Abstract
Background
In 2015, herpes simplex virus 1 (HSV-1)-derived talimogene laherparepvec (T-VEC) was the first oncolytic virus approved by the US Food and Drug Administration as a therapeutic agent for cancer treatment. However, its antitumor application is limited to local treatment of melanoma, and there is a lack of understanding of the mechanisms underlying the regulation of HSV-1 replication in cancer cells and the associated antitumor immunity. We hypothesized that increasing the replication capacity of HSV-1 in tumor cells would enhance the antitumor effect of this virus.
Methods
We systematically identified IFN-stimulated genes induced by HSV-1 by performing functional screens and clarified the mechanism by which BACH1 acts against HSV-1. Then, we tested the effect of BACH1 deficiency on immunogenic cell death induced by HSV-1. Furthermore, we investigated the antitumor effect of BACH1 deficiency on HSV-1 in MCA205 and B16 murine tumor models.
Results
We identified eight IFN-stimulated genes (ISGs) controlling HSV-1 replication, among which BTB and CNC homology 1 (BACH1) suppressed HSV-1 replication by inhibiting the transcription of ICP4, ICP27, and UL39. Loss of Bach1 function not only increased HSV-1 proliferation but also promoted HSV-1-induced cell apoptosis, HMGB1 secretion, and calreticulin exposure in tumor cells. More importantly, hemin, an FDA-approved drug known to downregulate BACH1, significantly enhanced HSV-1-mediated antitumor activity with increased T lymphocyte infiltration at the tumor site.
Conclusions
Our studies uncovered a novel antiviral activity of BACH1 and provided a new strategy for improving the clinical efficiency of the oncolytic virus HSV-1.
Keywords: IFN stimulated genes, Bach1, HSV-1, Hemin, Antitumor immunity
Subject terms: Immunotherapy, Tumour virus infections
Introduction
The immune system plays a vital role in the formation of tumors, and a variety of immunotherapies have been developed [1, 2]. The emergence of immunotherapy, especially immune checkpoint inhibitors, has completely changed the approach to cancer treatment. For some cancers, anti-PD-1/PD-L1 antibodies have become the preferred treatment, as they promote T lymphocyte-mediated elimination of tumors [3–6]. However, anti-PD-1/PD-L1 antibodies have no significant effects on patients with low lymphocyte infiltration in the tumor microenvironment [7, 8]. Therefore, oncolytic viruses that change the tumor microenvironment and promote lymphocyte infiltration have become a promising therapy [9, 10].
Oncolytic viruses selectively replicate in and lyse tumor cells, promoting the presentation of tumor antigens by antigen-presenting cells and the release of damage- or pathogen-associated molecular patterns [11]. Various oncolytic viruses, including herpes simplex virus, vaccinia, adenovirus, reovirus, vesicular stomatitis virus, poliovirus, measles, and coxsackie virus, have been tested in clinical trials; in particular, HSV-1 can induce local and distant antitumor immunity in melanoma patients [12–14]. In 2015, the FDA approved the oncolytic herpes virus talimogene laherparepvec (T-VEC), which lacks ribonucleotide reductase and expresses granulocyte-macrophage colony-stimulating factor (GM-CSF), as a first-in-class agent for metastatic melanoma. T-VEC works in two ways: direct lysis of neoplastic cells and stimulation of a tumor antigen-specific adaptive immune response [15, 16]. In a phase 3 clinical trial, the response rate in the T-VEC treatment group was 16%, which needs to be improved [17]. There are two potential reasons for the low efficacy of T-VEC in patients: neutralizing anti-HSV-1 antibodies and innate anti-HSV-1 resistance mechanisms that control HSV-1 replication and dissemination. Therefore, further studies are needed to improve the efficacy of T-VEC.
Type I interferons (IFNs) play an important role in antiviral responses by binding IFN receptors (IFNARs), resulting in the induction of IFN-stimulated gene (ISG) transcription [18, 19]. ISGs have diverse effects on different viruses mediated through distinct mechanisms. CH25H and IFITMs inhibit viral entry, PARP12 promotes the degradation of the Zika virus NS1 and NS3 proteins, and TETHERIN prevents the release of virions from infected cells [20–24]. There is evidence that HSV-1 is sensitive to IFN, and some ISGs that act against HSV-1, such as CH25H and IRF1, have been identified [21, 25]. Herein, we identified BACH1 as an inhibitor of HSV-1 replication. BACH1 belongs to the basic region leucine zipper (bZip) family, whose members bind to promoters and thus regulate the transcription of target genes [26]. BACH1 plays important roles in diverse cellular processes, such as the cell cycle, differentiation, the oxidative stress response, heme homeostasis, ferroptosis, and cancer metastasis [27–32]. The intracellular level of the BACH1 protein is regulated by heme, which stimulates the ubiquitination and proteasomal degradation of BACH1 [33].
To systematically identify ISGs induced against HSV-1, we performed a functional screen and identified eight ISGs with antiviral effects on HSV-1. We validated that BACH1 inhibited the replication of HSV-1 by repressing the transcription of ICP4, ICP27, and UL39. Furthermore, Bach1 deficiency enhanced immunogenic cell death and antigen presentation induced by HSV-1. Finally, Bach1 depletion or degradation by hemin increased the antitumor effect of HSV-1, which highlighted the clinical relevance of our findings. This study provides a new strategy for improving the clinical efficacy of the oncolytic virus T-VEC.
Results
Screening of interferon-inducible genes that inhibit HSV-1 replication
In a previous study, we performed a microarray analysis of IFN-α- and IFN-γ-stimulated murine bone marrow-derived macrophages and identified 288 IFN-stimulated genes (ISGs) [34]. To identify the specific ISGs with activity against HSV-1, HEK293T cells were transfected with the individual ISGs in triplicate for 36 h and then infected with HSV-1 coexpressing a luciferase reporter gene (HSV-1-Luc) for 12 h. Then, we lysed the cells and measured HSV-1 luciferase activity. IRF1, which has been identified to act against HSV-1, was used as a positive control for the screen [25]. Normalized results for the measured luciferase activity were obtained by comparison with infected cells that were cotransfected with the control gene GFP, because it has been reported that GFP does not influence HSV-1 replication [35]. Thirteen genes that could inhibit HSV-1-Luc production by ~60% were identified in the screening results (Fig. 1A). The median HSV-1 luciferase activity was 104% in cells transfected with GFP (Fig. 1B). Next, we verified these candidate genes in independent experiments and confirmed that the eight genes still inhibited luciferase activity (Fig. 1C). Similar results were found when we measured the genome copy number and plaque-forming units (pfu) of HSV-1 (Fig. 1D, E). Given these results, we found that eight ISGs inhibited the replication of HSV-1. Among these genes, BACH1 has not been previously reported to have antiviral activity. Therefore, we chose to further examine the anti-HSV-1 role of BACH1.
Fig. 1.
