SUMMARY
Homogalacturonan (HG), the most abundant pectic glycan, functions as a cell wall structural and signaling molecule essential for plant growth, development and response to pathogens. HG exists as a component of pectic homoglycans, heteroglycans and glycoconjugates. HG is synthesized by members of the GALACTURONOSYLTRANSFERASE (GAUT) family. UDP-GalA-dependent homogalacturonan:galacturonosyltransferase (HG:GalAT) activity has previously been demonstrated for GAUTs 1, 4, 11 and the GAUT1:GAUT7 complex. Here we show that GAUTs 10, 13 and 14 are also HG:GalATs and that GAUTs 1, 10, 11, 13, 14 and 1:7 synthesize polymeric HG in vitro. Comparison of the in vitro HG:GalAT specific activities of the heterologously-expressed proteins demonstrates GAUTs 10 and 11 with lowest, GAUT1 and GAUT13 moderate, and GAUT14 and the GAUT1:GAUT7 complex highest HG:GalAT activity. GAUT13 and GAUT14 are also shown to de novo synthesize (initiate) HG synthesis in the absence of exogenous HG acceptors, an activity previously demonstrated for GAUT1:GAUT7. The rate of de novo HG synthesis by GAUT13 and GAUT14 is similar to their acceptor dependent HG synthesis, in contrast to GAUT1:GAUT7 whose de novo synthesis occurred at much lower rates than acceptor-dependent synthesis. The results suggest a unique role for de novo HG synthesis by GAUTs 13 and 14. The reducing end of GAUT13-de novo-synthesized HG has covalently attached UDP, indicating that UDP-GalA serves as both a donor and acceptor substrate during de novo HG synthesis. The functional significance of unique GAUT HG:GalAT catalytic properties in the synthesis of different pectin glycan or glycoconjugate structures is discussed.
Keywords: Pectin, plant cell wall, biosynthesis, GAUT, galacturonosyltransferase, glycosyltransferase, polysaccharide synthesis, biosynthesis, initiation, galacturonan, Arabidopsis thaliana
INTRODUCTION
Pectins are one of the most complex classes of polysaccharides found in nature. They contain over a dozen unique monosaccharides and require the activity of at least 67 different transferases to synthesize the diversity of pectic structures (Mohnen, 2008). A wide range of pectin-associated functions within plant cells and in the plant body exist (Yang & Anderson, 2020). For example, they function in cell-cell adhesion (Bouton et al., 2002), cell signaling and defense (Ferrari et al., 2013), cell expansion and plant growth (Peaucelle et al., 2012; Peaucelle et al., 2015; Voxeur & Höfte, 2016; Haas et al., 2020), and in cell wall recalcitrance to deconstruction (Francocci et al., 2013; Biswal et al., 2015; Biswal et al., 2018a; Biswal et al., 2018b).
Traditional analyses of pectin structure have required chemical or enzymatic extraction of cell wall fractions to enrich for individual pectic polysaccharides and pectin glycan regions (also referred to as domains). The most abundant pectic glycan, homogalacturonan (HG), is composed of a linear backbone of d-GalA residues connected via α−1,4 linkages and accounts for the majority (~65%) of easily extractable pectin (Willats et al., 2001; Mohnen, 2008). HG can be synthesized in vitro as a homopolymer with a high degree of polymerization (DP) of greater than 500 GalA residues (Amos et al., 2018), yet it is unclear if this form of HG exists as an independent polymer unattached to other glycans in vivo (Fig. 1C, i). HG is known to be covalently linked via the backbone to more complex pectic glycans including rhamnogalacturonan-I (RG-I) and rhamnogalacturonan-II (RG-II) in vivo (Ishii & Matsunaga, 2001; Nakamura et al., 2002; Coenen et al., 2007) (Fig. 1C, ii-iii). RG-I and RG-II account for ~20-35% and 10%, respectively, of easily extractable pectin from plant cell walls (Mohnen, 2008). RG-I contains a disaccharide backbone of d-GalA and l-rhamnose (Rha) connected in an [−4-α-d-GalA-1-2-α-l-Rha-1-] repeating sequence. The backbone is substituted with side branches containing arabinose and galactose (McNeil et al., 1980; Mohnen, 2008). RG-II is composed of an HG backbone substituted with at least four different monosaccharide or oligosaccharide side branches (O'Neill et al., 2004; Ndeh et al., 2017). Finally, HG domains are known to be covalently associated with cell wall arabinogalactan proteins such as APAP1 (Tan et al., 2013) (Fig. 1C, iv). Despite multiple efforts to isolate individual pectic domains and polymers from different plant species and tissue types, the difficulty of defining the structural properties of HG due to the presence of these complex pectic hetero- and glycoconjugate polymeric forms continues to be recognized (Zdunek et al., 2021).
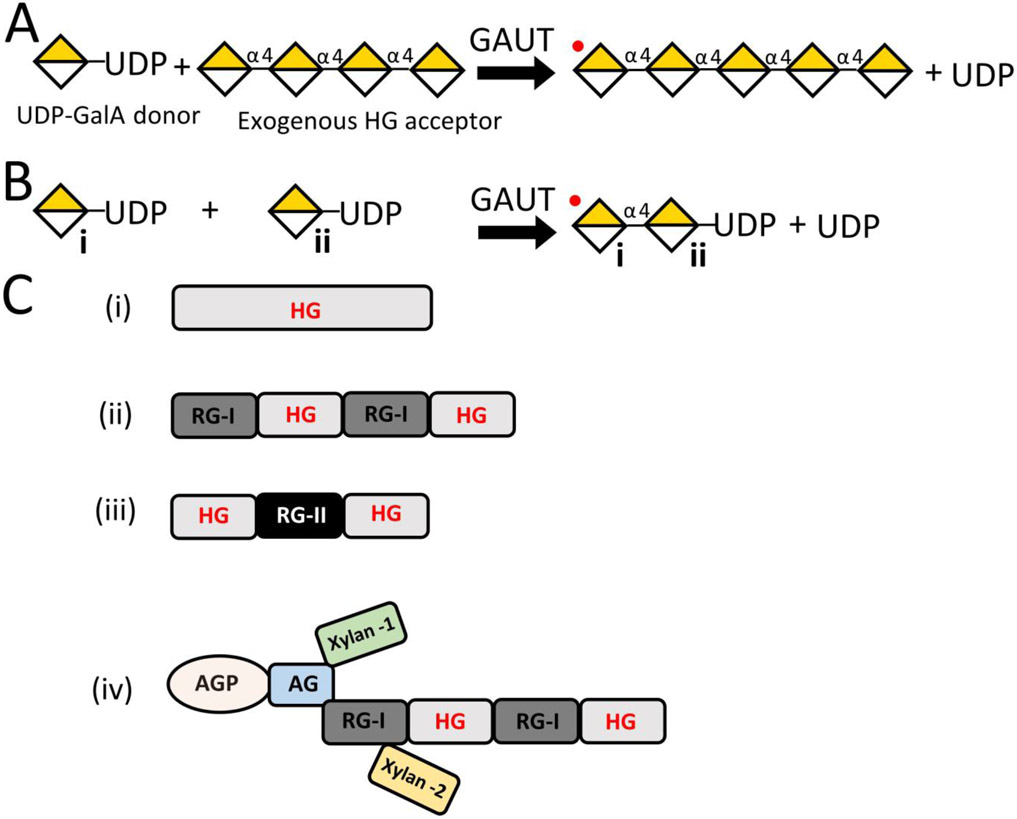
Figure 1. Schematic representation of homogalacturonan (HG) biosynthesis mechanisms and HG polymers that are synthesized in vitro or recovered from plant sources.
(A) Depiction of homogalacturonan galacturonosyltransferase (HG:GalAT) activity in the presence of UDP-GalA and exogenous HG acceptor. Multiple GAUTs are able to elongate exogenous HG acceptors (in this example, an acceptor length with a degree of polymerization of four) at the non-reducing end with GalA residues (yellow and white diamond) in the presence of UDP-GalA nucleotide sugar donor (Sterling et al., 2006; Atmodjo et al., 2011; Amos et al., 2018; Biswal et al., 2018a; Voinicuic et al., 2018). An HG homoglycan is generated that can be further glycosylated by addition of GalA to the non-reducing end (denoted by a red dot). (B) Depiction of HG:GalAT activity in the presence of UDP-GalA only. When no HG acceptor is accessible, data reported here show that at least some GAUTs can use UDP-GalA as an acceptor to synthesize and elongate HG, resulting in a polymeric HG product with UDP on its reducing end. The non-reducing end (denoted by a red dot) is available for subsequent GalA addition. This de novo synthesis activity has previously been detected for the GAUT1:GAUT7 complex but the presence of UDP on the reducing end could not be verified (Amos et al., 2018). In this paper we show that UDP is retained on the reducing end in product synthesized by GAUT13. (C) Depiction of HG or HG-containing structures either synthesized in vitro or recovered from plant cell walls. (i) Polymeric HG with a degree of polymerization > 500 can be synthesized in vitro (Amos et al., 2018), yet its existence in the cell wall as an exclusive polysaccharide, with no covalent linkages to other pectins, remains uncertain. HG isolated from cell walls has been shown to exist covalently connected via its backbone to (ii) RG-I, (iii) RG-II or (iv) as a component of the proteoglycan, APAP1 (Ishii & Matsunaga, 2001; Nakamura et al., 2002; Coenen et al., 2007; Tan et al., 2013).
One of the poorly understood aspects of in vivo HG biosynthesis includes accurate measurements of the length and location of HG in the wall. Measurements of HG enriched from various plant species and tissues identified polymeric chains with a degree of polymerization (DP) ranging from ~70-320 (Thibault et al., 1993; Ralet et al., 2008; Yapo, 2009; Round et al., 2010). Because the extended acid hydrolysis treatments typically used to remove neutral sugars from the more complex forms of pectin also reduce the length of the HG backbone (Round et al., 2010), it is likely that most measurements have underestimated the in vivo chain lengths of HG. Therefore, further investigation into the enzymes and mechanisms utilized for in vivo HG biosynthesis is vital.
HG biosynthesis is catalyzed by members of the GALACTURONOSYLTRANSFERASE (GAUT) family (Sterling et al., 2006), a family of proven and putative homogalacturonan:galacturonosyltransferases (HG:GalATs). Arabidopsis GAUT1 was the first member of this family shown to have HG:GalAT activity following heterologous expression in Human Embryonic Kidney (HEK293) cells (Sterling et al., 2006). In vivo, GAUT1 is truncated beyond its transmembrane domain (TMD) but is localized to the Golgi apparatus via disulfide-linkage with GAUT7 to form the GAUT1:GAUT7 complex (Atmodjo et al., 2011). In addition to GAUT1 and the GAUT1:GAUT7 complex (Sterling et al., 2006; Atmodjo et al., 2011; Amos et al., 2018), in vitro HG:GalAT activity has also been reported for GAUT4 expressed in Nicotiana benthamiana cells (Biswal et al., 2018a) and GAUT11 expressed in HEK293 cells (Voiniciuc et al., 2018).
The GAUTs are one of the largest non-cellulosic plant cell wall biosynthetic glycosyltransferase families in Arabidopsis with broad representation across the plant kingdom, suggesting a functional significance to the expansion of this family (Sterling et al., 2006; Yin et al., 2010; Atmodjo et al., 2013; Biswal et al., 2018a; Amos & Mohnen, 2019). While some of the Arabidopsis gaut mutants have mild or no observable phenotypes compared to wild-type, others exhibit severe defects or cannot be recovered (Bouton et al., 2002; Orfila et al., 2005; Persson et al., 2007; Caffall et al., 2009; Wang et al., 2013; Hao et al., 2014; Lund et al., 2020). Recently, GAUT5 and GAUT6 have been shown to serve as alternative anchors for GAUT1 in planta and may function as part of HG:GalAT complexes similar to the GAUT1:GAUT7 complex (Lund et al., 2020). These observations suggest that while there may be some functional redundancy within the family, at least some members have specialized and critical functions in the plant. Based on the prior results showing that several of the GAUTs have HG:GalAT activity or membrane anchoring functions in HG:GalAT complexes, we hypothesize that all GAUT family members are involved in HG biosynthesis.
The in vivo functions and enzyme activities of the full GAUT family are not yet understood. One way to determine their activities is via heterologous expression and assay of purified proteins for biosynthetic catalytic activity either individually or as protein complexes. The HG biosynthetic activities of GAUTs 1, 4, 11 and 1:7 were previously demonstrated by incubation of the heterologously expressed enzymes in the presence of UDP-GalA and exogenous HG acceptors (Fig. 1A). However, more recently the GAUT1:GAUT7 complex was shown to also de novo synthesize HG in the absence of exogenous acceptors when incubated with UDP-GalA in vitro (Amos et al., 2018) (Fig. 1B). Enzymes that synthesize polysaccharides in the absence of a starting oligosaccharide or polysaccharide acceptor using only an activated sugar (e.g., nucleotide-sugar) as a glycosyl residue substrate are defined as having de novo synthesis activity (Illingworth et al., 1961). Such de novo initiation of polysaccharide biosynthesis has been demonstrated for some bacterial (DeAngelis & White, 2002) and plant (Mukerjea & Robyt, 2013; Amos et al., 2018) glycosyltransferases. Because HG is elongated by the addition of GalA onto the non-reducing end of acceptor oligosaccharides, the simplest mechanism for de novo synthesis would be the non-reducing end elongation of a UDP-GalA molecule, resulting in a series of oligosaccharides that retain UDP on the reducing end. Such HG products could be distinguished from HG products produced by elongation of exogenous HG oligogalacturonide acceptors (Fig. 1, A and B) by the retention of UDP-GalA on the reducing end. Although a polymeric de novo synthesis product of GAUT1:GAUT7 has been detected (Amos et al., 2018), short-chain oligosaccharide products were not isolated at that time in sufficient quantities for chemical analysis. While non-reducing end de novo synthesis has been shown for several other polysaccharides, a general mechanism has not yet been demonstrated (DeAngelis & White, 2002; Chavaroche et al., 2011; Fiebig et al., 2014a; Fiebig et al., 2014b).
