Abstract
Glucose-6-phosphate dehydrogenase (G6PD) deficiency is a common X-linked enzyme disorder associated with hemolytic anemia after exposure to fava beans or certain medications. Activity testing is the gold standard for detecting G6PD deficiency; however, this test is affected by various hematologic parameters. Clinical G6PD genotyping is now included in pharmacogenetic arrays and clinical sequencing efforts and may be reconciled with activity results.
Patients (n=1,391) enrolled on an institutional pharmacogenetic testing protocol underwent clinical G6PD genotyping for 164 G6PD variants. An algorithm accounting for known interferences with the activity assay is proposed. We developed clinical decision support alerts to inform prescribers when high-risk medications were prescribed, warning of gene-drug interactions and recommending therapy alteration.
Of 1,391 patients with genotype results, 1,334 (95.9%) patients were predicted to have normal G6PD activity, 30 (2.1%) were predicted to have variable G6PD activity, and 27 (2%) were predicted to have deficient G6PD activity. Of the 417 patients with a normal genotype and an activity result, 415 (99.5%) had a concordant normal G6PD phenotype. Of the 21 patients with a deficient genotype and an activity result, 18 (85.7%) had a concordant deficient activity result. Genotyping reassigned phenotype in 5 patients with discordant genotype and activity results: 3 switched from normal to deficient, and 2 switched from deficient to normal.
G6PD activity and genotyping are two independent testing methods which can be used in conjunction to assign a more informed G6PD phenotype than either method alone.
Keywords: Glucose-6-phosphate-dehydrogenase (G6PD) deficiency, G6PD genotype, G6PD enzyme activity, pharmacogenomics, pharmacogenetics
Introduction
Glucose-6-phosphate-dehydrogenase (G6PD) deficiency is one of the most common enzyme disorders, affecting as much as 5% of the world’s population.[1] The G6PD gene is located on the X chromosome; therefore, males inherit one G6PD allele and females inherit two G6PD alleles. The population frequency of G6PD deficiency varies by sex and ancestry, affecting males more than females. It is more prevalent among individuals of African, South-East Asian, and Mediterranean descent [2]. G6PD reduces nicotinamide adenine dinucleotide phosphate (NADPH) and is the sole defense mechanism of red blood cells (RBC) against oxidative stress. People who are G6PD deficient are particularly susceptible to hemolytic events during infections or after exposure to fava beans and certain medications [3] such as rasburicase [4].
Identification of patients with G6PD deficiency can be achieved by two different methods: G6PD activity testing and G6PD genotyping [5]. The current gold standard for assigning G6PD phenotype is by activity; however, this test is not without limitations [6]. G6PD activity test results are affected by various hematologic parameters which require an assessment for interfering factors before interpretation. Four known interferences include: 1) critically low hemoglobin, 2) recent RBC transfusion, 3) elevated reticulocyte count, and 4) elevated white blood cell (WBC) count [7–9]. Thus, for many groups of patients (those with cancer, anemia, infections), G6PD activity test results may not be reliable.
The gene coding for G6PD is highly polymorphic, with over 200 known variant alleles; [10] however, the A-(202A_376G) variant accounts for the vast majority of low function alleles in African American populations [2, 11]. With growing use of clinical whole exome and whole genome sequencing, as well as pharmacogenetic arrays, G6PD genotype results will be increasingly available for patients. In cases where G6PD activity test results have also been generated, it is necessary to reconcile G6PD genotyping with any measured phenotypes. We herein describe how two laboratory tests (G6PD genotyping and G6PD activity) can be used in combination to assign G6PD phenotype. We also describe algorithms to assess for possible interferences with the G6PD activity test and to incorporate genotype to assign a more informed G6PD phenotype, especially in those with abnormal hematologic parameters, thereby properly guiding medication selection and improving patient safety.
