The authors identify a molecular signature of breast tumor–associated high endothelial venules, which correlate with presence of tertiary lymphoid structures and survival, suggesting the signature may provide information for treatment selection and determining prognosis.
Abstract
High endothelial venules (HEV) are specialized post-capillary venules that recruit naïve lymphocytes to lymph nodes. HEVs are essential for the development of adaptive immunity. HEVs can also develop in tumors where they are thought to be important for recruiting naïve T cells and B cells into the tumors and locally enhancing antitumor immunity by supporting the formation of tertiary lymphoid structures. Herein, we used comparative transcriptome analysis of human breast cancer to investigate genes differentially expressed between tumor-associated HEVs and the rest of the tumor vasculature. Tumor vessels highly expressing HEV-upregulated genes, such as the homeobox gene MEOX2 and the tetraspanin gene TSPAN7, were associated with extensive infiltration of T and B cells and the occurrence of tertiary lymphoid structures, which is known to predict therapeutic responses to immune-checkpoint inhibitors. Moreover, high transcript counts of these genes in clinical tumor specimens were associated with a significant survival benefit in advanced breast cancer. The molecular signature of HEVs identified herein may be useful for guiding immunotherapies and provides a new direction for investigating tumor-associated HEVs and their clinical significance.
See related Spotlight by Gallimore, p. 371.
Introduction
The development of immune-checkpoint inhibitors has revolutionized cancer treatment as these drugs have remarkable efficacy in some patients. However, only a subset of patients respond to these therapies at present. Overcoming this limitation has become an important challenge in cancer treatment today. A substantial body of clinical evidence indicates that abundant tumor infiltration by effector T cells as well as B cells is a prerequisite for successful immune-checkpoint inhibition therapy (1–6). However, these lymphocytes are largely excluded from many patients’ tumors (7–9). Both T cells and B cells are recruited from the blood circulation, suggesting a critical role of the tumor vasculature in antitumor immunity.
High endothelial venules (HEV) are post-capillary venules specialized for recruiting naïve T cells and B cells from the circulation into secondary lymphoid organs such as lymph nodes, tonsils, and Peyer's patches. Thus, HEVs function as the entry gates for these lymphocytes (10, 11). HEVs are also found at sites of chronic inflammation (12–14) and in malignant tumors (15–17). In many tumors, there are lymph node–like structures called tertiary lymphoid structures (TLS) inside or at the periphery (18). TLS are composed of clusters of accumulating immune cells that contain T cells, B cells, and dendritic cells surrounding HEVs. It is thought that the HEVs serve as gateways for the recruitment of T cells and B cells to the TLS, and that once recruited, naïve lymphocytes become activated locally by tumor antigens. The high density of TLS in tumors correlates with favorable prognosis in various types of cancers, suggesting the existence of antitumor immunity within TLS (16, 18–20). Data from three clinical studies show that the presence of B-cell clusters in TLS predicts a significant response to anti–PD-1 therapy (4–6). Studies of different cancer types have revealed lymphocyte infiltration specifically at HEV-rich areas of the tumors and a strong association between HEV density and T-cell/B-cell densities in these tumors (15, 21). Mature dendritic cells were also reported to be present around these HEVs. High HEV density correlates with favorable clinical parameters such as tumor regression and low invasion levels (15, 21). These clinical observations suggest tumor HEVs have a crucial role in lymphocyte recruitment that is necessary for successful cancer immunotherapy. Supporting this idea, increased HEV formation correlates with improved responses to anti–PD-L1 therapy in a mouse mammary tumor model (22).
The genes responsible for the formation of tumor-associated HEVs are largely unknown. Endothelial cells (EC) of HEVs typically exhibit a tall, plump shape (hence the name “high” endothelial venules), and they are morphologically distinct from the thin, flat ECs typical of other blood vessels. The ECs of HEVs are functionally adapted for facilitating leukocyte entry. Therefore, it is conceivable that the HEVs that develop in tumors express a set of genes that are different from the genes expressed by the rest of the blood vessels in the tumor vasculature. In this study, we tested this hypothesis by comparing the transcriptomes of HEVs and non-HEVs in clinical breast cancer specimens and identified a molecular signature for HEVs within the tumor vasculature.
Materials and Methods
Clinical breast cancer specimens
Informed written consent was obtained from all participants involved in the study, and all clinical investigations were conducted in line with the principles of the Declaration of Helsinki. The use of the cancer specimens was approved by the Institutional Review Board (IRB protocol# 2005-077) at the National Cancer Center Hospital, Japan. Tumor tissue samples were obtained from 101 patients, ages 38 to 71, with untreated stage I–III breast cancer, invasive ductal adenocarcinoma, during surgical resection of the tumors in 2017. The samples were immediately fixed with formalin and paraffin embedded for archiving. Histologic sections of these samples were analyzed.
Immunostaining of tumor and lymphoid tissues
Human tonsils were obtained from tonsillectomies through the Cooperative Human Tissue Network (CHTN) Southern Division, Department of Pathology at Duke University. Human lymph node FFPE sections were purchased from BioChain (cat. #T223416, Lot. C304112). IHC of breast tumor tissues, tonsils, and lymph nodes was performed using Leica BOND RX (Leica Biosystems), Bond Polymer Refine Detection, and Bond Polymer Refine Red Detection. The following primary antibodies were used to detect immune cells: CD8 (4B11, Leica PA0183), CD20 (L26, Leica PA0200), CD3 (LN10, Leica PA0553), Foxp3 (Novus Biological, NB100-39002), and DC-LAMP (1010E1.01, Novus DDX0191P-100). For staining HEVs, we used BLOXALL (Vector Laboratories, SP-6000) blocking solution, MECA-79 (BioLegend 120802, 1:100 dilution), MECA-79 Alexa Fluor 488 conjugate (Santa Cruz Biotech, sc-19602 AF488, 1:100), ImmPRESS-AP Goat Anti-Rat IgG (Vector Laboratories, MP-5444), ImmPACT Vector Red Alkaline Phosphatase (AP) Substrate (Vector Laboratories, SK-5105), and VECTOR Hematoxylin (Vector Laboratories, H-3401). Other antibodies used were specific for TSPAN7 (Sigma-Aldrich AMAB90624 and Thermo Fisher, MA5-24593), ANKRD53 (Sigma-Aldrich, HPA049574), MEOX2 (Sigma-Aldrich, HPA053793), ZNF280C (Sigma-Aldrich, HPA051494), MAdCAM-1 (LSBio, LS-C313201-100), UNC5B (Thermo Fisher, PA5-67631), SEMA3G (Abcam, ab197108), POSTN (Proteintech, 19899–-1-AP), CD31 (Leica Biosystems, PA0250, or Novus Biologicals, NB100-2284), collagen IV (LSBio, LS-B16212-0.1), and laminin (Novus Biological, NB300-144SS). Purified Rabbit Polyclonal Isotype Ctrl Antibody (BioLegend, 910801) and mouse IgG1 isotype control antibody (Cell Signaling Technology, 5415) were used for control staining. F2 antibody to detect the sialyl Lewis X carbohydrate structure was described previously (23). The following Alexa Fluor–conjugated secondary antibodies were obtained from Thermo Fisher Scientific: Alexa Fluor 555 goat anti-rabbit IgG (A21429), Alexa Flour 488 anti-mouse IgG (A11029), Alexa Fluor 647 anti-mouse IgG (A21236), and Alexa Fluor 555 anti-mouse IgG (A32727).
For immunofluorescence staining, FFPE specimens were deparaffinized, and antigen retrieval was performed at 120°C for 20 seconds and 90°C for 15 seconds by Decloaking Chamber (BioCare Medical, DC2002). Citrate Buffer pH 6 (Sigma-Aldrich, C9999) was used for antigen retrieval for TSPAN7, ANKRD53, MEOX2, ZNF280c, UNC5B, SEMA3G, POSTN, MADCAM-1, DC-LAMP, and F2 antibody staining. EDTA Buffer pH 8.5 (Sigma-Aldrich, E1161) was used for collagen IV, Laminin, and MAdCAM-1 antibody staining.
HEVs, CD3, CD8, CD20, and dendritic cells were identified in tumor sections by immunostaining, and the slides were scanned by Aperio Digital Pathology Slide Scanner. HEVs and immune cells were quantified in the scanned images using HALO software with Object Colocalization v1.3 module and CytoNuclear v2.0.5, respectively (Indica Labs). TSPAN7+ or MEOX2+ area was quantified with Area Quantification v2.13. The entire tumor area or region of interest (ROI) was analyzed in each tumor section. Image deconvolution of colorimetric staining was carried out using Deconvolution v1.0.0. module (Indica Labs). Fluorescence pictures were taken by Nikon ECLIPS 90i or Nikon ECLIPS Ti. Confocal images were taken by Nikon A1R confocal microscope.
Laser-capture microdissection and RNA isolation
FFPE breast tumor specimens were sectioned at 7 µm thickness, prepared on membrane slides (ZEISS, 415190-9041-000), and kept at 4°C. The sections were deparaffinized by 5 minutes in xylene two times, and 30 seconds each of 100%, 95%, 70% ethanol, and distilled water. For antigen retrieval, tumor sections were treated with sodium citrate buffer solution pH 4 at 70°C for 15 minutes (ACS HybEZTM Hybridization system). The sections were further incubated for 15 minutes with Animal-Free Blocker (Vector Laboratories, SP-5030) diluted with 200 µL BLOXALL, a peroxidase and AP blocking solution (Vector Laboratories, SP-6000) containing 10 µL of RNase inhibitor (Protector RNase inhibitor, Roche, 03335402001) at room temperature. Anti-CD31 (Novus Bio, NB100–2284) or MECA-79 (BioLegend, 120802) antibody was diluted at 1:20 in 200 µL blocking buffer containing 10 µL of RNase inhibitor and applied to the tissue sections for 20 minutes. After rinsing with PBS for 30 seconds, the secondary antibody (ImmPRESS-AP Goat Anti-Rat IgG, Vector Laboratories, MP-5444) was applied for 15 minutes. The slides were then washed in PBS for 1 minute and treated with ImmPACT Vector Red Alkaline Phosphatase (AP) Substrate (Vector Laboratories, SK-5105) for 2 minutes. The slides were dehydrated by a series of 3 seconds each of PBS, and 100% ethanol 2 times. Immediately after quick drying with compressed air, the slides were used for laser-capture microdissection.
To avoid RNA degradation, one tumor section was stained at a time, and one session of laser-capture microdissection was performed for up to 2 hours. MECA-79 or CD31-positive objects were captured either under bright-field or fluorescence microscopy (ZEISS PALM MicroBeam) with a 20× objective lens. The catapulted tissues were collected into AdhesiveCap opaque (Zeiss). The HEV endothelium was isolated from TLS-rich tumors. The non-HEV endothelium was isolated from either TLS-rich or TLS-free tumors and analyzed separately. Four cases of breast cancer were examined for each tumor type. The total captured tissue area was 220,000–260,000 µm2 for all three types of endothelium.