In this study, we hypothesized that additional GAUTs are HG:GalATs and that at least some may also synthesize HG in the absence of exogenous HG acceptors. To test this hypothesis, we heterologously expressed the GAUTs in Human Embryonic Kidney (HEK293) cells individually and in pairwise combination to identify new GAUTs with GalAT elongation and de novo activities and compared the characteristics of these activities with other family members. We also tested for evidence of enhanced enzyme activity upon in vitro expression of GAUT:GAUT pairs in an attempt to identify undiscovered GAUT complexes.
RESULTS
Expression of Arabidopsis GAUT gene family in HEK293 cells reveals that N-terminally truncated GAUTs 1, 10, 11, 13 and 14 are expressed at high levels as soluble proteins.
Recovery of purified proteins from a heterologous expression source makes possible the unambiguous assignment of glycosyltransferase (GT) enzyme activity (Amos & Mohnen, 2019). Based on previous success with the expression of GAUT1, GAUT11 and the GAUT1:GAUT7 complex in Human Embryonic Kidney (HEK293) cells (Sterling et al., 2006; Amos et al., 2018; Voiniciuc et al., 2018), the HEK293 system (Moremen et al., 2018) was used to express the remaining members of the GAUT gene family to determine if other GAUTs were also HG:GalATs. This strategy involved cloning of truncated GAUT coding regions into the mammalian expression vectors: pGEn1-DEST, which contains a signal sequence, 8x histidine tag (8xHis), StrepII and TEV cleavage sites; and pGEn2-DEST, which contains a signal sequence, 8xHis, Avi tag, GFP, and TEV cleavage site (Moremen et al., 2018) (Fig. S1). The pGEn2-DEST construct was used for all individual expression and co-expression studies, and the pGEn1-DEST for pairwise expression studies.
All GAUTs, except for GAUT2, contain a predicted N-terminal transmembrane domain (TMD; www.aramemnon.com) (Sterling et al., 2006). Constructs were generated for each GAUT coding region to enable production of proteins that could be secreted from HEK293 cells into the medium. This involved removal of the TMD by truncation downstream from the predicted TMD just N-terminal to the first charged residue as described (Moremen et al., 2018) and insertion of the truncated cDNA region into the pGEn2-DEST HEK293 expression vector (Fig. S1, Table S1). All GAUTs, except for GAUT2, were cloned and expressed in small scale (20 ml) HEK293 cultures as described above. In addition, Arabidopsis GAUT1 exists in a truncated form with proteolytic cleavage between amino acid residues 167 and 168 (GAUT1Δ167) (Atmodjo et al., 2011). This construct can be expressed and secreted at measurable levels from HEK293 cells (Amos et al., 2018), and thus, was expressed along with the GAUT1Δ42 construct generated using the strategy as outlined above in order to compare the efficacy of the different construct designs for expression of the GAUT1 protein. GAUT2 was omitted from this GAUT gene expression study due to the inability to PCR-amplify the cDNA from multiple Arabidopsis tissue sources and to the lack of available full-length cDNA clones. In addition to having a weakly predicted TMD compared to the other GAUTs, GAUT2 transcript is not expressed at measurable levels in Arabidopsis and is predicted to be a non-functional truncation homolog of GAUT1 (Caffall et al., 2009).
The production of sufficient amounts of GAUT protein to support detailed studies of enzyme activity required that each GAUT be folded properly, expressed at high levels, secreted into the medium as a soluble protein, and purified in an enzymatically active form. The level of expression (Table 1, Cells & Medium) and secretion into the medium (Table 1, Medium) for each GAUT was measured by the total fluorescence associated with the N-terminal GFP tag domain of the recombinant protein. A standard cutoff of 100 fluorescence units (F.U.) in the medium was chosen as the baseline level of expression of GAUT protein necessary for further studies. Based on this criterion, both the GAUT1Δ42 and GAUT1Δ167 constructs and GAUTs 10Δ39, 11Δ39, 13Δ61 and 14Δ61 were produced at sufficient levels (Table 1) for purification and enzyme activity assays.
Table 1. Expression and secretion of individual GAUTs in HEK293 cells.
The number after the Δ represents the amino acid residue truncation site at which each GAUT construct was truncated to remove its putative TMD or to provide the in vivo cleavage site (GAUT1) prior to insertion into the pGEn2-DEST vector for expression in HEK293 cells. Synthesis in the cells and secretion into the medium of each GAUT protein was monitored by fluorescence of the N-terminal domain GFP tag. Values are total gross fluorescence units and represent the average ± SEM of the designated GAUT protein expression from at least two independent 20 mL HEK293 cell cultures.
| GAUT | Cells & Medium Fluorescence | Medium Fluorescence |
|---|---|---|
| 1Δ42† | 379 ± 144 | 105 ± 2 |
| 1Δ167† | 639 ± 34 | 366 ± 31 |
| 3Δ28 | 164 ± 9 | 69 ± 3 |
| 4Δ29 | 27 ± 5 | 18 ± 1 |
| 5Δ31 | 129 ± 42 | 50 ± 11 |
| 6Δ31 | 32 ± 2 | 19 ± 0 |
| 7Δ43 | 204 ± 60 | 59 ± 20 |
| 8Δ43 | 30 ± 5 | 19 ± 0 |
| 9Δ50 | 242 ± 112 | 35 ± 8 |
| 10Δ39† | 953 ± 111 | 372 ± 3 |
| 11Δ39† | 1128 ± 187 | 795 ± 82 |
| 12Δ59 | 302 ± 100 | 46 ± 6 |
| 13Δ61† | 718 ± 52 | 245 ± 23 |
| 14Δ61† | 715 ± 67 | 296 ± 30 |
| 15Δ70 | 380 ± 47 | 35 ± 3 |
These constructs were selected for purification from the HEK293 medium
Co-expression of Arabidopsis GAUT gene family in HEK293 cells provides no evidence for novel GAUT:GAUT complexes
The GAUT1:GAUT7 complex exists in vivo and can be heterologously expressed in HEK293 cells and purified from HEK293 culture medium as an active protein complex (Atmodjo et al., 2011; Amos et al., 2018). In an effort to determine if any other GAUTs might function as protein complexes, all the truncated Arabidopsis GAUTs (with the exception of GAUT2) were expressed in pairwise combination in HEK293 cells (Fig. 2, Table S2). The goal was to determine whether, upon co-expression, evidence for the existence of GAUT:GAUT protein complexes, other than the well characterized GAUT1:GAUT7 complex, could be obtained. For example, we hypothesized that some of the GAUTs, such as GAUT7, that expressed poorly on their own (Table 1) would have increased expression once co-expressed with another GAUT member, such as GAUT1 (Fig. 1, Table S2).
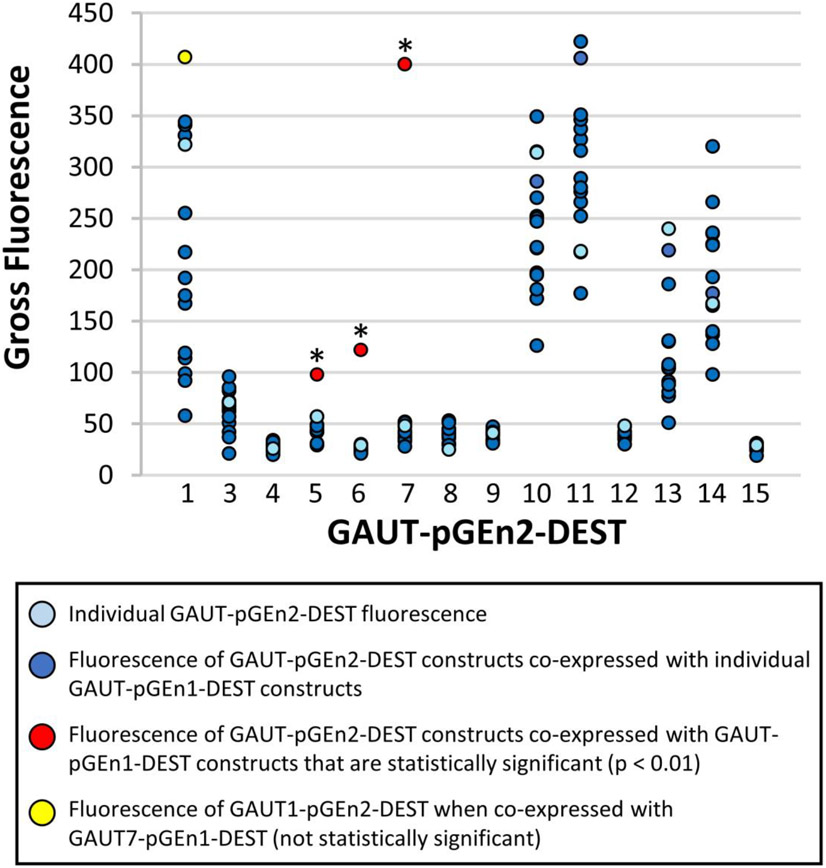
Figure 2. Secretion of pairwise expressed GAUT constructs in HEK293 cells.
Each GAUT, except for GAUT2, was truncated beyond its transmembrane domain and cloned into pGEn1-DEST and pGEn2-DEST vectors for HEK293 expression (as described in Table 1). Each GAUT-pGEn2 construct (x-axis) was expressed in pairwise combination with each GAUT-pGEn1 construct at a DNA total concentration of 3 μg/ml culture volume and the total gross medium fluorescence was measured (dark blue points). Fluorescence was measured exclusively from the expression and secretion of the GAUT-pGEn2 construct (the construct containing a GFP domain) due to the pGEn1 partner construct not containing a GFP domain. As controls, each GAUT was additionally expressed individually at the same concentration as the pGEn2 partner in all pairwise combinations (1.5 μg/ ml culture volume, light blue points). GAUT1 pGEn2 + GAUT7 pGEn1 resulted in the highest level of secretion of GAUT1 (yellow point) and GAUTs 5-7 pGEn2 + GAUT1 pGEn1 yielded the highest levels of secretions for these members (red points). Asterisks indicate those pairwise combinations for which an outlier value of gross fluorescence was detected in each set of pairwise expressed GAUTs (p < 0.01). Significance was determined by comparing calculated Grubb’s statistics in each expression set to published critical values (Table S2) (Grubbs, 1969).
In addition to the pGEn2 GFP-GAUT fusion vector constructs used for expression of individual GAUT proteins, each GAUT was also cloned into the pGEn1-DEST expression vector which does not contain the GFP fusion tag. GAUT-pGEn1 and GAUT-pGEn2 constructs were co-expressed in reciprocal combinations, such as GAUT1-pGEn2 + GAUT7-pGEn1 and GAUT1-pGEn1 + GAUT7-pGEn2, to determine whether enhanced expression was observed for unique partner pairs. The amount of total secreted GAUT protein produced was measured as the GFP fluorescence of the GAUT-pGEn2 partner in the medium of 20 mL HEK293 cultures. Each set of individual GAUT pGEn2 constructs co-expressed in pairwise combination with the pGEn1 construct partners, was then analyzed for evidence of co-expressed GAUT combinations that yielded a significant increase in expression, compared to single vector controls (Table S2). These analyses were performed to provide preliminary evidence for the formation of in vitro GAUT:GAUT complexes, in addition to GAUT1:GAUT7 (Table S2). The only outliers detected using this statistical method were increased expression of GAUT 5-pGEn2, GAUT6-pGEn2 and GAUT7- pGEn2 vectors when co-expressed with the GAUT1-pGEn1 vector, complexes already shown to exist in planta (Atmodjo et al., 2011; Lund et al., 2020). Therefore, due to a lack of evidence for any other putative GAUT:GAUT complexes using this approach, we focused the remainder of our efforts on analyzing the enzymatic activities and products produced by GAUTs 1, 10, 11, 13, 14 and the GAUT1:GAUT7 complex.
GAUTs 1, 10, 11, 13, 14 and 1:7 synthesize polymeric HG
The GAUT1:GAUT7 complex is the most extensively studied HG:GalAT to date, in part due to its ability to be highly expressed and purified from HEK293 cultures (Amos et al., 2018). GAUTs 1Δ42, 1Δ167, 10, 11, 13, 14, and 1:7 were expressed in large HEK293 cultures and purified by separation over a nickel resin column using fast protein liquid chromatography. As GAUT1Δ42 had poor protein yield from both large and small scale cultures it was not further analyzed. The purification of the remainder of the GAUTs, including GAUT1Δ167, yielded sufficient protein to support enzymatic assays for characterization of the GAUTs and the products they synthesize.