Materials and Methods
G6PD phenotype assigned based on activity
G6PD activity testing was performed as an in-house clinical laboratory test with a same day turnaround for a subset of patients who underwent G6PD genotyping. G6PD enzyme activity was measured in erythrocytes using a quantitative spectrophotometric assay, [12, 13] and tests were ordered as part of clinical screening, generally for those in whom rasburicase might be needed (e.g. newly diagnosed leukemia patients) or for those with anemia. Whole blood samples were collected in EDTA-containing tubes. Because a high WBC count may result in an artefactually elevated G6PD activity result, samples with a WBC count greater than 100 × 103 cells/mm3 had the buffy coat removed prior to assaying to minimize the potential for interference from G6PD content contributed by WBCs. Samples were prepared with Lyse reagent and assayed immediately on a Cobas® 6000 c501 analyzer (Roche Diagnostics, Indianapolis, IN).
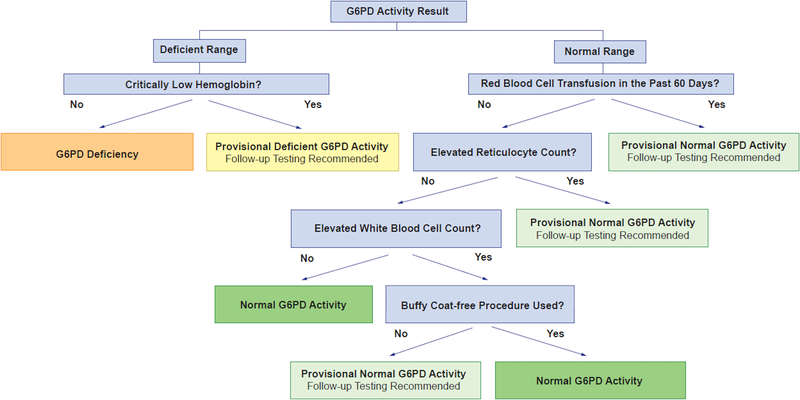
A result below the lower limit of normal (<6.3 units/g Hgb for the St. Jude Children’s Research Hospital analytical assay) was in the deficient range and a problem list entry of G6PD deficiency was placed into the electronic health record; other results were considered normal. Because the G6PD activity test can be affected by hematologic parameters, G6PD activity results were assessed for interferences prior to phenotype assignment (Fig. 1). Provisional deficient activity was assigned when a G6PD activity result was in the deficient range but in the setting of a critically low hemoglobin (<7 g/dL at St. Jude). This is because G6PD activity will be low if the hemoglobin concentration and RBC content of the sample are low. Provisional normal activity was assigned when a G6PD activity result was in the normal range but in the setting of an RBC transfusion within the past 60 days [14], elevated reticulocyte count (>0.085 × 106 cells/mm3 at St. Jude) [8], or WBC count greater than 100 × 103 cells/mm3 and the buffy coat-free procedure was not used. Follow-up testing was recommended when a provisional phenotype was assigned (Fig. 1). Follow-up testing included G6PD genotype or a repeat activity test when known interferences were no longer present.
FIGURE 1:
G6PD activity interference assessment algorithm.
G6PD phenotype assigned based on genotype
G6PD genotyping was performed for patients enrolled on our preemptive pharmacogenetic testing protocol (PG4KDS-www.stjude.org/pg4kds) [15] from September 2017 to June 2020. The primary objective of PG4KDS is to preemptively genotype all eligible patients receiving treatment for active disease at St. Jude Children’s Research Hospital to guide medication prescribing. Genotyping was performed using the PharmacoScan™ assay (Thermo Fisher Scientific, Waltham, MA) which interrogates 164 G6PD variants, including the A-(202A_376G) variant (Supplemental Table S1).
