Abstract
Purpose
CD8 cytotoxic T cells (CTLs) play a critical role in the clearance of virally infected cells. SARS-CoV-2-specific CD8 T cells and functional CTLs in natural infections and following COVID-19 vaccine in primary antibody deficiency (PAD) have not been reported. In this study, we evaluated T cell response following COVID-19 or COVID-19 mRNA vaccination in patients with PADs by assessing SARS-CoV-2 tetramer-positive CD8 T cells and functional CTLs.
Methods
SARS-CoV-2-specific CD8 and functional CTLs were examined in a patient with X-linked agammaglobulinemia (XLA) and a patient with common variable immunodeficiency (CVID) following COVID-19 infection, and in 5 patients with CVID and 5 healthy controls 1 month following 2nd dose of COVID-19 mRNA vaccine (Pfizer-BioNTech). Cells were stained with SARS-CoV-2 spike protein–specific tetramers, and for functional CTLs (CD8+ CD107a+ granzyme B+ perforin+), with monoclonal antibodies and isotype controls and analyzed by flow cytometry.
Results
SARS-CoV-2-specific tetramer + CD8 T cells and functional CTLs in the patient with XLA following COVID-19 infection were higher, as compared to healthy control subject following COVID-19 infection. On the other hand, SARS-CoV2-tetramer + CD8 T cells and functional CTLs were lower in CVID patient following COVID19 infection as compared to healthy control following COVID-19 infection. SARS-CoV2-tetramer + CD8 T cells and functional CTLs were significantly lower in SARS-CoV2-naive CVID patients (n = 10) following vaccination when compared to SARS-CoV-2-naive healthy vaccinated controls (n = 10).
Conclusions
CVID is associated with reduced SARS-CoV-2-specific CD8 T cells and functional CTLs in both natural SARS-CoV-2 infection and in response to SARS-CoV-2 mRNA vaccine, whereas natural infection in XLA is associated with a robust SARS-CoV-2-specific CD8 and functional CTL responses.
Keywords: CTLs, SARS-CoV-2-specific CD8 T cells, XLA, CVID, Pfizer/BNT vaccine
Introduction
Immunity reduces susceptibility to infection, reduces infectiousness, and reduces pathology. The magnitude of these components of immune efficacy depends on the levels of antibody and T cell immunity. However, over time, the levels of antibody and T cells wane. High levels of immunity are required to prevent individuals from getting infected (sterilizing immunity). If there is insufficient immunity to prevent infection, partial immunity can nonetheless allow the individual to mount a more rapid response, which typically results in more rapid control of the infection, and consequently lowers transmission and pathology. Introduction of vaccines has resulted in a significant decline in both COVID-19 cases and particularly in mortality. Neutralizing antibodies play an important role in defense against transmission from SARS-CoV-2 infection [1–8]. COVID-19 mRNA vaccines are 94–95% effective in preventing severe disease that is associated with the presence of significant amount of neutralizing antibodies [9–11]. A number of patients with primary antibody deficiencies do not make specific antibodies against SARS-CoV-2; however, some of these patients when infected with SARS-CoV-2 had either mild clinical course of COVID-19 or those hospitalized and with no comorbidity risks recover [12–20]. This would suggest that T cells may play an important role in determining the clinical course of SARS-CoV-2 infection. Generally, neutralizing antibodies prevent viral entry to target cells and, therefore, play an important role in the prevention of spread of infection, whereas cytotoxic T cells (CTLs) and natural killer cells kill virus-infected target cells, therefore containing and clearing the infection. Several studies have reported SARS-CoV-2-specific T cell responses in various clinical spectra of COVID-19; however, there are limited data on CD8 T cells and CTLs [21–30]. It is unclear if the immune responses to natural SARS-CoV-2 infection in antibody deficiency diseases are different from those from vaccination. A few studies have examined immune responses in inborn errors of immunity [31]. However, in none of these studies, SARS-CoV-2-specific tetramer-positive CD8 T cells and functional CTLs were evaluated. In this study, we analyzed SARS-CoV-2-specific tetramer-positive CD8 T cells and functional CD8 CTLs in patients with XLA and CVID who were infected with SARS-CoV-2, and compared them with SARS-CoV-2-naive CVID patient and healthy controls following vaccination with two doses of Pfizer-BioNTech COVID-19 vaccine to determine the difference between natural infection and vaccine-induced T cell responses and difference in immune response to vaccine between SARS-CoV-2-naive CVID patients and healthy controls. Our data show that following SARS-CoV-2 infection, XLA patient developed robust CD8 T cell responses, whereas CVID patient following SARS-CoV-2 infection had poor response as compared to healthy control following SARS-CoV-2 infection. Furthermore, SARS-CoV-2-naive CVID patients following immunization with 2 doses of Pfizer vaccine had significantly lower SARS-CoV-2-specific CD8 T cells and functional CTLs as compared to healthy controls following vaccination.