Screening of interferon-inducible genes that inhibit HSV-1 replication. A HEK293T cells were transfected with individual ISGs for 36 h and then infected with HSV-1 for 12 h. The cells were lysed, and luciferase activity was measured. Each dot represents an ISG, and its effect on HSV-1 luciferase activity was normalized to that of control cells transfected with GFP. Values represent the mean of triplicates. B Histogram of all ISGs. Their effects on HSV-1-Luc were normalized to HSV-1-Luc in control cells transfected with GFP. C Selected ISGs from the screen were verified in three independent experiments and normalized to the control. Values represent the mean ± SEM. D and E The inhibitory effects of ISGs selected from previous screens against HSV-1-Luc (C) were measured by RT–PCR (D) and a plaque assay (E). Values represent the mean ± SEM of three independent experiments
Deficiency of BACH1 leads to susceptibility to HSV-1 infection in vitro
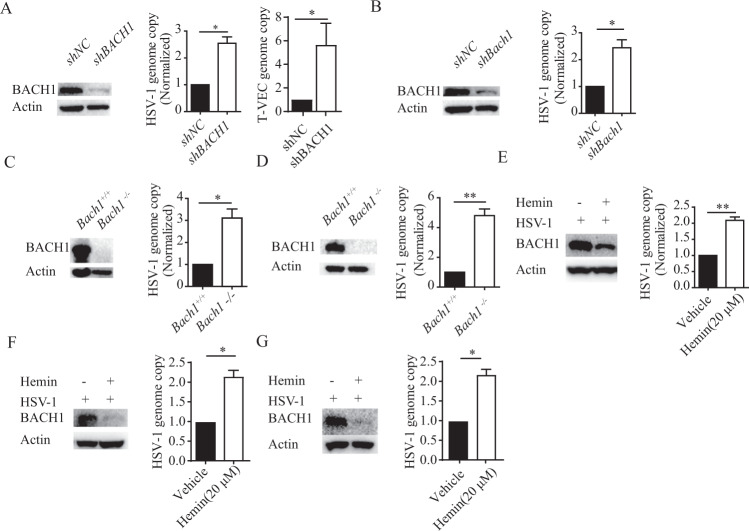
To further characterize the function of BACH1, we generated HEK293T and primary embryonic fibroblast (MEF) cell lines with stable Bach1 knockdown using small hairpin RNA (shRNA) sequences against BACH1 (Fig. 2A, B). Then, HEK293T cells with Bach1 knockdown were infected with HSV-1 or T-VEC. At 24 h post-infection, we found that the replication of HSV-1 and T-VEC was increased significantly (Fig. 2A). Similar results were found in MEFs with Bach1 knockdown (Fig. 2B) and MCA205 fibrosarcoma cells and B16 melanoma cells in which Bach1 was knocked out by a CRISPR–Cas9 approach (Fig. 2C, D). These results suggested that BACH1 played an important role in controlling HSV-1 replication.
Fig. 2.
Loss of BACH1 function increases the replication of HSV-1. A and B HEK293T cells (A) and MEFs (B) with shRNA-mediated stable knockdown of BACH1 were confirmed by western blotting. These stable knockdown cells were infected with HSV-1 and T-VEC (0.25 MOI), and then the genome copies of HSV-1 and T-VEC were measured by qRT–PCR at 24 hpi. C and D MCA205 cells (C) and B16 cells (D) with stable Bach1 knockout were confirmed by western blotting. These stable knockout cells were infected with HSV-1 (0.25 MOI), and the genome copies of HSV-1 were measured by qRT–PCR at 24 hpi. E–G HEK293T cells, MCA205 cells (F) and B16 cells (G) treated with 20 μM hemin or vehicle were infected with HSV-1 (0.25 MOI), and the expression level of BACH1 and genome copies of HSV-1 were measured by western blotting and qRT–PCR, respectively, at 24 hpi. Values represent the mean ± SEM of three independent experiments. *P < 0.05, **P < 0.01 by Student’s t test
BACH1 is degraded by the proteasome when free intracellular heme is high [33]. Hemin is a derivative of heme and can be used as an alternative agent to decrease the expression level of BACH1. We added 20 μM hemin to HEK293T, MCA205 fibrosarcoma or B16 melanoma cells and infected them with HSV-1. Then, the expression level of BACH1 was measured by western blotting. As shown in Fig. 2E-G, the BACH1 protein level was decreased in hemin-treated cells. To test the effect of hemin on HSV-1 replication, HEK293T cells, MCA205 fibrosarcoma cells and B16 melanoma cells were incubated with 20 μM hemin and infected with HSV-1. HSV-1 replication was increased with hemin treatment compared to control treatment. These results indicated that hemin increased the replication of HSV-1 by promoting BACH1 degradation (Fig. 2E, G).
BACH1 represses the transcription of ICP4, ICP27, and UL39
As a transcription factor, BACH1 usually interacts with MAFK to suppress the transcription of target genes, such as HO-1 [36]. We investigated whether BACH1-mediated inhibition of HSV-1 replication was dependent on MAFK. We knocked down MAFK in HEK293T cells with shRNA sequences against MAFK (Fig. S1A) and found that HSV-1 replication in the MAFK-knockdown cells was similar to that in control cells (Fig. S1B). Furthermore, the MAFK-knockdown cell line was transfected with GFP and BACH1 plasmids and then infected with HSV-1. BACH1 overexpression still inhibited HSV-1 replication in the MAFK-knockdown cell line (Fig. S1C). These results indicated that BACH1 inhibited the replication of HSV-1 in a MAFK-independent manner.
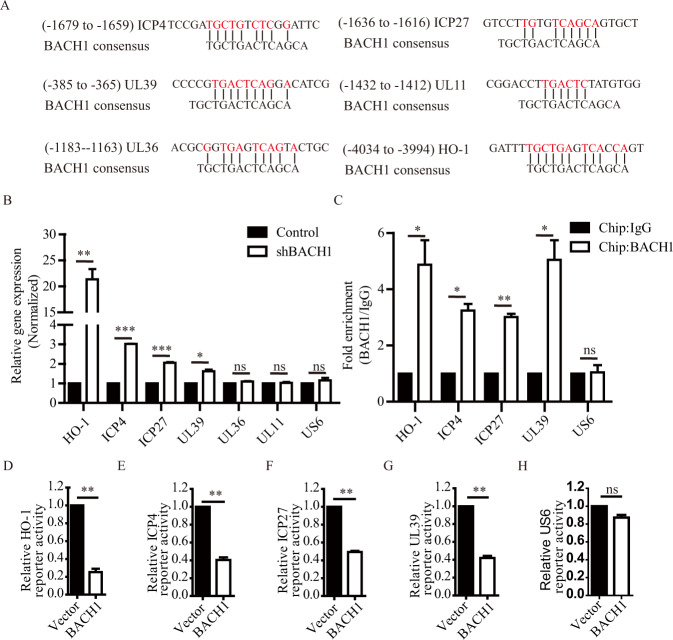
Next, we investigated whether BACH1 inhibited the replication of HSV-1 by repressing the transcription of viral genes. We performed a search for BACH1-binding sequences in the genome of HSV-1 strain F (GenBank accession no. GU734771.1) and found five viral gene promoters (ICP4, ICP27, UL11, UL36, and UL39) containing potential BACH1-binding sequences (Fig. 3A). To study whether BACH1 represses the expression of these genes, we detected the mRNA expression levels of these genes in shNC and shBACH1 HEK293T cells infected with HSV-1 and found that knockdown of BACH1 increased the expression of HO-1, ICP4, ICP27, and UL39 but not that of UL11, UL36, or US6; US6 does not contain a BACH1-binding sequence (Fig. 3B). Next, we confirmed increased BACH1 occupancy on the HO-1, ICP4, ICP27, and UL39 promoters but not on the UL11, UL36, and US6 promoters in HEK293T cells stably expressing BACH1 (Fig. 3C and Fig. S2). To further identify whether BACH1 suppressed the transcription of these genes via these promoter interactions, the promoter regions containing the BACH1-binding sequence were cloned into luciferase reporter vectors, and overexpression of BACH1 significantly decreased the luciferase activity driven by the HO-1, ICP4, ICP27 or UL39 promoter (Fig. 3D–G) but failed to alter the luciferase activity driven by the US6 promoter (Fig. 3H). These results suggested that BACH1 inhibited the replication of HSV-1 by repressing the transcription of ICP4, ICP27, and UL39.
Fig. 3.