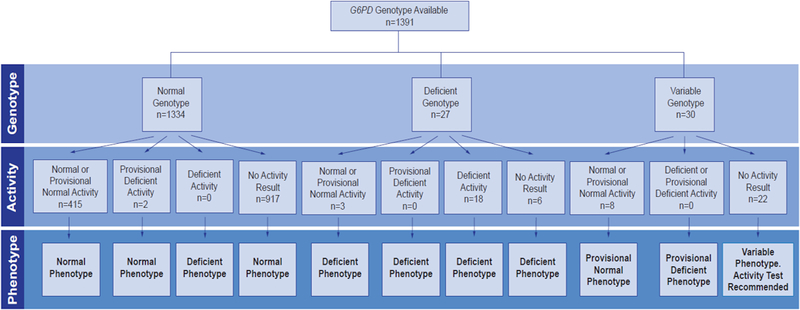
Phenotype assignment from genotype differed for male and female patients [16] and was consistent with the phenotype assignment outlined in the Clinical Pharmacogenetics Implementation Consortium guideline for rasburicase and G6PD [5]. G6PD alleles were categorized using the World Health Organization classification method according to enzyme activity [17] with class I, II, and III alleles (e.g., A-(202A_376G), A-(968C_376G), Asahi, and Kalyan- Kerala variants) consistent with deficient G6PD enzyme activity and class IV alleles (e.g., A [18] and Mira d’Aire variants and the wildtype B allele) consistent with normal G6PD enzyme activity. Males with one deficient G6PD allele (class I-III) and females with two deficient alleles were assigned a G6PD deficient phenotype. Heterozygous females, with one deficient allele (class I-III) and one normal allele (class IV), were assigned a variable G6PD phenotype. Patients with only normal alleles (class IV) were assigned a normal G6PD phenotype [5]. G6PD phenotype was assigned from genotype alone for patients who did not have a G6PD activity result available in the medical record; however, for females with a predicted variable phenotype, a recommendation was made to obtain an activity test before a high-risk medication was prescribed (Fig. 2).
FIGURE 2:
G6PD phenotype assignment.
G6PD phenotype based on genotype and activity
The procedures used for assigning G6PD phenotype based on genotype and clinical activity test are illustrated in Figure 2. For those with normal or deficient genotype, if an activity measure was present, their concordance was assessed. The results were considered concordant if the activity result was in the expected range predicted by genotype (i.e. ≥ 6.3 units/g Hgb for patients with a normal genotype and <6.3 units/g Hgb for patients with a deficient genotype); others were considered discordant. In patients with a normal or deficient genotype we estimated the sensitivity when using the patient’s activity result to predict G6PD phenotype.
Normal G6PD phenotype assignment
Patients with no observed deficient alleles by genotype and either a normal activity, provisional normal activity, or provisional deficient activity result were assigned a normal G6PD phenotype (Fig. 2).
Deficient G6PD phenotype assignment
Patients with a deficient G6PD genotype, regardless of activity, were assigned a G6PD deficiency phenotype. In addition, patients with no observed deficient alleles by genotype but deficient by activity were assigned a G6PD deficiency phenotype, due to the possibility that the patient may have had a deficient G6PD allele not interrogated on the genotyping assay. All patients assigned a deficient phenotype had a G6PD deficiency problem list entry added to their electronic health record.
Variable G6PD phenotype assignment
Due to X chromosome inactivation, heterozygous females may exhibit G6PD activity ranging from normal to deficient, and G6PD activity may change throughout their lifetime [13, 14]. For this reason, genotype alone cannot predict G6PD phenotype in heterozygous females. In such cases, a variable phenotype status was assignedand a G6PD activity test was recommended [5] if a result was not already present in the electronic health record. Even those with activity results were assigned a provisional phenotype based on activity, to reflect the potential for a change in G6PD activity and phenotype in the future (Fig. 2). Patients assigned a provisional deficient phenotype had a G6PD deficiency problem list entry added to their electronic health record.