Materials and Methods
Subjects
A patient with XLA and a patient with CVID with mild-to-moderate COVID-19 infection, 10 SARS-CoV-2-naive CVID patients, and 10 healthy controls following two doses of Pfizer-BioNTech COVID-19 vaccine were evaluated. The patient with XLA was a 70-year-old man who had been on IgG replacement therapy since age 3 years. His laboratory evaluation revealed absent circulating B cells, undetectable IgA and IgM, and a pathogenic mutation in BTK gene. He also had bronchiectasis. He developed COVID-19 disease in early 2021 manifested as mild pneumonia and recovered completely without any specific treatment or hospitalization. The patients with CVID were diagnosed according to European Society for Immunodeficiencies and Pan-American Group for Primary Immunodeficiency criteria of low IgG and low IgA/IgM, and impaired response to vaccine [32]. No gene mutation studies were performed. One of the patients with CVID, a 67-year-old woman with infection-only phenotype and untested genotype, developed a mild COVID-19 disease presenting as severe fatigue and mild pneumonia that also resolved without any complications or hospitalization. Blood samples were collected 29 days following COVID-19 infection in XLA patient, and 32 days following COVID-19 infection in CVID patient, and prior to next immunoglobulin infusion. Blood samples from healthy control were drawn 30 days following COVID-19 infection. Blood samples were drawn from 10 SARS-CoV-2-naive CVID patients 4–5 weeks (mean = 31.8 days; median 31.5 days) and 10 SARS-CoV-2-naive healthy controls 4–6 weeks (mean = 32.4 days, median 31.0 days) following 2nd dose Pfizer-BioNTech COVID-19 vaccine. Blood samples from healthy controls were obtained from UCI Institute for Clinical and Translational Science (ICTS). Patients with PAD were from UCI Immunology clinics. Demographic data on study patients are shown in Table 1. CD3+, CD4+, and CD8+ T cells in COVID-19-infected patients were within normal ranges for healthy laboratory controls of institutional CLIA-certified laboratory.
Table 1.
Clinical and immunological characteristics of antibody-deficient patients
| Patients | Age/gender | IgRT | COVID-19 IgG antibodies | Associated conditions |
|---|---|---|---|---|
| SARS-CoV-2-infected (RT-PCR +) and not vaccinated | ||||
| XLA* | 70/M | IVIG | Negative | Bronchiectasis |
| CVID | 67/F | SCIG | Negative | Hashimoto’s thyroiditis, asthma |
| HC | 72/F | NA | Positive | NA |
| SARS-CoV-2-naive and vaccinated | ||||
| CVID | 65/M | IVIG | Negative | DM II, allergic rhinitis |
| CVID | 76/M | IVIG | Positive | None |
| CVID | 56/M | EFI# | Positive | DM II, asthma, rhinitis, adrenal, insufficiency |
| CVID | 49/F | IVIG | Positive | Hashimoto’s thyroiditis |
| CVID | 72/F | IVIG | Negative | Allergic rhinitis |
| CVID | 56/F | SCIG | Negative | Allergic rhinitis |
| CVID | 58/M | IVIG | Positive | Hypothyroidism |
| CVID | 74/F | IVIG | Negative | ITP, allergic rhinitis |
| CVID | 59/F | EFI | Negative | None |
| CVID | 48/F | SCIG | Negative | Hypothyroidism |
| Patients (age range) 48–76 (4 M/6 F) mean—61.3, median—58.6 | ||||
| Controls (age range) 44–77 (6 M/4 F) mean = 60.3, median = 59.0 | ||||
IgRT immunoglobulin replacement therapy, EFI# enzyme-facilitated immunoglobulin, SCIG subcutaneous immunoglobulin, HC healthy controls, DM II, type II diabetes mellitus, ITP idiopathic thrombocytopenia, NA not applicable
*XLA was diagnosed with BTK mutation (BTK c.1085A > G (p.His362Arg) hemizygous pathogenic)
Antibodies and Reagents
Antibodies used included CD8 PerCP (clone SK1), granzyme B Alexa647 (clone GB11), Perforin FITC (clone dG9), and CD107a PE (clone H4A3) all from BioLegend (San Diego), and HLA-A*02:01 SARS-CoV-2 Spike Glycoprotein Tetramer PE (YLQPRTFLL) from MBL International (Woburn, MA).
Methods
SARS-CoV-2-Specific Tetramer-Positive CD8 T Cells
Cells were analyzed by the following technique: 200 μL blood was mixed with 5 μL CD8PerCP monoclonal antibody and 10 μL HLA-A*0201 spike Tetramer PE (HLA-A*02:01 SARS-CoV-2 Spike Glycoprotein Tetramer YLQPRTFLL), vortexed gently and incubated for 30 min at room temperature protected from light. Red blood cells were lysed using 1 mL of Lyse Reagent supplemented with 0.2% formaldehyde fixative reagent per tube. Tubes were centrifuged at 150 × g for 5 min and supernatants were removed. Three milliliters of FACS buffer was added, and tubes were centrifuged at 150 × g for another 5 min. Cell pellets were resuspended in 500 μL of phosphate-buffered saline (PBS) and 0.1% formaldehyde and stored at 4 °C for 1 h in the dark prior to analysis by flow cytometry.
