Abstract
Keloids are an abnormal fibroproliferative wound-healing disease with a poorly understood pathogenesis, making it difficult to predict and prevent this disease in clinical settings. Identifying disease-specific signatures at the molecular and cellular levels in both the blood circulation and primary lesions is urgently needed to develop novel biomarkers for risk assessment and therapeutic targets for recurrence-free treatment. There is mounting evidence of immune cell dysregulation in keloid scarring. In this study, we aimed to profile keloid scar tissues and blood cells and found that downregulation of cytotoxic CD8+ T cells is a keloid signature in the peripheral blood and keloid lesions. Single-cell RNA sequencing revealed that the NKG2A/CD94 complex was specifically upregulated, which might contribute to the significant reduction in CTLs within the scar tissue boundary. In addition, the NKG2A/CD94 complex was associated with high serum levels of soluble human leukocyte antigen-E (sHLA-E). We subsequently measured sHLA-E in our hospital-based study cohort, consisting of 104 keloid patients, 512 healthy donors, and 100 patients with an interfering disease. The sensitivity and specificity of sHLA-E were 83.69% (87/104) and 92.16% (564/612), respectively, and hypertrophic scars and other unrelated diseases exhibited minimal interference with the test results. Furthermore, intralesional therapy with triamcinolone combined with 5-fluorouracil drastically decreased the sHLA-E levels in keloid patients with better prognostic outcomes, while an incomplete reduction in the sHLA-E levels in patient serum was associated with higher recurrence. sHLA-E may effectively serve as a diagnostic marker for assessing the risk of keloid formation and a prognostic marker for the clinical outcomes of intralesional treatment.
Keywords: Keloid, Biomarker, T cell immunology
Subject terms: Biomarkers, Translational immunology
Introduction
Keloids are a dermal fibroproliferative disease affecting 4.5–16% of the human population [1–3]. They continue to grow for many years and rarely regress spontaneously [4, 5]. Keloids are associated with cosmetic disfigurement, itching, pain, and considerable psychological distress [6, 7]. However, accurate prediction, reliable prevention, and effective treatment of keloids are difficult to achieve [8]. Only 81% of clinically diagnosed keloid cases have been proven to be accurately diagnosed [9]. Surgical excision alone has a recurrence rate from 70 to 100% [10]. To date, no diagnostic biomarker or targeted therapeutic agent for keloids has been proven to be effective [11]. This limitation is probably attributable to the substantial heterogeneity of this disease due to variable racial susceptibilities, local tissue tension or infection, and various endocrine factors, among other factors [3, 12, 13]. Equally, there is limited insight into the cellular and molecular basis of keloid lesion-related abnormalities and systemic dysregulation [14, 15]. Published host gene expression profiling studies relying on whole white blood cells to characterize predisposing genes have not revealed the underlying mechanism and, hence, cannot be used for targeted therapy development [11, 16]. Therefore, identifying disease-specific signatures at the molecular and cellular levels in both the blood circulation and primary lesions is urgently needed to develop novel biomarkers for risk assessment and therapeutic targets for recurrence-free treatment.
Strong genetic factors underlie the disease susceptibility of individuals to keloids [17]. Genome-wide association studies in sporadic keloid populations have identified single-nucleotide polymorphisms in multiple chromosomal regions [16, 18]. In addition, a pedigree analysis of keloid patients demonstrated possible autosomal dominant inheritance with incomplete penetrance [19]. Several candidate genes have been proposed using linkage analysis, including TNFAIP6 at 2q23, EGFR at 7p11, and the SMAD genes and PIAS at 18q21, but none of these genes have been determined to be true predisposition genes [20, 21]. Since inflammation plays a critical role in keloid pathogenesis, large-scale case–control studies have been performed to identify human leukocyte antigen (HLA) polymorphisms associated with keloid risk [22, 23]. The alleles found in keloid disease in ethnic Han Chinese individuals include HLA-DQA1*0104 and HLA-DQB1*0501, which were associated with increased risk, and HLA-DQA1*0501, HLA-DQB1*0201, and HLA-DQB1*0402, which were associated with decreased risk. These data suggest that the associations of HLA alleles with keloids underlie a potential mechanism of MHC-related abnormal fibrosis. However, the exact mechanism by which these alleles affect keloid pathogenesis and their potential clinical utility in risk prediction remains to be elucidated.
The mechanisms underlying keloid formation and pathogenesis are poorly defined and understood. However, keloid formation can be attributed to fibroblast abnormalities. Recent lineage tracing and single-cell transcriptome sequencing have revealed that skin fibroblasts consist of distinct subpopulations arising from different lineages with different roles in determining the dermal architecture in skin development and repair [24–26]. Another study combining site-specific in situ microdissection and gene expression profiling of keloid tissues found distinct gene signatures in clear keloid regions, highlighting the morphological heterogeneity within the keloid scar [27]. That study demonstrated keloid dermal gene signatures and supported the roles of dermal fibroblasts in extracellular matrix deposition and all phases of wound healing. In contrast, there is mounting evidence showing immune cell dysregulation in keloid scarring; several T cells, B cells, and mast cells infiltrate the lesion site compared with normal skin [28, 29]. Highly potent macrophages were also found in keloid tissue, promoting the differentiation of FoxP3+ Treg cells [30]. Upregulation of proinflammatory genes in fibroblasts, as revealed by microarray data, may cause keloid formation and recurrence after surgical resection [31, 32]. These genes include interleukin-1, interleukin-6, tumor necrosis factor, and transforming growth factor-β, which are activated by the hyperactive nuclear factor-κB signaling pathway in keloid tissues and fibroblasts [33]. Altogether, the dysregulated immune microenvironment of keloid lesions warrants further in-depth investigation.
In this study, we aimed to examine T cell subtypes in tissue specimens from keloid patients and normal scar and skin samples from healthy donors and to further survey granzyme B+CD8+ cytotoxic T cells (CTLs) using single-cell RNA sequencing (scRNA-seq). We uncovered a novel signaling mechanism involved in keloid pathogenesis and recurrence by detailed analysis of the lineages of CTLs in keloid tissues and defined a new pathogenic factor in the circulation via intralesional therapy. We present a novel biomarker and simple assay method to reliably predict keloid risk and monitor disease recurrence.
Materials and methods
This study was conducted according to the tenets of the Declaration of Helsinki (1964) and its successive modifications and was approved by the institutional review board of the Ninth People’s Hospital (Shanghai Jiao Tong University School of Medicine). Informed consent was obtained from the participants before sample collection and analysis.
Sample collection
In this study, peripheral blood and tissue samples were collected from various cohorts for a plethora of analyses. A flow chart was drawn to better elucidate the study design and designate the various cohorts (Fig. S1). Normal skin, normal scar/hypertrophic scar, and keloid tissues were collected from patients who underwent blepharoplasty and scar revision surgeries. In addition, blood samples were collected from healthy donors and patients into EDTA-containing and coagulation-promoting tubes.
This study comprised two healthy donor cohorts and an interference case cohort that were used for blood sample collection. The first healthy donor cohort (n = 36) included subjects who underwent cosmetic surgery. The exclusion criteria for this cohort were as follows: (1) a history of hypertension, diabetes, tuberculosis, hepatitis, asthma, or allergic rhinitis; (2) a history of drug therapy; (3) no incidence of influenza, trauma, surgery, or toothache within 1 year; and (4) normal scars within 2 years. The second healthy donor cohort (n = 512) included subjects who underwent physical examinations and had no organic disease diagnosed. The interference case cohort (n = 100), as listed in Table 2, was from the Department of Plastic and Reconstructive Surgery and the Department of General Surgery.
Table 2.
Detailed soluble human leucocyte antigen-E level in cases of interference
| Cases of Interference (n = 100) | |
|---|---|
| Disease (No.) | Serum sHLA-E (ng/ml, mean ± SD) |
| Malignant dermatofibrosarcoma protuberans (n = 4) | 1.24 ± 0.59 |
| Basal cell carcioma (n = 4) | 0.50 ± 0.46 |
| Hypertrophic scar (n = 26) | 1.37 ± 1.38 |
| Hemorrhoidal bleeding (n = 6) | 2.30 ± 1.88 |
| Bile duct/Gallbladder carcinoma (n = 4) | 1.03 ± 0.66 |
| Gaslrointestinal perforation (n = 19) | 2.37 ± 1.79 |
| Hepatoma (n = 1) | 8.88 |
| Anal canal carcinoma (n = 1) | 0.39 |
| Anal fistula (n = 2) | 1.98 ± 2.04 |
| Crissum abscess (n = 4) | 4.46 ± 6.51 |
| Obstructive jaundice (n = 3) | 0.71 ± 0.29 |
| Mixed hemorrhoids (n = 14) | 2.16 ± 2.10 |
| Crohn disease (n = 4) | 1.62 ± 1.22 |
| Appendicular abscess (n = 1) | 12.06 |
| Pelvic tumor (n = 3) | 5.15 ± 1.72 |
| Pancreatic carcinoma (n = 1) | 0.07 |
| Colorectal carcinoma (n = 3) | 0.86 ± 1.15 |
Three patient cohorts were included for blood sample collection. The first keloid cohort (n = 34) included patients who underwent intralesional treatments three times; blood samples were collected from these patients before (n = 34) and after (n = 16) treatments [34]. Patients were excluded if (1) they had previously received any kind of treatment; (2) they had no incidence of influenza, trauma, surgery history, or toothache within 1 year; or (3) their keloid lesions had any kind of complication, such as infection or ulceration.