Nickel resin purified GAUTs 1Δ167, 10, 11, 13 and 14 and the GAUT1:GAUT7 complex were first analyzed under non-reducing SDS PAGE to determine whether each GAUT exists primarily as a monomer or a protein complex under these conditions, and to identify any truncation or proteolysis products resulting during purification. The bulk of the GAUT 10, 11, 13 and 14 proteins electrophoresed primarily as monomers under non-reducing conditions (Fig. 3, third bracket), although each showed some level of high molecular weight (HMW) complex formation (Fig. 3, second bracket). The GAUT1:GAUT7 complex electrophoresed as the expected HMW protein complex (Amos et al., 2018), while GAUT1 electrophoresed as the expected mixture of HMW complexed GAUT1 (~150 kDa) and truncation products (Amos et al., 2018). Truncation products were observed for each of the GAUTs, however the small truncation or proteolysis degradation products were most abundant for GAUT1 (Fig. 3, bottom bracket), confirming the prior observation of relatively large amounts of small truncation products when GAUT1 is expressed in the absence of GAUT7 (Amos et al., 2018).
Figure 3. Non-reducing SDS-PAGE of purified GAUTs and the GAUT1:GAUT7 complex.
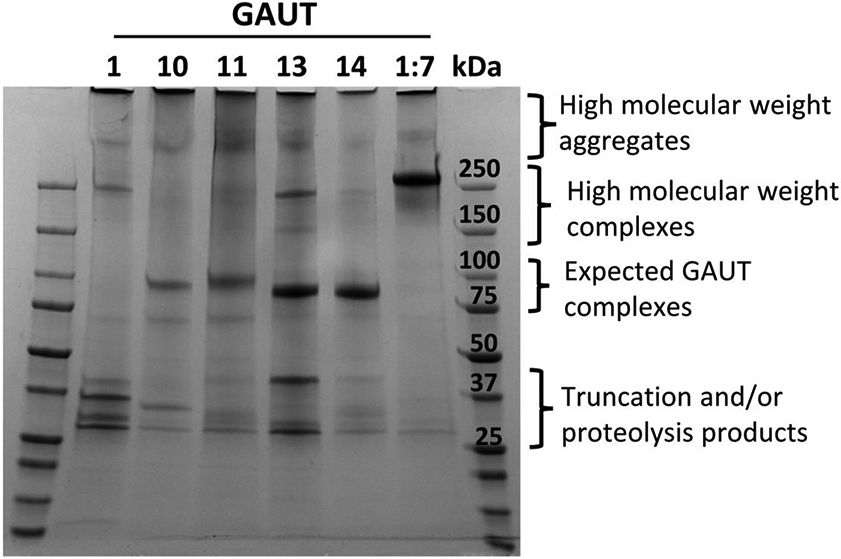
SDS-PAGE of GAUTs 1Δ167, 10Δ39, 11Δ39, 14Δ61 and 1Δ167:7Δ43 purified from HEK293 cultures via passage over a nickel column. The purified proteins (4 μg each) were separated by SDS-PAGE under non-reducing conditions and stained with Coomassie Brilliant Blue. Brackets indicate regions of different GAUT quaternary, aggregate and fragmented structure, specifically, high molecular weight aggregates, complexes, monomers, and truncation/proteolysis products. Expected masses of each GAUT with their N-terminal tags: 92.6 kDa (GAUT1 monomer), ~200-250 kDa (active GAUT1 homocomplex) (Amos et al., 2018), 99.6 kDa (GAUT7) (Amos et al., 2018), 91.2 kDa (GAUT10), 91.1 kDa (GAUT11) (Voiniciuc et al., 2018), 88.1 kDa (GAUT13), 87.8 kDa (GAUT14), and >250 kDa (GAUT1:GAUT7 complex) (Amos et al., 2018).
Each purified GAUT was assayed for HG:GalAT activity. GAUTs 1, 10, 11, 13 and 14 were incubated at 0.5, 4 or 24 h at different enzyme concentrations (0.2 μM – 5 or 10 μM) in reaction buffer containing the UDPase apyrase (E.C. 3.6.1.5) (20 mU/μL), 1 mM UDP-GalA and an HG acceptor of DP of 11 (10 μM), an acceptor size shown to be effective for maximum HG:GalAT activity by the GAUT1:GAUT7 complex (Amos et al., 2018) (Fig. 4, Fig. S2). The products formed by each GAUT were separated based on their relative charge and size via high percentage PAGE and visualized with alcian blue and silver staining. These conditions were selected to compare the sizes of HG products generated over time and to determine whether the GAUTs synthesized similar or unique size ranges of HG glycans. Commercial apyrase was added to the reactions to alleviate potential inhibition of catalysis by UDP product accumulation. We observed that the sizes of HG products formed by the GAUTs, especially by GAUT14, were influenced by the inclusion of apyrase (Fig. S3), indicating that the accumulation of UDP does inhibit GAUT activity.
Figure 4. Comparison of the sizes of HG synthesized by GAUTs 1, 10, 11, 14 and 1:7 in 4 hreactions containing HG DP11 exogenous acceptor at the enzyme concentrations indicated.
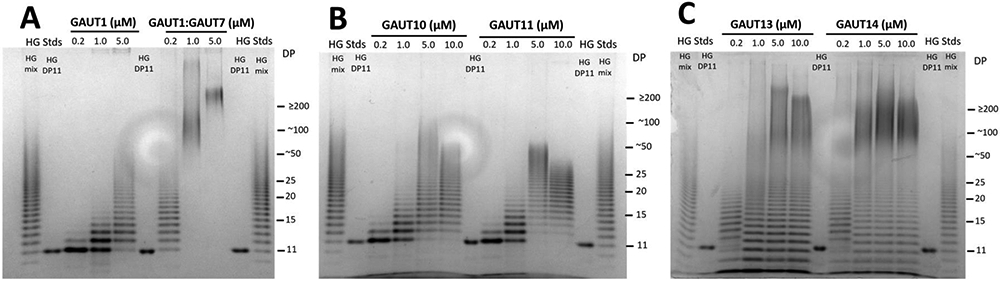
Sizes of HG products synthesized by each GAUT and the GAUT1:GAUT7 complex in 4 h reactions containing apyrase, the respective GAUT (0.2 – 5/10 μM), DP11 HG acceptor (10 μM) and UDP-GalA (1 mM) (GAUT1 and GAUT1:GAUT7, panel A; GAUT10 and GAUT11, panel B; GAUT13 and GAUT14, panel C). Products separated via PAGE over a 30% polyacrylamide gel and detected with alcian blue and silver staining. The first two, last two and the middle lane in each figure are HG standards: HG oligosaccharide mix enriched for DP 7-23, HG DP11.
Each of the GAUTs tested under these conditions were able to synthesize polymeric (DP ≥ 50) HG. The results showed that at the 4 h and 24 h time points, GAUT1 at a high enzyme concentration synthesized a product with a maximum DP of ~50-100 (Fig. 4A, Fig. S2 panel D). In comparison, GAUT1:GAUT7 quickly synthesized high molecular weight (HMW) (DP ≥ 200) at 30 min at high enzyme concentrations (Fig. S2 panel A), confirming the previously demonstrated high enzyme activity of this protein complex compared to GAUT1 alone (Amos et al., 2018). Over time, GAUT10 and GAUT11 synthesized polymeric HG at higher enzyme concentrations (Fig. 4B, Fig. S2 panels B and E). GAUT13 and GAUT14 synthesized HMW HG already at early time points at high enzyme concentrations (Fig. 4C, Fig. S2 panels C and F), suggesting that these GAUTs have high rates of HG biosynthesis. Taken together, the results support the hypothesis that all GAUTs are HG:GalATs but also demonstrate that individual GAUTs may differ in their rate of HG biosynthesis and/or in their use of multiple HG biosynthetic mechanisms in vitro.
GAUTs 13 and 14 de novo synthesize HG at high rates
During the analysis of HG product synthesized by the different GAUTs, we also noted that HG oligomers of a size smaller than the starting DP11 acceptor itself were generated by GAUT13 and GAUT14. This led to the question of whether the small HG oligomers were the result of de novo synthesis as previously described for the GAUT1:GAUT7 complex (Amos et al., 2018). To test this, GAUTs 13 and 14 were assayed for their ability to synthesize polymeric HG via both acceptor elongation and de novo biosynthesis mechanisms (Fig. 1A and B). GAUTs 13 and 14 were incubated under standard reaction conditions with UDP-GalA (1 mM) in the presence (+) and absence (−) of the HG DP11 acceptor (10 μM) for 5 min to 4 h (Fig. 5). At 5 min, only products elongated from the HG DP11 acceptor were the primary products observed, consistent with the acceptor-dependent HG:GalAT activity of these GAUTs. At 30 min, the reactions without exogenous HG DP11 acceptor displayed a range of apparent HG oligomers, both smaller and larger than the DP 11 acceptor. Some of these reaction products migrated at a slightly different position relative to those elongated using the HG DP11 acceptor, forming an offset banding pattern compared to the elongation products (Fig. 5). These results suggested a structural difference in the de novo reaction products compared to the exogenous acceptor-elongated products. Analysis of the 4 h reactions showed that the de novo synthesized products appeared to be the major polymeric HG products formed, based on the predominant emergence of the offset banding pattern. These results suggest that de novo synthesis is one of the primary biosynthetic mechanisms utilized by GAUT13 and GAUT14.
Figure 5. Comparison of products synthesized by GAUT13 and GAUT14 in the absence and presence of HG DP11 acceptor.
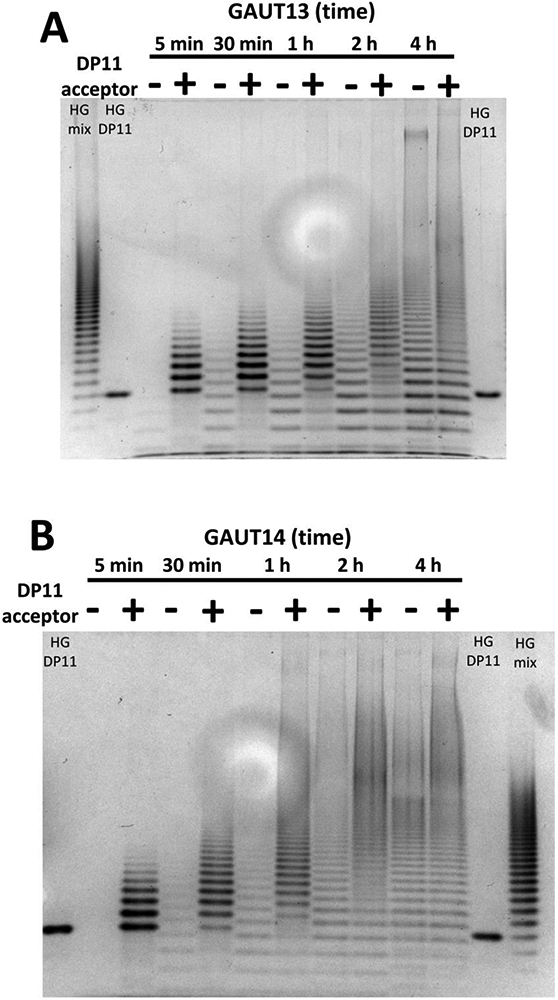
Time course (5 min, 30 min, 1 h, 2 h, 4 h) of products produced by GAUT13 (panel A) and GAUT14 (panel B) (1.0 μM each) incubated in reaction buffer containing apyrase, UDP-GalA (1 mM) and with (+) or without (−) HG DP11 acceptor (10 μM). Products were separated by PAGE over a 30% polyacrylamide gel and detected with alcian blue and silver staining. HG standards are HG oligomer mix enriched for DPs 7-23 and HG DP11. Only faint de novo reaction products (without HG DP11 acceptor) are visible in the first five min of the GAUT13 and GAUT14 reactions while substantive elongation of HG DP11 acceptor is observed. As time progresses, the de novo synthesis products become the most abundant products observed.
