Abstract
Purpose:
Loss of transforming growth factor β (TGFβ) signaling increases error-prone alternative end-joining (alt-EJ) DNA repair. We previously translated this mechanistic relationship as TGFβ and alt-EJ gene expression signatures, which are anti-correlated across cancer types. A score, βAlt, representing anti-correlation predicts patient outcome in response to genotoxic therapy. Here we sought to verify this biology in live specimens and additional datasets.
Experimental Design:
Human head and neck squamous cell (HNSC) carcinoma explants were treated in vitro to test whether the signatures report TGFβ signaling, indicated by SMAD2 phosphorylation, and unrepaired DNA damage, indicated by persistent 53BP1 foci after irradiation or olaparib. A custom NanoString assay was implemented to analyze the signatures’ expression in explants. Each signature gene was then weighted by its association with functional responses to define a modified score, βAltw, that was retested for association with response to genotoxic therapies in independent datasets.
Results:
Most genes in each signature were positively correlated with the expected biological response in tumor explants. Anticorrelation of TGFβ and alt-EJ signatures measured by Nanostring was confirmed in explants. βAltw was significantly (P<0.001) better than βAlt in predicting overall survival in response to genotoxic therapy in TCGA pancancer patients and in independent HNSC and ovarian cancer patient datasets.
Conclusion:
Association of the TGFβ and alt-EJ signatures with their biological response validates TGFβ competency as a key mediator of DNA repair that can be readily assayed by gene expression. The predictive value of βAltw supports its development to assist in clinical decision-making.
Keywords: TGFβ, DNA repair, alternative end-joining, biomarkers, carcinoma, genotoxic therapy
INTRODUCTION
Of its myriad roles, TGFβ maintenance of genomic stability is among the least studied. TGFβ regulates the expression or function of key DNA repair proteins encoded by ATM (ataxia telangiectasia mutated) (1), BRCA1 (breast cancer 1 gene) (2,3), FANCD2 (Fanconi anemia complementation group D2) (4) and LIG4 (DNA ligase 4) (5), which are necessary for maintenance of genomic integrity (reviewed in (6)). Inhibition or loss of TGFβ signaling decreases canonical DNA repair by homologous recombination (HRR) and non-homologous end-joining (NHEJ), which sensitizes cells to genotoxic treatments (1,3,5). Faulty DNA repair is a hallmark of cancer, and specific repair defects can provide the basis for response to precision therapies (7). Failure of HRR or NHEJ can increase the use of less-efficient repair by alternative end-joining (alt-EJ, also called microhomology-mediated end-joining) (8–11). Because inhibition of TGFβ signaling increases sensitivity to DNA damage by radiation or chemotherapy in preclinical models of breast, brain, lung and head and neck cancer (3,12–15), identifying TGFβ signaling defects in cancer may present a specific therapeutic opportunity (16).
Patients with head and neck squamous cell carcinoma (HNSC) associated with human papilloma virus (HPV) etiology exhibit a striking sensitivity to standard genotoxic therapy with cisplatin and radiotherapy (17). In addition to degrading p53 and RB proteins, HPV also blocks TGFβ by targeting its receptors and signal transduction (18). We have previously demonstrated that loss of TGFβ signaling competency in HPV-positive cancer is the mechanism by which canonical DNA double strand break repair shifts from HRR to alt-EJ. This repair pathway choice is recapitulated by blocking TGFβ signaling in HPV-negative cells (3). Consistent with compromised DNA repair upon loss of functional TGFβ signaling, cases of HPV-negative HNSC from The Cancer Genome Atlas (TCGA) characterized by low expression of TGFβ target genes were also associated with better overall survival (OS) following standard of care chemoradiation compared to those with high expression of TGFβ target genes (3). TGFβ regulation of genes involved in the DNA damage response identified in HNSC was also evident in glioblastoma cell lines and glioblastoma specimens in the TCGA (16).
We translated the mechanistic relationship between TGFβ and DNA repair into functional gene expression signatures consisting of TGFβ targets and genes necessary for alt-EJ (16). The TGFβ signature consists of 50 genes that were reciprocally regulated in MCF10A treated for 7 days with TGFβ or a small molecule inhibitor of the TGFβ type I receptor kinase (3,19). The 36-gene alt-EJ competency signature was curated from the literature and a screen of DNA damage repair gene using the EJ2GFP reporter (16,20,21). To classify patients according to the relationship between their TGFβ and alt-EJ transcriptional profiles, we defined a score, βAlt, to convey the relative expression of these signatures in each tumor. A high βAlt score, indicative of high alt-EJ and low TGFβ gene expression, correlates with specific mutational signatures, genomic instability characteristics, and better patient outcome in response to genotoxic treatment (16).
Given that both gene signatures were derived from in vitro studies of human cell lines, the non-transformed breast cell line MCF10A in the case of TGFβ target genes (19) and U2OS osteosarcoma cells for alt-EJ components (22), here we sought to validate that each signature reflects its respective biology in cancer specimens. We designed a custom NanoString panel to test each gene’s functional association with biological response, which also facilitated application to retrospective analysis of archival specimens. Through functional assays in HNSC tumor explants, targeted gene expression analyses, and integrative data modeling we demonstrate the biological coherence of the signatures, and subsequently redefined an optimized score, termed βAltw, that was validated in independent datasets. The results confirm that TGFβ regulation of DNA damage repair is an important biomarker of outcome after genotoxic treatment with RT or platinum chemotherapy. The developed tools and methods can be implemented to predict patient response.
METHODS
Explant cultures
HNSC tumor tissue and ovarian cancer specimen collection from patients who consented in writing under the US Common rule and was reviewed and approved by the University of California, San Francisco (UCSF) Institutional Review Board. Establishment of HNSC PDX and tumor collection were reviewed and approved by the UCSF Institutional Animal Care and Use Committee. Explants were established on floating rafts from HNSC PDX from immunocompromised mice (n=15) and patient primary tumors (n=22) collected during surgery as described previously (3). Characteristics of patients donating primary tissues are shown in Supplemental Table S1. Specimens were kept in DMEM and transported on ice. Tissues were dissected and one portion was frozen in liquid nitrogen for RNA extraction, and the remainder was portioned into approximately 1 mm3 fragments for explant cultures. Some explants were irradiated with 5 Gy at 48 hr; others were treated at 24 hr with 10 μm olaparib for 24 hours. All were harvested 5 hours after treatment. Specimens were embedded in OCT (Sigma Aldrich) and frozen on ethanol/dry ice bath. Blocks were kept at −80°C before cryosectioning.
Immunofluorescence and microscopy
Staining and visualization of 53BP1 DNA damage foci, indicative of unrepaired DNA damage, was assessed in treated explants and pSMAD2 was assayed in untreated explants as previously described (3). In brief, cryosections were fixed with 4% paraformaldehyde for 15 minutes and then permeabilized with 0.5% TritonX-100 followed by blocking with the supernatant of 0.5% casein stirred in PBS. Sections were incubated with antibodies to 53BP1 (Bethyl CAT#A700-011, 1:500) or SMAD2 phosphorylated on serine 465/467 at (Cell Signaling CAT#3108, 1:200) at 4°C overnight in a humidified chamber. After three rinses with PBS, sections were incubated for 1 hour with secondary donkey anti-rabbit IgG (Alexa Fluor 488/555, Invitrogen) or donkey anti-mouse IgG (Alexa Fluor 488/555, Invitrogen). Cell nuclei were counterstained with 4’,6-diamidino-2-phenylindole, dihydrochloride (DAPI). Slides were mounted in Vectashield (Sigma). A 40X objective with 0.95 numerical aperture was used on a Zeiss Axiovert equipped with epifluorescence. In-home developed macros in the opensource platform Fiji-ImageJ (NIH, Bethesda, MA) were used for image analyses of 8-bit images for each channel of fluorescence. The DAPI channel was used to generate the region of interest and 5 or more images per sample were randomly taken based on nuclear dye alone. At least 100 cells were analyzed for each sample. For analyses of radiation induced foci, spontaneous foci from sham-treated controls were subtracted unless otherwise noted.