Functional Cytotoxic CD8 T Cells
Functional cytotoxic CD8 T cells (CTLs) were analyzed by the following technique: 200 µ blood samples were incubated for 30 min with CD8PerCP and CD107a PE (a degranulation marker) for surface staining; lysed, fixed, and permeablized by Fix Perm buffer (BD biosciences, San Diego, CA); and then incubated with granzyme B AL647 and Perforin FITC monoclonal antibodies (MLB International, Woburn, MA) and appropriate isotype control.
All fluorescent minus one (FMO) controls and isotype controls were stained and fixed by 2% paraformaldehyde for flow cytometry. Cells were acquired by BD FACS Celesta (Becton-Dickenson, San Jose, CA) equipped with BVR laser. Forward and side scatters and singlets were used to gate and exclude cellular debris. Thirty thousand cells were acquired and analyzed using FLOWJO software (Ashland, OR).
Gating Strategy
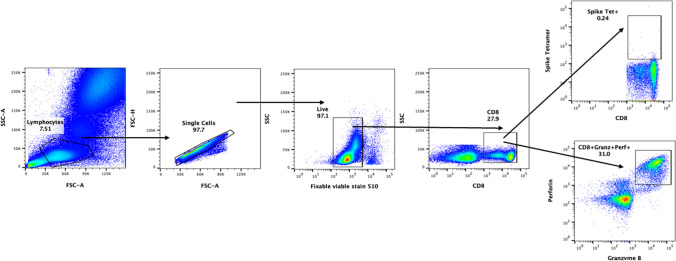
Representative pseudocolor plot was used for gating strategy; gated lymphocytes were analyzed for singlet, live, and CD8 expressing cells. These CD8+ cells were further analyzed for granzyme B and perforin expression, and for SARS-CoV-2 spike protein–specific tetramer-positive cells (Fig. 1).
Fig. 1.
Gating strategy. Representative pseudocolor plot shows gating strategy. Gated lymphocytes were analyzed for singlet, live, and CD8 expressing cells. These CD8+ cells were further analyzed for granzyme B and perforin expression, and for SARS-CoV-2 spike protein–specific tetramer-positive cells
Data are expressed as percent of total CD8+-gated (100%) T cells. Total numbers of SARS-CoV-2 spike protein–specific tetramer-positive cells and functional CTLs were calculated by multiplying percentage with absolute CD8+ T cell counts in the peripheral blood divided by 100.
Statistical analysis was performed by unpaired t-test using GraphPad Prism version 9.2.0 for Windows, GraphPad Software, San Diego, CA, USA A value of P < 0.05 is considered significant.
Results
SARS-CoV-2 Spike Protein–Specific Tetramer-Positive CD8+ T Cells and Functional Cytotoxic CD8 T Cells in XLA and CVID Patient Following COVID-19 Infection
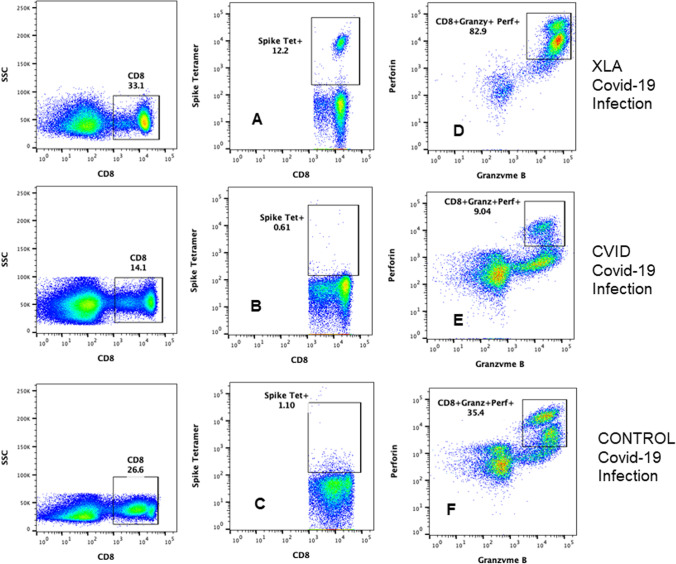
Data for SARS-CoV-2-specific tetramer-positive CD8+ T cells and functional cytotoxic CD8 T cells are shown in Fig. 2. Absolute CD8+ T cell counts in peripheral blood in XLA patient was 265, CVID patient 230, and healthy control was 310. SARS-CoV-2 spike protein–specific tetramer-positive CD8 T cells following SARS-CoV-2 infection were increased (12.2%; total # 32.33) in XLA patient (A) and decreased (0.61%; total # 1.4) in CVID patient (B) following SARS-CoV-2 infection (B) as compared to healthy control (1.10%; total # 3.41) following SARS-CoV-2 infection (C). Functional CD8 CTLs (CD8+ CD107a+ granzyme B+ perforin+) were increased (82.9%; total # 219.68) in XLA patient following SARS-CoV-2 infection (D), decreased (9.04%; total # 21.62) in SARS-CoV-2-infected CVID patient (E), as compared to healthy control (35.4%; total # 109.74) following SARS-CoV-2 infection (F).
Fig. 2.
