Abstract
Background:
Two acute respiratory distress syndrome (ARDS) subphenotypes with distinct clinical and biological features and differential treatment responses have been identified using latent class analysis (LCA) in seven individual cohorts. To facilitate bedside identification of subphenotypes, clinical-classifier models using readily available clinical variables have been described in five randomized-controlled trials. Performance of these models in observational cohorts of ARDS is unknown.
Methods:
We evaluated the performance of machine learning clinical-classifier models for assigning ARDS subphenotypes in two observational cohorts of ARDS: EARLI (n=335) and VALID (n=452), with LCA-derived subphenotype as the gold-standard. We also assessed model performance in EARLI using data automatically extracted from the electronic health record (EHR). In LUNG SAFE (n=2813), a multinational observational ARDS cohort, we applied the model to determine the prognostic value of the subphenotypes and tested their interaction with PEEP strategy, with mortality as the dependent variable.
Findings:
The clinical-classifier models had an area under receiver operating characteristic curve (AUC) of 0·92 (95% CI: 0·90–0·95) in EARLI and 0·88 (0·84–0·91) in VALID. Model performance was comparable when using exclusively EHR-derived predictors. In LUNG SAFE, 90-day mortality was higher in the Hyperinflammatory subphenotype (57% [414/725] vs. 33% [694/2088]; p<0·0001). There was a significant treatment interaction with PEEP strategy and ARDS subphenotype (p=0·041), with lower mortality in the high PEEP group in the Hyperinflammatory subphenotype, following similar patterns to those observed in prior analyses of the ALVEOLI trial.
Interpretation:
Classifier models using clinical variables alone can accurately assign ARDS subphenotypes in observational cohorts. Application of these models can provide valuable prognostic information and may inform management strategies for personalised treatment, including application of PEEP, once prospectively validated.
Funding:
National Institutes of Health (PS: GM142992, CSC: HL140026, LBW: HL103836, HL135849), European Society of Intensive Care Medicine.
Introduction
The acute respiratory distress syndrome (ARDS) remains a highly prevalent cause of acute respiratory failure, resulting in high morbidity and mortality.1 Yet, potentially as a consequence of underlying heterogeneity, few therapeutic options have proven to be beneficial in randomized controlled trials (RCTs).2–4 Two discrete biological subphenotypes have been identified using latent class analysis (LCA) in five RCTs and two observational cohorts, totaling over 4,000 patients.5–8 The two subphenotypes have distinct clinical and biological features, divergent outcomes, and in three RCTs, differential treatment responses were observed.
Although accurate parsimonious models for subphenotype identification have been developed, these models are reliant on measurement of protein biomarkers (e.g., interleukin (IL)-6, IL-8, soluble tumor necrosis factor receptor (sTNFR)-1, Protein C).9 The limited availability of real-time assays for these biomarkers represents a barrier to the clinical implementation and rapid identification of the subphenotypes.5,10 Recently, machine learning classification algorithms utilizing routinely available clinical variables have shown promise in identifying LCA-derived subphenotypes in RCT cohorts of ARDS.10 Their performance in unselected populations of ARDS patients, where patient heterogeneity may be even greater and where comparatively higher mortality is observed, is unknown.11 A critical step towards clinical application of these models is their validation in observational and representative populations of ARDS patients, particularly since it is these unselected, “real-world” patients in whom the models would be used to screen for enrollment in future RCTs.
The primary objective of this study was to validate machine learning classifier models that use readily available clinical data in observational cohorts of ARDS. Secondary objectives were (1) to evaluate model performance in an observational cohort of patients with predictor-variables automatically extracted from the electronic health record (EHR) and (2) to evaluate the clinical utility for prognostication and seeking differential responses to positive end-expiratory pressure (PEEP) strategy of ARDS subphenotypes derived using the clinical-classifier models in a large multinational observational cohort of ARDS.
Methods
Study populations
Details of the RCT cohorts used for model development are described in prior studies.10,12–15 Two observational cohorts of ARDS, Early Assessment of Renal and Lung Injury (EARLI, n=335) and Validating Acute Lung Injury markers for Diagnosis (VALID, n=452), served as independent validation cohorts for the models. EARLI is an ongoing prospectively enrolled cohort of patients admitted to UCSF Medical Center and Zuckerberg San Francisco General Hospital Intensive Care Units (ICUs). Study participants were identified in the Emergency Department upon request for admission to the ICU. For this analysis, patients were selected from EARLI if they were deemed to have ARDS as defined by the American-European Consensus Conference (AECC) criteria on either Day 1 or 2 of the study and included patients recruited between 2008–2018.16 Details of the study protocol have been previously published.17 VALID is an ongoing prospectively enrolled cohort of patients admitted to Vanderbilt University Medical Center ICU; details of the study protocol have been previously published.18 Study participants were enrolled in the study on the morning of the second day of admission to a medical, surgical, trauma, or cardiovascular ICU. Patients were selected from VALID for inclusion in this analysis if they were deemed to have ARDS as defined by AECC criteria on the first or second day of ICU admission and included patients were recruited between 2008–2016. Patients with trauma-related ARDS were excluded given biological and clinical differences (e.g., lower burden of inflammation and lower age-adjusted mortality) from patients with non-trauma ARDS and our previous work suggesting the subphenotyping schema is most valid in patients with non-trauma ARDS.8,19 The AECC definition was used because enrollment in both cohorts started prior to development of the Berlin definition and because patients continued to be enrolled using both definitions.20 The described strategy allowed capture of more patients for analysis. Further, the LCA-derived subphenotypes have been validated in these cohorts using the AECC definition with similar subphenotypes identified as when using the Berlin definition.8 Both cohorts include comprehensive demographic, clinical, and biomarker data from the day (or day prior to) of ARDS diagnosis that were manually collected by trained research coordinators, as well as clinical outcome data including ventilator-free days (VFD) and hospital mortality.
The Large Observational Study to Understand the Global Impact of Severe Acute Respiratory Failure (LUNG SAFE, n=2813) was a large, multinational, multicenter, prospectively enrolled cohort of patients admitted to 459 ICUs across 50 countries from February to March 2014; details of the study protocol have been previously published.1 Study participants were enrolled on the first day that acute hypoxemic respiratory failure criteria were satisfied. Patients were selected from LUNG SAFE for inclusion in this analysis if they were deemed to have ARDS as defined by Berlin criteria on the first or second day of study enrollment.20 The LUNG SAFE study was conducted after the description of the Berlin definition hence its use in this cohort. This cohort includes demographic, clinical, and respiratory data from the day patients were enrolled into the study and at pre-specified intervals until ICU discharge or death (see Supplement). All study cohorts were approved by the Institutional Review Board at each participating hospital.
