Abstract
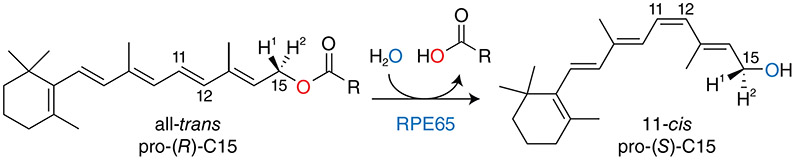
Vertebrate vision critically depends on an 11-cis-retinoid renewal system known as the visual cycle. At the heart of this metabolic pathway is an enzyme known as retinal pigment epithelium 65 kDa protein (RPE65), which catalyzes an unusual, possibly biochemically unique, reaction consisting of a coupled all-trans-retinyl ester hydrolysis and alkene geometric isomerization to produce 11-cis-retinol. Early work on this isomerohydrolase demonstrated its membership to the carotenoid cleavage dioxygenase superfamily and its essentiality for 11-cis-retinal production in the vertebrate retina. Three independent studies published in 2005 established RPE65 as the actual isomerohydrolase instead of a retinoid-binding protein as previously believed. Since the last devoted review of RPE65 enzymology appeared in this journal, major advances have been made in a number of areas including our understanding of the mechanistic details of RPE65 isomerohydrolase activity, its phylogenetic origins, the relationship of its membrane binding affinity to its catalytic activity, its role in visual chromophore production for rods and cones, its modulation by macromolecules and small molecules, and the involvement of RPE65 mutations in the development of retinal diseases. In this article, I will review these areas of progress with the goal of integrating results from the varied experimental approaches to provide a comprehensive picture of RPE65 biochemistry. Key outstanding questions that may prove to be fruitful future research pursuits will also be highlighted.
Keywords: retinal pigment epithelium, visual cycle, isomerase, isomerohydrolase, non-heme iron enzyme, photoreceptors, inhibitor, carbocation
1. Introduction and Scope
The retinal pigment epithelium 65 kDa protein (RPE65) is a critical component of the vertebrate visual system, playing a non-redundant role in the visual cycle metabolic pathway that regenerates visual chromophore, 11-cis-retinal, for rod and cone visual pigments. Within this pathway, RPE65 catalyzes the hallmark trans-cis isomerization reaction, taking all-trans-retinyl esters as substrates and converting them, through hydrolysis and alkene isomerization, into 11-cis-retinol. Owing to this dual catalytic activity, RPE65 is known as an isomerohydrolase, although terms such as retinoid isomerase and retinol isomerase are also in use. Interest in this enzymatic activity can be traced back to the late 1800’s with the work of Boll and Kühne on the mechanism of visual pigment regeneration in the frog retina (Boll, 1876; Kuhne, 1878). The involvement of vitamin A derivatives in visual pigment photochemistry was elucidated by George Wald and his colleagues who also first described a basic sketch of the visual cycle (Hubbard and Wald, 1952; Wald, 1933, 1968). Work during the 1980s and 90s biochemically established the existence of an isomerase system in the RPE that operates on a retinoid in the alcohol oxidation state (Bernstein et al., 1987; Bernstein and Rando, 1986; Canada et al., 1990; Stecher et al., 1999), which was later determined to be all-trans-retinyl ester (Moiseyev et al., 2003; Trehan et al., 1990). However, it was not until the late 1990’s, through studies on Rpe65 knockout mice (Redmond et al., 1998), that an essential role for RPE65 in the isomerohydrolase step of the visual cycle began to be revealed. Controversies regarding its precise function in the visual cycle were laid to rest by three studies published in 2005 demonstrating that RPE65 is the actual isomerohydrolase (Jin et al., 2005; Moiseyev et al., 2005; Redmond et al., 2005) as opposed to a mere retinyl ester-binding protein. For further details of the early history of research on RPE65 and the visual cycle in general, the reader is referred to prior reviews on this subject (Kiser and Palczewski, 2010; Ripps, 2008; Wright et al., 2015).
The RPE65 protein is also of major clinical interest owing to its involvement in retinal diseases as well as its identification as a target for pharmacologic inhibition. RPE65 was one of the first genes to be linked to the development of recessive Leber congenital amaurosis (LCA) and non-syndromic recessive retinitis pigmentosa (RP) (Gu et al., 1997; Marlhens et al., 1997), which triggered early efforts to develop a gene therapy for these conditions (Acland et al., 2001). These efforts culminated in the FDA-approval of the AAV-based RPE65 gene therapy (the first such approval for an inherited disease), voretigene neparvovec-rzyl, for the treatment of LCA/RP resulting from biallelic loss of function RPE65 gene mutations (Russell et al., 2017). For more details on the topic of RPE65 gene therapy, the reader is referred to several recent articles (e.g. Garafalo et al., 2020; Maguire et al., 2021).
Interestingly, the normal operation of RPE65 within the visual cycle pathway was shown to be critical for susceptibility of the retina to light damage (Grimm et al., 2000; Wenzel et al., 2001) and has been hypothesized to be a driver of disease progression in Stargardt disease and possibly age-related macular degeneration (reviewed in Travis et al., 2007). These findings have prompted efforts from several academic and industrial laboratories to develop therapeutic RPE65 inhibitors. Although the ultimate clinical utility of these inhibitors remains to be seen, they have proven to be valuable tools for understanding the enzymology of RPE65 as well as its physiological functions in the retina as described in detail later in this article.
Since the last devoted review of the RPE65 protein appeared in this journal in 2010 (Kiser and Palczewski, 2010), major advances have been made in understanding nearly all aspects of RPE65 structure and function. In this article, I will provide an update on progress made in understanding RPE65 biochemistry and enzymology, its phylogenetic origins, its pharmacology and role in visual cycle physiology, and its involvement in retinal disease with an emphasis on studies published within the last decade.
2. Overview of the Classical Visual Cycle
To provide a context for RPE65 functionality in the retina, I will provide a brief overview of the vertebrate visual cycle. For more details, the reader is referred to prior comprehensive reviews (Kiser et al., 2014; Lamb and Pugh, 2004, 2006; Saari, 2012).
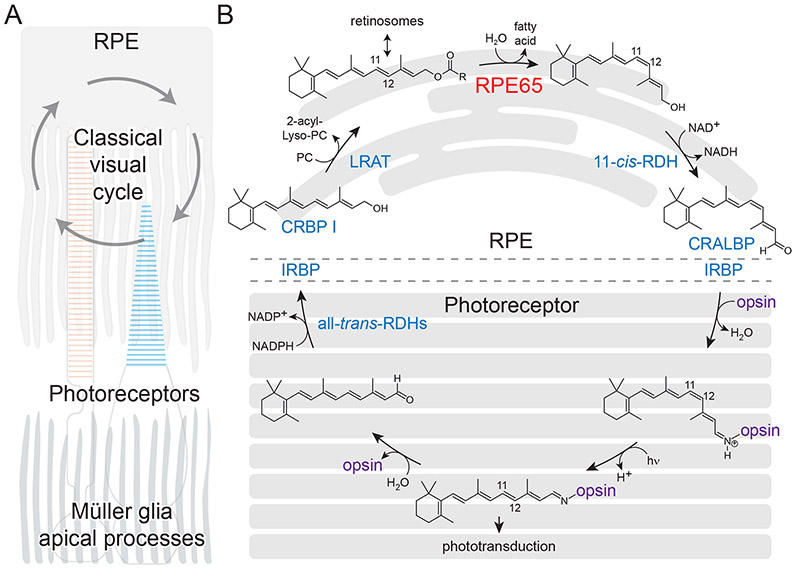
The mechanism of initial light detection and the continuous regeneration of this detection system within the vertebrate eye constitute a conserved, vitamin A-dependent biochemical system known as the visual cycle or retinoid cycle. The cycle begins when light passing through the photoreceptor layer of the retina is absorbed by opsin-based visual pigments, which include the rod pigment rhodopsin and the cone opsin pigments. Common to all of these photoreceptive molecules is the vitamin A-derived visual chromophore 11-cis-retinal, which is covalently attached via a Schiff-base linkage to an interior lysyl residue of the opsin moiety (Palczewski, 2006; Zhong et al., 2012). Light absorption by the chromophore results its photoisomerization to an all-trans configuration, which triggers conformational changes in the opsin necessary for initiation of phototransduction (Hofmann et al., 2009). The light-activated visual pigment hydrolytically decays, potentially after passing through additional photointermediates, to yield free all-trans-retinal, which must be rapidly cleared and replaced with fresh 11-cis-retinal in order for vision to remain continuous and to avoid the intrinsically toxic effects of free retinal or constitutive opsin signaling activity. The recycling of all-trans-retinal back into 11-cis-retinal is accomplished through several steps catalyzed or otherwise facilitated by enzymes, transporters and retinoid-binding proteins located in photoreceptors and the adjacent retinal pigment epithelium (RPE) (Figure 1).
Figure 1. Overview of the classical visual cycle.
A). Anatomic arrangement of the outer retina showing the operation of the visual cycle between photoreceptor outer segments (red and blue for rods and cones, respectively) and the RPE. Also shown are the apical processes of Müller glia cells, which play non-classical roles in the delivery of visual chromophore to photoreceptors, particularly cones. B) Details of the classical visual cycle. Protein components of the pathway are shown in blue with the exception of RPE65 and opsin. Non-standard abbreviations are as follows: CRALBP, cellular retinaldehyde-binding protein; CRBP, cellular retinol-binding protein; IRBP, interphotoreceptor retinoid-binding protein; LRAT, lecithin:retinol acyltransferase; PC, phosphatidylcholine; RDH, retinol dehydrogenase; RPE65, retinal pigment epithelium 65 kDa. Panel B is modified and used with permission from (Kiser et al., 2014).
Briefly, all-trans-retinal released from bleached visual pigments is reduced within the photoreceptor outer segments by retinol dehydrogenase 8 (RDH8), among other all-trans-RDHs, and trafficked to the RPE where it is esterified by lecithin:retinol acyltransferase (LRAT) (Batten et al., 2004). The resulting all-trans-retinyl esters (mainly palmitoyl esters) can be stored in lipid droplet-like organelles called retinosomes or further processed (Imanishi et al., 2004a). The “hallmark” trans-cis isomerization step of the renewal phase of the visual cycle is catalyzed by RPE65 (Jin et al., 2005; Moiseyev et al., 2005; Redmond et al., 2005). Specifically, RPE65 converts all-trans-retinyl esters, through a concurrent hydrolysis and isomerization reaction, into 11-cis-retinol and a free fatty acid. This reaction is biochemically notable for its involvement of two distinct chemical transformations, isomerization and hydrolysis, which has led to RPE65 being termed an isomerohydrolase reflecting its dual catalytic function. The 11-cis-retinol formed by RPE65 is subsequently oxidized within the RPE by 11-cis-retinol dehydrogenase enzymes (RDH5 and other 11-cis-RDHs) (Jang et al., 2001) and then shuttled back to the photoreceptor outer segment to regenerate ground-state visual pigments, thus completing the cycle.
3. RPE65 Phylogenetics: A Carotenoid Cleavage Enzyme Turned Isomerohydrolase
RPE65 belongs to a superfamily of enzymes known as carotenoid cleavage dioxygenases (CCDs), a discovery made following the cloning of the first functionally characterized CCD enzyme from Zea mays, known as viviparous-14 (VP14) (Schwartz et al., 1997). Realization of this homologous relationship of RPE65 to a known carotenoid-metabolizing enzyme (Tan et al., 1997) together with the visual cycle blockade found in Rpe65−/− mice (Redmond et al., 1998) provided the first clues as to the function of RPE65 in retinoid metabolism.
CCDs are broadly distributed in nature, involved in diverse physiological processes, and likely have an ancient history in generating retinal chromophore for both type 1 and type 2 opsin proteins (Ernst et al., 2014; Giuliano et al., 2003; Wyss, 2004; Zhong et al., 2012). These enzymes are characterized by a conserved 7-bladed β-propeller fold and a set of His/Glu residues that coordinate an Fe(II) prosthetic group (Kloer et al., 2005). As is evident from their name, CCDs typically catalyze oxygenolysis of carotenoids at specific alkene sites, although some cleave the non-aromatic alkene bond of phenylpropanoids such as resveratrol and isoeugenol (Kamoda and Saburi, 1993; Kiser, 2019). This catalytic activity is dependent on ferrous iron as shown in the first biochemical characterization of Zea mays VP14 (Schwartz et al., 1997).
Besides RPE65, most vertebrate genomes encode two additional CCDs, which are known as beta carotene dioxygenases (BCO) 1 and 2 (von Lintig et al., 2021) (Figure 2). These enzymes catalyze the cleavage of carotenoids or apocarotenoids by O2 at their 15-15’ and 9-10 (and/or 9’-10’) double bonds, respectively. In marked contrast, the isomerohydrolase reaction catalyzed by RPE65 does not appear to require an O2 co-substrate and is non-redox in nature. This intriguing change in catalytic activity, which is likely to have been a key event in the development of a visual system capable of operating in scotopic conditions (Lamb, 2013), has prompted efforts by different groups to understand when RPE65 first appeared during evolution.
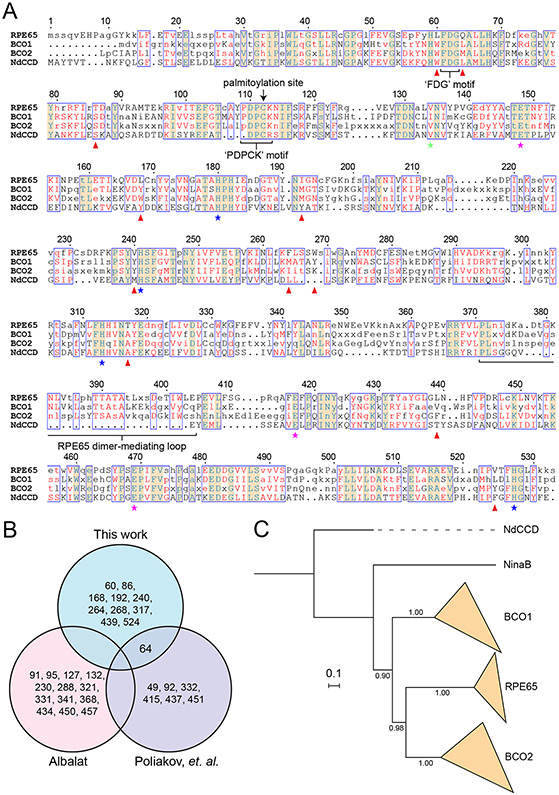
Figure 2. Sequence and phylogenetic relationship of RPE65 to carotenoid cleavage dioxygenase.
A) An alignment of RPE65, BCO1, and BCO2 consensus sequences along with the sequence of a metazoan-like archaeal CCD, NdCCD. Residues making up the conserved His/Glu iron coordination motif are marked with blue and red stars, respectively. The green star indicates the location of the residue that occludes one of the potential iron coordination sites. Red arrowheads indicate ‘RPE65-specific’ residues as defined in the main text. Consensus sequences were generated using the program Cons. Consensus residues were assigned based on the default plurality rule. Consensus residues are shown in uppercase if the corresponding column in the alignment consisted of all positively matched residues according to the BLOSUM62 scoring matrix. The alignment was generated using Clustal Omega (Sievers et al., 2011) and displayed using Espript3 (Gouet et al., 2003). With a few exceptions, CCD sequences from the following organisms were used to generate the consensus sequences: Bos taurus (cow), Mus musculus (mouse), Homo sapiens (human), Canis lupus familiaris (dog), Ornithorhynchus anatinus (platypus), Xenopus tropicalis (frog), Sus scrofa (pig), Felis catus (cat), Chelonia mydas (green sea turtle), Petromyzon marinus (Sea lamprey), Latimeria chalumnae (coelacanth), Gallus gallus (chicken), Eptatretus stoutii (hagfish). Hagfish BCO1 and BCO2 sequences were unavailable for use in consensus sequence generation. Additionally, the platypus BCO2 sequence was not used for consensus sequence generation owing to its questionable accuracy. B). Venn diagram comparison of residues identified as being “RPE65-specific” in different studies. C). Phylogeny of metazoan CCDs generated using MrBayes (Ronquist and Huelsenbeck, 2003). NdCCD was used as an outgroup sequence to root the tree. The scale bar indicates the average number of substitutions per site. Numbers along the bipartitions are posterior probabilities estimated by Markov Chain Monte Carlo using the Jones substitution matrix and assuming invgamma among-site rate variation. The majority rule consensus tree is shown.
It was clear early on that RPE65 is universally found in all jawed vertebrates and is a highly conserved protein both in terms of amino acid sequence identity as well as sequence length. Phylogenetic studies conducted in the late 2000’s suggested the presence of an RPE65 ortholog in the tunicate Ciona intestinalis (Ci-RPE65) (Takimoto et al., 2006; Takimoto et al., 2007), which is an invertebrate organism but with several proto-vertebrate characteristics (Corbo et al., 2001; Delsuc et al., 2006). These findings consequently suggested that the visual cycle developed in advance of rod photoreceptors, which are thought to have originated in the vertebrate last common ancestor during the Cambrian era (Asteriti et al., 2015). An important limitation of this investigation was its lack of direct functional demonstration of isomerohydrolase activity in this supposed RPE65 ortholog. Additionally, Ci-RPE65 exhibited only marginally greater sequence identity to human RPE65 (36.5%) than to human BCO1 (36.3%), in sharp contrast to the highly conserved nature of vertebrate RPE65 orthologs.
The issue of RPE65 evolutionary origins was reexamined a few years later by two different groups. A phylogenetic analysis carried out by Albalat revealed that Ci-RPE65 did not group among vertebrate RPE65 sequences (Albalat, 2012). Poliakov, Redmond, and colleagues, using similar phylogenetic methods, arrived at the same conclusion that Ci-RPE65 is not a true RPE65 ortholog (Poliakov et al., 2012). These investigators also directly showed that Ci-RPE65 is in fact a carotenoid cleaving enzyme rather than an isomerohydrolase. A later analysis of cephalochordate genomes also did not reveal any potential RPE65 orthologs, strongly suggesting that RPE65 was absent in the chordate last common ancestor (Poliakov et al., 2017). Poliakov and colleagues went on to demonstrate the presence of functional RPE65 and LRAT orthologs in the sea lamprey (Petromyzon marinus), which belongs to the cyclostome vertebrate lineage that diverged from jawed vertebrates about 550 Ma (Miyashita et al., 2019; Poliakov et al., 2012). Recently, Dong and Allison have demonstrated that Pacific Hagfish (Eptatretus stoutii), belonging to the other major lineage of jawless vertebrates, also express RPE65 within their retinal (non-pigmented) epithelial cells (Dong and Allison, 2021). Taken together, these studies have shown that RPE65 activity characteristic of the classical visual cycle is an innovation unique to the vertebrate lineage. However, it remains unclear whether RPE65 and the other classical visual cycle components evolved in advance of, or contemporaneously with, rhodopsin and rod photoreceptors. Additionally, the exact number and sequence of gene duplications and subsequent subfunctionalization and neofunctionalization events that occurred at the base of the vertebrate lineage remains unclear (Poliakov et al., 2020). For example, phylogenetic analyses conducted to date have not unambiguously resolved the relationships between the vertebrate CCD clades, which may suggest that these three lineages arose close in time from a common ancestral sequence, possibly through whole genome duplication(s) that occurred early in vertebrate evolution (Dehal and Boore, 2005).
To gain insights into the molecular changes that enabled neofunctionalization of the RPE65 gene lineage, Albalat and Poliakov and colleagues used two different approaches to identify RPE65-specific residues that could confer differential activity relative to BCO1/BCO2. Albalat identified 12 such residues based on their conservation in RPE65 orthologs relative to BCO1/BCO2 proteins and their involvement in RPE65-associated RP/LCA. Poliakov and colleagues found 7 residues based on a divergence comparison to BCO2 proteins. Most of the residues identified in the latter study are found outside of the active site pocket leading the authors to speculate that changes at these positions indirectly fine-tune active site residues to allow for RPE65-specific functions. Residues identified by Albalat are mostly located within the beta-propeller core or clustered on the outside of the helical cap near the membrane binding surface. Notably, the agreement between these studies on the exact residues that are adapted for RPE65 functions is poor, likely reflecting the different methodologies and specific sequences used to make the determinations (Figure 2B).
RPE65-specific residues and those common to vertebrate CCDs in general can also be identified by considering the shared and derived character states among the three groups of vertebrate CCDs. Figure 2A presents an alignment of the consensus sequences for RPE65, BCO1, and BCO2 along with a structurally characterized metazoan-like archaeal CCD from Nitrosotalea devanaterra (NdCCD) (Daruwalla et al., 2020) included as an outgroup sequence to polarize the character set (Figure 2C). In this representation, capital letters in the consensus sequences indicate highly or absolutely conserved residues (i.e. all positive matches of the consensus residue within the alignment column based on the BLOSUM62 matrix) across diverse gnathostomes and cyclostomes, lower case letters indicate incompletely conserved consensus residues, and “x” represents a position that fails to meet the plurality rule for consensus residue assignment.
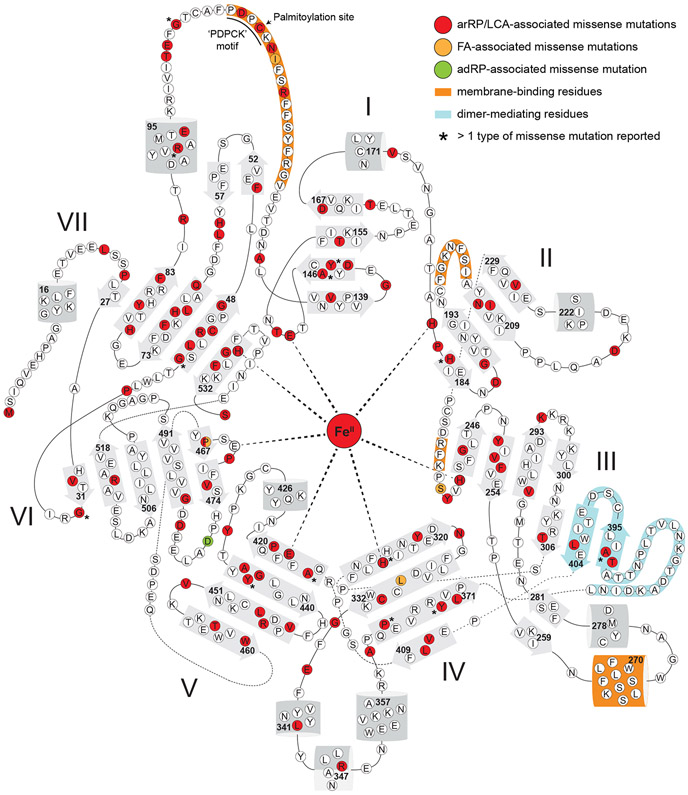
RPE65 shares a number of conserved sequence motifs with its carotenoid-cleaving paralogs. These include: 1) the 4-His/3-Glu iron binding motif (blue and magenta stars in Figure 2A, respectively) that unites the CCD superfamily, 2) a highly conserved ‘FDG’ motif (Poliakov et al., 2009) that is positioned near the iron center, 3) the ‘PDPC(K)’ motif (Poliakov et al., 2017) conserved in metazoan CCDs and partially conserved in NdCCD, which constitutes the initial region of a long flexible sequence believed to help mediate membrane binding (Hamel et al., 1993b; Kiser et al., 2009), and 4) an extended chordate-specific sequence that mediates dimerization in RPE65 (Kiser et al., 2012).
The alignment also reveals a number of character states that are specific to RPE65 (i.e. RPE65 synapomorphies). Here, an ‘RPE65-specific’ character is defined as one that is i. absolutely conserved in one state among the examined RPE65 orthologs, ii. highly or absolutely conserved (i.e. meeting the capitalization criteria described above) in states different from that of RPE65 in both BCO1 and BCO2 orthologs with no instances of the RPE65 character state present in the BCO1/BCO2 sequences used for the analysis, and iii. present in a state other than the one found in RPE65 in NdCCD. Of note, NinaB was not used as the outgroup sequence for this analysis since it also possesses C11-C12 isomerase activity, which may reflect convergent evolution of the NinaB and RPE65 sequences to achieve this common activity. This simple but stringent definition pinpoints characters under strong selective pressure for differential functions (catalytic or otherwise) between RPE65 and BCO1/BCO2 proteins. The RPE65 synapomorphies that meet this definition are Leu60, Gln64, Thr86, Leu168, Val240, Phe264, Trp268, Thr317, Leu439, Val524 (red triangles in Figure 2A). Alignments including other invertebrate CCD sequences such as Ci-RPE65 or Ci-BCO1 showed that the identified RPE65-specific characters are robust to the choice of outgroup, with the possible exception of Leu168 where the equivalent site in Ci-BCO1 is also found in the Leu character state. At least some of these RPE65-specific character states are likely to underlie the unique substrate specificity and isomerization capacity found in the RPE65 lineage. The structural and functional relevance of these residues will be further considered in Sections 5.5 and 6.