GAUT13 initiates HG biosynthesis by elongating UDP-GalA and producing HG with UDP-GalA on the reducing end
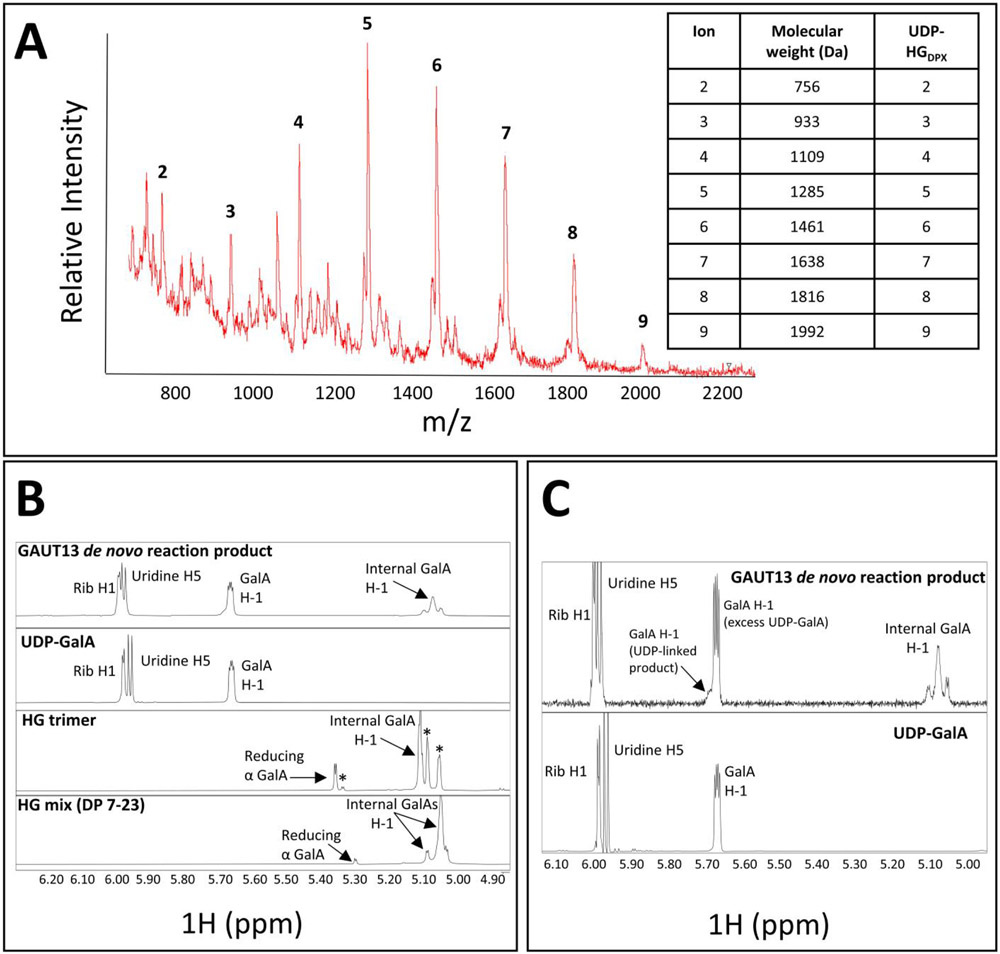
The high de novo synthesis activity of GAUT13 resulted in a population of HG chains with sufficient de novo synthesis product to support structural characterization of the reducing ends of the de novo synthesized HG product. HG synthesized in the absence of exogenous acceptor in 3h-GAUT13 reactions was purified by size-exclusion chromatography and analyzed by MALDI-TOF MS and NMR (Fig. 6). MALDI TOF MS identified masses expected for HG of DP 2-9 containing an additional mass of 386 Da corresponding to UDP. These results were consistent with retention of UDP-GalA on the reducing end of the synthesized product and with UDP-GalA serving as the initiating primer for HG synthesis (Fig. 6A). There were no ion peaks detected for HG oligomers that contain free reducing ends (Table S3). These results were confirmed by NMR analysis in which no free reducing GalA species was detected (Fig. 6B-D) while a clear resonance for UDP and an associated anomeric GalA H1 resonance were detected (Fig. 6C). Taken together the results indicate a GalA species conjugated to UDP and provide evidence for the presence of UDP at the reducing terminus of each de novo synthesized HG chain.
Figure 6. MS and NMR analysis of GAUT13 de novo synthesized products.
(A) MALDI-TOF MS of products synthesized by GAUT13 (1.1 μM) incubated in reaction buffer with UDP-GalA (1 mM) for 3 h and subsequently separated by size exclusion chromatography (SEC) (LH-20 resin). Data shown are for a SEC fraction of the HG enriched for HG of DP 2-9. The observed masses are consistent with HG chains containing one UDP (386 Da, see table inset). (B) Products from (A) (upper panel) analyzed by 1H NMR and standards of UDP-GalA (second panel), HG DP3 (third panel), and HG mix (DP 7-23) (fourth panel). Peak assignments are: Ribose H1 (~6.0 ppm), Uridine H5 (~5.95 ppm), conjugated anomeric GalA H1 (~5.65 ppm), free reducing α GalA (~5.3-5.35 ppm), and internal GalA (~5-.05-5.10 ppm). The HG trimer contained trace levels of contaminants, indicated by asterisks. The GAUT13 de novo reaction products contained no free reducing α GalA species, while the trimer and HG mix had free reducing α GalA species. (C) Zoomed in region of NMR spectra in (B) of the GAUT13 de novo reaction products and UDP-GalA standard to show the 5.65-5.75 ppm region containing the conjugated anomeric GalA H1. The shoulder present in the GAUT13 de novo reaction products at ~5.7 ppm is indicative of multiple GalA species that are conjugated to UDP, which is absent in the major peak generated from UDP-GalA alone.
GAUTs 1, 10, 11, 13, 14 and 1:7 have different acceptor dependent specific activities under identical HG:GalAT assay conditions; GAUTs 13 and 14 have high rates of HG de novo synthesis
The observation of different size ranges of polymeric HG synthesized by the GAUTs at different time points and enzyme concentrations led us to hypothesize that some GAUTs have higher rates of HG:GalAT activity than others. Thus, the HG:GalAT specific activity of the individual GAUTs 1, 10, 11, 13, 14 and the GAUT1:GAUT7 complex were compared under standard identical reaction conditions. A radioactive assay measuring incorporation of [14C]GalA from UDP-[14C]GalA into product was used to obtain a quantitative measure of enzyme activity (Sterling et al., 2005). The standard reaction conditions chosen were 100 μM UDP-GalA, 10 μM HG DP 7-23 oligosaccharide mix in reaction buffer with variable enzyme amounts (Amos et al., 2018; Voiniciuc et al., 2018). HG:GalAT activity was calculated in the linear region of the generated progress curves (Fig. S4). Normalization of activity to input quantity of enzyme yielded specific activity values for each of the GAUTs and enabled direct comparison. A concentration of 100 μM UDP-GalA was used because this is the maximum estimate of physiological UDP-GalA in multiple Arabidopsis tissues (Rautengarten et al., 2014; Rautengarten et al., 2017).
The results showed that the GAUTs exhibit a range of catalytic rates. GAUT14 had the highest specific activity of the singly expressed GAUTs, followed by GAUT13, GAUT1, GAUT10 and GAUT11, however, all exhibited lower specific activity than the GAUT1:GAUT7 complex (Table 2). In addition, the HG:GalAT specific activity of GAUT1 alone was 11-fold lower than the GAUT1:GAUT7 complex. These results confirm that GAUT7 acts not only as an anchor for GAUT1 in the Golgi membrane (Atmodjo et al., 2011), but also contributes to GAUT1 stability and/or activity (Amos et al., 2018). Overall, the GAUTs with higher specific activities synthesized larger HG polymers during extended reaction times (Fig. S2 panels D-F, Table 2).
Table 2. Specific activities of acceptor dependent GalAT activities by truncated GAUTs 1, 10, 11, 13, 14 and 1:7 and comparison of de novo GAUT13 and GAUT14 GalAT specific activities.
GAUTs 1Δ167, 10Δ39, 11Δ39, 13Δ61, 14Δ61 and GAUT1Δ167:GAUT7Δ43 were expressed and the resulting proteins purified from the HEK293 medium (see methods). HG:GalAT progress curves were generated under standard reaction conditions in reaction buffer containing added acceptor (100 μM UDP-GalA, 10 μM HG oligosaccharide acceptor mix, DP 7-23) and the specific activity (pmol GalA transferred min−1 mg−1 GAUT) of each GAUT was calculated at linear time points. GAUTs 13 and 14 were additionally assayed as described above but without added exogenous acceptor (i.e., only in the presence of 100 μM UDP-GalA) to calculate their de novo HG:GalAT biosynthetic rates. Values represent average ± SEM from at least two independent progress curves for each GAUT.
| GAUT | Acceptor Dependent HG:GalAT Specific Activity (pmol GalA transferred min−1 mg−1) |
de novo HG:GalAT Specific Activity |
|---|---|---|
| 1Δ167 | 584 ± 58 | |
| 10Δ39 | 43 ± 3 | |
| 11Δ39 | 39 ± 3† | |
| 13Δ61 | 958 ± 20 | 1124 ± 152 |
| 14Δ61 | 3541 ± 470 | 1039 ± 364 |
| 1Δ167:7Δ43 | 6330 ± 145 |
The specific activities of GAUT13 and GAUT14 for de novo synthesis in the absence of exogenous HG acceptors were also calculated (Fig. 7, Table 2) to compare the rate of de novo synthesis to that for acceptor dependent synthesis. The results showed that both GAUT13 and GAUT14 have high rates of de novo synthesis activity, comparable to their acceptor-dependent rates (Table 2). The GAUT13 de novo synthesis progress curve was almost identical to the acceptor dependent progress curve (Fig. 7), except for an initial lag in activity. GAUT14 also showed an initial lag in de novo synthesis. Such lags in de novo synthesis are associated with initiation phases of polymer synthesis (Levengood et al., 2011; Amos et al., 2018; Fiebig et al., 2018). The results support the hypothesis that de novo synthesis of HG is a major function of GAUT13 and GAUT14.
Figure 7. Acceptor dependent and de novo GalAT progress curves of GAUT13 and GAUT14.
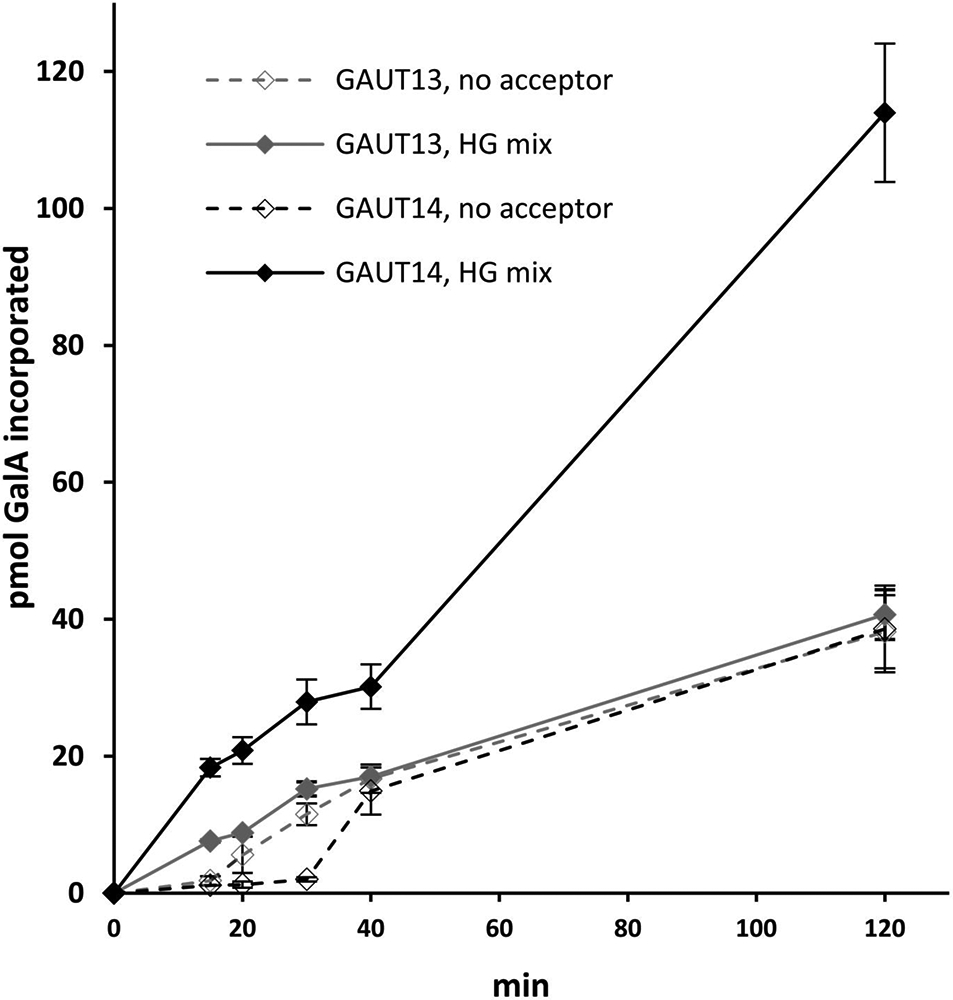
GAUT13 (grey lines) and GAUT14 (black lines) (200 nM each) were incubated with a mixture of UDP-GalA and UDP-[14C]GalA (100 μM total) and either with (solid lines) or without (dashed lines) HG acceptor (10 μM). Incorporated GalA was detected by an HG:GalAT filter assay (see methods) and pmol GalA incorporated from the linear region of the curve (between 15 and 40 min for both GAUT13 and GAUT14 without acceptor) was used to calculate specific activity as pmol GalA incorporated min−1 mg−1 protein. Data points represent average ± SEM of at least two independent progress curves.