Total RNA was isolated from the samples using NucleoSpin totalRNA FFPE (Clontech 740969.10). The concentration of isolated total RNA was estimated by smear analysis using an Agilent 2100 Bioanalyzer system, and 5 ng of RNA was used for RNA-seq analysis of each endothelium type. RNA quality was validated by DV200% (percentage of RNA fragments larger than 200 nucleotide-length). RNA samples that yielded DV200% between 42% and 71% were used for library generation.
Library generation
To construct libraries for RNA sequencing (RNA-seq), we used 5 ng total RNA from each microdissected endothelium type from each tumor sample. RNA amplification from FFPE samples was carried out by Single Primer Isothermal Amplification method using NuGEN TRIO RNA-seq Library Preparation kit (Part #0506). The quality and quantity of the libraries were analyzed using the Agilent Bioanalyzer (Agilent Technologies) and Kapa Biosystems qPCR (Sigma-Aldrich). Multiplexed libraries were pooled, and single-end 50 base-pair sequencing was performed on the Illumina HiSeq 2500.
RNA-seq analysis of samples obtained through laser-capture microdissection
Sequence reads were subjected to a variety of pre-and post-alignment quality control measures before being mapped against the reference genome, hg19, using STAR-2.5.3a. Gene-level quantification was determined using HTSeq 0.6.1 by summation of raw counts of reads aligned to the region associated with each gene. The average read count was 47 million reads. Differential expression analysis was performed using DESeq2_1.6.3 with batch correction. Principal component analysis (PCA) was conducted by using significantly differentially expressed genes in the comparison between the ECs of three different vessel types: HEV versus non-HEV vessels in TLS-rich or TLS-free tumors. A heat map was generated for the top 1,000 significantly differentially expressed genes between ECs of the three vessel types.
Comparison of our finding with previous transcriptome analyses of mouse HEV (24–26) was carried out as follows. The single-cell RNA-seq data of mouse lymph node ECs reported by Brulois and colleagues (24) was downloaded from the Gene Expressed Omnibus (GEO) database through the accession number GSE140348. Cell-type clusters and differentially expressed genes (Padj < 0.05) were identified using the FindClusters (resolution = 0.6) and FindMarkers functions of Seurat (V3.2.0) with the default setting. The differentially expressed genes were compared with those of human cancer HEVs from our study. For the comparison with single-cell RNA-seq data reported by Veerman and colleagues (25), the list of genes differentially expressed between homeostatic high endothelial cells (hHEC) and blood EC was downloaded. The genes with P < 0.05 statistical significance were used for comparison. For the comparison with microarray data reported by Lee and colleagues (26), a list of genes that have 1.5-fold or larger difference in the level of expression between high endothelial cells and capillary ECs (CAP EC) was analyzed. The differentially expressed genes commonly found in these previous mouse studies and in the current study were identified by using the LOOKUP function in Microsoft Excel.
Analysis of Human Protein Atlas data
IHC images publicly available at the Human Protein Atlas (proteinatlas.org; ref. 27) were examined to evaluate the expression of proteins of interest in human lymph node as well as breast cancer. Digital histology images of the following protein staining were evaluated: TSPAN7 antibody ID#HPA003140 (lymph nodes n = 3, breast cancer n = 11), MEOX2 antibody ID# HPA053793 (lymph nodes n = 3, breast cancer n = 11), ANKRD53 antibody ID# HPA049574 (lymph node n = 1, breast cancer n = 10), ZNF280C antibody ID # HPA051494 (lymph node n = 2, breast cancer n = 11), UNC5B antibody ID #HPA076687 (lymph node n = 2, breast cancer n = 10), POSTN antibody ID #HPA012306 (lymph node n = 2, breast cancer n = 11), ZFHX4 antibody ID #HPA023837 (lymph node n = 3, breast cancer n = 12), and SEMA3G antibody ID #HPA001761 (lymph node n = 2, breast cancer n = 12).
TCGA data and survival analysis
Genomic Data Commons Data Portal (GDC; https://portal.gdc.cancer.gov) was used to download RNA-seq data and relevant clinical data from The Cancer Genome Atlas Breast Invasive Carcinoma (TCGA-BRCA) data set. Data from 263 patients with stage III or IV breast cancer were analyzed to determine the correlation between survival and the expression of the genes of interest. Patients were classified into two groups based on the fragments per kilobase of transcript per million mapped reads (FPKM) value of each gene, and survival curves were plotted using Kaplan–Meier method as described before (28). Briefly, an FPKM cutoff was set for grouping patients using all FPKM values from the 20th to 80th percentiles. Significant differences between the survival outcomes of the high and low FPKM groups were examined, and the FPKM cutoff yielding the lowest log-rank P value was selected. The analysis was performed using R package survival (https://cran.r-project.org/web/packages/survival/index.html). A correlation matrix of HEV-associated genes and T-cell/B-cell markers was created using the data of 263 stage III and IV breast cancer patients in the TCGA database. Correlations were calculated with the Pearson method using the R stats package.
Statistical analysis for immunostaining
Differential analysis for two-group comparison was performed by Student t test using GraphPad Prism8 for quantitative immunostaining analyses.
Data availability statement
The raw data from the RNA-seq analyses were generated at the Genomic Core of the Sanford Burnham Prebys Medical Discovery Institute. Data analyses were conducted at the Biostatistics and Bioinformatics core of the Moffitt Cancer Center. The RNA-seq data are deposited in NCBI's Gene-Expression Omnibus and are accessible through GEO Series accession number GSE196703.
Results
HEVs are found in breast cancer exhibiting TLS
HEVs are typically found in the TLS that develop in malignant tumors. To investigate the relationship between tumor-associated HEVs and the immune environment in breast cancer, we analyzed 101 archived specimens of untreated invasive breast cancer for the presence of TLS. Histologic examination identified TLS in 49 of the 101 specimens. TLS were found in all subtypes of breast cancer we examined (luminal, HER2+, and triple-negative) regardless of hormone receptor expression, tumor grade, and tumor stage (Supplementary Table S1), but were found more frequently in hormone receptor–negative and triple-negative tumors. In addition, TLS were found more frequently in grade 3 tumors than in grade 1 or grade 2 tumors. The frequency of TLS formation did not appear to be affected by the cancer stage.
We further analyzed the 10 specimens with the most abundant TLS (TLS-rich tumors) and the 10 specimens with no TLS (TLS-free tumors). The pathologic evaluation of these tumors is reported with patient ID numbers in Supplementary Table S2. We then determined the density of HEVs in each sample. HEVs specifically express peripheral node addressin (PNAd), which can be detected using the MECA-79 antibody that binds the carbohydrate epitope 6-sulfo N-acetyllactosamine on extended core 1 O-glycans (29, 30). Immunostaining of FFPE sections for MECA-79 found MECA-79+ HEVs only in the TLS-rich specimens (Fig. 1A and B). We observed considerable heterogeneity in the patterns of MECA-79 staining in tumor HEVs compared with lymph node and tonsil HEVs (Supplementary Fig. S1). The ECs of both lymph node and tonsil HEVs showed prominent accumulation of PNAd (MECA-79) on the luminal surface, whereas weak staining was detected on the abluminal side of lymph node HEVs, indicating polarity. In contrast, MECA-79 staining was found on the luminal and basal sides as well as in the cytoplasm of tumor HEV ECs, whereas some tumor HEVs exhibited mainly luminal MECA-79 staining. We also observed diffused staining of MECA-79 around some tumor HEVs, which may be due to the shedding of PNAd. The basement membrane components, laminin and collagen IV, were detected in tumor HEVs as well as in lymph node and tonsil HEVs (Supplementary Fig. S1).
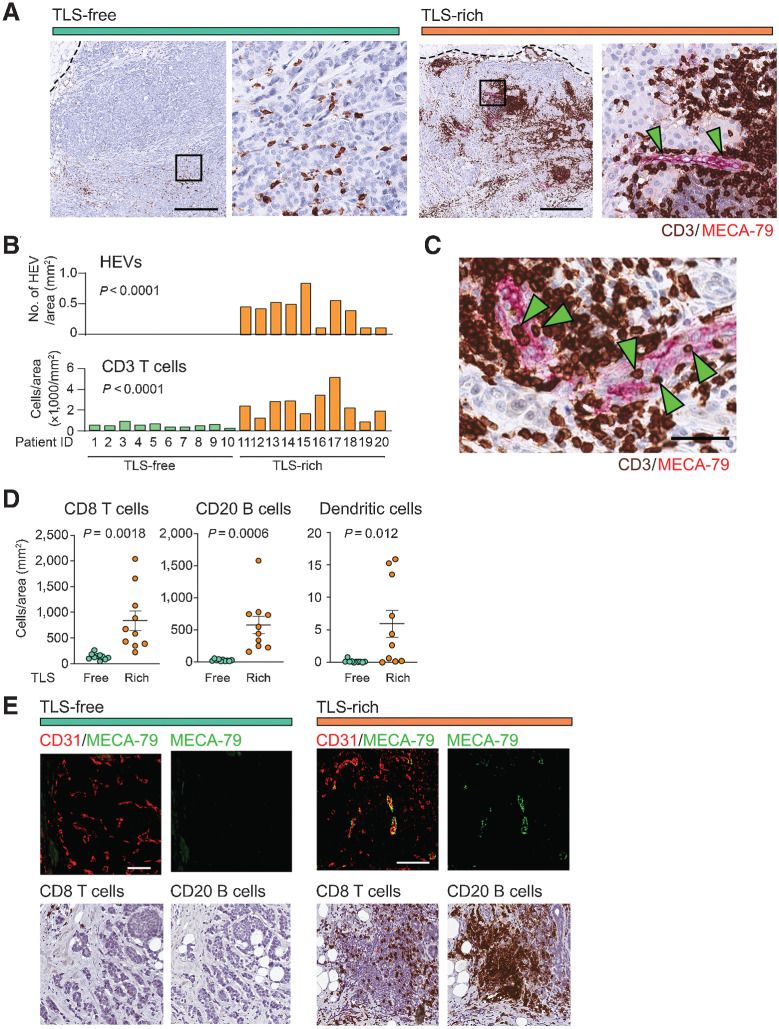
Figure 1.