NanoString assay
A custom NanoString panel was used consisting of 50 genes induced by chronic TGFβ, 36 genes necessary for execution of alt-EJ (16), and 12 housekeeping genes. Total RNA was extracted from samples using the TRIzol reagent (Invitrogen) and the miRNAeasy Mini Kit (Qiagen) for frozen HNSC or ovarian cancer samples. Formalin-fixed paraffin embedded ovarian cancer specimens were reviewed and approved by the Institutional Review Board at L’Hospitalet del Llobregat under the Declaration of Helsinki. RNA was prepared from Formalin-fixed paraffin embedded ovarian cancer sections using the RNeasy FFPE Kit (Qiagen) according to manufacturer’s instructions. 250 ng of total RNA from each sample were hybridized following the manufacturer’s protocol (NanoString Technologies, Seattle, Washington, USA). Gene expression was quantified using the standard nCounter methodology with multiplexed color-coded probe pairs (23). The raw expression data was processed and normalized using the nSolver software (NanoString). Normalized counts for HNSC and ovarian specimens were separately log2 transformed and mean-centered per gene by converting into z-scores.
Bioinformatic analyses
Gene expression heatmaps were constructed with unsupervised hierarchical clustering using the R package ComplexHeatmap (24). Euclidean distance was used as the similarity metric and the Ward.D2 method as the between-cluster distance metric. The gene correlation matrix was created by computing the Pearson correlation coefficient (rp) between the expression of every pair of TGFβ and alt-EJ signature genes, using the R package corrplot (https://www.rdocumentation.org/packages/corrplot). Genes were displayed based on the weight of their contribution to the first principal component of the gene expression profiles.
Weighted gene co-expression networks were built using the R package ggraph (https://ggraph.data-imaginist.com). Each gene was represented as a node and was colored by its signature. An edge between two genes corresponded to the expression correlation (rp); and rp > 0.007 was used as a cutoff for edge generation. Layout force-directed algorithm from Fruchterman-Reingold was applied for network construction. To identify central genes in the network, the weighted centrality degree of each gene was calculated considering the number and weight of the edges connecting to any other gene of the same signature.
The βAltw score
To compute βAltw, signature genes were weighted using biological data generated from the HNSC explants based on: (1) a centrality degree within the corresponding signature calculated based on the gene co-expression network analysis after rescaling it into a 0-0.5 range; (2) the strength of the positive correlation (Spearman correlation coefficient, rs) between expression and the corresponding biologic measurement (frequency of cells with pSMAD2-positive nuclei for the genes from the TGFβ signature or cells with 5 or more 53BP1 foci indicative of unrepaired DNA damage for the genes from the alt-EJ signature); and (3) the strength of the negative correlation (rs multiplied by −1) between expression and the other biologic measurement (53BP1 for the genes from the TGFβ signature and pSMAD2 for the genes from the alt-EJ signature).
The weight of each TGFβ gene (Fig. S4) was calculated as follows:
Likewise, the weight of each alt-EJ gene (Fig. S4) was calculated as:
Next, a factor (Fig. S4) was assigned to each gene based on its weight with the following formula:
Taking this into account, the TGFβ and alt-EJ weighted expression scores were calculated for each tumor as the sum of the expression of the genes from each signature multiplied by their factors:
The βAltw score conveys in one value the relative expression of both signatures in each tumor and is computed as follows:
Datasets
Gene expression data of the TCGA-pancancer cohort was downloaded from the Genomic Data Commons portal from the file EBPlusPlusAdjustPANCAN_IlluminaHiSeq_RNASeqV2.geneExp.tsv. The downloaded gene expression values were trimmed mean of M values normalized, log2 transformed and mean-centered per gene by converting them into z-scores. Primary solid tumor samples were analyzed. Glioblastoma samples categorized as ‘neural’ were excluded (25), as were mislabeled pancreatic cancer samples (26). Overall survival, tumor stage, and age from all TCGA patients whose standard of care would include genotoxic RT and/or chemotherapy based on their cancer type and stage (n=4597) were obtained from the dataset “TCGA-CDR-SupplementalTableS1.xlsx” from Genomic Data Commons in November 2020 (16).
Gene expression of the TCGA-HNSC cohort was downloaded from the dataset “TCGA.HNSC.sampleMap/HiSeqV2” of the University of California Santa Cruz Xena platform using the R package UCSCXenaTools in November 2020, excluding those patients whose primary curative treatment had been surgery so that the remaining (n=419) were likely to have received genotoxic treatment with RT and/or chemotherapy. Gene expression had been measured by RNA-seq with the platform IlluminaHiSeq_RNASeqV2 and values had been RSEM normalized and log2(x+1) transformed. The downloaded gene expression values were mean-centered per gene by converting them into z-scores for primary tumor samples. Overall survival, tumor stage, and age from TCGA-HNSC patients were obtained from the dataset “TCGA-CDR-SupplementalTableS1.xlsx” from Genomic Data Commons in November 2020. Information about HPV status from the TCGA-HNSC tumors was downloaded from cBioPortal in November 2020 from the project “Head and Neck Squamous Cell Carcinoma (TCGA, PanCancer Atlas)”. TCGA-HNSC patient treatment information was downloaded from the University of California Santa Cruz Xena platform using the R package UCSCXenaTools (27).
The GSE41613 HNSC dataset (n=97) and the GSE26712 ovarian cancer dataset (n=185) were downloaded from the Gene Expression Omnibus in March and April 2021, respectively, using the R package GEOquery. In GSE41613, gene expression had been measured with the platform GPL570 and normalized into log2 gcRMA signal. In GSE26712, gene expression had been measured with the platform GPL96 and normalized into RMA signal value. For genes with multiple probes, the average expression of the probes was calculated. The downloaded gene expression values were mean-centered per gene by converting them into z-scores.
Gene expression data from the NCI-60 cell lines (n=60) was downloaded from cBioPortal in April 2021 and the surviving fraction after 2 Gy (SF2) of the cell lines was obtained from the literature based on reported clonogenic assays (28).
Statistical Analyses
Kaplan-Meier survival curves, as defined in TCGA (29), GEO GSE26712 (30), and GSE41613 (31), were generated for βAltw score tertile 1 versus 3 via R package ‘survminer’ and the multivariable Cox regressions were performed using the R package ‘survival’ as indicated. Adjusted hazard ratios (HR) with corresponding 95% confidence interval (CI) were reported. The Cox proportional hazards assumption was assessed graphically with the ‘coxph’ function via the Schoenfeld residuals, and there was no evidence of PH violation. All statistical comparisons were two-sided and considered as statistically significant at P < 0.05.
To compare the performance of the βAltw score and the original βAlt in terms of predicting OS after genotoxic treatment, we followed a similar methodology as described (32). The TCGA-pancancer dataset of patients whose standard of care would include genotoxic RT and/or chemotherapy (n=4597) was randomly split into 500 surrogate datasets using the bootstrapping resampling method with the R package ‘boot’ to calculate the original βAlt score and the βAltw score. The OS HR calculated according to each score for the top and bottom tertile. The resulting HR obtained with the βAltw score and the original βAlt were compared using a paired T-test.