The second keloid patient cohort comprised blood samples collected from 10 patients before intralesional treatment: three of the patients had influenza within 1 month, and the remaining seven had infections in their keloid lesions. Blood samples were also collected from the third keloid patient cohort (n = 106) before intralesional treatment. Patients who had received any kind of prior treatment were excluded from these two cohorts. A total of 61 out of 102 patients whose lesions had a flat appearance after treatment or no obvious remission (patients requested therapy suspension) agreed to donate blood samples immediately before intralesional therapy suspension. These 61 patients were then followed up for 12 months to monitor the status of their lesions. The diagnostic criteria for keloid relapse were as follows: (1) >2.0 mm growth in any axis, as shown in photographs sent to doctors after in-home measurement of the major and minor lesion axes using a ruler and (2) obvious lesion regrowth noticed by the patients. All keloid patients were diagnosed by Dr. H.X. and reassessed by Dr. Y.Z. based on the following criteria: scar-like structures resulting from wound healing and a protruding skin surface that had infiltrated beyond the original damaged area, accompanied by itching, pain, and other discomfort symptoms.
Intralesional treatment
Five hundred milliliters of 2.5% 5-fluorouracil (FU) was added to 1 mL of 1% triamcinolone and then mixed with 4 mL of 2% lidocaine. The total mixture, at a dose of 0.2 mL/cm, was injected intralesionally three times for each keloid at an interval of 4 weeks [3, 34].
The Vancouver Scar Scale (VSS) [35, 36] was used to evaluate the therapeutic effects of keloid intralesional treatment. Before the first treatment and 1 month after each therapy, keloid lesions were assessed using the VSS.
PBMC isolation for flow cytometry
Human peripheral blood mononuclear cells (PBMCs) were isolated using a Ficoll-Hypaque (GE17-1440-02; GE Healthcare) gradient. Cryopreserved PBMCs were rested overnight at 37 °C and 5% CO2 (RPMI-1640 medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin), washed, and stained for flow cytometric analysis.
Fluorochrome-conjugated mAbs directed against CD3 (HIT3a), CD4 (SK3), CD8 (SK1), CD94 (HP-3D9), CD56 (B159), granzyme B (GB11), LAG3 (T47–530), CD161 (DX12), PD-1 (MIH4), T-bet (O4–46), NKG2A (131411), IFN-γ (B27), Ki-67 (B56), and Bcl-2 (Bcl-2/100) were purchased from BD Pharmingen. Anti-FoxP3 (PCH101), anti-granulysin (DH2), and anti-KLRG1 (13F12F2) were purchased from Thermo Fisher. A mAb against homeobox only protein (HOPX) (aa39–67) was purchased from Lifespan. Sequential staining steps were performed using a fixable live/dead reagent, FC blocking controls, surface marker-specific antibodies, fixation/permeabilization reagents, and intranuclear/intracellular marker-specific antibodies; staining was assessed on an LSRFortessa flow cytometer (BD Biosciences) and analyzed using FlowJo 10 software (FlowJo LLC) [37–39].
Immunohistochemistry (IHC)
To detect the tissue expression of the HLA-E and HLA-G proteins in the skin, normal scars, and keloids, IHC was performed with anti–HLA-E (SC-51621) (1:200) (Santa Cruz) and anti–HLA-G (ab283260) (1:200) (Abcam) [40].
Opal multicolor IHC (mIHC) analysis of a tissue microarray
Tissue microarrays were produced based on the pathology reports of each tissue [41]. For multicolor immunohistochemistry (mIHC) staining, multiplex IHC antibodies against CD3, CD4, CD8, CD56, granzyme B, vimentin, NKG2A, and HLA-E were optimized and applied sequentially. In contrast, the spectral library was built based on single-stained slides [42]. Subsequent multiplex immunofluorescence staining and multispectral imaging of the proteins of interest were performed on one tissue microarray slide each using the PANO 7-plex IHC Kit (cat. 0004100100, Panovue, Beijing, China). Briefly, the slide was deparaffinized for 10 min in xylene three times, followed by dehydration in 100% ethanol, 95% ethanol, 85% ethanol, and 75% ethanol for 5 min each. After a 5-min rinse in distilled water, the slide was pretreated with 100 mL of antigen retrieval solution (citric acid solution, pH 6.0/pH 9.0) in a microwave oven (45 s at 100% power, then 15 min on 20% power) and washed with 1× TBST buffer. The slide was then blocked in 10% blocking solution for 10 min and stained with a primary antibody for 1 h at room temperature. After washing the slide with 1× TBST for 3 min twice, the slide was incubated with a polymer HRP–anti-mouse/rabbit IgG secondary antibody for 10 min at RT. The slide was then incubated with a tyramide (TSA)-conjugated fluorophore (TSA Fluorescence Kits, Panovue, Beijing, China) at a 1:100 dilution for 10 min at RT. Finally, the TSA was aspirated, and the slide was washed with 1× TBST for 3 min twice for staining with the next primary antibody. For every additional marker in the multiplex immunofluorescence assay [43], the aforementioned procedure was repeated. After staining for all antigens, nuclei were counterstained with 4-6-diamidino-2-phenylindole (DAPI; Sigma–Aldrich, D9542). Detailed information on the primary antibodies used for mIHC is summarized in Table S1.
To obtain multispectral images, each slide was scanned using the Polaris System (PerkinElmer, Waltham, MA), which captures the fluorescence spectra at 20-nm wavelength intervals from 420 to 720 nm with identical exposure times, and the scanned images were combined to build a single-stack image with high contrast and accuracy.
Single-cell RNA-seq and analysis
scRNA-seq was performed on a chromium platform using single-cell expression 5′ v2 profiling chemistry (10× Genomics) combined with cell hashing [44]. HTO-labeled cells from donors were pooled equally and then washed twice with RPMI-1640 medium immediately before loading on a 10x controller. Complementary DNA amplification and library construction were conducted according to the manufacturer’s protocol, supplemented with additional steps for HTO barcode amplification. Libraries were sequenced to a depth of ~50,000 reads per cell using a NovaSeq S2 (Illumina). Reads were aligned to the GRCh38 reference genome using Cellranger v2.1 (10× Genomics), and hashed cells were demultiplexed using the CITE-seq count tool (https://github.com/Hoohm/CITE-seq-Count).
Single-cell data were preprocessed using the Scanpy package (https://github.com/theislab/scanpy) [45]. First, cells with fewer than 100 nonzero expression genes were filtered out. MT genes, ERCC genes, and genes expressed in less than ten cells were removed. Gene expression values were normalized in two steps. Then, cell-level normalization was performed, during which each gene’s expression level in each cell was divided by the total gene expression level in the cell, multiplied by 10,000, and then transformed to an add-one natural logarithm scale. Furthermore, gene-level normalization was performed, during which the cell-level normalized values for each gene were standardized by subtracting the mean and dividing by the standard deviation across all cells expressing the gene. Then, highly variable genes were selected using the filter_genes_dispersion function of the Scanpy package. Clustering was performed using the top 1000 highly variable genes using the DESC [46] algorithm with the resolution set at 0.2.
Clustering results were visualized using t-distributed stochastic neighbor embedding (t-SNE) projections on the latent embeddings provided by the DESC. To further identify the cell type composition of each cluster, differential expression analysis of each cluster was performed using the Wilcoxon rank-sum test. Genes with a false discovery rate (FDR)–adjusted p value less than 0.05 were selected as differentially expressed genes. To specifically identify genes that distinguished the ICU-SEP and ICU-NoSEP populations, differentially expressed genes were filtered for those with an in-group fraction >0.4 and outgroup fraction <0.6. Consensus nonnegative matrix factorization analysis was performed as previously described [47]. To ensure that no subject-specific or batch-specific modules were analyzed, only gene programs with a mean usage of >50 across all subjects were included for further analysis.
Western blotting
Briefly, cells were lysed in 1× SDS–PAGE loading buffer (50 mM Tris-HCl at pH 6.8, 2% [w/v] SDS, 0.1% [w/v] BPB, and 10% [v/v] glycerol) containing 50 mM β-glycerophosphate, and the lysates were subjected to SDS–PAGE. The separated proteins were transferred to polyvinylidene difluoride membranes for western blotting using enhanced chemiluminescence detection reagents (Merck Millipore, Billerica, MA, USA). Antibodies recognizing HLA-E (ab2216) (1:1000), HLA-G (ab283260) (1:1000), and β-actin (ab8226) (1:2000) were purchased from Abcam. Antibodies against the matrix metalloproteinase proteins (MMPs) MMP1 (DF6325) (1:1000), MMP2 (AF0577) (1:1000), MMP3 (DF6334) (1:1000), MMP7 (AF0218) (1:1000), MMP8 (AF5446) (1:1000), MMP9 (AF5228) (1:1000), and MMP13 (AF5355) (1:1000) were purchased from Affinity. An antibody recognizing the FLAG epitope (F1804) (1:2000) was purchased from Sigma–Aldrich.
Cell transfection
Human dermal fibroblasts were isolated from healthy donors and cultured in 24-well plates. At 75% to 90% confluency, the cells were transiently transfected with an MMP9-3×FLAG expression vector (Genomeditech (Shanghai) Co., Ltd.) using Lipofectamine 3000 reagent (Invitrogen) according to the manufacturer’s instructions. Recombinant MMP9 was evaluated by western blotting at 72 h after transfection.
Sandwich ELISA for the detection of soluble human histocompatibility leukocyte antigen (sHLA)-E and sHLA-G
ELISA kits for detecting human major histocompatibility complex class I (MHCE/HLA-E) and major histocompatibility complex class I G (MHCG/HLA-G) were purchased from Elabscience Biotechnology Co., Ltd. (China) and used in accordance with the manufacturer’s instructions. Briefly, purified anti-human HLA-E or HLA-G antibodies were used to coat the wells of microtiter plates. Standard and serum samples with HLA-E or HLA-G (in duplicate) were added to the wells. The plates were subsequently incubated at 37 °C for 30 min. After three rounds of washing, an HRP-labeled anti-HLA-E or anti-HLA-G antibody was added and incubated for another 15 min at 37 °C. The addition of a sulfuric acid solution terminated the reaction, and the absorbance was measured at 450 nm. The concentration of human HLA-E or HLA-G was then determined by comparing the OD of the samples to the corresponding standard curve. Standard wells were also set up in the ELISA plates, and the concentrations of the HLA-E or HLA-G antigen were 20, 10, 5, 2.5, 1.25, 0.63, and 0.31 ng/mL. Blank wells without any sample or the HRP-conjugate reagent were established separately [48].