IHC analyses of tumor HEVs and immune-cell infiltration. A, Immunostaining of TLS-rich and TLS-free tumors for MECA-79 (magenta) and CD3 (dark brown) was performed to determine the abundance of HEVs and T cells. The dotted line shows the edge of each tumor. Arrowheads indicate the MECA-79+ HEVs. The higher magnification of each left panel (black square) is shown on the right. Scale bar, 500 µm. B, HEV and CD3+ T-cell densities were quantified in specimens of 10 patients for each tumor type (TLS-rich and TLS-free). C, Immunostaining of CD3 and MECA-79. Green arrowheads indicate several CD3+ cells on or in the endothelial wall of HEVs. Scale bar, 50 µm. D, Comparison of CD8+ T-cell, CD20+ B-cell, and DC-LAMP+ dendritic-cell densities between the two tumor groups analyzed in B. Error bar; SEM. E, Immunofluorescence detection of HEVs associated with lymphocyte clusters in TLS-rich tumors. Consecutive sections of the same tumors were stained for CD31/MECA-79 (immunofluorescence), CD8, or CD20 (dark brown) to orient the positions of immune cells and blood vessels. A representative TLS is shown here for TLS-rich tumors. CD31, EC marker (red). Scale bar, 100 µm. P value was calculated by Student t test.
Tumor HEVs were observed together with significant infiltration of CD3+ T cells in all 10 cases of TLS-rich breast cancer (Fig. 1A and B). We also observed many CD3+ T cells trapped on or within the endothelial wall of tumor HEVs, suggesting that the extravasation of T cells is taking place through the HEVs (Fig. 1C). The overall abundance of CD3+ T cells was greater in the TLS-rich tumors compared with the TLS-free tumors, which exhibited only sporadic T cells without clustering (Fig. 1A and B). IHC of the same sets of tumors for CD8+ T cells, CD20+ B cells, and DC-LAMP+ dendritic cells, with costaining using MECA-79, demonstrated that the infiltration of these immune cells was significantly greater in TLS-rich tumors than in TLS-free tumors: CD8+ T-cell density, 836 versus 135 cells/mm2; B-cell density, 577 versus 24 cells/mm2; dendritic-cell density 6.63 versus 0.16 cells/mm2 (Fig. 1D and E). These results suggest tumor-associated HEVs have an important role in shaping the tumor immune landscape.
Gene-expression profiling of tumor-associated HEVs
We hypothesized that HEVs in tumors express a unique set of genes, one that is distinct from the genes expressed by non-HEV tumor vessels, and that the expression of these genes confers the specialized function of HEVs as lymphocytes’ gateways to the tumor interior. To identify such genes, we compared the transcriptome of HEV versus non-HEV tumor vessels in clinical breast cancer specimens. The endothelium of each vessel type was isolated by laser-capture microdissection from tumor sections following the identification of MECA-79+ HEVs in TLS and MECA-79–CD31+ non-HEV vessels outside TLS by immunofluorescence staining. The differentially expressed genes in ECs were determined by RNA-seq. Four cases of breast cancer were examined for each tumor blood vessel type (HEVs from TLS-rich tumors and non-HEV vessels from TLS-rich tumors or TLS-free tumors). Each of the four breast cancer cases provided over 220,000 µm2 of microdissected tissue of the HEV or non-HEV endothelium, which were individually used for RNA-seq analyses. Several random areas in TLS outside of HEVs (mostly containing lymphocytes) were also captured by LCM to provide negative control samples representing non-EC types. The RNA-seq yielded 60,658 total transcripts, of which 20,296 encoded genes. The analysis of these genes demonstrated distinct gene-expression patterns between ECs of HEV and non-HEV tumor vessels (Fig. 2A). PCA of the top 1,000 expressed genes showed a clear separation of the HEV cluster from the clusters of non-HEV tumor vessels and non-EC types (Fig. 2B), corroborating that ECs of intratumoral HEVs express a distinct set of genes. In addition, we found a marked difference in the gene-expression patterns between the non-HEV vessels of TLS-rich tumors and TLS-free tumors, suggesting that different blood vessel phenotypes develop in these tumor types (Fig. 2A and B).
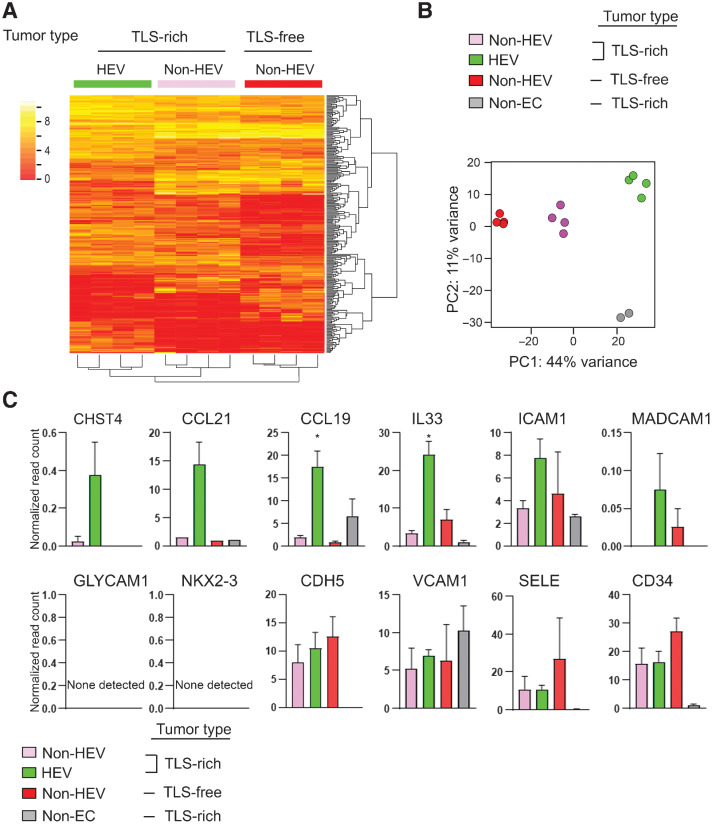
Figure 2.
Comparative transcriptome analysis of HEV vs. non-HEV tumor vessels. Tumors of four breast cancer patients exhibiting abundant TLS (TLS-rich tumors) and four exhibiting no TLS (TLS-free tumors) were examined. TLS-rich tumors were stained for MECA-79 to identify ECs of HEVs, and these ECs were isolated by laser-capture microdissection. ECs of non-HEV vessels were identified by CD31 staining in the HEV-free areas of each tumor type and isolated by laser-capture microdissection. RNA-seq was conducted for all three types of ECs. Non-EC: the areas outside of HEVs (mostly containing lymphocytes of TLS) were also examined in the same way to evaluate potential cell contamination during the laser-capture microdissection process. A, A heat map of the top 1,000 differentially expressed genes between the three different types of ECs. B, PCA shows clear separations of gene-expression patterns between different EC types as well as TLS lymphocytes (non-EC cells). C, Expression patterns of known HEV-associated genes in different EC types. Y-axis, normalized read count × 1/100. Data are presented as means ± SEM. *, P < 0.05.
It is known that CHST4 and IL33 are upregulated in HEVs of lymph nodes (31, 32). CHST4 is a sulfotransferase responsible for the synthesis of the MECA-79 antigen/L-selectin ligand, 6-sulfo-sLex motif (32). IL33, originally known as NF-HEV, is an IL1-like cytokine that also acts as a nuclear factor and is preferentially expressed in HEVs of secondary lymphoid organs (33, 34). Our RNA-seq analysis of the ECs from the different tumor vessel types showed that IL33 was significantly upregulated in tumor HEVs compared with non-HEV tumor vessels (Fig. 2C). There was also a trend of CHST4 upregulation (Fig. 2C). CCL21 and CCL19 are chemokines that facilitate the recruitment of CCR7+ leukocytes such as naïve/memory T cells and dendritic cells (31). These chemokines are abundant in lymph nodes and predominantly produced by fibroblastic reticular cells (35, 36). It has been reported that CCL21 is also expressed by HEVs in mouse lymph nodes (37). We detected high expression of CCL21 and CCL19 in HEVs in human breast cancer (Fig. 2C). GLYCAM1 is reported to be expressed in mouse but not human lymph node HEVs (25, 38). NKX2-3 is a transcription factor highly expressed in HEVs of Payer's patches and mesenteric lymph nodes in mice (26). However, a previous study showed a lack of NKX2-3 expression in mouse peripheral lymph node HEV transcriptome (25). Our RNA-seq analysis showed that neither GLYCAM1 nor NKX2-3 was expressed in intratumoral HEVs of breast cancer (Fig. 2C). A tendency was observed for higher expression of ICAM1 in HEVs than in non-HEV tumor vessels. CD34, E-selectin (SELE), and VE-cadherin (CDH5) were expressed in all three types of tumor vessels. Taken together, some known marker genes of HEVs of secondary lymphoid organs were expressed by tumor HEVs, but other HEV-associated genes were absent in these vessels. These results demonstrate the feasibility of the laser-capture microdissection–RNA-seq method for identifying different gene-expression patterns in HEVs and non-HEV tumor vessels.
Identification of novel HEV-associated genes
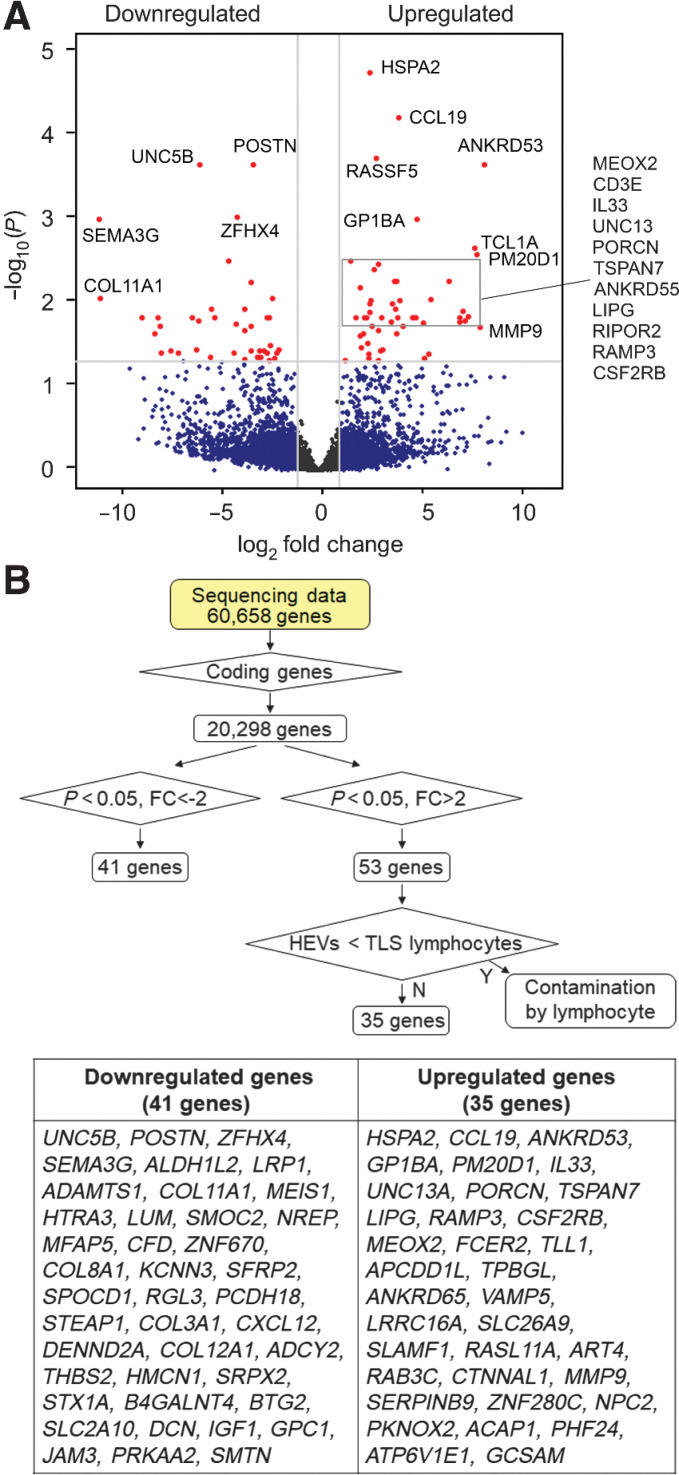
We further analyzed our laser-capture microdissection–RNA-seq data to characterize the gene-expression signature of intratumoral HEVs. The gene-expression profiles of non-HEV tumor vessels from TLS-rich and TLS-free tumors were combined and compared with the profile of HEVs from TLS-rich tumors. Genes with statistically significant >2-fold difference in expression level were identified (Fig. 3A). Because lymphocytes are recruited through HEVs, there may be lymphocytes captured in some laser-dissected HEV samples. Moreover, it is known that HEVs hold transmigrating lymphocytes in the vessel wall (39, 40); therefore, these lymphocytes could cause cell contamination in the laser-capture microdissection process and the subsequent gene-expression analysis. To eliminate data due to lymphocyte contamination, we looked at the expression profile of the non-EC cell types, which mostly consist of lymphocytes (Fig. 3B). We then identified the putative HEV-upregulated genes (genes on the right side of the volcano plot) that are expressed higher in the non-EC samples than in the HEV samples (Fig. 3B). TCF7, TCL1A, and CD3E, for example, met the criteria for such genes. We confirmed that the proteins encoded by these three genes were not expressed in human lymph node HEVs according to IHC data available at the Human Protein Atlas (Supplementary Fig. S2). These genes were eliminated from the final list as potential contaminants (Fig. 3B). After this data processing, we identified 35 statistically significantly upregulated and 41 statistically significantly downregulated genes in HEVs compared with non-HEV tumor vessels. This represents the gene-expression signature of intratumoral HEVs (Fig. 3B).