Model Development and Validation
All models were trained to predict the Hyperinflammatory phenotype. Of the machine learning clinical-classifier models described in the original study,10 we used the two best performing models to validate in this study, with a parsimonious (“vitals and labs”) model comprising only of vital signs and laboratory values serving as the primary model. As the secondary model, we used a “full feature” model comprising all the predictors in the primary model, with the addition of ventilatory variables and demographics. The “vitals and labs” model served as the primary model because it was less complex (fewer predictors), constituted exclusively of physiological predictors, was one of the most accurate in the original study, and was the most generalizable model.
In both EARLI and VALID, due to missing predictors, the original “vitals and labs” and “full feature” models could not be validated directly. In EARLI, the “vitals and labs” model had no predictors missing and the “full feature” model had one predictor missing (minute ventilation). In VALID, there was one predictor missing for the “vitals and labs” model (glucose) and three predictors missing (tidal volume, glucose, and body mass index) for the “full feature” model. To simplify the analysis, we developed new “vitals and labs” and “full feature” models comprising of common predictors available for each model in both EARLI and VALID from the original predictors. A final list of variables used in each model is described in Table S1.
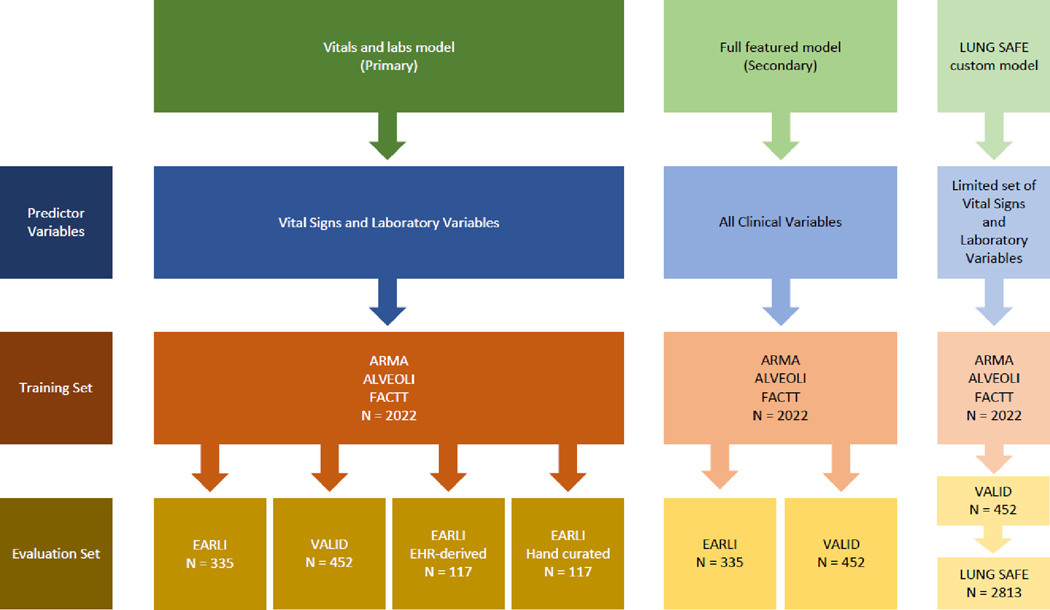
A schematic of the analysis plan is presented in Figure 1. For model development, we used a gradient-boosted machine algorithm, XGBoost: Extreme Gradient Boosting (version 1.3.2.1). In brief, gradient-boosted machines utilize an ensemble of multiple decision trees, where trees added sequentially to the model to attempt to correct the classification error of previous trees in the ensemble. We utilized 10-fold cross validation and hyperparameter tuning using a grid search to tune and optimize the models using the training set (see Supplement), recapitulating our prior approach.10 All models were developed using a training set comprised of a combination of three RCT cohorts, ARMA, ALVEOLI, and FACTT (n=2022), and model performance was tested externally in SAILS (n=745). The models output a continuous probability specifying the likelihood of classification to the Hyperinflammatory subphenotype for each patient. To evaluate the validity of these two new models in relation to the models developed in the original study,10 we compared the probabilities generated by corresponding new and original models using Pearson’s correlation coefficient.
Figure 1. Schematic of analysis plan.
The models were originally trained in ARMA, ALVEOLI and FACTT (n = 2022) and tested in SAILS (n=745), which were all randomised controlled trials. The models were validated in two observational cohorts: EARLI (n=335) and VALID (n=452). A custom (“LUNG SAFE”) model using a limited set of predictor variables was developed to evaluate the clinical utility of ARDS subphenotypes in LUNG SAFE (n=2813) a large multinational observational cohort of ARDS. The optimal probability cutoff for the “LUNG SAFE” model determined by first evaluating the model in VALID (n=452).
Next, performance of these models was evaluated independently in EARLI (n=335) and VALID (n=452). Validation cohorts were kept isolated from the training and testing procedures. LCA-derived subphenotypes served as the reference standard for model training, testing, and validation. The procedure for handling missing data is detailed in the Supplement and the missingness for predictors in each validation cohort are presented in Table S2. Overall model performance in EARLI and VALID was evaluated by (a) calculating the area under the receiver operating characteristic curve (AUC) with confidence intervals (estimated using 2000 stratified bootstrap replicates); and (b) generating calibration plots. For each model, class was assigned using a probability cutoff of 0·5 to report on accuracy, sensitivity, and specificity of subphenotype assignments. As with our prior work, we additionally performed sensitivity analysis using cutoffs of 0·3, 0·4, 0·6, and 0·7. Once patients were assigned subphenotypes, we evaluated differences in protein biomarkers and clinical outcomes (e.g., mortality and ventilator-free days).
Model validation in EHR-derived cohort
Patients in the EARLI cohort were identified in the UCSF’s electronic health record, Epic (Epic Systems Corp.). Patients enrolled before implementation of the UCSF EHR in 2012 were excluded, as were patients admitted at San Francisco General Hospital, due to the Epic EHR being implemented at this institution after the study period (post 2018). All vital signs and laboratory values from each participant’s admission encounter were queried using SQL and downloaded from Epic Clarity, a data warehouse and relational database that stores the majority of clinical data within Epic. Additionally, we queried usage of intravenous vasoactive agents and incorporated this into the cohort as a binary variable. The most extreme values (e.g., highest heart rate or lowest serum bicarbonate level) observed ± 12 hours of ARDS diagnosis were extracted. We used this “EHR-derived EARLI cohort” to identify ARDS subphenotypes using the “vitals and labs” model. Model performance was evaluated using the same procedures described above with LCA-derived subphenotype serving as the reference standard. For comparison, we evaluated model performance for the same patients identified in the EHR cohort but using vital signs and labs collected manually during the original EARLI prospective study enrollment that were used or the original LCA.
Model evaluation in LUNG SAFE
In LUNG SAFE, due to limited data collection, only a small selection of predictor variables was available for modelling (Table S1). A custom clinical-classifier model, comprising only these variables, was developed using the same procedure described above (training: ARMA, ALVEOLI, and FACTT, testing: SAILS). As LCA-derived subphenotypes were not known in LUNG SAFE and the model contained a sparse set of predictor variables, a priori, we first sought the best probability cutoff to assign class in VALID (an observational cohort) to optimize classification accuracy. Optimal cutoff in VALID was determined based on the tradeoff between sensitivity and specificity (i.e., Youden index).21 EARLI was not used to determine cutoff due to some of the LUNG SAFE variables not being available. For sensitivity analysis, we additionally evaluated model results in LUNG SAFE across a range of probability cutoffs.
