Abstract
Antiretroviral therapy suppresses HIV replication but leaves a population of infected CD4+ T cells with integrated proviruses. While most of these proviruses contain defects, such as deletions, some intact proviruses persist and can reinitiate viral replication. In this issue of the JCI, Duette, Hiener, and colleagues performed a tour de force proviral landscape analysis on clinical samples collected over many years with in vitro functional assays. The researchers showed that effector memory CD4+ T cells provide partial sanctuary to intact proviruses from CD8+ T cells and this was associated with superior Nef-mediated MHC-I downregulation relative to less mature CD4+ T cell populations. This finding implicates differential immunoevasion as a cell-intrinsic property, influencing proviral persistence, and highlights Nef as a therapeutic target.
Collecting needles from 96 haystacks
The vast majority (92%–98%) of the HIV-infected cells that persist with antiretroviral therapy (ART) contain proviruses with deletions, premature stop codons, or other defects that preclude production of infections viruses (1). An outstanding question for the field is whether these defective proviruses have a role in HIV persistence or ongoing inflammation (2, 3). In this issue of the JCI, Duette, Hiener, and colleagues contribute to this area of research by showing expression of viral proteins from proviral sequences, even those from sequences with large deletions (4). Nonetheless, because only intact proviruses can initiate viral rebound when treatment is interrupted, their characterization and quantification are paramount to HIV cure research (5) and are the main focus of the current study (4).
The group behind the current study previously developed a full-length individual proviral sequencing (FLIPS) assay, with a paired bioinformatics pipeline, to distinguish intact from different forms of defective proviruses (6). As this method is cost and labor intensive, other researchers have devised and implemented multiplex droplet-based PCR methods to selectively quantify intact proviruses (7, 8); however, scalability comes at the cost of sequence-level resolution. Duette, Hiener, and colleagues achieved an impressive feat; they applied their intensive FLIPS assay to a large scale, to T cells that had been sorted into four maturational phenotypes (naive [Tn]; central memory [Tcm]; transitional memory [Ttm]; and effector memory [Tem]) across a cohort of 24 participants with a wide range of years on ART (4).
This effort paid off. The resulting data showed unequal distributions of intact versus defective proviruses as a function of the T cell maturation state, confirming and extending previous observations (9, 10). However, it was the deep dive into sequence-level data that yielded clues, and then evidence, revealing a mechanism underlying how Tem cells maintain a particularly rich reservoir of intact proviruses while Tcm cells show provirus depletion in a progressive manner. The results implicate ongoing pressure by HIV-specific CD8+ cytotoxic T cells, also called cytotoxic T lymphocytes (CTLs), in shaping these proviral landscapes and a particular ability of Tem cells to evade these CTLs.
Ongoing CTL pressure and implications for therapeutic strategies
HIV infection elicits a robust virus-specific CD8+ T cell response, which suppresses viral replication to varying degrees — in part by killing infected cells. In a typical untreated infection, this response helps reduce viral load by a few orders of magnitude from an acute infection peak to a chronic set point. In rare cases, however, exceptional control is achieved, with elite controllers maintaining undetectable viral loads without the aid of ART (ref. 11). This potent antiviral activity raises a question: Why are HIV-specific CD8+ T cells unable to eliminate the rare infected cells that persist once viral replication is abrogated by ART?
The main paradigm has been that this reservoir of infected cells maintains a state of viral latency and, thus, is invisible to HIV-specific CD8+ T cells. It follows that reactivating HIV with latency-reversing agents would be required to engage CD8+ T cells in elimination of reservoirs (12, 13). A series of studies, however, challenge the completeness of this model by providing evidence in support of ongoing interactions between HIV-specific CD8+ T cells and viral reservoirs in individuals on ART (14–16).