4. RPE65 Cellular Biology
4.1. Retinal Cell Specificity of RPE65 Expression
Consistent with its assigned name, the expression of RPE65 has been found to be restricted to the RPE in many studies (Bavik et al., 1992; Hamel et al., 1993a; Hemati et al., 2005; Jacobson et al., 2007; Kiser, 2010; Nicoletti et al., 1995; Redmond, 2009; Seeliger et al., 2001). However, RPE65 expression has also been reported in cone photoreceptors at both the mRNA (Ma et al., 1998) and protein (Tang et al., 2011a; Tang et al., 2011b; Znoiko et al., 2002) levels, which led to speculation that RPE65 may be involved in cone-specific visual cycle pathways. This hypothesis was directly tested in a study by Kolesnikov and colleagues using transgenic mice expressing RPE65 under control of a cone-specific promoter (Kolesnikov et al., 2018). Despite achieving high-level transgenic expression of RPE65 within cones, electrophysiological recordings from these mice did not reveal any augmentation of cone photoresponses and in fact showed a slight delay in dark adaptation. The authors concluded that RPE65 does not serve a functional role in cone-specific visual chromophore regeneration pathways.
All studies reporting RPE65 expression in cones relied on the ‘PETLET’ rabbit polyclonal antibody (Redmond and Hamel, 2000), which was generated against residues 150-164 of the human/bovine RPE65 sequence. Although specificity of this antibody for RPE65 was reported (Redmond and Hamel, 2000; Znoiko et al., 2002), the fact that it is polyclonal raises the possibly that it could recognize non-RPE65 antigens. Using monoclonal RPE65 antibodies, we and others have not observed the presence of RPE65 in cone photoreceptors of mixed BL6/129 or BALB/c mice (Hemati et al., 2005; Kiser, 2010). It was suggested that RPE65 expression level in cones is dependent on the mouse strain used for experiments, with C57BL/6 and BALB/c mice having the highest and lowest (non-detectable) cone expression, respectively (Tang et al., 2011b). However, this idea is in conflict with the original study describing cone photoreceptor RPE65 expression, which reported strong RPE65 signals in the cones of BALB/c mice (Znoiko et al., 2002). Additionally, the cone photoreceptor reactivity in these studies exhibited variable localization in different species, which is unexpected for an enzyme with a putatively conserved function in the retina (Tang et al., 2013).
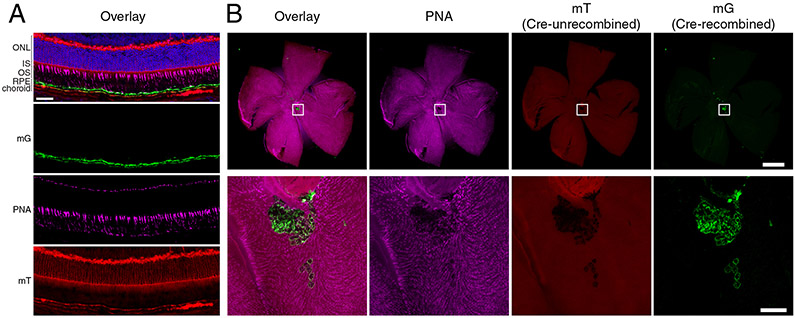
Given the conflicting immunohistochemical data on RPE65 expression in the neural retina, it is informative to consider results from orthogonal methods that inform on RPE65 expression in cone photoreceptors. In particular, the availability of retinal gene expression data obtained by single-cell RNA sequencing (RNAseq) has helped clarify the distribution of RPE65 in the retina. Studies in both humans (Cowan et al., 2020) and mice (Hoang et al., 2020; Voigt et al., 2019) have shown that RPE65 is expressed in virtually all RPE cells at a relatively high level whereas expression in cones was not above background levels. A novel Rpe65CreERT2 mouse line has also provided information on Rpe65 expression in the retina (Choi et al., 2021). In this mouse line, an inducible Cre recombinase (CreERT2) is co-translationally expressed with Rpe65 under control of the native Rpe65 promoter. We crossed this mouse with the mT/mG Cre reporter strain (Muzumdar et al., 2007), which undergoes an irreversible switch in expression from red fluorescence protein (mT) to green fluorescence protein (mG) following Cre-mediated LoxP recombination. This reporter system concept, which is widely used in the neuroscience community (Guenthner et al., 2013), thus serves as a sensitive test for Rpe65 promoter activity, since Cre expression driven by the native promoter at any time during the induction period leads to permanent fluorescent marking of the cell. Consistent with the RNAseq data mentioned above, we did not observe fluorescent marking of cone photoreceptors in any of the retinal sections or wholemounts examined (Figure 3).
Figure 3. Absence of Rpe65 promoter activity in cone photoreceptors.
A) Retinal cryo-section from a Rpe65CreERT2 mT/mG+/− mouse treated with tamoxifen to induce Cre activity. Green fluorescence (mG) shows cells where the Rpe65 promoter was active resulting in Cre activity at the mT/mG locus. Red fluorescence (mT) shows cells lacking Cre activity. The sections were co-stained with DAPI (blue) and peanut agglutinin (PNA, magenta) to demarcate nuclei and cones, respectively. The composite fluorescence signal is shown at the top along with the individual channels. Co-localization of the PNA and mG signals was not observed. Conversely, the mG signal did colocalize with ezrin, a marker of RPE cell apical cell processes (Huang et al., 2009), as described in (Choi et al., 2021). The scale bar represents 50 μm. B) A retinal flatmount from an Rpe65CreERT2 mT/mG+/− mouse treated with tamoxifen to induce Cre activity. The flatmount was co-stained with PNA to allow visualization of cone photoreceptors and imaged with its RPE-associated side facing the camera. The composite fluorescence signal is shown to the left along with the individual channels. Magnified images corresponding to the indicated boxes are shown below each wholemount image. The green fluorescent and PNA signals did not colocalize demonstrating that Cre recombinase in not expressed in the cone photoreceptors of this animal model. The green fluorescence shown in the magnified images originated from RPE cells that remained attached to the neural retina during the dissection procedure. The scale bar in the whole retina image represents 1000 μm. That in the bottom zoomed image represents 100 μm. Used with permission from (Choi et al., 2021).
Considered together, the current data strongly suggest that RPE65 expression in the eye is confined to the RPE. The variously localized signals associated with cone photoreceptors in prior immunohistochemical studies may be the result of off-target antigen recognition by the polyclonal antibody preparation used in those studies.
4.2. Subcellular Localization and Dynamics
Within the RPE, the RPE65 protein is localized to the cytoplasm of the cell body and is excluded from the apical processes (Huang et al., 2009). RPE65 has long been understood to associate with RPE microsomal membranes derived from smooth endoplasmic reticulum (Bavik et al., 1992; Hamel et al., 1993a) although the exact nature of this interaction has been a highly contentious subject (reviewed in Kiser and Palczewski, 2010). Data available to date indicate that the intrinsic affinity of RPE65 for membranes arises from 1) hydrophobic and cationic patches on its surface (described in detail in Section 5.4) and 2) cysteine palmitoylation at position 112 which provides an additional hydrophobic anchor to the protein (Kiser et al., 2009; Kiser and Palczewski, 2010; Takahashi et al., 2009; Uppal et al., 2019a; Uppal et al., 2019b). Notably, cysteine palmitoylation at the equivalent site in BCO2 has also been reported (Uppal et al., 2020).
In many databases (e.g. OMIM), RPE65 is annotated as existing in membrane-bound and soluble forms (mRPE65 and sRPE65) which differ in their palmitoylation status. This nomenclature was introduced by an influential (but now largely disproven) study purporting that RPE65 membrane localization and activity are controlled through a “palmitoylation switch” mechanism (Xue et al., 2004). Under the assumption that these two forms of RPE65 are physiological, it was previously speculated that “sRPE65” could play a role in mobilizing retinyl esters or retinol from retinosomes delivering them for metabolism in the smooth ER (Imanishi et al., 2004b; Lamb and Pugh, 2004; Lyubarsky et al., 2005). This hypothesis is appealing considering the fact that retinyl esters massively accumulate in Rpe65−/− mice (Redmond et al., 1998) driven by the continual influx of retinol from the circulation (Qtaishat et al., 2003), which is suggestive of a mobilization defect. However, fluorescence microscopy (Imanishi et al., 2004a) and proteomic/lipidomic (Orban et al., 2011) studies of bovine RPE retinosomes revealed that RPE65 does not appreciably associate with the retinosomes, contrary to the idea that it might bind and extract retinoids from this organelle. It thus remains an outstanding question why retinyl esters accumulate at such high levels in Rpe65−/− mice (Imanishi et al., 2004a; Redmond et al., 1998), and to a lesser extent in Rpe65 hypomorphs (Li et al., 2015; Sheridan et al., 2017).
A related unresolved question is how stored retinyl esters are eventually returned or reformed in the smooth ER for metabolism by RPE65. Studies on traditional lipid droplets containing triglycerides or cholesterol esters have identified three pathways for liberation of the stored lipids (Welte and Gould, 2017): 1) surface-associated hydrolases that extract and cleave core lipids thus releasing them into the cytosol (Zimmermann et al., 2004), 2) autophagy of the storage bodies (lipophagy) in which endocytic acid hydrolases mediate release of the lipid (Singh et al., 2009), and 3) direct release of core lipids into the organelle membranes via contact sites (Olzmann and Carvalho, 2019). Since RPE65 involvement as a direct lipid droplet hydrolase appears to be ruled out by the data described above, its involvement in retinosome homeostasis may be more indirect. For example, the processing of retinyl esters by RPE65 and subsequent protected shuttling of 11-cis-retinoid by other visual cycle components could be critical for limiting retinoid access to the powerful acyltransferase activity of LRAT (Figure 1) thus allowing their efflux or elimination from the RPE or retina.
Despite the flaws of the “palmitoylation switch” study, the ideas that RPE65 palmitoylation and localization are dynamically regulated have received support from a few studies over the past decade. Recent studies by Uppal and colleagues have revisited the issue of RPE65 palmitoylation using mass spectrometry-based approaches (Uppal et al., 2019a). These investigators found that dynamic palmitoylation of RPE65 at Cys112 plays a crucial role in its proper localization to endoplasmic membranes with the extent of palmitoylation being affected by the activity of LRAT. However, it was shown in prior work that treatment of native bovine RPE microsomal membranes with the thioester-cleaving compound, hydroxylamine, does not appreciably solubilize RPE65 (Golczak et al., 2010). Additionally, it was shown that loss of LRAT activity does not impact RPE65 membrane association (Jin et al., 2007). A potential unifying explanation for these collective findings is that Cys112 palmitoylation could be involved in the initial trafficking of RPE65 to the endoplasmic reticulum membrane but is less essential for maintenance of RPE65 membrane affinity, which instead could be mediated primarily by its intrinsic structural elements. Uppal and colleagues also found evidence for RPE65 palmitoylation at Cys146, although RPE65 crystal structures show this residue being buried within a rigid region of the protein and hence poorly accessible for an acyl transfer reaction.
Another study examined the role of the Usher syndrome 1B-associate myosin motor protein, MYO7A, on RPE65 expression and localization (Lopes et al., 2011). The authors found that wild-type mice exhibit a change in RPE65 localization upon light exposure. Specifically, RPE65 is localized throughout the RPE cell body under dark conditions but relocates to the central, smooth endoplasmic reticulum-rich, portion of the RPE cell upon light exposure. This relocation was not observed for Myo7a-mutant mice. Moreover, RPE65 levels were significantly lower in Myo7a-mutant mice as compared to wild-type controls, leading to elevations in all-trans-retinyl ester levels as well as resistance to light-induced retinal damage. The authors suggested that altered RPE65 trafficking in Myo7a-mutant mice may target the protein for degradation. MYO7A is known to play a key role in the transport of membranous structures including melanosomes and phagosomes and it has been proposed that its function may extend to transport of smooth ER membrane structures, similar to the function of MYO5 (Williams and Lopes, 2011). Further study on the precise subcellular structures and mechanisms involved in this light-dependent RPE65 translocation phenomenon are needed to elaborate its function.
5. Structural Biochemistry of RPE65
5.1. Overview
The crystal structure of RPE65 was first resolved in 2009 revealing the details of its active site structure and oligomeric state (Kiser et al., 2009). Like other CCD superfamily members, RPE65 adopts a 7-bladed β-propeller fold capped on its top face by a mostly alpha-helical dome that covers the catalytic iron center and forms the substrate binding pocket. Notable features of the RPE65 structure include 1) an iron center coordinated by a 4-His/3-Glu motif with marked structural similarity to carotenoid-cleaving CCDs, 2) the presence of a hydrophobic/cationic patch on the protein exterior that is believed to mediate the binding of RPE65 to phospholipid membranes, 3) a predominantly hydrophobic tunnel leading from the hydrophobic patch into the interior of the protein forming the presumed active site, and 4) the formation of a dimeric assembly mediated by a sequence insertion uniformly present in vertebrate CCDs (Figure 2). These features have served as foci for further study by crystallography and complementary structural approaches as detailed in the next section. It is notable that to date only RPE65 obtained from a native source, bovine RPE microsomes, has been amenable to crystallization despite efforts by our laboratory, and presumably other laboratories, to crystallize heterologously expressed protein. This experimental restriction has prevented direct mutagenesis studies that are typically performed to elucidate structure-function relationships. To help circumvent this difficulty, our laboratory has focused on using information from RPE65 homologs and chimeras to provide insights into the roles of specific residues in RPE65 catalysis as well as the structural effects of pathogenic mutations (see Section 11).
5.2. RPE65 Molecular Architecture and Iron Center
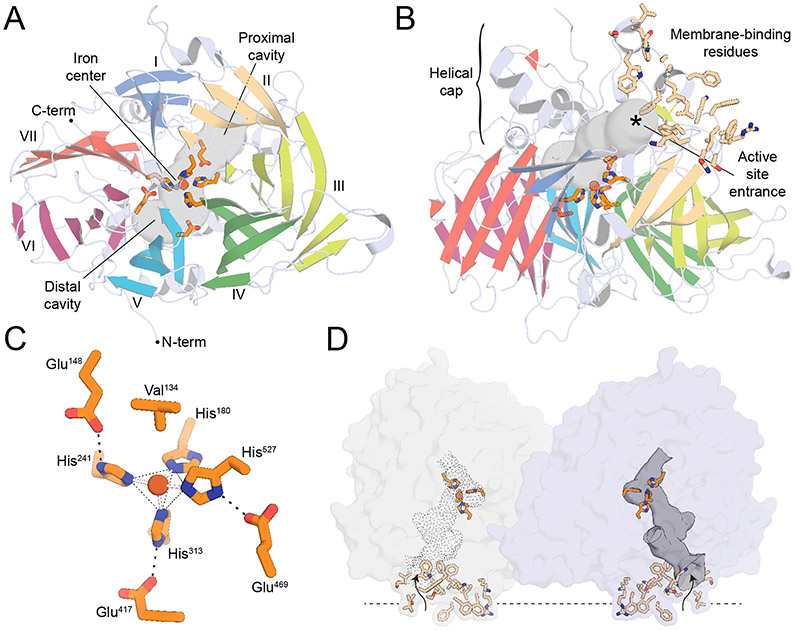
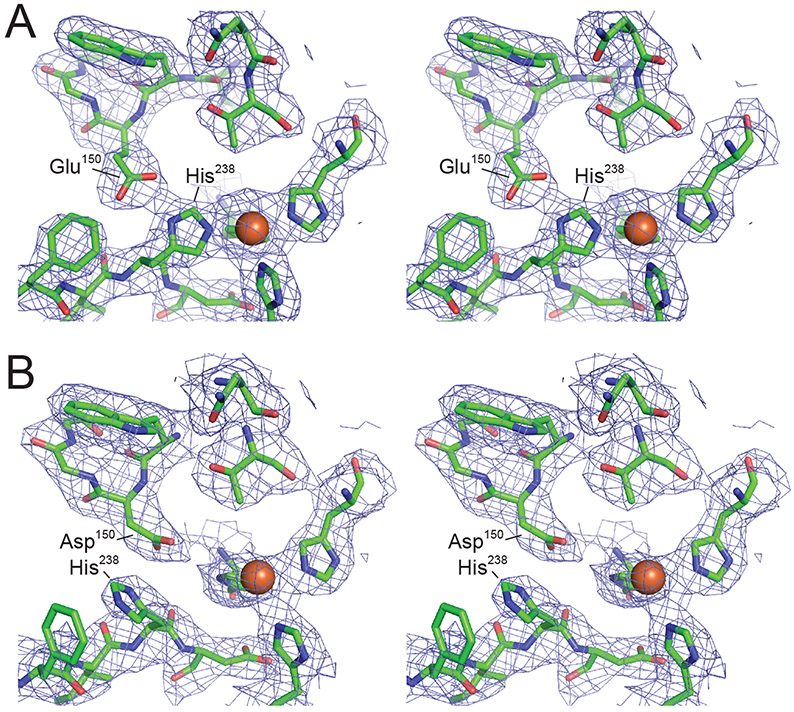
The original RPE65 crystal structure determination in space group P65 revealed that RPE65 adopts a 7-bladed β-propeller fold with single-strand extensions on blades VI and VII and a two-strand extension on blade III (Kiser et al., 2009) (Figure 4A). Sitting on top of this beta propeller core is a group of 7 alpha-helices, 2 short beta-sheets and other irregular secondary structure elements that together form the membrane-binding surface and catalytic active site of the enzyme (Figure 4B). Like many other non-heme iron proteins, RPE65 likely acquires its iron cofactor from buffered intracellular iron stores (Foster et al., 2014), which are abundant in the RPE (He et al., 2007). The iron center sits at the interface of these two structural regions being coordinated by the strictly conserved 4-His/3-Glu motif introduced in Section 3. The inner sphere is composed of the 4 His residues (180, 241, 313, and 527), with the latter three forming hydrogen bonding interactions with Glu residues 148, 417, 469, which form an anionic outer sphere (Figure 4C). A noteworthy feature of the coordination system is that each blade of the propeller structure contributes exactly one element to the iron coordination sphere. The primary sphere His ligands thus occupy four positions within the iron coordination sphere with a geometry that can be described as an incomplete octahedral or square/trigonal pyramidal structure. The iron center geometry and coordination state is further influenced by the nearby Val134 side chain which is oriented with one of its methyl groups positioned close (~4.6 Å) to the iron trans to His313, effectively blocking coordination at this site. While the presence of a non-protein ligand bound to iron trans to His527 was apparent from the original RPE65 structure determinations, its identify was initially ambiguous.
Figure 4. Overview of the RPE65 protein structure.
A) The RPE65 crystal structure viewed down its beta-propeller axis. The propeller blades are marked with Roman numerals. The tripartite active site cavity is delineated with a grey surface. The iron ion (red-brown sphere) is shown along with its 4-His/3-Glu coordination motif (orange sticks). B) Orthogonal view of the RPE65 structure displaying the helical cap on the top surface of the beta propeller. A cluster of hydrophobic and positively charged residues that mediate membrane binding are shown as wheat-colored sticks. C) Structure of the RPE65 iron center. D). Structure of the RPE65 dimeric assembly showing the localization of the membrane binding elements to a common face of the dimer and their close proximity to the active site openings (indicated by curvy arrows).
The RPE65 iron center has been further investigated by X-ray absorption spectroscopy (XAS) which revealed a 5-coordinate (or distorted 6-coordinate) FeII center with an average Fe-His bond length of 2.15 Å (Kiser et al., 2012). The presence of FeII in the native enzyme structure is consistent with prior biochemical work showing a dependence of RPE65 activity on ferrous iron (Moiseyev et al., 2006). Crystals structures and XAS data from highly diverse RPE65 homologs have shown that the structure of the iron center is highly conserved across the entire superfamily (Daruwalla et al., 2020; Kloer et al., 2005; Messing et al., 2010; Sui et al., 2017). Consequently, it has become clear that the isomerohydrolase activity of RPE65 did not arise from a fundamental change to the iron center structure. The XAS data together with later crystallographic studies have shown that the iron center is frequently found in complex with the carboxylate group of a palmitate ligand, indicating that the iron center has been repurposed in RPE65 to bind and polarize the ester moiety of retinyl ester substrates, described in more detail in Section 5.5. This observation implies that other CCDs could also display hydrolase activity upon encounter of an appropriate substrate, although such activity remains to be demonstrated.
The identification of alternative RPE65 crystallization conditions has facilitated more recent detailed investigations into its structure. The originally identified crystallization conditions produced a high percentage of merohedrally twinned crystals of limited value for investigating novel structural features (Golczak et al., 2010). Recently, we have found new, highly reproducible conditions that eliminate the twinning susceptibility and routinely give rise to crystals diffracting to < 2 Å resolution (Blum et al., 2021). Additionally, novel crystallization conditions have helped resolve previously poorly ordered regions of the protein. For example, crystallization of RPE65 under alkaline conditions with low molecular weight polyethylene glycols revealed an alternative N-terminal conformation with the chain packed against a cusp formed by the fifth blade of the beta-propeller (Zhang et al., 2015). The improved electron density for this region showed an apparent loss of the initiator Met residue and its replacement with an acetyl group, thus indicating that RPE65 is subject to N-terminal methionine excision with subsequent acetylation of the following Ser residue, both of which are common protein modifications (Giglione et al., 2004; Ree et al., 2018).
5.3. Dimeric Structure
In all crystal structures of RPE65 determined to date, the protein is found as a dimeric assembly mediated primarily by the vertebrate-associated two-strand extension of the third propeller blade (Figure 4D). The dimer has a number of properties that suggest it is relevant in vivo. First, it is two-fold symmetrical and symmetrical assemblies are known to be the rule in physiological homooligomeric protein complexes (Goodsell and Olson, 2000). Second, the dimeric arrangement places the membrane-binding residues and active site entrances on the same face of the assembly as required for it to be functionally significant. Third, the dimer forms independent of crystallization conditions or space group symmetry which argue against it being a crystallization artifact. Finally, computational analyses of the ~1,563 Å2 dimer interface with the PISA (Krissinel and Henrick, 2007) and EPPIC (Bliven et al., 2018) servers both indicate the dimeric structure is thermodynamically stable. Functionally, the presumed enhanced membrane affinity of the dimer as compared to the monomer could promote the necessary extraction of retinyl ester substrates thus enhancing catalytic activity. Such parallel dimer arrangements are commonly observed for monotopic membrane proteins (Allen et al., 2019; Bracey et al., 2004).
A prior size-exclusion chromatography (SEC) experiment suggested that detergent-solubilized RPE65 is monomeric in solution (Kiser et al., 2009). However, in retrospect, too much weight was probably given to this experiment given the propensity of SEC to produce inaccurate absolute molecular weight estimates. Future SEC coupled with multi-angle light scattering (SEC-MALS) or analytical ultracentrifugation experiments are required to provide an accurate assessment of the RPE65’s oligomeric state in solution. Additional studies will be required to confirm whether the dimeric assembly exists under in vivo conditions. Knowledge of the RPE65 oligomeric state could help explain some of its biochemical properties and contribute to our understanding of the pathology associated with a dominant-acting RPE65 mutation (see Section 11.3).