BACH1 suppressed the transcription of the viral genes ICP4, ICP27, and UL39 by binding to their promoters. A Alignment of the BACH1-binding site and promoter sequences of the ICP4, ICP27, UL39, UL11, UL36, and HO-1 genes. B qRT–PCR analyses of RNA from shNC and shBACH1 HEK293T cells infected with HSV-1-Luc for 12 hpi. C BACH1 enrichment in the promoter regions of the ICP4, ICP27, and UL39 genes assayed by a ChIP-qRT–PCR assay (compared with control IgG binding). HEK293T cells stably transfected with a plasmid containing BACH1 and infected with HSV-1-Luc for 24 h. The BACH1-DNA complex was precipitated with an anti-BACH1 antibody or control rabbit IgG. The precipitated DNA was subjected to qRT–PCR. D–H Luciferase activities driven by the HO-1 (D), ICP4 (E), ICP27 (F), UL39 (G) and US6 (H) promoters in HEK293T cells stably transfected with plasmids containing BACH1 (BACH1) or the empty vector (Vector). Values represent the mean ± SEM of three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant; Student’s t test
Loss of Bach1 function promotes the antitumor activity of HSV-1
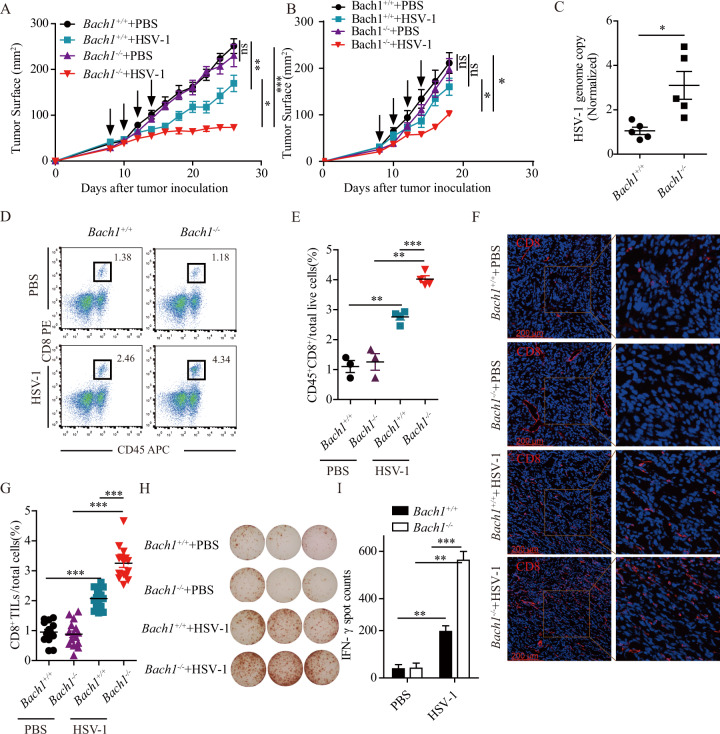
As an oncolytic virus, HSV-1 can generate systemic antitumor immune responses [37]. Therefore, we anticipated that Bach1 deficiency would further enhance the antitumor activity of HSV-1 by increasing the replication of HSV-1. As shown in Fig. 4A, Bach1-deficient MCA205 fibrosarcoma tumors did not show a difference in tumor growth compared to control cells in the absence of HSV-1 infection, while a growth delay was detected when MCA205 fibrosarcoma tumors were infected with HSV-1. More importantly, the sizes of Bach1−/− MCA205 fibrosarcoma tumors injected with HSV-1 were significantly decreased. Similar results were found in B16-F10 cells (Fig. 4B). However, injection of HSV-1 into B16-F10 tumors did not significantly decrease tumor growth, which may be due to the differences between MCA205 fibrosarcoma cells and B16 cells, and the reasons need to be further studied.
Fig. 4.
Loss of Bach1 function enhances the oncolytic effect of HSV-1. A and B Bach1+/+ and Bach1−/− MCA205 cells (A) and B16 cells (B) were injected subcutaneously (s.c.) into the right flank of naive C57BL/6 mice. When the tumor size reached approximately 25 mm2, the tumors were treated by intratumoral injection of 50 μL of 5 × 107 pfu/mL HSV-1 or vehicle every other day for a total of four times (arrows). Tumor growth was monitored 3–4 times per week. Each group contained at least five mice. Quantitative data are reported as the mean ± SEM; *P < 0.05, **P < 0.01, ***P < 0.001 by the Mann–Whitney U test. C Twenty-four hours after the first HSV-1 injection, we extracted genomic DNA from MCA205 tumors, which was used to quantify HSV-1 genomic DNA by qRT–PCR. Each group contained five mice, and quantitative data are reported as the mean ± SEM; *P < 0.05 by Student’s t test. D–I Twenty-four hours after the last HSV-1 injection, MCA205 tumors were harvested and processed to detect the indicated proteins by flow cytometry (D and E), immunofluorescence microscopy (F and G) and IFN-γ ELISpot (H and I). D and E Representative flow cytometry graphs showing the percentages of tumor-infiltrating CD45+CD8+ T cells (D) and statistical data (E). Each group contained at least three mice, and quantitative data are reported as the mean ± SEM; **P < 0.01, ***P < 0.001 by Student’s t test. F and G Representative immunofluorescence staining images of CD8+ T cells (F) and statistical data (G). Each group contained four mice, and quantitative data are reported as the mean ± SEM; ***P < 0.001 by Student’s t test. H and I Quantification of IFN-γ T cells from MCA205 tumors represented by images of IFN-γ T cells (H) and statistical data (I). Each group contained three mice, and quantitative data are reported as the mean ± SEM; **P < 0.01, ***P < 0.001 by Student’s t test
To determine whether Bach1 deficiency increased HSV-1 replication in the above mouse tumor model, Bach1−/− and Bach1+/+ MCA205 fibrosarcoma cells were injected subcutaneously into the right flank of immunocompetent C57BL/6 mice. When the tumor sizes reached ~5 × 5 mm2, 50 μL of 5 × 107 pfu HSV-1 or vehicle was injected into the tumors. At 24 h post-injection, the tumors were harvested, and the genomic DNA of HSV-1 was extracted to determine the HSV-1 genome copy number by qPCR. As shown in Fig. 4C, Bach1 deficiency still increased HSV-1 replication in the mouse tumor model.
HSV-1-mediated inhibition of tumor growth is usually accompanied by the infiltration of T lymphocytes. We therefore analyzed tumor-infiltrating T lymphocytes in our mouse model. We harvested vehicle-treated Bach1+/+ MCA205 fibrosarcoma tumors, HSV-1-treated Bach1+/+ MCA205 fibrosarcoma tumors, vehicle-treated Bach1−/− MCA205 fibrosarcoma tumors, and HSV-1-treated Bach1−/− MCA205 fibrosarcoma tumors at 24 h after the last treatment with HSV-1. Through FACS analysis, we found no difference in CD45+CD8+ T lymphocyte infiltration between the vehicle-treated Bach1+/+ and Bach1−/− MCA205 fibrosarcoma tumors. However, CD45+CD8+ T lymphocyte infiltration was increased in HSV-1-treated Bach1+/+ MCA205 fibrosarcoma tumors compared with vehicle-treated Bach1+/+ MCA205 fibrosarcoma tumors. More importantly, Bach1 deficiency in MCA205 fibrosarcoma cells forming tumors infected with HSV-1 further increased the infiltration of CD45+CD8+ T lymphocytes (Fig. 4D and E). Similar results were found by IFC and ELISpot analyses (Fig. 4F–I). These experiments demonstrated that Bach1 deficiency in the context of HSV-1 infection further augmented T lymphocyte infiltration and antitumor activity.
Bach1 deficiency promotes immunogenic cell death and antigen presentation induced by HSV-1
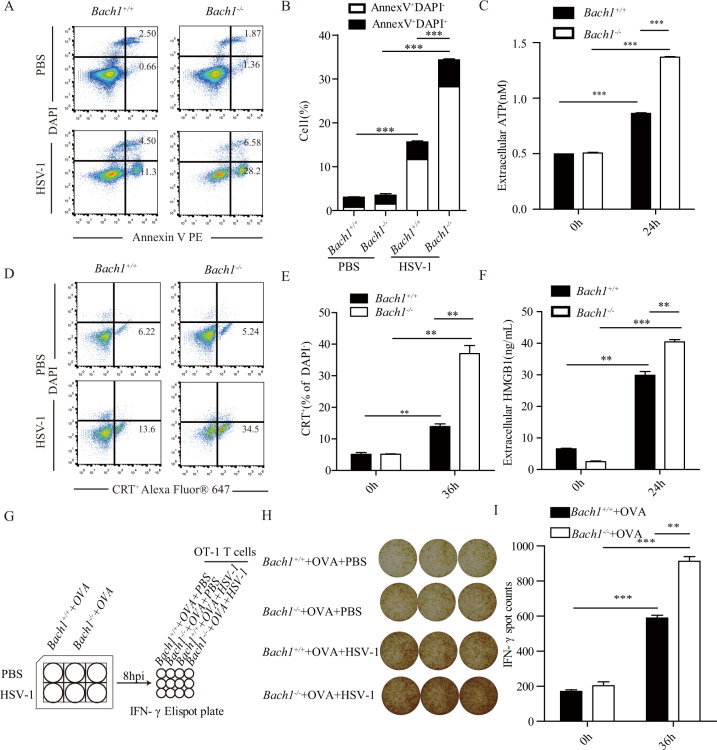
To further study how Bach1 deficiency increases T lymphocyte infiltration induced by HSV-1, we first examined the effect of Bach1 deficiency on HSV-1-induced apoptosis and found that HSV-1-induced cell apoptosis was significantly enhanced in Bach1−/− MCA205 cells compared with Bach1+/+ MCA205 cells (Fig. 5A, B). Then, we detected a series of hallmarks of immunogenic cell death and found that Bach1 deficiency strongly increased HSV-1-induced ATP release, calreticulin exposure and HMGB1 secretion (Fig. 5C–F). In addition, we sought to examine the effect of Bach1 deficiency on antigen presentation induced by HSV-1 with an OT-I mouse model in which OT-I CD8+ T cells can specifically recognize an ovalbumin peptide and become activated. We found that HSV-1 infection significantly increased the secretion of IFN-γ from OT-I CD8+ T cells in the presence of OVA-expressing Bach1−/− MCA205 cells compared with OVA-expressing Bach1+/+ MCA205 cells (Fig. 5G–I). These experiments demonstrated that Bach1 deficiency enhanced immunogenic cell death and antigen presentation induced by HSV-1.