Figure 3.
Gene-expression signature of intratumoral HEVs. The RNA-seq data generated as outlined for Fig. 2 were further analyzed to identify differentially expressed genes. A, Volcano plot of >2-fold upregulated or downregulated genes in ECs of intratumoral HEVs compared with non-HEV blood vessels of TLS-rich and TLS-free tumors combined. Red dots, genes with adjusted P values (Padj) < 0.05. B, Flowchart of RNA-seq data processing for the identification of differentially upregulated or suppressed genes and a list of 35 upregulated and 41 downregulated genes in intratumoral HEVs compared with non-HEV tumor vessels. FC, fold change; Y, Yes; N, No.
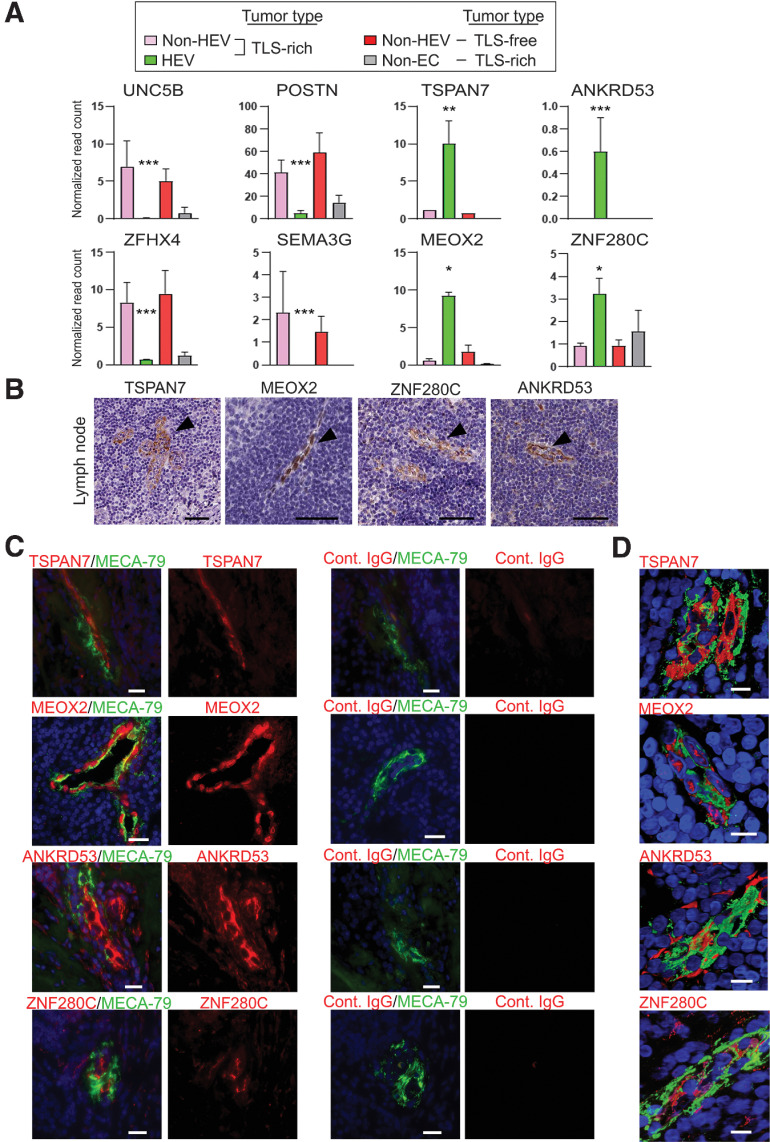
We analyzed the expression of these 76 genes in the IHC data available at the Human Protein Atlas. We found that four of the genes we found to be upregulated in tumor HEVs, MEOX2, TSPAN7, ANKRD53, and ZNF280C (Fig. 4A), were also expressed in HEVs of human lymph nodes (Fig. 4B). We then confirmed the expression of these proteins in HEVs of TLS-rich breast cancer by costaining of MECA-79 (Fig. 4C and D). We examined 10 cases of breast cancer for each tumor type to confirm consistency among patients with the same tumor type. The expression patterns of TSPAN7 and MEOX2 in the TLS-rich and TLS-free breast cancer specimens were further confirmed by IHC staining (Supplementary Fig. S3A–S3C). The majority of MECA-79+ HEVs were costained with TSPAN7 and MEOX2: 96.1% (±1.9 SEM) of MECA-79+ vessels were TSPAN7+, and 95.7% (±2.0 SEM) were MEOX2+. The coexpression of these two proteins in MECA-79+ HEVs was further demonstrated by triple-immunofluorescence staining (Supplementary Fig. S3D). Among the 41 genes significantly downregulated in intratumoral HEVs compared with non-HEV tumor vessels were UNC5B, POSTN, SEMA3G, and ZFHX4 (Fig. 4A). The protein expression of these genes was not detected in tumor-associated HEVs or normal lymph node HEVs, but they were readily detected in non-HEV tumor vessels (Supplementary Fig. S4A and S4B). These results suggest that the expression pattern of these genes—high expression of MEOX2, TSPAN7, ANKRD53, and ZNF280C and the lack or minimal expression of UNC5B, POSTN, SEMA3G, and ZFHX4—represents a molecular signature of HEVs.
Figure 4.
Unique gene-expression pattern of intratumoral HEVs. A, Normalized read counts of eight individual genes are shown. The data were generated using the RNA-seq data generated as outlined for Fig. 2. Adjusted P values for the comparison between the three tumor vessel types (n = 4 for each EC type, n = 2 for non-EC). Y-axis, normalized read count × 1/100. Data are presented as means ± SEM. *, Padj < 0.05; **, Padj < 0.01; ***, Padj < 0.001; error bar: SEM. B, Expression of the proteins encoded by the four genes in A in human lymph nodes is shown in the images obtained from the Human Protein Atlas. Arrowheads, lymph node HEVs. Scale bar, 50 µm. C, Immunofluorescence staining of breast cancer TLS to validate the protein expression of four genes that were found upregulated in intratumoral HEVs by RNA-seq analysis (A). MECA-79 (green), TSPAN7, MEOX2, ANKRD53, or ZNF280C (red), Hoechst nuclear staining (blue). MECA-79+ vessels stained with each isotype-matched control IgG are shown on the right. Ten tumor sections were examined. Scale bar, 50 µm. D, Confocal microscopy to analyze costaining of HEVs with MECA-79 (green) and TSPAN7, MEOX2, ANKRD53, or ZNF280C (red) in 3-D images reconstructed from ≥45 Z-sections. Hoechst (blue). MEOX2 is a transcription factor and shows nuclear staining. Scale bar, 10 µm.
MEOX2 (also known as GAX) is a homeobox gene encoding a transcription factor that suppresses the angiogenic activity of ECs (41–43). Because HEV formation appears to require suppression of angiogenic activation of ECs (22), MEOX2 is an interesting candidate marker for HEVs. Tetraspanins (TSPAN) are known to regulate transendothelial migration of leukocytes (44). Upregulation of TSPAN7 in ECs may be involved in diapedesis of lymphocytes. ANKRD53 is a poorly characterized ankyrin repeat-containing protein (45). ZNF280C is a putative zinc finger transcription factor with an unknown function. On the other hand, UNC5B, POSTN, and SEMA3G encode known angiogenesis activators/regulators (46–48). ZFHX4 is a homeodomain-zinc finger transcription factor involved in neural and muscle differentiation (49) that also regulates the maintenance of tumor-initiating stem cells in glioblastoma (50).
Comparison with previous transcriptome analyses of HEVs
Next, we compared our data with data previously obtained by three independent studies of mouse lymph node HEVs. Lee and colleagues investigated differential gene expression between HEVs and capillary blood vessels in mouse lymph nodes using microarray expression profiling (26), identifying 570 upregulated and 465 downregulated genes in HEVs compared with capillary vessels (Supplementary Fig. S5). The study by Veerman and colleagues determined differentially expressed genes using single-cell RNA-seq (25). This study identified 1,189 upregulated and 368 downregulated genes in HEVs compared with capillary vessels (Supplementary Fig. S5). The study by Brulois and colleagues also used single-cell RNA-seq. According to their data set, which is available at the GEO data repository, there were 235 upregulated and 309 downregulated genes in HEVs (24). We compared these data with the 35 upregulated and 41 downregulated genes from our laser-capture microdissection—RNA-seq analysis. This comparison identified two genes (TSPAN7 and SERPINB9) commonly upregulated and two genes (SEMA3G and CXCL12) commonly downregulated in all four studies (Supplementary Fig. S5B). In this study, we also identified 58 differentially expressed genes (26 upregulated and 32 downregulated genes) in HEV versus non-HEV tumor vessels that were not observed in the studies of mouse lymph node (Supplementary Fig. S5B). These genes included CCL19, ANKRD53, ZNF280C, and ZFHX4. The upregulation of MEOX2 was observed in the microarray analysis (26) and one of the single-cell RNA-seq analyses (24) of mouse lymph node HEVs.
