Keywords: Birds, Diplostomidae, Fibricola, mammals, molecular phylogeny, Neodiplostomum
Abstract
Fibricola and Neodiplostomum are diplostomid genera with very similar morphology that are currently separated based on their definitive hosts. Fibricola spp. are normally found in mammals, while Neodiplostomum spp. typically parasitize birds. Previously, no DNA sequence data was available for any member of Fibricola. We generated nuclear ribosomal and mtDNA sequences of Fibricola cratera (type-species), Fibricola lucidum and 6 species of Neodiplostomum. DNA sequences were used to examine phylogenetic interrelationships among Fibricola and Neodiplostomum and re-evaluate their systematics. Molecular phylogenies and morphological study suggest that Fibricola should be considered a junior synonym of Neodiplostomum. Therefore, we synonymize the two genera and transfer all members of Fibricola into Neodiplostomum. Specimens morphologically identified as Neodiplostomum cratera belonged to 3 distinct phylogenetic clades based on mitochondrial data. One of those clades also included sequences of specimens identified morphologically as Neodiplostomum lucidum. Further study is necessary to resolve the situation regarding the morphology of N. cratera. Our results demonstrated that some DNA sequences of N. americanum available in GenBank originate from misidentified Neodiplostomum banghami. Molecular phylogentic data revealed at least 2 independent host-switching events between avian and mammalian hosts in the evolutionary history of Neodiplostomum; however, the directionality of these host-switching events remains unclear.
Introduction
Fibricola Dubois, 1932 (Diplostomidae Poirier, 1886) is a small genus of diplostomid digeneans distributed in North and South America, Africa, Asia and Australia (Barker, 1915; Bisseru, 1957; Seo et al., 1964; Kifune and Uyema, 1982; Cribb and Pearson, 1993; Niewiadomska, 2002; Lima et al., 2013). Members of Fibricola are often reported in ecological and parasite survey studies, most commonly from their frog second intermediate hosts (e.g. Ulmer, 1970; Premvati and Bair, 1979; Gillilland and Muzzall, 1999; Goldberg and Bursey, 2001; Goldberg et al., 2001; Bolek and Coggins, 2003; Richardson, 2013; Weinstein et al., 2019). Although most members of Fibricola are known to parasitize intestines of mammals, some Fibricola species have also been reported from crocodilians (Bisseru, 1957; Dubois, 1982).
In contrast to Fibricola spp., the currently accepted members of the morphologically similar Neodiplostomum Railliet, 1919 parasitize intestines of birds with few exceptions. The majority of Neodiplostomum spp. known from mammals were collected in the Old World and originally placed into Fibricola based on their parasitism in mammals (e.g. Neodiplostomum seoulensis (Seo, Rim and Lee, 1964) and Neodiplostomum minor (Dubois, 1936)), and later transferred to Neodiplostomum (Seo et al., 1964; Cribb and Pearson, 1993; Hong and Shoop, 1994). Notably, Neodiplostomum vaucheri Dubois, 1983, described from the frog-eating big-eared woolly bat Chrotopterus auritus Peters in Peru, was the only member of Neodiplostomum from mammals originally assigned into the genus (Dubois, 1983). Noteworthily, N. seoulensis has been reported from humans in Korea (Huh et al., 1994). Despite the general trends of parasitism in different groups of definitive hosts (birds vs mammals), adult Neodiplostomum spp. and Fibricola spp. are remarkably morphologically similar. In the most recent detailed taxonomic revision of the group, Niewiadomska (2002) admitted that the two genera lack consistent morphological differences that can be used to reliably distinguish one from another. Although Niewiadomska (2002) retained the traditional generic and subfamily status of Neodiplostomum and Fibricola, she emphasized that the resolution of the real relationship between these genera needs to be supported by both morphological and molecular evidence.
Currently, DNA sequences are available for six species of Neodiplostomum (Woodyard et al., 2017; Heneberg et al., 2020; Lee et al., unpublished results), but DNA sequence data from morphologically identified adult Fibricola specimens are lacking. Herein, we provide partial sequences of the nuclear small ribosomal subunit (18S), internal transcribed spacer region (ITS1, 5.8S, ITS2) and large ribosomal subunit (28S) rRNA genes as well as a fragment of the mitochondrial cytochrome c oxidase subunit 1 (cox1) mtDNA gene for Fibricola cratera (Barker and Noll, 1915) and Fibricola lucidum (La Rue and Bosma, 1927) from mammals as well as five nominal species of Neodiplostomum and an unidentified Neodiplostomum species from a bird. We use newly generated and previously available DNA sequences to examine the phylogenetic interrelationships of Fibricola and Neodiplostomum species and re-evaluate their systematics.
Materials and methods
Adult specimens belonging to genera Fibricola and Neodiplostomum were collected from a variety of mammalian and avian definitive hosts as well as amphibian intermediate hosts in North and South America (Table 1). Live digeneans removed from hosts were briefly rinsed in saline, killed with hot water and preserved in 80% ethanol. Dead digeneans were immediately preserved in 80% ethanol. Specimens for light microscopy were stained with aqueous alum carmine and mounted permanently according to Lutz et al. (2017). Specimens were measured using an Olympus® BX53 microscope (Olympus America, Center Valley, Pennsylvania, USA) equipped with a digital imaging system. Voucher specimens are deposited in the collection of the Harold W. Manter Laboratory (HWML), University of Nebraska State Museum, Lincoln, NE, USA and the Museo de Zoología, Pontificia Universidad Católica del Ecuador (QCAZI), Quito, Ecuador. Due to the inconsistent reporting of morphological characteristics in the descriptions of Neodiplostomum spp., we re-measured type and voucher specimens of Neodiplostomum americanum Chandler and Rausch, 1947, Neodiplostomum banghami Penrod, 1947 and Neodiplostomum reflexum Chandler and Rausch, 1947 (syn. Neodiplostomum delicatum Chandler and Rausch, 1947) for comparison with specimens collected in the current study. Type and voucher specimens were borrowed for our study from the Natural History Museum of Geneva and the Smithsonian Institution Museum of Natural History. We use the terms prosoma and opisthosoma as explained by Achatz et al. (2019a) and Tkach et al. (2020).
Table 1.
Hosts, geographic origin, GenBank and museum accession numbers of Neodiplostomum (syn. Fibricola) spp. used in this study
| Digenean taxa | Host species | Geographic origin | Museum number | Accession numbers | |
|---|---|---|---|---|---|
| Ribosomal | cox1 | ||||
| Neodiplostomum cf. cratera 1 n. comb.* | Didelphis virginiana | Arkansas, USA | HWML-216754 | — | OL770020 |
| Neodiplostomum cf. cratera 2 n. comb.* | Didelphis virginiana | California, USA | HWML-216765 | OL799069, OL799070 (ITS1), OL770124, OL770125 (ITS2) | OL770021–OL770023 |
| Neodiplostomum cf. cratera 2 n. comb.* | Procyon lotor | California, USA | — | OL799097 (28S) | OL770024 |
| Neodiplostomum cf. cratera 3 n. comb.* | Didelphis virginiana | Mississippi, USA | HWML-216755 | OL799071 (18S–28S) | OL770025 |
| Neodiplostomum cf. cratera 3 n. comb.* | Lithobates pipiens | North Dakota, USA | — | OL799098 (28S) | OL770026 |
| Neodiplostomum cf. cratera 3 n. comb.* | Procyon lotor | Minnesota, USA | HWML-216766 | OL799072, OL799073 (ITS2–28S) | OL770027, OL770028 |
| Neodiplostomum cf. cratera 3 n. comb.* | Neogale vison | Minnesota, USA | HWML-216767 | OL799074 (18S–ITS2) | OL770029 |
| Neodiplostomum cf. cratera 3 n. comb.* | Taxidea taxus | North Dakota, USA | — | OL799099 (28S) | OL770030 |
| Neodiplostomum cf. lucidum* | Didelphis virginiana | Arkansas, USA | HWML-216752, HWML-216753 | OL799075 (18S–28S) | OL770031, OL770032 |
| Neodiplostomum cf. lucidum* | Didelphis virginiana | Mississippi, USA | — | — | OL770033–OL770037 |
| Neodiplostomum cf. lucidum* | Didelphis virginiana | Nebraska, USA | HWML-216768 | OL799076 (18S–28S) | OL770038, OL764381 |
| Neodiplostomum cf. lucidum* | Didelphis virginiana | North Carolina, USA | HWML-216769 | OL799100, OL799101 (28S) | OL770039, OL770040 |
| Neodiplostomum cf. lucidum* | Lithobates catesbeianus | Mississippi, USA | — | OL799077, OL799078 (ITS1–28S) | OL770041, OL770042 |
| Neodiplostomum cf. lucidum* | Procyon lotor | California, USA | HWML-216770 | OL799102 (28S) | OL770043 |
| Neodiplostomum microcotyle | Busarellus nigricollis | Pantanal, Brazil | HWML-216771 | OL799079 (18S–28S) | OL770044 |
| N. microcotyle | Buteogallus urubitinga | Pantanal, Brazil | HWML-216772 | — | OL770045 |
| Neodiplostomum americanum | Accipiter cooperii | North Dakota, USA | HWML-216773 | OL799080 (18S), OL770126 (ITS1–28S) | OL770046 |
| N. americanum | Bubo virginianus | Arkansas, USA | HWML-216774 | OL799103 (28S) | OL770047 |
| N. americanum | Bubo virginianus | Mississippi, USA | HWML-216756, HWML-216757, HWML-216760 | OL799081–OL799083 (ITS region) | OL770048–OL770050 |
| N. americanum | Nerodia fasciata | Mississippi, USA | — | OL799084 (ITS1–28S) | OL770051 |
| N. americanum | Strix varia | Mississippi, USA | — | OL799085 (ITS region) | OL770052 |
| N. americanum | Thalasseus maximus | Mississippi, USA | — | OL799086 (ITS1–28S) | OL770053 |
| Neodiplostomum banghami | Falco columbarius | North Dakota, USA | — | OL799087 (18S–28S) | OL770054 |
| N. banghami | Lithobates sylvatica | North Dakota, USA | — | OL799104 (28S) | OL770055 |
| N. banghami | Thamnophis sirtalis | North Dakota, USA | — | OL799105 (28S) | OL770056 |
| Neodiplostomum reflexum | Bubo virginianus | North Dakota, USA | HWML-216775 | OL799106 (28S) | OL770057 |
| N. reflexum | Bubo virginianus | Mississippi, USA | HWML-216759 | OL799088 (ITS region) | OL770058 |
| N. reflexum | Buteo jamaicensis | North Dakota, USA | — | OL799089 (18S–28S) | OL770059 |
| N. reflexum | Buteo jamaicensis | Mississippi, USA | — | OL799090 (ITS region) | OL770060 |
| N. reflexum | Strix varia | Mississippi, USA | HWML-216758, HWML-216761–216763 | OL799091 (18S–28S), OL799092–OL799094 (ITS region) | OL770061–OL770064 |
| Neodiplostomum vaucheri | Trachops cirrhosus | Ecuador | QCAZI 264292 | OL799095 (18S–28S), OL799107, OL799108 (28S) | OL770065–OL770067 |
| Neodiplostomum sp. VVT1 | Bubo virginianus | North Dakota, USA | HWML-216776 | OL799096 (18S–28S) | OL770068 |
HWML, Harold W. Manter Laboratory; QCAZI, Museo de Zoología, Pontificia Universidad Católica del Ecuador.