Recent advances have uncovered that, with time on ART, the landscape of HIV proviruses becomes increasingly restricted to those that are either too defective to produce antigen or those that are located in genomic regions that are unfavorable to transcription (e.g., gene deserts; ref. 16). While this skewing occurs slowly, some elite controllers appear to have vanquished all but the above proviruses (17) — scenarios that some have hailed as spontaneous cures. The authors of these studies imply that CD8+ T cells most likely exert the selective pressure that shapes these landscapes. This premise is supported by studies demonstrating that the kinetics and functional profiles of HIV-specific CD8+ T cells in ART-treated individuals reflect ongoing recognition of infected cells. T cells targeting the early gene product Nef appeared to be disproportionately sensitive to this stimulation (14, 15), perhaps reflecting a limited window of time for recognition, where late gene products (e.g., Gag) are expressed only after Nef-mediated loss of surface MHC-I has occurred.
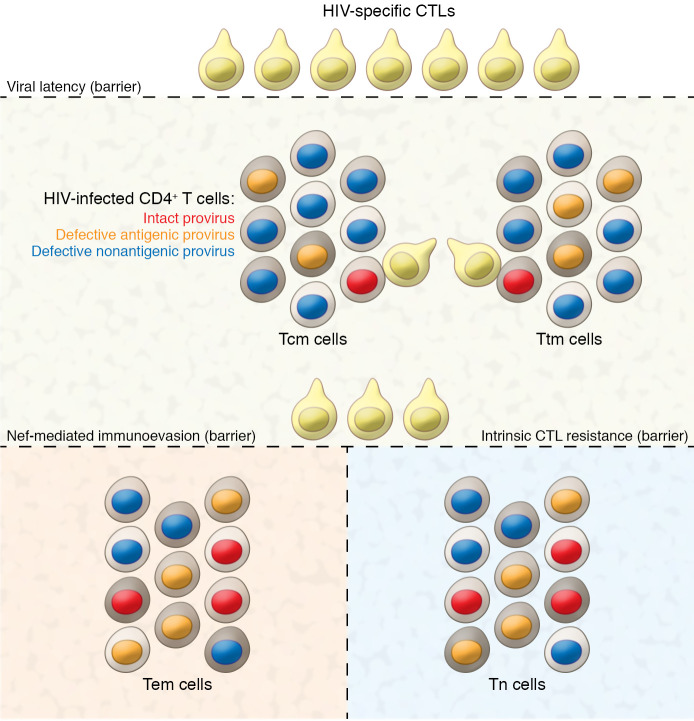
In the current issue of the JCI, Duette, Hiener, and colleagues add considerably to the evidence supporting ongoing surveillance of infected cells from participants on ART and highlight the potential of an alternative modality of therapeutic intervention (4). They showed progressive loss of proviruses possessing CD8+ T cell epitopes over many years of ART, providing direct evidence that these cells exert ongoing selective pressure. The unexpected finding that the ongoing selective pressure was restricted to Tcm cells, alongside the observation that proviral sequences encoding Nef were preferentially maintained in Tem cells, led to the fascinating finding that the degree of HIV-Nef–mediated MHC-I surface loss varied by host cell maturational status (Tcm < Ttm < Tem). The authors, thus, proposed a model whereby their initial observation of enriched frequencies of intact proviruses in Tem cells reflected superior Nef-mediated immunoevasion in these cells (Figure 1). They inferred that therapeutic strategies to inhibit Nef may expose the HIV reservoirs in Tem cells to the same CD8+ T cell pressure reflected by Tcm cells and, thus, lead to reductions in HIV reservoirs even without therapeutic latency reversal. This model is an attractive prospect in light of the ongoing challenges in developing safe and effective latency-reversing therapeutic agents. While we are not aware of any clinical stage Nef-inhibitor drugs, preclinical programs have shown promise and highlight the potential for clinical use (18).
Figure 1. Interactions between HIV-infected cells of different maturational phenotypes and CTLs from participants on ART.
Viral latency is a substantial but imperfect barrier to CTL engagement. Effector memory and naive CD4+ T cells possess the additional barriers of superior Nef-mediated immunoevasion and cell-intrinsic CTL resistance, respectively, which allow for higher frequencies of intact proviruses to persist in Tem and Tn cells. Additional layers of diversity exist within these maturational subsets (depicted by cell membrane shading), and further dissecting each subset in relation to immunoevasion/resistance is a key frontier.