Code Availability
Data Availability
All data in the article that are not from open-access datasets are available from the corresponding author upon request.
RESULTS
TGFβ and alt-EJ signatures associate with biological readouts of TGFβ and DNA repair
Gene expression of TGFβ and alt-EJ signatures was assessed by a targeted NanoString custom panel in 15 HNSC patient-derived xenografts (PDX) and 22 HNSC patients’ primary tumors from which we then established explants (Fig. 1A). Unsupervised hierarchical clustering of the HNSC tumors and their transcriptomic phenotype clustered genes from both signatures into two major clades characterized by low TGFβ and high alt-EJ or high TGFβ and low alt-EJ gene expression (Fig. 1B), reproducing the anti-correlation we reported using RNAseq data from TCGA (16). As reported therein, most HPV-positive samples gathered in the dendrogram arm with low TGFβ and high alt-EJ.
Figure 1. Evaluation of TGFβ and alt-EJ gene expression signatures in HNSC tumors.

(A) Schematic illustration of 15 PDX and 22 primary HNSC tumors collected for NanoString or immunostaining assays. (B) Unsupervised hierarchical clustering based on the expression (high, yellow, low, blue) of TGFβ (pink) and alt-EJ (green) signature genes using the NanoString custom platform and RNA extracted from the 37 HNSC samples HPV positive (purple), negative or not determined (grey) are indicated with the tissue origin from PDX (red) or primary patient specimens (light blue) at the bottom.
NanoString assays are well-suited to retrospective studies (33). To test how the panel reports on the TGFβ and alt-EJ pathways in archival specimens, RNA was extracted from 18 snap-frozen and 22 formalin-fixed paraffin tissue sections of ovarian cancer. Unsupervised hierarchical clustering of the NanoString signatures revealed two clades characterized by low TGFβ and high alt-EJ or high TGFβ and low alt-EJ genes expression (Fig. S1A). Duplicates (indicated by red bars) were adjacent, demonstrating the reproducibility of gene expression measured by NanoString.
We next sought to validate the biological significance of the TGFβ signature by assessing pathway activity by measuring phosphorylated SMAD2 (pSMAD2, Fig. 2A). DNA repair proficiency was assessed 5 hours after irradiation with 5 Gy by quantifying persistent 53BP1 foci (34) (Fig. 2B). The mean expression of the TGFβ signature genes was significantly correlated with the percentage of pSMAD2 positive cells (rs = 0.45, P = 0.0067), supporting that the signature is indicative of TGFβ signaling competency (Fig. 2C). Likewise, the mean expression of the alt-EJ signature was positively correlated with the unrepaired DNA damage (rs = 0.51, P = 0.03), supporting the alt-EJ signature as one of less efficient DNA repair (Fig. 2D).
Figure 2. TGFβ signaling status associates with DNA repair proficiency.

(A) Percentage of pSMAD2 positive cells in HNSC explants (n=35). Representative images of a pSMAD2-low sample (*) and a pSMAD2-high sample (+). (B) Percentage of HNSC cells with 5 or more 53BP1 foci at 5 hours after 5 Gy irradiation (n=19). Representative images of a HNSC sample with few cells with residual 53BP1 foci (*) and a sample with high residual 53BP1 foci (+). (C) Correlation of percent nuclear pSMAD2 positive cells with the expression of TGFβ signature genes calculated from NanoString data in the same sample (n=18). (D) Percentage of 53BP1 foci positive cells after irradiation correlated with the expression of the alt-EJ signature genes from the same HNSC sample. (E) Anti-correlation of percent nuclear pSMAD2 positive cells with percent 53BP1 positive cells after irradiation in the same panel of HNSC explants. (F) Anti-correlation of TGFβ signature with alt-EJ signature genes’ expression in the same panel of HNSC explants. (G) Percentage of 53BP1 foci positive cells are positively correlated with alt-EJ score from olaparib treated HNSC explants (n=19). (H) Anti-correlation of TGFβ signature score with percentage of 53BP1 foci positive cells from olaparib treated HNSC explants. Spearman correlations were performed for r and P values. Trend lines (blue) from linear regression are shown. Purple bars or dots are HPV-positive samples.
The percentage of pSMAD2 and 53BP1 positive cells were negatively correlated across all specimens (Fig. 2E, rs = −0.5, P = 0.037), confirming the correlation between robust TGFβ signaling and competent DNA damage repair (3). Consistent with our prior study, HPV-positive specimens had fewer pSMAD2 positive cells and more cells with 53BP1 foci indicative of unrepaired DNA damage and TGFβ and alt-EJ signatures were negatively correlated (rs = −0.41, P = 0.013; Fig. 2F).
Olaparib, an FDA approved poly-adenosine diphosphate-ribose polymerase (PARP) inhibitor, can cause DSB in cells that are HRR deficient. Our prior work indicated that TGFβ loss or inhibition decreases HRR and increases sensitivity to PARP inhibition (3). To further test this relationship, we treated explants with olaparib for 24 hours and examined 53BP1 foci after 5 hours of recovery. As for irradiated explants, unrepaired DNA damage marked by 53BP1 foci in olaparib treated explants was positively correlated with alt-EJ signature expression (rs = 0.59, P = 0.01; Fig. 2G). 53BP1 foci were anti-correlated with mean expression of the TGFβ signature (rs = −0.61, P = 0.005) and the percent pSMAD2 positive cells (rs = −0.65, P = 0.004; Fig. 2H). Thus, tumors with low TGFβ signaling indicated by pSMAD2 expression or TGFβ signature expression have more unrepaired damage following PARP inhibition.
To evaluate the relevance of each TGFβ and alt-EJ gene, we next assessed the expression of each gene per sample as a function of the percentage of pSMAD2 and 53BP1 positive cells (Fig. 3A,B). Most TGFβ genes were positively correlated with the frequency of pSMAD2 positive cells (37/50) and negatively correlated with 53BP1 positive cells (36/50), whereas most alt-EJ genes were positively correlated with residual DNA damage marked by 53BP1 (30/36) and negatively correlated with pSMAD2 (31/36).
Figure 3. Association of TGFβ and alt-EJ genes with functional biological readouts and gene-to-gene correlations.
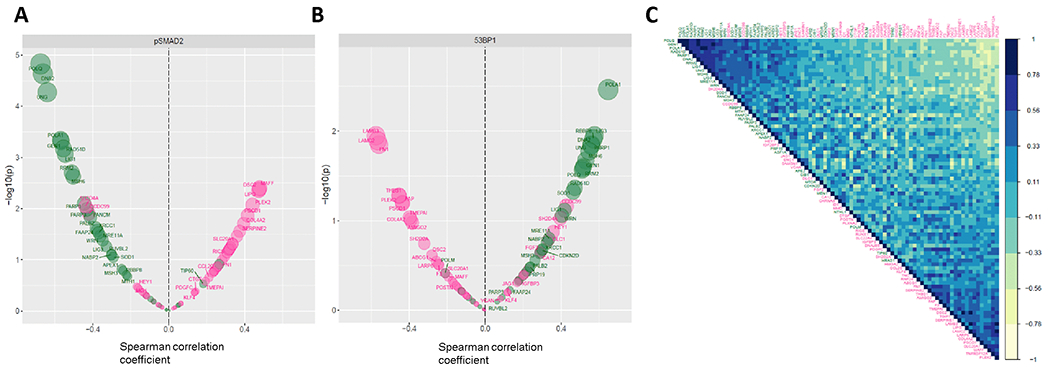
Volcano plots showing the statistical significance (Y axis, −log10 P value) of the correlation (X axis, rs) between the expression of the individual TGFβ (pink) and alt-EJ (green) signature genes and the percentage of pSMAD2 positive cells (A) or the percentage of cells with 53BP1 foci (B) in the panel of HNSC tumor explants. (C) Gene correlation matrix showing the rp between the expression of each pair of TGFβ (pink) and alt-EJ (green) signature genes in the HNSC tumor explants (yellow = negative rp; blue = positive rp). Genes are displayed in first principal component order.