GAUTs 1, 10, 11, 13, 14 and 1:7 are HG-specific GalATs
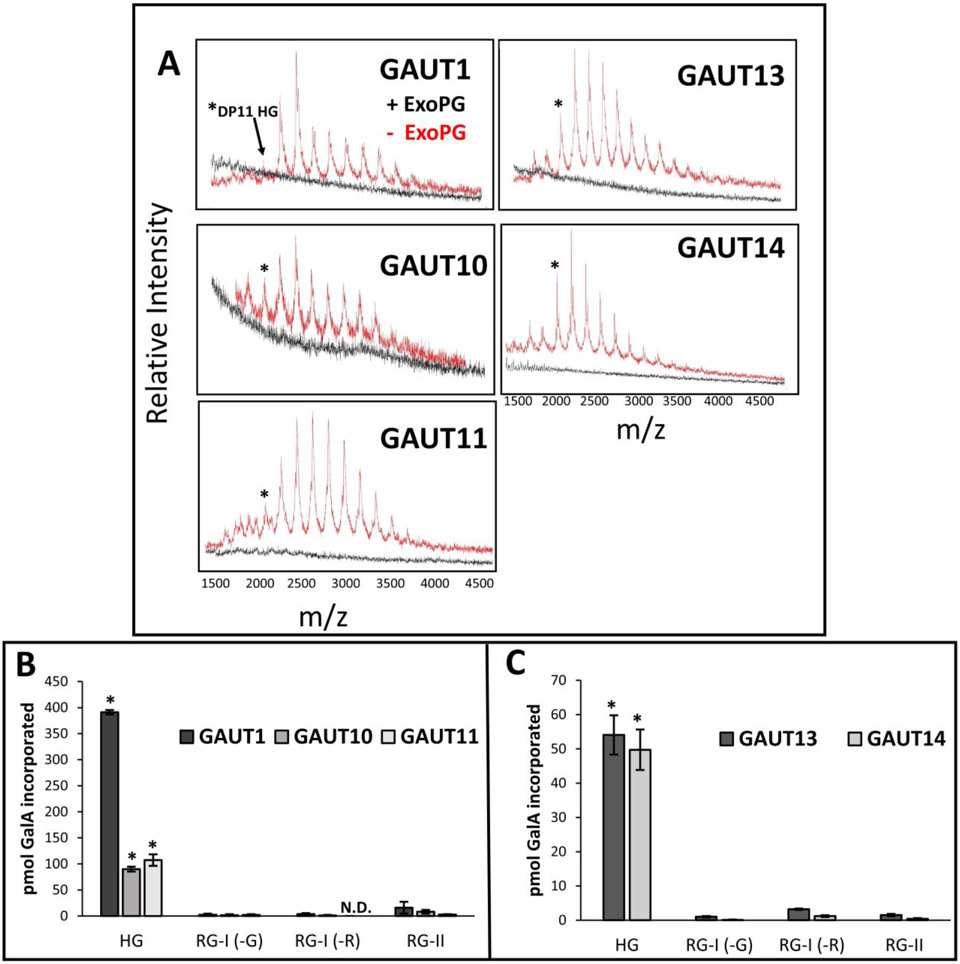
Previous studies of GAUT1 (Sterling et al., 2006), GAUT11 (Voiniciuc et al., 2018) and the GAUT1:GAUT7 complex (Amos et al., 2018) demonstrated that the products synthesized onto the HG acceptor by these GAUTs were α1,4-linked HG due to their sensitivity to hydrolysis by polygalacturonase. Here the products synthesized by GAUTs 1, 10, 11, 13 and 14 were also tested for their sensitivity to hydrolysis by exopolygalacturonase (ExoPG). As shown in Fig. 8A, treatment with ExoPG hydrolyzed the HG products produced, confirming that GAUTs 1, 10, 11, 13 and 14 are HG:GalATs.
Figure 8. Sensitivity of products synthesized by GAUTs 1, 10, 11, 13 and 14 to exopolygalacturonase and pectin acceptor specificities.
(A) MALDI-TOF MS of HG:GalAT products produced by GAUTs 1, 10, 11, 13 and 14 and sensitivity of the products to hydrolysis by exopolygalacturonase (ExoPG). GAUT1 (10 μg, 2.7 μM), GAUT10 (10 μg, 5.5 μM), GAUT11 (18 μg, 9.9 μM), GAUT13 (2 μg, 1.1 μM) and GAUT14 (2 μg, 1.1 μM) were incubated in reaction buffer with 1 mM UDP-GalA and 100 μM GalA11-2AB acceptor (asterisk) for 24 h and subsequently mixed with ExoPG buffer only (red spectra) or ExoPG buffer plus active ExoPG (2.4 mU) (black spectra) for ≥ 6h. Enzyme amounts were chosen so that the initial 24 h spectra showed ion masses up to a DP 20 to efficiently demonstrate the loss of these signals upon exopolygalacturonase treatment. The loss of ion mass signals associated with HG of various DPs after incubation with active ExoPG demonstrates that these GAUTs are HG:GalATs. (B) Products produced by GAUT1Δ167, GAUT10 and GAUT11 using diverse pectic exogenous acceptors. Each GAUT (3 μM each) was incubated in reaction buffer for 4.5 h containing a mixture of UDP-GalA and UDP-[14C]GalA (50 μM total) and one of four alternative pectic acceptors (10 μM each): homogalacturonan (HG) oligogalacturonides DP 7-23, rhamnogalacturonan-I (RG-I) DP 6-24 with GalA (-G) or Rha (-R) at the nonreducing end, rhamnogalacturonan-II (RG-II) monomer. Precipitated products were detected by scintillation counting. Data are average ± SEM of two independent assays (n = 3). ND = not detected. Asterisks indicate those acceptor substrates for which significant levels of GalA incorporation was detected for each of GAUT1, GAUT10 and GAUT11 (p < 0.01). Significance was determined by one-way ANOVA followed by Tukey’s multiple comparison test in R for each individual GAUT. (C) Products produced by GAUT13 and GAUT14 using diverse pectic exogenous acceptors. Incorporation of [14C]GalA by GAUT13 (200 nM, 90 min), GAUT14 (200 nM, 120 min) onto the indicated pectic acceptors. Each GAUT was incubated in reaction buffer containing UDP-[14C]GalA (5 μM) and one of four alternative pectic acceptors (10 μM each) as described above in (B). Data are average ± SEM of at least two independent assays (n ≥ 3). ND = not detected. Asterisks indicate those acceptor substrates for which significant levels of HG product formation was detected for each of GAUT13 and GAUT14 (p < 0.05). Significance was determined by one-way ANOVA followed by Tukey’s multiple comparison test in R for each individual GAUT.
It has been shown here and in previous studies that GAUTs can transfer GalA from UDP-GalA onto HG acceptors (Sterling et al., 2006; Atmodjo et al., 2011; Amos et al., 2018; Biswal et al., 2018a). However, two other pectins (RG-I and RG-II) in the plant cell wall contain GalA and have covalently-linked HG attached to their backbones which could potentially also be acceptor substrates utilized by the GAUTs. To test this, we determined whether GAUTs 1, 10 or 11 transferred GalA onto the HG backbone of RG-II (O'Neill et al., 2004; Ndeh et al., 2017) or onto the alternating disaccharide repeat backbones of RG-I with either GalA or Rha at the non-reducing end (Renard et al., 1997; Mutter et al., 1998), via incubation of each GAUT in reaction buffer in the presence of radiolabeled UDP-GalA donor substrate (UDP-[14C]GalA) and either HG, RG-I or RG-II acceptors (Fig. 8B). Following the reactions, the acceptors and products were precipitated and the transfer of GalA onto the acceptors was detected by measurement of the amount of [14C]GalA incorporation into product. The GAUTs were incubated in the presence of 50 μM UDP-GalA and 10 μM pectin acceptor to facilitate maximum elongation of each acceptor, as GAUTs 1, 10 and 11 had low or moderate HG biosynthetic rates (Table 2). However, when these conditions were used to assay GAUT13 and GAUT14, large amounts of incorporated [14C]GalA were obtained for all acceptors tested, likely due to the inherent de novo activities of these enzymes. Therefore, GAUT13 and GAUT14 were re-assayed for their acceptor specificities using modified reaction conditions with limiting amounts (5 μM) of UDP-[14C]GalA. All of the GAUTs tested were found to preferentially use HG as an acceptor, with minimal transfer of GalA onto the additional pectin acceptors (Fig. 8C). Similar results demonstrating preferential addition to HG acceptors were also previously observed for the GAUT1:GAUT7 complex purified from Arabidopsis microsomal membranes (Atmodjo et al., 2011). Taken together, these results showed that all GAUTs tested are HG-specific GalATs.
DISCUSSION
In vitro homogalacturonan:galacturonosyltransferase (HG:GalAT) activity has been demonstrated for heterologously expressed and purified Arabidopsis GAUTs 1, 10, 11, 13, 14 and 1:7
Prior to the work reported here, only Arabidopsis GAUTs 1, 4, 11 and the GAUT1:GAUT7 complex had been shown to have HG:GalAT activity (Sterling et al., 2006; Atmodjo et al., 2011; Amos et al., 2018; Biswal et al., 2018a; Voiniciuc et al., 2018). Furthermore, GAUTs 5, 6 and 7 were associated with HG:GalAT activity by their abilities to anchor GAUT1 to the Golgi (Atmodjo et al., 2011; Lund et al., 2020). The HG:GalAT activity of the other members of the GAUT family remained untested or unverified and no comparisons of the sizes of products generated by the different GAUTs had been reported. Here the generation of GAUT constructs containing a fluorescent GFP domain allowed for facile screening for GAUT protein expression and secretion in HEK293 cells. This system supported both the expression of individual GAUTs and the co-expression of multiple GAUTs to test for HG:GalT activity and for evidence of GAUT:GAUT complex formation in addition to the GAUT1:GAUT7 complex. The expression screen identified individual GAUTs1Δ42, 1Δ167, 10Δ39, 11Δ39, 13Δ61 and 14Δ61 as capable of being expressed and secreted from the HEK293 system at measurable levels. Upon purification from small and large scale HEK293 cell cultures, each of these GAUTs, with the exception of GAUT1Δ42 for which sufficient protein could not be purified, were shown to have HG:GalAT activity (Fig. 4, Fig. S2, Fig. 8A), thereby increasing to nine the number of GAUTs shown to have HG:GalAT activity or to be part of HG:GalAT enzyme complexes.
Expression of the GAUTs in pairwise combination, with the goal of identifying GAUTs for which co-expression significantly increased GAUT protein expression above the level achieved by individual GAUT expression, identified only three GAUTs for which co-expression significantly increased GAUT expression (Fig. 2, Table S2). Only GAUTs 5, 6 and 7 co-expressed with GAUT1 were shown to increase the expression of another GAUT protein in a statistically significant manner above the level obtained by individual GAUT expression, supporting the known interactions between these members (Atmodjo et al., 2011; Amos et al., 2018; Lund et al., 2020). No other GAUT:GAUT combination showed significant changes in secretion upon co-expression. These data suggest that the other HG:GalATs, GAUT10, GAUT11, GAUT13 and GAUT14, likely do not form abundant and stable GAUT:GAUT complexes, in comparison to the proven GAUT1:GAUT7 complex.
GAUTs 3, 4, 8, 9, 12, and 15 were not successfully expressed as proteins using the defined HEK293 strategy reported here, thus, their proposed HG:GalAT activity awaits confirmation. Several approaches could be explored to facilitate expression of the remaining GAUTs as soluble proteins. Modified construct design to vary the site of sequence truncation C-terminal to the TMD may be worth investigation. Notably, the original identification of GAUT1 HG:GalAT activity by Sterling and colleagues (Sterling et al., 2006) was based on the recovery of HG:GalAT activity from GAUT1Δ41 immunoprecipitated from the HEK293 cell medium and confirmed by both depletion of HG:GalAT activity in Arabidopsis suspension cultures upon incubation with an anti-GAUT1 antibody and recovery of HG:GalAT activity in the immunoadsorbed fractions. The relatively low level of HG:GalAT activity recovered in the heterologously expressed GAUT1 in the original paper (Sterling et al., 2006), and the lack of detectable activity from the GAUT1Δ42 construct generated in this study, may have been due to the sites chosen for truncation following the TMD. Strategies to test multiple truncation cloning sites as well as codon optimization for expression in mammalian cells may be avenues to explore for future expression of the remaining GAUTs as functional proteins.
Significance of the identified in vitro HG:GalAT activities for in vivo function of the GAUTS
This study establishes that all GAUTs that expressed as soluble proteins in vitro in HEK293 cells synthesized HG (Fig. 8A), supporting the hypothesis that all GAUTs are likely HG:GalATs or members of HG:GalAT complexes (Atmodjo et al., 2011; Atmodjo et al., 2013; Biswal et al., 2018a). Specifically, we show that all the expressed GAUTs, GAUTs 1, 10, 11, 13, 14 and the GAUT1:GAUT7 complex synthesize polymeric (DP ≥ 50) HG in vitro. The demonstration that GAUT1 can synthesize high molecular weight HG with a DP 50-100 was surprising since a prior study showed that the size of product generated by GAUT1 was limited to HG of DP ~30-50 (Amos et al., 2018). The finding here that GAUT1 synthesized larger MW products may be due to the greater enzyme concentration or specific GAUT1 enzyme preparation used (Fig. 3A, Fig. S2 upper and lower panels A). Interestingly, under the same standard conditions, some GAUTs, such as GAUTs 13 and 14, had greater in vitro HG:GalAT activity rates than other individually expressed GAUTs, and thus, synthesized high molecular weight HG relatively rapidly (Fig. 4, Fig. S2, Table 2). Conversely, other GAUTs, such as GAUTs 10 and 11, had substantially lower HG:GalAT activities.