Spotlight on intrinsic properties of reservoir-harboring cells
HIV persistence on ART takes the form of long-lived cells (predominately CD4+ T cells) with genome-integrated proviruses, which undergo clonal expansions and contractions driven by incompletely understood forces (19–21). Until recently, however, consideration of the role of cell-intrinsic properties in HIV persistence has been largely restricted to (a) whether the host cell is resting versus activated (the former being more conducive to viral latency, ref. 22), and (b) the proliferative capacity of the host cell (which, in rare cases, is influenced by the proviral integration site itself; ref. 23). More recent studies have uncovered prosurvival characteristics that allow for infected cells to persist by resisting virus- and/or CD8+ T cell–mediated cytopathicity, such as overexpression of BCL-2 and BCL-XL (24–28). Through their demonstration of differential Nef-mediated MHC-I downmodulation, Duette, Hiener, and colleagues further enriched this line of inquiry by uncovering another dimension of heterogeneity influencing proviral persistence: differential virus-mediated immunoevasion as a function of cell-intrinsic properties (4). While studies into the mechanism of superior Nef-medicated immunoevasion in Tem cells are warranted, the authors allude to this resulting from higher general HIV gene product expression in Tem cells compared with that from less mature cells. It is an interesting case of counterintuition that cells expressing more HIV may end up less susceptible to CTLs, as a result of superior expression of a viral immunoevasion factor.
Further heterogeneity and plasticity of CD4+ T cells
HIV establishes infection across diverse CD4+ T cell populations early on during acute infection, with a preference for memory versus naive subsets, and this pattern persists after ART initiation (29). The ability of HIV to draw benefit from properties of individual host cells begins to move into focus as a powerful force favoring HIV persistence when one considers the tremendous heterogeneity and plasticity of CD4+ T cells. Duette, Hiener, and colleagues approached their investigation at the level of maturational phenotype, an important and logical starting point (4). However, there are multiple additional layers of heterogeneity for future studies to explore. Several distinct lineages of CD4+ T cells can be derived from Tn cells, including Th1, Th2, Th17, Th9, Th22, and Treg populations, which can then each progress along the maturational pathways highlighted in the study by Duette, Hiener, and colleagues. Thus, while Nef-mediated MHC-I sequestration is generally superior in Tem cells versus Tcm cells, it would be interesting to see the breakdown by lineage, for example, by comparing Tem Tregs with Th2 Tcm cells. Indeed, the study by Duette, Hiener, and colleagues reflects considerable heterogeneity in Nef function even within each maturational stage, which further subsetting may resolve (4).
A previous study by Lee et al. applied a similar proviral sequencing methodology to CD4+ T cells sorted into Th1, Th17, Th2, or Th9 lineages and found that Th1 cells were relatively enriched for intact versus defective proviruses (30). However, these lineages were not subdivided by maturational phenotype. While a comprehensive assessment of HIV persistence in relation to maturational phenotype, layered on top of lineage, would be daunting, the findings of Lee et al., together with those of Duette, Hiener, and colleagues, suggest some logical inroads (4). Deeper dives into CD4+ T cell heterogeneity can now also benefit from pairing higher-throughput methods for distinguishing intact from defective proviruses, with the application of more information-rich sequencing methods to targeted populations. The study by Duette, Hiener, and colleagues provides a model for how insights gained from such investigations can have direct implications for therapeutic strategies.
An orthogonal approach to delineating heterogeneity within CD4+ T cells relies on the basis of clonality, where cells bearing the same TCR rearrangement or having an identical HIV provirus can be assigned as members of an expanded clone. To avoid having large expanded clones dominate the proviral landscape, and having the properties of the expanded clones supersede differences between maturational subsets, Duette, Hiener, and colleagues collapsed the clones by only counting one instance of identical proviruses, which serves as a marker of proliferation of infected cells (4). Their findings, however, also raise the question of whether cell-intrinsic properties relevant to persistence may vary from one clone to another, i.e., does the degree of Nef-mediated immunoevasion also track with the clonotype analogously with the maturational state? The plasticity of CD4+ T cells comes into play in tackling these questions, as expanded clones can span phenotypically diverse populations (21). However, recognizing this plasticity alongside the diversity of infected cells is fundamental to understanding what we now know to be incredibly dynamic reservoirs of HIV and intimately tied to normal CD4+ T cell biology.