Once LUNG SAFE patients were classified into subphenotypes, we compared clinical outcomes, resolution of ARDS, prevalence of underlying chronic diseases, and ventilatory/respiratory variables stratified by model-assigned subphenotype. Building on prior work showing differential subphenotype responses to PEEP, we evaluated the interaction between subphenotype allocation and PEEP in LUNG SAFE.5 In order to create two groups with substantially different levels of PEEP usage, patients were classified into tertiles according to their mean PEEP over days 1–3. The top tertile was labelled as “high PEEP” and bottom tertile as “low PEEP,” with the middle tertile excluded from analysis. A logistic regression model was created with the interaction term of PEEP-group and subphenotype as an independent variable and 90-day mortality as the dependent variable. As sensitivity analyses, we tested for differences in mortality, VFDs, and PEEP treatment interaction for a range of probability cutoffs. We also tested for subphenotype treatment interaction with PEEP-groups derived when using quintiles instead of tertiles. Further sensitivity analyses included testing treatment interaction of PEEP-groups with subgroups of ARDS severity as stratified by (a) PaO2/FiO2 and (b) Sequential Organ Failure Assessment (SOFA) score (see Supplement for details).
Statistical Analysis
Differences in outcomes between subphenotypes were tested using Pearson’s chi-squared test. Between-group differences were tested using Student’s t-test and Wilcoxon rank-sum test, depending on variable distribution. For differences in outcomes between ARDS subphenotypes, we also computed odds ratios for mortality and rank biserial correlations for VFDs. The Wald test was used to test for significance of the interaction term in the logistic regression models. All analyses were done using R (version 4.03) and RStudio interface (version 1.4.1106). The codes used for analysis can be found on our group’s GitHub page, available at https://github.com/Calfee-Sinha-PrecisionCriticalCareLab.
Role of funders
The funder of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Results
Baseline patient characteristics for the training set, both validation cohorts (EARLI and VALID), and LUNG SAFE are shown in Table S3.
The top ten most important features for the “vital and labs” and “full feature” models in the training dataset are shown in Figures S1A and S1B respectively and in line with prior models.10 In SAILS, the probabilities of subphenotype assignment generated by the new models developed for use in this study were highly correlated to probabilities generated by our previously described models: “vital and labs” model (r = 0·97, p <0·0001, Figure S2A) and “full feature” model (r = 0·95, p <0·0001; Figure S2B).10
Model evaluation in observational cohorts
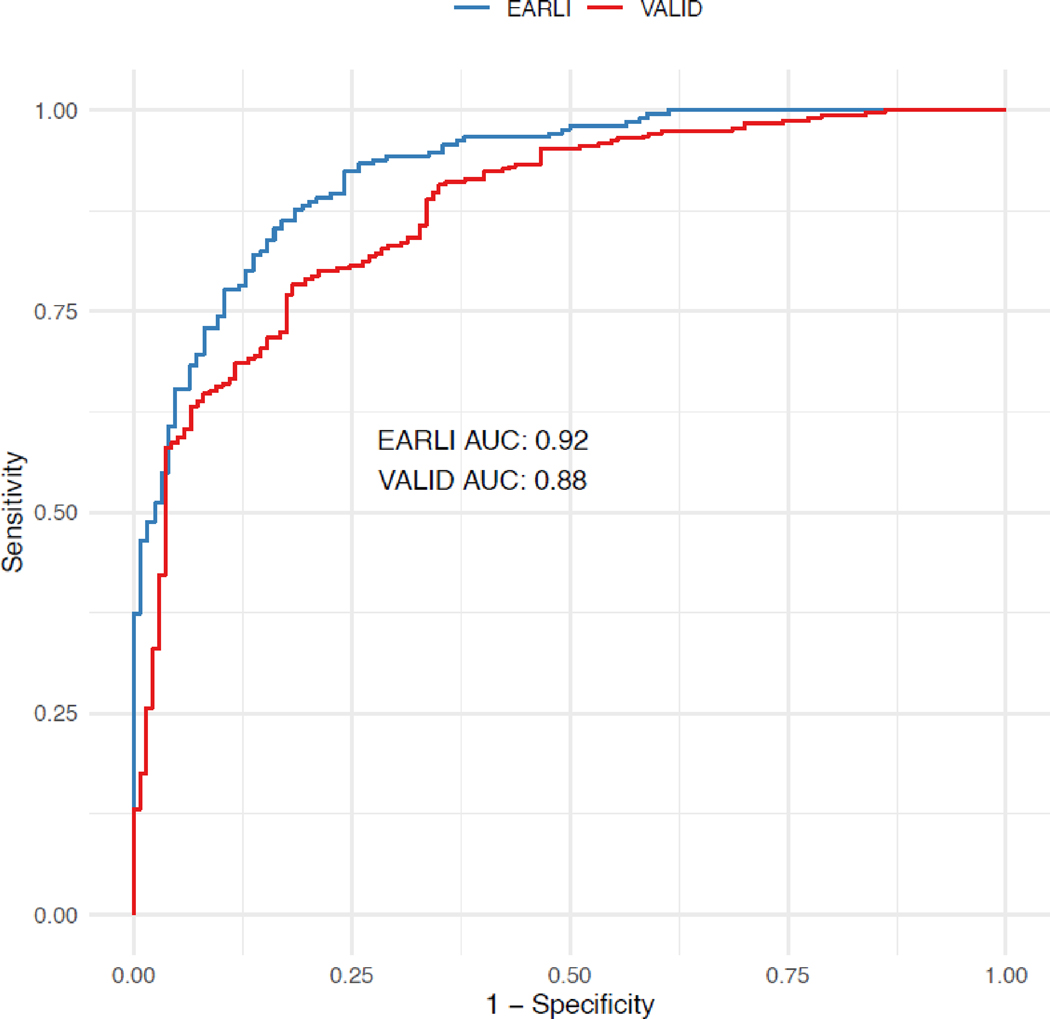
The “vitals and labs model” had an AUC of 0·92 (95% CI: 0·90 – 0·95) in EARLI and 0·88 (95% CI: 0·84 – 0·91) in VALID (Figure 2). Model sensitivity, specificity, and accuracy when using a probability cut-off of 0·5 are reported in Table 1, and over a range of probability cut-offs in Table S4. The calibration plot for the model in both cohorts is presented in Figure S3A and S3B.
Figure 2. Receiver operating characteristic (ROC) curve for primary (“vital and labs”) model in EARLI (n=335) and VALID (n=452).
AUC = Area under the ROC curve. EARLI AUC = 0.92; VALID AUC = 0.88.