The HG:GalAT activities of GAUTs 13 and 14 (Fig. 4, Fig. 5, Fig. S2, Table 2), including their high HG:GalAT in vitro acceptor dependent and de novo synthesis activity, along with the large size of the in vitro-synthesized HG suggests that these GAUTs may synthesize high molecular weight HG in vivo. GAUT13 and GAUT14 share 94% amino acid sequence identity and have been shown to function redundantly in pollen tube formation (Wang et al., 2013). Single gaut13 and gaut14 mutants are viable, while homozygous double mutants are not recoverable due to pollen tube swelling (Wang et al., 2013). In an RNA-Seq study of Arabidopsis tissues across multiple developmental stages, GAUT13 and GAUT14 were the most highly expressed GAUTs in anthers and pollen (Klepikova et al., 2015). These prior observations, along with the demonstrated HG:GalAT activities and sizes of the synthesized HG products reported here, support the hypothesis that GAUT13 and GAUT14 are pollen tube localized GAUTs that synthesize high molecular weight HG necessary for proper pollen tube development and maintenance.
The modest activity of GAUT10 and GAUT11 identified here could indicate a function for these GAUTs in synthesizing HG at a relative low rate or small size, or rather, that they require different reaction conditions for optimal activity, for example, different acceptors. GAUT10 is most highly expressed in seeds, roots and flowers (Klepikova et al., 2015), and thus, may synthesize HG with specific functions in these tissues. GAUT11 has previously been shown to be an HG:GalAT functioning in the synthesis of seed mucilage (Caffall et al., 2009; Voiniciuc et al., 2018), a polysaccharide abundant in RG-I. gaut11 mutants have reduced seed mucilage compared to WT seeds (Caffall et al., 2009) and the sugar composition of gaut11 seed mucilage shows reductions in GalA, Rha and Xyl (Caffall et al., 2009; Voiniciuc et al., 2018). These observations have led to the hypothesis that GAUT11 synthesizes HG necessary for the synthesis of RG-I in the mucilage (Voiniciuc et al., 2018). The demonstration here that GAUTs 10 and 11 are HG:GalATs with relative low activity suggests that they may serve specialized functions in these tissues, for example, the synthesis of short HG domains associated with RG-I-rich mucilage.
Mechanisms of HG biosynthesis by the GAUTs
In addition to the GAUT1:GAUT7 complex, GAUT13 and GAUT14 have now been shown to synthesize HG de novo, i.e. in the absence of exogenous HG acceptor (Fig. 1A and B, Fig. 5). It was previously hypothesized that during HG initiation and elongation by GAUT1:GAUT7, the UDP associated with the priming UDP-GalA was either retained at the reducing end of the HG, or rather, that the reaction was primed by UDP-GalA followed by cleavage of the UDP (Amos et al., 2018). However, because only large polymeric de novo synthesized reaction products could be recovered in the in vitro GAUT1:GAUT7 reactions, those hypotheses were not able to be tested due to insufficient reducing end signal amidst the high internal GalA residue signals in the HMW HG product (Amos et al., 2018). In the present study, the high de novo synthesis activity of GAUT13 and GAUT14 allowed us to modify the in vitro synthesis conditions to facilitate size-specific production of relatively short HG chains. The production of short HG products enabled a test of the hypothesis that UDP-GalA is present on the reducing end of de novo synthesized HG chains. Specifically, after production of de novo synthesized product by GAUT13, the HG was separated by size-exclusion chromatography and analyzed by mass spectrometry and NMR spectroscopy, revealing that UDP was covalently attached at the reducing end of the synthesized HG. These results show that de novo HG synthesis by GAUT13 (Fig. 6) occurs via use of UDP-GalA as a primer and support the hypothesis that de novo synthesis of HG by GAUT1:7, GAUT14 and possibly other GAUTs occurs in a similar manner. It remains unresolved at the present time whether appreciable de novo synthesis of HG polymers occurs in vivo with the use of UDP-GalA as an initiating acceptor or whether this mode of polymer synthesis is only a product of the in vitro reaction conditions. Further analysis of in vivo HG polymer products will be required to resolve this issue.
Relevance of results for the Multiple HG Domain Hypothesis
The results reported here show that multiple GAUTs are HG:GalATs that synthesize polymeric HG via acceptor elongation and de novo synthesis. A question that arises is why plant cells have so many enzymes that synthesize the same glycan. The results indicating that different GAUTs synthesize HG at different rates are consistent with the hypothesis that individual GAUTs synthesize HG with unique structural or functional roles in different HG-containing polymers (Atmodjo et al., 2011; Atmodjo et al., 2013; Biswal et al., 2018a; Biswal et al., 2018b).
At the time GAUT1 was discovered, the diversity of pectic polymers was thought to be primarily HG, RG-I and RG-II (Sterling et al., 2006). Therefore, the identification of GAUT1 as an HG:GalAT belonging to a large 15-member family was unexpected. However, since then, multiple studies have provided support for the hypothesis that different GAUTs synthesize different HG glycans. For example, knockdown expression of the main GAUT4 homologs in poplar and switchgrass revealed that GAUT4 synthesizes the bulk of HG and HG associated with RG-II (Biswal et al., 2018a) in these species. Furthermore, the identification of pectic proteoglycans that contain HG as well as xylan (Tan et al., 2013) indicates that at least one GAUT must synthesize HG associated with this proteoglycan. Although differences in enzyme catalytic properties of GTs with the same catalytic function do not, by themselves, indicate the polymer into which the HG glycan resides, the unique enzymatic rates for the different GAUTs and the ability of at least some of the GAUTs to both de novo initiate and elongate preexisting HG acceptors is consistent with the hypothesis that the unique catalytic properties may enable the different GAUTs to synthesize HG in unique HG-containing polymers. The challenge for the future will be to determine the in vivo polymer(s) into which the HG synthesized by each of the GAUTs resides.
EXPERIMENTAL PROCEDURES
Cloning and expression of the Arabidopsis GAUT gene family in single and pairwise fashion in HEK293 Cells
Cloning, expression and purification methods were performed as previously described for GAUT11Δ39 (Voiniciuc et al., 2018), with modification as described below. Protein sequences of each GAUT were analyzed for their putative transmembrane domains (TMD) using the AramTmCon tool from the ARAMEMNON database (Schwacke et al., 2003) (http://aramemnon.botanik.uni-koeln.de/). Primers were designed to amplify the coding DNA sequence of each GAUT truncated up to the first charged residue beyond the predicted TMD (Table S1). Truncated GAUT constructs were PCR-amplified either from whole 7-day-old Arabidopsis seedling cDNA or TAIR cDNA clones (Table S1) (www.arabidopsis.org). A second round of PCR amplification was performed using the attB F and R universal primers (Table S1) and the attB PCR products were cloned into the Gateway pDONR221 entry vector using the Gateway BP Clonase II Enzyme (ThermoFisher) according to the manufacturer’s instructions. JM109 competent cells were transformed and colonies selected as previously described (Voiniciuc et al., 2018). GAUT-pDONR221 constructs were sequence-confirmed using universal M13F and M13RpUC primers (Macrogen) and a GAUT specific internal primer (Table S1).
Sequence-confirmed GAUT-pDONR221 plasmids were isolated and recombined into pGEn1-DEST and pGEn2-DEST HEK293 expression vectors using the Gateway LR Clonase II Enzyme (ThermoFisher) and confirmed with pGEn1/2 forward and reverse and GAUT specific internal primers (Table S1) (Moremen et al., 2018; Voiniciuc et al., 2018). Glycerol stocks of all sequence-confirmed constructs were stored at −80°C until further use. Plasmids for expression in HEK293 cells were amplified in JM-109 competent cells and isolated using Invitrogen PureLink HiPure Plasmid Filter Maxiprep or Gigaprep kits (ThermoFisher) as previously described (Voiniciuc et al., 2018).
Transfection of sterile GAUT pGEn2-DEST DNA into HEK293 cells (Freestyle 293-F cells; ThermoFisher) was done at a total DNA concentration of 3 μg mL−1 total cell culture volume (20 mL cultures for initial screen; 400-1000 mL for large scale purification) with 9 μg mL−1 polyethyleneimine (linear 25-kD polyethyleneimine; Polysciences) as described previously (Moremen et al., 2018). For pairwise GAUT expression, transfection was performed with 1.5 μg DNA mL−1 total cell culture volume for each construct (3 μg mL−1 combined DNA amount). For expression of the individual GAUTs in the pairwise study (Table S2, no partner), only 1.5 μg mL−1 of the pGEn2-DEST construct was used.
Purification of GAUT protein from HEK293 cultures
Six days after transfection with the DNA constructs, the HEK293 cells were separated from the medium by centrifugation, and the resulting clarified medium was filtered through a 5-μm nylon filter. Total GFP fluorescence (cells + medium) and secreted GFP fluorescence (medium only) was measured on a SpectraMAX GeminiXS microplate reader (Molecular Devices, Sunnyvale, CA) using an excitation wavelength of 450 nm and emission wavelength of 515 nm (Moremen et al., 2018). GAUT1Δ167, GAUT10Δ39, GAUT11Δ39, GAUT13Δ61, GAUT14Δ61 and GAUT1Δ67:GAUT7Δ43 were purified from large-scale HEK293 cultures (400 - 1000 mL) using an ÄKTA FPLC system equipped with a 1 or 5-mL His-Trap HP column (GE Healthcare) as previously described (Voiniciuc et al., 2018). To verify the purity of the protein preparations, an aliquot of each (4 μg) was separated by non-reducing SDS-PAGE and the proteins were visualized by staining of the gel with Coomassie Brilliant Blue.
HG:GalAT enzyme assays of GAUTs
For quantitative enzyme assays using radiolabeled UDP-[14C]GalA, GAUT1Δ167 (2.5 μM), GAUT10Δ39 (2.2 μM), GAUT13Δ61 (200 nM), GAUT14Δ61 (200 nM) and GAUT1Δ167:GAUT7Δ43 (400 nM) were incubated at 30°C in individual 30 μL reactions containing reaction buffer (50 mM HEPES, pH 7.2, 0.05% (w/v) BSA, 0.125 mM MnCl2) with HG mix (DP 7-23) (10 μM), UDP-GalA (95 μM) (CarboSource Services) and UDP-[14C]GalA (5 μM), except for GAUT10 and GAUT11 which used 80 μM UDP-GalA and 20 μM UDP-[14C]GalA. De novo reactions for GAUT13 and GAUT14 were performed as above, but without the inclusion of HG mix. Incorporated GalA was detected by the HG:GalAT filter assay (Sterling et al., 2005) and GalA incorporation plotted to determine linear time points for use in calculations of specific activity. The amount of pmol GalA incorporated was calculated by subtraction of T0 CPM values from the overall CPM values at each time point and converted to pmol GalA incorporated using the UDP-[14C]GalA specific activity of 249 mCi/mmol. T0 samples were prepared by addition of 5 μL NaOH (400 mM) to the sample prior to addition of the enzyme. HG mix (DP 7-23) was prepared as previously described (Doong & Mohnen, 1998). UDP-[14C]GalA was synthesized from UDP-[14C]GlcA (Perkin Elmer) as previously described (Liljebjelke et al., 1995). After plotting the HG:GalAT progress curves for each GAUT, linear time points were identified and these were used to calculate the specific activity of each GAUT under the above conditions. Specific activity is reported in pmol GalA transferred min−1 mg−1 enzyme.
The sensitivity of HG:GalAT reaction products to exopolygalacturonase (ExoPG) (EC 3.2.1.67) (specific activity 15 U/mg, 1U = 1 μmol GalA produced/min) was detected as described (Voiniciuc et al., 2018). HG:GalAT reactions (20 μL) containing GAUT1 (10 μg, 2.7 2.7 μM, based on dimer mass), GAUT10 (10 μg, 5.5 μM), GAUT11 (18 μg, 9.9 μM), GAUT13 (2 μg, 1.1 μM) or GAUT14 (2 μg, 1.1μM) in reaction buffer, 100 μM GalA11X-2AB and 1 mM UDP-GalA were analyzed at time zero and after a 24-h incubation using a Bruker LT mass spectrometer as described previously (Amos et al., 2018; Voiniciuc et al., 2018). Enzyme amounts were chosen to obtain a product size of a DP ~20 to be used as substrates for digestion with purified ExoPG. After 24 h, the reactions (10 μL) were mixed with 1 M sodium acetate buffer, pH 4.2 (2 μL), and 2 M acetic acid (10 μL). Two aliquots (11 μL each) were made, with one aliquot given active ExoPG (2.4 mU/μL, 1 μL) and the other water only. The reactions were incubated ≥ 6 h at 30°C, and the products were analyzed using the Bruker LT mass spectrometer. ExoPG was purified from Aspergillus tubengensis as described (Voiniciuc et al., 2018). The GalA11X-2AB acceptor was generated by labeling HG of DP 11 with the fluorescent probe 2-aminobenzamide on the reducing end as previously described, dialyzed against water (four changes) and recovered by lyophilization (Ishii, 2002). The homogenous HG DP 11 acceptor was generated by partial digestion of polygalacturonic acid with endopolygalacturonase and purified by HPAEC-PAD as described previously (Doong & Mohnen, 1998).