*Species previously considered to be within Fibricola.
For comparative purposes, specimens of the following species have been examined from the collection of the Natural History Museum, London (NHM): Neodiplostomum australiense (Dubois, 1937) from Australia (co-types, NHM 1950.12.6.18–22), Neodiplostomum ramachandrani (Betterton, 1976) from Malaysia (paratypes: NHM 1979.8-3.36, 44–46; NHM 1976.4.21.74; vouchers: NHM 1976.8.4.7–8) and Neodiplostomum spathula (Creplin, 1829) from Minnesota (vouchers: NHM 1975.1.7.35-42).
Genomic DNA was extracted from either fragments (in case of larger specimens) or whole individuals of each species according to the methods described by Tkach and Pawlowski (1999). An approximately 1800 bp long fragment at the 5′ end of the 18S rRNA gene and a 1300 bp long fragment at the 5′ end of the 28S rRNA gene were amplified by polymerase chain reactions (PCRs) in a T100™ thermal cycler (Bio-Rad, Hercules, CA, USA). The 18S fragment was amplified using the forward primer WormA (5′–GCG AAT GGC TCA TTA AAT CAG–3′) and the reverse primer WormB (5′–ACG GAA ACC TTG TTA CGA CT–3′), whereas 28S was amplified using the forward primer digL2 (5′–AAG CAT ATC ACT AAG CGG–3′) and the reverse primer 1500R (5′–GCT ATC CTG AGG GAA ACT TCG–3′) (Littlewood and Olson, 2001; Tkach et al., 2003). In addition, fragments of the ribosomal internal transcribed spacer region (ITS1 + 5.8S + ITS2) were amplified for some of the studied taxa using the forward primer ITSf (5′–CGC CCG TCG CTA CTA CCG ATT G–3′) and the reverse primer 300R (5′–CAA CTT TCC CTC ACG GTA CTT G–3′) (Littlewood and Olson, 2001; Snyder and Tkach, 2007). A fragment of the mitochondrial cox1 gene was amplified using the previously published forward primer Dipl_Cox_5′ (5′-ACK TTR GAW CAT AAG CG-3′) and the reverse primer Dipl_Cox_3′ (5′-WAR TGC ATN GGA AAA AAA CA–3′) (Achatz et al., 2021b). For a subset of taxa collected in Mississippi and Arkansas (USA), the molecular methods described by Woodyard et al. (2017) were used.
An ExoSAP-IT PCR clean-up enzymatic kit from Affymetrix (Santa Clara, California, USA) was used to clean-up the PCR products following the manufacturer's protocol. PCR products were cycle-sequenced directly using BrightDye® Terminator Cycle Sequencing Kit chemistry (MCLAB, San Francisco, California, USA), alcohol precipitated and run on an ABI 3130 automated capillary sequencer (Life Technologies, Grand Island, New York, USA). PCR primers were used for sequencing of 18S, 28S and cox1 genes as well as the ribosomal ITS region. In addition, internal forward primer 18S-8 (5′-GCA GCC GCG GTA ATT CCA GC-3′) and internal reverse primer WB1 (5′-CTT GTT ACG ACT TTT ACT TCC-3′) were used for sequencing of the 18S fragment; internal forward primer DPL600F (5′-CGG AGT GGT CAC CAC GAC CG-3′) and internal reverse primer DPL700R (5′-CAG CTG ATT ACA CCC AAA G-3′) were used for sequencing of 28S PCR reactions; internal forward primer d58F (5′-GCG GTG GAT CAC TCG GCT CGT G-3′ was used to sequence the ITS region (Littlewood and Olson, 2001; Kudlai et al., 2015; Achatz et al., 2019d). Contiguous sequences were assembled using Sequencher version 4.2 software (GeneCodes Corp., Ann Arbor, Michigan, USA). Our newly generated sequences are deposited in GenBank (Table 1).
Phylogenetic interrelationships among the members of Fibricola and Neodiplostomum and other members of the Diplostomoidea Poirier, 1886 were analysed using the 18S, 28S and cox1 sequence data, in part, to match the data published by Heneberg et al. (2020). Interrelationships among the members of the genus-level clades of Fibricola/Neodiplostomum were studied using two cox1 datasets based on the presence of two distinct clades of Neodiplostomum as observed from the results of our analyses of 18S and 28S as well as the suprageneric analysis of cox1 data. Sequences were aligned with the assistance of ClustalW as implemented in MEGA7 (Kumar et al., 2016); alignments were trimmed to the length of the shortest respective sequence. Cyathocotyle prussica Mühling, 1896 was used as the outgroup for the analysis of 18S and Strigea strigis (Schrank, 1788) was used as the outgroup for the suprageneric analysis of cox1 to be comparable with the analysis of Heneberg et al. (2020). Suchocyathocotyle crocodili (Yamaguti, 1954) was selected as the outgroup for the 28S analysis based on the topology presented by Achatz et al. (2019d). On the basis of the results of the broader phylogenetic analyses (see ‘Results’ section), we opted to not use an outgroup in the phylogenetic analyses of interrelationships within the clade uniting Fibricola spp. with the majority of Neodiplostomum spp. clade based on cox1 data, because of the high level of genetic divergence between the members of this clade and other diplostomoidean taxa.
The 18S alignment included newly generated sequences of Fibricola spp. (n = 3) and Neodiplostomum spp. (n = 5) as well as previously published sequences of Neodiplostomum spp. (n = 3) and other members of the Diplostomidae (n = 21). The 28S alignment included newly generated sequences of Fibricola (n = 3) and Neodiplostomum spp. (n = 5) along with a previously published sequence of N. americanum. The 28S analysis also included previously published sequences of members of the Diplostomidae (n = 17), the Proterodiplostomidae Dubois, 1936 (n = 2) and Strigeidae Railliet, 1919 (n = 12). The suprageneric cox1 alignment included new sequences of Fibricola (n = 2) and Neodiplostomum spp. (n = 6) as well as previously published sequences of Neodiplostomum spp. (n = 7). This alignment also included previously published sequences of other members of the Diplostomidae (n = 15). The cox1 alignment limited to the members of the Fibricola/Neodiplostomum clade (clade I) included 21 newly generated sequences. The second cox1 alignment limited to the second clade of Neodiplostomum species (clade II) included nine newly generated sequences and nine previously published sequences.
Independent phylogenetic analyses were conducted using Bayesian inference (BI) as implemented in MrBayes Ver. 3.2.6 software (Ronquist and Huelsenbeck, 2003). The general time-reversible model with estimates of invariant sites and gamma distributed among site variation (GTR + I + G) was identified as the best-fitting nucleotide substitution model for the datasets using MEGA7 (Kumar et al., 2016). BI analyses of 18S, 28S and cox1 of the Diplostomidae were carried out with the following settings: Markov chain Monte Carlo chains run for 3 000 000 generations with a sample frequency of 1000, log-likelihood scores were plotted and only the final 75% of trees were used to produce the consensus trees. The cox1 analyses of clades I and II were carried out with identical settings, but sequence data were analysed as codons. The number of generations was considered sufficient when the s.d. value reduced well below 0.01. Due to limited representation, the ITS region sequences were not used for phylogenetic inference; however, we provided a pairwise sequence comparison for all isolates that have a complete ITS1 + 5.8S + ITS2 fragment sequenced.
Results
Molecular phylogenies
To maintain continuity and consistency in presenting and discussing our results, we are stating herein that we consider Fibricola to be a junior synonym of Neodiplostomum (see results of 18S, 28S and suprageneric cox1 analyses and discussion below). We refer to F. cratera and F. lucidum as Neodiplostomum cratera n. comb. (Barker and Noll, 1915) and Neodiplostomum lucidum La Rue and Bosma, 1927, respectively, throughout the remainder of the text. Justification for the synonymization is provided in the discussion.