T-cell and B-cell clusters surround tumor vessels expressing HEV signature genes
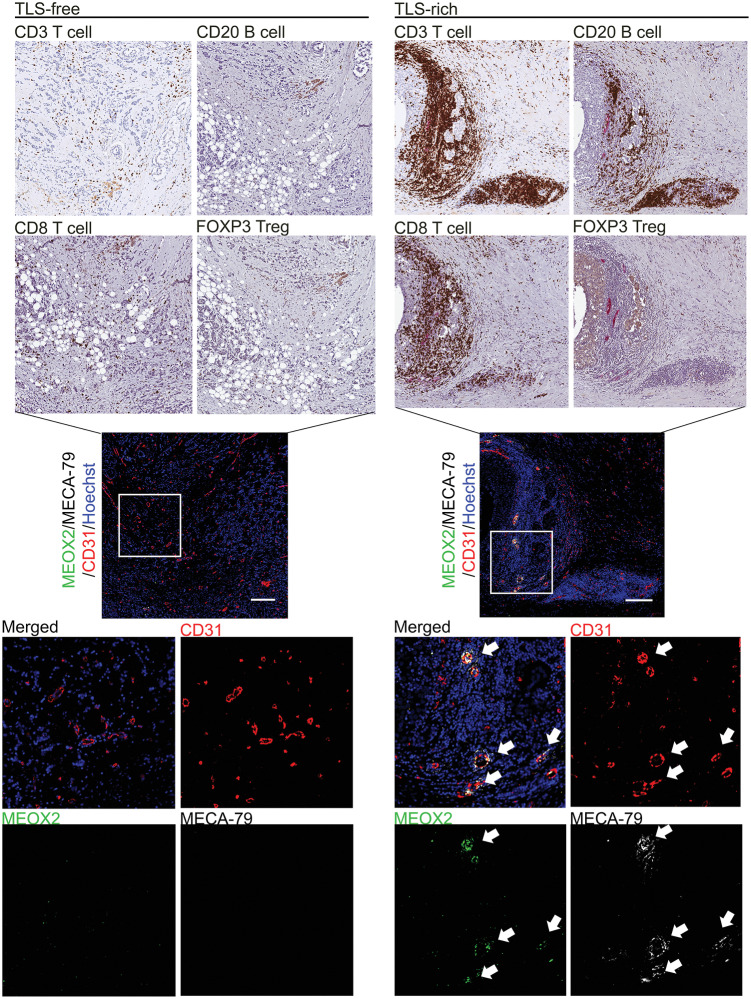
To understand the immune landscape surrounding the blood vessels expressing the HEV-associated genes we identified, we conducted immunostaining of breast cancer tissues for different lymphocyte markers and determined their spatial relationship with the tumor blood vessels expressing these genes. We examined 10 breast cancer specimens for each tumor type to confirm the consistency among the patients with the same tumor type (Supplementary Table S2). We found that MECA-79+ HEVs coexpressing TSPAN7, MEOX2, ANKRD53, and ZNF280C were associated with extensive accumulation of CD3+ and CD8+ T cells as well as B cells in TLS (Figs. 5 and 6A; Supplementary Figs. S6 and S7). In contrast, Foxp3+ Tregs were rarely detected around these vessels in TLS of breast cancer, but they were readily detected in the tonsil, a secondary lymphoid organ (Fig. 5; Supplementary Figs. S6–S8). In the lymph node, cytokines derived from dendritic cells are crucial for the development and function of HEVs (51). In our tumor study, a higher density of dendritic cells was observed in TLS compared with the tumor area outside of TLS (Supplementary Fig. S9), consistent with the idea that cytokines from dendritic cells, such as lymphotoxin β, play an important role in the formation and/or maintenance of tumor-associated HEVs and TLS.
Figure 5.
Immunostaining of MEOX2 identifies tumor HEVs associated with TLS in human breast cancer. IHC staining of different lymphocyte markers was conducted in serial tumor sections to visualize infiltration of T cells, B cells, and Tregs in TLS-free and TLS-rich breast cancer (brown staining, top). Tumor specimens of 10 patients were examined for each tumor type (TLS-rich and TLS-free). These tumor specimens were from the same patients assessed in Fig. 1. Tumor sections were also costained with MECA-79 antibodies (white) to highlight HEVs. The consecutive sections of the same tumors were analyzed by immunofluorescence of MEOX2, MECA-79, and CD31 (center and bottom). Hoechst, nuclear staining (blue). The data shown are for 1 patient representative of each tumor type. Scale bar, 200 µm. Areas in the white squares are enlarged in the bottom plots.
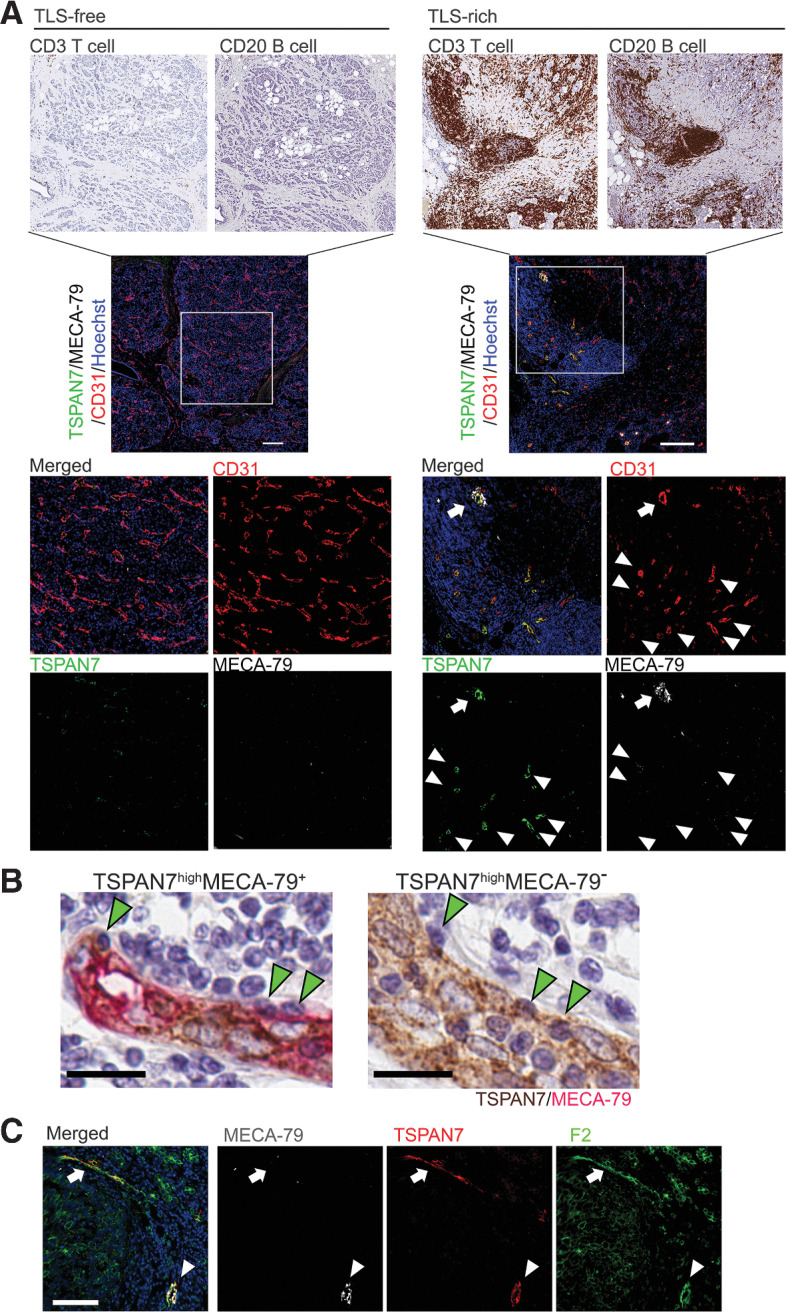
Figure 6.
TSPAN7+MECA-79– tumor vessels are associated with lymphocyte clustering and found in TLS. A, IHC staining of different lymphocyte markers was conducted in serial tumor sections to visualize infiltration of T cells, B cells, and Tregs in TLS-free and TLS-rich breast cancer. Tumor specimens of 10 patients were examined for each tumor type (TLS-rich and TLS-free). These tumor specimens were from the same patients assessed in Fig. 1. The consecutive sections of the same tumors were analyzed by immunofluorescence for TSPAN7, MECA-79, and CD31 (lower panels). TSPAN7+MECA-79+ double-positive HEVs (arrows) and TSPAN7+MECA-79– vessels (arrowheads) are both localized in TLS. For the IHC image of the TLS-free tumor, a lower magnification of Fig. 1E is shown. The data shown are for 1 patient representative of each tumor type. Scale bar, 200 µm. B, Immunostaining of MECA-79 (red) and TSPAN7 (brown) in a TLS-rich breast cancer specimen. Leukocytes engrafted in the endothelial wall of TSPAN7highMECA-79+ and TSPAN7highMECA-79– blood vessels are shown. Scale bar, 20 µm. C, Immunostaining of TSPAN7, MECA-79, and sLeX (by F2 antibody) in TLS-rich tumor sections. Arrows show TSPAN7+MECA-79– vessels detected by the F2 antibody. Arrowhead, TSPAN7+MECA-79+ vessels. A representative image of four TLS-rich tumor samples is shown. Scale bar, 100 µm.
We observed that there were TSPAN7+ vessels that were MECA-79low or MECA-79–. These blood vessels were highly enriched in TLS compared with the stroma or tumor areas of breast cancer specimens (Supplementary Fig. S10). TSPAN7highMECA-79– vessels were more abundant than MECA-79+ vessels in TLS, and they were closely associated with clusters of T cells and B cells (Fig. 6A). In addition, we observed the presence of leukocytes engrafted in the endothelium of these vessels, similar to what was observed for TSPAN7highMECA-79+ HEVs, suggesting that leukocyte transmigration is taking place across the vessel wall (Fig. 6B; Supplementary Fig. S11). The sialyl Lewis X (sLex) tetrasaccharide structure is an L-selectin ligand that allows blood vessels to recruit lymphocytes. sLex is a part of the MECA-79 epitope, but it can also be in glycan configurations that do not contain the MECA-79 epitope (52). Therefore, we assessed sLex expression in TSPAN7+MECA-79+ and TSPAN7+MECA-79– vessels in breast cancer TLS. Both types of vessels were strongly stained by the F2 antibody that recognizes sLex (23), suggesting that these vessels can facilitate lymphocyte homing and recruitment (Fig. 6C).