Table 1. Model performance metrics of clinical classifier models.
Model validation in EARLI (n=335) and VALID (n=452) cohorts, for both the primary (“vitals and labs”) model and the secondary (“full featured”) model (demographics, vital signs, laboratory values, and ventilator parameters), using a probability cutoff of 0·5 for subphenotype assignments.
| Cohort | Model | AUC | Accuracy | Sensitivity | Specificity | Classifier model-derived Hyperinflammatory Subphenotype | LCA-derived Hyperinflammatory Subphenotype |
|---|---|---|---|---|---|---|---|
| EARLI n=335 | Vitals and labs | 0·92 (0·90 – 0·95) | 0·84 | 0·85 | 0·84 | 41% (139/335) | 37% (124/335) |
| Full features | 0·92 (0·89 – 0·95) | 0·85 | 0·80 | 0·88 | 37% (124/335) | ||
| VALID n=452 | Vitals and labs | 0·88 (0·84 – 0·91) | 0·80 | 0·66 | 0·86 | 30% (134/452) | 30% (137/452) |
| Full features | 0·87 (0·84 – 0·90) | 0·81 | 0·64 | 0·89 | 27% (123/452) |
Abbreviations: Area Under Receiver Operating Characteristic Curve (AUC with 95% confidence intervals), LCA: latent class analysis.
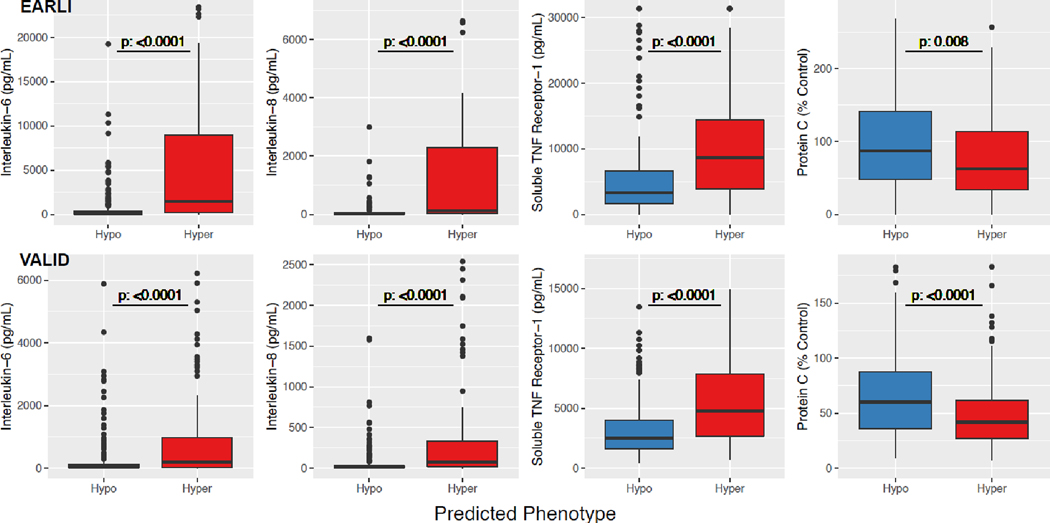
In both cohorts, the Hyperinflammatory subphenotype identified by “vitals and labs” models had significantly higher levels of interleukin-6 (IL-6), interleukin-8 (IL-8), and soluble TNF receptor-1 (sTNFR1), and lower levels of Protein C (Figure 3). The Hyperinflammatory phenotype was associated with higher in-hospital mortality and fewer ventilator-free days (Table 2). Clinical outcomes for the subphenotypes in both cohorts over a range of probability cutoffs were similar to those using a cutoff ≥ 0·5 (Table S5).
Figure 3. Differences in protein biomarkers in ARDS subphenotypes.
ARDS subphenotypes were identified by the “vitals and labs” model using a probability cut-off of 0.5. Differences in biomarker data are presented in EARLI (n=335) and VALID (n=452). Y-axis was limited to aid better data visualization. Consequently, in EARLI, 9, 10, 13, and 4 observations were censored, and in VALID, 13, 16, 17, and 3 observations were censored for Interleukin-6, Interleukin-8, Soluble tumor necrosis factor (TNF) receptor-1, and Protein C, respectively.
In both EARLI and VALID, the “full feature” model had similar model performance metrics (Figure S4, Tables 1 and S4) and differences in biomarkers (Figure S5) and clinical outcomes (Table S6) between the subphenotypes as the “vitals and labs” model.
Model validation in EHR-derived cohort
117 patients from the EARLI cohort were identified in the UCSF EHR. Baseline patient characteristics along with feature missingness are shown in Table S7. The “vitals and labs” model using EHR-derived data had an AUC of 0·88 (95% CI: 0·81 – 0·94) compared to an AUC of 0·92 (95% CI: 0·88 – 0·97; Figure S6) using hand-curated variables for the same patients. Clinical outcomes in subphenotypes assigned using EHR-derived data were similar to those derived using hand-curated data (Table S8).
Clinical-classifier model in LUNG SAFE
When first evaluated in SAILS and VALID, the LUNG SAFE classifier model resulted in an AUC of 0·93 (0·91 – 0·95) and 0·87 (0·83 – 0·90) respectively. In VALID, the model had the highest Youden index at a probability cutoff of 0·4 (Table S9). This probability cutoff was used to classify subphenotypes in LUNG SAFE.
