australian elapids. Mol Biol Evol. 2005;22(9):1853–64. [DOI] [PubMed] [Google Scholar]
- 84. Earl STH, Birrell GW, Wallis TP, et al. Post-translational modification accounts for the presence of varied forms of nerve growth factor in Australian elapid snake venoms. Proteomics. 2006;6(24):6554–65. [DOI] [PubMed] [Google Scholar]
- 85. McGlothlin JW, Chuckalovcak JP, Janes DE, et al. Parallel evolution of tetrodotoxin resistance in three voltage-gated sodium channel genes in the garter snake Thamnophis sirtalis. Mol Biol Evol. 2014;31(11):2836–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Yin W, Wang Z, Li Q, et al. Evolutionary trajectories of snake genes and genomes revealed by comparative analyses of five-pacer viper. Nat Commun. 2016;7(1):doi: 10.1038/ncomms13107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Perry BW, Card DC, McGlothlin JW, et al. Molecular adaptations for sensing and securing prey and insight into amniote genome diversity from the garter snake genome. Genome Biol Evol. 2018;10(8):2110–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Schield DR, Perry BW, Pasquesi GIM, et al. Applications of genomics and related technologies for studying reptile venoms. In: Handbook of Venoms and Toxins of Reptiles. 2nd ed. CRC Press; 2021. [Google Scholar]
- 89. Thamnophis elegans isolate rThaEle1, whole genome shotgun sequencing project. https://www.ncbi.nlm.nih.gov/assembly/GCF_009769535.1/, 2019. [Google Scholar]
- 90. International Human Genome Sequencing Consortium . Finishing the euchromatic sequence of the human genome. Nature. 2004;431(7011):931–45. [DOI] [PubMed] [Google Scholar]
- 91. Seppey M, Manni M, Zdobnov EM, et al. , eds. Gene Prediction: Methods and Protocols. New York, NY: Springer; 2019. [Google Scholar]
- 92. Jauhal AA, Newcomb RD.. Assessing genome assembly quality prior to downstream analysis: N50 versus BUSCO. Mol Ecol Resour. 2021;21(5):1416–21. [DOI] [PubMed] [Google Scholar]
- 93. Edwards R, Amos T, Tang J, et al. Pseudodiploid pseudo-long-read whole genome sequencing and assembly of Pseudonaja textilis (eastern brown snake) and Notechis scutatus (mainland tiger snake). F1000Res. 2018:doi: 10.7490/f1000research.1115550.1. [DOI] [Google Scholar]
- 94. Le TS, Yang F-J, Lo Y-H, et al. Non-Mendelian assortment of homologous autosomes of different sizes in males is the ancestral state in the Caenorhabditis lineage. Sci Rep. 2017;7(1):12819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Birdsell JA. Integrating genomics, bioinformatics, and classical genetics to study the effects of recombination on genome evolution. Mol Biol Evol. 2002;19(7):1181–97. [DOI] [PubMed] [Google Scholar]
- 96. de Koning APJ, Gu W, Castoe TA, et al. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011;7(12):e1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Liehr T. Repetitive elements in humans. Int J Mol Sci. 2021;22(4):2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Benjamini Y, Speed TP.. Summarizing and correcting the GC content bias in high-throughput sequencing. Nucleic Acids Res. 2012;40(10):e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Figueroa A, McKelvy AD, Grismer LL, et al. A species-level phylogeny of extant snakes with description of a new Colubrid subfamily and genus. PLoS One. 2016;11(9):e0161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Harvey PH, Pagel MD. The Comparative Method in Evolutionary Biology. Oxford University Press; 1991. [Google Scholar]
- 101. Whelan S, Liò P, Goldman N.. Molecular phylogenetics: state-of-the-art methods for looking into the past. Trends Genet. 2001;17(5)262–72. [DOI] [PubMed] [Google Scholar]
- 102. Huelsenbeck JP, Rannala B. Phylogenetic methods come of age: testing hypotheses in an evolutionary context. Science. 2021;276(5310):227–32. [DOI] [PubMed] [Google Scholar]
- 103. Fry BG, Vidal N, Norman JA, et al. Early evolution of the venom system in lizards and snakes. Nature. 2006;439(7076):584–8. [DOI] [PubMed] [Google Scholar]
- 104. Casewell NR, Wüster W, Vonk FJ, et al. Complex cocktails: the evolutionary novelty of venoms. Trends Ecol Evol. 2013;28(4):219–29. [DOI] [PubMed] [Google Scholar]
- 105. Fry BG, Scheib H, van der weerd L, et al. Evolution of an arsenal: structural and functional diversification of the venom system in the advanced snakes (Caenophidia). Mol Cell Proteomics. 2008;7(2):215–46. [DOI] [PubMed] [Google Scholar]