Clinical Decision Support Alerts
Clinical decision support (CDS) alerts were based on the presence of G6PD deficiency in the problem list or on the absence of a G6PD activity test result coupled with prescribing a high-risk G6PD medication [19–21]. A comprehensive list of medications contraindicated in patients with G6PD deficiency lacks universal consensus and remains controversial. This led to the development of an institution-specific two-tiered list of high-risk medications to avoid or to use with caution in patients with G6PD deficiency. This list was originally approved by our institution’s Pharmacogenetics Oversight Committee, Antimicrobial Utilization and Improvement Committee, and Pharmacy and Therapeutics Committee in 2014 and continues to be updated based on evidence in the literature. Medications likely to cause hemolytic anemia were categorized to avoid in G6PD deficiency, whereas medications that may cause hemolytic anemia were categorized to use with caution in patients with G6PD deficiency. The complete list of medications that constituted the avoid and caution lists are detailed in Supplemental Figs. S1A and S1B and can be viewed at www.stjude.org/g6pd.
Ancestry
Genomic ancestry was assigned from the PharmacoScan™ assay; allele frequencies from 1000 genome populations (EUR, AFR, AMR, EAS and SAS) were used as references. We selected 1054 ancestry informative markers on the array with allele frequencies that differed at least 5% between any two populations. A likelihood method based on reference frequencies was then used to infer the admixture of population for each patient [22]. Patients were then classified into ancestry groups using the following criteria: Whites had European ancestry >90%, Blacks had African ancestry >70%, Asians had eastern or south Asian ancestry >90%, Hispanics had Native American ancestry greater than 10% and higher Native American ancestry than any other non-European ancestry.
Results
G6PD genotype results were obtained in 1,391 patients. Observed G6PD genotypes by ancestry and sex are presented in Table 1. The median age was 8.3 years (range: 54 days to 38 years); 89% were patients treated on an oncology service, 10% were patients treated on a hematology service (primarily with a sickle cell disease diagnosis), and 1% were patients treated on an infectious disease service.
TABLE 1.
Observed G6PD genotypes
| Normal Phenotype | Deficient Phenotype | Variable Phenotype | ||
|---|---|---|---|---|
| Race | Genotype, n (%) | Genotype, n (%) | Genotype, n (%) | |
| Female | White, non-Hispanic (n=355) | B/B, 354 (99.7) | 0 (0) | A-(202A_376G)/B, A/Asahi, 1 (0.3) |
| White, Hispanic (n=61) | B/B, 59 (96.8) A/B, 1 (1.6) |
0 (0) | A-(202A_376G)/B, A/Asahi, 1 (1.6) | |
| Black (n=137) | B/B, 58 (42.3) A/B, 40 (29.2) A/A, 7 (5.1) |
A-(202A_376G)/A-(202A_376G), 4 (3.0) A-(202A_376G)/A-(968C_376G), 1 (0.7) A-(968C_376G)/A-(968C_376G), 1 (0.7) |
A-(202A_376G)/B, A/Asahi, 20 (14.6) A-(202A_376G)/A, 6 (4.4) |
|
| Multiracial (n=59) | B/B, 50 (84.7) A/B, 6 (10.2) B/Mira d’Aire, 1 (1.7) |
0 (0) | A-(202A_376G)/B, A/Asahi, 2 (3.4) | |
| Asian (n=11) | B/B, 11 (100) | 0 (0) | 0 (0) | |
| Total females (n=623) | 587 (94.2) | 6 (1) | 30 (4.8) | |
| Male | White, non-Hispanic (n=447) | B, 447 (100) | 0 (0) | NA |
| White, Hispanic (n=52) | B, 52 (100) | 0 (0) | NA | |
| Black (n=159) | B, 105 (66) A, 36 (22.7) |
A-(202A_376G), 18 (11.3) | NA | |
| Multiracial (n=89) | B, 80 (89.9) A, 7 (7.9) |
A-(202A_376G), 2 (2.2) | NA | |
| Asian (n=21) | B, 20 (95.2) | Kalyan-Kerala, 1 (4.8) | NA | |
| Total males (n=768) | 747 (97.3) | 21 (2.7) | NA | |
| Total patients | Total (n=1391) | 1334 (95.9) | 27 (2) | 30 (2.1) |
B designates no variant was observed and is considered the wild-type allele; A designates the c.376G SNP was observed alone; A-(202A_376G) designates the c.202A and c.376G SNPS were observed on the same allele; A-(202A_376G)/B, A/Asahi designates the c.202A and c.376G SNPS were observed but it could not be determined if the SNPS were on the same or separate alleles; A-(968C_376G) designates the c.968C and c.376G SNPS were observed on the same allele, Mira d’Aire designates the c.1048C SNP was observed; Kalyan-Kerala designates the c.949A SNP was observed. Race was determined based on PharmacoScan™ assay result.