Fig. 5.
Bach1 deficiency promotes immunogenic cell death and antigen presentation induced by HSV-1. A and B Bach1+/+ and Bach1−/− MCA205 cells were infected with HSV-1 at a multiplicity of infection of 1 or left uninfected for 48 h, and apoptosis was assessed through staining with Annexin V and DAPI. Representative flow cytometry graphs are shown in (A), and a quantitative summary is shown in (B). C Bach1+/+ and Bach1−/− MCA205 cells were infected with HSV-1 at a multiplicity of infection of 1 or left uninfected for 24 h, and the supernatants were harvested. Extracellular ATP was quantified with the ENLITEN ATP Assay System. D and E Bach1+/+ and Bach1−/− MCA205 cells were infected with HSV-1 at a multiplicity of infection of 1 or left uninfected for 36 h, and calreticulin exposure was detected by staining with an anti-CRT antibody plus DAPI. Representative flow cytometry graphs are shown in (D), and a quantitative summary is shown in (E). F Bach1+/+ and Bach1−/− MCA205 cells were infected with HSV-1 at a multiplicity of infection of 1 or left uninfected for 24 h, and the supernatants were harvested. The amounts of HMGB1 were measured by HMGB1 enzyme-linked immunosorbent assay (ELISA). G–I Bach1+/+ and Bach1−/− MCA205 OVA-expressing cells were infected with HSV-1 for 8 h at a multiplicity of infection of 1, and the cells were harvested and mixed with OT1 cells. Then, IFN-γ was quantified. A schematic diagram is shown in (G). Representative images of IFN-γ+ T cells are shown in (H), and statistical data are shown in (I). Values represent the mean ± SEM of three independent experiments. **P < 0.01, ***P < 0.001 by Student’s t test
Combination treatment with hemin and HSV-1 significantly activates the antitumor immune response
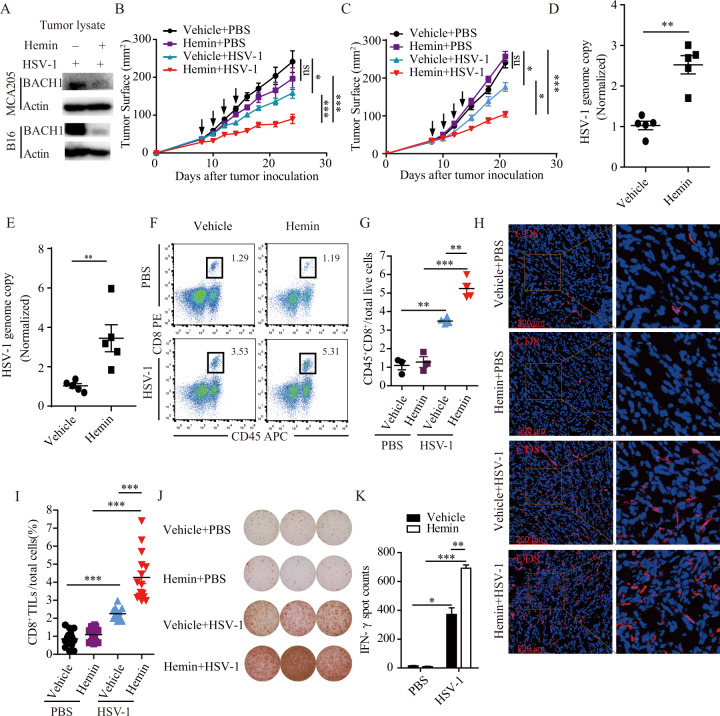
Hemin, an FDA-approved drug, has been shown to degrade BACH1 in vitro. To test its effect on BACH1 in vivo, we injected MCA205 fibrosarcoma or B16 cells subcutaneously into C57BL/6 mice and treated the mice with or without hemin daily by intraperitoneal injection. When the tumor size reached ~5 × 5 mm2, 50 μL of 5 × 107 pfu HSV-1 was injected into the tumors. At 24 h post-injection, the tumors were harvested. As shown in Fig. 6A, the expression level of BACH1 was reduced in tumors treated with hemin.
Fig. 6.
Combination treatment with hemin and HSV-1 further activates the antitumor immune response. A Representative western blot showing the relative expression of BACH1 in tumor lysates treated with hemin or vehicle and infected with HSV-1. Mice were injected with MCA205 cells and B16 cells subcutaneously and treated with 50 mg/kg hemin daily by intraperitoneal injection. 50 μL of 5 × 107 pfu/mL HSV-1 was injected when the tumor size reached ~25 mm2. Twenty-four hours after the HSV-1 injection, the tumors were harvested, and the expression levels of BACH1 were determined by western blotting. The experiments were repeated twice with similar results. B and C Tumor growth of MCA205 cell (B) and B16 cell (C) xenografts treated with 50 mg/kg hemin daily by intraperitoneal injection and/or 50 μL of 5 × 107 pfu/mL HSV-1 or vehicle every other day for a total of four times (arrows) by intratumoral injection beginning when the tumor size reached ~25 mm2. Tumor growth was monitored 3–4 times per week. Each group contained at least five mice, and the results are representative of two independent experiments. Quantitative data are reported as the mean ± SEM; *P < 0.05, **P < 0.01, ***P < 0.001 by the Mann–Whitney U test. D and E Twenty-four hours after the first HSV-1 injection, we extracted genomic DNA from mice injected with MCA205 cells and B16 cells and quantified HSV-1 genomic DNA by RT–PCR. Each group contained five mice, and quantitative data are reported as the mean ± SEM; *P < 0.05 by Student’s t test. F–K Twenty-four hours after the last HSV-1 injection, tumors from mice injected with MCA205 cells were harvested and processed to detect the indicated proteins by flow cytometry (F and G), immunofluorescence microscopy (H and I) and IFN-γ ELISpot (J and K). F and G Representative flow cytometry graphs of the percentages of tumor-infiltrating CD45+CD8+ T cells (F) and statistical data (G). Each group contained at least three mice, and quantitative data are reported as the mean ± SEM; **P < 0.01, ***P < 0.001 by Student’s t test. H and I Representative immunofluorescence staining images of CD8+ T cells (H) and statistical data (I). Each group contained four mice, and quantitative data are reported as the mean ± SEM; ***P < 0.001 by Student’s t test. J and K Quantification of IFN-γ+ T cells from MCA205 tumors. Each well represents one mouse tumor (J), and statistical data are shown (K). Data are represented as the mean ± SEM; *P < 0.05, **P < 0.05, ***P < 0.001
We then investigated the effect of hemin on antitumor activity induced by HSV-1. As shown in Fig. 6B, MCA205 fibrosarcoma cells treated with hemin did not show a significant difference in growth compared to control-treated cells. However, when hemin was combined with HSV-1 to treat MCA205 fibrosarcoma cells, hemin significantly enhanced the tumor-suppressive effects of HSV-1. Similar results were found for B16-F10 cells (Fig. 6C).