5.4. Interaction with the Membrane
RPE65 is well known to bind phospholipid membranes and is best thought of as a monotopic integral membrane protein based on its preferential extraction with detergents and the requirement of phospholipid membranes for its activity (Golczak et al., 2010; Nikolaeva et al., 2009). The original structural studies of RPE65 identified a patch of surface-exposed hydrophobic and positively charged amino acid side chains located near the entrance to the active site tunnel of the protein that could serve to anchor the protein to the smooth ER membrane. Similar hydrophobic patches have been found in all carotenoid-cleaving CCD structures determined to date whereas their sizes and constituent residues are variable (Daruwalla et al., 2020; Kloer et al., 2005; Messing et al., 2010). In RPE65, the surface-exposed hydrophobic patch is formed by residues 109-126, 196-202, 234-236, and 261-272, although their precise conformations were originally not well determined owing to disorder and consequent weak electron density support. It is notable that the detergent-solubilized RPE65 used for the original crystallization study was catalytically inactive, which may have been caused, at least in part, by detergent-induced destabilization of the membrane-binding regions.
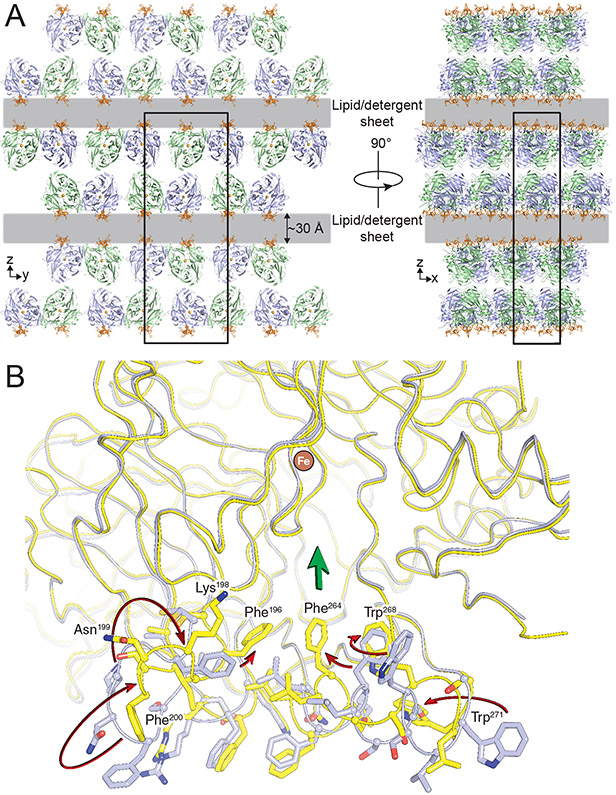
To gain further insights into the roles of the membrane-binding residues in supporting RPE65 activity, we crystallized a catalytically active form of RPE65 obtained directly from bovine RPE65 microsomes solubilized with low levels of detergent (Kiser et al., 2012). The resulting lipid embedded crystals featured an unusual crystal packing arrangement in which sheets of RPE65 molecules were separated by a ~20–30 Å gap extending in two dimensions throughout the crystal with no direct protein–protein contacts between sheets (Figure 5A). These gaps were postulated to be filled with mixed micelles or bilayer-like lipid–detergent sheets lacking defined electron density owing to their fluidity. The arrangement of RPE65 molecules in the unit cell with their membrane-binding surfaces facing the lipid/detergent-filled gap is consistent with this interpretation. In this native-like membrane environment, the membrane-binding residues were well ordered and exhibited novel conformations that likely support the catalytic function of the protein. As compared with the originally determined RPE65 structure, residues 196–202 were flipped by ~90° allowing them to adopt a more ideal β-sheet structure packing against blade II of the β-propeller (Figure. 5B). This conformation positions the Phe196 side chain deeper into the active-site tunnel. Additionally, residues 263–271 adopted an unwound conformation in the lipid-embedded structure, resulting in loss of their alpha-helical structure found in the original structure and positioning of the Phe264 and Trp268 side chains close to the active-site entrance. This loss of alpha-helical structure is consistent with circular dichroism spectroscopy studies that found similar structural differences in membrane-bound vs. detergent-solubilized RPE65 (Nikolaeva et al., 2011).
Figure 5. Molecular packing and conformational difference observed in lipid-embedded RPE65 crystals.
A) Orthogonal views down the crystallographic axes demonstrate well-packed RPE65 sheets separated by 20-30 Å gaps (grey layers) that extend in two dimensions and contain lipid-detergent mixed-micelle sheets. The membrane binding surface of RPE65 (orange) faces the lipid-filled sheet. B) Conformational differences in the membrane-binding surface and active site entrance observed in lipid-embedded RPE65 crystals and delipidated RPE65 crystals. Arrows pointing from the delipidated (grey) to the lipid-embedded (yellow) structures are shown to help illustrate the structural differences. Phe196, Phe264 and Trp268 in the lipid-embedded structure form a continuous aromatic surface near the active site entrance (denoted by the broad green arrow). Used with permission from (Kiser et al., 2012).
An analysis of the RPE65-membrane interaction with the OMP server (Lomize et al., 2012) predicts that the protein is embedded in the lipid bilayer at a depth of ~4 Å with the dimer axis oriented parallel to the membrane normal. As described in more detail in Section 5.5., this membrane penetration is likely essential to allow the extraction of retinyl esters from the membrane in the correct orientation.
The structure of residues 109-126 remained poorly defined in the lipid-embedded RPE65 crystal, although a stretch of weak electron density originating from these residues that runs parallel to the membrane surface suggests this region could be appropriately positioned to contribute to RPE65 membrane binding. Indeed, the ‘PDPC(K)’ sequence motif whose Cys112 residue is palmitoylated as described in Section 4.2., constitutes the N-terminal region of this loop providing further evidence that this sequence is in contact with the membrane. It was previously suggested that residues 109-126 could adopt an amphipathic α-helical structure with a prominent hydrophobic face that would promote membrane binding (Hamel et al., 1993b; Kiser et al., 2009). Notably, this region has never been clearly resolved in any structure of RPE65 reported to date, despite the use of a wide range of crystallization conditions and alternative detergents. These findings suggest that residues 109-126 could be intrinsically disordered. In support of this proposal, we recently determined the crystal structure of an archaeal CCD enzyme containing a homologous sequence with strong identity to that of sequence 109-126 of RPE65 and found it to be poorly ordered despite the protein having been crystallized in the absence of detergents (Daruwalla et al., 2020). Alternatively, residues 109-126 may become helical only when embedded in a lipid membrane. AlphaFold (Jumper et al., 2021) and RosettaFold (Baek et al., 2021) both model residues 115-124 as an alpha-helix but with very low confidence, again suggesting that this region may be unstructured or highly mobile. The identification of NdCCD as an RPE65 homolog with a mostly conserved ‘PDPC(K)’ loop that is amenable to mutagenesis and heterologous expression may provide an ideal model system to further examine the structure and membrane-binding roles of this nebulous sequence element.
5.5. How Retinyl Esters Bind to the RPE65 Active Site to be Cleaved and Isomerized
The active site of RPE65 is composed of an elongated tunnel that begins at the membrane-binding surface extending along the junction between the beta propeller domain and the helical cap (membrane-proximal region) past the iron center, ending in a curved pocket within the protein interior (distal region). This tunnel also serves a second function in providing the passageway for retinyl ester substrate to be extracted from the lipid bilayer. Because the iron center is located deep within the protein, the retinyl ester must be desorbed from the membrane into the enzyme to be processed, which necessitates penetration of the enzyme into the acyl core of the lipid bilayer (Forneris and Mattevi, 2008). The RPE65 active site tunnel is well suited to promote retinyl ester partitioning from the membrane owing to its predominantly hydrophobic character. The diameter of this tunnel is too narrow to allow the ester to enter the active site in a bent, hairpin conformation but is of an appropriate length (~30 Å) to accommodate the retinyl palmitate substrate in an extended conformation (also ~30 Å). The binding of retinyl ester substrate in an extended conformation is also consistent with the close relationship of RPE65 to BCO1 and BCO2, which must be able to accommodate long and mainly linear (apo)carotenoid molecules in their active sites. However, it remained ambiguous, following the initial RPE65 structure determination, whether it is the retinoid moiety or the acyl chain that enters the protein first and thus how the substrate is oriented in the active site during catalysis. Multiple attempts by us to obtain RPE65 in complex with retinoids failed likely due to the very poor aqueous solubility of these compounds.
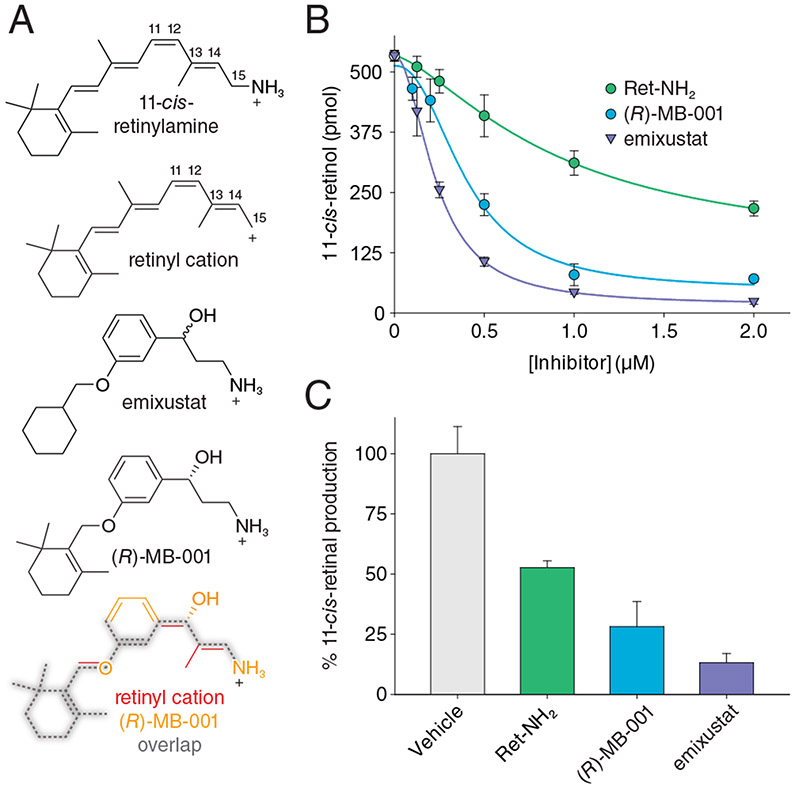
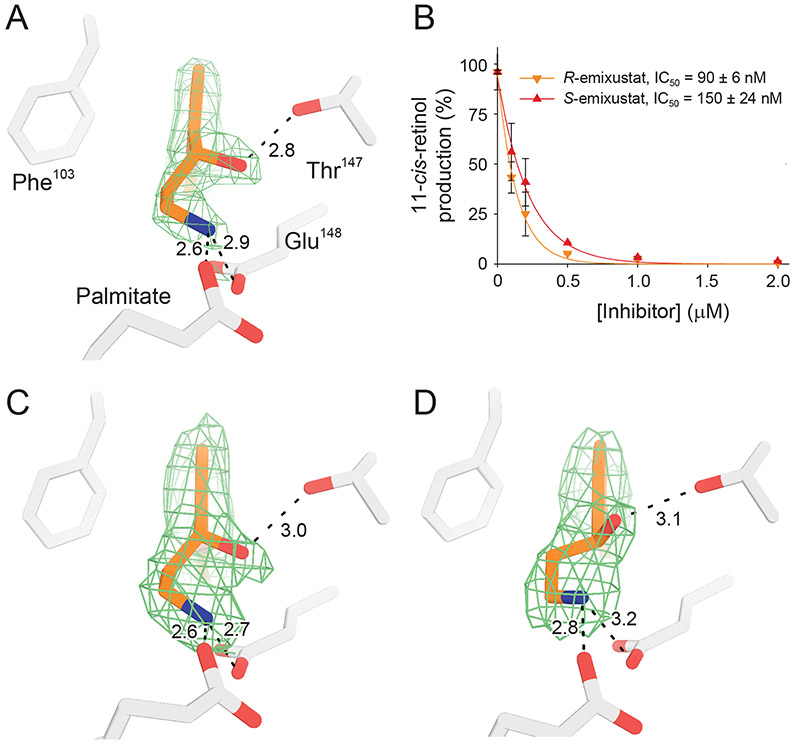
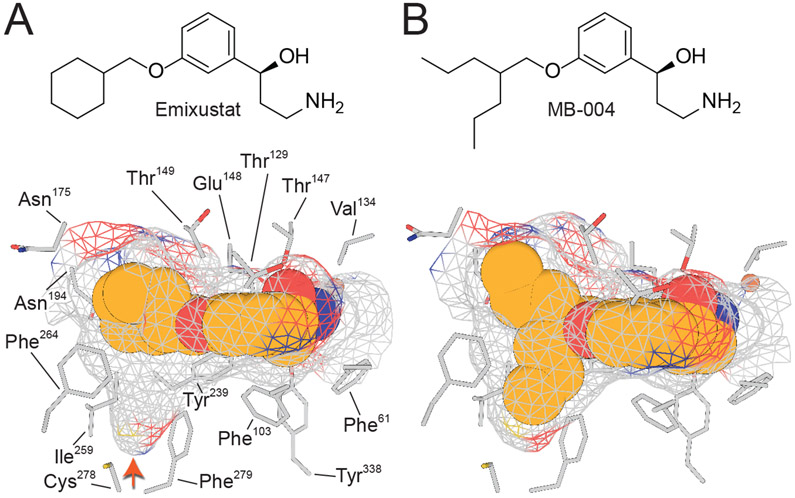
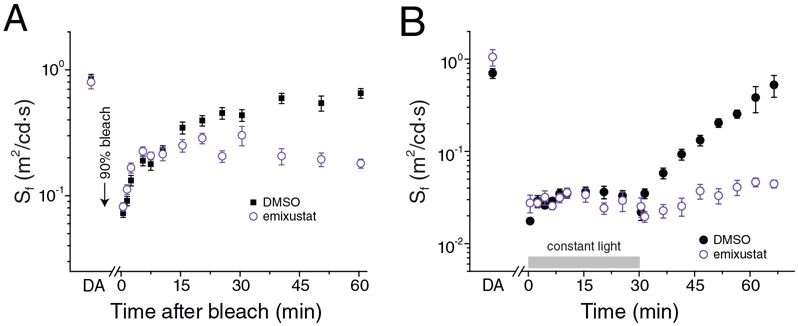
A resolution to the substrate orientation question came from structural studies of RPE65 in complex with the retinoid mimetic compounds, emixustat and MB-001, where both the retinoid mimetic as well as a co-purified palmitate molecule were simultaneously located within the RPE65 active site (Kiser et al., 2015). Emixustat is a non-retinoid inhibitor of RPE65 that was developed as a potential therapeutic for various retinal diseases (see Section 9 for further details). Despite its non-retinoid nature, emixustat exhibits strong correspondence to the retinoid backbone structure consistent with it having been derived from retinylamine, which is a rationally design transition state inhibitor of the isomerohydrolase based on its hypothesized carbocation intermediate (Golczak et al., 2005b) (see section 9.1.). Emixustat retains the positively charged primary amine functionality of retinylamine but its backbone structure is effectively one methylene group shorter than retinylamine making it an even closer analog of the putative C15 retinyl cation intermediate (Figure 6A). To restore greater retinoid character to this molecule, we substituted the terminal cyclohexyl moiety with a beta-ionone ring to give a molecule known as MB-001 (Figure 6A). Both emixustat and MB-001 have much greater water solubility as compared to retinoids and exhibit high affinity for the RPE65 active site as shown by their greater in vitro RPE65 inhibitory activity (Figure 6B and Table 1), which are the attributes that allowed their successful use for structural studies. Both emixustat and MB-001 are also more effective visual cycle suppressors than retinylamine in vivo (Figure 6C).
Figure 6. Structural and activity relationships among retinoids and non-retinoid RPE65 inhibitors.
A) Comparison of the chemical structures of C15 retinyl cation to retinylamine, emixustat, and MB-001 demonstrate the close relatedness of the latter two compound to the retinoid carbon backbone, indicating their ability to serve as retinoid mimetics for structural studies. B) Inhibitory effects of retinylamine, emixustat, and MB-001 on retinoid isomerase activity in vitro. Bovine RPE microsomes were used as the enzyme source for this assay. C) Inhibitory effects of retinylamine, emixustat, and MB-001 on visual chromophore regeneration in mice. Modified and used with permission from (Kiser et al., 2015).
Table 1.
Select RPE65 inhibitors and their in vitro potencies
| Compound name |
Chemical Structure |
In vitro IC50 towards RPE65 |
Reference |
|---|---|---|---|
| 13-cis-retinoic acid |

|
>100 μM | (Golczak et al., 2008) |
| all-trans-retinylamine |
|
0.65-2.03 μM | (Golczak et al., 2008) (Kiser et al., 2015) |
| Emixustat |

|
R: 91-175 nM (native RPE microsomes) or 4.4 nM (heterologously expressed RPE65) S: 150 nM |
(Bavik et al., 2015; Kiser et al., 2017; Zhang et al., 2015) |
| MB-001 |

|
396 nM | (Kiser et al., 2015) |
| MB-004 |

|
106 nM | (Kiser et al., 2017) |
| Compound 24 |

|
50 nM | (Blum et al., 2021) |
| Triacsin C |
|
500 nM | (Eroglu et al., 2016) |
| PBN |

|
61-100 μM | (Mandal et al., 2011; Poliakov et al., 2011) |
| CU239 |

|
6 μM | (Shin et al., 2018) |
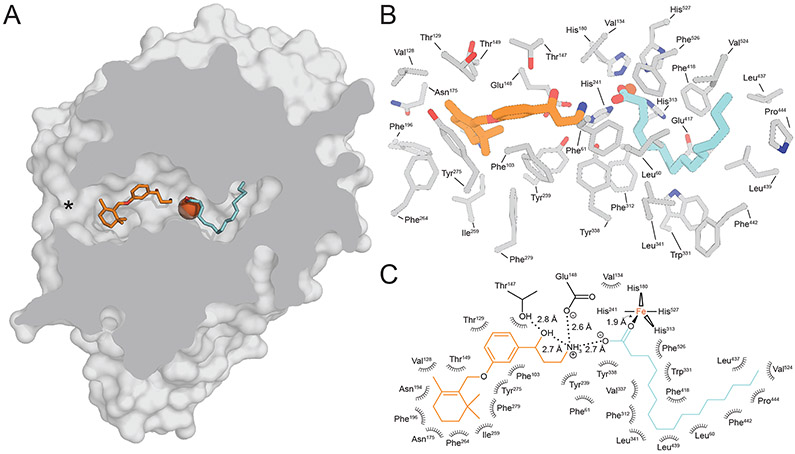
We observed that these retinoid mimetics bind within the proximal region of the active site whereas the palmitate molecule was found in the distal cavity with its carboxylate moiety forming a coordinate bond with the FeII center (Figure 7). In silico retinoid docking experiments were also consistent with this substrate orientation (Kiser et al., 2015). Based on this model, the acyl moiety of the ester is extracted out of the membrane and then “snakes” its way through the active site to reach its final catalytically relevant binding site within the distal pocket. The substrate binding process could be explained through the hydrophobic effect where displacement of water from within the hydrophobic distal cavity by the retinyl ester acyl chain could provide an energetic force to drive substrate abstraction from the membrane. The higher affinity binding of all-trans-retinyl palmitate to RPE65 in comparison to the corresponding hexanoate and acetate esters are consistent with the notion that the acyl chain plays an important role in driving substrate uptake (Maiti et al., 2005). This model further predicts that retinyl esters with acyl chains longer than palmitate are expected to be poorer substrates due to potential misalignment of the retinyl ester with respect to the catalytic groups. These ideas could be further tested by examining the in vitro isomerohydrolase activity of RPE65 towards a series of retinyl ester substrates of varying acyl chain length (Nikolaeva et al., 2009).
Figure 7. Structure of RPE65 in complex with the retinoid-mimetic MB-001 and palmitate.
A) Cut-away view of the RPE65 active site showing the binding sites for MB-001 (orange sticks) and palmitate (cyan sticks) within the proximal and distal regions of the active site cavity defined with respect to the active site opening at the membrane binding surface (marked by an asterisk). The iron ion is shown as a red-brown sphere. B) Detailed view of the residues forming the different regions of the active site cavity. C) Two-dimensional representation of the MB-001/palmitate interaction with the RPE65 active site. Modified and used with permission from (Kiser et al., 2015).
The carboxylate moiety of the palmitate ligand interacts with the iron center via a ~2 Å monodentate coordinate bond giving the iron a 5-coordinate trigonal bipyramidal structure, consistent with the XAS studies described in Section 5.2. This interaction provided direct structural support for the previously hypothesized role of iron acting to promote the retinyl ester hydrolysis step of the isomerohydrolase reaction. Hence, it appears that the iron center was repurposed, essentially without structural modification, from its original role as a dioxygen-activating cofactor in alkene-cleaving CCDs to a Lewis acid catalyst in RPE65 enzymes. Such use of redox-active metals for Lewis acid function is by no means unprecedented in nature (Valdez et al., 2014).
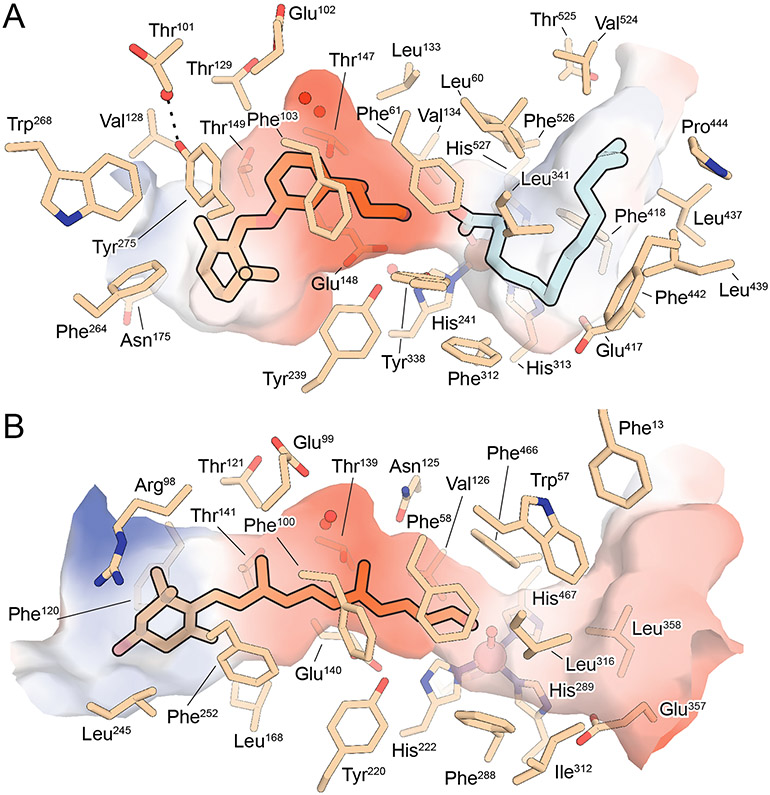
The proximal active site has two obvious features that could enable it to promote retinoid isomerization (Figure 8A). First, the cavity exhibits a pronounced bent shape, which is expected to promote the torsional movements of the retinoid necessary for it to change configuration from trans to cis. Indeed, in silico retinoid docking experiments showed that if the polyene double bond planarity restraints are artificially removed, the retinoid prefers to dock into the active site in an 11-cis-like conformation indicating that steric factors are important for promoting isomerization of the C11-C12 bond (Kiser et al., 2015). This idea was further supported by structure determination of NdCCD in complex with an apocarotenoid product of catalysis (Daruwalla et al., 2020). The proximal active site region of NdCCD, where the bound apocarotenoid is located, has striking similarities to that of RPE65, but is linear in structure as required to accommodate the all-trans-apocarotenoid substrate (Figure 8B). Comparison of the two active site structures revealed a key role for Tyr275 in generating the bent geometry observed for RPE65, versus the homologous Phe252 residue in NdCCD, which adopts a different side chain conformation as required for a linear substrate binding pocket.
Figure 8. Comparison of the active sites between RPE65 and the metazoan-like, apocarotenoid-cleaving CCD, NdCCD.
A) Structure RPE65 in complex with MB-001 (orange sticks) and palmitate (cyan sticks) (PDB accession code 4RSE). Residues the vicinity of the bound ligands are shown as wheat-color sticks. The iron ion is shown as a brown sphere and waters are red spheres. The electrostatic surface in the vicinity of the bound ligands is shown with red and blue representing negative and positive electrostatic potential, respectively. B) Structure of NdCCD in complex with an apocarotenoid product (orange sticks). Note the conserved negative electrostatic potential found within the retinoid/apocarotenoid binding sites of both proteins as well as the presence of buried water molecules in the central region of both active sites. Also note the conformational difference between Tyr275 in RPE65 and Phe252 in NdCCD that produces significantly different shapes near the substrate entrance as well the presence of the ‘RPE65-specific’ residues, Phe264 and Trp268, near Tyr275, which also contribute to the active site geometry. Modified and used with permission from (Daruwalla et al., 2020).