SARS-CoV-2 spike protein–specific tetramer-positive CD8 T cells and functional CTLs following SRAS-CoV-2 infection. Data are shown in Fig. 1. CD8+-gated T cells were further characterized for SARS-CoV-2 spike protein–specific tetramer-positive CD8 T cells. SARS-CoV-2-specific CD8 T cells in XLA patient (A) were increased, and decreased in CVID patient (B), as compared to healthy control (C) following SARS-CoV-2 infection. Cytotoxic CD8 T lymphocytes (CTLs) are CD8+- and CD107a+-gated cell that were further characterized for the expression of granzyme B and perforin. CD8+ CD107a+ granzyme B+ perforin+ CTLs in XLA (D) were increased, and reduced in CVID patient (E) as compared to healthy control (F) following SARS-CoV-2 infection
SARS-CoV-2 Spike Protein–Specific Tetramer-Positive CD8+ T Cells and Functional Cytotoxic CD8 T Cells in SARS-CoV-2-Naive Vaccinated Patients
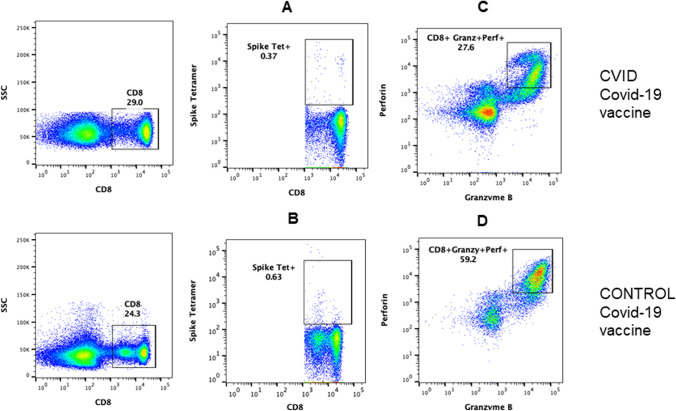
A representative flow cytograph of SARS-CoV-2 spike protein–specific tetramer-positive CD8+ T cells and functional CTLs is in shown in Fig. 3. SARS-CoV-2-naive CVID patient following vaccination had significantly decreased SARS-CoV-2 spike protein–specific tetramer-positive CD8+ T cells (A) and functional CD8 CTLs (C) as compared to SARS-CoV-2 spike protein–specific tetramer-positive CD8+ T cells (B) and functional CTLs (D) in SARS-CoV-2-naive COVID-19-vaccinated healthy control (Fig. 4).
Fig. 3.
SARS-CoV-2 spike protein–specific tetramer-positive CD8 T cells and functional CTLs in SRAS-CoV-2 infection-naive vaccinated patients. CD107a+ granzyme B+ and perforin+ CD8 T cells. A representative cytoflourograph is shown in Fig. 2. SARS-CoV-2 spike protein–specific tetramer-positive CD8 T cells were decreased in CVID patient (A), as compared to healthy control (B) following 2 doses of vaccine. CD8+ CD107a+ granzyme B+ perforin+ CTLs were reduced in CVID patient (C) as compared to healthy control (D) following 2 doses of Pfizer/BioNTech vaccine
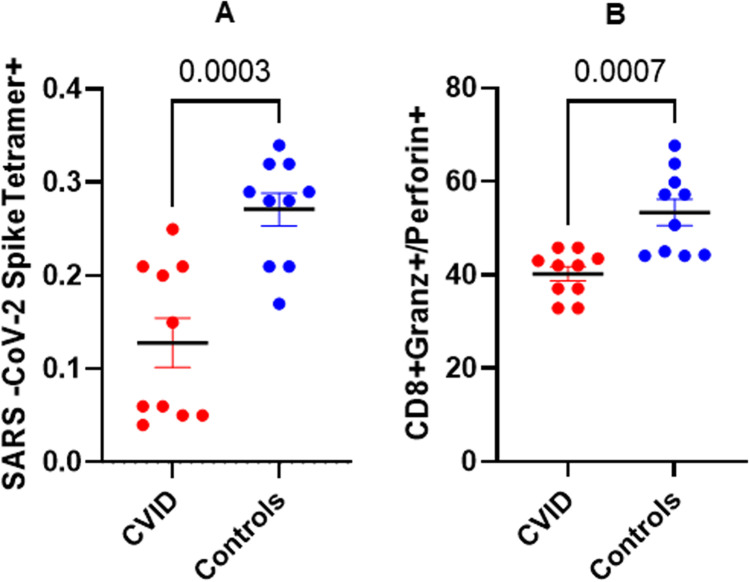
Fig. 4.