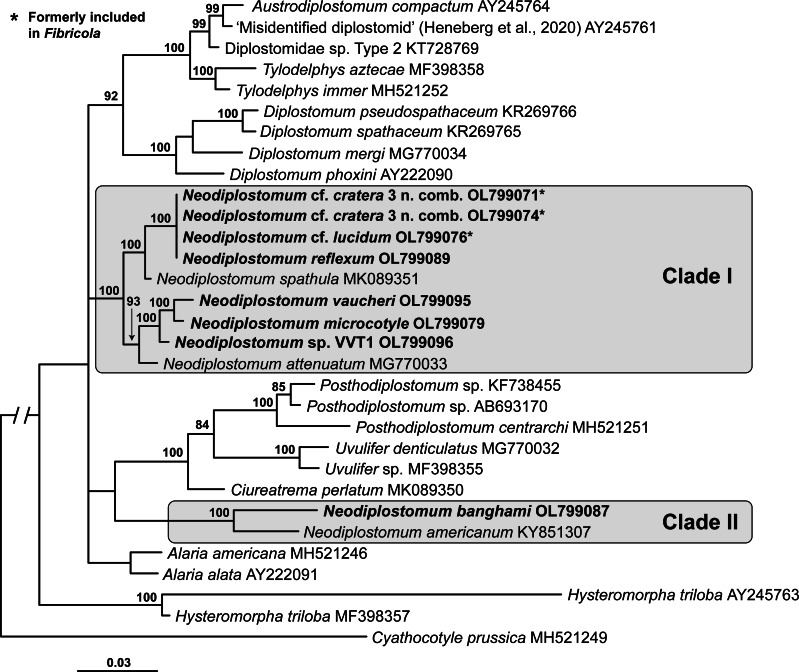
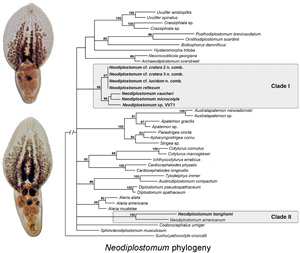
The 18S alignment was 1619 bp long; 22 bases were excluded from the analysis due to ambiguous homology. The phylogenetic tree resulting from the BI analysis of 18S (Fig. 1) demonstrated a similar topology to that presented by Heneberg et al. (2020). Neodiplostomum spp. were positioned in two distinct clades within a larger polytomy of diplostomids. Clade I (100% supported) of Neodiplostomum spp. contained Neodiplostomum cf. cratera 3 (Barker and Noll, 1915) (former type-species of Fibricola; see discussion below), Neodiplostomum cf. lucidum La Rue and Bosma, 1927, Neodiplostomum spathula (Creplin, 1829) (former type-species of Conodiplostomum Dubois, 1937) + Neodiplostomum attenuatum (Linstow, 1906) + Neodiplostomum microcotyle Dubois, 1937 + N. reflexum + N. vaucheri + Neodiplostomum sp. VVT1. Clade II (100% supported) of Neodiplostomum spp. only contained N. americanum + N. banghami.
Fig. 1.
Phylogenetic interrelationships among the Diplostomidae including Neodiplostomum (syn. Fibricola) based on BI analysis of partial 18S rRNA gene sequences. Members of Neodiplostomum are indicated by the shaded rectangles. BI posterior probability values lower than 80% are not shown. The new sequences are indicated in bold. The scale bar indicates the number of substitutions per site. GenBank accession numbers are provided after the names of species.
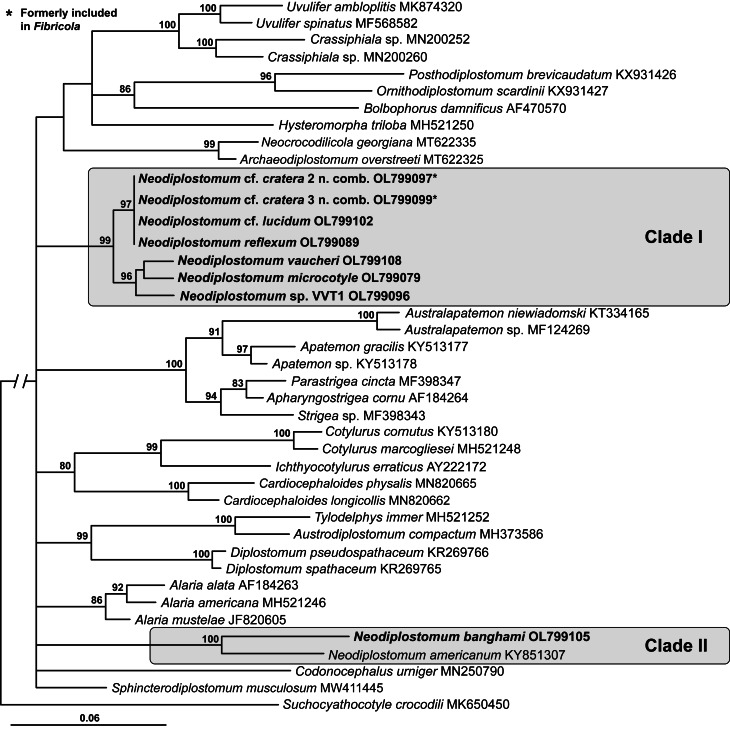
The 28S alignment was 1135 bp long; three bases were excluded from the analysis due to ambiguous homology. The phylogenetic tree resulting from the BI analysis of 28S demonstrated the non-monophyly of the Diplostomidae and Strigeidae and monophyly of the Proterodiplostomidae (Fig. 2), similar to previous molecular phylogenetic analyses of the Diplostomoidea (e.g. Blasco-Costa and Locke, 2017; Hernández-Mena et al., 2017; Achatz et al., 2019b, 2019c, 2022, 2020, 2021a; Queiroz et al., 2020; Tkach et al., 2020; Locke et al., 2021). All sequences of taxa/lineages representing Fibricola formed a 99% supported clade (clade I) with four Neodiplostomum species: N. microcotyle, N. reflexum and N. vaucheri as well as unidentified species-level lineage Neodiplostomum sp. VVT1. This clade was separated into two strongly supported sub-clades. The first sub-clade (97%) included sequences of Fibricola from mammals + N. reflexum from birds. The second sub-clade (96%) contained Neodiplostomum sp. VVT1 from great horned owl Bubo virginianus (Gmelin) + a weakly supported assemblage of [N. microcotyle + N. vaucheri]. Clade II of Neodiplostomum spp. (100% supported) contained N. americanum + N. banghami (Fig. 2).
Fig. 2.
Phylogenetic interrelationships among the diplostomoidean taxa including Neodiplostomum (syn. Fibricola) based on BI analysis of partial 28S rRNA gene sequences. Members of Neodiplostomum are indicated by the shaded rectangles. BI posterior probability values lower than 80% are not shown. The new sequences generated are indicated in bold. The scale bar indicates the number of substitutions per site. GenBank accession numbers are provided after the names of species.
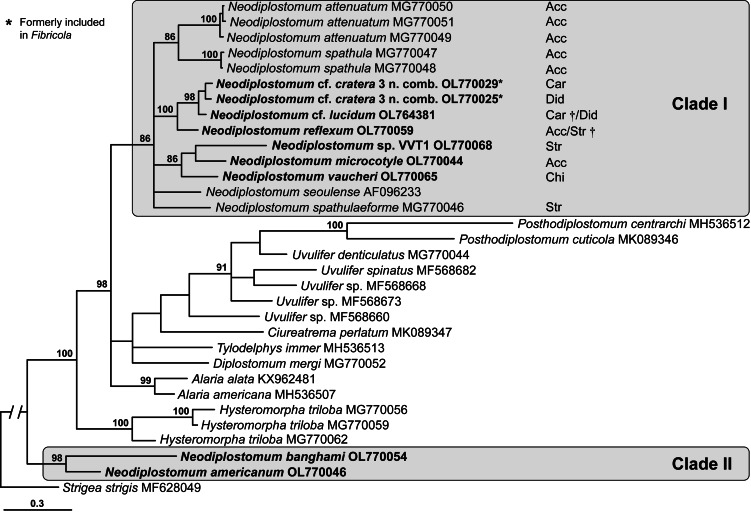
The suprageneric cox1 alignment was 285 bp long; the alignment length was limited by the short length of sequences published by Heneberg et al. (2020). Similar to the 18S and 28S analyses, Neodiplostomum taxa were split among the two clades (Fig. 3). Clade I (86% supported) consisted of a large polytomy with a poorly resolved internal topology (Fig. 3). The polytomy consisted of Neodiplostomum spathulaeforme (Brandes, 1888) (type-species of Neodiplostomum) + Neodiplostomum seoulense + a 100% supported clade of [N. reflexum + N. cf. cratera 3 + N. cf. lucidum] + an 88% supported clade of [N. vaucheri + N. microcotyle + Neodiplostomum sp. VVT1] + an 86% supported clade of [N. attenuatum + N. spathula] (Fig. 3).
Fig. 3.
Phylogenetic interrelationships among the Diplostomidae including 16 members of Neodiplostomum (syn. Fibricola) based on BI analysis of partial cox1 mtDNA gene sequences. Members of Neodiplostomum are indicated by the shaded rectangles. BI posterior probability values lower than 80% are not shown. The new sequences generated are indicated in bold. The scale bar indicates the number of substitutions per site. GenBank accession numbers are provided after the names of species. The orders of definitive hosts are provided after GenBank accession numbers for Neodiplostomum spp. in clade I. Abbreviations for orders of definitive host: Acc, Accipitriformes; Car, Carnivora; Chi, Chiroptera; Did, Didelphimorphia; Str, Strigiformes. †We also sequenced additional conspecific isolates collected from additional orders of definitive hosts.
On the basis of their phylogenetic position in the 18S, 28S and suprageneric cox1 analyses, N. microcotyle, N. reflexum, N. vaucheri and Neodiplostomum sp. VVT1 were included in the focused cox1 analysis together with former Fibricola spp. (clade I in Figs 1–3). This alignment was 456 bp long; three bases (one codon) were excluded from the analysis as an indel. The internal branch topology of the resulting tree (Fig. 4) was somewhat different and better resolved than in the 18S and 28S analyses. Neodiplostomum microcotyle + Neodiplostomum sp. VVT1 from B. virginianus + N. vaucheri formed a strongly (100%) supported clade separate from the 100% supported clade of N. reflexum + former Fibricola lineages. Neodiplostomum microcotyle was positioned as a sister group to a weakly supported clade of Neodiplostomum sp. VVT1 + N. vaucheri. All sequences of N. reflexum formed a 100% supported, long-branch clade as a sister clade to a weakly supported clade containing sequences of former Fibricola cf. cratera 3 (Fig. 4). The remaining sequences of former Fibricola formed two clades. One of them was weakly (82%) supported and included N. cf. cratera 1 (formerly F. cf. cratera 1) and specimens that were morphologically identified as N. cf. lucidum (formerly F. cf. lucidum) (Fig. 4). The other was a 100% supported clade of N. cf. cratera 2 (formerly F. cf. lucidum).