G6PD phenotype assignment based on genotype alone
Nine hundred forty-five patients had a genotype result without a G6PD activity result and were assigned a G6PD phenotype based on genotype alone. Of these, 917 patients (97%) were assigned a normal G6PD phenotype, 6 (1%) were assigned a deficient G6PD phenotype, and 22 (2%) patients were assigned a variable G6PD phenotype (Fig. 2).
Of the 917 patients assigned a normal phenotype based on genotype alone, no variant was observed in 465 males and 378 females, consistent with the wild-type genotype result of B and B/B (class IV or normal activity), respectively, 32 males were hemizygous for the class IV A variant, 36 females were heterozygous and 5 females were homozygous for the class IV A variant, and 1 female was heterozygous for the class IV Mira d’Aire variant.
Of the 6 patients assigned a deficient G6PD phenotype based on genotype alone, 5 males were hemizygous for the class III A-(202A_376G) variant and 1 male was hemizygous for the class III Kalyan-Kerala variant.
Of the 22 female patients assigned a variable phenotype based on genotype alone, in 18 patients heterozygous genotypes at the single nucleotide polymorphisms (SNPs) which constitute the A-(202A_376G) variant were observed, a finding consistent with an A-(202A_376G)/B or A/Asahi genotype depending on whether the SNPs are present on the same or separate alleles, respectively; and the other 4 patients had the A-(202A_376G)/A variants.
G6PD phenotype based on genotype and activity
Four hundred forty-six patients had a G6PD activity result in the medical record at the time the genotype result was obtained. Of these, 8 female heterozygous patients (2 with A-(202A_376G)/A genotype, and 6 with A-(202A_376G)/B, A/Asahi genotype) had a normal or provisional normal activity result and were assigned a provisional normal G6PD activity phenotype (Fig. 2).
Four hundred seventeen patients with a normal genotype had activity test results: 415 patients had a concordant normal G6PD activity phenotype and 2 patients had discordant results. Of the 2 with discordant results, 1 was a male with a class IV A variant and 1 was a male with no observed variants, but both had provisional deficient activity (due to critically low hemoglobin concentrations of <5 g/dL at the time the G6PD sample was drawn) and both were subsequently assigned a normal phenotype (Fig. 2). Of the 415 patients with concordant normal genotype and activity results, no variant was observed in 238 males and 154 females, consistent with the wild-type genotype results of B and B/B (class IV), respectively; 10 males were hemizygous for the class IV A variant, 11 females were heterozygous and 2 females were homozygous for the class IV A variant.
Twenty-one patients with deficient genotype (15 males hemizygous and 4 females homozygous for the A-(202A_376G) variant, 1 female homozygous for the A-(968C_376G) variant, and 1 female compound heterozygous for the A-(202A_376G) and A-(968C_376G) variants) had activity test results; 18 patients had a concordant deficient activity result and were assigned a G6PD deficiency phenotype; 3 patients had discordant results, with normal or provisional normal G6PD activity results, and were subsequently reassigned a G6PD deficiency phenotype (Fig. 2). Of these 3 patients with possible discordances, one patient had been assigned a provisional normal activity because he had received an RBC transfusion in the 60-day period prior to the G6PD activity being drawn; one patient had the G6PD activity sample drawn in the setting of reticulocytosis (0.2647 × 106 cells/mm3); and one patient experienced hemolysis after receiving rasburicase despite having a normal activity result. For this final patient, there was a lack of documentation about RBC transfusion status prior to her malignancy diagnosis.