Cumulative data for SARS-CoV-2-specific CD8 T cells and functional CTLs in SARS-CoV-2-naive CVID patients and controls following 2 doses of vaccine. Ten SARS-CoV-2-naive CVID patients and 10 healthy controls received 2 doses of Pfizer vaccine and blood samples were simultaneously analyzed 4–6 weeks following 2nd dose. Data show that SARS-CoV-2 spike protein–specific tetramer-positive CD8 T cells are significantly reduced (P < 0.0007) in SARS-CoV-2-naive CVID patients as compared to SARS-CoV-2-naive healthy controls (A) following vaccination. Functional CTL CD8 T cells were also significantly lower (P < 0.0003) in SARS-CoV-2-naive CVID patients as compared to SARS-CoV-2-naive healthy subjects (B)
Discussion
A growing number of studies have been published on both humoral and cellular immune responses in various clinical stages of SARS-CoV-2 infection [21–30]. Furthermore, studies investigating vaccine responses predominantly focused on the production of specific antibodies while very few studies examined the T cell response evaluated by SARS-CoV-2-specific tetramer-positive CD8 T cells and functional CTLs [21–30]. Several reports around the world on mild-to-moderate COVID-19 in patients with primary antibody deficiencies suggested an important role of T cells in modifying severity of the disease [12–19, 31, 33, 34]. However, data on specific immune responses to COVID-19 in patients with inborn errors of immunity (IEI) are limited [31, 33, 34]. In addition, there have been no studies evaluating SARS-CoV-2-specific CD8 T cells and functional CTLs in patients with IEI.
One of the limitations of our study is that in natural SARS-CoV-2 infection group, we have only one patient with XLA and CVID, and a healthy control. A study of larger cohort of patients and healthy controls with natural infection is needed to draw a definitive conclusion. Several investigators have studied immune responses in mild-to-moderate COVID-19 disease. Ni et al. reported SARS-CoV-2-specific T and B cell responses in COVID-19 convalescent individuals [2]. They observed a correlation between neutralizing antibody titers and T cell responses. However, CTLs or tetramer-positive CD8 T cells were not reported. Rha et al., using cytokine secretion assays combined with MHC class I multimer staining, revealed that the proportion of interferon-γ (IFNγ)-producing cells was significantly lower among SARS-CoV-2-specific CD8 T cells than those specific to influenza A virus in COVID-19 convalescent individuals [35]. Peng et al. studied SARS-CoV-2-activated CD4 and CD8 T cells using pentamers in convalescent individuals following COVID-19 [36]. They observed increased membrane and nucleocapsid-specific CD8 T cells as compared to spike protein–specific CD8 T cells in mild disease; no difference was observed in CD107+ CD8+ T cells between mild and severe disease. These investigators did not examine granzyme and perforin markers. Steiner et al. examined SARS-CoV-2 peptide pool (spike and nucleocapsid)–stimulated T cells in previously healthy SARS-CoV-2 antibody-positive and antibody-negative convalescent COVID-19 patients, who had mild disease [34]. Using cytokine polyfunctionality in CD4 T cells and CD8+ CD137+ T cell criteria of T cell responses, they observed no difference in T cell responses among seropositive and seronegative COVID-19 patients, suggesting a good response in both groups. In peptide-stimulated SARS-Cov-2-specific T cell responses, peripheral blood mononuclear cells are stimulated with multiple peptides for CD4 and CD8 T cells for 20–24 h and expression of activation marker is assayed. In this assay, T cells cross-reactive to exposure to previous corona virus may also be activated. Similar to the case reported by Peng et al., our XLA patient with SARS-CoV-2 infection had increased number of SARS-CoV-2-specific tetramer-positive CD8 T cells as compared to SARS-CoV-2 infection healthy control [36]. However, in contrast to the Peng et al. [36], where there was no difference in CD8+ CD107+ cells, we observed increased functional CTLs (CD8+ CD107a+ perforin+ granzyme B+). In contrast, the patient with CVID who had COVID-19 infection had decreased CTLs as well as decreased SARS-CoV-2-specific tetramer-positive CD8 T cells as compared to healthy subject who had COVID-19 infection. These data demonstrate distinctly different SARS-CoV-2-specific CD8 and CTL responses in XLA and CVID following mild-to-moderate COVID-19 infection as compared to healthy controls. Since clinical course in both XLA patient and CVID patient was mild and both cases were SARS-CoV-2 spike IgG antibody–negative, CD8 T cell responses were remarkably different suggesting that additional immune responses (i.e., NK cells, unconventional T cells) may also play a role the clinical outcome of SARS-CoV-2 infection. Chen and Wherry suggested that robust clonal expansion of CD8+ T cells may be associated with milder disease or recovery [21]. Jouan et al. reported that CD69+ iNKT and CD69+ MAIT cells may be predictive of clinical course and disease severity [37]. Carcetti et al. observed high frequency of NK cells was associated with asymptomatic and mild SARS-CoV-2 infection [38].
Our XLA patient with COVID-19 displayed very high SARS-CoV-2 tetramer-positive CD8 T cells that may suggest a compensatory mechanism for lack of antibody response to change the clinical course of the disease. This is similar to the observations of CD8 T cells in patients with hematological malignancies with impaired humoral immunity. Bange et al. [39], in a large cohort of COVID-19 patients with hematological malignancies with impaired B cell responses, preserved CD8 T cell response which were associated with lower viral load and mortality. They also reported that depletion of B cells with anti-CD20 therapy was not associated with increased mortality compared to other hematological cancer, when adequate CD8 T cells were present.