Fig. 4.
Phylogenetic interrelationships among the 20 members of Neodiplostomum clade I based on BI analysis of partial cox1 mtDNA gene sequences. BI posterior probability values lower than 80% are not shown. The new sequences generated are indicated in bold. The scale bar indicates the number of substitutions per site. GenBank accession numbers are provided after the names of species.
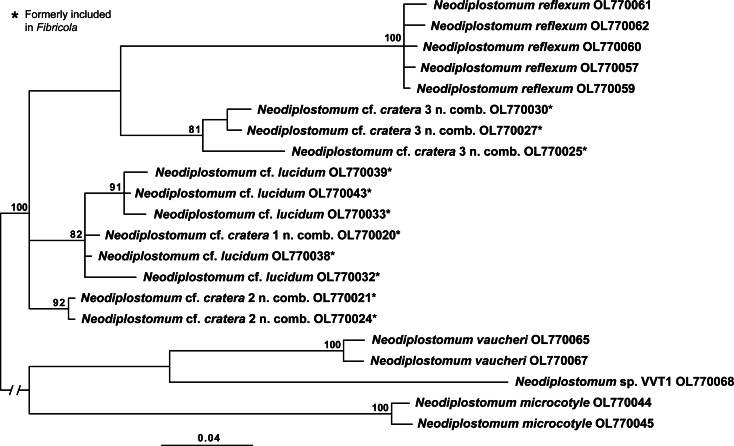
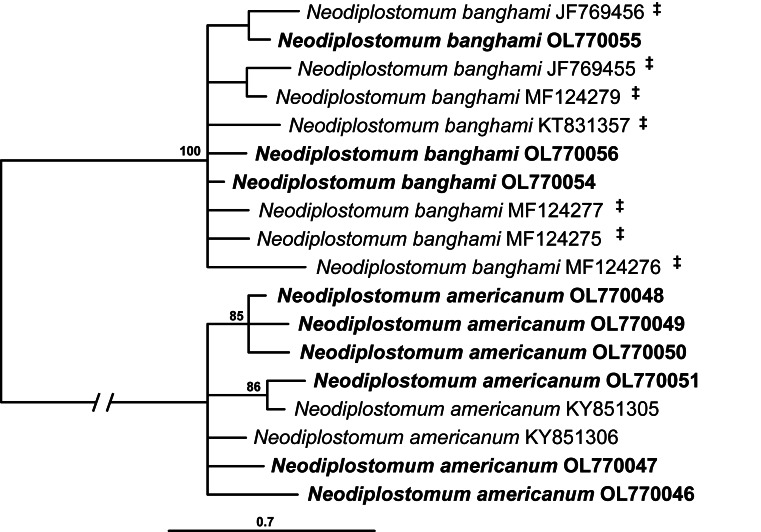
The cox1 alignment of the second Neodiplostomum clade (clade II in Figs 1–3) was 366 bp long. The sequences of N. americanum and N. banghami formed separate 100% supported clades (Fig. 5).
Fig. 5.
Phylogenetic interrelationships among the two species of Neodiplostomum clade II based on BI analysis of partial cox1 mtDNA gene sequences. BI posterior probability values lower than 80% are not shown. The new sequences generated are indicated in bold. The scale bar indicates the number of substitutions per site. GenBank accession numbers are provided after the names of species. ‡Isolates previously identified as Neodiplostomum americanum in GenBank.
Genetic variation
Taxa included in clade I demonstrated a low interspecific divergence in 18S sequences (0–1.1%). No differences were detected among the 18S sequences of N. cf. cratera 3 (multiple sequences), N. cf. lucidum and N. reflexum; Neodiplostomum cf. cratera 3 vs N. vaucheri, N. cf. lucidum vs N. vaucheri and N. reflexum vs N. vaucheri had the greatest level of interspecific divergence in 18S among Neodiplostomum spp. in clade I. Neodiplostomum americanum and N. banghami, members of clade II, differed by 1.6% between their 18S sequences. No intraspecific variation was detected among the 18S sequences of N. cf. cratera 3 n. comb., N. reflexum and N. americanum. Complete pairwise comparisons of 18S sequences are provided in Table 2.
Table 2.
Pairwise comparisons of partial sequences of the 18S rDNA among Neodiplostomum (syn. Fibricola) species included in this study based on a 1602 bp long alignment
| (1) OL799074 |
(2) OL799071 |
(3) OL799076 |
(4) OL799089 |
(5) MK089351 |
(6) MG770033 |
(7) OL799096 |
(8) OL799079 |
(9) OL799095 |
(10) KY851307 |
(11) OL799087 |
|
|---|---|---|---|---|---|---|---|---|---|---|---|
| (1) Neodiplostomum cf. cratera 3 n. comb. OL799074* | — | 0% | 0% | 0% | 0.4% | 0.8% | 0.9% | 1% | 1.1% | 2.9% | 3.2% |
| (2) Neodiplostomum cf. cratera 3 n. comb. OL799071* | 0 | — | 0% | 0% | 0.4% | 0.8% | 0.9% | 1% | 1.1% | 2.9% | 3.2% |
| (3) Neodiplostomum cf. lucidum OL799076* | 0 | 0 | — | 0% | 0.4% | 0.8% | 0.9% | 1% | 1.1% | 2.9% | 3.2% |
| (4) Neodiplostomum reflexum OL799089 | 0 | 0 | 0 | — | 0.4% | 0.8% | 0.9% | 1% | 1.1% | 2.9% | 3.2% |
| (5) Neodiplostomum spathula MK089351 | 6 | 6 | 6 | 6 | — | 0.6% | 0.6% | 0.7% | 0.9% | 2.6% | 2.9% |
| (6) Neodiplostomum attenuatum MG770033 | 13 | 13 | 13 | 13 | 9 | — | 0.4% | 0.6% | 0.7% | 2.7% | 2.9% |
| (7) Neodiplostomum sp. VVT1 OL799096 | 14 | 14 | 14 | 14 | 10 | 7 | — | 0.2% | 0.4% | 2.8% | 3% |
| (8) Neodiplostomum microcotyle OL799079 | 16 | 16 | 16 | 16 | 12 | 9 | 4 | — | 0.2% | 2.7% | 3% |
| (9) Neodiplostomum vaucheri OL799095 | 18 | 18 | 18 | 18 | 14 | 11 | 6 | 4 | — | 2.9% | 3.1% |
| (10) Neodiplostomum americanum KY851307 | 46 | 46 | 46 | 46 | 42 | 43 | 45 | 44 | 46 | — | 1.6% |
| (11) Neodiplostomum banghami OL799087 | 52 | 52 | 52 | 52 | 47 | 46 | 48 | 48 | 49 | 26 | — |
Percentage differences are given above the diagonal and the number of variable nucleotide positions is given below the diagonal. Taxa previously included in Fibricola are denoted by *.
The interspecific divergence in 28S sequences of Neodiplostomum spp. (clade I) was similar to differences among the 18S sequences (0–1.2%). No differences were detected among the 28S sequences of N. cf. cratera 2 and 3 n. comb., N. cf. lucidum and N. reflexum. The unidentified Neodiplostomum sp. VVT1 from B. virginianus vs N. cf. cratera 2 and 3 n. comb., Neodiplostomum sp. VVT1 vs N. cf. lucidum and Neodiplostomum sp. VVT1 vs N. reflexum had the greatest level of interspecific divergence in 28S (1.2%) among Neodiplostomum spp. in clade I. In contrast, the overall interspecific variability among the members of Neodiplostomum spp. in clade II was greater; N. americanum and N. banghami were 3.7% divergent in the sequenced 28S fragment. Notably, no intraspecific variation in sequences of 28S was detected in any of the Neodiplostomum taxa with multiple isolates included in the analysis. Complete pairwise comparisons of 28S sequences are provided in Table 3.
Table 3.
Pairwise comparisons of partial sequences of the 28S rDNA among Neodiplostomum (syn. Fibricola) species included in this study based on a 1176 bp long alignment
| (1) OL799097 |
(2) OL799071 |
(3) OL799102 |
(4) OL799089 |
(5) OL799079 |
(6) OL799108 |
(7) OL799096 |
(8) KY851307 |
(9) OL799105 |
|
|---|---|---|---|---|---|---|---|---|---|
| (1) Neodiplostomum cf. cratera 2 n. comb. OL799097* | — | 0% | 0% | 0% | 1.1% | 1.1% | 1.2% | 5% | 5.8% |
| (2) Neodiplostomum cf. cratera 3 n. comb. OL799071* | 0 | — | 0% | 0% | 1.1% | 1.1% | 1.2% | 5% | 5.8% |
| (3) Neodiplostomum cf. lucidum OL799102* | 0 | 0 | — | 0% | 1.1% | 1.1% | 1.2% | 5% | 5.8% |
| (4) Neodiplostomum reflexum OL799089 | 0 | 0 | 0 | — | 1.1% | 1.1% | 1.2% | 5% | 5.8% |
| (5) Neodiplostomum microcotyle OL799079 | 13 | 13 | 13 | 13 | — | 0.9% | 1% | 5.8% | 6.2% |
| (6) Neodiplostomum vaucheri OL799108 | 13 | 13 | 13 | 13 | 10 | — | 1.1% | 5.4% | 6.2% |
| (7) Neodiplostomum sp. VVT1 OL799096 | 14 | 14 | 14 | 14 | 12 | 13 | — | 5.5% | 5.6% |
| (8) Neodiplostomum americanum KY851307 | 59 | 59 | 59 | 59 | 68 | 64 | 65 | — | 3.7% |
| (9) Neodiplostomum banghami OL799105 | 68 | 68 | 68 | 68 | 73 | 73 | 66 | 43 | — |
Percentage differences are given above the diagonal and the number of variable nucleotide positions is given below the diagonal. Taxa previously included in Fibricola are denoted by *.