- 106. Hargreaves AD, Swain MT, Hegarty MJ, et al. Restriction and recruitment—gene duplication and the origin and evolution of snake venom toxins. Genome Biol Evol. 2014;6(8):2088–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Reyes-Velasco J, Card DC, Andrew AL, et al. Expression of venom gene homologs in diverse python tissues suggests a new model for the evolution of snake venom. Mol Biol Evol. 2015;32(1):173–83. [DOI] [PubMed] [Google Scholar]
- 108. Casewell NR, Huttley GA, Wüster W. Dynamic evolution of venom proteins in squamate reptiles. Nat Commun. 2012;3(1):doi: 10.1038/ncomms2065. [DOI] [PubMed] [Google Scholar]
- 109. Gibbs HL, Mackessy SP.. Functional basis of a molecular adaptation: prey-specific toxic effects of venom from Sistrurusrattlesnakes. Toxicon. 2009;53(6):672–9. [DOI] [PubMed] [Google Scholar]
- 110. Romero IG, Ruvinsky I, Gilad Y.Comparative studies of gene expression and the evolution of gene regulation. Nat Rev Genet. 2012;13:505–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Shapiro MD, Marks ME, Peichel CL, et al. Genetic and developmental basis of evolutionary pelvic reduction in threespine sticklebacks. Nature. 2004;428(6984):717–23. [DOI] [PubMed] [Google Scholar]
- 112. Margres MJ, McGivern JJ, Wray KP, et al. Linking the transcriptome and proteome to characterize the venom of the eastern diamondback rattlesnake (Crotalus adamanteus). J Proteomics. 2014;96:145–58. [DOI] [PubMed] [Google Scholar]
- 113. Schwartz TS, Tae H, Yang Y, et al. A garter snake transcriptome: pyrosequencing, de novo assembly, and sex-specific differences. BMC Genomics. 2010;11(1):doi: 10.1186/1471-2164-11-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Hofmann EP, Rautsaw RM, Strickland JL, et al. Comparative venom-gland transcriptomics and venom proteomics of four Sidewinder Rattlesnake (Crotalus cerastes) lineages reveal little differential expression despite individual variation. Sci Rep. 2018;8(1):doi: 10.1038/s41598-018-33943-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Zheng Y, Wiens JJ.. Combining phylogenomic and supermatrix approaches, and a time-calibrated phylogeny for squamate reptiles (lizards and snakes) based on 52 genes and 4162 species. Mol Phylogenet Evol. 2016;94:537–47. [DOI] [PubMed] [Google Scholar]
- 116. Feiner N. Accumulation of transposable elements in Hox gene clusters during adaptive radiation of Anolis lizards. Proc Biol Sci. 2016;283(1840):20161555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Platt RN, Vandewege MW, Ray DA.. Mammalian transposable elements and their impacts on genome evolution. Chromosome Res. 2018;26(1-2):25–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Yin W, Wang Z, Li Q, et al. Evolutionary trajectories of snake genes and genomes revealed by comparative analyses of five-pacer viper. Nat Commun. 2016;7(1):doi: 10.1038/ncomms13107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Barlow A, Pook CE, Harrison RA, et al. Coevolution of diet and prey-specific venom activity supports the role of selection in snake venom evolution. Proc Biol Sci. 2009;276(1666):2443–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Daltry JC, Wüster W, Thorpe RS.. Diet and snake venom evolution. Nature. 1996;379(6565):537–40. [DOI] [PubMed] [Google Scholar]
- 121. Pawlak J, Mackessy SP, Fry BG, et al. Denmotoxin, a three-finger toxin from the Colubrid snake Boiga dendrophila (mangrove catsnake) with bird-specific activity. J Biol Chem. 2006;281(39):29030–41. [DOI] [PubMed] [Google Scholar]
- 122. Zancolli G, Calvete JJ, Cardwell MD, et al. When one phenotype is not enough: divergent evolutionary trajectories govern venom variation in a widespread rattlesnake species. Proc Biol Sci. 2019;286(1898):20182735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Holding ML, Biardi JE, Gibbs HL.. Coevolution of venom function and venom resistance in a rattlesnake predator and its squirrel prey. Proc Biol Sci. 2016;283(1829):20152841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Holding ML, Margres MJ, Rokyta DR, et al. Local prey community composition and genetic distance predict venom divergence among populations of the northern Pacific rattlesnake (Crotalus oreganus). J Evol Biol. 2018;31(10):1513–28. [DOI] [PubMed] [Google Scholar]
- 125. Fry BG. From genome to “venome”: molecular origin and evolution of the snake venom proteome inferred from phylogenetic analysis of toxin sequences and related body proteins. Genome Res. 2005;15(3):403–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Fry BG, Roelants K, Champagne DE, et al. The toxicogenomic multiverse: convergent recruitment of proteins into animal venoms. Annu Rev Genomics Hum Genet. 2009;10(1):483–511. [DOI] [PubMed] [Google Scholar]