To determine whether hemin treatment increased HSV-1 replication in the mouse models, C57BL/6 mice were injected with MCA205 cells or B16 cells and treated with hemin daily by intraperitoneal injection. When the tumor size reached ~5 × 5 mm2, 50 μL of 5 × 107 pfu HSV-1 or vehicle was injected into the tumors. The tumors were harvested at 24 h after HSV-1 injection for genomic DNA extraction, and the HSV-1 genome copy number was measured by qPCR. As shown in Fig. 6D, E, hemin treatment increased HSV-1 replication in the mouse models.
We also wanted to know whether this tumor growth inhibition was accompanied by changes in T lymphocyte infiltration. Through FACS analysis, we found that HSV-1 infection significantly increased CD45+CD8+ T lymphocyte infiltration in MCA205 fibrosarcoma tumor tissues and that hemin treatment further enhanced CD45+CD8+ T lymphocyte infiltration in MCA205 fibrosarcoma tumors infected with HSV-1 (Fig. 6F, G). Similar results were found by IFC and ELISpot analyses (Fig. 6H–K). From these results, we concluded that hemin treatment could enhance CD45+CD8+ T lymphocyte infiltration and antitumor activity induced by HSV-1.
Discussion
There are various immunotherapeutic approaches for cancer treatment, such as anti-PD-1/PD-L1, CAR-T cell, and oncolytic virus therapies [1]. Among these, oncolytic virus therapy has become a promising therapeutic approach because it can alter the tumor microenvironment [9, 10]. Talimogene laherparepvec (T-VCE) is the first-in-class oncolytic agent that was approved in 2015 by the US Food and Drug Administration for metastatic melanoma. In a clinical study involving four European countries, it was reported that 26 (55.3%) patients had no remaining injectable lesions after receiving T-VEC as their first-line therapy [38]. At the Netherlands Cancer Institute, a total of 93 patients with stage IIIB-IVM1a melanoma were treated with T-VEC between December 2016 and January 2020, and 51% of the patients achieved a durable response [39]. However, for those who did not respond to T-VEC treatment, we speculated that one of the reasons might be low replication of the virus in the tumors. Therefore, it is important to improve the replication capability of this oncolytic virus in tumors to further increase treatment efficacy. In this study, we not only identified BACH1 as a novel antiviral ISG that can inhibit HSV-1 replication by repressing the transcription of ICP4, ICP27, and UL39 but also, more importantly, demonstrated that hemin, an FDA-approved drug with the ability to degrade BACH1, significantly enhanced the HSV-1-mediated antitumor immune response.
Our functional screens of 288 ISGs identified 8 ISGs with antiviral activity in HSV-1 infection. Some of these ISGs such as IRF1, CH25H, IFITM3, and IFIH1 have been reported to suppress the replication of HSV-1. However, others such as BIRC6, TMEM171, and PEX26 have not been shown to inhibit HSV-1 replication [21, 22, 25, 40]. Therefore, further studies are needed to elucidate the mechanisms by which these ISGs inhibit HSV-1. T-VEC, which is used in the clinic, does not contain the ICP34.5 gene, which can interact with TBK-1 and result in the inhibition of IRF3 activation [41]. Therefore, some other ISGs that have activity against T-VEC but not HSV-1 may have been omitted in our screening experiments.
As a transcriptional regulator, BACH1 usually interacts with MAFK to suppress gene expression [26], especially in the contexts of oxidative stress and heme homeostasis, in which the BACH1 and MAFK complex binds to the promoter of target genes, such as HO-1. After binding to the target gene promoter, BACH1 suppresses gene transcription. In our study, we found that the BACH1-mediated inhibition of HSV-1 replication was independent of MAFK. Furthermore, we demonstrated that BACH1 bound to the promoters of HSV-1 viral genes such as ICP4, ICP27, and UL39 and repressed gene expression, which led to reduced HSV-1 replication. However, our studies could not exclude the involvement of other targets of BACH1 in HSV-1 replication.
Recently, it was reported that Nectin-1 expression increased the susceptibility of malignant melanoma to an oncolytic herpes simplex virus in vitro and in vivo by promoting the entry of the oncolytic herpes simplex virus and could serve as a biomarker for T-VEC-induced melanoma regression [42]. In our studies, we also demonstrated that BACH1 deficiency could enhance HSV-1-mediated antitumor activity. One possible mechanism underlying this enhanced antitumor activity is the increased HSV-1 replication in Bach1−/− tumors compared with that in Bach1+/+ tumors. In addition, we also observed increased apoptosis, ATP release, calreticulin exposure, HMGB1 secretion and antigen presentation in Bach1−/− MCA205 cells compared to Bach1+/+ MCA205 cells after HSV-1 infection. Furthermore, loss of BACH1 function increased the infiltration of T lymphocytes, including IFN-γ-positive T cells, after intratumoral injection of HSV-1. It is possible that these events are all interrelated and together change the tumor microenvironment, resulting in stronger antitumor immunity. However, the increased population of infiltrating T lymphocytes may not contain only tumor-specific T cells. Some of these cells may be virus-specific T cells. Therefore, further studies are needed to determine the antigen specificity of these infiltrating T cells. In the B16 mouse model, we found that HSV-1 infection did not significantly reduce tumor size. This may be the result of a difference between tumors, but the specific mechanism is currently unclear.
Recently, combining oncolytic viruses with other immunotherapies, such as an anti-PD-1 antibody or anti-CTLA4 antibody, was shown to significantly increase the efficacy of the oncolytic viruses [37, 43, 44]. This is because oncolytic virotherapy can increase T cell infiltration, but reactive expression of PD-1 inhibits the function of these T lymphocytes, and this inhibition can be overcome by using an anti-PD-1 antibody. After oncolytic virotherapy and anti-PD-1 therapy are combined, tumor antigen-specific T lymphocytes are further stimulated, leading to an improved antitumor response. Our studies have validated the possibility of using two FDA-approved drugs, hemin and T-VEC, as a combination therapy against cancers. Hemin can induce the nuclear export of BACH1 and promote BACH1 ubiquitination and proteasomal degradation [33, 45]; therefore, it can regulate the expression levels of BACH1. When we treated HEK293T cells with hemin, the levels of BACH1 decreased, and the replication of HSV-1 increased. More importantly, when we treated mice with hemin combined with HSV-1, significant inhibition of tumor growth was observed, and T lymphocyte infiltration was also increased. However, in our study, we used HSV-1 instead of T-VEC. Future studies are needed to determine the effect of combined treatment with hemin and T-VEC on tumors in mouse models and, more importantly, the clinical applicability of this combination therapy.
Materials and methods
Cell lines, virus, and reagents
MEFs were isolated from Day 13.5 to 14.5 Balb/c mouse embryos. HEK293T, Vero and MCA205 fibrosarcoma cells were cultured in Dulbecco’s modified Eagle’s medium containing L-glutamine (DMEM) (Gibco). B16-F10 cells were cultured in RPMI 1640 medium (Gibco). All the media were supplemented with 10% fetal bovine serum (Gibco) and 100 U/mL penicillin–streptomycin (Gibco). All cell lines were incubated at 37 °C in 5% CO2. HSV-1 (strain F) was propagated in Vero cells and titrated as described previously [46]. T-VEC was purchased from ORIENGENE (Beijing, China). Hemin (MedChemExpress, HY-19424) was dissolved in 20 mM NaOH, and the solution was adjusted to pH 7.5. Then, the hemin solution was filtered using 0.22 μm filters for in vitro assays and mouse treatments.
HSV-1 screening
HEK293T cells were plated in a 96-well plate treated with poly-L-lysine solution (Sigma, P4707). Individual ISG expression plasmids were transfected with Lipo3000 (Invitrogen, L3000015) for 36 h. Cells were infected with HSV-1-Luc at a multiplicity of infection (MOI) of 0.25 for 1 h and lysed in 1× passive lysis buffer (Promega, E1941) at 12 hpi. Then, luciferase activity was measured with a firefly luciferase substrate kit (Promega, E1960).
Detection of the HSV-1 genome copy number
HEK293T cells were plated in 24-well plates treated with poly-L-lysine solution. Individual ISG expression plasmids were transfected with Lipo3000 for 36 h. Cells were infected with HSV-1-Luc at a MOI of 0.25 for 1 h. Genomic DNA was extracted at 24 hpi using a TIANamp Genomic DNA Kit (TIANGEN, DP304-03), and the HSV-1 genome copy number was quantified with qPCR. To quantify the HSV-1 genome copy number in tumors, HSV-1 was intratumorally injected into tumors. After 24 h of HSV-1 infection, the tumors were harvested. Genomic DNA was extracted using a TIANamp Genomic DNA Kit, and the HSV-1 genome copy number was quantified with qPCR.