Statistical analysis
Data were analyzed using GraphPad Prism 8.0 software (GraphPad Prism Software Inc., San Diego, CA) and are presented as the mean ± SD unless otherwise specified. The correlation between time duration and flow cytometry data was assessed using linear regression analyses. Comparisons between two groups were performed with a 2-tailed Student’s t test if the data were normally distributed or with the Mann–Whitney U unpaired test if the populations were not normally distributed. For three or more groups, one-way ANOVA with the corresponding Tukey test for multiple comparisons was used for data with a normal distribution, and the Kruskal–Wallis test with Dunn’s test was used for multiple comparisons of data with a nonnormal distribution. Statistical significance was set at P < 0.05 [39, 49].
Results
Downregulation ofdisease-specific CTLs and subtypes in keloid scar tissue
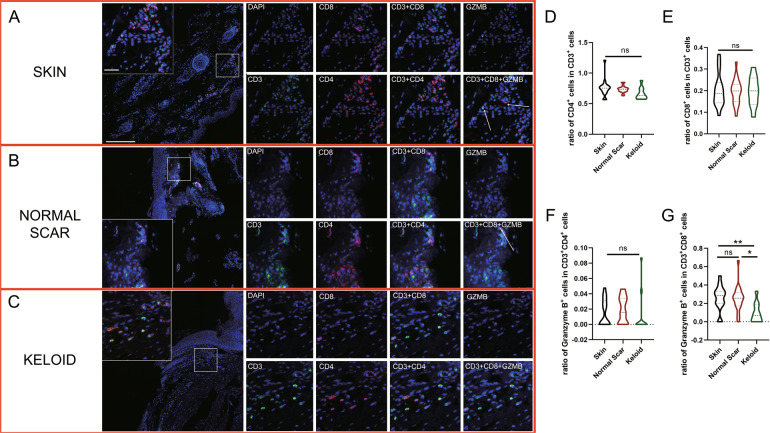
We performed opal mIHC staining to delineate different T cell subtypes in healthy skin, normal scar, and keloid tissues. Notably, the number of CD3+ T cells was much lower in the keloid tissues than in the skin and normal scar tissues (Fig. 1A–C) due to inefficient infiltration by CD3+ T cells. The ratios of CD3+CD4+/CD3+, CD3+CD8+/CD3+, and granzyme B+CD4+/CD4+ T cells exhibited no statistically significant differences among the skin, normal scar, and keloid tissues (Fig. 1D–F). However, compared to the skin and normal scar tissues, the keloid tissues exhibited a significantly lower ratio of granzyme B+CD8+/CD8+ T cells (Fig. 1G), suggesting a diminished CTL level in the scar tissues.
Fig. 1.
Characterization of infiltrating T cells in keloid tissues. Opal multicolor immunohistochemistry (IHC) analysis of CD3, CD8, CD4, and granzyme B revealed the infiltration patterns of different types of T cells in A skin, B normal scar, and C keloid tissues. The ratios of D CD3+CD4+/CD3+, E CD3+CD8+/CD3+, and F Granzyme B+CD4+/CD4+ T cells were not significantly different. G A smaller ratio of Granzyme B+CD8+/CD8+ T cells was observed in keloid tissues. Genomic DNA was stained with DAPI. GZMB: granzyme B; ns: no significant difference; *P < 0.05; **P < 0.01
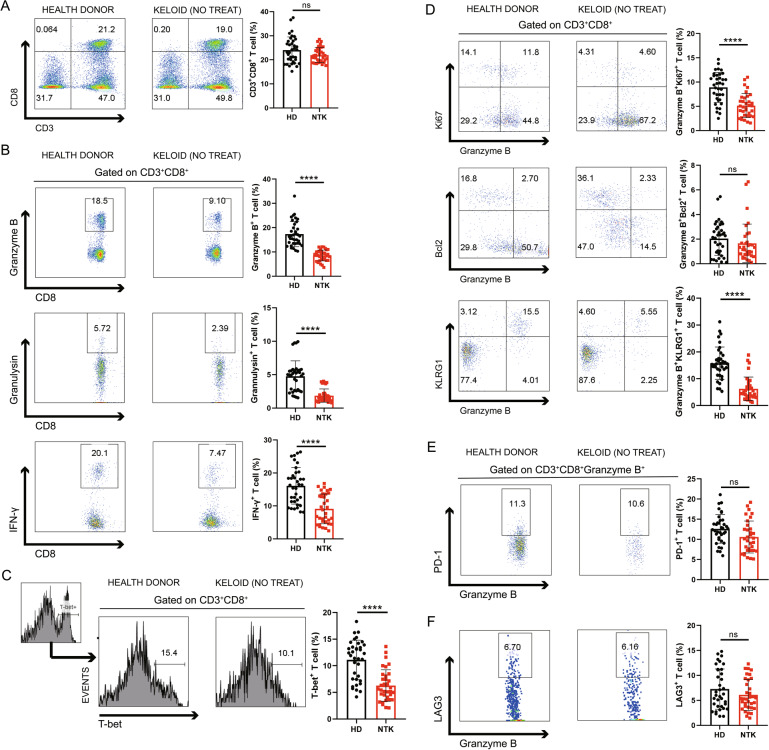
To determine whether the number of CTLs is also altered in patient blood, we assessed the levels of various cytokines and functional markers using PBMCs from 34 keloid patients without any therapeutic treatment and 36 healthy donors. As expected, the percentage of CD3+CD8+ T cells was not significantly different between the healthy donors and keloid patients (Fig. 2A). However, the proportions of granzyme B-, granulysin-, and IFN-γ-producing CTLs were significantly decreased in the keloid patients (P < 0.0001) (Fig. 2B), as corroborated by the downregulated T-bet expression shown in Fig. 2C (P = 0.0002). No significant differences were found for the other cell types assessed, namely, CD3+CD4+ T cells, CD3+CD4+FoxP3-granzyme B+ effector T cells, and CD3+CD4+FoxP3+ regulatory T cells (Fig. S2A–C). Other protein markers of CTLs, such as Ki67 and KLRG1, were decreased in the CD3+CD8+granzyme B+ CTL fraction (P < 0.0001), while Bcl-2 remained virtually unchanged (Fig. 2D).
Fig. 2.
Profiles of peripheral cytotoxic CD8+ T cells in keloid patients. Peripheral blood mononuclear cells (PBMCs) from 36 healthy donors and 34 keloid patients without any treatments or other diseases were analyzed by flow cytometry. A The proportions of CD3+CD8+ T cells showed no significant difference, while the relative frequencies of B Granzyme B-, Granulysin- and IFN-γ-producing CTLs were significantly decreased in the keloid patients. These reductions were associated with C downregulated T-bet expression. D Ki67 and KLRG1 were lower in the CTL fraction, while Bcl-2 remained virtually the same. E, F The expression levels of PD-1 and LAG3 showed no differences. HD: health donor; NTK: nontreated keloid; ns: no significant difference; ****P < 0.001
To clarify whether inhibitory receptors [50] are involved in the CTL reduction, PD-1 and LAG3 expression were analyzed, and their expression displayed no significant changes (Fig. 2E, F). To enhance our understanding of the CTL reduction, 10 keloid patients, among whom three had influenza within 1 month and seven had infections in their keloid lesions, were further evaluated. Granzyme B expression was markedly elevated in these 10 patients (Fig. S2D). Hence, downregulation of CTLs, both in the circulatory system and in local scar tissue, emerged as a keloid-specific cellular signature. However, CTLs in keloid patients could still respond to immune stimuli, similar to those in normal patients.
NKG2A/CD94-induced downregulation of CTLs
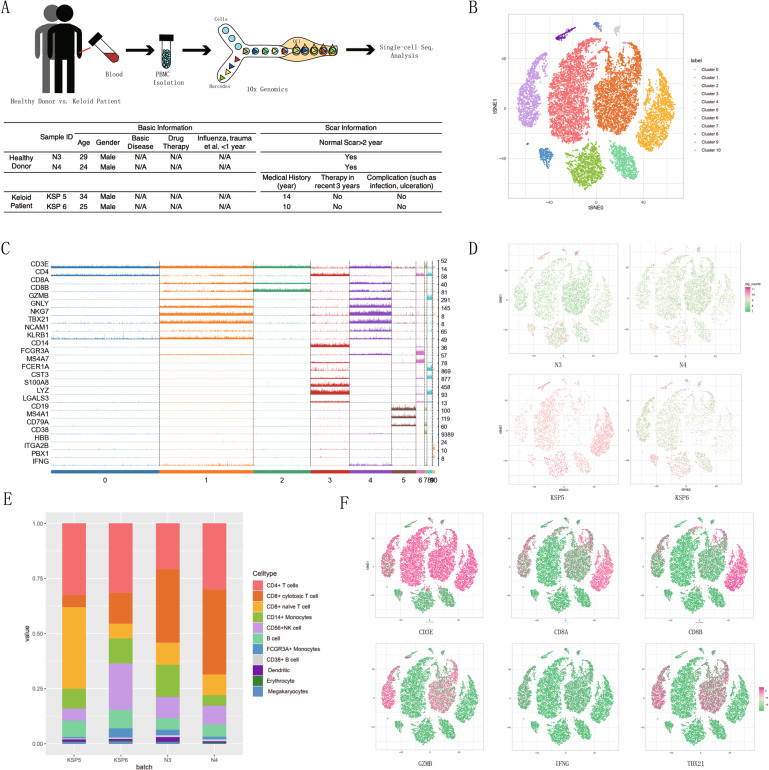
To understand the mechanism underlying the CTL reduction, we performed scRNA-seq analysis of PBMCs from two keloid patients and two normal controls using a 5′ tag RNA-seq approach (Fig. 3A). After filtering and normalization, we performed unsupervised deep embedding clustering [46] and visualized 11 cell clusters with t-SNE (Fig. 3B). Based on lineage marker gene expression (Fig. 3C), we defined the 11 clusters as CD4+ T cells (CD3E and CD4), CTLs (CD3E, CD8A, CD8B, GZMB, GNLY, and TBX21), CD8+ naive T cells (CD3E, CD8A, and CD8B), CD14+ monocytes (CD14 and FCGR3A), CD56+ NK cells (NCAM1, NKG7, and FCGR3A), B cells (CD19, MS4A1, and CD79A), FCGR3A+ monocytes (FCGR3A), CD38+ B cells (CD19, CD38, and CD79A), dendritic cells (FCER1A), erythrocytes (HBB), and megakaryocytes (ITGA2B). Importantly, the number of Cluster 1-CTLs was lower in the keloid patients than in the healthy donors (Fig. 3D, E). The expression levels of lineage markers, namely, CD3E, CD8A, CD8B, GZMB, IFNG, and TBX21, were visualized on the 2D t-SNE space to further verify the Cluster 1-CTL identity (Fig. 3F). Although the scRNA-seq data were obtained from a limited number of samples, the results of the scRNA-seq analysis were generally consistent with the aforementioned findings.