Finally, while blood samples can yield important insights into how host cell properties relate to the proviral landscapes that they harbor, it is important to stay mindful of the fact that cells of different maturational states will move through distinctive anatomical compartments of the body, e.g., Tn and Tcm cells migrate back to lymphoid tissue, whereas Tem cells generally circulate through peripheral tissues. These environments can both alter the states of these cells and expose these populations to differential selective pressures, such as exposure to CTLs and NK cells. Tissue-based and in situ studies will thus be important complements to those based on peripheral blood as we work toward a more comprehensive understanding of these aspects of HIV persistence.
In summary, Duette, Hiener, and colleagues used HIV proviral landscape analysis in different T cell subsets and found that Nef expression, both from intact and defective proviruses, promotes immune evasion from CD8+ T cell killing in this highly proliferative cell population. This study highlights the power of HIV proviral landscape analysis, which required sequence analysis to reveal the effect of viral gene expression on HIV persistence and immune surveillance.
Acknowledgments
LL and RBJ are supported by the National Institute of Allergy and Infectious Diseases of the NIH under awards UM1AI164565 (to RBJ), R01AI165301, UM1AI164562, R01AI147845, and R01AI131798 (to RBJ). Awards UM1AI164565 and UM1AI164562 were also supported by the National Institute of Diabetes and Digestive and Kidney Diseases, the National Institute of Neurological Disorders and Stroke, the National Institute on Drug Abuse, and the National Heart, Lung, and Blood Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Version 1. 04/01/2022
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2022, Leyre et al. This is an open access article published under the terms of the Creative Commons Attribution 4.0 International License.
Reference information: J Clin Invest. 2022;132(7):e158872. https://doi.org/10.1172/JCI158872.
Contributor Information
Louise Leyre, Email: lol4001@med.cornell.edu.
R. Brad Jones, Email: rbjones@med.cornell.edu.
References
- 1.Ho YC, et al. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell. 2013;155(3):540–551. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Imamichi H, et al. Defective HIV-1 proviruses produce viral proteins. Proc Natl Acad Sci U S A. 2020;117(7):3704–3710. doi: 10.1073/pnas.1917876117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pollack RA, et al. Defective HIV-1 proviruses are expressed and can be recognized by cytotoxic T lymphocytes, which shape the proviral landscape. Cell Host Microbe. 2017;21(4):494–506. doi: 10.1016/j.chom.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duette G, et al. The HIV-1 proviral landscape reveals that Nef contributes to HIV-1 persistence in effector memory CD4+ T cells. J Clin Invest. 2022;132(7):154422. doi: 10.1172/JCI154422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davey RT, Jr, et al. HIV-1 and T cell dynamics after interruption of highly active antiretroviral therapy (HAART) in patients with a history of sustained viral suppression. Proc Natl Acad Sci U S A. 1999;96(26):15109–15114. doi: 10.1073/pnas.96.26.15109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hiener B, et al. Amplification of near full-length HIV-1 proviruses for next-generation sequencing. J Vis Exp. 2018(140):58016. doi: 10.3791/58016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruner KM, et al. A quantitative approach for measuring the reservoir of latent HIV-1 proviruses. Nature. 2019;566(7742):120–125. doi: 10.1038/s41586-019-0898-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kinloch NN, et al. HIV-1 diversity considerations in the application of the Intact Proviral DNA Assay (IPDA) Nat Commun. 2021;12(1):165. doi: 10.1038/s41467-020-20442-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Venanzi Rullo E, et al. Persistence of an intact HIV reservoir in phenotypically naive T cells. JCI Insight. 2020;5(20):e133157. doi: 10.1172/jci.insight.133157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hiener B, et al. Identification of genetically intact HIV-1 proviruses in specific CD4+ T cells from effectively treated participants. Cell Rep. 2017;21(3):813–822. doi: 10.1016/j.celrep.2017.09.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walker BD. Elite control of HIV Infection: implications for vaccines and treatment. Top HIV Med. 2007;15(4):134–136. [PubMed] [Google Scholar]