During embryonic development, immature HEVs express mucosal addressin cell adhesion molecule-1 (MAdCAM-1; ref. 53). We analyzed MAdCAM-1 expression in the TSPAN7+MECA-79– vessels to see whether these vessels are similar to the developmentally immature HEVs. The majority of these vessels were MAdCAM-1–, suggesting that they are different from early HEVs in the developing lymph node. In contrast, some HEVs of the adult lymph node were MAdCAM-1+ (Supplementary Fig. S12), as reported in a single-cell analysis in mice (25).
Expression of MEOX2 and TSPAN7 correlates with prolonged survival in breast cancer
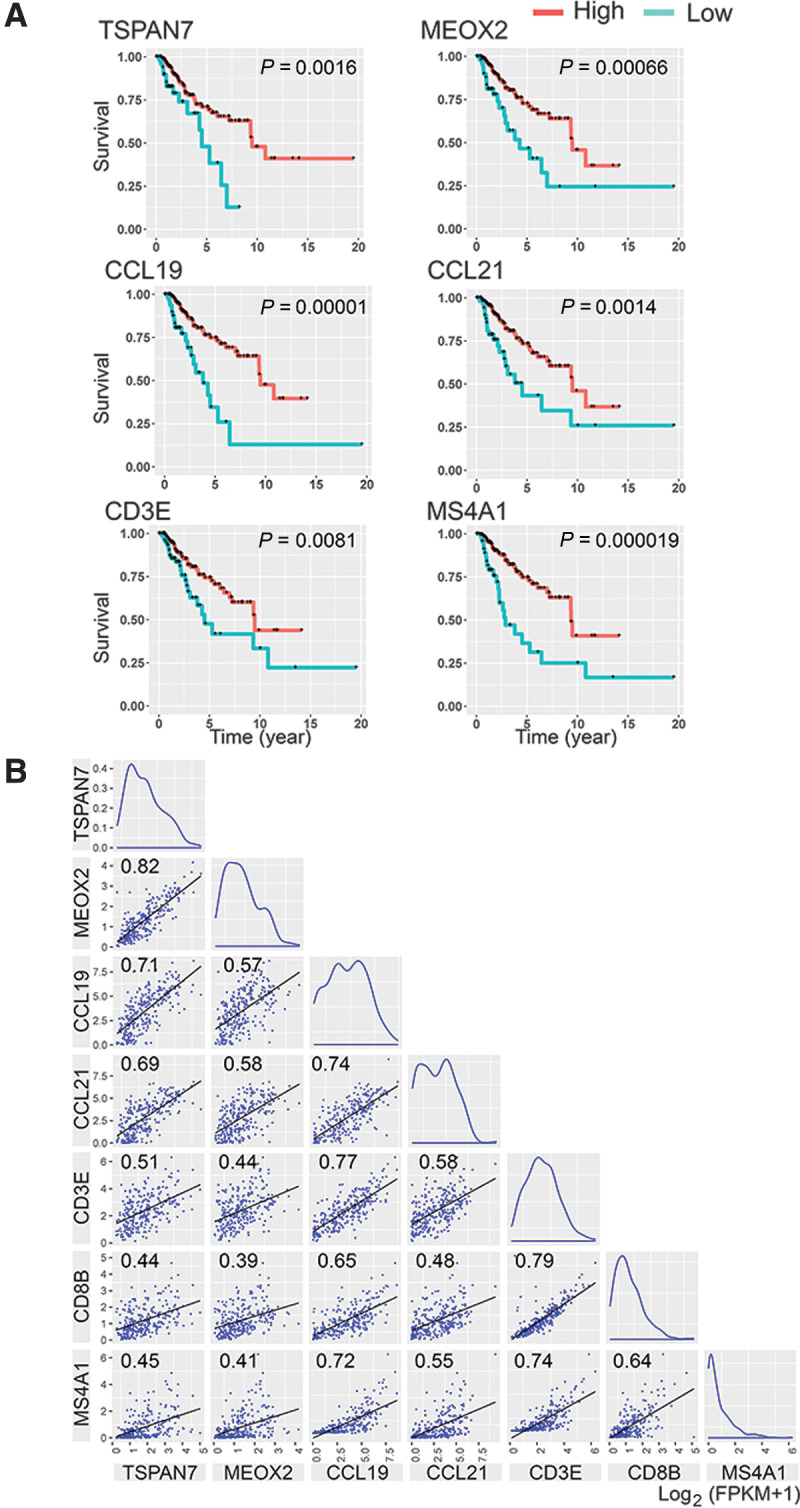
Molecular markers to detect TLS-associated blood vessels in RNA from a patient's tumor may be useful for prognostic purposes. To explore this possibility, we analyzed TCGA data to determine the relationship between breast cancer survival and expression of TSPAN7 or MEOX2. We chose these two genes because we observed high expression of them in the ECs of TLS-associated MECA-79+ and MECA-79– blood vessels, whereas minimal or no expression was found in cancer cells or other stromal cells in breast cancer (Figs. 5 and 6; Supplementary Fig. S10). IHC of several cases of breast cancer presented in the Human Protein Atlas database also indicated no staining of TSPAN7 and MEOX2 in cancer cells and stromal cells supporting this idea. We classified the TCGA survival data of stage III and IV breast cancer (n = 263) into two patient groups based on the abundance of TSPAN7 or MEOX2 transcript determined by bulk RNA-seq analysis. We found a significant survival advantage for the TSPAN7high and MEOX2high patient groups compared with the TSPAN7low and MEOX2low groups (Fig. 7A). The 5-year survival of the TSPAN7high patient group was 71%, compared with 48% for the TSPAN7low group (Fig. 7A). Likewise, the 5-year survival of the MEOX2high group was 72%, compared with 48% for the MEOX2low group (Fig. 7A). Eight-year survival of the TSPAN7high group and MEOX2high group was 63% and 64%, respectively, compared with 13% and 25% for their respective low-expressing groups. These findings suggest that TSPAN7 and MEOX2 could serve as prognostic markers for breast cancer survival. High transcript counts of other HEV-associated genes found in breast cancer, CCL19 and CCL21, also correlated with similar improved breast cancer survival (Fig. 7A). Similar shifts of survival curve were found in the patient group with high transcript counts of CD3E or MS4A1 (CD20 gene), correlating the T-cell or B-cell abundance in tumors with prolonged survival (Fig. 7A).
Figure 7.
Prolonged breast cancer survival is associated with high transcript counts of HEV signature genes in the tumor. A, The survival of 263 stage III and IV breast cancer patients was analyzed using data from TCGA. The survival data were classified into two patient groups based on the abundance of transcript of the gene of interest in each patient's tumor. The gene-expression cutoff for the patient classification was determined as the FPKM value of the gene that yields maximal separation of survival curves between the two patient groups at the lowest P value. The cutoff FPKM for each gene was: TSPAN7 = 0.88, MEOX2 = 0.52, CCL19 = 2.07, CCL21 = 0.96, CD3E = 2.3, and MS4A1 = 0.06. B, A correlation matrix of HEV-associated genes and lymphocyte markers. The correlation matrix of five HEV-associated genes and T-cell/B-cell markers was created using the bulk RNA-seq data of 263 breast cancer patients in TCGA database. The correlation coefficient of each comparison is indicated in the graph. The X and Y axes indicate mRNA levels of the genes described as log2 (FPKM + 1). P < 1×10–8 for all correlations.
We next used the TCGA data to examine whether the abundance of the transcripts of these genes in tumor RNA correlated with the abundance of T cells and B cells. Plotting the transcript counts of two genes in each tumor sample onto X–Y axes demonstrated a strong linear correlation between TSPAN7 and MEOX2 (correlation coefficient: 0.823), consistent with the observation that both genes are upregulated in HEVs (Fig. 7B). In addition, these two genes also correlated with other tumor HEV-upregulated genes, CCL19 and CCL21 (correlation coefficient: 0.713 and 0.686 with TSPAN7, respectively; 0.574 and 0.583 with MEOX2, respectively). TSPAN7 and MEOX2 also showed statistically significant correlations with CD3E, CD8B, and MS4A1 (0.507, 0.438, and 0.446, respectively), suggesting correlations with the T-cell and B-cell abundance in each tumor (Fig. 7B). These results are consistent with our histologic observations that the expression of MEOX2 and/or TSPAN7 in tumor vessels coincided with T-cell and B-cell accumulation around these vessels (Figs. 5 and 6). CCL19 and CCL21 also correlated with T-cell and B-cell markers (Fig. 7B). In particular, CCL19 strongly correlated with CD3E and MS4A1. On the other hand, CHST4, which is the gene responsible for the MECA-79 epitope, showed no correlation with tumor HEV markers or lymphocyte markers, likely due to its low FPKM values detected in the tumors. Thus, we conclude that CHST4 is not a suitable marker for predicting HEV formation from tumor RNA analyses.
Discussion
At present, the molecular characteristics of tumor-associated HEVs are largely unknown. In this study, we took an unbiased approach of comparative transcriptome analysis for identifying the gene-expression signature of these blood vessels. We found genes that were differentially regulated between ECs of TLS-associated HEVs and ECs of non-HEV tumor blood vessels outside TLS. Among 35 genes significantly upregulated in TLS-associated HEVs, at least four genes, TSPAN7, MEOX2, ANKRD53, and ZNF280C, were also found to be upregulated in HEVs of the tonsil and lymph nodes compared with capillary vessels in these secondary lymphoid organs. Immunostaining of the corresponding proteins detected HEVs within or closely associated with TLS in clinical specimens of invasive breast cancer.
TSPAN7 is a member of the tetraspanin family of proteins, which compartmentalize plasma membrane microdomains to regulate various cellular signaling and functions (54, 55). In addition, tetraspanins are involved in tension control of the plasma membrane (56) and endocytosis of viral particles (57). ECs express several tetraspanins including TSPAN5, TSPAN17, CD9, CD63, CD81, and CD151. Lymphocyte migration across the blood vessel wall during diapedesis involves a dynamic regulation of the EC plasma membrane. Tetraspanins are known to contribute to the formation of adhesive platforms during transendothelial migration of leukocytes (44). The finding that TSPAN7 is highly upregulated in TLS-associated blood vessels suggests a role for this tetraspanin in diapedesis of lymphocytes and the function of these blood vessels as the gateways for lymphocyte recruitment. The homeobox gene MEOX2 encodes a transcription factor that negatively regulates the angiogenic activity of ECs (41) and upregulates the expression of cyclin kinase inhibitor p21WAF1/CIP1 in ECs (43). On the other hand, the expression of MEOX2 is negatively regulated by an epithelial–mesenchymal transition mediator ZEB2 in ECs (42). It is thought that suppression of angiogenic stimuli in the tumor microenvironment is important for HEV formation, as exemplified by the observations that VEGF receptor-2 inhibition promotes HEV formation in mouse mammary tumors (22), and tumor vascular normalization is associated with TLS formation in pancreatic cancer (16). This idea is consistent with our observation that an antiangiogenic transcription factor, MEOX2, is highly upregulated in tumor HEVs.