Fig. 3.
Single-cell RNA-sequencing (scRNA-seq) analysis of PBMCs from keloid patients and healthy individuals. A Processing pipeline for blood samples used for scRNA-seq analysis (healthy donors: n = 2; keloid patients: n = 2). B After filtering and normalization, t-distributed stochastic neighbor embedding (t-SNE) analysis revealed 11 cell clusters (Clusters 0–10). C Trackplot showing the expression of lineage marker genes to define the clusters in (E). D t-SNE analysis and E a bar plot showing the distribution patterns of different cell clusters in the 4 samples. F Lineage markers including CD3E, CD8A, CD8B, GZMB, IFNG, and TBX21 are visualized in the 2D t-SNE space to confirm the Cluster 1-CTL identity
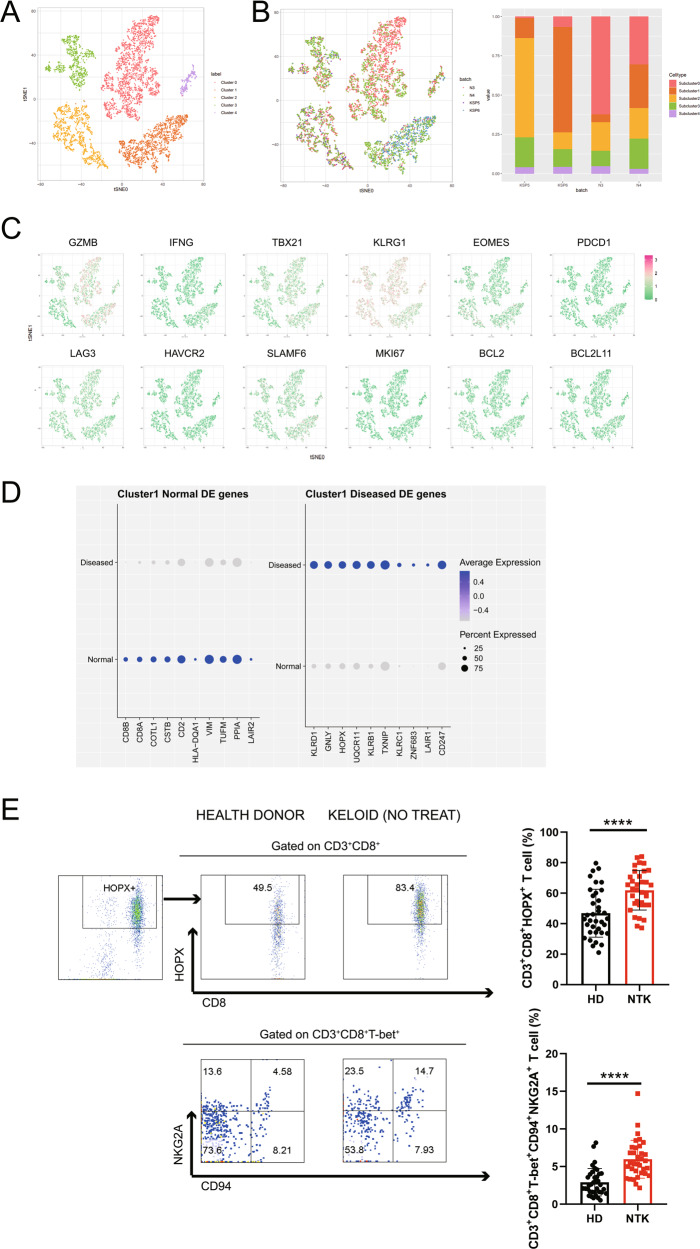
To investigate the mechanisms underlying the CTL reduction, Cluster 1-CTL was split into five subclusters (subclusters 0–4) and visualized using t-SNE (Fig. 4A). The distribution pattern in the t-SNE plot and the bar plot demonstrated a collapsed subcluster 0 in the four keloid samples (Fig. 4B). Subsequently, we examined 12 functionally related genes including GZMB, IFNG, TBX21, KLRG1, EOMES, PDCD1, LAG3, HAVCR2, SLAMF6, MKI67, BCL2, and BCL2L11 (Fig. 4C). Subcluster 0 was strongly enriched with marker genes such as GZMB, TBX21, and KLRG1, which represented decreasing CTLs. Then, we analyzed the top 10 differentially expressed genes (either higher in healthy donors or keloid patients, 20 genes in total; Fig. 4D) between 34 keloid patients and 36 healthy donors. In addition to specific markers for CTLs, such as CD8A, CD8B, CSTB, and GNLY, the numbers of CD3+CD8+HOPX+ T (HOPX encoded by the HOPX gene) and CD3+CD8+T-bet+CD94+NKG2A+ T (CD94 and NKG2A encoded by KLRD1 and KLRC1, respectively) cells were found to be significantly higher in PBMCs from the keloid patients (Fig. 4E). The pattern of mIHC staining for DAPI, CD8, granzyme B, and NKG2A revealed that in keloid tissue, the number of CD8+ granzyme B+ cells was decreased and more CD8+NKG2A+ cells were observed (Fig. S3). After regrouping the keloid samples according to disease duration (Fig. S4A), with 2 years as the cutoff, NKG2A/CD94 complex expression increased as the disease progressed (Fig. S4A, B); a weak correlation was identified by regression analysis (R2 < 0.4; Fig. S4C). In conclusion, the observed CTL downregulation might be attributed to the upregulation of HOPX, particularly NKG2A/CD94, in specific subtypes of T cells in keloid patients.
Fig. 4.
Identification of subtypes in Cluster 1-CTL based on scRNA-seq data. A Cluster 1-CTL could be further clustered into 5 subclusters (Subclusters 0–4) and visualized using t-SNE. B Merged t-SNE and bar plots showing the distributions of derived cell types from the four samples. C t-SNE revealed 12 functionally related genes (GZMB, IFNG, TBX21, KLRG1, EOMES, PDCD1, LAG3, HAVCR2, SLAMF6, MKI67, BCL2, and BCL2L11). D Top 10 differentially expressed genes between health donors and keloid patients (either higher in healthy donors or vice versa, 20 genes in total). E Based on analysis of PBMCs from 36 healthy donors and 34 keloid patients without any treatment or disease, the fractions of CD3+CD8+HOPX+ T cells (HOPX encoded by the HOPX gene) and CD3+CD8+T-bet+CD94+NKG2A+ T cells (CD94 and NKG2A encoded by KLRD1 and KLRC1, respectively) were significantly elevated in the keloid patients. HD: healthy donor; NTK: nontreated keloid; ****P < 0.001
Restoration of CTLs in keloid patients by intralesional therapy
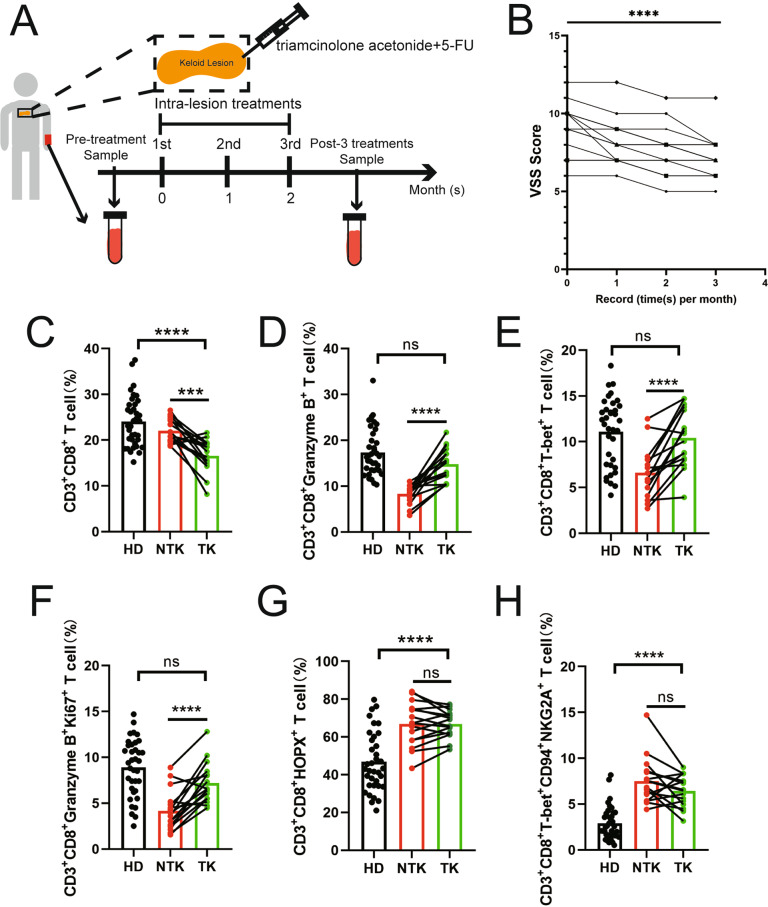
To further investigate how the NKG2A/CD94 complex is regulated, we followed 16 keloid patients without any prior treatments or complications after treatment to evaluate the profiles of CTLs before and after three rounds of combined intralesional therapy with triamcinolone+5-FU (Fig. 5A). The treatment was highly effective, as assessed based on VSS scores recorded monthly pre- and post-intralesional therapy administration (Fig. 5B). As expected, the number of CD3+CD8+ T cells was significantly decreased post-treatment due to triamcinolone inhibition (Fig. 5C). Interestingly, the proportion of CTLs was restored to normal levels (Fig. 5D), as the fractions of both CD3+CD8+T-bet+ T cells and Ki67+ CTLs recovered from the pretreatment levels (Fig. 5E, F). The upregulation of CTLs as a positive response to intralesional injection of triamcinolone and 5-FU supported the critical role for CTLs in keloid pathogenesis. However, the expression level of HOPX, particularly NKG2A/CD94, remained unchanged post-treatment (Fig. 5G, H). Thus, the mechanism underlying CTL regulation remains unclear.