Figure 4. Refinement of the TGFβ and alt-EJ signatures by weighting the functional relevance of each gene.
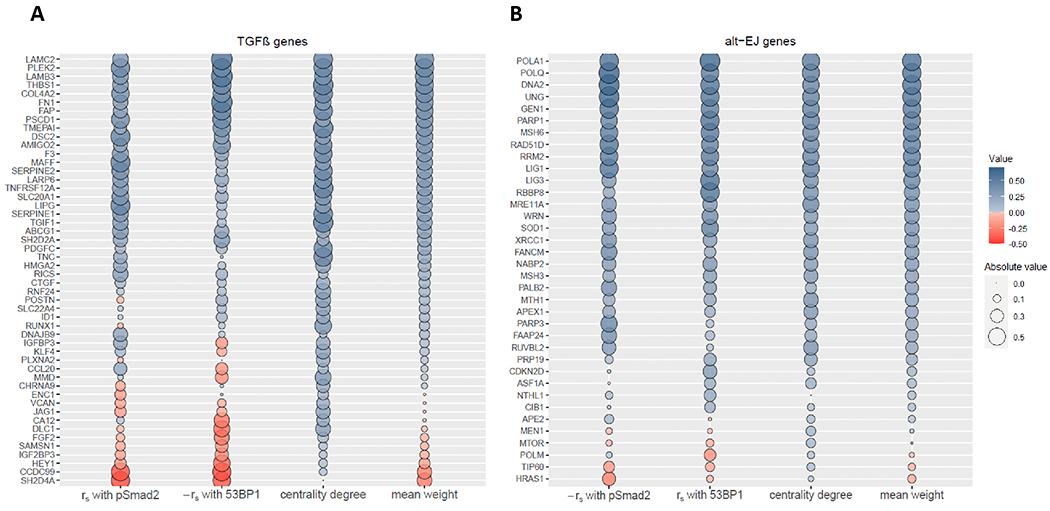
Relative weight of the TGFβ signature genes (A) and the alt-EJ signature genes (B) according to the strength of their association with the frequency of pSMAD2 positive cells (column 1, rs for TGFβ genes and −rs for alt-EJ genes), the strength of their association with the frequency of 53BP1 positive cells (column 2, −rs for TGFβ genes and rs for alt-EJ genes), and their centrality degree (column 3) in the HNSC tumor explants. The fourth column represents the mean of the other three columns. The dot size indicates the absolute value, and the color indicates its direction (blue, positive; red, negative).
Next, the Pearson correlation coefficient (rp) was computed between expression values of each pair of genes to construct a correlation matrix (Fig. 3C). Endorsing the premise that each signature embodies a distinct functional pathway, most genes from the same signature were highly correlated. Conversely, gene pairs between the signatures were found to be commonly anti-correlated, consistent with TGFβ suppression of alt-EJ genes (16). We then constructed TGFβ and alt-EJ gene correlation networks in the HNSC specimens to identify “hub” genes (i.e., genes with a relatively high number of positive correlations, expected to be functionally more relevant within the signature) and calculated the weighted centrality degree of each component (Fig. S2A,B). Similar networks were computed from the NanoString analysis of the ovarian cancer specimens (Fig. S2C,D). Notably, functional ranks of the genes based on their association with pSMAD2 and 53BP1 cellular readouts and network centrality were found to be similar (Fig. 4).
Association between gene ranks and clinical outcomes
The previously defined βAlt score was calculated on single-sample gene set enrichment analysis (ssGSEA) of the TGFβ and alt-EJ signatures that reports their anti-correlation and is associated with response to genotoxic cancer therapy (16). A high βAlt score represented specimens in which the expression of TGFβ target genes was low and expression of alt-EJ genes was high, whereas a low βAlt score indicated the opposite. To test whether the above evidence of differences in the strength of association between biology and specific genes in each signature improved the translational relevance of the signatures, we calculated βAlt score using only the top-ranked 15 genes or the bottom-ranked 15 genes from each signature for HNSC patients from TCGA, excluding those whose primary curative treatment had been surgery (n=419). Survival was significantly associated with a βAlt score calculated using only the top 15 genes (P = 0.0093, HR = 0.62, 95% CI 0.43-0.89; Fig. S3A), whereas one calculated using the bottom 15 genes was not (P = 0.49, HR = 0.89, 95% CI 0.63-1.24; Fig. S3B). This suggests that the genes with the greatest weight based on the biology and centrality exhibited by the HNSC tumor explants are also the most clinically relevant.
Given that 11 TGFβ signature genes and 4 alt-EJ signature genes were negatively weighted (Fig. 4), we examined whether excluding these genes would increase βAlt prognostic power compared to the original score. Notably, both the original (P = 0.00099, hazard ratio = 0.53, 95% CI 0.37-0.78; Fig. S3C) and shortened βAlt scores (P = 0.0036, hazard ratio = 0.59, 95% CI 0.41-0.84; Fig. S3D) were significantly associated with HNSC patient OS (P = 0.0009, HR = 0.53, 95% CI 0.37-0.78 versus P = 0.0036, HR = 0.59, 95% CI 0.41-0.84). Hence, we concluded that removing low weighted genes does not improve βAlt prognostic capacity.
Development of the βAltw score and validation of its predictive value
We next developed a model, termed βAltw, based on the sum of gene expression levels weighted by their estimated functional relevance (Methods). To compare the performance of the βAltw score to the original βAlt score we used pancancer patients in TCGA whose standard of care would include radiotherapy and/or genotoxic chemotherapy based on their cancer type and stage (n=4597) (16) and the bootstrap resampling method to split the dataset into 500 surrogate datasets (35). Patients in the top tertile had significantly longer OS compared to those in the bottom tertile using either the original βAlt score (Fig. 5A, B) or the βAltw score (Fig. 5C, D). The βAltw score (HR = 0.61, 95% CI 0.54-0.68) between the resampled patient tertile were significantly lower (P <0.0001) than those calculated using the original βAlt score (HR = 0.64, 95% CI 0.57-0.73), indicating superiority of the βAltw score as a biomarker of prognosis in response to genotoxic treatment.
Figure 5. Comparison of βAlt and βAltw as predictors of pancancer OS after genotoxic treatment.