Acceptor substrates tested in enzyme specificity assays were HG mix (DP 7–23), RG-I-R (RG-I with Rhamnose on the non-reducing end) and RG-I-G (RG-I with Galacturonic Acid on the non-reducing end) oligomers (DP 6–26), and RG-II monomer from wine, all of which were prepared or obtained as previously described (Atmodjo et al., 2011) (gift from Malcolm O’Neill). These reactions contained the respective pectin acceptor (10 μM) and UDP-[14C]GalA (5 μM for GAUT13 and GAUT14; 50 μM for GAUT1, GAUT10, GAUT11) in the above reaction buffer. The reaction products formed by GAUT10 (3 μM, 4.5 h), GAUT10 (3 μM, 4.5 h), GAUT11 (3 μM, 4.5 h), GAUT13 (200 nM, 90 min) and GAUT14 (200 nM, 120 min), were precipitated, unincorporated [14C]GalA removed, and the incorporated [14C]GalA detected by scintillation counting as described (Doong & Mohnen, 1998).
Synthesis and analysis of products synthesized by GAUTs including polyacrylamide gel electrophoresis (PAGE) with alcian blue and silver staining
For PAGE analysis of the GAUT reaction products, reactions (≥30 uL) were performed in reaction buffer with each GAUT (0.2 – 10 μM) for the designated amounts of time (0.5, 4, 24 h) with UDP-GalA (1 mM) and either with (for acceptor elongation activity) or without (for de novo synthesis) 10 μM DP11 HG acceptor at 30°C. All synthesis reactions analyzed by PAGE contained added potato apyrase (Sigma-Aldrich) (20 mU/μL) unless otherwise noted. To prevent loss of enzyme activity due to freeze-thaw, the apyrase was diluted to 200 mU/uL in 10 uL aliquots in 50 mM HEPES, pH 7.2 and frozen at −20°C. Fresh aliquots were used for each assay at calculated volumes to obtain the final working concentration.
Separation of the HG:GalAT reaction products formed by the GAUTs was performed by electrophoresis of the products on a high-percentage (30%) polyacrylamide gel as described (Amos et al., 2018). After the termination of each reaction by boiling, samples were centrifuged (14,000xg for 5 min) and 10 uL of the supernatant was mixed with 2 uL 6X loading dye (0.63 M Tris, pH 6.8, 0.05% phenol red, 50% glycerol). HG mix (DP 7-23) (500 ng) and 50 ng of DP11 HG were separated on the gels to serve as size standards. After separation, the gels were fixed in 40% MeOH and 10% acetic acid for 20 min. Gels were stained with freshly made 0.2% (w/v) Alcian Blue (Sigma) in 40% EtOH solution for ≥20 min and washed in ddH2O with gentle rocking until background stain was removed. Finally, the gels were stained using the silver stain kit and associated protocol (Bio-Rad) and terminated by the addition of 5% (v/v) acetic acid. Gels were imaged on a ChemiDoc XRS+ imager using the silver stain option on the ImageLab software (BioRad).
Preparation and analysis of GAUT13 de novo reaction products
GAUT13 de novo reaction products from 3 h reactions were generated in multiple 600 μL reactions containing 1.1 μM GAUT11 and 1 mM UDP-GalA in reaction buffer containing 50 mM HEPES, pH 7.2, 0.05% (w/v) BSA, and 0.125 mM MnCl2. No apyrase was present in the reactions. After the reaction, the final volume was adjusted to 1 mL with dH2O and frozen. The reaction products were fractionated using Sephadex LH-20 size exclusion resin (HxL) (GE LifeSciences) and ten fractions were collected (22 mL total) with the following volumes (mL) (6, 4, 2, 2, 1, 1, 1, 1, 2, 2). Fractions were frozen, lyophilized and resuspended in 100 μL dH2O. Fractions were analyzed by PAGE as described above (2 μL sample in 8 μL water and 2 μL dye) and the resulting gel was stained with alcian blue and silver staining. The HG products were present in fractions 2-4. MALDI-TOF analysis of fractions 2-4 revealed that fraction 2 had the highest signal with the maximum size of HG product being DP ~11/12. The products in Fraction 2, UDP-GalA (CarboSource Services), HG DP3 (HG trimer) (Sigma-Aldrich) and HG mix (DP 7-23, see above) were analyzed by 1H NMR using an 800 MHz Bruker Avance III spectrometer (Hedenström et al., 2008).
ACCESSION NUMBERS
Uniprot accession numbers for GAUTs 1-15 are provided in Table S1.
Supplementary Material
Table S1. Cloning and sequencing primers of Arabidopsis GAUTs used for expression in HEK293 cells.
Table S3. Expected molecular weights of HG oligomers with no UDP attachment.
Table S2. Outlier Analysis of Pairwise Expressed GAUTs in HEK293 Cells.
Figure S1. pGEn1-DEST and pGEn2-DEST HEK293 expression vectors used for individual and pairwise heterologous expression of the GAUTs.
Figure S2. Comparison of the sizes of HG synthesized by GAUTs 1, 10, 11, 14 and 1:7 in the presence of HG DP11 exogenous acceptor after 30 min and 24 h at various enzyme concentrations.
Figure S3. Effect of apyrase on HG:GalAT activities of GAUTs 1, 10 and 14.
Figure S4. Progress curves for GAUT1Δ167, 10Δ39, 13Δ61, 14Δ61 and 1Δ167:7Δ43 acceptor-dependent HG:GalAT activities.
SIGNIFICANCE.
Three new GAUTs (10, 13 and 14) are shown to be HG:GalATs. Comparison of products synthesized in vitro by GAUTs 1, 1:7, 10, 11, 13 and 14 under identical reaction conditions shows that all can synthesize polymeric HG. GAUTs 13 and 14 also de novo synthesize HG at rates comparable to their acceptor-dependent activities. The results support the hypothesis that different GAUTs synthesize homogalacturonan with unique structural and/or functional roles in vivo.
ACKNOWLEDGEMENTS
We thank Dr. Breanna Urbanowicz and Dr. Pradeep Prabhakar for technical support on FPLC based separation of the GAUTs, Dr. Zachary Wood for guidance on outlining the enzyme kinetic experiments, Dr. Malcolm O’Neill for technical support on the MALDI-TOF MS and providing RG-II monomer, Dr. Bryon Donohoe for helpful discussions on Golgi dimensions, and the members of the Mohnen laboratory, including Dr. Ajaya K. Biswal, Paul J. Glatz, Erica Chandler, for training and helpful research discussions. We also recognize the technical assistance provided by Graf Exum, Caleb Lavigne and Shelby Everett for assistance in cloning the GAUT constructs and generating the ExoPG. This work was supported primarily by BioEnergy Science Center (BESC) Grant DE-PS02-06ER64304 and partially by the Center for Bioenergy Innovation (CBI). The research was also partially funded by the Department of Energy–funded Center for Plant and Microbial Complex Carbohydrates Grant DE-SC0015662 (DE-FG02-93ER20097) and National Institutes of Health Grants P41GM103390 and P01GM107012. The BioEnergy Science Center and the Center for Bioenergy Innovation are United States Department of Energy Bioenergy Research Centers supported by the Office of Biological and Environmental Research in the Department of Energy’s Office of Science. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest with the contents of this article.
DATA AVAILABILITY STATEMENT
All relevant data can be found within the manuscript and its supporting materials.
REFERENCES
- Amos RA, Pattathil S, Yang JY, Atmodjo MA, Urbanowicz BR, Moremen KW, & Mohnen D (2018) A two-phase model for the non-processive biosynthesis of homogalacturonan polysaccharides by the GAUT1:GAUT7 complex. Journal of Biological Chemistry, 293, 19047–19063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos RA, & Mohnen D (2019) Critical review of plant cell wall matrix polysaccharide glycosyltransferase activities verified by heterologous protein expression. Frontiers in Plant Science, 10, 915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atmodjo MA, Sakuragi Y, Zhu X, Burrell AJ, Mohanty SS, Atwood III JA, Orlando R, Scheller HV, & Mohnen D (2011) Galacturonosyltransferase (GAUT)1 and GAUT7 are the core of a plant cell wall pectin biosynthetic homogalacturonan:galacturonosyltransferase complex. Proceedings of the National Academy of Sciences of the United States of America, 108, 20225–20230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atmodjo MA, Hao Z, & Mohnen D (2013) Evolving views of pectin biosynthesis. Annual Review of Plant Biology, 64, 747–779. [DOI] [PubMed] [Google Scholar]
- Biswal AK, Hao Z, Pattathil S, Yang X, Winkeler K, Collins C, Mohanty SS, Richardson EA, Gelineo-Albersheim I, Hunt K, Ryno D, Sykes RW, Turner GB, Ziebell A, Gjersing E, Lukowitz W, Davis MF, Decker SR, Hahn MG, & Mohnen D (2015) Downregulation of GAUT12 in Populus deltoides by RNA silencing results in reduced recalcitrance, increased growth and reduced xylan and pectin in a woody biofuel feedstock. Biotechnology for Biofuels, 8, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal AK, Atmodjo MA, Li M, Baxter HL, Yoo CG, Pu Y, Lee YC, Mazarei M, Black IM, Zhang JY, Ramanna H, Bray AL, King ZR, LaFayette PR, Pattathil S, Donohoe BS, Mohanty SS, Ryno D, Yee K, Thompson OA, Rodriguez M Jr., Dumitrache A, Natzke J, Winkeler K, Collins C, Yang X, Tan L, Sykes RW, Gjersing EL, Ziebell A, Turner GB, Decker SR, Hahn MG, Davison BH, Udvardi MK, Mielenz JR, Davis MF, Nelson RS, Parrott WA, Ragauskas AJ, Stewart CN Jr., & Mohnen D (2018a) Sugar release and growth of biofuel crops are improved by downregulation of pectin biosynthesis. Nature Biotechnology, 36, 249–257. [DOI] [PubMed] [Google Scholar]
- Biswal AK, Atmodjo MA, Pattathil S, Amos RA, Yang X, Winkeler K, Collins C, Mohanty SS, Ryno D, Tan L, Gelineo-Albersheim I, Hunt K, Sykes RW, Turner GB, Ziebell A, Davis MF, Decker SR, Hahn MG, & Mohnen D (2018b) Working towards recalcitrance mechanisms: increased xylan and homogalacturonan production by overexpression of GAlactUronosylTransferase12 (GAUT12) causes increased recalcitrance and decreased growth in Populus. Biotechnology for Biofuels, 11, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouton S, Leboeuf E, Mouille G, Leydecker M-T, Talbotec J, Granier F, Lahaye M, Höfte H, & Truong HN (2002) QUASIMODO1 encodes a putative membrane-bound glycosyltransferase required for normal pectin synthesis and cell adhesion in Arabidopsis. The Plant Cell, 14, 2577–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffall KH, Pattathil S, Phillips SE, Hahn MG, & Mohnen D (2009) Arabidopsis thaliana T-DNA mutants implicate GAUT genes in the biosynthesis of pectin and xylan in cell walls and seed testa. Molecular Plant, 2, 1000–1014. [DOI] [PubMed] [Google Scholar]
- Chavaroche AA, van den Broek LA, Springer J, Boeriu C, & Eggink G (2011) Analysis of the polymerization initiation and activity of Pasteurella multocida heparosan synthase PmHS2, an enzyme with glycosyltransferase and UDP-sugar hydrolase activity. Journal of Biological Chemistry, 286, 1777–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coenen GJ, Bakx EJ, Verhoef RP, Schols HA, & Voragen AGJ (2007) Identification of the connecting linkage between homo- or xylogalacturonan and rhamnogalacturonan type I. Carbohydrate Polymers, 70, 224–235. [Google Scholar]
- DeAngelis PL, & White CL (2002) Identification and molecular cloning of a heparosan synthase from Pasteurella multocida type D. Journal of Biological Chemistry, 277, 7209–7213. [DOI] [PubMed] [Google Scholar]
- Doong RL, & Mohnen D (1998) Solubilization and characterization of a galacturonosyltransferase that synthesizes the pectic polysaccharide homogalacturonan. The Plant Journal, 13, 363–374. [Google Scholar]
- Ferrari S, Savatin DV, Sicilia F, Gramegna G, Cervone F, & Lorenzo GD (2013) Oligogalacturonides: plant damage-associated molecular patterns and regulators of growth and development. Frontiers in Plant Science, 4, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiebig T, Berti F, Freiberger F, Pinto V, Claus H, Romano MR, Proietti D, Brogioni B, Stummeyer K, Berger M, Vogel U, Costantino P, & Gerardy-Schahn R (2014a) Functional expression of the capsule polymerase of Neisseria meningitidis serogroup X: a new perspective for vaccine development. Glycobiology, 24, 150–158. [DOI] [PubMed] [Google Scholar]