Second, the proximal region exhibits an overall anionic electrostatic potential along with strategically placed Phe103 and Thr147 residues that form a constriction within the binding pocket precisely at the location where the C11-C12 bond is expected to reside in the RPE65-retinyl ester Michaelis complex. These residues are arranged on opposite sides of the tunnel and are positioned appropriately within the negatively charged environment to stabilize a putative retinyl cation intermediate formed during the isomerohydrolase reaction. This proposal is supported by previous mutagenesis studies, which showed that Phe103 and Thr147 are key determinants of 11-cis versus 13-cis isomerization selectivity (Chander et al., 2012; Redmond et al., 2010; Takahashi et al., 2014). Interestingly, Phe103 and Thr147 are highly conserved residues in BCO enzymes as well as NdCCD (Figure 2) and the crystal structure of NdCCD revealed that this enzyme also exhibits strong negative electrostatic potential in its proximal active site cavity (Figure 8B) (Daruwalla et al., 2020). This structural and electrostatic similarity support the idea that the reactions catalyzed by RPE65 and its carotenoid-cleaving relatives could involve a common cationic reaction intermediate (Poliakov et al., 2020) that is stabilized electrostatically (Warshel et al., 2006). It also suggests that CCDs in the metazoan/metazoan-like clade have an intrinsic capacity to acts as alkene isomerases – a capacity that manifests only if the active site geometry is conducive to such a transformation. For example, the insect CCD NinaB is predicted to have a proximal active site structure similar to that of RPE65 and has independently acquired C11-C12 trans-cis isomerization activity coupled with C15-C15’ alkene cleavage activity (Oberhauser et al., 2008). BCO1 is also capable of simultaneously cleaving and isomerizing 9-cis-beta-carotene to form all-trans-retinal as the dominant product as opposed to the 1:1 mixture of 9-cis and all-trans-retinals expected for a pure double bond cleavage reaction (Maeda et al., 2011; Nagao and Olson, 1994).
The partially conserved structure of the retinoid/(apo)carotenoid binding site in RPE65 and its carotenoid-cleaving relatives raises the question of what distinguishing active site adaptations enable the unique substrate specificity and catalytic activity of RPE65. Insights into this question can be gained be considering the locations of the RPE65-characteristic residues delineated in Section 3. A majority of these residues are seen to cluster near the opening of the active site cavity as well as within the palmitoyl-binding pocket of the distal cavity. In the proximal cavity, these residues, which include Val240, Phe264, and Trp268, are major contributors to formation of the beta-ionone binding site nearby the critical Tyr275 residue mentioned above. Indeed, Tyr275 nearly meets the definition of an RPE65-specific character, missing it only due incomplete conservation at the equivalent position in the BCO2 clade. Ile192 is adjacent to Val240 and its specialization is likely a consequence of co-evolution with Val240. Again, these molecular adaptations appear to serve the purpose of generating an appropriately shaped active-site cavity to promote the trans-cis isomerization reaction. Notably, these residues are among those that were observed to adopt differing conformations between the inactive and active RPE65 structures (see Section 5.4) providing further support for their functional importance (Kiser et al., 2012). Within the distal binding pocket, a cluster consisting of Leu60, Leu439, and Val524 engage in van der Waals interactions with the bound palmitate ligand. In many cases, these aliphatic residues are substituted for aromatic residues in the carotenoid-cleaving enzymes likely reflecting the differing binding requirements of an aliphatic acyl chain and a carotenoid polyene. Gln64 does not directly line the active site but instead flanks, along with Leu60, the highly conserved ‘FDG’ motif described in the Section 3. It is tempting to speculate that specialization of its flanking residues could in some way confer RPE65-specific function to the ‘FDG’ motif, although a comparison of the RPE65 and NdCCD structures does not reveal any major structural differences for this sequence. Thr86 is located adjacent to Gln64 and its characteristic state in RPE65 again likely reflects co-evolution with Gln64. The remaining two RPE65-characteristic residues, Leu168 and Thr317, are more distantly removed from the active site and their relevance to RPE65 catalytic activity is not immediately clear.
6. Current View of the Isomerohydrolase Reaction Mechanism
The fundamental aspects of the isomerohydrolase reaction were worked out through elegant isotope labeling experiments, many of which were conducted prior to RPE65 being identified as the responsible catalyst (Kiser and Palczewski, 2010). The results of these studies are briefly summarized in Figure 9. A finding established early on and confirmed by several groups is that the oxygen atom bonded to C15 of the retinyl ester is lost during the isomerohydrolase reaction (Law and Rando, 1988; McBee et al., 2000; Redmond et al., 2010). This is accompanied by an inversion of the pro-chirality of C15 upon addition of the nascent hydroxyl moiety. Later experiments using 18O-labeled water demonstrated that the hydroxyl group found in the 11-cis-retinol product originates from bulk water (Kiser et al., 2009). Thus, the reaction strictly speaking is consistent with the definition of a hydrolysis reaction (with coupled geometric isomerization) as defined by the Enzyme Commission (EC) classification system:
with Aiso denoting an isomerized form of the A moiety of the substrate molecule, in this case the retinoid, and B denoting the carboxyl leaving group. The nature of this hydrolysis reaction is, however, rather different from a typical biological ester hydrolysis which involves cleavage of the acyl-oxygen bond. Instead it is the C15 alkyl-oxygen bond that is cleaved in the isomerohydrolase reaction (Figure 9). While apparently rare in biological systems, O-alkyl cleavage is well established in non-biological organic chemistry and generally occurs when the positive charge that develops on the alkyl carbon can be delocalized or otherwise stabilized. Such a situation is relevant to the retinyl ester as a C15 retinyl cation can be resonance stabilized through delocalization within the polyene pi electron system. Indeed, it is well known that retinyl esters undergo O-alkyl cleavage reactions in the presence of acids to form retinyl cation intermediates, e.g. the Carr-Price reaction (Blatz and Pippert, 1968a). Based on these similarities, it was proposed that the retinyl cation could be a key intermediate in the isomerohydrolase reaction that possesses the reduced bond order necessary for the geometric isomerization reaction to take place (Kiser et al., 2014). Additionally, the thermodynamically uphill trans-cis isomerization has been proposed to be driven by the energy released by ester hydrolysis (Deigner et al., 1989). However, it is important to note that the efficiency of 11-cis-retinol production is dependent on the presence of retinoid-binding proteins suggesting that product release from the enzyme may in fact be rate-limiting (McBee et al., 2000; Saari et al., 2001; Stecher et al., 1999; Winston and Rando, 1998).
Figure 9. Summary of isotope-labeling studies performed on the RPE65-catalyzed isomerohydrolase reaction.
Note the unusual cleavage of the C15-O bond as opposed to acyl bond cleavage more commonly observed in biological ester hydrolysis reactions. Used with permission from (Kiser et al., 2015).
Other intermediates besides the retinyl cation have also been proposed. These include a retinyl radical-cation (Poliakov et al., 2011) and a retinyl-nucleophile adduct (Deigner et al., 1989). The latter was ruled out based on the absence of cysteine or other suitably nucleophilic side chains in the RPE65 active site cavity. The former intermediate was proposed based on the observed inhibition of RPE65 activity by free radical spin trap compounds such as α-Phenyl-N-tert-butylnitrone (PBN) (see Section 9.1). Although such an intermediate cannot be completely ruled out, the PBN spin adduct was not spectroscopically verified, and it remains less clear how such an intermediate could be formed as compared to the straightforward mechanism of carbocation generation that accompanies O-alkyl hydrolysis. Additionally, it is known that RPE65 is subject to inhibition by diverse hydrophobic molecules (Stecher and Palczewski, 2000) and the relatively high concentration of PBN necessary for effective RPE65 inhibition is in line with a presumably low affinity binding to RPE65 (Poliakov et al., 2011) (Table 1).
The fact that RPE65 belongs to an oxygenase superfamily raises the possibility that its catalytic mechanism could involve O2, even though such a role is not apparent from consideration of the isotope labeling results described above or the non-redox nature of the overall reaction. We examined this possibility by carrying out the isomerohydrolase reaction in O2-depleted and O2-saturated buffers using Synechocystis apocarotenoid oxygenase (SynACO) as an internal oxygen-dependent control (Sui et al., 2015). As expected, SynACO activity was markedly impaired following argon-purging but was rescued and in fact augmented by reintroduction of O2 into the reaction buffer. By contrast, when RPE65 activity was measured following the same deoxygenation treatment, the amount of 11-cis-retinol formed was only marginally reduced (~14%), and the reduced activity was not restored by reintroduction of O2 into the reaction mixture. Moreover, O2 supplementation of the reaction mixture without prior deoxygenation actually depressed RPE65 activity by ~30% possibly due to oxidation of the iron cofactor, which is required to be in its ferrous form to be catalytically competent (Moiseyev et al., 2006). Considered together with prior biochemical data (Kiser et al., 2009; Law and Rando, 1988; McBee et al., 2000; Redmond et al., 2010), these results strongly indicate that RPE65 does not depend on O2 to exert its catalytic activity.
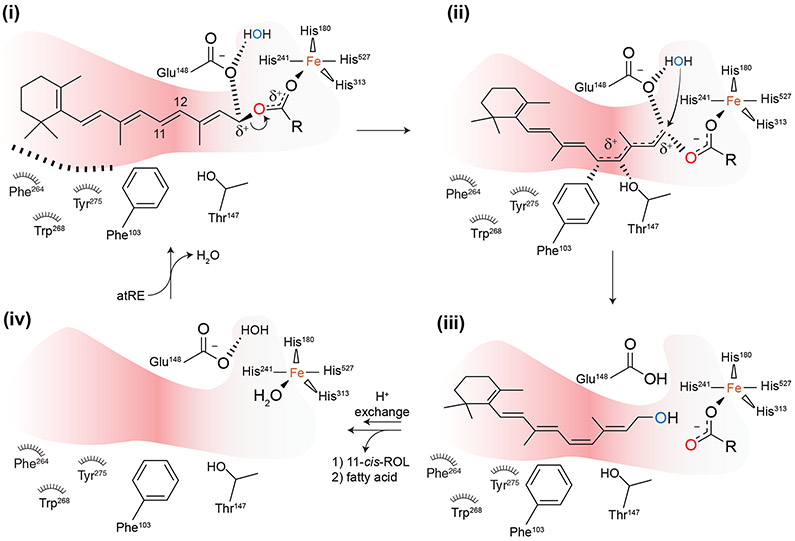
Knowledge of the mode of retinyl ester binding to the RPE65 active site has allowed a detailed mechanistic hypothesis of the isomerohydrolase reaction to be developed that reconciles much of the structural and biochemical data on RPE65 obtained to date. The proposed reaction follows an AAL1 SN1 mechanism (Smith and March, 2001) with the carbocation intermediate undergoing geometric trans-cis isomerization (Figure 10). Given the observed curved shape of the proximal active site, conferred in part by the RPE65-characteristic residues and Tyr275 described in Section 5.5, the retinoid moiety likely binds in a conformation in which steric factors promote trans-cis isomerization. The ester functionality binds close to the iron allowing its carbonyl oxygen to engage in a coordinate bond, which polarizes the ester making it a good leaving group. The developing positive charge on C15 during ester dissociation is likely stabilized by the Glu148 carboxylate group as mimicked by the observed interaction of this residue with the emixustat/MB-001 primary amine functionality. Delocalization of the retinyl cation onto the C11 atom is facilitated by the overall negative electrostatic potential of the proximal active site together with specific aromatic-cation and dipolar interactions involving Phe103 and Thr147, which as mentioned earlier, are well positioned to stabilize the carbocation intermediate at the retinoid C11 atom. This stabilization lowers the C11-C12 bond order allowing molecular rotation to a cis-like configuration. Repositioning of the C15 atom facilitates nucleophilic attack of nearby water molecule, which, following a proton transfer, generates the nascent 11-cis-retinol molecule. Given that the RPE65 active site is effectively a cul-de-sac, the 11-cis-retinol product must first dissociate back into the membrane followed by palmitic acid.
Figure 10. Proposed mechanism of the RPE65-catalyzed isomerohydrolase reaction.
Individual steps are described in Section 6 of the main text. Modified and used with permission from (Kiser et al., 2015).
Although the retinyl cation appears to be the most plausible intermediate in comparison to others that have been proposed to date, it still remains hypothetical and requires spectroscopic validation. Given the presumed transient nature of the intermediate, more detailed studies will likely require the use of model proteins that can be produced in large quantities necessary for stopped-flow or rapid freeze-quench absorbance (Blatz and Pippert, 1968a), fluorescence (Blatz and Pippert, 1968b), NMR (Kildahl-Andersen et al., 2003) or other advanced spectroscopic techniques.
7. Role of RPE65 in the Biosynthesis of Meso-xeaxanthin
The macula region of the primate retina exhibits a characteristic yellow color that is due to the accumulation of three different carotenoid species: lutein, zeaxanthin, and meso-zeaxanthin (Arunkumar et al., 2020). These compounds are believed to improve visual performance and protect the retina from blue light damage (Krinsky et al., 2003). Because primates, like other metazoan species, lack the ability to synthesize carotenoids de novo, these compounds must be obtained from dietary sources. Dietary sources of lutein and zeaxanthin are well known and include green leafy vegetables, mangos, cantaloupe, corn, and eggs among many others (Krinsky et al., 2003). By contrast, meso-zeaxanthin, an achiral diastereomer of zeaxanthin, has been suggested not to be present in significant amounts in foods commonly consumed by humans, although this idea is not without controversy (Nolan et al., 2013). It has been suggested that meso-zeaxanthin can be derived from zeaxanthin or lutein via either oxidation/reduction or isomerization mechanisms, respectively, but the enzymes potentially involved in these pathways had remained elusive (Krinsky et al., 2003).
In 2017, Shyam, Bernstein and colleagues reported that RPE65 possesses a second enzymatic activity, catalyzing a lutein double bond migration to form meso-zeaxanthin (Shyam et al., 2017). These investigators showed that overexpression of RPE65 in mammalian cell cultures conferred an ability to convert lutein but not zeaxanthin into meso-zeaxanthin, although the catalytic efficiency was apparently very low relative to the canonical isomerohydrolase activity of RPE65. The authors also studied the role of RPE65 in forming meso-zeaxanthin in vivo by injecting an emixustat-like RPE65 inhibitor (ACU-5200) in the yolk sac of a developing chicken embryo at embryonic days 17 and 19. The authors observed a dose-dependent reduction in meso-zeaxanthin levels in the RPE/choroid of the drug-treated embryos. Future research devoted to replicating and extending these studies will be important to help evaluate the physiological relevance of this meso-zeaxanthin biosynthetic pathway and the catalytic mechanism by which it is achieved.
8. RPE65 Protein Interactions
In some of the earliest studies on RPE65, it was recognized that this protein tends to form complexes with other proteins within the smooth endoplasmic reticulum (Bavik et al., 1992). One of the best characterized is the interaction of RPE65 with the RDH5 dehydrogenase enzyme that functions just downstream of RPE65 in the visual cycle pathway (Golczak et al., 2010; Hemati et al., 2005; Simon et al., 1995; Trudel et al., 2006). This biochemical relationship suggests that the RPE65-RDH5 interaction is functionally important for efficient transfer of retinoids and adequately rapid generation of 11-cis-retinal. In addition to RDH5, RPE65 has also been documented to form complexes with CRALBP and RGR-opsin (Bhattacharya et al., 2002; Golczak et al., 2010). Elucidation of the functional importance of these interactions has recently taken on an elevated importance given the renewed interest in the roles these proteins play in visual chromophore production for cone photoreceptors (Kolesnikov et al., 2021; Morshedian et al., 2019; Zhang et al., 2019).
Another set of proteins that RPE65 has been shown to interact with or otherwise be modulated by are the fatty acid transport proteins (FATPs) belonging to the SLC27 protein family (DiRusso et al., 2005). A yeast two-hybrid screen identified the cytosolic C-terminal sequence of FATP1 as a direct RPE65 binding partner (Guignard et al., 2010). FATP1 is expressed in the RPE and localizes to the smooth ER where RPE65 resides suggesting the interaction could be physiological. FATP1 inhibited the isomerohydrolase activity of RPE65, which could be mediated by a direct interaction or through its modulation of retinyl ester composition. The role of FATP1 in modulating RPE65 activity was further studied in loss- and gain-of-function mouse models. Fatp1−/− mice exhibited attenuated ERG b-wave recovery after photobleaching compared to wild-type mice but had no detectable difference in visual cycle kinetics (Chekroud et al., 2012). Interestingly, overexpression of human FATP1 in the RPE of mice caused visual cycle kinetic changes and altered ocular retinoid profiles, including an overaccumulation of all-trans-retinyl esters within retinosomes, along with an increased susceptibility to light-induced retinal degeneration (Cubizolle et al., 2017). FATP1 is known to be involved in the formation of lipid droplets (Xu et al., 2012), which may explain the increased retinosome content in these mice (Olzmann and Carvalho, 2019). The increased susceptibility to retinal phototoxicity in FATP1-overexpressing mice is somewhat unexpected given that FATP1 was previously found to inhibit RPE65 activity, which would be expected to be protective against light-induced retinal degeneration (Grimm et al., 2000). This light damage susceptibility was explained by the 25% greater number photoreceptors and 35% longer photoreceptor outer segments in the FATP1-overexpressing mice as compared to wild-type controls, which provides a greater amount of rhodopsin to mediate light damage (Cubizolle et al., 2017).
FATP4 has also been characterized in detail by Jin and colleagues for its ability to modulate RPE65 activity. This protein was identified as an RPE65 suppressor through cDNA library screening (Li et al., 2013). Parenthetically, the fatty acid elongation enzyme ELOVL1 was also identified as a modulator in this study although it hasn’t been characterized in further detail to date. FATP4, like FATP1, is expressed in the RPE and partially co-localizes with RPE65 in the smooth ER. FATP4 was also shown to be the most abundant FATP expressed in the RPE, at least at the mRNA level. Fatp4−/− mice exhibited more rapid visual cycle kinetics, which enhanced their susceptibility to light-induced retinal degeneration. Interestingly, the enhancement of visual cycle activity conferred by loss of FATP4 activity could be leveraged to augment visual function and cone photoreceptor viability in a mouse model of LCA2 that harbors the loss-of-function Arg91Trp RPE65 mutation, suggesting that FATP4 inhibition could be a viable approach to improving vision in individuals with RPE65-associated retinopathy (Li et al., 2020). Unlike FATP1, FATP4 is not known to directly interact with RPE65. Instead, it was suggested that FATP4 could compete with RPE65 for all-trans-retinyl palmitate binding or generate products such as all-trans-retinyl lignocerate that could compete with all-trans-retinyl palmitate for binding to RPE65 but not serve as viable substrates. Long acyl chain retinyl esters, such as retinyl lignocerate, generated by FATP4 are unlikely to serve as viable RPE65 substrates given the active site constraints mentioned in Section 5.5. However, these compounds could still accumulate in storage droplets, similar to the retinosome accumulation that was observed in the FATP1 overexpression mouse model, which would sequester the retinoid outside of the circulating visual cycle pool and reduce visual chromophore production.
The apparently transient and low-affinity interactions of RPE microsomal proteins with RPE65 together with the sensitivity of RPE65 activity to detergents needed for purification of putative complexes have hampered efforts to characterize the structures of these complexes at high resolution. The advent of detergent-free solubilization methods using styrene maleic acid co-polymer (SMA), a molecule capable of forming native lipid nanodisks (Dorr et al., 2016), may provide a method to purify RPE65 in its native protein and lipid environment (Sander et al., 2021).
9. The Pharmacology of RPE65 Inhibition
9.1. Overview of RPE65 Inhibitors Characterized to Date
Efforts to develop inhibitors of RPE65 have their origin in two distinct research goals. The earliest studies aimed to develop chemical biology tools to aid in understanding the biochemistry of the retinoid isomerohydrolase reaction and the physiology of RPE65 function particularly as it relates to cone photoreceptor activity (Golczak et al., 2005b; Muniz et al., 2009). Following the discovery that RPE65 activity is correlated with the formation of retinal lipofuscin, a heterogenous mixture of retinal-derived compounds believed to drive progression of certain retinal diseases (Katz and Redmond, 2001; Kim et al., 2004), efforts were made to develop RPE65 inhibitors for potential therapeutic use that could protect against retinal disease (Maeda et al., 2006; Sieving et al., 2001). It is noteworthy that a major bisretinoid component of retinal lipofuscin, A2E, has been shown to directly inhibit RPE65, which may underlie some of the visual impairment experienced by individuals with Stargardt disease (Moiseyev et al., 2010).
The earliest efforts focused on retinoids and related isoprenoid compounds (Golczak et al., 2005b; Law and Rando, 1989; Maiti et al., 2006) (Table 1). Of these, retinylamine exhibited the most potent and efficacious RPE65 inhibition both in vitro and in vivo (Golczak et al., 2008) and has served as an important chemical biology tool for study of visual cycle biology (Babino et al., 2015; Schonthaler et al., 2007). Retinylamine was rationally designed to act as a transition-state inhibitor of RPE65 based on the hypothesis that the retinoid isomerohydrolase reaction occurs through a retinyl cation intermediate (Golczak et al., 2005b; McBee et al., 2000). Notably, this compound is subject to amidation by LRAT (Golczak et al., 2005a) and also interacts with cellular retinol-binding protein (Silvaroli et al., 2016) and possibly other retinoid-interacting proteins, which may have been factors that initially obscured the identification of RPE65 as a molecular target of retinylamine (Golczak et al., 2005b).
Retinylamine was later licensed by Acucela Inc. and used as a basis molecule to develop the non-retinoid RPE65 inhibitor, emixustat, discussed earlier in Section 5.5 (Bavik et al., 2015). Although not a retinoid, emixustat retains substantial structural similarity to retinylamine including its core carbon backbone and analogously placed primary amine functional group (Figure 6). However, emixustat is a substantially more potent RPE65 inhibitor than retinylamine both in vitro and in vivo (Kiser et al., 2015; Zhang et al., 2015) (Figure 6 and Table 1). Emixustat and its derivatives remain the best characterized RPE65 inhibitors developed to date and will be discussed in detail in the next sections.
Other RPE65 inhibitors that are chemically unrelated to retinoids have been discovered over the past decade (Table 1). α-phenyl-N-tert-butylnitrone (PBN) and its derivatives are free radical spin trap molecules that inhibit RPE65, albeit relatively weakly, and protect against light-induced retinal degeneration at least in part by slowing visual cycle kinetics (Chander et al., 2012; Mandal et al., 2011; Poliakov et al., 2011; Stiles et al., 2015). CU239, a ureidothiazole-containing compound identified through in silico crystal structure docking, is a moderately potent competitive RPE65 inhibitor that also exhibits in vivo activity in suppressing visual cycle activity and protecting against light-induced retinal degeneration (Shin et al., 2018). Triacsin C, a linear nitrosohydrazine-containing molecule and established inhibitor of fatty acyl:CoA ligases, was found to be another moderately potent competitive inhibitor RPE65. The non-polar tail of triacsin C may bind to the distal region of the RPE65 active site, similar to the linear detergent C8E6 (Blum et al., 2021), with its nitroso group coordinated to the iron center, thereby blocking entry of retinyl ester substrates.
9.2. Molecular Mechanism of RPE65 Inhibition by Emixustat and its Derivatives
For clarity of discussion, it is helpful to divide the emixustat molecule into three regions: 1) the gamma-hydroxyamine functionality, 2) the central phenyl ring, and 3) the methoxycyclohexyl moiety. The contribution of each of these regions to RPE65 active site affinity has been studied extensively by in vitro and in vivo RPE65 activity assays as well as X-ray crystallography. As described in Section 5.5., emixustat binds in the proximal region of the RPE65 active site cavity nearest to the active site opening.