Thus, of 446 patients with both activity and genotype measures, having a genotype result changed G6PD status in 5 patients (1.1%): status changed from provisional deficient to normal in 2 patients and from normal or provisional normal to G6PD deficient in 3 patients. In 4 of the 5 cases, abnormal hematologic parameters at the time of the G6PD activity measure could explain the unreliable activity measure, and in the fifth case, drug-induced anemia confirmed a genotype-based diagnosis of G6PD deficiency missed by the activity measure.
In 173 females, the sensitivity of the activity test was 83.33% (35.88–99.58%) and in 265 males the sensitivity was 86.67% (59.54–98.34%).
Interpretive consults and clinical decision support alerts
An interpretive pharmacogenetics consult placed in the medical record assigned G6PD phenotype based on genotype alone or based on genotype and activity if an activity result was already available. Examples of interpretive consults which assign G6PD phenotype from G6PD genotype and activity are provided in Supplemental Figs. S2A and S2B.
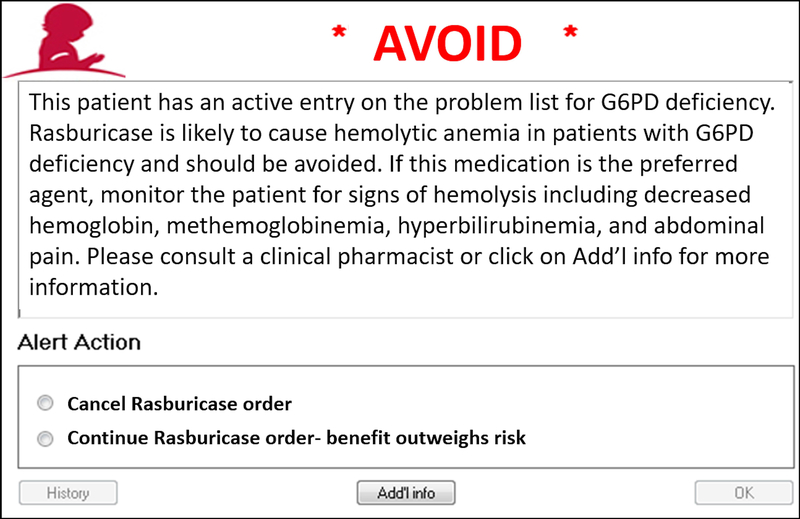
Four CDS alerts were implemented in the electronic health record. For patients with G6PD deficiency, when a high-risk medication is prescribed, a CDS alert is presented to the prescriber, recommending avoiding the drug (Fig. 3) or using caution with the drug (Supplemental Fig. S3A). Additionally, for patients without a G6PD genotype or G6PD activity result, when a high-risk medication is prescribed, a CDS alert is presented to the prescriber with a recommendation to order a G6PD activity test to assign a G6PD status (Supplemental Fig. S3B). Similarly, when a high-risk medication is prescribed to a patient with variable G6PD phenotype, an alert is presented to the prescriber with a recommendation to order a G6PD activity test to assess for G6PD deficiency (Supplemental Fig. S3C).
FIGURE 3:
Clinical decision support alert for medications to avoid in patients with G6PD deficiency.
Discussion
Our results highlight the need to assess for interferences when interpreting G6PD activity results to prevent a potentially incorrect G6PD phenotype assignment. Patients incorrectly phenotyped by G6PD activity alone in our study included those with severe anemia, reticulocytosis, recent red blood cell transfusion, and new leukemia diagnoses. These patients were correctly categorized by G6PD genotyping. Additionally, for some patients (e.g., those with sickle cell disease or those with anemia) G6PD activity results may be uninterpretable due to abnormal hematologic parameters, making G6PD genotype the only available method to confirm G6PD deficiency and prevent adverse effects, such as hemolytic anemia, from high-risk medications.