Plaque assay
In total, 2 × 105 Vero cells were seeded in 12-well plates. Supernatants from HSV-1-infected cells were serially diluted and used to infect the Vero cells for 1 h. The cells were then covered with DMEM containing 1% low-melting agarose (VWR LIFE SCIENCE, 0815-100 G) and 2% FBS. Viral plaques were counted after 72 h.
Construction of stable cell lines
The shRNA sequences targeting human BACH1 (forward: 5′-GCCTTAATGACCAGCGGAAGA; reverse: 3′-TCTTCCGCTGGTCATTAAGGC), human MAFK (forward: 5′-GCTCAGCGATGATGAGCTGGT; reverse: 3′-ACCAGCTCATCATCGCTGAGC) and mouse Bach1 (forward: 5′ GGAACTGACAAGATCCGAACT; reverse: 3′-AGTTCGGATCTTGTCAGTTCC) were cloned into LV2N (U6/Puro) (Gene Pharma). These plasmids were cotransfected with the plasmids pMD2.G (Addgene, 12259) and psPAX2 (Addgene, 12260) for 48 h to produce lentiviruses. Then, the lentiviruses were used to infect target cells. After 48 h of infection, the cells were selected with puromycin (0.3 µg/mL) for 10 days. To construct knockout cell lines, sgRNA sequences targeting mouse Bach1 (forward: 5′-CACCGGCGGTTCCGAGCCCACCGCT; reverse: 3′- AAACAGCGGTGGGCTCGGAACCGCC) were cloned into the vector LentiCRISPR v2 (Addgene 52961). The plasmid was cotransfected with the plasmids pMD2.G (Addgene 12259) and psPAX2 (Addgene 12260) for 48 h to produce lentiviruses. Then, the lentiviruses were used to infect target cells. After 48 h of infection, the cells were selected with puromycin (0.4 µg/mL) for 8 days. Single clones were isolated by FACS (Becton Dickinson, San José, CA, USA) and confirmed by western blotting. To construct a BACH1-overexpressing cell line, the BACH1 open reading frame was cloned into the vector pMXsIG-IgkFLAG and transfected into HEK293T cells. After transfection for 48 h, BACH1-overexpressing cells were sorted by fluorescence-activated cell sorting.
Immunoblotting
Cells and tumors were lysed with RIPA buffer supplemented with a protease inhibitor cocktail (Roche, 04693132001) at 4 °C and quantified with a BCA Protein Assay Kit (Beyotime, P0011). After quantification, 30 µg of protein was separated on 10% SDS–PAGE gels. The separated proteins were transferred to PVDF membranes. Then, the membranes were blocked with 5% nonfat milk in TBST and incubated with antibodies against BACH1 (Abcam, ab124919), MAFK (Immunoway, YN2091), actin (Cell Signaling Technology, 8457 S) and tubulin (Cell Signaling Technology, 2146 S). The secondary antibody used for western blotting was anti-rabbit IgG (Cell Signaling Technology, 7074 S). Target proteins were visualized with ECL (Tanon, 180-5001) and scanned with a ChemiDoc XRS + system (Bio–Rad, USA).
Cell death, CRT exposure, ATP release, and HMGB1 secretion
Cell lines were seeded and infected with HSV-1 for the indicated times. Early- or late-stage apoptosis was analyzed with DAPI (BBI Life Sciences, 564907) and Annexin V-PE (BD Bioscience, 556421) staining following the manufacturer’s instructions. Calreticulin exposure was quantified by surface staining with an Alexa Fluor® 647-conjugated rabbit anti-Calreticulin antibody (Abcam, ab196159). Extracellular ATP was quantified with the ENLITEN ATP Assay System (catalog FF2000, Promega). For the HMGB1 secretion assay, supernatants were collected and detected with an HMGB1 ELISA kit (IBL, ST51011).
Antigen presentation
Cell lines were seeded and infected with HSV-1 for the indicated times. Then, 106 cells/microwell were seeded in ELISpot plates precoated with an anti-IFN-γ capture antibody (BD Biosciences, 551881) and cultured for 18–24 h at 37 °C in a 5% CO2 humidified incubator. After the coculture period, the plates were washed and incubated with a biotinylated anti-mouse IFNγ detection antibody (BD Biosciences, 551881) for 2 h at RT. Next, the plates were incubated with an enzyme conjugate for 1 h at RT, followed by substrate reaction. Finally, the plates were washed to stop the reaction and dried, followed by scanning and counting with CTL ImmunoSpot® S6 Analyzers (LLC, OH, USA).
Real-time quantitative PCR
Total RNA was isolated with NucleoZol (MN, 740404.200) according to the manufacturer’s instructions, and cDNA was synthesized with a One Step PrimeScript RT–PCR kit (TaKaRa, 6110 A). Real-time PCR was carried out using a LightCycler 96 (Roche) and SYRB Green qPCR mix (Bimake.cn, B21202). The primer pairs used are shown in Table S1.
Construction of promoter reporter plasmids
The primers used to amplify the promoters of HO-1, ICP4, ICP27, UL39, and US6 are shown in Table S1. Amplified products were cloned into the luciferase reporter vector pGL4 (Addgene, E6721) using standard molecular cloning techniques.
Promoter reporter analysis
HEK293T cells stably transfected with pMXsIG-IgkFLAG-BACH1 or pMXsIG-IgkFLAG were transfected with 500 ng of plasmids containing a promoter reporter and 20 ng of Renilla luciferase-expressing plasmid (pRL-TK; Promega) using Lipofectin 3000 reagent (Invitrogen). The cells were lysed at 48 h after transfection, and luciferase activities were assayed using a Dual-Glo assay kit (Promega). For each lysate, the firefly luciferase activity was normalized to the Renilla luciferase activity to assess the transfection efficiency.
ChIP-PCR assay
HEK293T cells stably transfected with pMXsIG-IgkFLAG-BACH1 were infected with HSV-1. At 24 h after infection, the cells were subjected to a ChIP assay using a method described previously [47, 48]. ChIP-DNA was immunoprecipitated with an anti-BACH1 (Abcam, ab245548) antibody or rabbit IgG (Cell Signaling Technology, 2729 S) and subjected to real-time quantitative PCR. The primers for ChIP-PCR are shown in Table S1.
Tumor model
Female C57BL/6 mice (aged between 6 and 8 weeks) were purchased from Beijing Vital River Company and were randomly assigned to the indicated groups. Bach1+/+ and Bach1−/− MCA205 cells and B16 cells were injected subcutaneously (s.c.) into the right flank of naive C57Bl/6 mice. When the tumor size reached ~25 mm2, the tumors were treated by intratumoral injection of 50 μL of 5 × 107 pfu/mL HSV-1 or vehicle every other day for a total of four times, and tumor growth was monitored with calipers and recorded as tumor size (mm2).
For the hemin-treated tumor model, MCA205 cells and B16 cells were injected subcutaneously (s.c.) into the right flank of naive C57BL/6 mice that were treated with 50 mg/kg hemin daily by intraperitoneal injection. When the tumor size reached ~25 mm2, the tumors were treated by intratumoral injection of 50 μL of 5 × 107 pfu/mL HSV-1 or vehicle every other day for a total of four times, and tumor growth was monitored with calipers and recorded as tumor size (mm2). All animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) of the Suzhou Institute of Systems Medicine (ISM-IACUC-0019-R).
Immune cell isolation and flow cytometry analysis
Tumors were harvested from mice 24 h after the fourth intratumoral injection of HSV-1 and digested into a single-cell suspension with 200 U/mL DNase I (Calbiochem, San Diego, CA, USA) and 0.4 Wünsch units/mL Liberase TL (Roche, Mannheim, Germany) at 37 °C for 30 min. The cell suspension was filtered through 70 μm cell strainers, followed by staining with fluorophore-labeled antibodies against vivid yellow (Invitrogen, L34959), CD8 (BioLegend, 100708), and CD45.2 (BioLegend, 109814) at 4 °C for 25 min. Then, the cells were detected on an Attune NxT flow cytometer (Thermo Fisher Scientific) and analyzed with FlowJo software (TreeStar, Inc., Ashland, OR, USA).