The interspecific divergence in ITS1 + 5.8S + ITS2 sequences of Neodiplostomum spp. (clade I) was greater than among the 18S and 28S sequences (0.2–6.6%). Up to 0.1% variation was detected among the ITS region sequences of N. cf. lucidum and N. reflexum. Neodiplostomum reflexum and N. vaucheri were the most divergent pairs of sequences among Neodiplostomum spp. in clade I. Up to 0.2% variation was detected among the N. cf. lucidum/cratera lineages. At the same time, the interspecific variability among the ITS region sequences from the two members of Neodiplostomum spp. in clade II was even greater (9.3–9.4%). Intraspecific variation in sequences of the ITS region was detected in N. americanum (up to 0.6%). The divergence between the members of clades I and II was much greater (13–16.7%). Complete pairwise comparisons of ITS region (ITS1 + 5.8S + ITS2) sequences are provided in Table 4.
Table 4.
Pairwise comparisons of ITS1 + 5.8S + ITS2 rDNA region among Neodiplostomum (syn. Fibricola) species included in this study based on a 1073 bp long alignment
| (1) OL799074 |
(2) OL799076 |
(3) OL799077 |
(4) OL799089 |
(5) OL799091 |
(6) OL799079 |
(7) OL799095 |
(8) OL799096 |
(9) OL799080 |
(10) OL799086 |
(11) OL799084 |
(12) OL799087 |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (1) Neodiplostomum cf. cratera 3 n. comb. VT OL799074* | — | 0.2% | 0.1% | 0.4% | 0.3% | 5.9% | 6.3% | 4.8% | 13% | 13.3% | 13.1% | 14.9% |
| (2) Neodiplostomum cf. lucidum OL799076* | 2 | — | 0.1% | 0.4% | 0.3% | 6% | 6.5% | 4.9% | 13.1% | 13.4% | 13.2% | 15% |
| (3) Neodiplostomum cf. lucidum OL799077* | 1 | 1 | — | 0.5% | 0.4% | 6% | 6.4% | 4.9% | 13.1% | 13.4% | 13.2% | 15% |
| (4) Neodiplostomum reflexum OL799089 | 4 | 4 | 5 | — | 0.1% | 6% | 6.5% | 4.9% | 13.1% | 13.4% | 13.2% | 15% |
| (5) Neodiplostomum reflexum OL799091 | 3 | 3 | 4 | 1 | — | 6.1% | 6.6% | 5% | 13.2% | 13.5% | 13.3% | 15.1% |
| (6) Neodiplostomum microcotyle OL799079 | 63 | 64 | 64 | 64 | 65 | — | 5.3% | 4.8% | 14.1% | 14.4% | 14.2% | 15.8% |
| (7) Neodiplostomum vaucheri OL799095 | 68 | 70 | 69 | 70 | 71 | 57 | — | 4.4% | 14.4% | 14.7% | 14.5% | 16.7% |
| (8) Neodiplostomum sp. VVT1 OL799096 | 52 | 53 | 53 | 53 | 54 | 51 | 47 | — | 13.2% | 13.5% | 13.3% | 15.5% |
| (9) Neodiplostomum americanum OL799080 | 140 | 141 | 141 | 141 | 142 | 151 | 155 | 142 | — | 0.6% | 0.1% | 9.4% |
| (10) Neodiplostomum americanum OL799086 | 143 | 144 | 144 | 144 | 145 | 154 | 158 | 145 | 6 | — | 0.5% | 9.4% |
| (11) Neodiplostomum americanum OL799084 | 141 | 142 | 142 | 142 | 143 | 152 | 156 | 143 | 1 | 5 | — | 9.3% |
| (12) Neodiplostomum banghami OL799087 | 160 | 161 | 161 | 161 | 162 | 169 | 179 | 166 | 101 | 101 | 100 | — |
Percentage differences are given above the diagonal and the number of variable nucleotide positions is given below the diagonal. Taxa previously included in Fibricola are denoted by *.
Interspecific differences of cox1 sequences among Neodiplostomum spp. of clade I, excluding the N. cf. cratera/N. cf. lucidum cluster, were in the range of 8.6–13.4%. Neodiplostomum vaucheri vs Neodiplostomum sp. VVT1 showed the lowest divergence (8.6–8.8%), whereas N. reflexum and N. microcotyle had the greatest divergence (12.7–13.4%). The N. cf. cratera/N. cf. lucidum cluster demonstrated up to 6.2% divergence among its members. The cox1 sequences of N. americanum and N. banghami (clade II) differed by 12.3–14.2%. With the exception of N. cf. cratera lineages, all Neodiplostomum spp. in clades I and II with more than a single sequence available showed no more than 1.6% intraspecific variation in cox1 sequences (Supplementary Tables S1 and S2). Complete pairwise comparisons of cox1 sequences of clade I Neodiplostomum taxa are provided in Supplementary Table S1 and clade II Neodiplostomum taxa in Supplementary Table S2.
Discussion
The systematic histories of Fibricola and Neodiplostomum are complex and tightly interwoven. Dubois (1932) originally established genus Fibricola for F. cratera described from muskrat Ondatra zibethicus (Linnaeus) by Barker (1915). Subsequently, Dubois (1937) added Fibricola minor Dubois, 1936 to the genus and noted an error in the topography of reproductive system organs in the original description of F. cratera. Dubois (1937) used parasitism in mammals along with confinement of the vitellarium to the prosoma as the justification for separation between Fibricola and Neodiplostomum which typically parasitizes birds and has vitellarium in both parts of the body. However, Dubois (1938) noted that the vitellarium of F. cratera may extend into the opisthosoma to the level of the anterior testis. Miller (1940) later described a second North American species of the genus, Fibricola laruei Miller, 1940, from raccoon Procyon lotor (Linnaeus) collected in Quebec.
A third member of the genus from North America, Fibricola texensis Chandler, 1942, was described based on specimens collected from P. lotor in Texas. The original description of the species reported its vitellarium extending to variable levels in the opisthosoma (Chandler, 1942). Additionally, Chandler (1942) noted that the vitellarium of F. laruei also extended into the opisthosoma, but only to the level of the vitelline reservoir situated between the testes. Zerecero (1943) subsequently described the fourth species of Fibricola from North America, Fibricola caballeroi Zerecero, 1943, collected from the brown, or Norway, rat Rattus norvegicus (Berkenhout) in Mexico.
Later, Dubois (1944) erected Theriodiplostomum Dubois, 1944 for F. texensis and N. lucidum from Virginia opossum Didelphis virginiana (Kerr) collected in Texas, based on vitellarium distributed in both the prosoma and opisthosoma and parasitism in mammals. Theriodiplostomum spp. were considered morphologically intermediate forms between Fibricola and Neodiplostomum (Dubois, 1944).
Chandler and Rausch (1946) described a fifth member of Fibricola in North America, Fibricola nana Chandler and Rausch, 1946, from American red squirrel Tamiasciurus hudsonicus (Erxleben) (syn. Sciurus hudsonicus) in Michigan. Importantly, Chandler and Rausch (1946) deemed the use of the distribution of vitellarium and parasitism in either mammals or birds not tenable for differentiation among the genera and rejected Theriodiplostomum. Read (1948) agreed with this decision and considered F. nana and F. laruei synonyms of F. cratera. Read (1948) proposed the tendency for greater concentration of vitelline follicles in the prosoma in members of Fibricola species as the main distinguishing character from Neodiplostomum spp.
Dubois and Rausch (1950) transferred the former Theriodiplostomum lucidum (La Rue and Bosma, 1927) to Fibricola. In contrast to the previous authors, Pearson (1959) viewed Fibricola as a subgenus of Neodiplostomum. Odening (1965) maintained Fibricola as a subgenus of Neodiplostomum based on similarities of larval morphology (i.e. the identical flame cell formula, 2[(1 + 1 + 1) + (1 + 1 + [1])] = 12).
Several Fibricola spp. were previously described from mammalian hosts outside of North America and later transferred to Neodiplostomum. For example, N. seoulensis, described from R. norvegicus collected in Korea, was originally included in Fibricola based, in part, on parasitism in mammals. Noteworthily, this species has been reported from humans in Korea (Huh et al., 1994). Hong and Shoop (1994) transferred this species into Neodiplostomum based on the morphology of adults and metacercariae. Similarly, Cribb and Pearson (1993) transferred three Fibricola spp. from Australian mammals into Neodiplostomum based on adult morphology.
Despite similarities in larval and adult morphology, Dubois (1970) rejected the placement of Fibricola as a subgenus of Neodiplostomum and insisted that the distribution of vitellarium and specificity to mammals were sufficient for separation between the two genera. In spite of his own statement, Dubois (1983) placed N. vaucheri collected from a chiropteran host into Neodiplostomum.
Although specificity to either mammalian or avian hosts has often been used as a distinguishing characteristic of Fibricola and Neodiplostomum species, some studies (e.g. Ulmer, 1955; Seo, 1989) demonstrated that Fibricola spp. can develop in avian hosts. Nevertheless, the most recent revision of the Diplostomoidea by Niewiadomska (2002) maintained Fibricola and Neodiplostomum as separate genera belonging to different subfamilies (the Alariinae and the Diplostominae Poirier, 1886, correspondingly) based on parasitism in either mammals or birds. Shoop (1989) provided an alternative hypothesis for the subfamily structure of the Diplostomidae; in his system, Fibricola was placed together with Neodiplostomum within the Neodiplostominae Shoop, 1989 based on morphology. Recently, Achatz et al. (2021c) rejected the use of subfamilies of the Diplostomidae based on morphological and molecular data. The molecular phylogeny presented by Achatz et al. (2021c) and other recent molecular phylogenetic studies (e.g. Hernández-Mena et al., 2017; Achatz et al., 2019c, 2022, in press; Queiroz et al., 2020) clearly do not support the system provided by Shoop (1989).