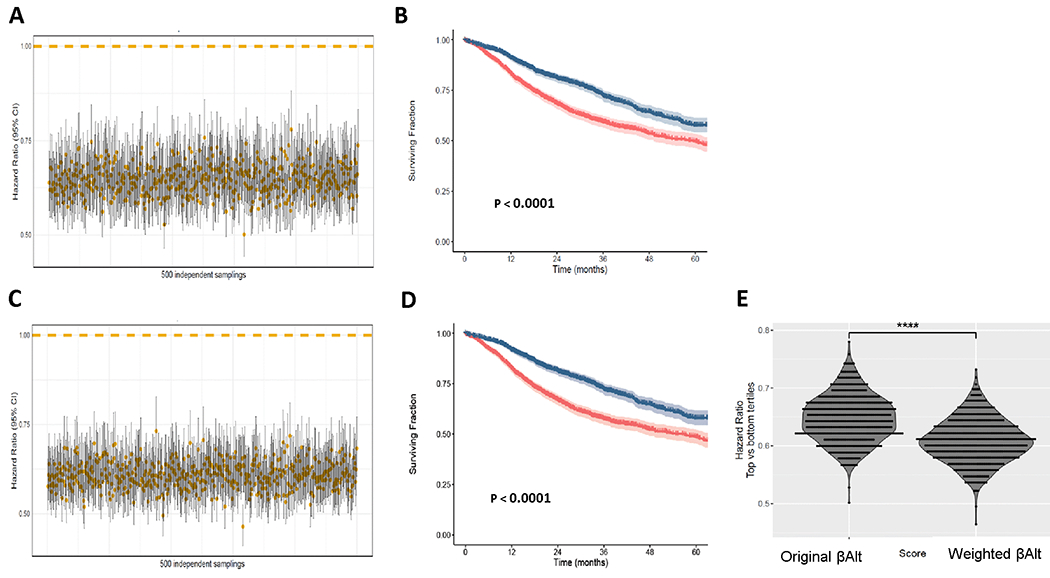
Cox regression analyses of 500 bootstrapping sets generated from the TCGA-pancancer dataset. (A) HR and 95% CI of the top vs bottom βAlt tertile of patients calculated for each sample set using a Cox model. (B) Kaplan-Meier overall survival curves of the top (blue) vs bottom (red) βAlt tertile in the TCGA-pancancer dataset. P values were calculated with log-rank tests here and in C. (C) HR and 95% CI of the top vs bottom βAltw tertile of patients calculated using a Cox model. (D) Kaplan-Meier overall survival curves of the top (blue) vs bottom (red) βAltw tertile in the TCGA-pancancer dataset. (E) Comparison of HR for βAlt and βAltw for the 500 test sets (paired T-test, **** P < 0. 0001).
The βAltw score is mechanistically based on TGFβ control of the response to DNA damage (16). To confirm this, we evaluated βAltw prediction of the radiosensitivity of NCI-60 pancancer cell lines (n=60). As expected, the weighted TGFβ and alt-EJ signatures were significantly anticorrelated (rs= −0.51, P < 0.0001; Fig. S5A). Radiation sensitivity, measured as the surviving fraction after exposure to 2 Gy (SF2), was significantly correlated with βAltw (rs = −0.36, P = 0.0046; Fig. S5B), which supports the functional validity of βAltw.
To further test the predictive power of βAltw we reanalyzed TCGA HNSC patients (n=419). As shown for the original βAlt (Fig. S3A), patients with a high βAltw score had significantly better OS compared to those with a low βAltw (P = 0.019, HR = 0.65, 95% CI 0.45-0.94; Fig. 6A). Although in a multivariable Cox regression adjusted for HPV status, age and stage the association of the βAltw score with OS compared as a function of tertile was not significant (P = 0.1099, HR = 0.697, 95% CI 0.45-1.08; table S2), it was significant as a continuous variable (P = 0.0250, HR = 0.995, 95% CI 0.99-0.99). To eliminate the potential impact of HPV or tumor location as confounding variables, we analyzed HPV negative oral squamous carcinoma patients from the GSE41613 dataset (n=97). Patients from the high βAltw tertile had significantly better cancer-specific survival than those from the low βAltw tertile (P = 0.015, HR = 0.30, 95% CI 0.11-0.84; Fig. 6B). Multivariable Cox regression analysis including age and stage maintained βAltw statistical significance (P = 0.0393, HR = 0.33, 95% CI 0.11-0.95; Supplemental table S2). These analyses support the predictive capacity of the βAltw score in HNSC patients.
Figure 6. The βAltw score predicts clinical outcomes after genotoxic therapy in independent HNSC and ovarian cancer datasets.
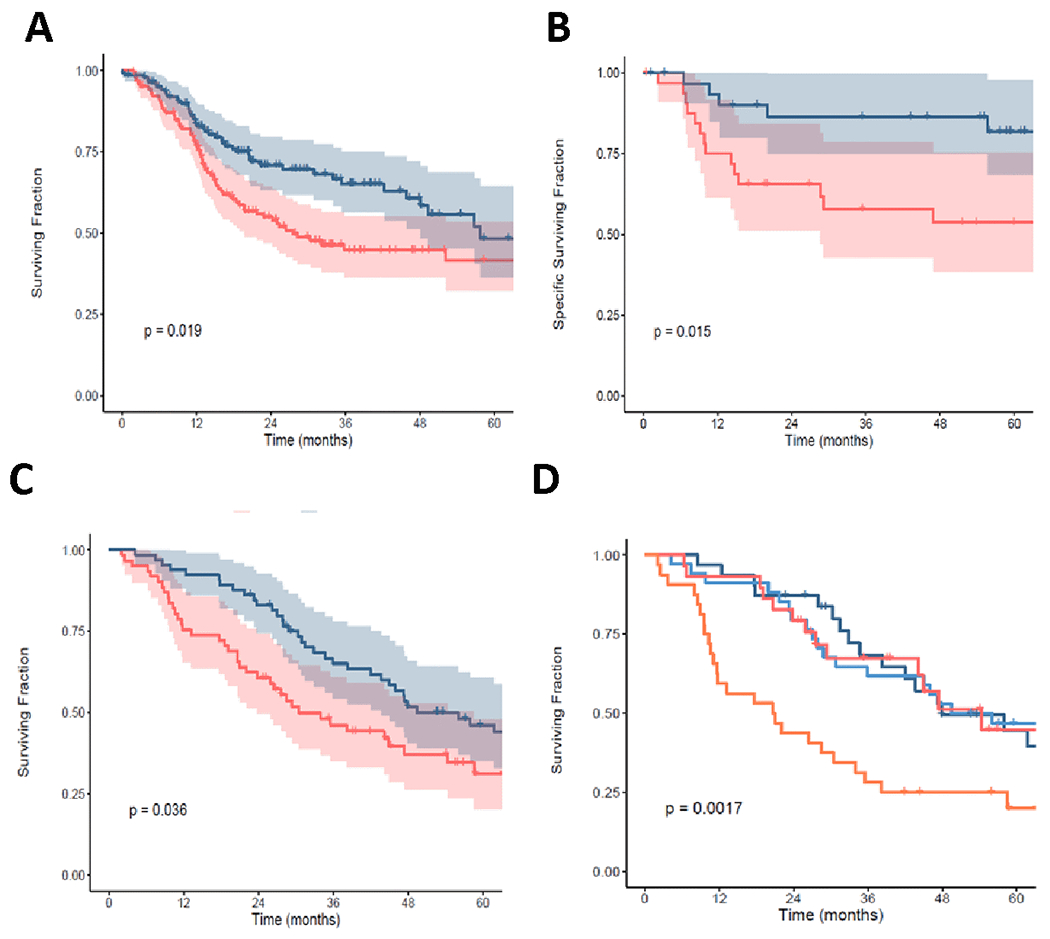
(A) βAltw top (blue) tertile is associated with better OS (log-rank, P = 0.019) compared to bottom (red) tertile of HNSC patients from TCGA. (B) βAltw top (blue) tertile is associated with better cancer-specific survival (log-rank, P = 0.015) compared to bottom (red) tertile of HPV-negative oral squamous carcinoma patients (GSE41613). (C) βAltw top (blue) tertile is associated with better OS (log-rank, P = 0.036) compared to bottom (red) tertile of naive stage II-III high grade OV carcinoma patients treated with adjuvant platinum chemotherapy (GSE26712). (D) The same population as in panel C classified by optimal (dark blue and dark red) versus suboptimal (light blue and orange) debulking status. βAltw top (light blue) tertile of suboptimally debulked patients is associated with better OS (log-rank, P = 0. 0017) compared to patients in the bottom (orange) tertile. In all plots, P values were calculated with log-rank tests.