Fig. 5.
Effects of triamcinolone+5-fluorouracil intralesional therapy on peripheral cytotoxic CD8+ T cells. A We analyzed PBMCs isolated from 16 keloid patients without any treatment or disease to profile the peripheral cytotoxic CD8+ T cell population pre- and post-treatment with 3 rounds of triamcinolone+5-FU intralesional treatment. B Vancouver Scar Scale (VSS) scores measured pre- and post-intralesional therapy with triamcinolone+5-FU. C Compared to 36 healthy donors, keloid patients showed significantly decreased numbers of CD3+CD8+ T cells after intralesional therapy. D The proportion of CTLs returned to a normal level, as demonstrated by the recovered fractions of both E CD3+CD8+T-bet+ T cells and F Ki67+ CTLs. G The HOPX and H NKG2A/CD94 expression levels remained unchanged before and after treatment. HD: healthy donor; NTK: nontreated keloid; TK: treated keloid; ns: no significant difference; ***P < 0.005; ****P < 0.001
Given that the NKG2A/CD94 complex is typically expressed on the surface of NK cells, we analyzed CD3−CD56+CD94+ NK cells and found that the number of this particular subtype was increased in keloid patients, which could not be reversed by intralesional injection therapy (Fig. S5A–C). In addition, the expression levels of the NKG2A/CD94 complex in NK cells (Fig. S5D) in nontreated keloid patients did not differ significantly from those in treated keloid patients (Fig. S5E, F). These results imply that the exact role of NKG2A/CD94 in the CTL reduction and restoration observed remains unknown.
Signaling transduction of the NKG2A/CD94-HLA-E axis for CTL regulation
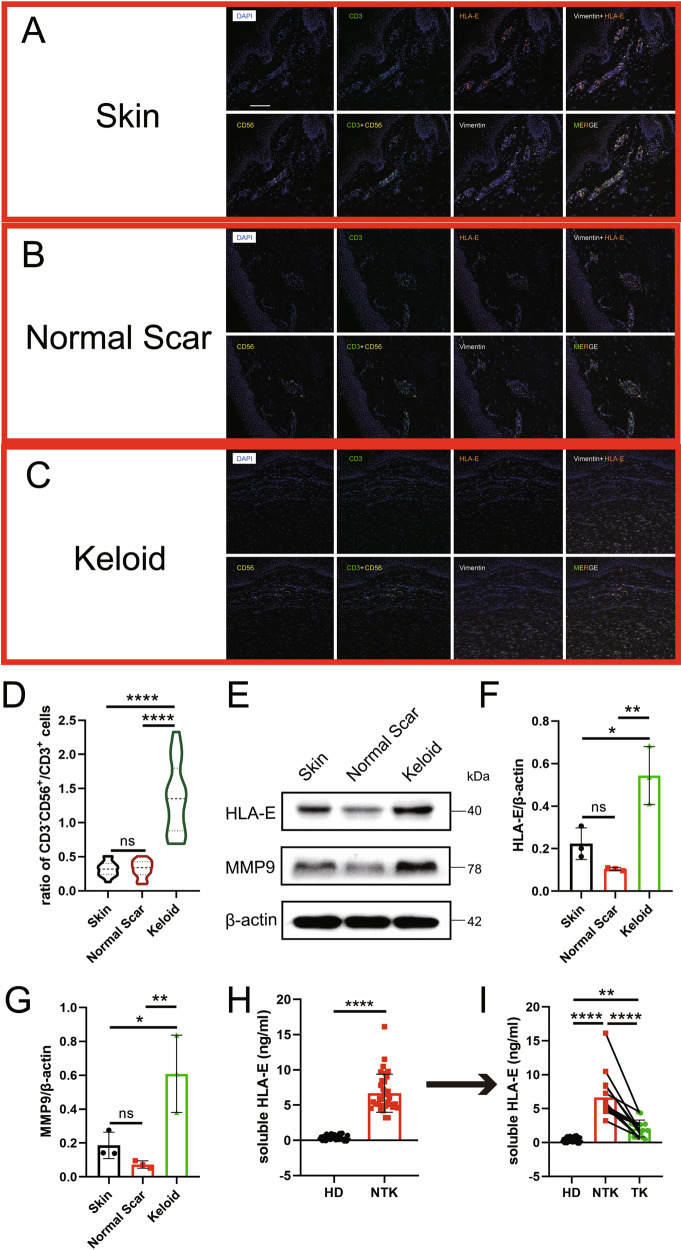
The NKG2A/CD94 complex binds specifically to the nonclassical MHC I molecule HLA-E in humans [51]. Therefore, opal mIHC was performed on skin, normal scar, and keloid tissue specimens to evaluate HLA-E expression in keloid patients. Consistent with the previous findings, we observed a decreased number of CD3+ T cells and an increased number of CD3−CD56+ NK cells in the keloid tissue compared to that in the skin and normal scar tissues (Fig. 6A–C), resulting in a significantly higher ratio of CD3−CD56+ NK cells/CD3+ T cells (Fig. 6D). Intriguingly, HLA-E was expressed mainly on vimentin+ fibroblasts in normal skin and scar tissues, but fibroblast-enriched keloid tissue did not show any HLA-E signal (Fig. 6A–C). However, higher HLA-E expression was observed in keloid tissues by western blot analysis (Fig. 6E, F). In contrast, HLA-G expression was significantly upregulated in keloid fibroblasts, as revealed by immunofluorescence staining and western blotting (Figs. S6 and S7). Since HLA-E and HLA-G can exist as soluble moieties (soluble HLA-E [sHLA-E] and soluble HLA-G [sHLA-G]) in biological fluids [52], we further investigated the amounts of sHLA-E and sHLA-G in serum samples and observed a dramatically increased level of sHLA-E in keloid patients (36 healthy donors and 34 keloid patients, Fig. 6H, P < 0.0001) but no significant changes in sHLA-G (13 healthy donors and 12 keloid patients, Fig. S9). Concomitantly, we examined the expression of mediator MMPs [53] and found that MMP1, MMP3, MMP8, MMP9, and MMP1,3 but not MMP2 or MMP7 were upregulated in keloid tissues (Figs. 6E, G, S7). Furthermore, a slight increase in HLA-E expression was observed due to ectopic expression of MMP9, a well-known mediator of HLA-E solubilization in human dermal fibroblasts (Fig. S8) [54]. Hence, we suggest that sHLA-E is the major form secreted by local keloid fibroblasts and then released into the circulatory system.
Fig. 6.
Abnormal expression of human leucocyte antigen-E (HLA-E) is involved in keloids. A–C By using opal multicolor immunohistochemistry (IHC), nuclei (DAPI), CD56, CD3, HLA-E, and vimentin were stained to identify NK cells and HLA-E expression in A skin, B normal scar, and C keloid tissues. The ratio of D CD3−CD56+/CD3 cells was significantly higher in keloids. E Western blot analysis (MMP9 and HLA-E) of fibroblasts from skin, normal scar and keloid tissues, and the expression was analyzed (F, G). H Serum samples (36 healthy donors vs. 34 keloid patients without any treatments or diseases) showed increased soluble HLA-E (sHLA-E) levels in the keloid patients. I The serum samples of 16 keloid patients followed over time showed that sHLA-E was significantly decreased after 3 rounds of triamcinolone+5-FU intralesional therapy. HD: healthy donor; NTK: nontreated keloid; TK: treated keloid; ns: no significant difference; **P < 0.01; ****P < 0.001
After three intralesional injections, sHLA-E was decreased to a level much lower than the pretreatment state (Fig. 6I), although this level was still higher than that of healthy donors. We subsequently expanded the study cohort to 104 keloid patients without prior treatment, 512 healthy donors, and 100 patients with an interfering disease. The level of serum sHLA-E was 6.34 ± 2.58 ng/mL in the keloid patients, significantly higher than the levels of 1.26 ± 1.00 ng/mL in the healthy donors and 2.05 ± 2.41 ng/mL in the patients with an interfering disease (Tables 1, 2). When the cutoff value was set at 3 ng/mL for use of sHLA-E as a marker to distinguish among the keloid patient, healthy control, and interference groups, the sensitivity was 83.69%, and the specificity was 93.36%, 94.73%, and 79.00% for keloid patients vs. healthy donors and interference patients, keloid patients vs. healthy donors, and keloid patients vs. interference patients, respectively (Table 1). Furthermore, we followed 61 keloid patients who received treatment >3 times in 12 months (triamcinolone combined with 5-FU). After therapy was suspended, 13 patients (21.31%) had keloid recurrence within 6 months. Their serum level of sHLA-E was 4.06 ± 1.70 ng/mL after the last therapy session, which was significantly higher than that of the other patients who did not experience recurrence within 6 months (1.73 ± 1.07 ng/mL, P < 0.0002; Table 1). Interestingly, recurrence occurred preferentially in female patients (62.50% vs. 30.77%; P = 0.02), indicating that sex differences may play a role in treatment response. Additionally, sHLA-E was capable of distinguishing keloids from hypertrophic scars (1.37 ± 1.38; P < 0.0001), malignant dermatofibrosarcoma protuberans (1.24 ± 0.59; P < 0.0001), basal cell carcinoma (0.50 ± 0.46; P < 0.0001), and a host of other interfering diseases (Table 2). These data suggested that the level of sHLA-E was inversely correlated with the numbers of CTLs in keloid patients in both the peripheral blood and keloid lesions. As the pathogenic process or remission phase proceeded, the level of sHLA-E was either upregulated or downregulated in lockstep with CTL downregulation or restoration.