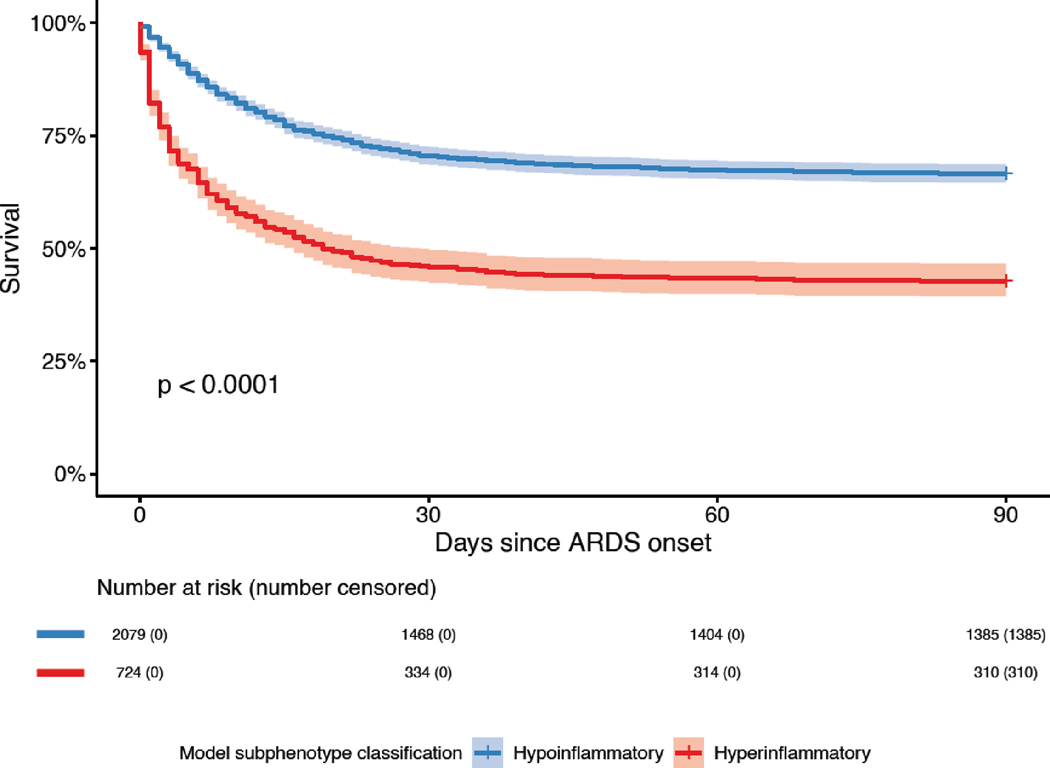
Using a cutoff of 0·4, 26% (725/2813) of patients in LUNG SAFE were classified in the Hyperinflammatory subphenotype. Mortality at day 90 in the Hyperinflammatory subphenotype was 57% (414/725) compared to 33% (694/2088) in the Hypoinflammatory group (p <0·0001). VFDs were significantly fewer in the Hyperinflammatory subphenotype (p <0·0001; Table 2). Survival between groups diverged at day 1 that was sustained over 90 days, with a significantly lower survival in the Hyperinflammatory group (Figure 4). The observed differences in mortality and VFDs were consistent across a range of probability cutoffs (Table S10).
Table 2. Clinical outcomes in the ARDS subphenotypes.
Mortality (count; percentage) and Ventilator Free Days (VFD; median and interquartile range) in the three observational cohorts of ARDS (EARLI, VALID, and LUNG SAFE). In EARLI (n=335) and VALID (n=452), outcomes are presented for the “vitals and labs” model, with a probability cutoff of 0·5 for subphenotype assignments. In LUNG SAFE (n=2813), outcomes are presented for a custom classifier model using a limited set of features and a probability cutoff of 0·4 for subphenotype assignments. Effect size was estimated using odds ratio for mortality and rank biserial correlation for VFD, with 95% confidence intervals.
| Cohort | Model | Outcome | Hypoinflammatory | Hyperinflammatory | Effect size | P value |
|---|---|---|---|---|---|---|
| EARLI n=335 | Vitals and labs | Mortality* | 29% (57/196) | 58% (80/139) | 3·3 (2·1 – 5·2) | <0·0001 |
| VFD | 24 (0 – 28) | 0 (0 – 24) | 0·30 (0·18 – 0·41) | <0·0001 | ||
| VALID n=452 | Vitals and labs | Mortality* | 27% (85/318) | 49% (66/134) | 2·7 (1·7 – 4·0) | <0·0001 |
| VFD | 21 (5 – 25) | 6 (0 – 22) | 0·31 (0·20 – 0·41) | <0·0001 | ||
| LUNG SAFE n=2813 | Custom feature set | Mortality† | 33% (694/2088) | 57% (414/725) | 2·7 (2·2 – 3·2) | <0·0001 |
| VFD | 15 (0 – 23) | 0 (0 – 19) | 0·23 (0·18 – 0·28) | <0·0001 |
P-value represent the Chi-squared test for mortality and Wilcoxon-rank test for VFD.
In Hospital Mortality;
90-day Mortality.
Figure 4. Survival curves of the two ARDS subphenotypes in LUNG SAFE (n=2813).
ARDS subphenotypes were identified by a custom clinical-classifier (“LUNG SAFE”) model using a probability cutoff of 0·4 to assign class. Abbreviations: Acute Respiratory Distress Syndrome (ARDS). P-value was calculated using the log-rank test.
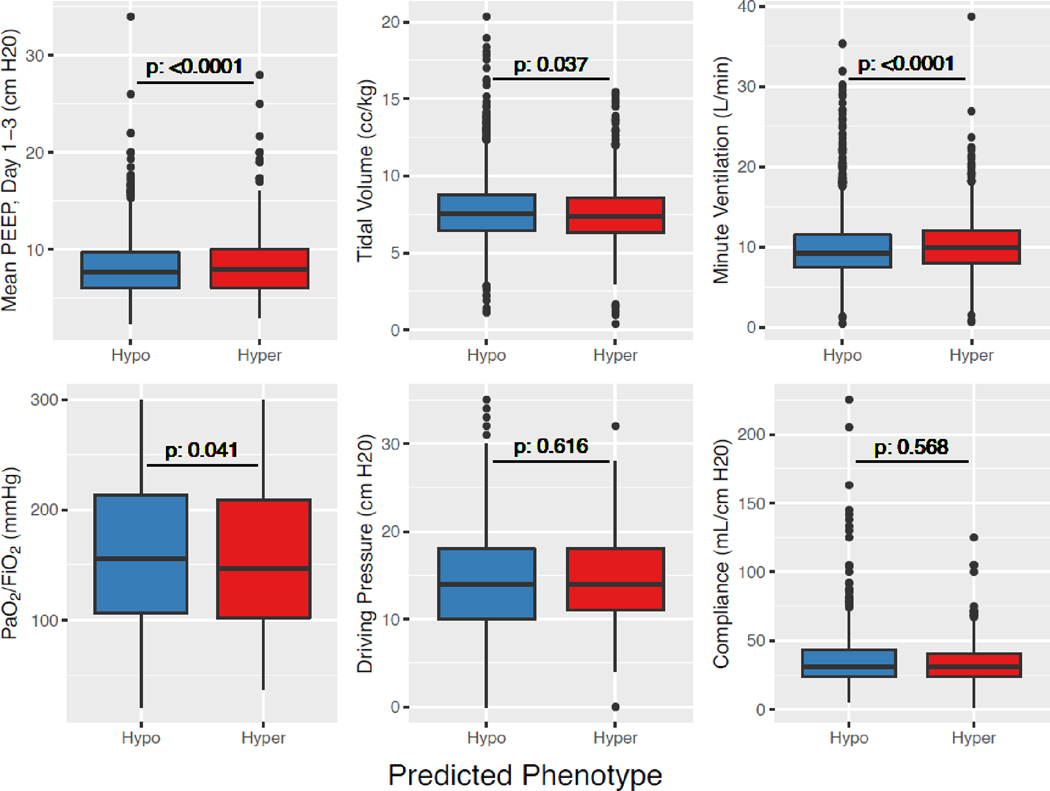
More patients in the Hypoinflammatory subphenotype had resolution of ARDS on day 2 (35%; 510/1447) compared to the Hyperinflammatory subphenotype (28%; 129/469; p = 0·0024), suggesting temporal stability of ARDS diagnosis in the latter. Prevalence of underlying chronic liver disease was significantly higher in the Hyperinflammatory subphenotype, whereas prevalence of chronic obstructive pulmonary disease (COPD) was lower (Table S11). Difference in respiratory variables, even among those that were statistically significant, were not clinically significant between the two subphenotypes (Figure 5).