Prior analysis of βAlt in TCGA showed a significant association with outcomes of ovarian cancer patients in which standard of care is genotoxic chemotherapy (16); thus we sought to test βAltw using ovarian cancer data from the GSE26712 dataset of naïve stage II-III high grade ovarian cancer patients treated with adjuvant platinum-based chemotherapy (n=185). Patients from the high βAltw tertile experience significantly longer OS than those from the low βAltw tertile (P = 0.036, HR = 0.64, 95% CI 0.42-0.97; Fig. 6C). In this dataset, lack of additional stage and age information precluded multivariable Cox regression analysis.
Patients with advanced stage ovarian cancer are first treated by extensive debulking surgery, followed by genotoxic platinum chemotherapy combined with paclitaxel. Patients who are optimally debulked have a substantially improved survival compared with patients who are left with bulky residual disease (36). Hence, we conducted a subset analysis of this dataset to determine whether the βAltw score is equally predictive of outcome as a function of debulking status. Comparison of the OS of the top and bottom βAltw tertile from optimally and suboptimally debulked patients showed that those with a high βAltw had substantially better outcomes (P = 0.0017; Fig. 6D) in response to standard of care treatment with platinum chemotherapy. Notably, sub-optimally debulked patients with a high βAltw had an equivalent OS as patients who were optimally debulked, underscoring the prognostic significance of the βAltw score in patients treated with genotoxic therapies.
DISCUSSION
The induction of DNA damage by radiotherapy or genotoxic chemotherapy is arguably still the most widely deployed cancer treatment approach. However, chemo/radioresistance constitutes a major obstacle to effective or personalized treatment. Thus, identifying novel predictive biomarkers is imperative to guide therapeutic decisions and improve cancer patients’ survival. In this study, we showed that TGFβ and alt-EJ reciprocal gene expression signatures, which are predictive of patient outcome in response to genotoxic therapy (16), are significantly correlated with their respective biology in HNSC explants. The TGFβ signaling signature was associated with the frequency of pSMAD2 positive cells whereas the alt-EJ signature was correlated with unrepaired DNA damage. These data formed the basis for weighting the signatures into a single score, termed βAltw, which was clinically validated in independent HNSC and ovarian cancer patient datasets.
Several insights were gained from this research. First, the strong correlation between biological response in the human HNSC explants treated in vitro together with the NanoString analysis of TGFβ and alt-EJ genes demonstrate that the signatures accurately report TGFβ signaling and functional responses to DNA damage, which further supports the extensive control of TGFβ signaling in DNA repair choice (16).
Second, the original βAlt biomarker used whole-transcriptome profiling, where the requirements for high-quality RNA or sufficient bulk tumor can impede widespread clinical adoption (37). Implementation of the custom NanoString panel is a cost-effective alternative to whole genome expression profiling and suited for analysis of archival tissues, allowing assessment of large patient cohorts for which only formalin-fixed, paraffin-embedded tissue is available. The technical advantages of this method were substantiated by the reproducibility of the gene expression in archival ovarian cancer replicates.
Third, comparing the performance of predicting pancancer patient survival after genotoxic treatment between the original βAlt and the βAltw demonstrates the superiority of βAltw and that it is a robust predictor of patient prognosis in response to genotoxic treatment, as was confirmed in independent HNSC and ovarian cancer datasets. In HNSC, appropriate therapy decision and stratification remain a major challenge (38). HPV positivity in patients with oropharyngeal carcinoma is a potent stratification factor that is associated with better survival due to increased sensitivity to radiation and platinum chemotherapy (17,39). HPV positive cancer lack TGFβ responsiveness, which impairs DNA repair by HRR and increases the use of error-prone alt-EJ (3). Inhibiting TGFβ in HPV negative HNSC also increases alt-EJ (3). That HPV is not the only means by which cancers become TGFβ incompetent was substantiated by the TCGA pancancer anti-correlation of these signatures (16). Here, we show that βAltw predictive capacity is maintained in HPV-negative HNSC patients independently of clinical characteristics including age and stage. Efforts are increasing to de-intensify standard cancer treatments in HPV positive patients (40) and likewise, identification of HPV negative patients resistant to genotoxic therapy is critical to enrich selection for clinical trials of intensified therapy. Thus, if further validated in prospective clinical trials, this biomarker could provide predictive and prognostic information to personalize therapies to the individual patient’s likelihood of response.
Fourth, the validation of the βAltw score in ovarian cancer patients treated with genotoxic therapy highlights fundamental tumor biology that has direct implications for prognosis. Ovarian cancer is the most lethal gynecologic malignancy in women worldwide (41). Ovarian cancer is sensitive to platinum-based chemotherapy and the current standard is carboplatin and paclitaxel in the first-line setting. Interestingly, βAltw predicts that loss of TGFβ signaling overcomes the survival disadvantage shown for sub-optimally debulked patients (30).
Moreover, the biology of alt-EJ points to additional therapeutic choices. Maintenance with poly(ADP-ribose) polymerase (PARP) inhibitors after platinum-based therapy is approved for ovarian cancer patients with BRCA1 or BRCA2 mutations but also provides significant benefit in patients with platinum-sensitive, relapsed, high-grade serous ovarian cancer (42). Repair by alt-EJ relies on polymerase θ (encoded by POLQ), for which clinically viable inhibitors were recently identified (43). Cells in which TGFβ signaling is blocked are sensitized to PARP inhibition and silencing POLQ increases response (3). Thus, targeting cancers using alt-EJ by inhibiting PARP or polymerase θ selectively kills cancer cells via synthetic lethality and spares TGFβ competent normal cells. Importantly, although TGFβ regulates HRR utilization via regulation of BRCA1, HRR deficiency is not required for alt-EJ to increase upon TGFβ inhibition (16). Hence, the anti-correlation of TGFβ and alt-EJ reported by βAlt is a clinically actionable basis by which to stratify patients.
This study has limitations. Although we validated the βAltw score in published datasets, this biomarker has not been tested prospectively in a clinical trial, which is a necessary step before widespread adoption of the score as a prognostic tool in the clinical setting. Also, the extent of genotoxic treatments given to patients in some public datasets, such as TCGA, is not annotated in individual detail, for which inferences on patient treatments had to be made based on the standard of care of each cancer type and stage. Additionally, gene expression levels do not necessarily reflect gene function, given that many genes are not regulated at the transcriptional level. Lastly, comparison of outcomes as a function of βAltw score tertile was used as an unbiased approach, but further studies are needed to determine the optimal cutoff for clinical application.
In summary, the TGFβ and alt-EJ transcriptomic signatures represent functional biological processes, and their anti-correlation provides important clinical insight. The βAltw score, or a similar means to assess this biology, may serve as a predictive biomarker for patients receiving genotoxic radiation or chemotherapy. The clinical utility of these signatures needs to be validated in a prospective clinical trial to determine if the βAlt score can provide sufficient predictive information to stratify and help guide patient management in both HNSC and ovarian cancer. If so, this mechanism-based score could assist in clinical decision making and enable more personalized cancer therapy for these patients.
Nonetheless, the biological validation of TGFβ and alt-EJ signatures, introduction of a custom gene set in a platform that can be used for retrospective analysis of existing specimens, and the development of the βAltw score support the prospective use of this biology to predict patient response to genotoxic therapies.
Supplementary Material
Translational Relevance.