The gamma-hydroxyamine moiety is located deepest within the active site cavity where it engages in electrostatic interactions with Glu148, Thr147, and an oxygen atom of an iron-bound palmitate carboxyl moiety. The gamma-hydroxyamine features a chiral center at the alcohol functional group, which has an R configuration in emixustat. Co-crystallization of racemic emixustat with RPE65 led to predominant binding of the R-isomer suggesting it is the more active isomer (Figure 11A). This idea was further evaluated by synthesis and activity testing of pure S and R isomers of emixustat, which revealed a roughly two-fold greater potency of the R-isomer (Zhang et al., 2015) (Figure 11B). Crystal structures of RPE65 in complex with the two isomers revealed somewhat more favorable hydrogen bonding interactions involving the gamma-hydroxyamine moiety for the R-isomer which may explain the affinity difference (Figure 11, C and D). Stronger inhibitory activity for the R-isomers of emixustat derivatives have also been observed (Blum et al., 2021). A molecule known as MB-002, in which the terminal amine is replaced with a hydroxyl group, is also an effective, albeit less potent, RPE65 inhibitor (Zhang et al., 2015). Interestingly, the RPE65 MB-002 co-crystal structure revealed that this compound can bind to both the proximal (S-isomer) and distal (R-isomer) regions of the active site cavity, although binding in the distal pocket may only occur at high MB-002 concentrations as used for the crystallization experiment.
Figure 11. Analysis of emixustat enantiomer binding to and inhibition of RPE65.
A) RPE65 crystals obtained in the presence of racemic emixustat show preferential binding of the (R) stereoisomer. B) In vitro RPE65 activity assays demonstrate a roughly two-fold greater potency of the R isomer of emixustat as compared to the S isomer. Crystal structures of RPE65 in complex with pure R-emixustat (C) and pure S-emixustat (D) demonstrate more a more favorable hydrogen bonding pattern for the former stereoisomer, thus explaining its greater inhibitory potency. The mesh in panels A, C, and D represents unbiased σA-weighted omit mFo-DFc electron density contoured at 3 RMSD. Dashed lines indicate hydrogen bonds with bond lengths given in Angstroms. Modified and used with permission from (Zhang et al., 2015).
The central aryl moiety is positioned at about the midpoint bend of the proximal active site region where it engages in van der Waals interactions with several nearby side chains and two solvent molecules as well as pi-pi interactions with Phe103 and Tyr275. Bulky substitutions at the aryl moiety not surprisingly greatly impair active site binding and weaken or abolish RPE65 inhibitory activity (Kiser et al., 2017). A fluorine scan of the aryl moiety showed that fluoro-substitution at the 4-position substantially improves inhibitory activity in vitro (Blum et al., 2021). Crystal structures of such derivatives in complex with RPE65 showed that the 4-fluoro moiety engages in a ~3 Å en face interaction with the Tyr275 aryl ring. Quantum chemical calculations suggest that the greater binding affinity of these derivatives arises from favorable van der Waals interactions between the fluoro and aryl moiety, as opposed to electrostatic effects found for some systems in which the aryl ring is electron deficient (Li et al., 2017).
The terminal methoxycyclohexyl moiety of emixustat resides near the active site entrance and weakly interacts with the protein as evidenced by the comparatively weak electron density support, higher B-factors, and multiple conformations observed for this moiety (Kiser et al., 2015; Zhang et al., 2015). The cyclohexyl binding region features an additional unused pocket that was targeted to help stabilize the terminal moiety and improve binding affinity (Figure 12A). A molecule known as MB-004, in which the cyclohexyl moiety of emixustat is replaced by a dipropanyl group, was found to be a highly potent RPE65 inhibitor and exerted strong in vivo visual cycle suppression (Kiser et al., 2017). The crystal structure of RPE65 in complex with MB-004 showed occupancy of a propanyl group within the unused pocket, which was well-resolved in the electron density maps, suggesting that it makes a substantial contribution to MB-004 affinity for the RPE65 active site (Figure 12B).
Figure 12. Comparison of emixustat and MB-004 binding to the RPE65 active site.
A) The cyclohexyl ring of emixustat is located close to a water-filled pocket within the membrane-proximal region of the active site pocket. Owing to its bulkiness, the cyclohexyl moiety is unable to occupy a nearby apolar pocket (red arrow). B) One half of the MB-004 dipropyl moiety occupies the aforementioned hydrophobic cavity forming non-bonded contacts with residues lining the pocket. The other half of this moiety resides in approximately the same position as the cyclohexyl moiety of emixustat. Ligands are shown as van der Waals spheres with carbon, nitrogen and oxygen atoms colored orange, blue and red, respectively. Used with permission from (Kiser et al., 2017).
10. Probing RPE65 Visual Cycle Physiology with Pharmacological Inhibitors
10.1. Introduction
The pharmacological inhibitors described in Section 9 have proven to be critical tools for understanding the physiological roles of RPE65 in the visual system. As compared to genetic knockout/knockdown approaches, pharmacological inhibitors in general are more readily applied in an acute fashion in adult animals, which minimizes the contribution of developmental, degenerative, or compensatory effects to the observed phenotype. The advent of potent and selective RPE65 inhibitors has enabled this pharmacological approach to be applied to the study of RPE65 function in various aspects of visual system physiology, including its role in cone photoreceptor function, inner retinal photoreception, and macular pigment biosynthesis (discussed in Section 7).
10.2. Role of RPE65 in Cone Photoreceptor Physiology
The question of RPE65’s role in support of cone photoreceptor-mediate vision is one that has a long history but was not fully resolved through genetic experimental approaches. It is well established that cone ERG responses are severely attenuated or extinguished in mice (Maeda et al., 2009; Redmond et al., 1998; Seeliger et al., 2001), dogs (Acland et al., 2005), and humans (Jacobson et al., 2007; Jacobson et al., 2009) with loss-of-function mutations in RPE65 and that cone visual pigments fail to form in the Nrl−/− Rpe65−/− mouse model (Feathers et al., 2008; Wenzel et al., 2007) in which the photoreceptor population is predominantly cone-like in contrast to the normal mouse retina which is rod dominated. Additionally, the enrichment of RPE65 expression within the portion of the RPE in contact with the cone-rich fovea (Jacobson et al., 2007) as well as the enhanced isomerohydrolase activity observed for RPE65 from the cone-dominant chicken as compared to orthologs from rod-dominant rodents (Moiseyev et al., 2008) are consistent with adaptive mechanisms whereby classical visual cycle activity can be adjusted based on the needs of the retinal photoreceptor population.
Conversely, there is abundant biochemical and electrophysiological evidence suggesting that the retina possesses non-RPE65-dependent mechanisms to support cone, and potentially rod, visual pigment regeneration that are based within Müller glia (Mata et al., 2002; Morshedian et al., 2019; Muniz et al., 2009; Wang et al., 2009), the RPE (Zhang et al., 2019) or the photoreceptors themselves (Frederiksen et al., 2021; Kaylor et al., 2017). As noted in Section 3, the emergence of ancestral bleaching cone pigments during chordate evolution likely preceded the appearance of RPE65 and the classical visual cycle by several million years which points to alternative chromophore regeneration pathway(s) that could be retained in extant species (Kusakabe et al., 2001; Lamb, 2013).
The role of continuous RPE65 activity in cone vision was first addressed by Schonthaler, von Lintig and colleagues by testing the impact of retinylamine treatment on photoreceptor function in the cone-dominated larval zebrafish retina 5 days post-fertilization (Schonthaler et al., 2007). These investigators showed that retinylamine reduced, but did not eliminate, 11-cis-retinal regeneration following a deep photobleach. Importantly, the effect of retinylamine was not augmented by simultaneous morpholino knockdown of RPE65a (the ohnolog expressed within the RPE), which confirmed the dose of retinylamine used in the study was sufficient to abolish RPE65 activity. Zebrafish are known to store 11-cis-retinyl esters in their RPE which could serve as a source of 11-cis-retinal under conditions where RPE65 is acutely inhibited. This hypothesis was investigated several years later by the same laboratory again using retinylamine to acutely block RPE65 activity (Babino et al., 2015). These investigators found that 11-cis-retinyl esters are consumed during exposure to bright light and resynthesized in an RPE65-dependent manner during dark adaptation. Thus, the residual visual chromophore production in zebrafish in the setting of acute pharmacological blockade of RPE65 activity can be attributed in part to the existence of storage forms of visual chromophore.
Analogous studies have been carried out in Gnat−/− mice, which lack rod photoresponses (Calvert et al., 2000), using emixustat-family compounds to acutely block RPE65 activity (Kiser et al., 2018). Following a brief step bleach of ~95% of the visual pigment, it was found that acute RPE65 inhibition with emixustat does not impact the initial phase of cone photoreceptor sensitivity recovery, which is believed to reflect intraretinal visual cycle activity, while the slower RPE-dependent phase was significantly delayed (Figure 13A). However, when the bleaching light exposure was presented over the course of 30 min where multiple cycles of bleaching and regeneration could occur, all phases of pigment regeneration were significantly delayed (Figure 13B). This delay suggested that the mouse retina contains storage form(s) of 11-cis-retinoid that can be mobilized to quickly regenerate cone pigment but are subject to depletion during extended bright light exposure. Because the normal mouse retina does not contain measurable 11-cis-retinyl esters as described above for zebrafish, 11-cis-retinoids bound to CRALBP were considered a potentially relevant source of visual chromophore. This hypothesis was further tested in Rlbp1+/− mice that have a ~50% reduction in CRALBP expression both in Müller glia and the RPE (Kolesnikov et al., 2021). With the classical visual cycle intact, there was no observable difference in cone photoreceptor dark adaptation following a strong photobleach between wild-type and Rlbp1+/− mice. However, acute blockade of RPE65 activity using MB-001 revealed a significant slowing of cone photoreceptor dark adaptation caused by CRALBP deficiency following either acute or extended light exposure bleaching conditions. These results demonstrated that holo-CRALBP plays a role in cone visual pigment regeneration independent of RPE65 function. This study also demonstrated that genetic ablation of CRALBP expression combined with acute pharmacological blockade of RPE65 with MB-001 was sufficient to essentially eliminate cone photoreceptor dark adaptation following a deep photobleach further highlighting the direct role RPE65 plays in supplying cone photoreceptors with visual chromophore.
Figure 13. Effects of acute RPE65 inhibition by emixustat on mouse cone dark adaptation in vivo.
A) The recovery of cone b-wave flash sensitivity following a 90% bleach was compared between vehicle- and emixustat-treated Gnat1−/− mice The late, RPE visual cycle-driven phase of cone dark adaptation was suppressed by emixustat. By contrast, the early, Müller cell-driven phase of the cone dark adaptation curve was unaffected indicating the presence of visual chromophore sources that are not acutely dependent on RPE65 activity. B) Impact of RPE65 inhibition by emixustat on in vivo Gnat1−/− mouse cone photoreceptor responses during and after prolonged exposure to background light. Cone b-wave sensitivity was monitored during a 30 min exposure to bright (300 cd/m2) white background light (gray bar), and then for 35 min in darkness. Compared to vehicle controls, cones from mice treated with emixustat were equivalently desensitized by the background light but had largely suppressed subsequent dark adaptation. Modified and used with permission from (Kiser et al., 2018)
The impact of acute inhibition of RPE65 activity with MB-001 on cone function was also studied in wild-type mice and cone-dominant ground squirrels (Kiser et al., 2018). Following three days of treatment with doses of MB-001 that are sufficient to abolish RPE65 activity, as measured by scotopic ERG (mice) or in vitro RPE65 activity assays using isolated RPE (ground squirrels), it was observed that cone dark adaptation was slowed but not abolished in both animal models. The ground squirrel in particular retained relatively rapid cone dark recovery as exhibited by the essentially normal cone ERG responses obtained 10 min after a strong photobleach.
The impact of RPE65 inhibition by emixustat on cone vision was further tested in cone-dominant zebrafish larvae through the use of optokinetic response (OKR) measurements, which assess visual reflexes (Ward et al., 2020). Ward, Kennedy and colleagues found that emixustat inhibited photopic, cone-based OKRs when fish were tested immediately following a dark adaptation period whereas responses following a prolonged light adaptation period were not affected by RPE65 inhibition. Interestingly, the sustained photopic response was achieved in the face of continued low levels of 11-cis-retinal caused by RPE65 inhibition, which is consistent with the ability of cones to mediate functional vision with a large percentage of their visual pigments bleached (Rodieck, 1998).
Taken together, these pharmacological data are consistent with the idea that continuous RPE65 activity is critical to ensure that cones receive adequate visual chromophore to support their normal function. At the same time, the data clearly demonstrate the availability of alternative visual chromophore sources for cones. These include the 11-cis-retinoid storage forms (11-cis-retinyl esters and/or holo-CRALBP depending on the species) that are generated at least in part by RPE65-dependent classical visual cycle activity as well as RPE65-independent mechanisms that seem, for the most part, to depend on light as the energy source to achieve trans-cis retinoid isomerization (Chen et al., 2001; Frederiksen et al., 2021; Kaylor et al., 2017; Morshedian et al., 2019; Ward et al., 2020; Zhang et al., 2019). However, all of these alternative chromophore synthesis pathways ultimately depend on RPE65 activity to function, as the phenotypes of animal models with RPE65 loss-of-function tell us (Acland et al., 2005; Redmond et al., 1998). As discussed in Section 4.2., this is likely due to the fact that an absence of RPE65 isomerohydrolase activity leaves the powerful retinyl ester synthase activity of LRAT unchecked resulting in the sequestration of ocular retinoids away from these alternative pathways.
10.3. Role of RPE65 in Melanopsin-Based Photoresponses
In addition to the photoreceptor visual pigments, the melanopsin protein located in intrinsically photosensitive retinal ganglion cells (ipRGCs) is another well characterized light-sensing opsin protein that depends on 11-cis-retinal for its function (Terakita, 2005). Melanopsin is important for entrainment of the circadian clock and also plays an important role, along with (mainly rod) photoreceptors, in the pupillary light response (Keenan et al., 2016; Lucas et al., 2003; Panda et al., 2003). The question has arisen as to the dependence of melanopsin on RPE65 and the classical visual cycle to obtain its chromophore. Again, pharmacological inhibitors of RPE65 activity have proven valuable tools to interrogate this question.
Tu, Van Gelder and colleagues studied the impact of RPE65 inhibition with retinylamine treatment or via genetic means on pupillary light responses in mouse models with or without an intact outer retina (Zhao et al., 2016). Whereas retinylamine treatment dramatically reduced PLR sensitivity in wild-type mice in which the PLR is driven by both rods and ipRGCs, it had little impact on PLRs from rd/rd mice in which rods and most cone degenerate. This finding suggested that the impact of acute RPE65 inhibition on PLRs in wild-type mice arose entirely from blockade of rod function while ipRGC responses were spared. This finding is consistent with the biochemical features of melanopsin as a bistable opsin capable of photic regeneration of its active state.
A related study by Zhao, Wong, and colleagues using 13-cis-retinoic acid and PBN to inhibit RPE65 (along with other components of the visual cycle) found that visual chromophore synthesized in the RPE does in fact play a role in melanopsin-based PLRs during intense illumination when the presumed rate of melanopsin bleaching outstrips its photoreversion capacity (Zhao et al., 2016). These findings were mirrored in a follow-up study using Rpe65−/− mice instead of PBN and 13-cis-retinoic acid, thus supporting a role for RPE65 in maintenance of melanopsin-based photoresponses under strong or prolonged illuminance (Harrison et al., 2021). This same study also showed that CRALBP present in the Müller glia plays an important role in transporting visual chromophore generated in the RPE across the retina to the ipRGCs and also likely serves as a storage pool of readily mobilized 11-cis-retinal as CRALBP is well known to concentrate in the Müller glia end-feet in close proximity to the RGCs (Saari et al., 2001). This study thus adds support to the idea that visual chromophore found within Müller cell-expressed holo-CRALBP has its origins at least in part in the RPE65-dependent visual cycle (Kiser et al., 2019).
10.4. Impact of RPE65 Inhibition by Emixustat in Human Subjects
The use of emixustat in clinical studies has provided a unique opportunity to observe the impact of selective, acute and chronic RPE65 inhibition on the function of the human visual system. Single doses of emixustat (from 2 to 75 mg) resulted in a graded, time-dependent suppression of rod ERG recovery following a strongly rod-suppressing photobleach while no detectable impact on cone photoreceptor ERG responses was observed (Kubota et al., 2012). Similar ERG findings have been reported with chronic dosing regimens and in various patient populations (Dugel et al., 2015; Kubota et al., 2014; Kubota et al., 2020). The lack of an effect on cone function may be due to either incomplete RPE65 blockade at the doses studied and/or the availability of cone-specific sources of visual chromophore that do not acutely depend on RPE65 activity. An interesting common side effect of emixustat treatment is abnormal color vision (dyschromatopsia). It was speculated that this effect could arise from a modulatory effect of suppressed rods on color vision pathways (Kubota et al., 2012).
As mentioned in the introduction to this section, partial suppression of RPE65 has been theorized to be therapeutically beneficial for retinopathies in which bisretinoid-based lipofuscin plays a role in the disease pathogenesis. Geographic atrophy is one such disorder where the impact of emixustat treatment was evaluated in a randomized, double-blind, placebo-controlled clinical study. Unfortunately, daily emixustat treatment for 2 years did not achieve its primary efficacy endpoint in slowing the annual growth rate of the total geographic atrophy area (Rosenfeld et al., 2018). This molecule continues to be studied in other diseases including Stargardt macular dystrophy (Kubota et al., 2020) and diabetic retinopathy (Kubota et al., 2019; Kubota et al., 2021).
11. Expanding Involvement and Mechanisms of RPE65 Mutations in Retinal Disease
11.1. Overview of RPE65-associated Retinal Disease
The involvement of RPE65 in retinal disease was discovered in 1997 by two groups that identified homozygous RPE65 mutations in patients with autosomal recessive retinitis pigmentosa (RP20, OMIM #613794) or Leber congenital amaurosis (LCA2, OMIM #204100) (Gu et al., 1997; Marlhens et al., 1997). Over the past decade, knowledge of pathogenic RPE65 mutations has expanded greatly. There are now nearly 200 distinct RPE65 mutations in the Human Gene Mutation Database including over 115 sites of missense mutations. These and additional pathogenic missense mutations listed in ClinVar as well as recent publications (Gao et al., 2021; Lopez-Rodriguez et al., 2021) are summarized in Figure 14. Our understanding of the pathogenic effects of these recessive mutations has been facilitated by the generation of mouse models including the Arg91Trp RPE65 mouse (Samardzija et al., 2008) and the Pro25Leu RPE65 mouse (Li et al., 2015), which feature variable degrees of visual cycle deficiency. Not limited to recessive RP/LCA, RPE65 mutations have also been linked to recessive fundus albipunctatus (a disease normally associated with RDH5 or RLBP1 mutations) (Schatz et al., 2011; Yang et al., 2017) and, in one case, to an atypical form of dominant RP (Bowne et al., 2011). In mice, the Rd12 mutation in RPE65 (Pang et al., 2005), which results in a premature stop site at codon 44, has been found to exert a semidominant negative effect on visual function possibly due to effects of the mutant transcript on translational machinery (Wright et al., 2014). A detailed description of the disease phenotypes in RPE65-associated RP/LCA patients is beyond the scope of this paper but has been expertly reviewed in recent publications (Cideciyan, 2010; Kiang et al., 2020; Lopez-Rodriguez et al., 2021; Tsang and Sharma, 2018; Verbakel et al., 2018). Additionally, several different approaches to the rescue of vision in RPE65-associated RP/LCA are currently under investigation, some of which being pharmacological bypass of the defective visual cycle (Kiang et al., 2020; Maeda et al., 2013), pharmacological stabilization of mutant RPE65 structure (Li et al., 2014; Li et al., 2016), recently FDA-approved RPE65 gene augmentation therapy (Russell et al., 2017), and gene editing approaches (Jo et al., 2019; Suh et al., 2021). Some of these approaches have recently been reviewed (Dias et al., 2018; Kiser and Palczewski, 2020; Roska and Sahel, 2018).
Figure 14. Two-dimensional topology diagram of RPE65 mapped with corresponding residues showing sites found to be mutated in retinal disease.
Dashed lines indicate residues involved in iron coordination.
A variety of mechanisms may be at play in causing RPE65 protein dysfunction depending on the specific mutation. As shown in Figure 14, most of the known functional regions of the RPE65 molecule have been identified as being mutated in retinal disease. Notably, the beta-propeller fold adopted by RPE65 likely makes this protein intrinsically sensitive to protein folding detects as complete folding is likely required before the protein achieves a stable structural state. Hence, some of the point mutants described in Figure 14 could cause protein dysfunction not only through distortion of the folded protein structure but also through alterations in folding kinetics which could lead to partially-folded, aggregation-prone (due to exposed beta sheet edges) intermediates that are subject to proteolytic degradation.
The lack of a method for crystallization of recombinantly produced RPE65 has complicated efforts to understand the detailed structural basis for how specific point mutation lead to protein dysfunction. This barrier has been circumvented through the use of an apocarotenoid-cleaving RPE65 homolog from Synechocystis (SynACO) as a model system that is amenable to high-resolution structural analysis. Below, I highlight two studies employing this model system that have provided insights into how both recessive- and dominant-acting point mutations cause alterations in the RPE65 structure.
11.2. Structural Impact of a Glu148Asp-equivalent Mutation Associated with LCA2
As described in Section 5.2., the iron-coordinating His and Glu residues of CCDs are highly structurally conserved amongst all members of the superfamily including RPE65 which enabled the use of SynACO as a model to study the impact of a second-sphere Glu148Asp mutation on the protein structure and iron coordination (Sui et al., 2016). This site was of particular interest since the structural role of the second sphere Glu residues was not well-established prior to this study and the associated pathogenic mutation (Hanein et al., 2004; Zernant et al., 2005) produced a charge-conserved substitution. Mutation of the corresponding Glu residue of SynACO (residue 150) to an Asp residue produced a protein that was expressed nearly as well as the wild-type protein and was readily crystallized, both of which suggesting that the mutation does not cause a global disruption of the protein structure. However, the catalytic efficiency of the mutant protein was reduced to ~9% of the wild-type enzyme, demonstrating that mutation is also detrimental within the context of the SynACO structure. The crystal structure of Glu150Asp SynACO was determined at 2.8 Å resolution. Electron density for the Asp150 side chain showed that it had flipped to a vacant site opposite the iron center relative to the normal positioning of the Glu150 side chain in the wild-type enzyme (Figure 15). In this conformation, no hydrogen bond was formed between Asp150 and His238 (equivalent to His240 in RPE65) resulting in disruption of coordination between His238 and the iron center. Although the His238 side chain was well resolved, it was oriented away from the metal, clearly demonstrating an iron coordination defect in the mutant protein. Interestingly, a Glu150Gln mutant, in which a hydrogen bond interaction with His238 was maintained, also showed drastically reduced catalytic activity accompanied by an iron coordination defect. This finding along with data from other biochemical studies on RPE65 (Nikolaeva et al., 2010; Takahashi et al., 2005), indicate that the negative charge of the second sphere Glu residues is critical for their role in supporting iron coordination by the primary sphere His residues for CCDs in general.
Figure 15.
Impact of a Glu148Asp-equivalent mutation on the structure of the CCD iron center. A) Stereo-view of the iron center of an RPE65 homolog, SynACO, showing the interaction of Glu150 (equivalent to Glu148 in RPE65) with the iron-coordinating His238 residue (equivalent to His241 in RPE65). B) The Glu150Asp mutation alters the conformation of His238 resulting in a loss of its interaction with the iron ion and lower iron occupancy in the active site. The blue mesh in both panels represents the final σA-weighted 2Fo-Fc maps contoured at 1 RMSD. Modified and used with permission from (Sui et al., 2016).
11.3. Structural Insights into the Mechanism of a Dominant-acting RPE65 Mutation
To date, there has been only one RPE65 mutation linked to dominant retinitis pigmentosa which gives rise to an Asp to Gly substitution at position 477 in the human sequence. This mutation was originally reported in an Irish family (Bowne et al., 2011) and has since been reported in several additional unrelated families (Hull et al., 2016; Jauregui et al., 2020; Jauregui et al., 2018). The mutation causes an atypical form of RP with features of choroideremia and variable penetrance (Kiang et al., 2020). Four different mouse models of this disease have been generated, none of which closely mimic the human disease phenotype (Choi et al., 2018; Kiang et al., 2020; Li et al., 2019; Shin et al., 2017).