Figure 5. Comparison of respiratory variables between the two ARDS subphenotypes in LUNG SAFE (n=2813).
ARDS subphenotypes were identified by a custom clinical-classifier (“LUNG SAFE”) model using a probability cutoff of 0·4 to assign class. Driving pressure is defined as the difference between plateau pressure and PEEP. Abbreviations: Positive End Expiratory Pressure (PEEP); Hyperinflammatory subphenotype (Hyper); Hypoinflammatory subphenotype (Hypo). P-value was calculated using either the t-test or Wilcoxon rank test depending on the distribution of the data.
When stratified into tertiles based on mean day 1 to 3 PEEP, median PEEP in the top tertile (“high PEEP”; n=992) was 11 cm H2O (10 – 12) and bottom tertile (“low PEEP”; n=943) was 5 cm H2O (5 – 6). Differences between the characteristics of the low- and high-PEEP groups can be found in Table S12. There was a significant interaction between PEEP subgroups and ARDS subphenotypes with 90-day mortality as the outcome; Hyperinflammatory subphenotype: “high PEEP” 54% [169/313] vs. “Low PEEP” 62% [127/205]; Hypoinflammatory subphenotype: “high PEEP” 34% [231/675] vs. “Low PEEP” 32% [233/734] (p = 0·041; Table 3). The differences in outcomes and treatment interaction were significant across a range of probability cutoffs (Table S13). The interaction term remained significant after adjusting the model for age and PaO2/FiO2 (p = 0·047). A sensitivity analysis using quintiles to define PEEP groups (with the middle quintile eliminated) also revealed significant treatment interactions (Table S14). Significant interactions with PEEP groups were not observed when the population was stratified by other measures of disease severity such as PaO2/FiO2 (p = 0·96) or SOFA score (p = 0·30; Table S15 and S16).
Table 3. Mortality at day 90 in PEEP-groups stratified by ARDS Subphenotypes in LUNG SAFE (n=2813).
ARDS Subphenotypes were assigned by a custom clinical classifier (“LUNG SAFE”) model using a probability cutoff of 0·4. PEEP subgroups were defined as “high PEEP” (n=992; median PEEP 11 cm H2O [10 – 12]) and “low PEEP” (n=943; median 5 cm H2O [5 – 6]) subgroups based on the mean PEEP over the first three days.
| Subphenotype | Mortality in Low PEEP group | Mortality in High PEEP group | P value |
|---|---|---|---|
| Hyperinflammatory | 62% (127/205) | 54% (169/313) | 0·041 |
| Hypoinflammatory | 32% (233/734) | 34% (231/675) |
P-value is for the interaction term of PEEP subgroups and ARDS subphenotypes with mortality as the dependent variable and was derived using the Wald test. Abbreviations: Positive End Expiratory Pressure (PEEP).
Discussion
In this study, we report that machine learning classifier models, using only readily available clinical variables as predictors, can accurately assign ARDS subphenotypes in observational cohorts. Our models consistently captured the rich biological information that define the LCA-derived subphenotyping schema, with marked differences in protein biomarkers between the two identified phenotypes. The models identified patients at high risk for adverse outcomes, including in the large multinational observational cohort (LUNG SAFE), where protein biomarker data were not available. Further, in LUNG SAFE, we observed differential responses to PEEP strategy by subphenotype, with higher PEEP associated with improved outcomes in the Hyperinflammatory subphenotype, similar to patterns previously identified in secondary analyses of the ALVEOLI trial.5 Finally, the “vitals and labs” model performed robustly even when utilizing clinical data extracted automatically from the EHR (as opposed to values obtained manually during study enrollment). Taken together, the models presented in these studies represent a substantial step towards translating ARDS subphenotypes into the clinical workflow. Pending prospective evaluation, these models may be valuable tools for prognostication and treatment stratification in future trials.
The utility of subphenotypes in ARDS is contingent on feasible bedside identification. Although point-of-care and real-time assays are being developed rapidly, they remain experimental.22 In the interim, or as an alternative, clinical-classifier models can be a useful adjunct. Clinical-classifier models to date have been validated only in retrospective secondary analyses of relatively uniform RCTs,10 which enroll typically only 5–10% of potentially eligible patients,11 thus limiting their routine application. By contrast, validation of these models in observational cohorts of all-comer patients with ARDS indicates that such classification algorithms can be reliably applied to more generalizable populations and could potentially be used to screen patients for eligibility for enriched RCTs. Embedding such models into the EHR would allow for bedside screening for and enrollment into prospective clinical trials to evaluate for prognostic or therapeutic differences among patients with ARDS. Moreover, such models could more easily capture temporal trends given the rich, abundant data stream in the ICU. By demonstrating the high-performance metrics of the models with EHR-derived data, our study serves as a proof of concept that EHR-embedded machine learning models are feasible for classifying ARDS subphenotypes. If validated prospectively, such EHR-embedded models could provide on-demand decision support for clinicians and/or clinical trials, while limiting disruption to clinical workflow by automatically incorporating clinical data into the models.
The implementation of these models in the clinical setting are, however, contingent on two factors. First, it must be prospectively demonstrated that the models can classify phenotypes robustly and consistently in real-time clinical scenarios in diverse settings. Prior to their clinical implementation, the models will need rigorous evaluation for their interaction with missing data frequently encountered in the real world setting of critical care. Second, it is imperative that a clear clinical utility of the subphenotypes is demonstrated prior to their EHR implementation. Based on its performance in our study, we would advocate the use of the “vital and labs” model for prospective evaluation in future studies. Interestingly, the clinical utility and divergent characteristics of the subphenotypes identified using the sparse model in LUNG SAFE would suggest that a model comprising of even fewer features than the “vitals and lab model” may classify with sufficient accuracy. The development and validation of such parsimonious models requires careful evaluation using the most important variables identified in the “vitals and labs” model, rather than sets of variables constrained by availability, such as in the LUNG SAFE model.
Though model performance was comparable between both observational cohorts, model performance in EARLI was marginally better compared to VALID, potentially due a variety of factors including the timing of enrollment into the studies. Patients were enrolled on the day of hospital admission in EARLI, whereas in VALID enrollment was on day two of ICU admission. Earlier study enrollment may have captured the most extreme physiological characteristics for each patient and higher classification into the Hyperinflammatory subphenotype, but without serial protein biomarker quantification and LCA classification, the temporal kinetics of the subphenotypes remain a key knowledge gap in the field. The longitudinal model performance metrics of the clinical-classifier model requires further studies.