Steiner et al. [34] reported decreased HCoV strains and SARS-CoV-2 cross-reactive T cells in unexposed patients with CVID. Kinoshita et al. [33] reported antibody and T cell responses specific to spike and nucleocapsid at day 33 and 76 cells following infection in 3 members in a family with varying degree of antibody deficiency, one patient each with CVID, hypogammaglobulinemia, and specific antibody deficiency who developed mild COVID-19, and in 2 unrelated CVID patients who also developed mild disease. All except one patient had detectable antibodies against NP and S proteins. All patients made CD4 T cell responses to S, NP. and M proteins; however, patients and control did not show CD8 T cell responses. CD107a expression was minimal and did not differ between patients and controls. In all these studies, cells were stimulated with peptides; however, no tetramers were used to examine SARS-CoV-2-specific T cells. In our study, we examine de novo tetramer-positive SARS-CoV-2-specific CD8 T cells. Castano-Jaramillo et al. [12] reported clinical course of 31 adult and pediatric patients with different inborn errors of immunity. Sixteen percent of patients died. Hospitalization and mortality did not differ between 7 XLA and 11 CVID patients.
Safety, efficacy, and immunogenicity of various COVID-19 vaccines have been reported [9–11, 31, 40, 41]. However, there are limited data on T cells, especially CTLs, from clinical trials of current vaccines. Anderson et al. [9] reported that in Moderna vaccine (SARS-CoV-2 mRNA-1273) clinical trial in adults who were 71 years and older, there was induction of TH1 CD4+ T cell response in response to S-specific peptide pool characterized by increased cytokines, IL-2 > TNF-α > IFNγ. TH-2 responses (IL-4, IL-13) were unaffected. Sahin et al. [11] also reported skewed TH1 responses in both CD4 and CD8 T cells induced with receptor binding domain (RBD) peptide. Data on vaccine response in PAD are even more limited, and no data have been reported for SARS-CoV-2-specific CD8 and functional CTLs. Recently, Salinas et al. [42] reported T and B cell responses in patients with XLA and CVID 1 week following SARS-CoV-2 BNT162b2 vaccine. They observed normal response in majority of XLA patients, whereas Hagin et al. [43] reported increased IFNγ-producing T cells in all 4 XLA patients, whereas 4 of 12 CVID patients did not make T cell responses to S peptide. No data for SARS-CoV-2-specific CD8 or functional CTLs were reported. Increased IFNγ-producing T cells following vaccination reported by Hagin et al. [43] are similar to marked increased SARS-CoV-2 spike protein–specific CD8 and functional CTLs in our XLA patient following SARS-CoV-2 infection. In our SARS-CoV-2-naive CVID patients following vaccination, we observed significantly decreased SARS-CoV-2-specific tetramer-positive CD8 cells and functional CTLs. This is similar to decreased IFNγ-producing T cells reported by Salinas et al. [42] and Hagin et al. [43].
In summary, patients with XLA appear to have increased SARS-CoV-2 spike protein–specific CD8 T cells and function CTL response, whereas CVID patients appear to have impaired CD8 T cell responses to SARS-CoV-2 infection. Furthermore, SARS-CoV-2-naive patients have impaired SARS-CoV-2 spike protein–specific CD8 T cells and functional CTL response to Pfizer-BioNTech vaccine as compared to healthy controls following vaccination. This may suggest a need for additional/frequent booster vaccination in patients with CVID.
Author Contribution
SG conceived the project and wrote and edited the manuscript. YY collected clinical and laboratory data from CVID patients and edited the manuscript. AS collected clinical and routine immunological laboratory data from XLA patient. HS performed the experiments. SA analyzed the data and supervised HS.
Funding
This study was supported by unrestricted funds of the Division of Basic and Clinical Immunology.
Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics Approval
This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Ethics Committee (Institutional Review Board [Human] of University of California at Irvine (HS: 2001–2073).
Consent to Participate
Informed consent was obtained from all individual participants included in the study.