Table 1.
Soluble human leucocyte antigen-E level in health donor, cases of interference and keloid patients
| Not Keloid | Keloid (No treat) (n = 104) | ||
|---|---|---|---|
| Health Donor (n = 512) | Cases of Interference (n = 100) | ||
| Age (years, mean ± SD) | 42.64 ± 13.87 (21.00–89.00) | 59.63 ± 18.00 (23.00–95.00) | 33.67 ± 12.44 (15.00–73.00) |
| Male sex (No.) | 42.77% (219) | 62.00% (62) | 40.38% (42) |
| Serum sHLA-E (ng/ml, mean ± SD) | 1.26 ± 1.00 (0.02–4.83) | 2.05 ± 2.41 (0.06–14.09) | 6.34 ± 2.58 (2.63–16.14) |
| Serum sHLA-E > 3 (ng/ml) | Not Keloid | Keloid | |
| Health Donor | Cases of Interference | ||
| True | 27 | 21 | 86 |
| False | 485 | 79 | 18 |
| Not Keloid vs Keloid (No treat) | Health Donor vs Keloid (No treat) | Cases of Interference vs Keloid (No treat) | |
| Sensitivity | 83.69% | n.a. | n.a. |
| Specificity | 92.16% | 94.73% | 79.00% |
| Keloid (Treated) (n = 61) | p value | ||
| Recurrence post-intralesional therapy suspended <6 months? | Yes (n = 13, 21.31%) | No (n = 48, 78.69%) | |
| Age (years, mean ± SD) | 32.92 ± 13.01 (20.00–62.00) | 37.13 ± 11.90 (18.00–68.00) | 0.27 |
| Male sex (No.) | 30.77% (4) | 62.50% (30) | 0.02 |
| Serum sHLA-E (ng/ml, mean ± SD)post last therapy | 4.06 ± 1.70 (1.81–6.97) | 1.73 ± 1.07 (0.46–4.51) | <0.0001 |
Discussion
Our work identified the downregulation of CTLs in the target tissue and blood circulation as a cellular signature of keloid patients. This signature raises the novel understanding that keloids are a systemic disease. The abnormal tissue microenvironment is likely driven by systemic dysfunction. Therefore, peripheral blood samples can potentially be used to comprehensively survey the host immune landscape. This study showed that CTLs and sHLA-E, in particular, can be used as biomarkers for the diagnosis and prediction of keloid risk. Measurement of serum sHLA-E levels can also help distinguish between keloid and early-stage hypertrophic scars, as well as certain types of carcinomas. For instance, the level of sHLA-E in malignant dermatofibrosarcoma protuberans, which appears to resemble keloids and often leads to misdiagnosis [55], is significantly lower than that in keloids. In addition, the sHLA-E level is correlated with keloid recurrence after intralesional therapy. Based on the post-treatment level of serum sHLA-E, keloids can be classified into N-type, in which the level of sHLA-E returns completely to normal, and H-type, in which the sHLA-E level remains stubbornly high. Hence, physicians can classify keloid patients as N- or H-type by measuring the serum level of sHLA-E to guide intralesional treatment. Furthermore, the sHLA-E signature can guide the use of a systemic therapeutic approach to improve disease prognosis when combined with topical therapy. For instance, a systemic approach can be used in future clinical trials for keloid treatment using monalizumab, a monoclonal antibody that has been reported to be a novel checkpoint inhibitor targeting the NKG2A/CD94 complex in cancer immunotherapy [56].
HOPX, a transcription cofactor, is induced by T-bet and upregulated in terminally differentiated effector/memory Th1 cells [57]. In addition, induced regulatory T cells (Tregs) require HOPX expression to suppress peripheral T cell function [58]. Therefore, the higher percentage of CD3+CD8+HOPX+ T cells in keloid patients than in healthy donors indicates that HOPX may drive CTLs to differentiate into abnormal lineages. NKG2A forms a heterodimer with CD94, known as the NKG2A/CD94 receptor complex, and is expressed on the surface of CD8+ T cells [59]. In addition, the NKG2A/CD94 complex acts as an immune checkpoint capable of suppressing immunoreactive NK and CD8+ T cell activity [59]. Van Montfoort et al. demonstrated that antibody blockade of NKG2A/CD94 on CD8+ T cells improved survival in certain solid tumor models [60]. The ligand of NKG2A/CD94 is HLA-E, which can be upregulated in cancer cells to transduce inhibitory signals to immunoreactive T cells, leading to poor outcomes [61]. In keloids, we found that HLA-E was secreted into the tissue microenvironment and circulation by enriched fibroblasts rather than remaining membrane-bound. We speculated that sHLA-E binds to NKG2A/CD94 to inhibit the functions of NK cells and CTLs and decrease the number of immunoreactive CTLs. Circulating sHLA-E molecules act as a reservoir of pathogenic stimuli, causing keloid regeneration after surgical resection.
Monthly intralesional triamcinolone injection is considered the first-line treatment for keloid lesions, with a response rate of 50–100% [8, 62]. Combination therapy with 5-FU results in a 92% reduction in lesion size compared to the 73% reduction in lesions treated with triamcinolone alone [63]. We observed that effective intralesional therapy (triamcinolone+5-FU) was mediated, at least in part, by restoring CTLs. Evidence suggests that triamcinolone can inhibit CD3+CD8+ T cells, which are abundant in keloid scars [64]. The overlapping cytolytic capabilities of CTLs and NK cells warrant closely regulated collaboration, but they also antagonize each other under certain conditions [65]. One important notion is that the upregulated NK cells in keloids might mediate CTL elimination [66]. Increased HLA-E expression has also been observed in the development of multiple sclerosis, an autoimmune disease [67]. HLA-E and HLA-G belong to the HLA-1b family and play roles in immune cell modulation through membrane-bound or soluble isoforms [53]. Our data indicate that HLA-E and HLA-G expression is enhanced in keloid tissues. HLA-E exists predominantly as sHLA-E, while HLA-G is found as the membrane-bound isoform due to the relatively low expression of MMP2, which mediates the membrane shedding of HLA-G [68], and enhanced HLA-E secretion is mediated by increased expression of MMP9 [54]. This may explain the significant CTL reduction and high recurrence rate, approximately 9–50%, after post-intralesional therapy [69]. Hence, selective and specific blockade of sHLA-E might be an effective therapy for keloids. However, HLA-E-restricted CD8+ T cells can also kill infected fibroblasts and may play a role complementary to that of conventional CTLs [70–72] in addition to transducing inhibitory signals via NKG2A/CD94 on immunoreactive T cells [61]; hence, sHLA-E might exert immunoregulatory or immunostimulatory activities under different circumstances. Thus, additional studies are required to address this question and gain further insight into keloid pathogenesis.
Keloid risk is difficult to predict, and a long time after surgery is often needed to recognize that a patient is susceptible to scar formation. Genome-wide association studies have identified several single-nucleotide polymorphisms associated with keloid risk [16, 18]. However, these genetic variants confer an increased risk to only a very small proportion of the population [18].
In this study, we provided, for the first time, a reliable immunoassay using sHLA-E in the serum as a single biomarker to diagnose patients with keloids and monitor keloid recurrence. In a hospital-based cohort, the single-target test showed robust performance characteristics with excellent accuracy; moreover, interfering diseases did not significantly affect the test outcome. The assay could also predict whether recurrence occurred after intraepithelial injections, assessing treatment efficacy. However, we could not validate test performance in an independent prospective cohort. This limitation remains to be addressed in the future by using powerful multicenter cross-validation studies in real-world clinical settings.
In conclusion, our findings suggest that sHLA-E may effectively serve as a diagnostic marker for assessing the risk of keloid formation and a prognostic marker for the clinical outcomes of intralesional treatment.
Supplementary information
Acknowledgements
The authors wish to acknowledge Prof. Min Cui from Huazhong Agricultural University (China) for her insightful assistance and Prof. Moubin Lin from Yangpu Hospital, Tongji University School of Medicine (China) for providing technical assistance. We would like to acknowledge the patients for their collaboration. This study was funded by the National Natural Science Foundation of China (Nos. 81772098, 81672247, and 82002064), Shanghai Sailing Program (No. 20YF1422700) and Shanghai Municipal Education Commission Gaofeng Clinical Medicine Grant Support (No. 20152227). The funders of the study had no role in the design or conduct of the study; the collection, management, analysis, and interpretation of the data; the preparation, review, or approval of the manuscript; or the decision to submit the manuscript for publication. We wish to thank the editors for providing editorial assistance.
Author contributions
HX, BL, YW, and YZ designed the study. HX, ZZ, and JH performed experiments and collected, analyzed, and interpreted data. JH and JS collected data and contributed to data interpretation. HX, HZ, XW, and BL wrote the manuscript. HX, ZZ, JH, and JS contributed equally to this work. All authors critically revised the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Heng Xu, Zhu Zhu, Jian Hu, Jiawei Sun.
Contributor Information
Heng Xu, Email: xuh1990@gmail.com.
Bin Li, Email: binli@shsmu.edu.cn.
Yixin Zhang, Email: zhangyixin6688@hotmail.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41423-021-00834-1.