- Fiebig T, Freiberger F, Pinto V, Romano MR, Black A, Litschko C, Bethe A, Yashunsky D, Adamo R, Nikolaev A, Berti F, & Gerardy-Schhn R (2014b) Molecular cloning and functional characterization of components of the capsule biosynthesis complex of Neisseria meningitidis serogroup A: toward in vitro vaccine production. Journal of Biological Chemistry, 289, 19395–19407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiebig T, Litschko C, Freiberger F, Bethe A, Berger M, & Gerardy-Schahn R (2018) Efficient solid-phase synthesis of meningococcal capsular oligosaccharides enables simple and fast chemoenzymatic vaccine production. Journal of Biological Chemistry, 293, 953–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francocci F, Bastianelli E, Lionetti V, Ferrari S, De Lorenzo G, Bellincampi D, & Cervone F (2013) Analysis of pectin mutants and natural accessions of Arabidopsis highlights the impact of de-methyl-esterified homogalacturonan on tissue saccharification. Biotechnology for Biofuels, 6, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubbs FE (1969) Procedures for detecting outlying observations in samples. Technometrics, 11, 1–21. [Google Scholar]
- Haas KT, Wightman R, Meyerowitz EM, & Peaucelle A (2020) Pectin homogalacturonan nanofilament expansion drives morphogenesis in plant epidermal cells. Science, 367, 1003–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Z, Avci U, Tan L, Zhu X, Glushka J, Pattathil S, Eberhard S, Sholes T, Rothstein GE, Lukowitz W, Orlando R, Hahn MG, & Mohnen D (2014) Loss of Arabidopsis GAUT12/IRX8 causes anther indehiscence and leads to reduced G lignin associated with altered matrix polysaccharide deposition. Frontiers in Plant Science, 5, 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedenström M, Wiklund S, Sundberg B, & Edlund U (2008) Visualization and interpretation of OPLS models based on 2D NMR data. Chemometrics and Intelligent Laboratory Systems, 92, 110–117. [Google Scholar]
- Illingworth B, Brown DH, & Cori CF (1961) The de novo synthesis of polysaccharide by phosphorylase. Proceedings of the National Academy of Sciences of the United States of America, 47, 469–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii T, & Matsunaga T (2001) Pectin polysaccharide rhamnogalacturonan II is covalently linked to homogalacturonan. Phytochemistry, 57, 969–974. [DOI] [PubMed] [Google Scholar]
- Ishii T (2002) A sensitive and rapid bioassay of homogalacturonan synthase using 2-aminobenzamide-labeled oligogalacturonides. Plant and Cell Physiology, 43, 1386–1389. [DOI] [PubMed] [Google Scholar]
- Kang B-H & Staehelin A (2008) ER-to-Golgi transport by COPII vesicles in Arabidopsis involves a ribosome-excluding scafflod that is transferred with the vesicles to the Golgi matrix. Protoplasma, 234, 51–64. [DOI] [PubMed] [Google Scholar]
- Klepikova AV, Logacheva MD, Dmitriev SE, & Penin AA (2015) RNA-seq analysis of an apical meristem time series reveals a critical point in Arabidopsis thaliana flower initiation. BMC Genomics, 16, 466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levengood MR, Splain RA, & Kiessling LL (2011) Monitoring processivity and length control of a carbohydrate polymerase. Journal of American Chemical Society, 133, 12758–12766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljebjelke K, Adolphson R, Baker K, Doong RL, & Mohnen D (1995) Enzymatic synthesis and purification of uridine diphosphate [14C] galacturonic acid: a substrate for pectin biosynthesis. Analytical Biochemistry, 225, 296–304. [DOI] [PubMed] [Google Scholar]
- Lund CH, Stenbæk A, Atmodjo MA, Rasmussen RE, Moller IE, Erstad SM, Biswal AK, Mohnen D, Mravec J, & Sakuragi Y (2020) Pectin synthesis and pollen tube growth in arabidopsis involves three GAUT Golgi-anchoring proteins: GAUT5, GAUT6, and GAUT7. Frontiers in Plant Science, 11, 585774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil M, Darvill A, & Albersheim P (1980) Structure of plant cell walls. X. Rhamnogalacturonan I, a structurally complex pectic polysaccharide in the walls of suspension-cultured sycamore cells. Plant Physiology, 66, 1128–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohnen D (2008) Pectin structure and biosynthesis. Current Opinion in Plant Biology, 11, 266–277. [DOI] [PubMed] [Google Scholar]
- Moremen KW, Ramiah A, Stuart M, Steel J, Meng L, Forouhar F, Moniz HA, Gahlay G, Gao Z, Chapla D, Wang S, Yang JY, Prabhakar PK, Johnson R, Rosa MD, Geisler C, Nairn AV, Seetharaman J, Wu SC, Tong L, Gilbert HJ, LaBaer J, & Jarvis DL (2018) Expression system for structural and functional studies of human glycosylation enzymes. Nature Chemical Biology, 14, 156–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukerjea R, & Robyt JF (2013) Tests for the mechanism of starch biosynthesis: de novo synthesis or an amylogenin primer synthesis. Carbohydrate Research, 372, 55–59. [DOI] [PubMed] [Google Scholar]
- Mutter M, Renard CMGC, Beldman G, Schols HA, & Voragen AG (1998) Mode of action of RG-hydrolase and RG-lyase toward rhamnogalacturonan oligomers. Characterization of degradation products using RG-rhamnohydrolase and RG-galacturonohydrolase. Carbohydrate Research, 311, 155–164. [DOI] [PubMed] [Google Scholar]
- Nakamura A, Furuta H, Maeda H, Takao T, & Nagamatsu Y (2002) Structural studies by stepwise enzymatic degradation of the main backbone of soybean soluble polysaccharides consisting of galacturonan and rhamnogalacturonan. Bioscience, Biotechnology, and Biochemistry, 66, 1301–1313. [DOI] [PubMed] [Google Scholar]
- Ndeh D, Rogowski A, Cartmell A, Luis AS, Baslé A, Gray J, Venditto I, Briggs J, Zhang X, Labourel A, Terrapon N, Buffetto F, Nepogodiev S, Xiao Y, Field RA, Zhu Y, O'Neil MA, Urbanowicz BR, York WS, Davies GJ, Abbott DW, Ralet MC, Martens EC, Henrissat B, & Gilbert HJ (2017) Complex pectin metabolism by gut bacteria reveals novel catalytic functions. Nature, 544, 65–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill MA, Ishii T, Albersheim P, & Darvill AG (2004) Rhamnogalacturonan II: structure and function of a borate cross-linked cell wall pectic polysaccharide. Annual Review of Plant Biology, 55, 109–139. [DOI] [PubMed] [Google Scholar]
- Orfila C, Sørensen SO, Harholt J, Geshi N, Crombie H, Truong HN, Reid JS, Knox JP, & Scheller HV (2005) QUASIMODO1 is expressed in vascular tissue of Arabidopsis thaliana inflorescence stems, and affects homogalacturonan and xylan biosynthesis. Planta, 222, 613–622. [DOI] [PubMed] [Google Scholar]
- Peaucelle A, Braybrook S, & Höfte H (2012) Cell wall mechanics and growth control in plants: the role of pectins revisited. Frontiers in Plant Science, 3, 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peaucelle A, Wightman R, & Höfte H (2015) The control of growth symmetry breaking in the Arabidopsis hypocotyl. Current Biology, 25, 1746–1752. [DOI] [PubMed] [Google Scholar]
- Persson S, Caffall KH, Freshour G, Hilley MT, Bauer S, Poindexter P, Hahn MG, Mohnen D, & Somerville C (2007) The Arabidopsis irregular xylem8 mutant is deficient in glucuronoxylan and homogalacturonan, which are essential for secondary cell wall integrity. The Plant Cell, 19, 237–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralet MC, Crépeau M-J, Lefébvre J, Mouille G, Höfte H, & Thibault J-F (2008) Reduced number of homogalacturonan domains in pectins of an Arabidopsis mutant enhances the flexibility of the polymer. Biomacromolecules, 9, 1454–1460. [DOI] [PubMed] [Google Scholar]
- Rautengarten C, Ebert B, Moreno I, Temple H, Herter T, Link B, Doñas-Cofré D, Moreno A, Saéz-Aguayo S, Blanco F, Mortimer JC, Schultink A, Reiter WD, Dupree P, Pauly M, Heazlewood JL, Scheller HV, & Orellana A (2014) The Golgi localized bifunctional UDP-rhamnose/UDP-galactose transporter family of Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America, 111, 11563–11568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautengarten C, Birdseye D, Pattathil S, McFarlane HE, Saez-Aguayo S, Orellana A, Persson S, Hahn MG, Scheller HV, Heazlewood JL, & Ebert B (2017) The elaborate route for UDP-arabinose delivery into the Golgi of plants. Proceedings of the National Academy of Sciences of the United States of America, 114, 4261–4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renard CMGC, Lahaye M, Mutter M, Voragen FGJ, & Thibault J-F (1997) Isolation and structural characterisation of rhamnogalacturonan oligomers generated by controlled acid hydrolysis of sugar-beet pulp. Carbohydrate Research, 305, 271–280. [DOI] [PubMed] [Google Scholar]
- Round AN, Rigby NM, MacDougall AJ, & Morris VJ (2010) A new view of pectin structure revealed by acid hydrolysis and atomic force microscopy. Carbohydrate Research, 345, 487–497. [DOI] [PubMed] [Google Scholar]
- Schwacke R, Schneider A, van der Graaff E, Fischer K, Catoni E, Desimone M, Frommer WB, Flügge UI, & Kunze R (2003) ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiology, 131, 16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterling JD, Lemons JA, Forkner IF, & Mohnen D (2005) Development of a filter assay for measuring homogalacturonan: α-(1,4)-galacturonosyltransferase activity. Analytical Biochemistry, 343, 231–236. [DOI] [PubMed] [Google Scholar]
- Sterling JD, Atmodjo MA, Inwood SE, Kolli VSK, Quigley HF, Hahn MG, & Mohnen D (2006) Functional Identification of an Arabidopsis pectin biosynthetic homogalacturonan galacturonosyltransferase. Proceedings of the National Academy of Sciences of the United States of America, 103, 5236–5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Eberhard S, Pattathil S, Warder C, Glushka J, Yuan C, Hao Z, Zhu X, Avci U, Miller JS, Baldwin D, Pham C, Orlando R, Darvill A, Hahn MG, Kieliszewski MJ, & Mohnen D (2013) An Arabidopsis cell wall proteoglycan consists of pectin and arabinoxylan covalently linked to an arabinogalactan protein. The Plant Cell, 25, 270–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault J-F, Renard CMGC, Axelos MAV, Roger P, & Crépeau M-J (1993) Studies of the length of homogalacturonic regions in pectins by acid hydrolysis. Carbohydrate Research, 238, 271–286. [Google Scholar]
- Voiniciuc C, Engle KA, Günl M, Dieluweit S, Schmidt MH, Yang JY, Moremen KW, Mohnen D, & Usadel B (2018) Identification of Key Enzymes for Pectin Synthesis in Seed Mucilage. Plant Physiology, 178, 1045–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voxeur A, & Höfte H (2016) Cell wall integrity signaling in plants: "To grow or not to grow that's the question". Glycobiology, 26, 950–960. [DOI] [PubMed] [Google Scholar]
- Wang L, Wang W, Wang YQ, Liu YY, Wang JX, Zhang XQ, Ye D, & Chen LQ (2013) Arabidopsis galacturonosyltransferase (GAUT) 13 and GAUT14 have redundant functions in pollen tube growth. Molecular Plant, 6, 1131–1148. [DOI] [PubMed] [Google Scholar]
- Willats WGT, McCartney L, Mackie W, & Knox JP (2001) Pectin: cell biology and prospects for functional analysis. Plant Molecular Biology, 47, 9–27. [PubMed] [Google Scholar]
- Yang Y, & Anderson CT (2020). Biosynthesis, localisation, and function of pectins in plants. In Kontogiorgos K (Ed.), Pectin: Technological and Physiological Properties. Switzerland: Springer Nature. [Google Scholar]
- Yapo BM (2009) Pineapple and banana pectins comprise fewer homogalacturonan building blocks with a smaller degree of polymerization as compared with yellow passion fruit and lemon pectins: Implication for gelling properties. Biomacromolecules, 10, 717–721. [DOI] [PubMed] [Google Scholar]
- Yin Y, Chen H, Hahn MG, Mohnen D, & Xu Y (2010) Evolution and function of the plant cell wall synthesis-related glycosyltransferase family 8. Plant Physiology, 153, 1729–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdunek A, Pieczywek PM, & Cybulska J (2021) The primary, secondary, and structures of higher levels of pectin polysaccharides. Comprehensive Reviews In Food Science And Food Safety, 20, 1101–1117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Cloning and sequencing primers of Arabidopsis GAUTs used for expression in HEK293 cells.
Table S3. Expected molecular weights of HG oligomers with no UDP attachment.
Table S2. Outlier Analysis of Pairwise Expressed GAUTs in HEK293 Cells.
Figure S1. pGEn1-DEST and pGEn2-DEST HEK293 expression vectors used for individual and pairwise heterologous expression of the GAUTs.
Figure S2. Comparison of the sizes of HG synthesized by GAUTs 1, 10, 11, 14 and 1:7 in the presence of HG DP11 exogenous acceptor after 30 min and 24 h at various enzyme concentrations.
Figure S3. Effect of apyrase on HG:GalAT activities of GAUTs 1, 10 and 14.
Figure S4. Progress curves for GAUT1Δ167, 10Δ39, 13Δ61, 14Δ61 and 1Δ167:7Δ43 acceptor-dependent HG:GalAT activities.
Data Availability Statement
All relevant data can be found within the manuscript and its supporting materials.