Immunofluorescence staining
Tumors were harvested from mice at 24 h after the fourth intratumoral injection of HSV-1 and fixed with 4% polyformaldehyde for 4 h at room temperature (RT) or overnight at 4 °C. The fixed tumor tissues were transferred to a 30% sucrose solution (in 1× PBS) for 24 h and embedded in OCT. Then, the tumor tissues were frozen at −20 °C. Frozen tissue sections (5 μm) were obtained with a CM1950 cryostat (Leica Biosystems) and then adhered to slides. For immunofluorescence staining, frozen sections were recovered to RT and washed three times. Then, the sections were fixed in 2% polyformaldehyde for 15 min and washed three times. After fixation, the sections were permeabilized with 0.5% Triton X-100 for 5 min and washed three times. After permeabilization, the sections were blocked with 10% FBS for 1 h and subsequently incubated with anti-CD8a antibodies (Abcam, ab217344) at 4 °C overnight. After three thorough washes, the sections were incubated with Alexa488-conjugated goat anti-rabbit IgG (Thermo Fisher Scientific, A-11008) for 1 h at RT. The sections were then washed three times and stained with Hoechst 33258 (Thermo Fisher Scientific, H3569) for 5 min at RT. Images were captured with a confocal microscope (Leica TCS SP8, installed at the Suzhou Institute of Systems Medicine, China) and analyzed with ImageJ software.
Enzyme-linked immunospot (ELISpot) assay
Tumors were harvested from mice at 24 h after the fourth intratumoral injection of HSV-1 and digested into a single-cell suspension. Then, 106 cells/microwell were seeded in ELISpot plates precoated with an anti-IFN-γ capture antibody (BD Biosciences, 551881) and cultured for 18–24 h at 37 °C in a 5% CO2 humidified incubator. After the coculture period, the plates were washed and incubated with a biotinylated anti-mouse IFNγ detection antibody (BD Biosciences, 551881) for 2 h at RT. Next, the plates were incubated with an enzyme conjugate for 1 h at RT, followed by substrate reaction. Finally, the plates were washed to stop the reaction and dried, followed by scanning and counting with CTL ImmunoSpot® S6 Analyzers (LLC, OH, USA).
Statistical analysis
Results are presented as the mean ± SEM. The significance of differences between groups was detected by an unpaired, two-tailed Student’s t test. Tumor growth curves were compared by the Mann–Whitney U test. Statistical analyses were performed using Prism 5 software (GraphPad) or Excel 2007 (Microsoft). A P value of <0.05 was considered statistically significant.
Supplementary information
Acknowledgements
This project was financially supported by the Chinese Academy of Medical Sciences Innovation Fund for Medical Sciences (2021-I2M-1-047 and 2019-I2M-5-049), National Science Funds of China (82073181, 81802870 and 82102371), Nonprofit Central Research Institute Fund of Chinese Academy of Medical Sciences (2020-PT310-006, 2019XK310002 and 2018TX31001), as well as NIH R01AI069120, AI158154, and AI140718 grants, the UCLA AIDS Institute, and UCLA David Geffen School of Medicine–Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research Award Program. H.Y. is supported by science funds from Jiangsu Province (BK20211554, BK20170407) and the Innovative and Entrepreneurial Team grant (2018-2021) from Jiangsu Province. L.L. is supported by Innovative and Entrepreneurial Doctor grant (2020-2022) from Jiangsu Province.
Author contributions
CP and QC contributed equally to this work. CP and QC performed the experiments. CP, XL, and LY analyzed data. CP, QC, YS, JR, LSC, HY and GC wrote the paper. QC, LL, LY, YC, JL, WL, MG, TS, XW, HF, and SC provided technical support. All authors have read and approved the final paper.
Competing interests
HY and CH have filed a patent related to the anti-HSV-1 activity of BACH1.
Footnotes
These authors contributed equally: Chaohu Pan, Qiaomei Cai.
Change history
5/13/2022
A Correction to this paper has been published: 10.1038/s41423-022-00860-7
Contributor Information
Jiayong Liu, Email: liujiayong_doc@163.com.
Heng Yang, Email: yhmyt@hotmail.com.
Genhong Cheng, Email: gcheng@mednet.ucla.edu.
Supplementary information
The online version contains supplementary material available at 10.1038/s41423-021-00824-3.
References
- 1.Mellman I, Coukos G, Dranoff G. Cancer immunotherapy comes of age. Nature. 2011;480:480–9. doi: 10.1038/nature10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science. 2011;331:1565–70. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 3.Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348:56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- 4.Fridman WH, Pages F, Sautes-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer. 2012;12:298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- 5.Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–4. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 6.Vesely MD, Kershaw MH, Schreiber RD, Smyth MJ. Natural innate and adaptive immunity to cancer. Annu Rev Immunol. 2011;29:235–71. doi: 10.1146/annurev-immunol-031210-101324. [DOI] [PubMed] [Google Scholar]
- 7.Herbst RS, Soria JC, Kowanetz M, Fine GD, Hamid O, Gordon MS, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515:563–7. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–71. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin CZ, Xiang GL, Zhu XH, Xiu LL, Sun JX, Zhang XY. Advances in the mechanisms of action of cancer-targeting oncolytic viruses. Oncol Lett. 2018;15:4053–60.. doi: 10.3892/ol.2018.7829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Vloten JP, Workenhe ST, Wootton SK, Mossman KL, Bridle BW. Critical Interactions between Immunogenic Cancer Cell Death, Oncolytic Viruses, and the Immune System Define the Rational Design of Combination Immunotherapies. J Immunol. 2018;200:450–8. doi: 10.4049/jimmunol.1701021. [DOI] [PubMed] [Google Scholar]
- 11.Russell SJ, Barber GN. Oncolytic Viruses as Antigen-Agnostic Cancer Vaccines. Cancer Cell. 2018;33:599–605. doi: 10.1016/j.ccell.2018.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng M, Huang J, Tong A, Yang H. Oncolytic viruses for cancer therapy: barriers and recent advances. Mol Ther Oncolytics. 2019;15:234–47. doi: 10.1016/j.omto.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ledford H. Cancer-fighting viruses win approval. Nature. 2015;526:622–3. doi: 10.1038/526622a. [DOI] [PubMed] [Google Scholar]
- 14.Kaufman HL, Kim DW, DeRaffele G, Mitcham J, Coffin RS, Kim-Schulze S. Local and distant immunity induced by intralesional vaccination with an oncolytic herpes virus encoding GM-CSF in patients with stage IIIc and IV melanoma. Ann Surg Oncol. 2010;17:718–30. doi: 10.1245/s10434-009-0809-6. [DOI] [PubMed] [Google Scholar]
- 15.Liu BL, Robinson M, Han ZQ, Branston RH, English C, Reay P, et al. ICP34.5 deleted herpes simplex virus with enhanced oncolytic, immune stimulating, and anti-tumour properties. Gene Ther. 2003;10:292–303. doi: 10.1038/sj.gt.3301885. [DOI] [PubMed] [Google Scholar]
- 16.Andtbacka RH, Ross M, Puzanov I, Milhem M, Collichio F, Delman KA, et al. Patterns of Clinical Response with Talimogene Laherparepvec (T-VEC) in Patients with Melanoma Treated in the OPTiM Phase III Clinical Trial. Ann Surg Oncol. 2016;23:4169–77. doi: 10.1245/s10434-016-5286-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andtbacka RH, Kaufman HL, Collichio F, Amatruda T, Senzer N, Chesney J, Delman KA, Spitler LE, Puzanov I, Agarwala SS, et al. Talimogene Laherparepvec Improves Durable Response Rate in Patients With Advanced Melanoma. J Clin Oncol. 2015;33:2780–8. doi: 10.1200/JCO.2014.58.3377. [DOI] [PubMed] [Google Scholar]