Consent for Publication
Consent form contains permission to publish data without identifier.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Long QX, Liu BZ, Deng HJ, Wu GC, Deng K, Chen YK, et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med. 2020;26(6):845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 2.Ni L, Ye F, Cheng ML, Feng Y, Deng YQ, Zhao H, et al. Detection of SARS-CoV-2-specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity. 2020;52(6):971–7 e3. doi: 10.1016/j.immuni.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siracusano G, Pastori C, Lopalco L. Humoral immune responses in COVID-19 patients: a window on the state of the art. Front Immunol. 2020;11:1049. doi: 10.3389/fimmu.2020.01049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wajnberg A, Amanat F, Firpo A, Altman DR, Bailey MJ, Mansour M, et al. Robust neutralizing antibodies to SARS-CoV-2 infection persist for months. Science. 2020;370(6521):1227–1230. doi: 10.1126/science.abd7728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woodruff MC, Ramonell RP, Nguyen DC, Cashman KS, Saini AS, Haddad NS, et al. Extrafollicular B cell responses correlate with neutralizing antibodies and morbidity in COVID-19. Nat Immunol. 2020;21(12):1506–1516. doi: 10.1038/s41590-020-00814-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao J, Yuan Q, Wang H, Liu W, Liao X, Su Y, et al. Antibody responses to SARS-CoV-2 in patients with novel coronavirus disease 2019. Clin Infect Dis. 2020;71(16):2027–2034. doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chvatal-Medina M, Mendez-Cortina Y, Patino PJ, Velilla PA, Rugeles MT. Antibody responses in COVID-19: a review. Front Immunol. 2021;12:633184. doi: 10.3389/fimmu.2021.633184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weisberg SP, Connors TJ, Zhu Y, Baldwin MR, Lin WH, Wontakal S, et al. Distinct antibody responses to SARS-CoV-2 in children and adults across the COVID-19 clinical spectrum. Nat Immunol. 2021;22(1):25–31. doi: 10.1038/s41590-020-00826-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson EJ, Rouphael NG, Widge AT, Jackson LA, Roberts PC, Makhene M, et al. Safety and immunogenicity of SARS-CoV-2 mRNA-1273 vaccine in older adults. N Engl J Med. 2020;383(25):2427–2438. doi: 10.1056/NEJMoa2028436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R, et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med. 2021;384(5):403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sahin U, Muik A, Derhovanessian E, Vogler I, Kranz LM, Vormehr M, et al. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature. 2020;586(7830):594–599. doi: 10.1038/s41586-020-2814-7. [DOI] [PubMed] [Google Scholar]
- 12.Castano-Jaramillo LM, Yamazaki-Nakashimada MA, O’Farrill-Romanillos PM, MuzquizZermeno D, Scheffler Mendoza SC, Venegas Montoya E, et al. COVID-19 in the context of inborn errors of immunity: a case series of 31 patients from Mexico. J Clin Immunol. 2021;41(7):1463–1478. doi: 10.1007/s10875-021-01077-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esenboga S, Ocak M, Akarsu A, Bildik HN, Cagdas D, Iskit AT, et al. COVID-19 in patients with primary immunodeficiency. J Clin Immunol. 2021;41(7):1515–1522. doi: 10.1007/s10875-021-01065-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ho HE, Mathew S, Peluso MJ, Cunningham-Rundles C. Clinical outcomes and features of COVID-19 in patients with primary immunodeficiencies in New York City. J Allergy Clin Immunol Pract. 2021;9(1):490–3 e2. doi: 10.1016/j.jaip.2020.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marcus N, Frizinsky S, Hagin D, Ovadia A, Hanna S, Farkash M, et al. Minor clinical impact of COVID-19 pandemic on patients with primary immunodeficiency in Israel. Front Immunol. 2020;11:614086. doi: 10.3389/fimmu.2020.614086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meyts I, Bucciol G, Quinti I, Neven B, Fischer A, Seoane E, et al. Coronavirus disease 2019 in patients with inborn errors of immunity: an international study. J Allergy Clin Immunol. 2021;147(2):520–531. doi: 10.1016/j.jaci.2020.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Milito C, Lougaris V, Giardino G, Punziano A, Vultaggio A, Carrabba M, et al. Clinical outcome, incidence, and SARS-CoV-2 infection-fatality rates in Italian patients with inborn errors of immunity. J Allergy Clin Immunol Pract. 2021;9(7):2904–6 e2. doi: 10.1016/j.jaip.2021.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quinti I, Lougaris V, Milito C, Cinetto F, Pecoraro A, Mezzaroma I, et al. A possible role for B cells in COVID-19? Lesson from patients with agammaglobulinemia. J Allergy Clin Immunol. 2020;146(1):211–3 e4. doi: 10.1016/j.jaci.2020.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shields AM, Burns SO, Savic S, Consortium UPC- Richter AG COVID-19 in patients with primary and secondary immunodeficiency: the United Kingdom experience. J Allergy Clin Immunol. 2021;147(3):870–5 e1. doi: 10.1016/j.jaci.2020.12.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soresina A, Moratto D, Chiarini M, Paolillo C, Baresi G, Foca E, et al. Two X-linked agammaglobulinemia patients develop pneumonia as COVID-19 manifestation but recover. Pediatr Allergy Immunol. 2020;31(5):565–569. doi: 10.1111/pai.13263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Z, John WE. T cell responses in patients with COVID-19. Nat Rev Immunol. 2020;20(9):529–536. doi: 10.1038/s41577-020-0402-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Candia P, Prattichizzo F, Garavelli S, Matarese G. T cells: warriors of SARS-CoV-2 infection. Trends Immunol. 2021;42(1):18–30. doi: 10.1016/j.it.2020.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guihot A, Litvinova E, Autran B, Debre P, Vieillard V. Cell-mediated immune responses to COVID-19 infection. Front Immunol. 2020;11:1662. doi: 10.3389/fimmu.2020.01662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le Bert N, Tan AT, Kunasegaran K, Tham CYL, Hafezi M, Chia A, et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584(7821):457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 25.Mathew D, Giles JR, Baxter AE, Oldridge DA, Greenplate AR, Wu JE, et al. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369(6508). [DOI] [PMC free article] [PubMed]