- 12.Deeks SG. HIV: shock and kill. Nature. 2012;487(7408):439–440. doi: 10.1038/487439a. [DOI] [PubMed] [Google Scholar]
- 13.Ward AR, et al. Immunological approaches to HIV cure. Semin Immunol. 2021;51:101412. doi: 10.1016/j.smim.2020.101412. [DOI] [PubMed] [Google Scholar]
- 14.Stevenson EM, et al. HIV-specific T cell responses reflect substantive in vivo interactions with antigen despite long-term therapy. JCI Insight. 2021;6(3):142640. doi: 10.1172/jci.insight.142640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomas AS, et al. T-cell responses targeting HIV Nef uniquely correlate with infected cell frequencies after long-term antiretroviral therapy. PLoS Pathog. 2017;13(9):e1006629. doi: 10.1371/journal.ppat.1006629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Einkauf KB, et al. Parallel analysis of transcription, integration, and sequence of single HIV-1 proviruses. Cell. 2022;185(2):266–282. doi: 10.1016/j.cell.2021.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang C, et al. Distinct viral reservoirs in individuals with spontaneous control of HIV-1. Nature. 2020;585(7824):261–267. doi: 10.1038/s41586-020-2651-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lv Y, et al. Research progress of HIV-1 Nef inhibitors. AIDS Rev. 2020;22(4):221–226. doi: 10.24875/AIDSRev.20000050. [DOI] [PubMed] [Google Scholar]
- 19.Hosmane NN, et al. Proliferation of latently infected CD4+ T cells carrying replication-competent HIV-1: potential role in latent reservoir dynamics. J Exp Med. 2017;214(4):959–972. doi: 10.1084/jem.20170193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cohn LB, et al. Clonal CD4+ T cells in the HIV-1 latent reservoir display a distinct gene profile upon reactivation. Nat Med. 2018;24(5):604–609. doi: 10.1038/s41591-018-0017-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gantner P, et al. Single-cell TCR sequencing reveals phenotypically diverse clonally expanded cells harboring inducible HIV proviruses during ART. Nat Commun. 2020;11(1):4089. doi: 10.1038/s41467-020-17898-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chun TW, et al. In vivo fate of HIV-1-infected T cells: quantitative analysis of the transition to stable latency. Nat Med. 1995;1(12):1284–1290. doi: 10.1038/nm1295-1284. [DOI] [PubMed] [Google Scholar]
- 23.Maldarelli F, et al. HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science. 2014;345(6193):179–183. doi: 10.1126/science.1254194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cummins NW, et al. Prime, shock, and kill: priming CD4 T cells from HIV patients with a BCL-2 antagonist before HIV reactivation reduces HIV reservoir size. J Virol. 2016;90(8):4032–4048. doi: 10.1128/JVI.03179-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ren Y, et al. BCL-2 antagonism sensitizes cytotoxic T cell-resistant HIV reservoirs to elimination ex vivo. J Clin Invest. 2020;130(5):2542–2559. doi: 10.1172/JCI132374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang SH, et al. Latent HIV reservoirs exhibit inherent resistance to elimination by CD8+ T cells. J Clin Invest. 2018;128(2):876–889. doi: 10.1172/JCI97555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ren Y, et al. Selective BCL-XL antagonists eliminate infected cells from a primary-cell model of HIV latency but not from ex vivo reservoirs. J Virol. 2021;95(15):e0242520. doi: 10.1128/JVI.02425-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang SH, et al. Have cells harboring the HIV reservoir been immunoedited? Front Immunol. 2019;10:1842. doi: 10.3389/fimmu.2019.01842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leyre L, et al. Abundant HIV-infected cells in blood and tissues are rapidly cleared upon ART initiation during acute HIV infection. Sci Transl Med. 2020;12(533):eaav3491. doi: 10.1126/scitranslmed.aav3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee GQ, et al. Clonal expansion of genome-intact HIV-1 in functionally polarized Th1 CD4+ T cells. J Clin Invest. 2017;127(7):2689–2696. doi: 10.1172/JCI93289. [DOI] [PMC free article] [PubMed] [Google Scholar]