- 18.Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8:559–68. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schoggins JW, Rice CM. Interferon-stimulated genes and their antiviral effector functions. Curr Opin Virol. 2011;1:519–25. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–5. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu SY, Aliyari R, Chikere K, Li G, Marsden MD, Smith JK, et al. Interferon-inducible cholesterol-25-hydroxylase broadly inhibits viral entry by production of 25-hydroxycholesterol. Immunity. 2013;38:92–105. doi: 10.1016/j.immuni.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith S, Weston S, Kellam P, Marsh M. IFITM proteins-cellular inhibitors of viral entry. Curr Opin Virol. 2014;4:71–77. doi: 10.1016/j.coviro.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li L, Zhao H, Liu P, Li C, Quanquin N, Ji X, et al. PARP12 suppresses Zika virus infection through PARP-dependent degradation of NS1 and NS3 viral proteins. Sci Signal. 2018;11:eaas9332. [DOI] [PMC free article] [PubMed]
- 24.Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451:425–30. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 25.Ru J, Sun H, Fan H, Wang C, Li Y, Liu M, Tang H. MiR-23a facilitates the replication of HSV-1 through the suppression of interferon regulatory factor 1. PLoS ONE. 2014;9:e114021. doi: 10.1371/journal.pone.0114021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oyake T, Itoh K, Motohashi H, Hayashi N, Hoshino H, Nishizawa M, Yamamoto M, Igarashi K. Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol Cell Biol. 1996;16:6083–95. doi: 10.1128/MCB.16.11.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang X, Guo J, Wei X, Niu C, Jia M, Li Q, Meng D. Bach1: function, regulation, and involvement in disease. Oxid Med Cell Longev. 2018;2018:1347969. doi: 10.1155/2018/1347969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hira S, Tomita T, Matsui T, Igarashi K, Ikeda-Saito M. Bach1, a heme-dependent transcription factor, reveals presence of multiple heme binding sites with distinct coordination structure. IUBMB Life. 2007;59:542–51. doi: 10.1080/15216540701225941. [DOI] [PubMed] [Google Scholar]
- 29.Nishizawa H, Matsumoto M, Shindo T, Saigusa D, Kato H, Suzuki K, Sato M, Ishii Y, Shimokawa H, Igarashi K. Ferroptosis is controlled by the coordinated transcriptional regulation of glutathione and labile iron metabolism by the transcription factor BACH1. J Biol Chem. 2020;295:69–82. doi: 10.1074/jbc.RA119.009548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wiel C, Le Gal K, Ibrahim MX, Jahangir CA, Kashif M, Yao H, et al. BACH1 Stabilization by Antioxidants Stimulates Lung Cancer Metastasis. Cell. 2019;178:330–45 e322. doi: 10.1016/j.cell.2019.06.005. [DOI] [PubMed] [Google Scholar]
- 31.Lee J, Yesilkanal AE, Wynne JP, Frankenberger C, Liu J, Yan J, et al. Effective breast cancer combination therapy targeting BACH1 and mitochondrial metabolism. Nature. 2019;568:254–8. doi: 10.1038/s41586-019-1005-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anderson NM, Simon MC. BACH1 Orchestrates Lung Cancer Metastasis. Cell. 2019;179:800. doi: 10.1016/j.cell.2019.09.032. [DOI] [PubMed] [Google Scholar]
- 33.Zenke-Kawasaki Y, Dohi Y, Katoh Y, Ikura T, Ikura M, Asahara T, Tokunaga F, Iwai K, Igarashi K. Heme induces ubiquitination and degradation of the transcription factor Bach1. Mol Cell Biol. 2007;27:6962–71. doi: 10.1128/MCB.02415-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu SY, Sanchez DJ, Aliyari R, Lu S, Cheng G. Systematic identification of type I and type II interferon-induced antiviral factors. Proc Natl Acad Sci USA. 2012;109:4239–44. doi: 10.1073/pnas.1114981109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y, Wang S, Zhu H, Zheng C. Cloning of the herpes simplex virus type 1 genome as a novel luciferase-tagged infectious bacterial artificial chromosome. Arch Virol. 2011;156:2267–72. doi: 10.1007/s00705-011-1094-9. [DOI] [PubMed] [Google Scholar]
- 36.Kitamuro T, Takahashi K, Ogawa K, Udono-Fujimori R, Takeda K, Furuyama K, et al. Bach1 functions as a hypoxia-inducible repressor for the heme oxygenase-1 gene in human cells. J Biol Chem. 2003;278:9125–33. doi: 10.1074/jbc.M209939200. [DOI] [PubMed] [Google Scholar]
- 37.Moesta AK, Cooke K, Piasecki J, Mitchell P, Rottman JB, Fitzgerald K, et al. Local Delivery of OncoVEX(mGM-CSF) Generates Systemic Antitumor Immune Responses Enhanced by Cytotoxic T-Lymphocyte-Associated Protein Blockade. Clin Cancer Res. 2017;23:6190–202. doi: 10.1158/1078-0432.CCR-17-0681. [DOI] [PubMed] [Google Scholar]
- 38.van Akkooi ACJ, Haferkamp S, Papa S, Franke V, Pinter A, Weishaupt C, et al. A Retrospective Chart Review Study of Real-World Use of Talimogene Laherparepvec in Unresectable Stage IIIB-IVM1a Melanoma in Four European Countries. Adv Ther. 2021;38:1245–62. doi: 10.1007/s12325-020-01590-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stahlie EHA, Franke V, Zuur CL, Klop WMC, van der Hiel B, Van de Wiel BA, Wouters M, Schrage YM, van Houdt WJ, van Akkooi ACJ. T-VEC for stage IIIB-IVM1a melanoma achieves high rates of complete and durable responses and is associated with tumor load: a clinical prediction model. Cancer Immunol Immunother. 2021;70:2291–300. doi: 10.1007/s00262-020-02839-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barral PM, Sarkar D, Su ZZ, Barber GN, DeSalle R, Racaniello VR, Fisher PB. Functions of the cytoplasmic RNA sensors RIG-I and MDA-5: key regulators of innate immunity. Pharm Ther. 2009;124:219–34. doi: 10.1016/j.pharmthera.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manivanh R, Mehrbach J, Knipe DM, Leib DA. Role of Herpes Simplex Virus 1 gamma34.5 in the Regulation of IRF3 Signaling. J Virol. 2017;91:e01156–17. [DOI] [PMC free article] [PubMed]
- 42.Schwertner B, Lindner G, Toledo Stauner C, Klapproth E, Magnus C, Rohrhofer A, et al. Nectin-1 Expression Correlates with the Susceptibility of Malignant Melanoma to Oncolytic Herpes Simplex Virus In Vitro and In Vivo. Cancers (Basel). 2021;13:3058. [DOI] [PMC free article] [PubMed]
- 43.Ribas A, Dummer R, Puzanov I, VanderWalde A, Andtbacka RHI, Michielin O, et al. Oncolytic Virotherapy Promotes Intratumoral T Cell Infiltration and Improves Anti-PD-1 Immunotherapy. Cell. 2017;170:1109–19 e1110. doi: 10.1016/j.cell.2017.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bommareddy PK, Shettigar M, Kaufman HL. Integrating oncolytic viruses in combination cancer immunotherapy. Nat Rev Immunol. 2018;18:498–513. doi: 10.1038/s41577-018-0014-6. [DOI] [PubMed] [Google Scholar]
- 45.Suzuki H, Tashiro S, Hira S, Sun J, Yamazaki C, Zenke Y, Ikeda-Saito M, Yoshida M, Igarashi K. Heme regulates gene expression by triggering Crm1-dependent nuclear export of Bach1. EMBO J. 2004;23:2544–53. doi: 10.1038/sj.emboj.7600248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xing J, Wang S, Lin F, Pan W, Hu CD, Zheng C. Comprehensive characterization of interaction complexes of herpes simplex virus type 1 ICP22, UL3, UL4, and UL20.5. J Virol. 2011;85:1881–6. doi: 10.1128/JVI.01730-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun HS, Hsiao KY, Hsu CC, Wu MH, Tsai SJ. Transactivation of steroidogenic acute regulatory protein in human endometriotic stromalcells is mediated by the prostaglandin EP2 receptor. Endocrinology. 2003;144:3934–42. doi: 10.1210/en.2003-0289. [DOI] [PubMed] [Google Scholar]
- 48.Chen KF, Lai YY, Sun HS, Tsai SJ. Transcriptional repression of human cad gene by hypoxia inducible factor-1alpha. Nucleic Acids Res. 2005;33:5190–8. doi: 10.1093/nar/gki839. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.