- 26.Mazzoni A, Salvati L, Maggi L, Capone M, Vanni A, Spinicci M, et al. Impaired immune cell cytotoxicity in severe COVID-19 is IL-6 dependent. J Clin Invest. 2020;130(9):4694–4703. doi: 10.1172/JCI138554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nelde A, Bilich T, Heitmann JS, Maringer Y, Salih HR, Roerden M, et al. SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nat Immunol. 2021;22(1):74–85. doi: 10.1038/s41590-020-00808-x. [DOI] [PubMed] [Google Scholar]
- 28.Weiskopf D, Schmitz KS, Raadsen MP, Grifoni A, Okba NMA, Endeman H, et al. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci Immunol. 2020;5(48). [DOI] [PMC free article] [PubMed]
- 29.Zhang F, Gan R, Zhen Z, Hu X, Li X, Zhou F, et al. Adaptive immune responses to SARS-CoV-2 infection in severe versus mild individuals. Signal Transduct Target Ther. 2020;5(1):156. doi: 10.1038/s41392-020-00263-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng HY, Zhang M, Yang CX, Zhang N, Wang XC, Yang XP, et al. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell Mol Immunol. 2020;17(5):541–543. doi: 10.1038/s41423-020-0401-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gupta S, Su H, Narsai T, Agrawal S. SARS-CoV-2-associated T-cell responses in the presence of humoral immunodeficiency. Int Arch Allergy Immunol. 2021;182(3):195–209. doi: 10.1159/000514193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conley ME, Notarangelo LD, Etzioni A. Diagnostic criteria for primary immunodeficiencies. Representing PAGID (Pan-American Group for Immunodeficiency) and ESID (European Society for Immunodeficiencies) Clin Immunol. 1999;93(3):190–7. doi: 10.1006/clim.1999.4799. [DOI] [PubMed] [Google Scholar]
- 33.Kinoshita H, Durkee-Shock J, Jensen-Wachspress M, Kankate VV, Lang H, Lazarski CA, et al. Robust antibody and T cell responses to SARS-CoV-2 in patients with antibody deficiency. J Clin Immunol. 2021;41(6):1146–1153. doi: 10.1007/s10875-021-01046-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steiner S, Sotzny F, Bauer S, Na IK, Schmueck-Henneresse M, Corman VM, et al. HCoV- and SARS-CoV-2 cross-reactive T cells in CVID patients. Front Immunol. 2020;11:607918. doi: 10.3389/fimmu.2020.607918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rha MS, Jeong HW, Ko JH, Choi SJ, Seo IH, Lee JS, et al. PD-1-expressing SARS-CoV-2-specific CD8(+) T cells are not exhausted, but functional in patients with COVID-19. Immunity. 2021;54(1):44–52 e3. doi: 10.1016/j.immuni.2020.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peng Y, Mentzer AJ, Liu G, Yao X, Yin Z, Dong D, et al. Broad and strong memory CD4(+) and CD8(+) T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat Immunol. 2020;21(11):1336–1345. doi: 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jouan Y, Guillon A, Gonzalez L, Perez Y, Boisseau C, Ehrmann S, Ferreira M, Daix T, Jeannet R, François B, Dequin PF, Si-Tahar M, Baranek T, Paget C. Phenotypical and functional alteration of unconventional T cells in severe COVID-19 patients. J Exp Med. 2020;217(12):e20200872. doi: 10.1084/jem.20200872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carsetti R, Zaffina S, Piano Mortari E, Terreri S, Corrente F, Capponi C, et al. Different innate and adaptive immune responses to SARS-CoV-2 infection of asymptomatic, mild, and severe cases. Front Immunol. 2020;11:610300. doi: 10.3389/fimmu.2020.610300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bange EM, Han NA, Wileyto P, Kim JY, Gouma S, Robinson J, et al. CD8(+) T cells contribute to survival in patients with COVID-19 and hematologic cancer. Nat Med. 2021;27(7):1280–1289. doi: 10.1038/s41591-021-01386-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xia S, Zhang Y, Wang Y, Wang H, Yang Y, Gao GF, et al. Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine, BBIBP-CorV: a randomised, double-blind, placebo-controlled, phase 1/2 trial. Lancet Infect Dis. 2021;21(1):39–51. doi: 10.1016/S1473-3099(20)30831-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y, Zeng G, Pan H, Li C, Hu Y, Chu K, et al. Safety, tolerability, and immunogenicity of an inactivated SARS-CoV-2 vaccine in healthy adults aged 18–59 years: a randomised, double-blind, placebo-controlled, phase 1/2 clinical trial. Lancet Infect Dis. 2021;21(2):181–192. doi: 10.1016/S1473-3099(20)30843-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salinas AF, Mortari EP, Terreri S, Quintarelli C, Pulvirenti F, Di Cecca S, et al. SARS-CoV-2 vaccine induced atypical immune responses in antibody defects: everybody does their best. J Clin Immunol. 2021;41(8):1709–1722. doi: 10.1007/s10875-021-01133-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hagin D, Freund T, Navon M, Halperin T, Adir D, Marom R, et al. Immunogenicity of Pfizer-BioNTech COVID-19 vaccine in patients with inborn errors of immunity. J Allergy Clin Immunol. 2021;148(3):739–749. doi: 10.1016/j.jaci.2021.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.