Heneberg et al. (2020) demonstrated the non-monophyly of Neodiplostomum and proposed Conodiplostomum to be a junior synonym of Neodiplostomum based on molecular phylogenies. Unfortunately, this solution did not remove the problem of the non-monophyly of Neodiplostomum. Members of Neodiplostomum consistently formed two distinct clades in our analyses (Figs 1–3). Currently, 18S and 28S sequences of N. spathulaeforme (type-species) are not available. The suprageneric analysis of shorter fragment of cox1 (Fig. 3) revealed a fairly well supported clade of Neodiplostomum (including N. spathulaeforme) + former Fibricola + the former type-species of Conodiplostomum (N. spathula). At the same time, the second well-supported clade of Neodiplostomum was positioned separately within this phylogeny (Fig. 3) and contained only N. americanum + N. banghami. Similar patterns related to the constituents of the two Neodiplostomum clades (e.g. the position of Fibricola within clade I) were strongly supported in 18S and 28S analyses (Figs 1 and 2). The position of the type-species of Neodiplostomum (N. spathulaeforme) in the suprageneric analysis of cox1 (Fig. 3) clearly indicates that taxa within clade I should be considered true Neodiplostomum.
On the basis of our examination of adult morphology (e.g. variable distribution of vitellarium in the prosoma and opisthosoma among and within Fibricola species) and previous studies of larval morphology (e.g. Odening, 1965), no morphological characters reliably support the status of Fibricola as an independent genus. Neodiplostomum reflexum from avian hosts and F. cratera lineages from mammals lack any differences among the sequences of 18S and 28S, which demonstrates the taxa to be congeneric. Molecular data demonstrate the lack of specificity to mammalian or avian definitive hosts within the Neodiplostomum + Fibricola clade. Therefore, we consider Fibricola to be a junior synonym of Neodiplostomum and transfer the constituent species of Fibricola into Neodiplostomum. Fibricola cratera and F. caballeroi are being transferred into Neodiplostomum as N. cratera n. comb. and Neodiplostomum caballeroi Zerecero, 1943, respectively. Notably, F. lucidum was originally described as N. lucidum; thus, this species is returned to its original genus. Below, we provide an amended diagnosis of Neodiplostomum based on the diagnosis by Niewiadomska (2002). Due to the lack of distinct morphological features differentiating Neodiplostomum spp. clade II from true Neodiplostomum (clade I), we temporarily retain the species from clade II within Neodiplostomum. We anticipate that future detailed studies of their morphology and/or life cycles will provide differentiating characters and may allow placement of the clade II members into a currently undescribed genus.
Neodiplostomum Railliet, 1919 (after Niewiadomska, 2002 with changes)
Diagnosis: Body distinctly bipartite; prosoma spatulate or oval; opisthosoma cylindrical or oval. Pseudosuckers absent. Oral and ventral suckers and pharynx present. Holdfast organ round or oval, with median slit. Testes of similar size, tandem; anterior usually asymmetrical; posterior symmetrical, often bilobed. Ovary reniform or ellipsoidal, pretesticular, median or submedian, situated close to borderline between prosoma and opisthosoma, rarely near middle of opisthosoma. Vitellarium may extend almost to intestinal bifurcation. Copulatory bursa small or large; genital cone absent; hermaphroditic duct opens directly into bursa. In avian and mammalian definitive hosts. Cosmopolitan. Metacercariae in amphibians; paratenic hosts reptilians and mammals. Cercariae with two pairs of pre- and paracetabular penetration glands; flame-cell formula 2[(1 + 1 + 1) + (1 + 1 + [1])] = 12. Type-species N. spathulaeforme (Brandes, 1888).
After the re-evaluation of the validity of Fibricola and its constituents in North America, 11 valid named species of Neodiplostomum are currently known from North America: N. cratera n. comb., N. lucidum and N. caballeroi n. comb. from mammals as well as Neodiplostomum accipitris Dubois and Rausch, 1948, N. attenuatum, Neodiplostomum centuri Dubois and Macko, 1972, Neodiplostomum isomegalocotyle Dubois and Macko, 1972, Neodiplostomum pearsoni Dubois, 1962, N. reflexum, N. americanum, N. banghami from birds (e.g. Dubois, 1968, 1982; Dubois and Macko, 1972; current data). As mentioned above, N. americanum and N. banghami are kept in Neodiplostomum only provisionally due to the lack of suitable differentiating morphological characters. The same may potentially apply to N. accipitris, N. centuri, N. isomegalocotyle and N. pearsoni for which sequence data are currently lacking.
Notably, our data revealed the presence of three genetically distinct lineages of digeneans morphologically corresponding to N. cratera in North America (Fig. 4). One of these lineages appeared in the clade with specimens morphologically corresponding to N. lucidum. Our adult specimens of N. cf. cratera collected from several mammalian hosts throughout the USA, morphologically conform to the original description of F. cratera by Barker (1915) from O. zibethicus collected in Nebraska and redescribed by Dubois (1937). Because this situation does not affect the main conclusions from the present phylogenetic study, we cautiously designate these forms as N. cf. cratera 1–3 and N. cf. lucidum. Although the cox1 sequences of N. cf. cratera 1 (GenBank: OL770020) were clearly conspecific to sequences of samples that morphologically correspond to N. cf. lucidum, the cox1 sequences of N. cf. cratera 1 and 2 differ from N. cf. cratera 3 by 4.6–6.2% of nucleotide positions (Supplementary Table S1). Currently, N. cratera and N. lucidum are differentiated based on the distribution of vitellarium (primarily in prosoma in N. cratera vs extending far into opisthosoma in N. lucidum) (e.g. Dubois, 1968). However, based on our data, it is clear that distribution of vitellarium cannot be used to distinguish between these species.
Interestingly, the morphology of samples in the cluster of N. cf. cratera 3 somewhat varied. Specimens collected in the northern USA (HWML-216766, 216767) were distinctly smaller than the sequenced specimen from Mississippi (HWML-216755). These morphologically distinct forms from geographically distant regions differed by 2.6–2.9% of nucleotide positions in cox1. It should be mentioned that the vertebrate hosts of these species have broad, overlapping distributions. Notably, F. laruei, a species synonymized with F. cratera (=N. cratera) by Read (1948), was originally described from Canada, relatively close to the area where we collected our specimens. The main characters differentiating F. laruei from F. cratera were the smaller body size and elliptical shape of the holdfast organ in the former species. The somewhat significant level of genetic divergence between the larger form from the south and smaller form from the north suggests that the validity of F. laruei may need to be re-visited. A definitive answer can be obtained only when DNA sequence data from the type territory of F. laruei (Quebec) become available and the question of morphological identity of N. cratera is resolved.
Our results clearly demonstrate that the sequences of N. americanum available in GenBank represent two distinct species (Tables 2 and 3, Fig. 3; Supplementary Table S2). Our specimens of N. americanum are conspecific with specimens previously published by Woodyard et al. (2017) based on partial sequences of 28S, the ITS region and cox1. Furthermore, our specimens and the material of Woodyard et al. (2017) conform to the original morphological description of N. americanum by Chandler and Rausch (1947). Our morphological examination of voucher specimens of adult N. americanum sequenced by Blasco-Costa and Locke (2017) revealed that the taxon was misidentified. The morphological characteristics of N. americanum sequenced and deposited by Blasco-Costa and Locke (2017) closely conform to those of N. banghami (Supplementary Table S3). Additionally, cox1 sequences of N. americanum published by Blasco-Costa and Locke (2017) are clearly conspecific with our sequences of N. banghami (Supplementary Table S2; Fig. 5).
Our cox1 phylogeny (Fig. 3) demonstrated at least two independent host-switching events between avian and mammalian hosts in the evolutionary history of Neodiplostomum. The clade of N. reflexum + a cluster of [N. cf. lucidum + N. cf. cratera] suggest a transition from avian definitive hosts (orders Accipitriformes Vieillot and Strigiformes Wagler) to a diversity of mammalian definitive hosts (orders Carnivora Bowdich and Didelphimorphia Gill). The position of N. vaucheri in a clade with N. microcotyle and Neodiplostomum sp. VVT1 confirmed the initial generic placement of this species by Dubois (1983) and revealed a transition to bats (order Chiroptera Blumenbach); additional data are needed to determine the directionality of the secondary host-switching event due to the lack of internal support within this clade. The bat in which N. vaucheri was found is known to feed on amphibians. This dietary overlap with more traditional hosts of Neodiplostomum spp. (birds of prey, carnivorous mammals) created conditions for host switching. It remains to be observed how DNA sequences from other former Fibricola species that parasitize mammals as adults (e.g. N. caballeroi n. comb.) and species from southeast Asia and Australia (e.g. N. australiense) will impact the current picture of the interrelationships of Neodiplostomum.
Similar to other previous molecular phylogenetic studies, in our analyses Neodiplostomum did not form a clade with other members of the formerly accepted Diplostominae (Figs 1–3) (e.g. Achatz et al., 2019b, 2021a, 2021c; Queiroz et al., 2020). Our results, along with other recent molecular phylogenetic studies (e.g. Blasco-Costa and Locke, 2017; Hernández-Mena et al., 2017; Locke et al., 2018; Sereno-Uribe et al., 2019; Achatz et al., 2020, 2021a, 2021b, 2021c, 2022, in press; Queiroz et al., 2020; Locke et al., 2021), strongly suggest that the most recently accepted subfamilies of the Diplostomidae cannot be considered valid. Achatz et al. (2021c) rejected the subfamily system of the Diplostomidae. The data presented in the current study and Achatz et al. (in press) further corroborate the decision by Achatz et al. (2021c). Re-evaluation of the systematics of the superfamily Diplostomoidea remains necessary, but is beyond the scope of the current study.