- 127. Conant GC, Wolfe KH.. Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet. 2008;9(12):938–50. [DOI] [PubMed] [Google Scholar]
- 128. Holland PW, Garcia-Fernàndez J, Williams NA, et al. Gene duplications and the origins of vertebrate development. Dev Suppl. 1994:125–33. [PubMed] [Google Scholar]
- 129. Calvete JJ, Pérez A, Lomonte B, et al. Snake venomics of Crotalus tigris: the minimalist toxin arsenal of the deadliest neartic rattlesnake venom. Evolutionary clues for generating a pan-specific antivenom against Crotalid Type II venoms. J Proteome Res. 2012;11(2):1382–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Charlesworth B, Charlesworth D.. Elements of Evolutionary Genetics. 1st ed. WH Freeman; 2010. [Google Scholar]
- 131. Casewell NR, Jackson TNW, Laustsen AH, et al. Causes and consequences of snake venom variation. Trends Pharmacol Sci. 2020;41(8):570–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Rautsaw RM, Hofmann EP, Margres MJ, et al. Intraspecific sequence and gene expression variation contribute little to venom diversity in sidewinder rattlesnakes (Crotalus cerastes). Proc Biol Sci. 2019;286(1906):20190810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Kosiol C, Anisimova M.. Selection acting on genomes. In: Anisimova M, ed. Evolutionary Genomics: Statistical and Computational Methods. New York, NY: Springer; 2019. [DOI] [PubMed] [Google Scholar]
- 134. Margres MJ, Bigelow AT, Lemmon EM, et al. Selection to increase expression, not sequence diversity, precedes gene family origin and expansion in rattlesnake venom. Genetics. 2017;206(3):1569–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Reyes-Velasco J, Card DC, Andrew AL, et al. Expression of venom gene homologs in diverse python tissues suggests a new model for the evolution of snake venom. Mol Biol Evol. 2015;32(1):173–83. [DOI] [PubMed] [Google Scholar]
- 136. Tattini L, D'Aurizio R, Magi A. Detection of genomic structural variants from next-generation sequencing data. Front Bioeng Biotechnol. 2015;3:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Massey DJ, Calvete JJ, Sánchez EE, et al. Venom variability and envenoming severity outcomes of the Crotalus scutulatus scutulatus (Mojave rattlesnake) from Southern Arizona. J Proteomics. 2012;75(9):2576–87. [DOI] [PubMed] [Google Scholar]
- 138. Pla D, Sanz L, Quesada-Bernat S, et al. Phylovenomics of Daboia russeliiacross the Indian subcontinent. Bioactivities and comparative in vivo neutralization and in vitro third-generation antivenomics of antivenoms against venoms from India, Bangladesh and Sri Lanka. J Proteomics. 2019;207:103443. [DOI] [PubMed] [Google Scholar]
- 139. Durban J, Sanz L, Trevisan-Silva D, et al. Integrated venomics and venom gland transcriptome analysis of juvenile and adult Mexican rattlesnakes Crotalus simus, C. tzabcan, and C. culminatus revealed miRNA-modulated ontogenetic shifts. J Proteome Res. 2017;16(9):3370–90. [DOI] [PubMed] [Google Scholar]
- 140. Laxme RRS, Khochare S, de Souza HF, et al. Beyond the ‘big four’: venom profiling of the medically important yet neglected Indian snakes reveals disturbing antivenom deficiencies. PLoS Negl Trop Dis. 2019;13(12):e0007899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Kimura M. Evolutionary rate at the molecular level. Nature. 1968;217(5129):624–6. [DOI] [PubMed] [Google Scholar]
- 142. Wright S. Evolution in Mendelian populations. Genetics. 1931;16(2):97–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Oguiura N, Collares MA, Furtado MFD, et al. Intraspecific variation of the crotamine and crotasin genes in Crotalus durissus rattlesnakes. Gene. 2009;446(1):35–40. [DOI] [PubMed] [Google Scholar]
- 144. Charlesworth B. Effective population size and patterns of molecular evolution and variation. Nat Rev Genet. 2009;10(3):195–205. [DOI] [PubMed] [Google Scholar]
- 145. Ludington AJ, Sanders KL.. Demographic analyses of marine and terrestrial snakes (Elapidae) using whole genome sequences. Mol Ecol. 2021;30(2):545–54. [DOI] [PubMed] [Google Scholar]
- 146. Ochoa A, Broe M, Moriarty Lemmon E, et al. Drift, selection and adaptive variation in small populations of a threatened rattlesnake. Mol Ecol. 2020;29(14):2612–25. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Blair Perry -- 1/18/2022 Reviewed
Matthew Holding -- 1/18/2022 Reviewed
Data Availability Statement
Not applicable.