ANKRD53 is an ankyrin repeat-containing protein (45), and ZNF280C is a putative zinc finger transcription factor. There is one report that ANKRD53 is a microtubule-associated protein involved in microtubule stabilization during mitosis (45). However, the functions of ANKRD53 and ZNF280C are both unknown otherwise, at present. Neither ANKRD53 nor ZNF280C was detected as an HEV-upregulated gene in the previous single-cell analyses or microarray analysis of mouse lymph node HEVs (24–26). In contrast, immunostaining of clinical breast cancer specimens and human lymph nodes both indicated the upregulation of these genes in HEVs. The roles of ANKRD53 and ZNF280C in HEVs warrant future studies.
We also identified 41 genes that were significantly downregulated in ECs of HEV in comparison with non-HEV tumor vessels. Some of these genes are known regulators of angiogenesis such as UNC5B, SEMA3G, and DCN. UNC5B is a vascular receptor of netrin that induces cell repulsion (46). SEMA3G is a class 3 semaphorin and a neuropilin ligand that regulates ECs and smooth muscle cell functions (48). DCN is a TGFβ-regulating proteoglycan decorin (58). The strong downregulation of these genes in HEVs suggests that repression of these genes in ECs may be necessary for the formation, maintenance, and/or function of HEVs. Compared with ECs of other venules, ECs of HEVs have markedly different cell morphology and are specialized for facilitating T-cell and B-cell extravasation. At present, the pattern of EC gene expression that allows tumor blood vessels to become HEVs or create new HEVs de novo is unknown. However, the gene-expression signature we identified in this study may hold a critical set of genes that need to be expressed or repressed. In this regard, it is interesting that we found transcription factors such as MEOX2, ZNF280C, ZFHX4, and ZNF670 were differentially regulated between HEVs versus non-HEV tumor vessels. These HEV-associated transcription factors may also control the expression of other HEV-associated genes. Future studies will determine the precise role of these genes in the regulation of HEVs.
At present, the detection of TLS or HEVs in patients’ tumors requires histologic examination. A molecular detection method would be useful for the prognostic assessment of cancer. Our study suggests that the abundance of TSPAN7 and MEOX2 transcripts in bulk tumor RNA from clinical specimens can be used for predicting breast cancer survival. Other marker genes we identified in tumor HEVs, such as CCL19, also correlated with breast cancer survival. The chemokines CCL19 and CCL21 are crucial for the recruitment of naïve T cells. However, a previous in situ hybridization study demonstrated the lack of CCL19 mRNA expression in HEVs of the tonsils (59), and there has been no clear evidence for CCL19 expression in lymph node HEVs (60). Instead, fibroblastic reticular cells provide CCL19 for T-cell chemotaxis in these lymphoid organs (61). Therefore, it is interesting that we found ECs of HEVs upregulate CCL19 expression in tumor TLS, presumably to promote naïve T-cell recruitment. Although CHST4 was upregulated in HEVs, its expression level was 25- to 40-fold lower than the levels of TSPAN7, MEOX2, and CCL19, and it is not suitable for a similar correlative analysis to predict cancer survival.
TSPAN7 is particularly interesting because high TSPAN7 protein expression was also found in many TLS-associated MECA-79– vessels. These blood vessels lack the MECA-79 epitope, the classic HEV marker PNAd, 6-sulfo N-acetyllactosamine on the extended core 1 O-glycan (30). However, the L-selectin ligand 6-sulfo sLex structure can also form on the core 2 O-glycan without the MECA-79 epitope (29, 30). We found that TSPAN7highMECA-79– vessels were positive for sLex expression, and they appeared to be actively recruiting T cells and B cells based on the extensive accumulation of lymphocytes associated with them and the presence of lymphocytes in the endothelial wall. These findings suggest heterogeneity of blood vessels in TLS that are capable of active lymphocyte recruitment. This is an important observation as clinical studies have demonstrated that the presence of B cell–rich TLS is the strongest prognostic factor for a high response rate to PD-1 blockade (4–6). In addition, studies in mice show that the induction of MECA-79+ (PNAd+) HEV-like blood vessels enhances tumor immunity (62, 63). However, the classic PNAd+ HEVs may not be the sole key players of the TLS vasculature. The precise role of the TLS-associated PNAd−TSPAN7high vessels in tumor immunity and cancer immunotherapy warrants further investigation.
Predicting the abundance of TLS-associated blood vessels from tumor RNA based on TSPAN7 and MEOX2 expression would give a considerable advantage in analyzing clinical specimens. Overall, our findings provide potential new prognostic molecular markers, which may be useful in selecting appropriate therapies for patients with breast cancer.
Supplementary Material
Acknowledgments
The Shared Resources and Research Support of Johns Hopkins All Children's Research Center provided Leica BOND RX for IHC. RNA-seq was performed at Sanford Burnham Prebys Medical Discovery Institute Genomic Core. The RNA-seq data analyses were conducted at the Biostatistics and Bioinformatics core of the Moffitt Cancer Center. This work was supported by the NIH, NCI (R01CA125255), Florida Department of Health (20B01), and the Florida Breast Cancer Foundation.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Immunology Research Online (http://cancerimmunolres.aacrjournals.org/).
Authors' Disclosures
M. Komatsu reports grants from NIH, Florida Department of Health, and Florida Breast Cancer Foundation during the conduct of the study; other support from Vascular Biosciences Pharmaceuticals outside the submitted work; in addition, M. Komatsu has a patent for US20190022170A1 granted to Vascular Biosciences. No disclosures were reported by the other authors.
Authors' Contributions
J. Sawada: Conceptualization, data curation, formal analysis, validation, investigation, visualization, methodology, writing–original draft. N. Hiraoka: Resources, formal analysis, investigation. R. Qi: Data curation, visualization. L. Jiang: Data curation, visualization. A.E. Fournier-Goss: Formal analysis, validation, investigation. M. Yoshida: Resources, investigation. H. Kawashima: Resources. M. Komatsu: Conceptualization, supervision, funding acquisition, project administration, writing–review and editing, formal analysis, investigation, visualization, methodology.
References
- 1. Sunshine J, Taube JM. PD-1/PD-L1 inhibitors. Curr Opin Pharmacol 2015;23:32–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lipson EJ, Forde PM, Hammers HJ, Emens LA, Taube JM, Topalian SL. Antagonists of PD-1 and PD-L1 in cancer treatment. Semin Oncol 2015;42:587–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cimino-Mathews A, Thompson E, Taube JM, Ye X, Lu Y, Meeker A, et al. PD-L1 (B7-H1) expression and the immune tumor microenvironment in primary and metastatic breast carcinomas. Hum Pathol 2016;47:52–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Helmink BA, Reddy SM, Gao J, Zhang S, Basar R, Thakur R, et al. B cells and tertiary lymphoid structures promote immunotherapy response. Nature 2020;577:549–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Petitprez F, de Reynies A, Keung EZ, Chen TW, Sun CM, Calderaro J, et al. B cells are associated with survival and immunotherapy response in sarcoma. Nature 2020;577:556–60. [DOI] [PubMed] [Google Scholar]
- 6. Cabrita R, Lauss M, Sanna A, Donia M, Skaarup Larsen M, Mitra S, et al. Tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature 2020;577:561–5. [DOI] [PubMed] [Google Scholar]
- 7. Buckanovich RJ, Facciabene A, Kim S, Benencia F, Sasaroli D, Balint K, et al. Endothelin B receptor mediates the endothelial barrier to T cell homing to tumors and disables immune therapy. Nat Med 2008;14:28–36. [DOI] [PubMed] [Google Scholar]
- 8. Joyce JA, Fearon DT. T cell exclusion, immune privilege, and the tumor microenvironment. Science 2015;348:74–80. [DOI] [PubMed] [Google Scholar]
- 9. Chiba T, Ohtani H, Mizoi T, Naito Y, Sato E, Nagura H, et al. Intraepithelial CD8+ T-cell-count becomes a prognostic factor after a longer follow-up period in human colorectal carcinoma: possible association with suppression of micrometastasis. Br J Cancer 2004;91:1711–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sawada J, Perrot CY, Chen L, Fournier-Goss AE, Oyer J, Copik A, et al. High endothelial venules accelerate naive T cell recruitment by tumor necrosis factor-mediated R-Ras upregulation. Am J Pathol 2021;191:396–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Anderson AO, Anderson ND. Studies on the structure and permeability of the microvasculature in normal rat lymph nodes. Am J Pathol 1975;80:387–418. [PMC free article] [PubMed] [Google Scholar]
- 12. Ruddle NH. High endothelial venules and lymphatic vessels in tertiary lymphoid organs: characteristics, functions, and regulation. Front Immunol 2016;7:491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shomer NH, Fox JG, Juedes AE, Ruddle NH. Helicobacter-induced chronic active lymphoid aggregates have characteristics of tertiary lymphoid tissue. Infect Immun 2003;71:3572–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shipman WD, Dasoveanu DC, Lu TT. Tertiary lymphoid organs in systemic autoimmune diseases: pathogenic or protective? F1000Res 2017;6:196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Martinet L, Garrido I, Filleron T, Le Guellec S, Bellard E, Fournie JJ, et al. Human solid tumors contain high endothelial venules: association with T- and B-lymphocyte infiltration and favorable prognosis in breast cancer. Cancer Res 2011;71:5678–87. [DOI] [PubMed] [Google Scholar]
- 16. Hiraoka N, Ino Y, Yamazaki-Itoh R, Kanai Y, Kosuge T, Shimada K. Intratumoral tertiary lymphoid organ is a favourable prognosticator in patients with pancreatic cancer. Br J Cancer 2015;112:1782–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Avram G, Sanchez-Sendra B, Martin JM, Terradez L, Ramos D, Monteagudo C. The density and type of MECA-79-positive high endothelial venules correlate with lymphocytic infiltration and tumour regression in primary cutaneous melanoma. Histopathology 2013;63:852–61. [DOI] [PubMed] [Google Scholar]
- 18. Hiraoka N, Ino Y, Yamazaki-Itoh R. Tertiary lymphoid organs in cancer tissues. Front Immunol 2016;7:244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Di Caro G, Bergomas F, Grizzi F, Doni A, Bianchi P, Malesci A, et al. Occurrence of tertiary lymphoid tissue is associated with T-cell infiltration and predicts better prognosis in early-stage colorectal cancers. Clin Cancer Res 2014;20:2147–58. [DOI] [PubMed] [Google Scholar]