Acknowledgements
The authors are grateful to Tim Driscoll for providing carcases of hawks and owls. The authors are grateful to Dr João B. Pinho (Universidade Federal de Mato Grosso, Cuiabá, Brazil) for his help in organizing the collection of the specimens used in this study and in obtaining collecting permits for avian hosts in Brazil. The authors also extend their gratitude to Dr James Flowers for providing some of the specimens used in this study. The authors also acknowledge Dr Isabel Blasco-Costa of the Natural History Museum of Geneva and Georgia Tschen, Katie Ahlfeld and Dr Anna Phillips of the Smithsonian Institution Museum of Natural History for sending voucher specimens.
Author contributions
TJA, EEP, ETW, TGR and VVT conceived and supervised the study. All authors involved in the collection of digeneans or their hosts. TJA, EEP, ETW, TGR, JRM and VVT conducted the morphological and molecular studies of the digeneans. TJA, JRM and VVT were responsible for writing and revising the manuscript. All authors read and approved the final manuscript.
Financial support
This study was supported by the National Science Foundation, USA (VVT, grant DEB-1120734) The University of North Dakota (TJA, Esther Wadsworth Hall Wheeler Award, Student Research Stipend and Summer Doctoral Fellowship; JRM, Student Research Stipend) and the American Society of Parasitologists (ETW, Willis A Reid, Jr. Student Research Grant). AF was supported by a postdoctoral fellowship (PNPD scholarship) from the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES). JRM was supported by the National Science Foundation (REU Site award number 1852459) and the National Institute of General Medical Sciences of the National Institutes of Health (Institutional Development Award (IDeA) grant number P20GM103442) and the University of North Dakota School of Medicine & Health Sciences. Examination of specimens deposited at the NHM was supported (VVT) by the SYNTHESYS+ (project: http://www.synthesys.info/) financed by European Community Research Infrastructure Action under the H2020 Integrating Activities Programme, project number 823827.
Ethical standards
All applicable institutional, national and international guidelines for the collecting, care and use of animals were followed.
Supplementary material
For supplementary material accompanying this paper visit https://doi.org/10.1017/S003118202100216X.
click here to view supplementary material
Data availability
The data supporting the findings of this study are available within the article and in the Supplementary materials. All newly generated sequences were deposited in the GenBank database under the following accession numbers: OL764381, OL770020–OL770068, OL770124–OL770126 and OL799069–OL799108.
Conflict of interest
The authors declare there are no conflicts of interest.
References
- Achatz TJ, Curran SS, Patitucci KF, Fecchio A and Tkach VV (2019a) Phylogenetic affinities of Uvulifer spp. (Digenea: Diplostomidae) in the Americas with description of two new species from Peruvian Amazon. Journal of Parasitology 105, 704–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achatz TJ, Dmytrieva I, Kuzmin Y and Tkach VV (2019b) Phylogenetic position of Codonocephalus Diesing, 1850 (Digenea, Diplostomoidea), an unusual diplostomid with progenetic metacercariae. Journal of Parasitology 105, 821–826. [PubMed] [Google Scholar]
- Achatz TJ, Pulis EE, Fecchio A, Schlosser IJ and Tkach VV (2019c) Phylogenetic relationships, expanded diversity and distribution of Crassiphiala spp. (Digenea, Diplostomidae), agents of black spot disease in fish. Parasitology Research 118, 2781–2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achatz TJ, Pulis EE, Junker K, Tran BT, Snyder SD and Tkach VV (2019d) Molecular phylogeny of the Cyathocotylidae (Digenea, Diplostomoidea) necessitates systematic changes and reveals a history of host and environment switches. Zoologica Scripta 48, 545–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achatz TJ, Pulis EE, González-Acuña D and Tkach VV (2020) Phylogenetic relationships of Cardiocephaloides spp. (Digenea, Diplostomoidea) and the genetic characterization of Cardiocephaloides physalis from Magellanic penguin, Spheniscus magellanicus, in Chile. Acta Parasitologica 65, 525–534. [DOI] [PubMed] [Google Scholar]
- Achatz TJ, Bell JA, Melo FTV, Fecchio A and Tkach VV (2021a) Phylogenetic position of Sphincterodiplostomum Dubois, 1936 (Digenea: Diplostomoidea) with description of a second species from Pantanal, Brazil. Journal of Helminthology 95, E6. [DOI] [PubMed] [Google Scholar]
- Achatz TJ, Brito ES, Fecchio A and Tkach VV (2021b) Description and phylogenetic position of a new species of Herpetodiplostomum from Phrynops geoffroanus in Brazil and a re-evaluation of Cheloniodiplostomum. Journal of Parasitology 107, 455–462. [DOI] [PubMed] [Google Scholar]
- Achatz TJ, Chermak TP, Martens JR, Pulis EE, Fecchio A, Bell JA, Greiman SE, Cromwell KJ, Brant SV, Kent ML and Tkach VV (2021c) Unravelling the diversity of the Crassiphialinae (Digenea: Diplostomidae) with molecular phylogeny and descriptions of five new species. Current Research in Parasitology & Vector-Borne Diseases 1, 100051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achatz TJ, Martens JR, Kostadinova A, Pulis EE, Orlofske SA, Bell JA, Fecchio A, Oyarzún-Ruiz P, Syrota YY and Tkach VV (2022) Molecular phylogeny of Diplostomum, Tylodelphys, Austrodiplostomum and Paralaria (Digenea: Diplostomidae) necessitates systematic changes and reveals a history of evolutionary host switching events. International Journal for Parasitology 52, 47–63. doi: 10.1016/j.ijpara.2021.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achatz TJ, Chermak TP, Martens JR, Woodyard ET, Rosser TG, Pulis EE, Weinstein SB, McAllister CT, Kinsella JM and Tkach VV (in press) Molecular phylogeny supports invalidation of Didelphodiplostomum and Pharyngostomoides (Digenea: Diplostomoidea) and reveals a Tylodelphys from mammals. Zoological Journal of the Linnean Society. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker F (1915) Parasites of the American muskrat (Fiber zibethicus). Journal of Parasitology 1, 184–195. [Google Scholar]
- Bisseru B (1957) On two new trematodes (Proterodiplostomidae) from an African crocodile, and a list of strigeid parasites from Africa. Journal of Helminthology 31, 85–102. [DOI] [PubMed] [Google Scholar]
- Blasco-Costa I and Locke SA (2017) Life history, systematics and evolution of the Diplostomoidea Poirier, 1886: progress, promises and challenges emerging from molecular studies. Advances in Parasitology 98, 167–225. [DOI] [PubMed] [Google Scholar]
- Bolek MG and Coggins JR (2003) Helminth community structure of sympatric eastern American toad, Bufo americanus, northern leopard frog, Rana pipiens and blue-spotted salamander, Ambystoma laterale, from southeastern Wisconsin. Journal of Parasitology 89, 673–680. [DOI] [PubMed] [Google Scholar]
- Chandler AC (1942) The morphology and life cycle of a new strigeid, Fibricola texensis, parasitic in raccoons. Transactions of the American Microscopical Society 61, 156–167. [Google Scholar]
- Chandler AC and Rausch R (1946) A study of strigeids from Michigan mammals with comments on the classification of mammalian strigeids. Transactions of the American Microscopical Society 65, 328–337. [Google Scholar]
- Chandler AC and Rausch R (1947) A study of strigeids from owls in North Central United States. Transactions of the American Microscopical Society 66, 283–292. [Google Scholar]
- Cribb TH and Pearson JC (1993) Neodiplostomum spratti n. sp. (Digenea: Diplostomidae) from Antechinus spp. (Arsupialia: Dasyuridae) in Australia, with notes on other diplostomids from Australian mammals. Systematic Parasitology 25, 25–35. [Google Scholar]
- Dubois G (1932) Revision des Hemistomes et étude de forms nouvelles. Bulletin de la Société des Sciences Naturelles de Neuchâtel 56, 375–412. [Google Scholar]
- Dubois G (1937) Sur quelques Strigidés. Revue Suisse de Zoologie 44, 391–396. [Google Scholar]
- Dubois G (1938) Monographie des Strigeida (Trematoda). Mémoires de la Société des Sciences Naturelles de Neuchâtel 6, 1–535. [Google Scholar]
- Dubois G (1944) A propos de la spécificité parasitaire des Strigeida. Bulletin de la Société des Sciences Naturelles de Neuchâtel 69, 5–103. [Google Scholar]
- Dubois G (1962) Sur quelques néodiplostomes (Trematoda: Strigeida). Bulletin de la Société Neuchâtaloise des Sciences Naturelles 85, 121–142. [Google Scholar]
- Dubois G (1968) Synopsis des Strigeidae et des Diplostomatidae (Trematoda). Mémoires de la Société des Sciences Naturelles de Neuchâtel 10, 1–258. [Google Scholar]
- Dubois G (1970) Les fondements de la taxonomie des Strigeata La Rue (Trematoda: Strigeida). Revue Suisse de Zoologie 77, 663–685. [PubMed] [Google Scholar]
- Dubois G (1982) Répertoire des synonymes récents de genres et d'espéces de la superfamille des Strigeoidea Railliet, 1919 (Trematoda). Bulletin de la Société des Sciences Naturelles de Neuchâtel 105, 163–183. [Google Scholar]
- Dubois G (1983) Quelques Strigeoidea (Trematoda) récoltés chez des oiseaux du Paraguay par la mission claude Weber, Automne 1983, du Muséum d'Histoire naturelle de Genève. Revue Suisse de Zoologie 92, 641–648. [Google Scholar]
- Dubois G and Macko JK (1972) Contribution à l’étude des Strigeata La Rue, 1926 (Trematoda: Strigeida) de Cuba. Annales de Parasitologie Humaine et Comparee 47, 51–75. [DOI] [PubMed] [Google Scholar]