Asp is conserved at position 477 in RPE65 from primates to sea lamprey, consistent with an important structural or functional role for this residue. Interestingly, Gly is found at a position equivalent to position 477 of human RPE65 in some CCD enzymes including human BCO1 and BCO2 as well as SynACO, indicating that Gly is tolerated for protein structure and function in the presence of a suitable surrounding sequence. Asp477 is found within a beta-turn element connecting strands 32 and 33 of the beta-propeller (Figure 16A). This loop is located distant from the active site, does not contribute to any known functional region of the protein. Hence, the pathogenicity of the Asp477Gly substitution was not obvious from its position within the three-dimensional structure. However, beta-turns can exert pivotal roles in protein folding suggesting that the Asp477Gly substitution could lead to a protein folding defect.
Figure 16. Location of the Asp477 residue within the RPE65 structure and creation of an RPE65/SynACO chimera system for structural study of the Asp477Gly mutation.
A) Location of the Asp477-containing loop within the three-dimensional structure of RPE65. B) Structure of the Asp477 loop region in RPE65/SynACO chimera (blue to green gradient) compared to bovine RPE65 (grey). Auxiliary substituted residues in the chimera and the corresponding residues in bovine RPE65 are colored light orange and grey, respectively. The Leu residue substituted at position 44 is omitted for clarity. The Asp477 position is marked by an asterisk. Top residue labeling refers to the ACO/RPE65 sequence while the numbers on bottom (in grey) refer to the native RPE65 sequence. Note the structural equivalence between the two proteins in this region. C) Comparison of the same loop region of the RPE65/SynACO chimera Asp477Gly mutant to the original chimera structure shows only modest structural differences indicating that the Gly substitution does not grossly destabilize the Asp477 loop region. Modified and used with permission from (Choi et al., 2018).
To probe the structural consequences of the substitution, we took advantage of the high structural similarity between RPE65 and SynACO near position 477 to design a chimeric protein in which residues 474-485 of RPE65 were grafted in place of the equivalent SynACO sequence together with point substitutions at 4 additional sites that appeared important to avoid steric clashes or maintain important interactions. The chimera and its Asp477Gly mutant expressed in E. coli as soluble proteins and were catalytically active indicating that the Asp477Gly substitution does not have a marked effect on protein folding.
To directly examine the degree of structural preservation of the Asp477 loop in RPE65 and the RPE65/SynACO chimera, we crystallized the chimera under the same conditions used for wild-type SynACO. The structure revealed a loop structure nearly identical to that of native RPE65 (Figure 16B). This finding provides key validation for use of the chimera to infer structural consequences of the RPE65 Asp477Gly mutation. We then crystallized the Asp477Gly chimera variant again under the same conditions used for wild-type SynACO. Strikingly, this single amino acid change resulted in the exclusive formation of two new crystal forms not observed in prior structural studies on SynACO. The overall structure of the Asp477Gly loop region was similar to that of the Asp477 chimera with a few notable differences including variability in the residue 477-containing beta-turn structure and alterations in the structures of a few nearby residues, which were likely due to a difference in crystal packing (Figure 16C). However, in both crystal forms, the electron density corresponding to the Asp477 loop and surrounding residues was well resolved indicating that the Asp477Gly substitution does not structurally destabilize this loop region.
Molecular packing in the Asp477Gly crystals differed in a striking way from that of the RPE65/SynACO chimera crystals. Whereas the Asp477-containing loop did not engage in protein–protein contacts in crystals of the original chimera, contacts involving this loop were integrally involved in the crystal packing of both new crystal forms (Figure 17). Considering both crystal forms, the Asp477Gly loop engages in four distinct protein–protein interactions involving predominantly hydrophobic contacts with a few H-bonds and salt bridges (Figure 17, B and D) and are among the major contact points responsible for crystal formation. In all four cases, multiple van der Waals interactions involving the Gly alpha carbon indicate that the absence of the Asp side chain crucially enabled formation of the protein–protein contacts. The Asp477Gly substitution thus produced a gain of protein–protein interaction potential by allowing the residue 477-containing loop to form contacts with diverse molecular surfaces. From this observation it was suggested that Asp477 in RPE65 could serve as a hydrophilic “gatekeeper” residue that protects the aggregation-prone 477 loop from engaging in unwanted protein-protein contacts by interrupting an otherwise continuous site of hydrophobicity. Indeed, it was observed that Asp477Gly RPE65 was targeted for ubiquitination and degradation presumably because the mutant loops allowed for novel interactions with E3 ubiquitin ligases. Other abnormal protein-protein interactions could drive the RPE cell pathology that is observed in patients with this dominant form of RP.
Figure 17. Involvement of the Asp477 loop in crystal packing of RPE65/SynACO chimera Asp477Gly mutant.
A) Molecular packing in the trigonal crystal form revealing an interaction of the Asp477Gly loop and surrounding residues (shown as van der Waals spheres) with the other molecule in the asymmetric unit (contact I), which in turn also interacts with a symmetry-related molecule through the region surrounding the Asp477Gly loop (contact II). B) Details of the interactions formed between the Asp477Gly loop and its interaction partners. Dashed lines indicate van der Waals contacts (width proportional to number of atomic contacts), solid blue lines indicate hydrogen bonds and solid red lines indicate salt bridges. C) Molecular packing in the monoclinic crystal form mediated by the Asp477Gly loop and surrounding residues. Contact III occurs between the two molecules of the asymmetric unit whereas contact IV involves a symmetry-related molecule. These two contacts are related by a slight rotation. D) Interactions formed between the Asp477Gly loop and residues from its interacting partners in the monoclinic crystal form. In all four contacts, the introduction of an Asp side chain at position 434 would result in steric clashes which explains why these two crystal forms were only observed for the Asp477Gly mutant. Used with permission from (Choi et al., 2018).
12. Conclusions and Future Directions
Substantial progress has been made over the past decade in understanding the many facets of RPE65 biology, biochemistry, pharmacology, and disease involvement. Nevertheless, there are several key areas that warrant additional research. First, the role of RPE65-containing protein complexes in the operation of the visual cycle deserves closer inspection now that sophisticated mass spectrometry and extraction methods have been developed. Understanding the structure of such complexes will likely help reveal how retinoids are efficiently trafficked between visual cycle enzymes and retinoid binding proteins. With renewed interest in the biology of RGR-opsin, it will be informative to further examine the functional role of its interaction with RPE65. The precise intermediates involved in RPE65 catalysis remain to be spectroscopically characterized. Opportunities to study such intermediates may arise with the development of new methods for high-yield expression of RPE65 or RPE65-like model enzymes. Concerning the structure of RPE65, the one region of the protein that remains unresolved is the PDPC(K)-containing loop that is involved in membrane binding and substrate recognition. It will be of interest to further study the structure of this region through the use of model systems or structural biology techniques besides X-ray crystallography. The therapeutic utility of visual cycle modulation has yet to be demonstrated although clinical trials involving emixustat for the treatment of Stargardt macular dystrophy or diabetic retinopathy are ongoing. Further research is also required to fully understand the pathogenesis of RP associated with the Asp477Gly substitution. One of the most exciting future areas of research is the use of gene-editing technologies to correct pathological RPE65 mutations as shown in proof-of-principle mouse studies (Suh et al., 2021). The next decade promises to bring further advances in our understanding of this all-important retinoid isomerohydrolase.
Highlights:
Relationships of RPE65 to carotenoid cleavage dioxygenases have been further elucidated.
The mechanism of the RPE65-catalyzed isomerohydrolase reaction has been clarified.
Potent small molecule and protein modulators of RPE65 activity have been developed and identified.
Physiological roles of RPE65 in the retina have been clarified through use of pharmacological modulators.
Knowledge of disease-causing RPE65 mutations and their pathogenic mechanisms has advanced.
Acknowledgements
I thank my lab members and collaborators for their dedication to advancing RPE65 research and Drs. Adriana Briscoe (UC Irvine) and Vladimir Kefalov (UC Irvine) for valuable comments on this manuscript. This work was supported by funding from the Department of Veterans Affairs (I01BX004939), the National Science Foundation (CHE-2107713), the National Institutes of Health (R24EY027283), and an unrestricted grant from Research to Prevent Blindness to the Department of Ophthalmology at UC Irvine. The views expressed in this article are those of the author and do not necessarily reflect the position or policy of the funding agencies or the United States government. The author declares no competing financial interests.
References
- Acland GM, Aguirre GD, Bennett J, Aleman TS, Cideciyan AV, Bennicelli J, Dejneka NS, Pearce-Kelling SE, Maguire AM, Palczewski K, Hauswirth WW, Jacobson SG, 2005. Long-term restoration of rod and cone vision by single dose rAAV-mediated gene transfer to the retina in a canine model of childhood blindness. Mol. Ther 12, 1072–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acland GM, Aguirre GD, Ray J, Zhang Q, Aleman TS, Cideciyan AV, Pearce-Kelling SE, Anand V, Zeng Y, Maguire AM, Jacobson SG, Hauswirth WW, Bennett J, 2001. Gene therapy restores vision in a canine model of childhood blindness. Nat. Genet 28, 92–95. [DOI] [PubMed] [Google Scholar]
- Albalat R, 2012. Evolution of the genetic machinery of the visual cycle: a novelty of the vertebrate eye? Mol. Biol. Evol 29, 1461–1469. [DOI] [PubMed] [Google Scholar]
- Allen KN, Entova S, Ray LC, Imperiali B, 2019. Monotopic Membrane Proteins Join the Fold. Trends Biochem. Sci 44, 7–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arunkumar R, Gorusupudi A, Bernstein PS, 2020. The macular carotenoids: A biochemical overview. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1865, 158617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asteriti S, Grillner S, Cangiano L, 2015. A Cambrian origin for vertebrate rods. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babino D, Perkins BD, Kindermann A, Oberhauser V, von Lintig J, 2015. The role of 11-cis-retinyl esters in vertebrate cone vision. FASEB J. 29, 216–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek M, DiMaio F, Anishchenko I, Dauparas J, Ovchinnikov S, Lee GR, Wang J, Cong Q, Kinch LN, Schaeffer RD, Millan C, Park H, Adams C, Glassman CR, DeGiovanni A, Pereira JH, Rodrigues AV, van Dijk AA, Ebrecht AC, Opperman DJ, Sagmeister T, Buhlheller C, Pavkov-Keller T, Rathinaswamy MK, Dalwadi U, Yip CK, Burke JE, Garcia KC, Grishin NV, Adams PD, Read RJ, Baker D, 2021. Accurate prediction of protein structures and interactions using a three-track neural network. Science 373, 871–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batten ML, Imanishi Y, Maeda T, Tu DC, Moise AR, Bronson D, Possin D, Van Gelder RN, Baehr W, Palczewski K, 2004. Lecithin-retinol acyltransferase is essential for accumulation of all-trans-retinyl esters in the eye and in the liver. J. Biol. Chem 279, 10422–10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bavik C, Henry SH, Zhang Y, Mitts K, McGinn T, Budzynski E, Pashko A, Lieu KL, Zhong S, Blumberg B, Kuksa V, Orme M, Scott I, Fawzi A, Kubota R, 2015. Visual Cycle Modulation as an Approach toward Preservation of Retinal Integrity. PLoS ONE 10, e0124940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bavik CO, Busch C, Eriksson U, 1992. Characterization of a plasma retinol-binding protein membrane receptor expressed in the retinal pigment epithelium. J. Biol. Chem 267, 23035–23042. [PubMed] [Google Scholar]
- Bernstein PS, Law WC, Rando RR, 1987. Biochemical characterization of the retinoid isomerase system of the eye. J. Biol. Chem 262, 16848–16857. [PubMed] [Google Scholar]
- Bernstein PS, Rando RR, 1986. In vivo isomerization of all-trans- to 11-cis-retinoids in the eye occurs at the alcohol oxidation state. Biochemistry 25, 6473–6478. [DOI] [PubMed] [Google Scholar]
- Bhattacharya SK, Wu Z, Miyagi M, West KA, Jin Z, Nawrot M, Saari JC, Crabb JW, 2002. Interactions of CRALBP with other visual cycle proteins. Invest. Ophthalmol. Vis. Sci 43, U1301–U1301. [Google Scholar]
- Blatz PE, Pippert DL, 1968a. Carbonium Ion of All-Trans-Retinyl Acetate . Spectroscopic Detection and Identification of Absorbing Species . Effect of Environment on Spectral Properties. J. Am. Chem. Soc 90, 1296–1300. [Google Scholar]
- Blatz PE, Pippert DL, 1968b. Fluorescence Spectrum of Retinylic Cation. Chem. Commun, 176–177. [Google Scholar]
- Bliven S, Lafita A, Parker A, Capitani G, Duarte JM, 2018. Automated evaluation of quaternary structures from protein crystals. PLoS Comput. Biol 14, e1006104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum E, Zhang J, Zaluski J, Einstein DE, Korshin EE, Kubas A, Gruzman A, Tochtrop GP, Kiser PD, Palczewski K, 2021. Rational Alteration of Pharmacokinetics of Chiral Fluorinated and Deuterated Derivatives of Emixustat for Retinal Therapy. J. Med. Chem 64, 8287–8302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boll F, 1876. Zur Anatomie and Physiologie der Retina. Monatsber. dk preuss. Akad. d. Wissensch. Berlin. Aus dem Jahre [Google Scholar]
- Bowne SJ, Humphries MM, Sullivan LS, Kenna PF, Tam LC, Kiang AS, Campbell M, Weinstock GM, Koboldt DC, Ding L, Fulton RS, Sodergren EJ, Allman D, Millington-Ward S, Palfi A, McKee A, Blanton SH, Slifer S, Konidari I, Farrar GJ, Daiger SP, Humphries P, 2011. A dominant mutation in RPE65 identified by whole-exome sequencing causes retinitis pigmentosa with choroidal involvement. Eur. J. Hum. Genet 19, 1074–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracey MH, Cravatt BF, Stevens RC, 2004. Structural commonalities among integral membrane enzymes. FEBS Lett. 567, 159–165. [DOI] [PubMed] [Google Scholar]
- Calvert PD, Krasnoperova NV, Lyubarsky AL, Isayama T, Nicolo M, Kosaras B, Wong G, Gannon KS, Margolskee RF, Sidman RL, Pugh EN Jr., Makino CL, Lem J, 2000. Phototransduction in transgenic mice after targeted deletion of the rod transducin alpha -subunit. Proc. Natl. Acad. Sci. U.S.A 97, 13913–13918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canada FJ, Law WC, Rando RR, Yamamoto T, Derguini F, Nakanishi K, 1990. Substrate specificities and mechanism in the enzymatic processing of vitamin A into 11-cis-retinol. Biochemistry 29, 9690–9697. [DOI] [PubMed] [Google Scholar]
- Chander P, Gentleman S, Poliakov E, Redmond TM, 2012. Aromatic residues in the substrate cleft of RPE65 protein govern retinol isomerization and modulate its progression. J. Biol. Chem 287, 30552–30559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekroud K, Guillou L, Gregoire S, Ducharme G, Brun E, Cazevieille C, Bretillon L, Hamel CP, Brabet P, Pequignot MO, 2012. Fatp1 deficiency affects retinal light response and dark adaptation, and induces age-related alterations. PLoS ONE 7, e50231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Hao W, Rife L, Wang XP, Shen D, Chen J, Ogden T, Van Boemel GB, Wu L, Yang M, Fong HK, 2001. A photic visual cycle of rhodopsin regeneration is dependent on Rgr. Nat. Genet 28, 256–260. [DOI] [PubMed] [Google Scholar]
- Choi EH, Suh S, Einstein DE, Leinonen H, Dong Z, Rao SR, Fliesler SJ, Blackshaw S, Yu M, Peachey NS, Palczewski K, Kiser PD, 2021. An inducible Cre mouse for studying roles of the RPE in retinal physiology and disease. JCI Insight 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi EH, Suh S, Sander CL, Hernandez CJO, Bulman ER, Khadka N, Dong Z, Shi W, Palczewski K, Kiser PD, 2018. Insights into the pathogenesis of dominant retinitis pigmentosa associated with a D477G mutation in RPE65. Hum. Mol. Genet 27, 2225–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cideciyan AV, 2010. Leber congenital amaurosis due to RPE65 mutations and its treatment with gene therapy. Prog. Retin. Eye Res 29, 398–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbo JC, Di Gregorio A, Levine M, 2001. The ascidian as a model organism in developmental and evolutionary biology. Cell 106, 535–538. [DOI] [PubMed] [Google Scholar]
- Cowan CS, Renner M, De Gennaro M, Gross-Scherf B, Goldblum D, Hou Y, Munz M, Rodrigues TM, Krol J, Szikra T, Cuttat R, Waldt A, Papasaikas P, Diggelmann R, Patino-Alvarez CP, Galliker P, Spirig SE, Pavlinic D, Gerber-Hollbach N, Schuierer S, Srdanovic A, Balogh M, Panero R, Kusnyerik A, Szabo A, Stadler MB, Orgul S, Picelli S, Hasler PW, Hierlemann A, Scholl HPN, Roma G, Nigsch F, Roska B, 2020. Cell Types of the Human Retina and Its Organoids at Single-Cell Resolution. Cell 182, 1623–1640 e1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubizolle A, Guillou L, Mollereau B, Hamel CP, Brabet P, 2017. Fatty acid transport protein 1 regulates retinoid metabolism and photoreceptor development in mouse retina. PLoS ONE 12, e0180148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daruwalla A, Zhang J, Lee HJ, Khadka N, Farquhar ER, Shi W, von Lintig J, Kiser PD, 2020. Structural basis for carotenoid cleavage by an archaeal carotenoid dioxygenase. Proc. Natl. Acad. Sci. U.S.A 117, 19914–19925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehal P, Boore JL, 2005. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 3, e314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deigner PS, Law WC, Canada FJ, Rando RR, 1989. Membranes as the energy source in the endergonic transformation of vitamin A to 11-cis-retinol. Science 244, 968–971. [DOI] [PubMed] [Google Scholar]
- Delsuc F, Brinkmann H, Chourrout D, Philippe H, 2006. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature 439, 965–968. [DOI] [PubMed] [Google Scholar]
- Dias MF, Joo K, Kemp JA, Fialho SL, da Silva Cunha A Jr., Woo SJ, Kwon YJ, 2018. Molecular genetics and emerging therapies for retinitis pigmentosa: Basic research and clinical perspectives. Prog. Retin. Eye Res 63, 107–131. [DOI] [PubMed] [Google Scholar]
- DiRusso CC, Li H, Darwis D, Watkins PA, Berger J, Black PN, 2005. Comparative biochemical studies of the murine fatty acid transport proteins (FATP) expressed in yeast. J. Biol. Chem 280, 16829–16837. [DOI] [PubMed] [Google Scholar]
- Dong EM, Allison WT, 2021. Vertebrate features revealed in the rudimentary eye of the Pacific hagfish (Eptatretus stoutii). Proc. Biol. Sci 288, 20202187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorr JM, Scheidelaar S, Koorengevel MC, Dominguez JJ, Schafer M, van Walree CA, Killian JA, 2016. The styrene-maleic acid copolymer: a versatile tool in membrane research. Eur. Biophys. J 45, 3–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugel PU, Novack RL, Csaky KG, Richmond PP, Birch DG, Kubota R, 2015. Phase ii, randomized, placebo-controlled, 90-day study of emixustat hydrochloride in geographic atrophy associated with dry age-related macular degeneration. Retina 35, 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst OP, Lodowski DT, Elstner M, Hegemann P, Brown LS, Kandori H, 2014. Microbial and animal rhodopsins: structures, functions, and molecular mechanisms. Chem. Rev 114, 126–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eroglu A, Gentleman S, Poliakov E, Redmond TM, 2016. Inhibition of RPE65 Retinol Isomerase Activity by Inhibitors of Lipid Metabolism. J. Biol. Chem 291, 4966–4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feathers KL, Lyubarsky AL, Khan NW, Teofilo K, Swaroop A, Williams DS, Pugh EN Jr., Thompson DA, 2008. Nrl-knockout mice deficient in Rpe65 fail to synthesize 11-cis retinal and cone outer segments. Invest. Ophthalmol. Vis. Sci 49, 1126–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forneris F, Mattevi A, 2008. Enzymes without borders: mobilizing substrates, delivering products. Science 321, 213–216. [DOI] [PubMed] [Google Scholar]
- Foster AW, Osman D, Robinson NJ, 2014. Metal preferences and metallation. J. Biol. Chem 289, 28095–28103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederiksen R, Morshedian A, Tripathy SA, Xu T, Travis GH, Fain GL, Sampath AP, 2021. Rod Photoreceptors Avoid Saturation in Bright Light by the Movement of the G Protein Transducin. J. Neurosci 41, 3320–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao FJ, Wang DD, Li JK, Hu FY, Xu P, Chen F, Qi YH, Liu W, Li W, Zhang SH, Chang Q, Xu GZ, Wu JH, 2021. Frequency and phenotypic characteristics of RPE65 mutations in the Chinese population. Orphanet J. Rare Dis 16, 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garafalo AV, Cideciyan AV, Heon E, Sheplock R, Pearson A, WeiYang Yu C, Sumaroka A, Aguirre GD, Jacobson SG, 2020. Progress in treating inherited retinal diseases: Early subretinal gene therapy clinical trials and candidates for future initiatives. Prog. Retin. Eye Res 77, 100827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giglione C, Boularot A, Meinnel T, 2004. Protein N-terminal methionine excision. Cell. Mol. Life Sci 61, 1455–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliano G, Al-Babili S, von Lintig J, 2003. Carotenoid oxygenases: cleave it or leave it. Trends Plant Sci. 8, 145–149. [DOI] [PubMed] [Google Scholar]
- Golczak M, Imanishi Y, Kuksa V, Maeda T, Kubota R, Palczewski K, 2005a. Lecithin:retinol acyltransferase is responsible for amidation of retinylamine, a potent inhibitor of the retinoid cycle. J. Biol. Chem 280, 42263–42273. [DOI] [PubMed] [Google Scholar]