References
- 1.Alhady SM, Sivanantharajah K. Keloids in various races. A review of 175 cases. Plast Reconstr Surg. 1969;44:564–6. doi: 10.1097/00006534-196912000-00006. [DOI] [PubMed] [Google Scholar]
- 2.Gauglitz GG, Korting HC, Pavicic T, Ruzicka T, Jeschke MG. Hypertrophic scarring and keloids: pathomechanisms and current and emerging treatment strategies. Mol Med. 2011;17:113–25. doi: 10.2119/molmed.2009.00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolfram D, Tzankov A, Pülzl P, Piza-Katzer H. Hypertrophic scars and keloids-a review of their pathophysiology, risk factors, and therapeutic management. Dermatol Surg. 2009;35:171–81. doi: 10.1111/j.1524-4725.2008.34406.x. [DOI] [PubMed] [Google Scholar]
- 4.van den Broek LJ, Limandjaja GC, Niessen FB, Gibbs S. Human hypertrophic and keloid scar models: principles, limitations and future challenges from a tissue engineering perspective. Exp Dermatol. 2014;23:382–6. doi: 10.1111/exd.12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chike-Obi CJ, Cole PD, Brissett AE. Keloids: pathogenesis, clinical features, and management. Semin Plast Surg. 2009;23:178–84. doi: 10.1055/s-0029-1224797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bijlard E, Kouwenberg C, Timman R, Hovius S, Busschbach J, Mureau M. Burden of keloid disease: a cross-sectional health-related quality of life assessment. Acta Derm Venereol. 2017;97:225–9. doi: 10.2340/00015555-2498. [DOI] [PubMed] [Google Scholar]
- 7.Lee SS, Yosipovitch G, Chan YH, Goh CL. Pruritus, pain, and small nerve fiber function in keloids: a controlled study. J Am Acad Dermatol. 2004;51:1002–6. doi: 10.1016/j.jaad.2004.07.054. [DOI] [PubMed] [Google Scholar]
- 8.Trace AP, Enos CW, Mantel A, Harvey VM. Keloids and hypertrophic scars: a spectrum of clinical challenges. Am J Clin Dermatol. 2016;17:201–23. doi: 10.1007/s40257-016-0175-7. [DOI] [PubMed] [Google Scholar]
- 9.Gulamhuseinwala N, Mackey S, Meagher P, Powell B. Should excised keloid scars be sent for routine histologic analysis? Ann Plast Surg. 2008;60:186–7. doi: 10.1097/SAP.0b013e318056d6cc. [DOI] [PubMed] [Google Scholar]
- 10.Andrews JP, Marttala J, Macarak E, Rosenbloom J, Uitto J. Keloids: the paradigm of skin fibrosis—pathomechanisms and treatment. Matrix Biol. 2016;51:37–46. doi: 10.1016/j.matbio.2016.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Limandjaja GC, Niessen FB, Scheper RJ, Gibbs S. The keloid disorder: heterogeneity, histopathology, mechanisms and models. Front Cell Dev Biol. 2020;8:360. doi: 10.3389/fcell.2020.00360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burd A, Huang L. Hypertrophic response and keloid diathesis: two very different forms of scar. Plast Reconstr Surg. 2005;116:150e–157e. doi: 10.1097/01.prs.0000191977.51206.43. [DOI] [PubMed] [Google Scholar]
- 13.Robles DT, Berg D. Abnormal wound healing: keloids. Clin Dermatol. 2007;25:26–32. doi: 10.1016/j.clindermatol.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 14.Seifert O, Mrowietz U. Keloid scarring: bench and bedside. Arch Dermatol Res. 2009;301:259–72. doi: 10.1007/s00403-009-0952-8. [DOI] [PubMed] [Google Scholar]
- 15.Marttala J, Andrews JP, Rosenbloom J, Uitto J. Keloids: animal models and pathologic equivalents to study tissue fibrosis. Matrix Biol. 2016;51:47–54. doi: 10.1016/j.matbio.2016.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakashima M, Chung S, Takahashi A, Kamatani N, Kawaguchi T, Tsunoda T, et al. A genome-wide association study identifies four susceptibility loci for keloid in the Japanese population. Nat Genet. 2010;42:768–71. doi: 10.1038/ng.645. [DOI] [PubMed] [Google Scholar]
- 17.Shih B, Bayat A. Genetics of keloid scarring. Arch Dermatol Res. 2010;302:319–39. doi: 10.1007/s00403-009-1014-y. [DOI] [PubMed] [Google Scholar]
- 18.Zhu F, Wu B, Li P, Wang J, Tang H, Liu Y, et al. Association study confirmed susceptibility loci with keloid in the Chinese Han population. PLoS One. 2013;8:e62377. doi: 10.1371/journal.pone.0062377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glass DA., 2nd Current understanding of the genetic causes of keloid formation. J Investig Dermatol Symp Proc. 2017;18:S50–S53. doi: 10.1016/j.jisp.2016.10.024. [DOI] [PubMed] [Google Scholar]
- 20.Yan L, Lü XY, Wang CM, Cao R, Yin YH, Jia CS, et al. [Association between p53 gene codon 72 polymorphism and keloid in Chinese population] Zhonghua Zheng Xing Wai Ke Za Zhi. 2007;23:428–30. [PubMed] [Google Scholar]
- 21.Marneros AG, Norris JE, Watanabe S, Reichenberger E, Olsen BR. Genome scans provide evidence for keloid susceptibility loci on chromosomes 2q23 and 7p11. J Invest Dermatol. 2004;122:1126–32. doi: 10.1111/j.0022-202X.2004.22327.x. [DOI] [PubMed] [Google Scholar]
- 22.Brown JJ, Ollier WE, Thomson W, Bayat A. Positive association of HLA-DRB1*15 with keloid disease in Caucasians. Int J Immunogenet. 2008;35:303–7. doi: 10.1111/j.1744-313X.2008.00780.x. [DOI] [PubMed] [Google Scholar]
- 23.Lu WS, Wang JF, Yang S, Xiao FL, Quan C, Cheng H, et al. Association of HLA-DQA1 and DQB1 alleles with keloids in Chinese Hans. J Dermatol Sci. 2008;52:108–17. doi: 10.1016/j.jdermsci.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 24.Driskell RR, Lichtenberger BM, Hoste E, Kretzschmar K, Simons BD, Charalambous M, et al. Distinct fibroblast lineages determine dermal architecture in skin development and repair. Nature. 2013;504:277–81. doi: 10.1038/nature12783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rinkevich Y, Walmsley GG, Hu MS, Maan ZN, Newman AM, Drukker M, et al. Skin fibrosis. Identification and isolation of a dermal lineage with intrinsic fibrogenic potential. Science. 2015;348:aaa2151. doi: 10.1126/science.aaa2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Philippeos C, Telerman SB, Oules B, Pisco AO, Shaw TJ, Elgueta R, et al. Spatial and Single-Cell Transcriptional Profiling Identifies Functionally Distinct Human Dermal Fibroblast Subpopulations. J Invest Dermatol. 2018;138:811–25. doi: 10.1016/j.jid.2018.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jumper N, Hodgkinson T, Paus R, Bayat A, Tangrea MA. Site-specific gene expression profiling as a novel strategy for unravelling keloid disease pathobiology. PLoS One. 2017;12:e0172955. doi: 10.1371/journal.pone.0172955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boyce DE, Ciampolini J, Ruge F, Murison MS, Harding KG. Inflammatory-cell subpopulations in keloid scars. Br J Plast Surg. 2001;54:511–6. doi: 10.1054/bjps.2001.3638. [DOI] [PubMed] [Google Scholar]
- 29.Bagabir R, Byers RJ, Chaudhry IH, Müller W, Paus R, Bayat A. Site-specific immunophenotyping of keloid disease demonstrates immune upregulation and the presence of lymphoid aggregates. Br J Dermatol. 2012;167:1053–66. doi: 10.1111/j.1365-2133.2012.11190.x. [DOI] [PubMed] [Google Scholar]
- 30.Jin Q, Gui L, Niu F, Yu B, Lauda N, Liu J, et al. Macrophages in keloid are potent at promoting the differentiation and function of regulatory T cells. Exp Cell Res. 2018;362:472–6. doi: 10.1016/j.yexcr.2017.12.011. [DOI] [PubMed] [Google Scholar]
- 31.Chen W, Fu X, Sun X, Sun T, Zhao Z, Sheng Z. Analysis of differentially expressed genes in keloids and normal skin with cDNA microarray. J Surg Res. 2003;113:208–16. doi: 10.1016/s0022-4804(03)00188-4. [DOI] [PubMed] [Google Scholar]
- 32.Dong X, Mao S, Wen H. Upregulation of proinflammatory genes in skin lesions may be the cause of keloid formation (Review) Biomed Rep. 2013;1:833–6. doi: 10.3892/br.2013.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Messadi DV, Le A, Berg S, Huang G, Zhuang W, Bertolami CN. Effect of TGF-beta 1 on PDGF receptors expression in human scar fibroblasts. Front Biosci. 1998;3:a16–22. doi: 10.2741/A246. [DOI] [PubMed] [Google Scholar]
- 34.Deng K, Xiao H, Liu X, Ogawa R, Xu X, Liu Y, et al. Strontium-90 brachytherapy following intralesional triamcinolone and 5-fluorouracil injections for keloid treatment: A randomized controlled trial. PLoS ONE. 2021;16:e0248799. doi: 10.1371/journal.pone.0248799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee YI, Kim J, Yang CE, Hong JW, Lee WJ, Lee JH. Combined therapeutic strategies for keloid treatment. Dermatol Surg. 2019;45:802–10. doi: 10.1097/DSS.0000000000001695. [DOI] [PubMed] [Google Scholar]
- 36.Hedayatyanfard K, Ziai SA, Niazi F, Habibi I, Habibi B, Moravvej H. Losartan ointment relieves hypertrophic scars and keloid: a pilot study. Wound Repair Regen. 2018;26:340–3. doi: 10.1111/wrr.12648. [DOI] [PubMed] [Google Scholar]
- 37.Akimova T, Zhang T, Negorev D, Singhal S, Stadanlick J, Rao A, et al. Human lung tumor FOXP3+ Tregs upregulate four “Treg-locking” transcription factors. JCI Insight. 2017;2:e94075. [DOI] [PMC free article] [PubMed]