The anti-correlation of TGFβ and alt-EJ transcriptomic signatures represents a mechanistic relationship and indicates important biological processes that provide important clinical insight. The βAltw score, or a similar means to assess this biology, may serve as a predictive biomarker for patients receiving genotoxic therapy in either HNSC or ovarian cancer. The clinical utility of these signatures needs to be further validated in a prospective clinical trial to determine if the βAltw score can provide sufficient predictive information to stratify and help guide patient management. If so, this mechanism-based score could enable more personalized cancer therapy to assist in clinical decision making.
Acknowledgements
The authors would like to thank William Chou, Trevor Jones, and Colin Foster at UCSF for technical support. The results presented here are partly based on data generated by the TCGA Research Network (https://www.cancer.gov/tcga), and we would like to express our gratitude to the TCGA consortia and their coordinators for the data provision and clinical information used in this study.
Funding
This research was supported by funding from the National Institutes of Health (R01 CA239235) and the UCSF Resource Allocation Program to MHBH and AA. Tissue processing and immunostaining was performed using the Helen Diller Family Comprehensive Cancer Center Pathology Shared Resource, supported by the National Cancer Institute of the National Institutes of Health under Award Number P30CA082103. The National Natural Science Foundation of China (Grant No. 82073007) to QL, the Generalitat de Catalunya (SGR 2017-449, CERCA program to IDIBELL) and Carlos III Institute of Health (ISCIII), funded by FEDER funds (PI21/01306) to MAP.
Conflicts of Interests
MHBH has or is a recipient of research grants paid to UCSF from Roche-Genentech, Varian, Inc. and Lilly, Inc. and consults for EMD-Serono, Varian, and Genentech. FG reports research grants to ICO from Varian, ELecta and Zeiss. SY reports research grants paid to UCSF from Genentech, Merck, Bristol Myers-Squibb, and BioMimetix, royalties from UpToDate and Springer, and honoria from ASTRO. MHBH, LQ and MAP have filed a patent application submitted by the University of California, San Francisco that covers use of the gene signatures. JP reports research grants paid to ICO from AstraZeneca. AA is co-founder of Tango Therapeutics, Azkarra Therapeutics, Ovibio Corporation, Kytarro and BioAI; consultant or Scientific Advisory Board Member for Bluestar, ProLynx, Earli, Cura, GenVivo, GSK; Ambagon, Phoenix Molecular Designs, Genentech, GLAdiator, Circle; a board member of Cambridge Science Corporation and Cytomyx; receives grant/research support from SPARC and AstraZeneca. IG, JHM, PH, AL and JC have nothing to report.
Abbreviations:
- alt-EJ
alternative end-joining
- TGFβ
transforming growth factor β
- HRR
homologous recombination repair
- NHEJ
non-homologous end-joining
- HNSC
head and neck squamous cell carcinoma
- OS
overall survival
- PFS
progression-free survival
- TCGA
The Cancer Genome Atlas
- HR
hazard ratio
- CI
confidence interval
- HR
hazard ratio
References
- 1.Kirshner J, Jobling MF, Pajares MJ, Ravani SA, Glick AB, Lavin MJ, et al. Inhibition of transforming growth factor-beta1 signaling attenuates ataxia telangiectasia mutated activity in response to genotoxic stress. Cancer research 2006;66(22):10861–9 doi 10.1158/0008-5472.CAN-06-2565. [DOI] [PubMed] [Google Scholar]
- 2.Martinez-Ruiz H, Illa-Bochaca I, Omene C, Hanniford D, Liu Q, Hernando E, et al. A TGFbeta-miR-182-BRCA1 axis controls the mammary differentiation hierarchy. Science signaling 2016;9(457):ra118 doi 10.1126/scisignal.aaf5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Q, Ma L, Jones T, Palomero L, Pujana MA, Martinez-Ruiz H, et al. Subjugation of TGFb Signaling by Human Papilloma Virus in Head and Neck Squamous Cell Carcinoma Shifts DNA Repair from Homologous Recombination to Alternative End Joining. Clin Cancer Res 2018;24(23):6001–14 doi 10.1158/1078-0432.ccr-18-1346. [DOI] [PubMed] [Google Scholar]
- 4.Chen J, Shukla V, Farci P, Andricovich J, Jogunoori W, Kwong LN, et al. Loss of the transforming growth factor-β effector β2-Spectrin promotes genomic instability. Hepatology 2017;65(2):678–93 doi 10.1002/hep.28927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim MR, Lee J, An YS, Jin YB, Park IC, Chung E, et al. TGFbeta1 Protects Cells from gamma-IR by Enhancing the Activity of the NHEJ Repair Pathway. Molecular cancer research : MCR 2015;13(2):319–29 doi 10.1158/1541-7786.mcr-14-0098-t. [DOI] [PubMed] [Google Scholar]
- 6.Liu Q, Lopez K, Murnane J, Humphrey T, Barcellos-Hoff MH. Misrepair in Context: TGFbeta Regulation of DNA Repair. Frontiers in oncology 2019;9:799 doi 10.3389/fonc.2019.00799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ceccaldi R, Rondinelli B, D’Andrea AD. Repair Pathway Choices and Consequences at the Double-Strand Break. Trends in cell biology 2016;26(1):52–64 doi 10.1016/j.tcb.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iliakis G, Murmann T, Soni A. Alternative end-joining repair pathways are the ultimate backup for abrogated classical non-homologous end-joining and homologous recombination repair: Implications for the formation of chromosome translocations. Mutation research Genetic toxicology and environmental mutagenesis 2015;793:166–75 doi 10.1016/j.mrgentox.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 9.Sallmyr A, Tomkinson AE. Repair of DNA double-strand breaks by mammalian alternative end-joining pathways. The Journal of biological chemistry 2018;293(27):10536–46 doi 10.1074/jbc.TM117.000375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sfeir A, Symington LS. Microhomology-Mediated End Joining: A Back-up Survival Mechanism or Dedicated Pathway? Trends in biochemical sciences 2015;40(11):701–14 doi 10.1016/j.tibs.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wood RD, Doublie S. DNA polymerase theta (POLQ), double-strand break repair, and cancer. DNA repair 2016;44:22–32 doi 10.1016/j.dnarep.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bouquet SF, Pal A, Pilones KA, Demaria S, Hann B, Akhurst RJ, et al. Transforming growth factor b1 inhibition increases the radiosensitivity of breast cancer cells in vitro and promotes tumor control by radiation in vivo. Clin Cancer Res 2011;17(21):6754–65 doi 10.1158/1078-0432.CCR-11-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hardee ME, Marciscano AE, Medina-Ramirez CM, Zagzag D, Narayana A, Lonning SM, et al. Resistance of glioblastoma-initiating cells to radiation mediated by the tumor microenvironment can be abolished by inhibiting transforming growth factor-beta. Cancer research 2012;72(16):4119–29 doi 10.1158/0008-5472.CAN-12-0546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang M, Herion TW, Timke C, Han N, Hauser K, Weber KJ, et al. Trimodal Glioblastoma Treatment Consisting of Concurrent Radiotherapy, Temozolomide, and the Novel TGF-β Receptor I Kinase Inhibitor LY2109761. Neoplasia (New York, NY) 2011. 13(6):537–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du S, Bouquet F, Lo C-H, Pellicciotta I, Bolourchi S, Parry R, et al. Attenuation of the DNA Damage Response by TGFβ Inhibitors Enhances Radiation Sensitivity of NSCLC Cells In Vitro and In Vivo Int J Radiat Oncol Biol Phys 2014;91(1):91–9 doi 10.1016/j.ijrobp.2014.09.026. [DOI] [PubMed] [Google Scholar]