- Golczak M, Kiser PD, Lodowski DT, Maeda A, Palczewski K, 2010. Importance of membrane structural integrity for RPE65 retinoid isomerization activity. J. Biol. Chem 285, 9667–9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golczak M, Kuksa V, Maeda T, Moise AR, Palczewski K, 2005b. Positively charged retinoids are potent and selective inhibitors of the trans-cis isomerization in the retinoid (visual) cycle. Proc. Natl. Acad. Sci. U.S.A 102, 8162–8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golczak M, Maeda A, Bereta G, Maeda T, Kiser PD, Hunzelmann S, von Lintig J, Blaner WS, Palczewski K, 2008. Metabolic basis of visual cycle inhibition by retinoid and nonretinoid compounds in the vertebrate retina. J. Biol. Chem 283, 9543–9554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodsell DS, Olson AJ, 2000. Structural symmetry and protein function. Annu. Rev. Biophys. Biomol. Struct 29, 105–153. [DOI] [PubMed] [Google Scholar]
- Gouet P, Robert X, Courcelle E, 2003. ESPript/ENDscript: Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 31, 3320–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm C, Wenzel A, Hafezi F, Yu S, Redmond TM, Reme CE, 2000. Protection of Rpe65-deficient mice identifies rhodopsin as a mediator of light-induced retinal degeneration. Nat. Genet 25, 63–66. [DOI] [PubMed] [Google Scholar]
- Gu SM, Thompson DA, Srikumari CR, Lorenz B, Finckh U, Nicoletti A, Murthy KR, Rathmann M, Kumaramanickavel G, Denton MJ, Gal A, 1997. Mutations in RPE65 cause autosomal recessive childhood-onset severe retinal dystrophy. Nat. Genet 17, 194–197. [DOI] [PubMed] [Google Scholar]
- Guenthner CJ, Miyamichi K, Yang HH, Heller HC, Luo L, 2013. Permanent genetic access to transiently active neurons via TRAP: targeted recombination in active populations. Neuron 78, 773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guignard TJP, Jin MH, Pequignot MO, Li SH, Chassigneux Y, Chekroud K, Guillou L, Richard E, Hamel CP, Brabet P, 2010. FATP1 Inhibits 11-cis Retinol Formation via Interaction with the Visual Cycle Retinoid Isomerase RPE65 and Lecithin:Retinol Acyltransferase. J. Biol. Chem 285, 18759–18768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel CP, Tsilou E, Harris E, Pfeffer BA, Hooks JJ, Detrick B, Redmond TM, 1993a. A developmentally regulated microsomal protein specific for the pigment epithelium of the vertebrate retina. J. Neurosci. Res 34, 414–425. [DOI] [PubMed] [Google Scholar]
- Hamel CP, Tsilou E, Pfeffer BA, Hooks JJ, Detrick B, Redmond TM, 1993b. Molecular cloning and expression of RPE65, a novel retinal pigment epithelium-specific microsomal protein that is post-transcriptionally regulated in vitro. J. Biol. Chem 268, 15751–15757. [PubMed] [Google Scholar]
- Hanein S, Perrault I, Gerber S, Tanguy G, Barbet F, Ducroq D, Calvas P, Dollfus H, Hamel C, Lopponen T, Munier F, Santos L, Shalev S, Zafeiriou D, Dufier JL, Munnich A, Rozet JM, Kaplan J, 2004. Leber congenital amaurosis: comprehensive survey of the genetic heterogeneity, refinement of the clinical definition, and genotype-phenotype correlations as a strategy for molecular diagnosis. Hum. Mutat 23, 306–317. [DOI] [PubMed] [Google Scholar]
- Harrison KR, Reifler AN, Chervenak AP, Wong KY, 2021. Prolonged Melanopsin-based Photoresponses Depend in Part on RPE65 and Cellular Retinaldehyde-binding Protein (CRALBP). Curr. Eye Res 46, 515–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Hahn P, Iacovelli J, Wong R, King C, Bhisitkul R, Massaro-Giordano M, Dunaief JL, 2007. Iron homeostasis and toxicity in retinal degeneration. Prog. Retin. Eye Res 26, 649–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemati N, Feathers KL, Chrispell JD, Reed DM, Carlson TJ, Thompson DA, 2005. RPE65 surface epitopes, protein interactions, and expression in rod- and cone-dominant species. Mol. Vis 11, 1151–1165. [PubMed] [Google Scholar]
- Hoang T, Wang J, Boyd P, Wang F, Santiago C, Jiang L, Yoo S, Lahne M, Todd LJ, Jia M, Saez C, Keuthan C, Palazzo I, Squires N, Campbell WA, Rajaii F, Parayil T, Trinh V, Kim DW, Wang G, Campbell LJ, Ash J, Fischer AJ, Hyde DR, Qian J, Blackshaw S, 2020. Gene regulatory networks controlling vertebrate retinal regeneration. Science 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann KP, Scheerer P, Hildebrand PW, Choe HW, Park JH, Heck M, Ernst OP, 2009. A G protein-coupled receptor at work: the rhodopsin model. Trends Biochem. Sci 34, 540–552. [DOI] [PubMed] [Google Scholar]
- Huang J, Possin DE, Saari JC, 2009. Localizations of visual cycle components in retinal pigment epithelium. Mol. Vis 15, 223–234. [PMC free article] [PubMed] [Google Scholar]
- Hubbard R, Wald G, 1952. Cis-trans isomers of vitamin A and retinene in the rhodopsin system. J. Gen. Physiol 36, 269–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull S, Mukherjee R, Holder GE, Moore AT, Webster AR, 2016. The clinical features of retinal disease due to a dominant mutation in RPE65. Mol. Vis 22, 626–635. [PMC free article] [PubMed] [Google Scholar]
- Imanishi Y, Batten ML, Piston DW, Baehr W, Palczewski K, 2004a. Noninvasive two-photon imaging reveals retinyl ester storage structures in the eye. J. Cell Biol 164, 373–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanishi Y, Gerke V, Palczewski K, 2004b. Retinosomes: new insights into intracellular managing of hydrophobic substances in lipid bodies. J. Cell Biol 166, 447–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson SG, Aleman TS, Cideciyan AV, Heon E, Golczak M, Beltran WA, Sumaroka A, Schwartz SB, Roman AJ, Windsor EA, Wilson JM, Aguirre GD, Stone EM, Palczewski K, 2007. Human cone photoreceptor dependence on RPE65 isomerase. Proc. Natl. Acad. Sci. U.S.A 104, 15123–15128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson SG, Aleman TS, Cideciyan AV, Roman AJ, Sumaroka A, Windsor EA, Schwartz SB, Heon E, Stone EM, 2009. Defining the residual vision in leber congenital amaurosis caused by RPE65 mutations. Invest. Ophthalmol. Vis. Sci 50, 2368–2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang GF, Van Hooser JP, Kuksa V, McBee JK, He YG, Janssen JJ, Driessen CA, Palczewski K, 2001. Characterization of a dehydrogenase activity responsible for oxidation of 11-cis-retinol in the retinal pigment epithelium of mice with a disrupted RDH5 gene. A model for the human hereditary disease fundus albipunctatus. J. Biol. Chem 276, 32456–32465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauregui R, Cho A, Oh JK, Tanaka AJ, Sparrow JR, Tsang SH, 2020. Phenotypic expansion of autosomal dominant retinitis pigmentosa associated with the D477G mutation in RPE65. Cold Spring Harb. Mol. Case Stud 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauregui R, Park KS, Tsang SH, 2018. Two-year progression analysis of RPE65 autosomal dominant retinitis pigmentosa. Ophthalmic Genet. 39, 544–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M, Li S, Moghrabi WN, Sun H, Travis GH, 2005. Rpe65 is the retinoid isomerase in bovine retinal pigment epithelium. Cell 122, 449–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M, Yuan Q, Li S, Travis GH, 2007. Role of LRAT on the retinoid isomerase activity and membrane association of Rpe65. J. Biol. Chem 282, 20915–20924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo DH, Song DW, Cho CS, Kim UG, Lee KJ, Lee K, Park SW, Kim D, Kim JH, Kim JS, Kim S, Kim JH, Lee JM, 2019. CRISPR-Cas9-mediated therapeutic editing of Rpe65 ameliorates the disease phenotypes in a mouse model of Leber congenital amaurosis. Sci. Adv 5, eaax1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Zidek A, Potapenko A, Bridgland A, Meyer C, Kohl SAA, Ballard AJ, Cowie A, Romera-Paredes B, Nikolov S, Jain R, Adler J, Back T, Petersen S, Reiman D, Clancy E, Zielinski M, Steinegger M, Pacholska M, Berghammer T, Bodenstein S, Silver D, Vinyals O, Senior AW, Kavukcuoglu K, Kohli P, Hassabis D, 2021. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamoda S, Saburi Y, 1993. Cloning, expression, and sequence analysis of a lignostilbene-alpha,beta-dioxygenase gene from Pseudomonas paucimobilis TMY1009. Biosci. Biotechnol. Biochem 57, 926–930. [DOI] [PubMed] [Google Scholar]
- Katz ML, Redmond TM, 2001. Effect of Rpe65 knockout on accumulation of lipofuscin fluorophores in the retinal pigment epithelium. Invest. Ophthalmol. Vis. Sci 42, 3023–3030. [PubMed] [Google Scholar]
- Kaylor JJ, Xu T, Ingram NT, Tsan A, Hakobyan H, Fain GL, Travis GH, 2017. Blue light regenerates functional visual pigments in mammals through a retinyl-phospholipid intermediate. Nat. Commun 8, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan WT, Rupp AC, Ross RA, Somasundaram P, Hiriyanna S, Wu Z, Badea TC, Robinson PR, Lowell BB, Hattar SS, 2016. A visual circuit uses complementary mechanisms to support transient and sustained pupil constriction. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiang AS, Kenna PF, Humphries MM, Ozaki E, Koenekoop RK, Campbell M, Farrar GJ, Humphries P, 2020. Properties and Therapeutic Implications of an Enigmatic D477G RPE65 Variant Associated with Autosomal Dominant Retinitis Pigmentosa. Genes (Basel) 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kildahl-Andersen G, Lutnaes BF, Krane J, Liaaen-Jensen S, 2003. Structure elucidation of polyene systems with extensive charge delocalization-carbocations from allylic carotenols. Org. Lett 5, 2675–2678. [DOI] [PubMed] [Google Scholar]
- Kim SR, Fishkin N, Kong J, Nakanishi K, Allikmets R, Sparrow JR, 2004. Rpe65 Leu450Met variant is associated with reduced levels of the retinal pigment epithelium lipofuscin fluorophores A2E and iso-A2E. Proc. Natl. Acad. Sci. U.S.A 101, 11668–11672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiser PD, 2010. STRUCTURAL AND BIOCHEMICAL STUDIES OF RPE65, THE RETINOID ISOMERASE OF THE VISUAL CYCLE. Case Western Reserve University School of Graduate Studies. [Google Scholar]
- Kiser PD, 2019. Alkene-cleaving carotenoid cleavage dioxygenases, in: Scott RA (Ed.), Encyclopedia of Inorganic and Bioinorganic Chemistry. John Wiley & Sons, Ltd. [Google Scholar]
- Kiser PD, Farquhar ER, Shi W, Sui X, Chance MR, Palczewski K, 2012. Structure of RPE65 isomerase in a lipidic matrix reveals roles for phospholipids and iron in catalysis. Proc. Natl. Acad. Sci. U.S.A 109, E2747–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiser PD, Golczak M, Lodowski DT, Chance MR, Palczewski K, 2009. Crystal structure of native RPE65, the retinoid isomerase of the visual cycle. Proc. Natl. Acad. Sci. U.S.A 106, 17325–17330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiser PD, Golczak M, Palczewski K, 2014. Chemistry of the retinoid (visual) cycle. Chem. Rev 114, 194–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiser PD, Kolesnikov AV, Kiser JZ, Dong ZQ, Chaurasia B, Wang LP, Summers SA, Hoang T, Blackshaw S, Peachey NS, Kefalov VJ, Palczewski K, 2019. Conditional deletion of Des1 in the mouse retina does not impair the visual cycle in cones. FASEB J. 33, 5782–5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiser PD, Palczewski K, 2010. Membrane-binding and enzymatic properties of RPE65. Prog. Retin. Eye Res 29, 428–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiser PD, Palczewski K, 2020. Pathways and disease-causing alterations in visual chromophore production for vertebrate vision. J. Biol. Chem 296, 100072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiser PD, Zhang J, Badiee M, Kinoshita J, Peachey NS, Tochtrop GP, Palczewski K, 2017. Rational Tuning of Visual Cycle Modulator Pharmacodynamics. J. Pharmacol. Exp. Ther 362, 131–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiser PD, Zhang J, Badiee M, Li Q, Shi W, Sui X, Golczak M, Tochtrop GP, Palczewski K, 2015. Catalytic mechanism of a retinoid isomerase essential for vertebrate vision. Nat. Chem. Biol 11, 409–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiser PD, Zhang J, Sharma A, Angueyra JM, Kolesnikov AV, Badiee M, Tochtrop GP, Kinoshita J, Peachey NS, Li W, Kefalov VJ, Palczewski K, 2018. Retinoid isomerase inhibitors impair but do not block mammalian cone photoreceptor function. J. Gen. Physiol 150, 571–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloer DP, Ruch S, Al-Babili S, Beyer P, Schulz GE, 2005. The structure of a retinal-forming carotenoid oxygenase. Science 308, 267–269. [DOI] [PubMed] [Google Scholar]
- Kolesnikov AV, Kiser PD, Palczewski K, Kefalov VJ, 2021. Function of mammalian M-cones depends on the level of CRALBP in Muller cells. J. Gen. Physiol 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesnikov AV, Tang PH, Kefalov VJ, 2018. Examining the Role of Cone-expressed RPE65 in Mouse Cone Function. Sci. Rep 8, 14201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krinsky NI, Landrum JT, Bone RA, 2003. Biologic mechanisms of the protective role of lutein and zeaxanthin in the eye. Annu. Rev. Nutr 23, 171–201. [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K, 2007. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol 372, 774–797. [DOI] [PubMed] [Google Scholar]
- Kubota R, Al-Fayoumi S, Mallikaarjun S, Patil S, Bavik C, Chandler JW, 2014. Phase 1, dose-ranging study of emixustat hydrochloride (ACU-4429), a novel visual cycle modulator, in healthy volunteers. Retina 34, 603–609. [DOI] [PubMed] [Google Scholar]
- Kubota R, Birch DG, Gregory JK, Koester JM, 2020. Randomised study evaluating the pharmacodynamics of emixustat hydrochloride in subjects with macular atrophy secondary to Stargardt disease. Br. J. Ophthalmol [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota R, Boman NL, David R, Mallikaarjun S, Patil S, Birch D, 2012. Safety and effect on rod function of ACU-4429, a novel small-molecule visual cycle modulator. Retina 32, 183–188. [DOI] [PubMed] [Google Scholar]
- Kubota R, Calkins DJ, Henry SH, Linsenmeier RA, 2019. Emixustat Reduces Metabolic Demand of Dark Activity in the Retina. Invest. Ophthalmol. Vis. Sci 60, 4924–4930. [DOI] [PubMed] [Google Scholar]
- Kubota R, Jhaveri C, Koester JM, Gregory JK, 2021. Effects of emixustat hydrochloride in patients with proliferative diabetic retinopathy: a randomized, placebo-controlled phase 2 study. Graefes Arch. Clin. Exp. Ophthalmol 259, 369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhne W, 1878. On the photochemistry of the retina and on visual purple. University Press, Cambridge. [Google Scholar]
- Kusakabe T, Kusakabe R, Kawakami I, Satou Y, Satoh N, Tsuda M, 2001. Ci-opsin1, a vertebrate-type opsin gene, expressed in the larval ocellus of the ascidian Ciona intestinalis. FEBS Lett. 506, 69–72. [DOI] [PubMed] [Google Scholar]
- Lamb TD, 2013. Evolution of phototransduction, vertebrate photoreceptors and retina. Prog. Retin. Eye Res 36, 52–119. [DOI] [PubMed] [Google Scholar]
- Lamb TD, Pugh EN Jr., 2004. Dark adaptation and the retinoid cycle of vision. Prog. Retin. Eye Res 23, 307–380. [DOI] [PubMed] [Google Scholar]
- Lamb TD, Pugh EN Jr., 2006. Phototransduction, dark adaptation, and rhodopsin regeneration the proctor lecture. Invest. Ophthalmol. Vis. Sci 47, 5137–5152. [DOI] [PubMed] [Google Scholar]
- Law WC, Rando RR, 1988. Stereochemical inversion at C-15 accompanies the enzymatic isomerization of all-trans- to 11-cis-retinoids. Biochemistry 27, 4147–4152. [DOI] [PubMed] [Google Scholar]
- Law WC, Rando RR, 1989. The molecular basis of retinoic acid induced night blindness. Biochem. Biophys. Res. Commun 161, 825–829. [DOI] [PubMed] [Google Scholar]
- Li P, Maier JM, Vik EC, Yehl CJ, Dial BE, Rickher AE, Smith MD, Pellechia PJ, Shimizu KD, 2017. Stabilizing Fluorine-pi Interactions. Angew. Chem. Int. Ed. Engl 56, 7209–7212. [DOI] [PubMed] [Google Scholar]
- Li S, Izumi T, Hu J, Jin HH, Siddiqui AA, Jacobson SG, Bok D, Jin M, 2014. Rescue of enzymatic function for disease-associated RPE65 proteins containing various missense mutations in non-active sites. J. Biol. Chem 289, 18943–18956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Lee J, Zhou Y, Gordon WC, Hill JM, Bazan NG, Miner JH, Jin M, 2013. Fatty acid transport protein 4 (FATP4) prevents light-induced degeneration of cone and rod photoreceptors by inhibiting RPE65 isomerase. J. Neurosci 33, 3178–3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Samardzija M, Yang Z, Grimm C, Jin M, 2016. Pharmacological Amelioration of Cone Survival and Vision in a Mouse Model for Leber Congenital Amaurosis. J. Neurosci 36, 5808–5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SH, Gordon WC, Bazan NG, Jin MH, 2020. Inverse correlation between fatty acid transport protein 4 and vision in Leber congenital amaurosis associated with RPE65 mutation. Proc. Natl. Acad. Sci. U.S.A 117, 32114–32123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Furhang R, Ray A, Duncan T, Soucy J, Mahdi R, Chaitankar V, Gieser L, Poliakov E, Qian H, Liu P, Dong L, Rogozin IB, Redmond TM, 2019. Aberrant RNA splicing is the major pathogenic effect in a knock-in mouse model of the dominantly inherited c.1430A>G human RPE65 mutation. Hum. Mutat 40, 426–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Yu S, Duncan T, Li Y, Liu P, Gene E, Cortes-Pena Y, Qian H, Dong L, Redmond TM, 2015. Mouse model of human RPE65 P25L hypomorph resembles wild type under normal light rearing but is fully resistant to acute light damage. Hum. Mol. Genet 24, 4417–4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomize MA, Pogozheva ID, Joo H, Mosberg HI, Lomize AL, 2012. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 40, D370–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes VS, Gibbs D, Libby RT, Aleman TS, Welch DL, Lillo C, Jacobson SG, Radu RA, Steel KP, Williams DS, 2011. The Usher 1B protein, MYO7A, is required for normal localization and function of the visual retinoid cycle enzyme, RPE65. Hum. Mol. Genet 20, 2560–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Rodriguez R, Lantero E, Blanco-Kelly F, Avila-Fernandez A, Merida IM, Pozo-Valero MD, Perea-Romero I, Zurita O, Jimenez-Rolando B, Swafiri ST, Riveiro-Alvarez R, Trujillo-Tiebas MJ, Salas EC, Garcia-Sandoval B, Corton M, Ayuso C, 2021. RPE65-related retinal dystrophy: Mutational and phenotypic spectrum in 45 affected patients. Exp. Eye Res, 108761. [DOI] [PubMed] [Google Scholar]
- Lucas RJ, Hattar S, Takao M, Berson DM, Foster RG, Yau KW, 2003. Diminished pupillary light reflex at high irradiances in melanopsin-knockout mice. Science 299, 245–247. [DOI] [PubMed] [Google Scholar]
- Lyubarsky AL, Savchenko AB, Morocco SB, Daniele LL, Redmond TM, Pugh EN Jr., 2005. Mole quantity of RPE65 and its productivity in the generation of 11-cis-retinal from retinyl esters in the living mouse eye. Biochemistry 44, 9880–9888. [DOI] [PubMed] [Google Scholar]
- Ma J, Xu L, Othersen DK, Redmond TM, Crouch RK, 1998. Cloning and localization of RPE65 mRNA in salamander cone photoreceptor cells. Biochim. Biophys. Acta 1443, 255–261. [DOI] [PubMed] [Google Scholar]
- Maeda A, Maeda T, Golczak M, Imanishi Y, Leahy P, Kubota R, Palczewski K, 2006. Effects of potent inhibitors of the retinoid cycle on visual function and photoreceptor protection from light damage in mice. Mol. Pharmacol 70, 1220–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda T, Cideciyan AV, Maeda A, Golczak M, Aleman TS, Jacobson SG, Palczewski K, 2009. Loss of cone photoreceptors caused by chromophore depletion is partially prevented by the artificial chromophore pro-drug, 9-cis-retinyl acetate. Hum. Mol. Genet 18, 2277–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda T, Dong Z, Jin H, Sawada O, Gao S, Utkhede D, Monk W, Palczewska G, Palczewski K, 2013. QLT091001, a 9-cis-Retinal Analog, Is Well-Tolerated by Retinas of Mice with Impaired Visual Cycles. Invest. Ophthalmol. Vis. Sci 54, 455–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda T, Perusek L, Amengual J, Babino D, Palczewski K, von Lintig J, 2011. Dietary 9-cis-beta,beta-carotene fails to rescue vision in mouse models of leber congenital amaurosis. Mol. Pharmacol 80, 943–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire AM, Bennett J, Aleman EM, Leroy BP, Aleman TS, 2021. Clinical Perspective: Treating RPE65-Associated Retinal Dystrophy. Mol. Ther 29, 442–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti P, Gollapalli D, Rando RR, 2005. Specificity of binding of all-trans-retinyl ester to RPE65. Biochemistry 44, 14463–14469. [DOI] [PubMed] [Google Scholar]
- Maiti P, Kong J, Kim SR, Sparrow JR, Allikmets R, Rando RR, 2006. Small molecule RPE65 antagonists limit the visual cycle and prevent lipofuscin formation. Biochemistry 45, 852–860. [DOI] [PubMed] [Google Scholar]
- Mandal MN, Moiseyev GP, Elliott MH, Kasus-Jacobi A, Li X, Chen H, Zheng L, Nikolaeva O, Floyd RA, Ma JX, Anderson RE, 2011. Alpha-phenyl-N-tert-butylnitrone (PBN) prevents light-induced degeneration of the retina by inhibiting RPE65 protein isomerohydrolase activity. J. Biol. Chem 286, 32491–32501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlhens F, Bareil C, Griffoin JM, Zrenner E, Amalric P, Eliaou C, Liu SY, Harris E, Redmond TM, Arnaud B, Claustres M, Hamel CP, 1997. Mutations in RPE65 cause Leber's congenital amaurosis. Nat. Genet 17, 139–141. [DOI] [PubMed] [Google Scholar]
- Mata NL, Radu RA, Clemmons RC, Travis GH, 2002. Isomerization and oxidation of vitamin a in cone-dominant retinas: a novel pathway for visual-pigment regeneration in daylight. Neuron 36, 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBee JK, Kuksa V, Alvarez R, de Lera AR, Prezhdo O, Haeseleer F, Sokal I, Palczewski K, 2000. Isomerization of all-trans-retinol to cis-retinols in bovine retinal pigment epithelial cells: dependence on the specificity of retinoid-binding proteins. Biochemistry 39, 11370–11380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing SA, Gabelli SB, Echeverria I, Vogel JT, Guan JC, Tan BC, Klee HJ, McCarty DR, Amzel LM, 2010. Structural insights into maize viviparous14, a key enzyme in the biosynthesis of the phytohormone abscisic acid. Plant Cell 22, 2970–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita T, Coates MI, Farrar R, Larson P, Manning PL, Wogelius RA, Edwards NP, Anne J, Bergmann U, Palmer AR, Currie PJ, 2019. Hagfish from the Cretaceous Tethys Sea and a reconciliation of the morphological-molecular conflict in early vertebrate phylogeny. Proc. Natl. Acad. Sci. U.S.A 116, 2146–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moiseyev G, Chen Y, Takahashi Y, Wu BX, Ma JX, 2005. RPE65 is the isomerohydrolase in the retinoid visual cycle. Proc. Natl. Acad. Sci. U.S.A 102, 12413–12418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moiseyev G, Crouch RK, Goletz P, Oatis J Jr., Redmond TM, Ma JX, 2003. Retinyl esters are the substrate for isomerohydrolase. Biochemistry 42, 2229–2238. [DOI] [PubMed] [Google Scholar]
- Moiseyev G, Nikolaeva O, Chen Y, Farjo K, Takahashi Y, Ma JX, 2010. Inhibition of the visual cycle by A2E through direct interaction with RPE65 and implications in Stargardt disease. Proc. Natl. Acad. Sci. U.S.A 107, 17551–17556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moiseyev G, Takahashi Y, Chen Y, Gentleman S, Redmond TM, Crouch RK, Ma JX, 2006. RPE65 is an iron(II)-dependent isomerohydrolase in the retinoid visual cycle. J. Biol. Chem 281, 2835–2840. [DOI] [PubMed] [Google Scholar]