- Dubois G and Rausch R (1950) Troisième contribution a l’étude des strigeides (Trematoda) Nord-Américains. Société Neuchâteloise des Sciences Naturelles 73, 19–50. [Google Scholar]
- Gillilland III MG and Muzzall PM (1999) Helminths infecting froglets of the Northern leopard frog (Rana pipiens) from Foggy Bottom Marsh, Michigan. Journal of the Helminthological Society of Washington 66, 73–77. [Google Scholar]
- Goldberg SR and Bursey C (2001) Intestinal helminths of four species of skinks (Mabuya) (Sauria: Scincidae) from Southern Africa. Onderstepoort Journal of Veterinary Research 68, 143–147. [PubMed] [Google Scholar]
- Goldberg SR, Bursey CR and Gergus EWA (2001) Helminth communities of subpopulations of the Pacific treefrog, Hyla regilla (Hylidae), from Baja California, México. Southwestern Naturalist 46, 223–230. [Google Scholar]
- Heneberg P, Sitko J and Těšínský M (2020) Paraphyly of Conodiplostomum Dubois, 1937. Parasitology International 76, 102033. [DOI] [PubMed] [Google Scholar]
- Hernández-Mena DI, García-Varela M and Pérez-Ponce de León G (2017) Filling the gaps in the classification of the Digenea Carus, 1863: systematic position of the Proterodiplostomidae Dubois, 1936 within the superfamily Diplostomoidea Poirier, 1886, inferred from nuclear and mitochondrial DNA sequences. Systematic Parasitology 94, 833–848. [DOI] [PubMed] [Google Scholar]
- Hong ST and Shoop WL (1994) Neodiplostomum seoulensis n. comb. (Trematoda: Neodiplostomidae). Journal of Parasitology 80, 660–663. [PubMed] [Google Scholar]
- Huh S, Lee SU and Huh SC (1994) A follow-up examination of intestinal parasitic infections of the army soldiers in Whachon-gun, Korea. Korean Journal of Parasitology 32, 61–63. [DOI] [PubMed] [Google Scholar]
- Kifune T and Uyema N (1982) Reports of Fukuoka University scientific expedition to Peru, 1976. Part 3. Taxonomical studies on trematodes from marsupials and rodents with records of two crabs. Medical Bulletin of Fukuoka University 9, 241–256. [Google Scholar]
- Kudlai O, Stunžėnas V and Tkach V (2015) The taxonomic identity and phylogenetic relationships of Cercaria pugnax and C. helvetica XII (Digenea: Lecithodendriidae) based on morphological and molecular data. Folia Parasitologica 62, 3. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G and Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33, 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima CR, Hoppe EGL, Tebaldi JH, Cruz BC, Barros Gomes AA and Nascimento AA (2013) Gastrintestinal helminths of Cerdocyon thous (Linnaeus, 1766, Smith, 1839) from the caatinga area of the Paraíba State, Brazil. Semina Ciências Agrárias 34, 2879–2888. [Google Scholar]
- Littlewood DTJ and Olson PD (2001) Small subunit rDNA and the Platyhelminthes: signal, noise, conflict and compromise. In Littlewood DTJ and Bray RA (eds), Interrelationships of Platyhelminthes. London, UK: Taylor & Francis, pp. 186–193. [Google Scholar]
- Locke SA, Van Dam AR, Caffara M, Pinto HA, López-Hernández D and Blanar CA (2018) Validity of the Diplostomoidea and Diplostomida (Digenea, Platyhelminthes) upheld in phylogenomic analysis. International Journal of Parasitology 48, 1043–1059. [DOI] [PubMed] [Google Scholar]
- Locke SA, Drago FB, López-Hernández D, Chibwana FD, Núñez V, Van Dam A, Achinelly MF, Johnson PTJ, Assis JCA, Melo AL and Pinto HA (2021) Intercontinental distributions, phylogenetic position and life cycles of species of Apharyngostrigea (Digenea, Diplostomoidea) illuminated with morphological, experimental, molecular and genomic data. International Journal for Parasitology 51, 667–683. [DOI] [PubMed] [Google Scholar]
- Lutz HL, Tkach VV and Weckstein JD (2017) Methods for specimen-based studies of avian symbionts. In Webster M (eds), The Role of Collections in Ornithology: The Extended Specimen. Studies in Avian Biology. Boca Raton, Florida, USA: CRC Press, pp. 157–183. [Google Scholar]
- Miller MJ (1940) A new trematode, Fibricola laruei, from the racoon in Canada. Canadian Journal of Research 18, 333–335. [Google Scholar]
- Niewiadomska K (2002) Family Diplostomidae Poirier, 1886. In Gibson DI, Jones A, Bray RA (eds), Keys to the Trematoda. Vol. I. CAB International and the Natural History Museum, London, pp. 167–196. [Google Scholar]
- Odening K (1965) Kie lebenszyklen der trematoden Neodiplostomum spathoides Dubois and N. attenuatum (V. Linstow) in Raum Berlin. Monatsberichte der Königlichen Preussische Akademie des Wissenschaften zu Berlin 7, 952–954. [Google Scholar]
- Pearson JC (1959) Observations on the morphology and life cycle of Strigea elegans Chandler & Rausch, 1947 (Trematoda: Strigeidae). Journal of Parasitology 45, 155–174. [PubMed] [Google Scholar]
- Penrod FW (1947) Neodiplostomum banghami, a new diplostomid strigeoidean trematode from an eagle. Transactions of the American Microscopical Society 66, 144–148. [PubMed] [Google Scholar]
- Premvati G and Bair TD (1979) Trematode parasites of the opossum, Didelphis virginiana, from Florida. Proceedings of the Helminthological Society of Washington 46, 207–212. [Google Scholar]
- Queiroz MS, López-Hernández D, Locke SA, Pinto HA and Anjos LA (2020) Metacercariae of Heterodiplostomum lanceolatum (Trematoda: Proterodiplostomidae) found in Leptodactylus podicipinus (Anura: Leptodactylidae) from Brazil: a morphological, molecular and ecological study. Journal of Helminthology 94, E66. [DOI] [PubMed] [Google Scholar]
- Read CP (1948) Strigeids from Texas mink with notes on the genus Fibricola Dubois. Transactions of the American Microscopical Society 67, 165–168. [Google Scholar]
- Richardson DJ (2013) Helminth parasites of the raccoon (Procyon lotor), Virginia opossum (Didelphis virginiana), and striped skunk (Mephitis mephitis) from Keith County, Nebraska. Transactions of the Nebraska Academy of Sciences and Affiliated Societies 33, 35–38. [Google Scholar]
- Ronquist F and Huelsenbeck JP (2003) MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. [DOI] [PubMed] [Google Scholar]
- Seo BS (1989) Comparative growth and development of the metacercariae of Fibricola seoulensis (Trematoda: Diplostomidae) in vitro, in vivo and on the chick chorioallantois. Korean Journal of Parasitology 27, 231–248. [DOI] [PubMed] [Google Scholar]
- Seo BS, Rim HJ and Lee CW (1964) Studies on the parasitic helminths of Korea I. Trematodes of rodents. Korean Journal of Parasitology 2, 20–26. [DOI] [PubMed] [Google Scholar]
- Sereno-Uribe AL, Andrade-Gómez L, Ostrowski de Núñez M, Pérez-Ponce de León GP and García-Varela M (2019) Assessing the taxonomic validity of Austrodiplostomum spp. (Digenea: Diplostomidae) through nuclear and mitochondrial data. Journal of Parasitology 105, 102–112. [PubMed] [Google Scholar]
- Shoop WL (1989) Systematic analysis of the Diplostomidae and Strigeidae (Trematoda). Journal of Parasitology 75, 21–32. [PubMed] [Google Scholar]
- Snyder SD and Tkach VV (2007) Neosychnocotyle maggiae, n. gen., n. sp. (Platyhelminthes: Aspidogastrea) from freshwater turtles in northern Australia. Journal of Parasitology 93, 399–403. [DOI] [PubMed] [Google Scholar]
- Tkach VV and Pawlowski J (1999) A new method of DNA extraction from the ethanol-fixed parasitic worms. Acta Parasitologica 44, 147–148. [Google Scholar]
- Tkach VV, Littlewood DTJ, Olson PD, Kinsella JM and Swiderski Z (2003) Molecular phylogenetic analysis of the Microphalloidea Ward, 1901 (Trematoda: Digenea). Systematic Parasitology 56, 1–15. [DOI] [PubMed] [Google Scholar]
- Tkach VV, Achatz TJ, Pulis EE, Junker K, Snyder SD, Bell JA, Halajan A and Melo FTV (2020) Phylogeny and systematics of the Proterodiplostomidae Dubois, 1936 (Digenea: Diplostomoidea) reflect the complex evolutionary history of the ancient digenean group. Systematic Parasitology 97, 409–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmer MJ (1955) Notes on the morphology and host–parasite specificity of Fibricola cratera (Barker and Noll, 1915) Dubois 1932 (Trematoda: Diplostomatidae). Journal of Parasitology 41, 456–466. [PubMed] [Google Scholar]
- Ulmer MJ (1970) Notes on rearing snails in the laboratory. In MacInnis AJ and Voge M (eds), Experiments and Techniques in Parasitology. San Francisco, USA: WH Freeman and Company, pp. 143–144. [Google Scholar]
- Weinstein SB, Van Wert JC, Kinsella JM, Tkach VV and Laffery KD (2019) Infection at an ecotone: cross-system foraging increases satellite parasites but decreases core parasites in raccoons. Ecology 100, e02808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodyard ET, Rosser TG and Griffin MJ (2017) New data on Neodiplostomum americanum Chandler and Rausch, 1947 (Digenea: Diplostomidae), in the great horned owl Bubo virginianus Gmelin, 1788 and the eastern screech owl Megascops asio Linnaeus, 1758 in Mississippi, USA. Parasitology Research 116, 2075–2089. [DOI] [PubMed] [Google Scholar]
- Zerecero C (1943) Algunos tremátodos de las ratas domésticas de la Ciudad de México. Anales del Instituto de Biología serie Zoología 14, 507–526. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For supplementary material accompanying this paper visit https://doi.org/10.1017/S003118202100216X.
click here to view supplementary material
Data Availability Statement
The data supporting the findings of this study are available within the article and in the Supplementary materials. All newly generated sequences were deposited in the GenBank database under the following accession numbers: OL764381, OL770020–OL770068, OL770124–OL770126 and OL799069–OL799108.