We describe the clinical implementation of G6PD genotype testing performed preemptively in the context of a multi-gene panel. This preemptive approach is relatively inexpensive on a per-gene basis, as the cost to test many genes is not much greater than the cost to test one gene. With this preemptive testing approach, the results are already in hand at the time of prescribing a high-risk drug, removing the time constraints of ordering and waiting for a test result. The majority of patients have at least one high-risk pharmacogenetic result returned from a multi-gene panel approach [23].
In the current cohort, adding G6PD genotype to G6PD activity resulted in a change in G6PD status in 1% of patients, relative to status previously assigned based on G6PD activity alone. Incorrect phenotype assignment from G6PD activity results had implications for both false negatives (1 of 3 incorrectly identified as normal received a high-risk drug and developed drug-induced hemolytic anemia), and for false positives (2 were incorrectly identified as deficient but were normal), these latter patients can have the diagnosis of G6PD deficiency removed from their problem list and receive medications without regard to G6PD status. This current report suggests that G6PD genotype is a reliable method for assigning G6PD phenotype, as all patients with deficient activity in our cohort were deficient by genotype, and discordances between genotype and activity were explained by interferences in the G6PD activity assay. The 83.3% sensitivity of the activity test in females and 86.7% sensitivity in males highlights the opportunity for a genotype result to improve identification of patients with G6PD deficiency, especially in those with abnormal hematologic parameters which may interfere with the activity test results and interpretation. G6PD activity testing is still recommended in heterozygous females assigned a variable phenotype prior to administration of a high-risk G6PD drug because their G6PD phenotype can potentially change throughout their lifetime.
G6PD activity testing is affected by various hematologic parameters. Despite these limitations and because of its relatively quick turnaround (i.e., same day at our institution) compared to G6PD genotyping (two weeks at our institution), G6PD activity has been the gold standard test for G6PD phenotype assignment. As G6PD genotype results are becoming increasingly available to clinicians through clinical sequencing or genotyping tests, we provide clinicians with a method to assign a G6PD phenotype using genotype alone and using genotype and activity results when both are available.
Supplementary Material
Acknowledgments
The authors would like to thank the St. Jude Children’s Research Hospital PG4KDS team for their assistance in implementing G6PD genotyping/testing as well as Amy Turner, Gunter Scharer, and Praful Aggarwal of RPRD Diagnostics for their work in providing G6PD genotyping. NIH Grants CA 21765, GM 115279 and ALSAC.
Funding: NIH Grants CA 21765, GM 115279 and ALSAC.
Footnotes
Conflict of interest: Authors report no conflict of interest
References
- 1.Nkhoma ET, Poole C, Vannappagari V, Hall SA, Beutler E. The global prevalence of glucose-6-phosphate dehydrogenase deficiency: a systematic review and meta-analysis. Blood Cells Mol Dis 2009;42:267–278. [DOI] [PubMed] [Google Scholar]
- 2.Cappellini MD, Fiorelli G. Glucose-6-phosphate dehydrogenase deficiency. Lancet 2008;371:64–74. [DOI] [PubMed] [Google Scholar]
- 3.Glucose-6-phosphate dehydrogenase deficiency. WHO Working Group. Bull World Health Organ 1989;67:601–611. [PMC free article] [PubMed] [Google Scholar]