- Moiseyev G, Takahashi Y, Chen Y, Kim S, Ma JX, 2008. RPE65 from cone-dominant chicken is a more efficient isomerohydrolase compared with that from rod-dominant species. J. Biol. Chem 283, 8110–8117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morshedian A, Kaylor JJ, Ng SY, Tsan A, Frederiksen R, Xu T, Yuan L, Sampath AP, Radu RA, Fain GL, Travis GH, 2019. Light-Driven Regeneration of Cone Visual Pigments through a Mechanism Involving RGR Opsin in Muller Glial Cells. Neuron 102, 1172–1183 e1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniz A, Betts BS, Trevino AR, Buddavarapu K, Roman R, Ma JX, Tsin AT, 2009. Evidence for two retinoid cycles in the cone-dominated chicken eye. Biochemistry 48, 6854–6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo LQ, 2007. A global double-fluorescent cre reporter mouse. Genesis 45, 593–605. [DOI] [PubMed] [Google Scholar]
- Nagao A, Olson JA, 1994. Enzymatic formation of 9-cis, 13-cis, and all-trans retinals from isomers of beta-carotene. FASEB J. 8, 968–973. [DOI] [PubMed] [Google Scholar]
- Nicoletti A, Wong DJ, Kawase K, Gibson LH, Yang-Feng TL, Richards JE, Thompson DA, 1995. Molecular characterization of the human gene encoding an abundant 61 kDa protein specific to the retinal pigment epithelium. Hum. Mol. Genet 4, 641–649. [DOI] [PubMed] [Google Scholar]
- Nikolaeva O, Moiseyev G, Rodgers KK, Ma JX, 2011. Binding to lipid membrane induces conformational changes in RPE65: implications for its isomerohydrolase activity. Biochem. J 436, 591–597. [DOI] [PubMed] [Google Scholar]
- Nikolaeva O, Takahashi Y, Moiseyev G, Ma JX, 2009. Purified RPE65 shows isomerohydrolase activity after reassociation with a phospholipid membrane. FEBS J. 276, 3020–3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaeva O, Takahashi Y, Moiseyev G, Ma JX, 2010. Negative charge of the glutamic acid 417 residue is crucial for isomerohydrolase activity of RPE65. Biochem. Biophys. Res. Commun 391, 1757–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan JM, Meagher K, Kashani S, Beatty S, 2013. What is meso-zeaxanthin, and where does it come from? Eye (Lond) 27, 899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberhauser V, Voolstra O, Bangert A, von Lintig J, Vogt K, 2008. NinaB combines carotenoid oxygenase and retinoid isomerase activity in a single polypeptide. Proc. Natl. Acad. Sci. U.S.A 105, 19000–19005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzmann JA, Carvalho P, 2019. Dynamics and functions of lipid droplets. Nat. Rev. Mol. Cell Biol 20, 137–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orban T, Palczewska G, Palczewski K, 2011. Retinyl ester storage particles (retinosomes) from the retinal pigmented epithelium resemble lipid droplets in other tissues. J. Biol. Chem 286, 17248–17258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palczewski K, 2006. G protein-coupled receptor rhodopsin. Annu. Rev. Biochem 75, 743–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S, Provencio I, Tu DC, Pires SS, Rollag MD, Castrucci AM, Pletcher MT, Sato TK, Wiltshire T, Andahazy M, Kay SA, Van Gelder RN, Hogenesch JB, 2003. Melanopsin is required for non-image-forming photic responses in blind mice. Science 301, 525–527. [DOI] [PubMed] [Google Scholar]
- Pang JJ, Chang B, Hawes NL, Hurd RE, Davisson MT, Li J, Noorwez SM, Malhotra R, McDowell JH, Kaushal S, Hauswirth WW, Nusinowitz S, Thompson DA, Heckenlively JR, 2005. Retinal degeneration 12 (rd12): a new, spontaneously arising mouse model for human Leber congenital amaurosis (LCA). Mol. Vis 11, 152–162. [PubMed] [Google Scholar]
- Poliakov E, Gentleman S, Chander P, Cunningham FX Jr., Grigorenko BL, Nemuhin AV, Redmond TM, 2009. Biochemical evidence for the tyrosine involvement in cationic intermediate stabilization in mouse beta-carotene 15, 15’-monooxygenase. BMC Biochem. 10, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliakov E, Gubin AN, Stearn O, Li Y, Campos MM, Gentleman S, Rogozin IB, Redmond TM, 2012. Origin and evolution of retinoid isomerization machinery in vertebrate visual cycle: hint from jawless vertebrates. PLoS ONE 7, e49975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliakov E, Parikh T, Ayele M, Kuo S, Chander P, Gentleman S, Redmond TM, 2011. Aromatic lipophilic spin traps effectively inhibit RPE65 isomerohydrolase activity. Biochemistry 50, 6739–6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliakov E, Soucy J, Gentleman S, Rogozin IB, Redmond TM, 2017. Phylogenetic analysis of the metazoan carotenoid oxygenase superfamily: a new ancestral gene assemblage of BCO-like (BCOL) proteins. Sci. Rep 7, 13192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliakov E, Uppal S, Rogozin IB, Gentleman S, Redmond TM, 2020. Evolutionary aspects and enzymology of metazoan carotenoid cleavage oxygenases. Biochim. Biophys. Acta Mol. Cell Biol. Lipids, 158665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qtaishat NM, Redmond TM, Pepperberg DR, 2003. Acute radiolabeling of retinoids in eye tissues of normal and rpe65-deficient mice. Invest. Ophthalmol. Vis. Sci 44, 1435–1446. [DOI] [PubMed] [Google Scholar]
- Redmond TM, 2009. Focus on Molecules: RPE65, the visual cycle retinol isomerase. Exp. Eye Res 88, 846–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmond TM, Hamel CP, 2000. Genetic analysis of RPE65: from human disease to mouse model. Methods Enzymol. 316, 705–724. [DOI] [PubMed] [Google Scholar]
- Redmond TM, Poliakov E, Kuo S, Chander P, Gentleman S, 2010. RPE65, visual cycle retinol isomerase, is not inherently 11-cis-specific: support for a carbocation mechanism of retinol isomerization. J. Biol. Chem 285, 1919–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmond TM, Poliakov E, Yu S, Tsai JY, Lu Z, Gentleman S, 2005. Mutation of key residues of RPE65 abolishes its enzymatic role as isomerohydrolase in the visual cycle. Proc. Natl. Acad. Sci. U.S.A 102, 13658–13663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmond TM, Yu S, Lee E, Bok D, Hamasaki D, Chen N, Goletz P, Ma JX, Crouch RK, Pfeifer K, 1998. Rpe65 is necessary for production of 11-cis-vitamin A in the retinal visual cycle. Nat. Genet 20, 344–351. [DOI] [PubMed] [Google Scholar]
- Ree R, Varland S, Arnesen T, 2018. Spotlight on protein N-terminal acetylation. Exp. Mol. Med 50, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripps H, 2008. The color purple: milestones in photochemistry. FASEB J. 22, 4038–4043. [DOI] [PubMed] [Google Scholar]
- Rodieck RW, 1998. The first steps in seeing. Sinauer Associates, Sunderland, Mass. [Google Scholar]
- Ronquist F, Huelsenbeck JP, 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. [DOI] [PubMed] [Google Scholar]
- Rosenfeld PJ, Dugel PU, Holz FG, Heier JS, Pearlman JA, Novack RL, Csaky KG, Koester JM, Gregory JK, Kubota R, 2018. Emixustat Hydrochloride for Geographic Atrophy Secondary to Age-Related Macular Degeneration: A Randomized Clinical Trial. Ophthalmology 125, 1556–1567. [DOI] [PubMed] [Google Scholar]
- Roska B, Sahel JA, 2018. Restoring vision. Nature 557, 359–367. [DOI] [PubMed] [Google Scholar]
- Russell S, Bennett J, Wellman JA, Chung DC, Yu ZF, Tillman A, Wittes J, Pappas J, Elci O, McCague S, Cross D, Marshall KA, Walshire J, Kehoe TL, Reichert H, Davis M, Raffini L, George LA, Hudson FP, Dingfield L, Zhu X, Haller JA, Sohn EH, Mahajan VB, Pfeifer W, Weckmann M, Johnson C, Gewaily D, Drack A, Stone E, Wachtel K, Simonelli F, Leroy BP, Wright JF, High KA, Maguire AM, 2017. Efficacy and safety of voretigene neparvovec (AAV2-hRPE65v2) in patients with RPE65-mediated inherited retinal dystrophy: a randomised, controlled, open-label, phase 3 trial. Lancet 390, 849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saari JC, 2012. Vitamin A metabolism in rod and cone visual cycles. Annu. Rev. Nutr 32, 125–145. [DOI] [PubMed] [Google Scholar]
- Saari JC, Nawrot M, Kennedy BN, Garwin GG, Hurley JB, Huang J, Possin DE, Crabb JW, 2001. Visual cycle impairment in cellular retinaldehyde binding protein (CRALBP) knockout mice results in delayed dark adaptation. Neuron 29, 739–748. [DOI] [PubMed] [Google Scholar]
- Samardzija M, von Lintig J, Tanimoto N, Oberhauser V, Thiersch M, Reme CE, Seeliger M, Grimm C, Wenzel A, 2008. R91W mutation in Rpe65 leads to milder early-onset retinal dystrophy due to the generation of low levels of 11-cis-retinal. Hum. Mol. Genet 17, 281–292. [DOI] [PubMed] [Google Scholar]
- Sander CL, Sears AE, Pinto AFM, Choi EH, Kahremany S, Gao F, Salom D, Jin H, Pardon E, Suh S, Dong Z, Steyaert J, Saghatelian A, Skowronska-Krawczyk D, Kiser PD, Palczewski K, 2021. Nano-scale resolution of native retinal rod disk membranes reveals differences in lipid composition. J. Cell Biol 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz P, Preising M, Lorenz B, Sander B, Larsen M, Rosenberg T, 2011. Fundus albipunctatus associated with compound heterozygous mutations in RPE65. Ophthalmology 118, 888–894. [DOI] [PubMed] [Google Scholar]
- Schonthaler HB, Lampert JM, Isken A, Rinner O, Mader A, Gesemann M, Oberhauser V, Golczak M, Biehlmaier O, Palczewski K, Neuhauss SC, von Lintig J, 2007. Evidence for RPE65-independent vision in the cone-dominated zebrafish retina. Eur. J. Neurosci 26, 1940–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz SH, Tan BC, Gage DA, Zeevaart JA, McCarty DR, 1997. Specific oxidative cleavage of carotenoids by VP14 of maize. Science 276, 1872–1874. [DOI] [PubMed] [Google Scholar]
- Seeliger MW, Grimm C, Stahlberg F, Friedburg C, Jaissle G, Zrenner E, Guo H, Reme CE, Humphries P, Hofmann F, Biel M, Fariss RN, Redmond TM, Wenzel A, 2001. New views on RPE65 deficiency: the rod system is the source of vision in a mouse model of Leber congenital amaurosis. Nat. Genet 29, 70–74. [DOI] [PubMed] [Google Scholar]
- Sheridan C, Boyer NP, Crouch RK, Koutalos Y, 2017. RPE65 and the Accumulation of Retinyl Esters in Mouse Retinal Pigment Epithelium. Photochem. Photobiol 93, 844–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y, Moiseyev G, Chakraborty D, Ma JX, 2017. A Dominant Mutation in Rpe65, D477G, Delays Dark Adaptation and Disturbs the Visual Cycle in the Mutant Knock-In Mice. Am. J. Pathol 187, 517–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y, Moiseyev G, Petrukhin K, Cioffi CL, Muthuraman P, Takahashi Y, Ma JX, 2018. A novel RPE65 inhibitor CU239 suppresses visual cycle and prevents retinal degeneration. Biochim. Biophys. Acta Mol. Basis Dis 1864, 2420–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyam R, Gorusupudi A, Nelson K, Horvath MP, Bernstein PS, 2017. RPE65 has an additional function as the lutein to meso-zeaxanthin isomerase in the vertebrate eye. Proc. Natl. Acad. Sci. U.S.A 114, 10882–10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG, 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieving PA, Chaudhry P, Kondo M, Provenzano M, Wu D, Carlson TJ, Bush RA, Thompson DA, 2001. Inhibition of the visual cycle in vivo by 13-cis retinoic acid protects from light damage and provides a mechanism for night blindness in isotretinoin therapy. Proc. Natl. Acad. Sci. U.S.A 98, 1835–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvaroli JA, Arne JM, Chelstowska S, Kiser PD, Banerjee S, Golczak M, 2016. Ligand Binding Induces Conformational Changes in Human Cellular Retinol-binding Protein 1 (CRBP1) Revealed by Atomic Resolution Crystal Structures. J. Biol. Chem 291, 8528–8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon A, Hellman U, Wernstedt C, Eriksson U, 1995. The Retinal-Pigment Epithelial-Specific 11-Cis Retinol Dehydrogenase Belongs to the Family of Short-Chain Alcohol Dehydrogenases. J. Biol. Chem 270, 1107–1112. [PubMed] [Google Scholar]
- Singh R, Kaushik S, Wang YJ, Xiang YQ, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ, 2009. Autophagy regulates lipid metabolism. Nature 458, 1131–U1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M, March J, 2001. March's advanced organic chemistry : reactions, mechanisms, and structure, March's advanced organic chemistry : reactions, mechanisms, and structure, 5th ed. Wiley, New York, pp. xviii, 2083 p. [Google Scholar]
- Stecher H, Gelb MH, Saari JC, Palczewski K, 1999. Preferential release of 11-cis-retinol from retinal pigment epithelial cells in the presence of cellular retinaldehyde-binding protein. J. Biol. Chem 274, 8577–8585. [DOI] [PubMed] [Google Scholar]
- Stecher H, Palczewski K, 2000. Multienzyme analysis of visual cycle. Methods Enzymol. 316, 330–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiles M, Moiseyev GP, Budda ML, Linens A, Brush RS, Qi H, White GL, Wolf RF, Ma JX, Floyd R, Anderson RE, Mandal NA, 2015. PBN (Phenyl-N-Tert-Butylnitrone)-Derivatives Are Effective in Slowing the Visual Cycle and Rhodopsin Regeneration and in Protecting the Retina from Light-Induced Damage. PLoS ONE 10, e0145305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh S, Choi EH, Leinonen H, Foik AT, Newby GA, Yeh WH, Dong Z, Kiser PD, Lyon DC, Liu DR, Palczewski K, 2021. Restoration of visual function in adult mice with an inherited retinal disease via adenine base editing. Nat. Biomed. Eng 5, 169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui X, Golczak M, Zhang J, Kleinberg KA, von Lintig J, Palczewski K, Kiser PD, 2015. Utilization of Dioxygen by Carotenoid Cleavage Oxygenases. J. Biol. Chem 290, 30212–30223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui X, Weitz AC, Farquhar ER, Badiee M, Banerjee S, von Lintig J, Tochtrop GP, Palczewski K, Hendrich MP, Kiser PD, 2017. Structure and Spectroscopy of Alkene-Cleaving Dioxygenases Containing an Atypically Coordinated Non-Heme Iron Center. Biochemistry 56, 2836–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui X, Zhang J, Golczak M, Palczewski K, Kiser PD, 2016. Key Residues for Catalytic Function and Metal Coordination in a Carotenoid Cleavage Dioxygenase. J. Biol. Chem 291, 19401–19412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Moiseyev G, Ablonczy Z, Chen Y, Crouch RK, Ma JX, 2009. Identification of a novel palmitylation site essential for membrane association and isomerohydrolase activity of RPE65. J. Biol. Chem 284, 3211–3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Moiseyev G, Chen Y, Ma JX, 2005. Identification of conserved histidines and glutamic acid as key residues for isomerohydrolase activity of RPE65, an enzyme of the visual cycle in the retinal pigment epithelium. FEBS Lett. 579, 5414–5418. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Moiseyev G, Ma JX, 2014. Identification of Key Residues Determining Isomerohydrolase Activity of Human RPE65. J. Biol. Chem 289, 26743–26751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takimoto N, Kusakabe T, Horie T, Miyamoto Y, Tsuda M, 2006. Origin of the vertebrate visual cycle: III. Distinct distribution of RPE65 and beta-carotene 15,15'-monooxygenase homologues in Ciona intestinalis. Photochem. Photobiol 82, 1468–1474. [DOI] [PubMed] [Google Scholar]
- Takimoto N, Kusakabe T, Tsuda M, 2007. Origin of the vertebrate visual cycle. Photochem. Photobiol 83, 242–247. [DOI] [PubMed] [Google Scholar]
- Tan BC, Schwartz SH, Zeevaart JA, McCarty DR, 1997. Genetic control of abscisic acid biosynthesis in maize. Proc. Natl. Acad. Sci. U.S.A 94, 12235–12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang PH, Buhusi MC, Ma JX, Crouch RK, 2011a. RPE65 Is Present in Human Green/Red Cones and Promotes Photopigment Regeneration in an In Vitro Cone Cell Model. J. Neurosci 31, 18618–18626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang PH, Kono M, Koutalos Y, Ablonczy Z, Crouch RK, 2013. New insights into retinoid metabolism and cycling within the retina. Prog. Retin. Eye Res 32, 48–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang PH, Wheless L, Crouch RK, 2011b. Regeneration of Photopigment Is Enhanced in Mouse Cone Photoreceptors Expressing RPE65 Protein. J. Neurosci 31, 10403–10411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terakita A, 2005. The opsins. Genome Biol. 6, 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis GH, Golczak M, Moise AR, Palczewski K, 2007. Diseases caused by defects in the visual cycle: retinoids as potential therapeutic agents. Annu. Rev. Pharmacol. Toxicol 47, 469–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trehan A, Canada FJ, Rando RR, 1990. Inhibitors of retinyl ester formation also prevent the biosynthesis of 11-cis-retinol. Biochemistry 29, 309–312. [DOI] [PubMed] [Google Scholar]
- Trudel E, Beaufils S, Renault A, Breton R, Salesse C, 2006. Binding of RPE65 fragments to lipid monolayers and identification of its partners by glutathione S-transferase pull-down assays. Biochemistry 45, 3337–3347. [DOI] [PubMed] [Google Scholar]
- Tsang SH, Sharma T, 2018. Leber Congenital Amaurosis. Adv. Exp. Med. Biol 1085, 131–137. [DOI] [PubMed] [Google Scholar]
- Uppal S, Liu T, Poliakov E, Gentleman S, Redmond TM, 2019a. The dual roles of RPE65 S-palmitoylation in membrane association and visual cycle function. Sci. Rep 9, 5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppal S, Poliakov E, Gentleman S, Redmond TM, 2019b. RPE65 Palmitoylation: A Tale of Lipid Posttranslational Modification. Adv. Exp. Med. Biol 1185, 537–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppal S, Rogozin IB, Redmond TM, Poliakov E, 2020. Palmitoylation of Metazoan Carotenoid Oxygenases. Molecules 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdez CE, Smith QA, Nechay MR, Alexandrova AN, 2014. Mysteries of metals in metalloenzymes. Acc. Chem. Res 47, 3110–3117. [DOI] [PubMed] [Google Scholar]
- Verbakel SK, van Huet RAC, Boon CJF, den Hollander AI, Collin RWJ, Klaver CCW, Hoyng CB, Roepman R, Klevering BJ, 2018. Non-syndromic retinitis pigmentosa. Prog. Retin. Eye Res 66, 157–186. [DOI] [PubMed] [Google Scholar]
- Voigt AP, Mulfaul K, Mullin NK, Flamme-Wiese MJ, Giacalone JC, Stone EM, Tucker BA, Scheetz TE, Mullins RF, 2019. Single-cell transcriptomics of the human retinal pigment epithelium and choroid in health and macular degeneration. Proc. Natl. Acad. Sci. U.S.A 116, 24100–24107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Lintig J, Moon J, Babino D, 2021. Molecular components affecting ocular carotenoid and retinoid homeostasis. Prog. Retin. Eye Res 80, 100864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wald G, 1933. Vitamin A in the retina. Nature 132, 316–317. [Google Scholar]
- Wald G, 1968. The molecular basis of visual excitation. Nature 219, 800–807. [DOI] [PubMed] [Google Scholar]
- Wang JS, Estevez ME, Cornwall MC, Kefalov VJ, 2009. Intra-retinal visual cycle required for rapid and complete cone dark adaptation. Nat. Neurosci 12, 295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward R, Kaylor JJ, Cobice DF, Pepe DA, McGarrigle EM, Brockerhoff SE, Hurley JB, Travis GH, Kennedy BN, 2020. Non-photopic and photopic visual cycles differentially regulate immediate, early, and late phases of cone photoreceptor-mediated vision. J. Biol. Chem 295, 6482–6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warshel A, Sharma PK, Kato M, Xiang Y, Liu H, Olsson MH, 2006. Electrostatic basis for enzyme catalysis. Chem. Rev 106, 3210–3235. [DOI] [PubMed] [Google Scholar]
- Welte MA, Gould AP, 2017. Lipid droplet functions beyond energy storage. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1862, 1260–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel A, Reme CE, Williams TP, Hafezi F, Grimm C, 2001. The Rpe65 Leu450Met variation increases retinal resistance against light-induced degeneration by slowing rhodopsin regeneration. J. Neurosci 21, 53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel A, von Lintig J, Oberhauser V, Tanimoto N, Grimm C, Seeliger MW, 2007. RPE65 is essential for the function of cone photoreceptors in NRL-deficient mice. Invest. Ophthalmol. Vis. Sci 48, 534–542. [DOI] [PubMed] [Google Scholar]
- Williams DS, Lopes VS, 2011. The many different cellular functions of MYO7A in the retina. Biochem. Soc. Trans 39, 1207–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston A, Rando RR, 1998. Regulation of isomerohydrolase activity in the visual cycle. Biochemistry 37, 2044–2050. [DOI] [PubMed] [Google Scholar]
- Wright CB, Chrenek MA, Feng W, Getz SE, Duncan T, Pardue MT, Feng Y, Redmond TM, Boatright JH, Nickerson JM, 2014. The Rpe65 rd12 allele exerts a semidominant negative effect on vision in mice. Invest. Ophthalmol. Vis. Sci 55, 2500–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright CB, Redmond TM, Nickerson JM, 2015. A History of the Classical Visual Cycle. Prog. Mol. Biol. Transl. Sci 134, 433–448. [DOI] [PubMed] [Google Scholar]
- Wyss A, 2004. Carotene oxygenases: a new family of double bond cleavage enzymes. J. Nutr 134, 246S–250S. [DOI] [PubMed] [Google Scholar]
- Xu N, Zhang SO, Cole RA, McKinney SA, Guo F, Haas JT, Bobba S, Farese RV Jr., Mak HY, 2012. The FATP1-DGAT2 complex facilitates lipid droplet expansion at the ER-lipid droplet interface. J. Cell Biol 198, 895–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L, Gollapalli DR, Maiti P, Jahng WJ, Rando RR, 2004. A palmitoylation switch mechanism in the regulation of the visual cycle. Cell 117, 761–771. [DOI] [PubMed] [Google Scholar]
- Yang G, Liu Z, Xie S, Li C, Lv L, Zhang M, Zhao J, 2017. Genetic and phenotypic characteristics of four Chinese families with fundus albipunctatus. Sci. Rep 7, 46285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zernant J, Kulm M, Dharmaraj S, den Hollander AI, Perrault I, Preising MN, Lorenz B, Kaplan J, Cremers FP, Maumenee I, Koenekoop RK, Allikmets R, 2005. Genotyping microarray (disease chip) for Leber congenital amaurosis: detection of modifier alleles. Invest. Ophthalmol. Vis. Sci 46, 3052–3059. [DOI] [PubMed] [Google Scholar]
- Zhang J, Choi EH, Tworak A, Salom D, Leinonen H, Sander CL, Hoang TV, Handa JT, Blackshaw S, Palczewska G, Kiser PD, Palczewski K, 2019. Photic generation of 11-cis-retinal in bovine retinal pigment epithelium. J. Biol. Chem 294, 19137–19154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Kiser PD, Badiee M, Palczewska G, Dong Z, Golczak M, Tochtrop GP, Palczewski K, 2015. Molecular pharmacodynamics of emixustat in protection against retinal degeneration. J. Clin. Invest 125, 2781–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Pack W, Khan NW, Wong KY, 2016. Prolonged Inner Retinal Photoreception Depends on the Visual Retinoid Cycle. J. Neurosci 36, 4209–4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong M, Kawaguchi R, Kassai M, Sun H, 2012. Retina, retinol, retinal and the natural history of vitamin A as a light sensor. Nutrients 4, 2069–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann R, Strauss JG, Haemmerle G, Schoiswohl G, Birner-Gruenberger R, Riederer M, Lass A, Neuberger G, Eisenhaber F, Hermetter A, Zechner R, 2004. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science 306, 1383–1386. [DOI] [PubMed] [Google Scholar]
- Znoiko SL, Crouch RK, Moiseyev G, Ma JX, 2002. Identification of the RPE65 protein in mammalian cone photoreceptors. Invest. Ophthalmol. Vis. Sci 43, 1604–1609. [PubMed] [Google Scholar]