- 16.Liu Q, Palomero L, Moore J, Guix I, Espín R, Aytés A, et al. Loss of TGFβ signaling increases alternative end-joining DNA repair that sensitizes to genotoxic therapies across cancer types. Science translational medicine 2021;13(580):eabc4465 doi 10.1126/scitranslmed.abc4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tan PF, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med 2010;363(1):24–35 doi 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nees M, Geoghegan JM, Munson P, Prabhu V, Liu Y, Androphy E, et al. Human papillomavirus type 16 E6 and E7 proteins inhibit differentiation-dependent expression of transforming growth factor-beta 2 in cervical keratinocytes. Cancer research 2000;60(15):4289–98. [PubMed] [Google Scholar]
- 19.Andarawewa KL, Erickson AC, Chou WS, Costes SV, Gascard P, Mott JD, et al. Ionizing radiation predisposes nonmalignant human mammary epithelial cells to undergo transforming growth factor b induced epithelial to mesenchymal transition. Cancer research 2007;67:8662–70. [DOI] [PubMed] [Google Scholar]
- 20.Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ Is a Mechanistically Distinct Pathway of Mammalian Chromosome Break Repair. Plos Genet 2008;4(6):e1000110 doi ARTN e1000110 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DiBiase SJ, Zeng ZC, Chen R, Hyslop T, Curran WJ Jr., Iliakis G. DNA-dependent protein kinase stimulates an independently active, nonhomologous, end-joining apparatus. Cancer research 2000;60(5):1245–53. [PubMed] [Google Scholar]
- 22.Howard SM, Yanez DA, Stark JM. DNA damage response factors from diverse pathways, including DNA crosslink repair, mediate alternative end joining. PLoS genetics 2015;11(1):e1004943 doi 10.1371/journal.pgen.1004943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nature biotechnology 2008;26(3):317–25 doi 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 24.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics (Oxford, England) 2016;32(18):2847–9 doi 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 25.Sidaway P Glioblastoma subtypes revisited. Nature Reviews Clinical Oncology 2017;14(10):587- doi 10.1038/nrclinonc.2017.122. [DOI] [PubMed] [Google Scholar]
- 26.Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell 2017;32(2):185–203.e13 doi 10.1016/j.ccell.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang S, Liu X. The UCSCXenaTools R package: a toolkit for accessing genomics data from UCSC Xena platform, from cancer multi-omics to single-cell RNA-seq. Journal of Open Source Software 2019;4(40):1627 doi 10.21105/joss.01627. [DOI] [Google Scholar]
- 28.Eschrich S, Zhang H, Zhao H, Boulware D, Lee JH, Bloom G, et al. Systems biology modeling of the radiation sensitivity network: a biomarker discovery platform. Int J Radiat Oncol Biol Phys 2009;75(2):497–505 doi 10.1016/j.ijrobp.2009.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 2018;173(2):400–16.e11 doi 10.1016/j.cell.2018.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bonome T, Levine DA, Shih J, Randonovich M, Pise-Masison CA, Bogomolniy F, et al. A Gene Signature Predicting for Survival in Suboptimally Debulked Patients with Ovarian Cancer. Cancer research 2008;68(13):5478–86 doi 10.1158/0008-5472.can-07-6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lohavanichbutr P, Méndez E, Holsinger FC, Rue TC, Zhang Y, Houck J, et al. A 13-Gene Signature Prognostic of HPV-Negative OSCC: Discovery and External Validation. Clinical Cancer Research 2013;19(5):1197–203 doi 10.1158/1078-0432.ccr-12-2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen EG, Wang P, Lou H, Wang Y, Yan H, Bi L, et al. A robust gene expression-based prognostic risk score predicts overall survival of lung adenocarcinoma patients. Oncotarget 2018;9(6):6862–71 doi 10.18632/oncotarget.23490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Veldman-Jones MH, Brant R, Rooney C, Geh C, Emery H, Harbron CG, et al. Evaluating Robustness and Sensitivity of the NanoString Technologies nCounter Platform to Enable Multiplexed Gene Expression Analysis of Clinical Samples. Cancer research 2015;75(13):2587–93 doi 10.1158/0008-5472.can-15-0262. [DOI] [PubMed] [Google Scholar]
- 34.Asaithamby A, Hu B, Chen DJ. Unrepaired clustered DNA lesions induce chromosome breakage in human cells. Proceedings of the National Academy of Sciences 2011;108(20):8293–8 doi 10.1073/pnas.1016045108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Castellanos-Martin A, Castillo-Lluva S, Saez-Freire Mdel M, Blanco-Gomez A, Hontecillas-Prieto L, Patino-Alonso C, et al. Unraveling heterogeneous susceptibility and the evolution of breast cancer using a systems biology approach. Genome biology 2015;16:40 doi 10.1186/s13059-015-0599-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Riester M, Wei W, Waldron L, Culhane AC, Trippa L, Oliva E, et al. Risk Prediction for Late-Stage Ovarian Cancer by Meta-analysis of 1525 Patient Samples. JNCI: Journal of the National Cancer Institute 2014;106(5) doi 10.1093/jnci/dju048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piskol R, Huw L, Sergin I, Kljin C, Modrusan Z, Kim D, et al. A Clinically Applicable Gene-Expression Classifier Reveals Intrinsic and Extrinsic Contributions to Consensus Molecular Subtypes in Primary and Metastatic Colon Cancer. Clin Cancer Res 2019;25(14):4431–42 doi 10.1158/1078-0432.ccr-18-3032. [DOI] [PubMed] [Google Scholar]
- 38.Wichmann G, Rosolowski M, Krohn K, Kreuz M, Boehm A, Reiche A, et al. The role of HPV RNA transcription, immune response-related gene expression and disruptive TP53 mutations in diagnostic and prognostic profiling of head and neck cancer. International journal of cancer 2015;137(12):2846–57 doi 10.1002/ijc.29649. [DOI] [PubMed] [Google Scholar]
- 39.Leemans CR, Snijders PJF, Brakenhoff RH. The molecular landscape of head and neck cancer. Nat Rev Cancer 2018;18(5):269–82 doi 10.1038/nrc.2018.11. [DOI] [PubMed] [Google Scholar]
- 40.Mehanna H, Robinson M, Hartley A, Kong A, Foran B, Fulton-Lieuw T, et al. Radiotherapy plus cisplatin or cetuximab in low-risk human papillomavirus-positive oropharyngeal cancer (De-ESCALaTE HPV): an open-label randomised controlled phase 3 trial. Lancet (London, England) 2019;393(10166):51–60 doi 10.1016/s0140-6736(18)32752-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matsumoto K, Nishimura M, Onoe T, Sakai H, Urakawa Y, Onda T, et al. PARP inhibitors for BRCA wild type ovarian cancer; gene alterations, homologous recombination deficiency and combination therapy. Jpn J Clin Oncol 2019;49(8):703–7 doi 10.1093/jjco/hyz090. [DOI] [PubMed] [Google Scholar]
- 42.Mirza MR, Monk BJ, Herrstedt J, Oza AM, Mahner S, Redondo A, et al. Niraparib Maintenance Therapy in Platinum-Sensitive, Recurrent Ovarian Cancer. New England Journal of Medicine 2016;375(22):2154–64 doi 10.1056/NEJMoa1611310. [DOI] [PubMed] [Google Scholar]
- 43.Helleday T Polθ inhibitors unchained. Nature Cancer 2021;2(6):581–3 doi 10.1038/s43018-021-00225-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data in the article that are not from open-access datasets are available from the corresponding author upon request.