- 20. Bruno TC. New predictors for immunotherapy responses sharpen our view of the tumour microenvironment. Nature 2020;577:474–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Martinet L, Le Guellec S, Filleron T, Lamant L, Meyer N, Rochaix P, et al. High endothelial venules (HEVs) in human melanoma lesions: major gateways for tumor-infiltrating lymphocytes. Oncoimmunology 2012;1:829–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Allen E, Jabouille A, Rivera LB, Lodewijckx I, Missiaen R, Steri V, et al. Combined antiangiogenic and anti-PD-L1 therapy stimulates tumor immunity through HEV formation. Sci Transl Med 2017;9:eaak9679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Matsumura R, Hirakawa J, Sato K, Ikeda T, Nagai M, Fukuda M, et al. Novel antibodies reactive with Sialyl Lewis X in both humans and mice define its critical role in leukocyte trafficking and contact hypersensitivity responses. J Biol Chem 2015;290:15313–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brulois K, Rajaraman A, Szade A, Nordling S, Bogoslowski A, Dermadi D, et al. A molecular map of murine lymph node blood vascular endothelium at single cell resolution. Nat Commun 2020;11:3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Veerman K, Tardiveau C, Martins F, Coudert J, Girard JP. Single-cell analysis reveals heterogeneity of high endothelial venules and different regulation of genes controlling lymphocyte entry to lymph nodes. Cell Rep 2019;26:3116–31. [DOI] [PubMed] [Google Scholar]
- 26. Lee M, Kiefel H, LaJevic MD, Macauley MS, Kawashima H, O'Hara E, et al. Transcriptional programs of lymphoid tissue capillary and high endothelium reveal control mechanisms for lymphocyte homing. Nat Immunol 2014;15:982–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ponten F, Jirstrom K, Uhlen M. The Human Protein Atlas–a tool for pathology. J Pathol 2008;216:387–93. [DOI] [PubMed] [Google Scholar]
- 28. Uhlen M, Zhang C, Lee S, Sjostedt E, Fagerberg L, Bidkhori G, et al. A pathology atlas of the human cancer transcriptome. Science 2017;357:eaan2507. [DOI] [PubMed] [Google Scholar]
- 29. Hiraoka N, Petryniak B, Nakayama J, Tsuboi S, Suzuki M, Yeh JC, et al. A novel, high endothelial venule-specific sulfotransferase expresses 6-sulfo sialyl Lewis(x), an L-selectin ligand displayed by CD34. Immunity 1999;11:79–89. [DOI] [PubMed] [Google Scholar]
- 30. Yeh JC, Hiraoka N, Petryniak B, Nakayama J, Ellies LG, Rabuka D, et al. Novel sulfated lymphocyte homing receptors and their control by a Core1 extension beta 1,3-N-acetylglucosaminyltransferase. Cell 2001;105:957–69. [DOI] [PubMed] [Google Scholar]
- 31. Ager A, May MJ. Understanding high endothelial venules: lessons for cancer immunology. Oncoimmunology 2015;4:e1008791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kawashima H, Petryniak B, Hiraoka N, Mitoma J, Huckaby V, Nakayama J, et al. N-acetylglucosamine-6-O-sulfotransferases 1 and 2 cooperatively control lymphocyte homing through L-selectin ligand biosynthesis in high endothelial venules. Nat Immunol 2005;6:1096–104. [DOI] [PubMed] [Google Scholar]
- 33. Baekkevold ES, Roussigne M, Yamanaka T, Johansen FE, Jahnsen FL, Amalric F, et al. Molecular characterization of NF-HEV, a nuclear factor preferentially expressed in human high endothelial venules. Am J Pathol 2003;163:69–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Carriere V, Roussel L, Ortega N, Lacorre DA, Americh L, Aguilar L, et al. IL-33, the IL-1-like cytokine ligand for ST2 receptor, is a chromatin-associated nuclear factor in vivo. Proc Nat Acad Sci USA 2007;104:282–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Miyasaka M, Tanaka T. Lymphocyte trafficking across high endothelial venules: dogmas and enigmas. Nat Rev Immunol 2004;4:360–70. [DOI] [PubMed] [Google Scholar]
- 36. Girard JP, Moussion C, Forster R. HEVs, lymphatics and homeostatic immune cell trafficking in lymph nodes. Nat Rev Immunol 2012;12:762–73. [DOI] [PubMed] [Google Scholar]
- 37. Gunn MD, Tangemann K, Tam C, Cyster JG, Rosen SD, Williams LT. A chemokine expressed in lymphoid high endothelial venules promotes the adhesion and chemotaxis of naive T lymphocytes. Proc Nat Acad Sci USA 1998;95:258–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dowbenko D, Andalibi A, Young PE, Lusis AJ, Lasky LA. Structure and chromosomal localization of the murine gene encoding GLYCAM 1. A mucin-like endothelial ligand for L selectin. J Biol Chem 1993;268:4525–9. [PubMed] [Google Scholar]
- 39. Mionnet C, Sanos SL, Mondor I, Jorquera A, Laugier JP, Germain RN, et al. High endothelial venules as traffic control points maintaining lymphocyte population homeostasis in lymph nodes. Blood 2011;118:6115–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Freemont AJ, Jones CJ. Light microscopic, histochemical and ultrastructural studies of human lymph node paracortical venules. J Anat 1983;136:349–62. [PMC free article] [PubMed] [Google Scholar]
- 41. Gorski DH, Leal AJ. Inhibition of endothelial cell activation by the homeobox gene Gax. J Surg Res 2003;111:91–9. [DOI] [PubMed] [Google Scholar]
- 42. Chen Y, Banda M, Speyer CL, Smith JS, Rabson AB, Gorski DH. Regulation of the expression and activity of the antiangiogenic homeobox gene GAX/MEOX2 by ZEB2 and microRNA-221. Mol Cell Biol 2010;30:3902–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen Y, Leal AD, Patel S, Gorski DH. The homeobox gene GAX activates p21WAF1/CIP1 expression in vascular endothelial cells through direct interaction with upstream AT-rich sequences. J Biol Chem 2007;282:507–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yeung L, Hickey MJ, Wright MD. The many and varied roles of tetraspanins in immune cell recruitment and migration. Front Immunol 2018;9:1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kim S, Jang CY. ANKRD53 interacts with DDA3 and regulates chromosome integrity during mitosis. Biochem Biophys Res Commun 2016;470:484–91. [DOI] [PubMed] [Google Scholar]
- 46. Navankasattusas S, Whitehead KJ, Suli A, Sorensen LK, Lim AH, Zhao J, et al. The netrin receptor UNC5B promotes angiogenesis in specific vascular beds. Development 2008;135:659–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liu Y, Li F, Gao F, Xing L, Qin P, Liang X, et al. Periostin promotes tumor angiogenesis in pancreatic cancer via Erk/VEGF signaling. Oncotarget 2016;7:40148–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kutschera S, Weber H, Weick A, De Smet F, Genove G, Takemoto M, et al. Differential endothelial transcriptomics identifies semaphorin 3G as a vascular class 3 semaphorin. Arterioscler Thromb Vasc Biol 2011;31:151–9. [DOI] [PubMed] [Google Scholar]
- 49. Hemmi K, Ma D, Miura Y, Kawaguchi M, Sasahara M, Hashimoto-Tamaoki T, et al. A homeodomain-zinc finger protein, ZFHX4, is expressed in neuronal differentiation manner and suppressed in muscle differentiation manner. Biol Pharm Bull 2006;29:1830–5. [DOI] [PubMed] [Google Scholar]
- 50. Chudnovsky Y, Kim D, Zheng S, Whyte WA, Bansal M, Bray MA, et al. ZFHX4 interacts with the NuRD core member CHD4 and regulates the glioblastoma tumor-initiating cell state. Cell Rep 2014;6:313–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Moussion C, Girard JP. Dendritic cells control lymphocyte entry to lymph nodes through high endothelial venules. Nature 2011;479:542–6. [DOI] [PubMed] [Google Scholar]
- 52. Ivetic A, Hoskins Green HL, Hart SJ. L-selectin: a major regulator of leukocyte adhesion, migration and signaling. Front Immunol 2019;10:1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mebius RE, Streeter PR, Michie S, Butcher EC, Weissman IL. A developmental switch in lymphocyte homing receptor and endothelial vascular addressin expression regulates lymphocyte homing and permits CD4+ CD3− cells to colonize lymph nodes. Proc Nat Acad Sci USA 1996;93:11019–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Termini CM, Gillette JM. Tetraspanins function as regulators of cellular signaling. Front Cell Dev Biol 2017;5:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Singethan K, Tetraspanins Jürgen SS.: Small transmembrane proteins with big impact on membrane microdomain structures. Commun Integr Biol 2008;1:11–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Navarro-Hernandez IC, Lopez-Ortega O, Acevedo-Ochoa E, Cervantes-Diaz R, Romero-Ramirez S, Sosa-Hernandez VA, et al. Tetraspanin 33 (TSPAN33) regulates endocytosis and migration of human B lymphocytes by affecting the tension of the plasma membrane. FEBS J 2020;287:3449–71. [DOI] [PubMed] [Google Scholar]
- 57. Florin L, Lang T. Tetraspanin assemblies in virus infection. Front Immunol 2018;9:1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yamaguchi Y, Mann DM, Ruoslahti E. Negative regulation of transforming growth factor-beta by the proteoglycan decorin. Nature 1990;346:281–4. [DOI] [PubMed] [Google Scholar]
- 59. Baekkevold ES, Yamanaka T, Palframan RT, Carlsen HS, Reinholt FP, von Andrian UH, et al. The CCR7 ligand elc (CCL19) is transcytosed in high endothelial venules and mediates T cell recruitment. J Exp Med 2001;193:1105–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Berendam SJ, Koeppel AF, Godfrey NR, Rouhani SJ, Woods AN, Rodriguez AB, et al. Comparative transcriptomic analysis identifies a range of immunologically related functional elaborations of lymph node associated lymphatic and blood endothelial cells. Front Immunol 2019;10:816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Link A, Vogt TK, Favre S, Britschgi MR, Acha-Orbea H, Hinz B, et al. Fibroblastic reticular cells in lymph nodes regulate the homeostasis of naive T cells. Nat Immunol 2007;8:1255–65. [DOI] [PubMed] [Google Scholar]
- 62. Peske JD, Thompson ED, Gemta L, Baylis RA, Fu YX, Engelhard VH. Effector lymphocyte-induced lymph node-like vasculature enables naive T-cell entry into tumours and enhanced anti-tumour immunity. Nat Commun 2015;6:7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Colbeck EJ, Jones E, Hindley JP, Smart K, Schulz R, Browne M, et al. Treg depletion licenses T cell-driven HEV neogenesis and promotes tumor destruction. Cancer Immunol Res 2017;5:1005–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data from the RNA-seq analyses were generated at the Genomic Core of the Sanford Burnham Prebys Medical Discovery Institute. Data analyses were conducted at the Biostatistics and Bioinformatics core of the Moffitt Cancer Center. The RNA-seq data are deposited in NCBI's Gene-Expression Omnibus and are accessible through GEO Series accession number GSE196703.