- 38.Klicznik MM, Morawski PA, Hollbacher B, Varkhande SR, Motley SJ, Kuri-Cervantes L, et al. Human CD4(+)CD103(+) cutaneous resident memory T cells are found in the circulation of healthy individuals. Sci Immunol. 2019;4:eaav8995 [DOI] [PMC free article] [PubMed]
- 39.Xu H, Dahiya S, Wang L, Akimova T, Han R, Zhang T, et al. Utility of IL-2 complexes in promoting the survival of murine orthotopic forelimb vascularized composite allografts. Transplantation. 2018;102:70–8. doi: 10.1097/TP.0000000000001852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tao R, de Zoeten EF, Ozkaynak E, Chen C, Wang L, Porrett PM, et al. Deacetylase inhibition promotes the generation and function of regulatory T cells. Nat Med. 2007;13:1299–307. doi: 10.1038/nm1652. [DOI] [PubMed] [Google Scholar]
- 41.Fang M, Li Y, Huang K, Qi S, Zhang J, Zgodzinski W, et al. IL33 promotes colon cancer cell stemness via JNK Activation and macrophage recruitment. Cancer Res. 2017;77:2735–45. doi: 10.1158/0008-5472.CAN-16-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ying L, Yan F, Meng Q, Yu L, Yuan X, Gantier MP, et al. PD-L1 expression is a prognostic factor in subgroups of gastric cancer patients stratified according to their levels of CD8 and FOXP3 immune markers. Oncoimmunology. 2018;7:e1433520. doi: 10.1080/2162402X.2018.1433520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McGuire HM, Shklovskaya E, Edwards J, Trevillian PR, McCaughan GW, Bertolino P, et al. Anti-PD-1-induced high-grade hepatitis associated with corticosteroid-resistant T cells: a case report. Cancer Immunol Immunother. 2018;67:563–73. doi: 10.1007/s00262-017-2107-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stoeckius M, Zheng S, Houck-Loomis B, Hao S, Yeung BZ, Mauck WM, et al. Cell hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol. 2018;19:224. doi: 10.1186/s13059-018-1603-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19:15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li X, Wang K, Lyu Y, Pan H, Zhang J, Stambolian D, et al. Deep learning enables accurate clustering with batch effect removal in single-cell RNA-seq analysis. Nat Commun. 2020;11:2338. doi: 10.1038/s41467-020-15851-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kotliar D, Veres A, Nagy MA, Tabrizi S, Hodis E, Melton DA, et al. Identifying gene expression programs of cell-type identity and cellular activity with single-cell RNA-Seq. Elife. 2019;8:e43803. [DOI] [PMC free article] [PubMed]
- 48.Xu YP, Wieten L, Wang SX, Cai Y, Olieslagers T, Zhang L, et al. Clinical significance of HLA-E genotype and surface/soluble expression levels between healthy individuals and patients with acute leukemia. Leuk Lymphoma. 2019;60:208–15. doi: 10.1080/10428194.2018.1474521. [DOI] [PubMed] [Google Scholar]
- 49.Llorens F, Hermann P, Villar-Piqué A, Diaz-Lucena D, Nägga K, Hansson O, et al. Cerebrospinal fluid lipocalin 2 as a novel biomarker for the differential diagnosis of vascular dementia. Nat Commun. 2020;11:619. doi: 10.1038/s41467-020-14373-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol. 2015;15:486–99. doi: 10.1038/nri3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Braud VM, Allan DS, O'Callaghan CA, Söderström K, D'Andrea A, Ogg GS, et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. 1998;391:795–9. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- 52.Goel R, Kabeerdoss J, Mohan H, Danda S, Jayaseelan V, Kumar TS, et al. Soluble-HLA-E: a follow up biomarker in Takayasu arteritis, independent of HLA-E genotype. Int J Rheum Dis. 2018;21:532–40. doi: 10.1111/1756-185X.13027. [DOI] [PubMed] [Google Scholar]
- 53.Pistoia V, Morandi F, Wang X, Ferrone S. Soluble HLA-G: are they clinically relevant? Semin Cancer Biol. 2007;17:469–79. doi: 10.1016/j.semcancer.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shwetank S, Date OS, Kim KS, Manjunath R, Lee YM. Infection of human endothelial cells by Japanese encephalitis virus: increased expression and release of soluble HLA-E. PLoS One. 2013;8:e79197. doi: 10.1371/journal.pone.0079197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ogawa R, Akaishi S, Hyakusoku H. Differential and exclusive diagnosis of diseases that resemble keloids and hypertrophic scars. Ann Plast Surg. 2009;62:660–4. doi: 10.1097/SAP.0b013e31817e9d67. [DOI] [PubMed] [Google Scholar]
- 56.Andre P, Denis C, Soulas C, Bourbon-Caillet C, Lopez J, Arnoux T, et al. Anti-NKG2A mAb is a checkpoint inhibitor that promotes anti-tumor immunity by unleashing both T and NK cells. Cell. 2018;175:1731–43 e13. doi: 10.1016/j.cell.2018.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Albrecht I, Niesner U, Janke M, Menning A, Loddenkemper C, Kuhl AA, et al. Persistence of effector memory Th1 cells is regulated by Hopx. Eur J Immunol. 2010;40:2993–3006. doi: 10.1002/eji.201040936. [DOI] [PubMed] [Google Scholar]
- 58.Hawiger D, Wan YY, Eynon EE, Flavell RA. The transcription cofactor Hopx is required for regulatory T cell function in dendritic cell-mediated peripheral T cell unresponsiveness. Nat Immunol. 2010;11:962–8. doi: 10.1038/ni.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haanen JB, Cerundolo V. NKG2A, a new kid on the immune checkpoint block. Cell. 2018;175:1720–2. doi: 10.1016/j.cell.2018.11.048. [DOI] [PubMed] [Google Scholar]
- 60.van Montfoort N, Borst L, Korrer MJ, Sluijter M, Marijt KA, Santegoets SJ, et al. NKG2A blockade potentiates CD8 T cell immunity induced by cancer vaccines. Cell. 2018;175:1744–55 e15. doi: 10.1016/j.cell.2018.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Levy EM, Bianchini M, Von Euw EM, Barrio MM, Bravo AI, Furman D, et al. Human leukocyte antigen-E protein is overexpressed in primary human colorectal cancer. Int J Oncol. 2008;32:633–41. [PubMed] [Google Scholar]
- 62.Gold MH, et al. Updated international clinical recommendations on scar management: part 1-evaluating the evidence. Dermatol Surg. 2014;40:817–24. doi: 10.1111/dsu.0000000000000049. [DOI] [PubMed] [Google Scholar]
- 63.Davison SP, Dayan JH, Clemens MW, Sonni S, Wang A, Crane A. Efficacy of intralesional 5-fluorouracil and triamcinolone in the treatment of keloids. Aesthet Surg J. 2009;29:40–6. doi: 10.1016/j.asj.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 64.Shimba A, Ikuta K. Control of immunity by glucocorticoids in health and disease. Semin Immunopathol. 2020;42:669–80. doi: 10.1007/s00281-020-00827-8. [DOI] [PubMed] [Google Scholar]
- 65.Uzhachenko RV, Shanker A. CD8(+) T lymphocyte and nk cell network: circuitry in the cytotoxic domain of immunity. Front Immunol. 2019;10:1906. doi: 10.3389/fimmu.2019.01906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pallmer K, Oxenius A. Recognition and regulation of T cells by NK cells. Front Immunol. 2016;7:251. doi: 10.3389/fimmu.2016.00251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Laroni A, Armentani E, Kerlero de Rosbo N, Ivaldi F, Marcenaro E, Sivori S, et al. Dysregulation of regulatory CD56(bright) NK cells/T cells interactions in multiple sclerosis. J Autoimmun. 2016;72:8–18. doi: 10.1016/j.jaut.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 68.Rizzo R, Trentini A, Bortolotti D, Manfrinato MC, Rotola A, Castellazzi M, et al. Matrix metalloproteinase-2 (MMP-2) generates soluble HLA-G1 by cell surface proteolytic shedding. Mol Cell Biochem. 2013;381:243–55. doi: 10.1007/s11010-013-1708-5. [DOI] [PubMed] [Google Scholar]
- 69.Hayashi T, Furukawa H, Oyama A, Funayama E, Saito A, Murao N, et al. A new uniform protocol of combined corticosteroid injections and ointment application reduces recurrence rates after surgical keloid/hypertrophic scar excision. Dermatol Surg. 2012;38:893–7. doi: 10.1111/j.1524-4725.2012.02345.x. [DOI] [PubMed] [Google Scholar]
- 70.Mazzarino P, Pietra G, Vacca P, Falco M, Colau D, Coulie P, et al. Identification of effector-memory CMV-specific T lymphocytes that kill CMV-infected target cells in an HLA-E-restricted fashion. Eur J Immunol. 2005;35:3240–7. doi: 10.1002/eji.200535343. [DOI] [PubMed] [Google Scholar]
- 71.Schulte D, Vogel M, Langhans B, Krämer B, Körner C, Nischalke HD, et al. The HLA-E(R)/HLA-E(R) genotype affects the natural course of hepatitis C virus (HCV) infection and is associated with HLA-E-restricted recognition of an HCV-derived peptide by interferon-gamma-secreting human CD8(+) T cells. J Infect Dis. 2009;200:1397–401. doi: 10.1086/605889. [DOI] [PubMed] [Google Scholar]
- 72.Hoare HL, Sullivan LC, Pietra G, Clements CS, Lee EJ, Ely LK, et al. Structural basis for a major histocompatibility complex class Ib-restricted T cell response. Nat Immunol. 2006;7:256–64. doi: 10.1038/ni1312. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.