Our findings in LUNG SAFE are consistent with prior studies suggesting that ARDS subphenotypes capture unique information compared to other metrics of ARDS severity, such as PaO2/FiO2 or SOFA score.5,6,10 In our analysis, a treatment interaction was observed between PEEP groups and the subphenotypes with differential responses. Notably, this treatment interaction was consistent with our previous secondary analysis of the ALVEOLI trial that tested the efficacy of high PEEP versus low PEEP in ARDS.5,13 In that analysis, as in this study, high PEEP was associated with improved survival in the Hyperinflammatory subphenotype and worse survival in the Hypoinflammatory subphenotype, albeit the effect size in the latter was clinically insignificant in both studies. The consistent findings across both these studies suggest that there may be value in evaluating PEEP strategies more formally in subphenotype-specific trials with treatment directed by subphenotype. Specifically, in future trials testing high-PEEP strategies, inclusion of the Hypoinflammatory phenotype may lead to a dilution of the effective sample size, rendering the detection of a significant effect less likely.
This study has several strengths. First, the models performed comparably across two observational cohorts with variable inclusion criteria, suggesting model generalizability. Second, the models were able to identify high-risk patients when utilizing inputs automatically extracted from the EHR, showing that biological “signal” can be accurately captured despite the “noise” associated with EHR-derived data. Third, the primary model performed well despite utilizing a parsimonious set of features (only vital signs and laboratory values). This approach could allow future EHR-embedded models to use the “most objective” inputs while excluding features that are dependent on epidemiological factors (e.g., race/ethnicity), or those which are harder to capture in the EHR (e.g., ARDS risk factors and ventilatory variables). The “vitals and labs” model also has the advantage of being potentially applicable in low- and middle-income countries where availability of emerging point-of-care protein biomarker quantification may not be feasible.22 Fourth, unlike our prior studies, this is the first time we have tested the performance of the clinical classifier models and shown the clinical value of ARDS subphenotypes in a cohort consisting of patients derived from low and middle income countries, suggesting their generalizability across healthcare systems.
This study also has several limitations. Due to a lack of availability of predictor variables, we were not able to validate the exact models developed in our prior study.10 The strong correlation of the probabilities generated by the models we presented in this study compared to models in our prior study would, however, suggest that these models are highly overlapping. It is noteworthy that the “full feature” model was trained with the race variable stratified as white and non-white, thereby limiting its generalisability and validity in populations with greater racial or ethnic diversity. However, the “vitals and labs” model without this data also performed well. The EHR-derived cohort was limited by a relatively small sample size and high missingness for some variables. In addition to limiting model validity, the observed missingness, specifically in the EHR cohort, highlights some of the challenges in applying such models prospectively and embedded in the EHR. In LUNG SAFE, fewer features were available, and the tolerance of these models for variable missingness or predictor variable parsimony requires further evaluation. There were several differences in the clinical baseline characteristics of EARLI, VALID, and LUNG SAFE. Most notably, ARDS risk factors, PaO2/FiO2, and bicarbonate levels were substantially different in LUNG SAFE, and taken together with the lack of a comparative gold-standard (LCA-derived subphenotype) to evaluate model performance, the findings of this portion of the study should be interpreted cautiously. Further, interpretation of the findings of treatment interaction with PEEP groups and subphenotypes should also be cautious given that these data are generated from observational data and level of PEEP was not randomly assigned; however, their concordance with our previous findings from randomized PEEP trials is noteworthy and suggests validity. Finally, to date, application of these models has been retrospective, and their validity in real-time clinical settings remains to be tested.
In summary, machine learning classifier models using readily available clinical data accurately assigned inflammatory subphenotypes in observational populations of ARDS. Additionally, the models performed robustly in an EHR-derived observational cohort, suggesting such models can be potentially embedded into an EHR. Finally, the model identified high-risk patients and a treatment interaction between PEEP and inflammatory subphenotype in a large observational cohort without a reference standard of LCA-derived classification, providing further support of the hypothesis that the effect of PEEP may differ in each subphenotype. Application of these models to identify subphenotypes can provide valuable prognostic information linked to distinct biological characteristics and may inform management strategies to test in future clinical trials.
Supplementary Material
Research in context.
Evidence before this study
Using latent class analysis (LCA), two biological acute respiratory distress syndrome (ARDS) subphenotypes – termed “Hypoinflammatory” and “Hyperinflammatory”– have been identified, with distinct clinical and biological features, outcomes, and differential responses to therapy. Clinical use of these subphenotypes, however, is limited by complexity and lack of point of care biomarker assays. To facilitate bedside identification of these subphenotypes, machine learning classifier models using only readily available clinical variables have been developed and validated using data from randomized controlled trials. Performance and clinical utility of these models in observational cohorts of ARDS is not known. No formal literature search was done for this study.
Added value of this study
The presented study demonstrates the validity of machine learning clinical-classifier models in accurately identifying ARDS subphenotypes in two observational cohorts. Differences in biomarkers and clinical outcomes in subphenotypes identified using these models were similar to those in LCA-derived subphenotypes. The models performed comparably when utilizing a dataset comprised of variables automatically extracted from the electronic health record (EHR), suggesting that EHR-embedded models may be feasible. When applied to a large multinational observational cohort of ARDS, the models identified patients at risk for adverse clinical outcomes. The models also identified a treatment interaction with PEEP and subphenotype, with lower mortality observed with higher PEEP in the Hyperinflammatory subphenotype, similar to patterns observed in secondary analyses of the ALVEOLI trial.
Implications of all the available evidence
LCA-derived phenotyping has recently shown promise in identifying homogenous subgroups within larger, heterogenous populations of ARDS. Clinical-classifier models using readily available clinical data can accurately identify these subphenotypes at the bedside and could facilitate prospective, subphenotype-specific trials in ARDS. Response to PEEP may differ on the basis of subphenotype.
Acknowledgements:
We thank Fabiana Madotto, James Anstey, and Nader Najafi for their contributions in data collection, cleaning, and analysis. We thank all patients and researchers who participated in the National Heart Lung and Blood Institute (NHLBI) ARDS Network trials from which data from this study were derived. These include the ALVEOLI, FACTT, and SAILS trials. We acknowledge the contributions of healthcare providers and research staff that enabled the successful completion of these trials. In addition, we thank the contributions of the Biological Specimen and Data Repository Information Coordinating Center of the NHLBI (BIOLINCC) that made the data and biological specimens available to do these studies. This manuscript was prepared using ALVEOLI, ARDSNET, and FACTT Research Materials obtained from the NHLBI Biologic Specimen and Data Repository Information Coordinating Center and does not necessarily reflect the opinions or views of the ALVEOLI, ARDSNET, FACTT or the NHLBI.