- 4.Robinson KM, Yang W, Haidar CE, Hankins JS, Jay DW, Kornegay N, et al. Concordance between glucose-6-phosphate dehydrogenase (G6PD) genotype and phenotype and rasburicase use in patients with hematologic malignancies. Pharmacogenomics J 2019;19:305–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Relling MV, McDonagh EM, Chang T, Caudle KE, McLeod HL, Haidar CE, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for rasburicase therapy in the context of G6PD deficiency genotype. Clin Pharmacol Ther 2014;96:169–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Domingo GJ, Satyagraha AW, Anvikar A, Baird K, Bancone G, Bansil P, et al. G6PD testing in support of treatment and elimination of malaria: recommendations for evaluation of G6PD tests. Malar J 2013;12:391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Echler G Determination of glucose-6-phosphate dehydrogenase levels in red cell preparations. Am J Med Technol 1983;49:259–262. [PubMed] [Google Scholar]
- 8.Lakomek M, Schröter W, De Maeyer G, Winkler H. On the diagnosis of erythrocyte enzyme defects in the presence of high reticulocyte counts. British J Haematol 1989;72:445–451. [DOI] [PubMed] [Google Scholar]
- 9.Morelli A, Benatti U, Lenzerini L, Sparatore B, Salomino F, Melloni E, et al. The interference of leukocytes and platelets with measurement of clucose-6-phosphate dehydrogenase activity of erythrocytes with low activity variants of the enzyme. Blood 1981;58:642–644. [PubMed] [Google Scholar]
- 10.Luzzatto L, Ally M, Notaro R. Glucose-6-phosphate dehydrogenase deficiency. Blood 2020;136:1225–1240. [DOI] [PubMed] [Google Scholar]
- 11.Robinson K, Yang W, Karol S, Kornegay N, Jay D, Cheng C, et al. No evidence that G6PD deficiency affects the efficacy or safety of daunorubicin in acute lymphoblastic leukemia induction therapy. Pediatr Blood Cancer 2019;66:e27681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kornberg A, Horecker BL. Glucose-6-Phosphate Dehydrogenase. IN Methods in Enzymology. 1955;1:323. [Google Scholar]
- 13.Lohr GW, Waller HD. Glucose-6-Phosphate Dehydrogenase. IN Methods of Enzymatic Analysis. 1974:636. [Google Scholar]
- 14.Liumbruno G, Bennardello F, Lattanzio A, Piccoli P, Rossetti G. Recommendations for the transfusion of red blood cells. Blood Transfus 2009;7:49–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffman JM, Haidar CE, Wilkinson MR, Crews KR, Baker DK, Kornegay NM, et al. PG4KDS: a model for the clinical implementation of pre-emptive pharmacogenetics. Am J Med Genet C Semin Med Genet 2014;166C:45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Standardization of procedures for the study of glucose-6-phosphate dehydrogenase. Report of a WHO Scientific Group. World Health Organ Tech Rep Ser 1967;366:1–53. [PubMed] [Google Scholar]
- 17.Minucci A, Moradkhani K, Hwang MJ, Zuppi C, Giardina B, Capoluongo E. Glucose-6-phosphate dehydrogenase (G6PD) mutations database: review of the “old” and update of the new mutations. Blood Cells Mol Dis 2012;48:154–165. [DOI] [PubMed] [Google Scholar]
- 18.Town M, Bautista JM, Mason PJ, Luzzatto L. Both mutations in G6PD A- are necessary to produce the G6PD deficient phenotype. Hum Mol Genet 1992;1:171–174. [DOI] [PubMed] [Google Scholar]
- 19.Youngster I, Arcavi L, Schechmaster R, Akayzen Y, Popliski H, Shimonov J, et al. Medications and Glucose-6-Phosphate Dehydrogenase Deficiency. Drug Saf 2010;33:713–726. [DOI] [PubMed] [Google Scholar]
- 20.Luzzatto L, Seneca E. G6PD deficiency: a classic example of pharmacogenetics with on-going clinical implications. Br J Haematol 2014;164:469–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.St. Jude Children’s Research Hospital. Glucose-6-Phosphate Dehydrogenase (G6PD) [Available from: http://stjude.org/g6pd.] Accessed 06/19/2021.
- 22.Bansal V, Libiger O. Fast individual ancestry inference from DNA sequence data leveraging allele frequencies for multiple populations. BMC Bioinformatics 2015;16:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Driest SL, Shi Y, Bowton EA, Schildcrout JS, Peterson JF, Pulley J, et al. Clinically Actionable Genotypes Among 10,000 Patients With Preemptive Pharmacogenomic Testing. Clin Pharmacol Ther 2014;95:423–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.