Declaration of interests: Dr. Churpek reports grants from NIH/NIDA (R01 DA051464), grants from DOD/PRMRP, W81XWH-21-1-0009, grants from NIH/NIA (R21 AG068720), grants from NIH/NIGMS (R01 GM123193), grants from NIH/ NIDDK (R01 DK126933), grants from EarlySense (Tel Aviv, Isreal), grants from NIH/NHLBI (R01 HL157262) outside the submitted work. In addition, Dr. Churpek has a patent Patent pending (ARCD. P0535US.P2) pending to University of Chicago related to clinical deterioration risk prediction algorithms for hospitalized patients. Dr. Sarma reports grants from National Heart, Lung, and Blood Institute during the conduct of the study. Dr. Matthay reports grants from Roche-Genentec, personal fees from Johnson and Johnson, personal fees from Novartis Pharmaceuticals, personal fees from Gilead Pharmaceuticals, and personal fees from Pliant Therapeutics, outside the submitted work. Dr. Ware reports grants from National Institutes of Health (US), during the conduct of the study; grants and personal fees from Boehringer Ingelheim, grants from Genentech, grants from CSL Behring, personal fees from Merck, personal fees from Citius, personal fees from Quark, and personal fees from Foresee, outside the submitted work. Dr. Laffey reports grants from European Society of Intensive Care Medicine, during the conduct of the study; personal fees from Glaxosmithkline, and personal fees from Baxter, outside the submitted work. Dr. Bellani reports grants and personal fees from Draeger Medical, personal fees from Ge Healthcare, personal fees from Hamilton Medical, and personal fees from Flowmeter SPA, outside the submitted work. Dr. Calfee reports grants from NIH, during the conduct of the study; grants and personal fees from Roche/Genentech, grants and personal fees from Bayer, personal fees from Quark Pharmaceuticals, personal fees from Gen1e Life Sciences, personal fees from Vasomune, and grants from Quantum Leap Healthcare Collaborative, outside the submitted work. The other authors report no disclosures.
Footnotes
Data Sharing: Data from these studies can be provided to others upon reasonable request on approval of a written request to Dr Pratik Sinha. Data from the National Heart Lung and Blood Institute were accessed through the BIOLINCC public repository.
References
- 1.Bellani G, Laffey JG, Pham T, et al. Epidemiology, Patterns of Care, and Mortality for Patients With Acute Respiratory Distress Syndrome in Intensive Care Units in 50 Countries. JAMA 2016; 315(8): 788–800. [DOI] [PubMed] [Google Scholar]
- 2.Thompson BT, Chambers RC, Liu KD. Acute Respiratory Distress Syndrome. N Engl J Med 2017; 377(6): 562–72. [DOI] [PubMed] [Google Scholar]
- 3.Wilson JG, Calfee CS. ARDS Subphenotypes: Understanding a Heterogeneous Syndrome. Crit Care 2020; 24(1): 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matthay MA, Arabi YM, Siegel ER, et al. Phenotypes and personalized medicine in the acute respiratory distress syndrome. Intensive Care Med 2020; 46(12): 2136–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calfee CS, Delucchi K, Parsons PE, et al. Subphenotypes in acute respiratory distress syndrome: latent class analysis of data from two randomised controlled trials. Lancet Respir Med 2014; 2(8): 611–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calfee CS, Delucchi KL, Sinha P, et al. Acute respiratory distress syndrome subphenotypes and differential response to simvastatin: secondary analysis of a randomised controlled trial. Lancet Respir Med 2018; 6(9): 691–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Famous KR, Delucchi K, Ware LB, et al. Acute Respiratory Distress Syndrome Subphenotypes Respond Differently to Randomized Fluid Management Strategy. Am J Respir Crit Care Med 2017; 195(3): 331–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sinha P, Delucchi KL, Chen Y, et al. Latent class analysis-derived subphenotypes are generalisable to observational cohorts of acute respiratory distress syndrome: a prospective study. Thorax 2021: thoraxjnl-2021–217158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sinha P, Delucchi KL, McAuley DF, O’Kane CM, Matthay MA, Calfee CS. Development and validation of parsimonious algorithms to classify acute respiratory distress syndrome phenotypes: a secondary analysis of randomised controlled trials. Lancet Respir Med 2020; 8(3): 247–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sinha P, Churpek MM, Calfee CS. Machine Learning Classifier Models Can Identify Acute Respiratory Distress Syndrome Phenotypes Using Readily Available Clinical Data. Am J Respir Crit Care Med 2020; 202(7): 996–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pais FM, Sinha P, Liu KD, Matthay MA. Influence of Clinical Factors and Exclusion Criteria on Mortality in ARDS Observational Studies and Randomized Controlled Trials. Respir Care 2018; 63(8): 1060–9. [DOI] [PubMed] [Google Scholar]
- 12.Acute Respiratory Distress Syndrome N, Brower RG, Matthay MA, et al. Ventilation with lower tidal volumes as compared with traditional tidal volumes for acute lung injury and the acute respiratory distress syndrome. N Engl J Med 2000; 342(18): 1301–8. [DOI] [PubMed] [Google Scholar]
- 13.Brower RG, Lanken PN, MacIntyre N, et al. Higher versus lower positive end-expiratory pressures in patients with the acute respiratory distress syndrome. N Engl J Med 2004; 351(4): 327–36. [DOI] [PubMed] [Google Scholar]
- 14.National Heart L, Blood Institute Acute Respiratory Distress Syndrome Clinical Trials N, Wiedemann HP, et al. Comparison of two fluid-management strategies in acute lung injury. N Engl J Med 2006; 354(24): 2564–75. [DOI] [PubMed] [Google Scholar]
- 15.National Heart L, Blood Institute ACTN, Truwit JD, et al. Rosuvastatin for sepsis-associated acute respiratory distress syndrome. N Engl J Med 2014; 370(23): 2191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernard GR, Artigas A, Brigham KL, et al. The American-European Consensus Conference on ARDS. Definitions, mechanisms, relevant outcomes, and clinical trial coordination. Am J Respir Crit Care Med 1994; 149(3 Pt 1): 818–24. [DOI] [PubMed] [Google Scholar]
- 17.Kangelaris KN, Prakash A, Liu KD, et al. Increased expression of neutrophil-related genes in patients with early sepsis-induced ARDS. Am J Physiol Lung Cell Mol Physiol 2015; 308(11): L1102–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ware LB, Koyama T, Zhao Z, et al. Biomarkers of lung epithelial injury and inflammation distinguish severe sepsis patients with acute respiratory distress syndrome. Crit Care 2013; 17(5): R253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calfee CS, Eisner MD, Ware LB, et al. Trauma-associated lung injury differs clinically and biologically from acute lung injury due to other clinical disorders. Crit Care Med 2007; 35(10): 2243–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Force ADT, Ranieri VM, Rubenfeld GD, et al. Acute respiratory distress syndrome: the Berlin Definition. JAMA 2012; 307(23): 2526–33. [DOI] [PubMed] [Google Scholar]
- 21.Youden WJ. Index for rating diagnostic tests. Cancer 1950; 3(1): 32–5. [DOI] [PubMed] [Google Scholar]
- 22.Sinha P, Calfee CS, Cherian S, et al. Prevalence of phenotypes of acute respiratory distress syndrome in critically ill patients with COVID-19: a prospective observational study. Lancet Respir Med 2020; 8(12): 1209–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.