Abstract
Clinical blood isolates from three sequential episodes of endocarditis occurring over a 6-month period in a child with a malformative cardiopathy were investigated. All isolates identified as Abiotrophia defectiva were resistant to erythromycin-clindamycin and to tetracycline-minocycline, due to the presence of sequences homologous to the erythromycin resistance gene ermB and to the tetracycline resistance gene tet(M), respectively. These resistance genes were located on a chromosomally borne composite Tn916-related transposon. These results demonstrate the involvement of conjugative transposons in the dissemination of antibiotic resistance in the genus Abiotrophia.
Abiotrophia defectiva and Abiotrophia adiacens, previously referred to as nutritionally variant streptococci Streptococcus defectivus and Streptococcus adjacens, respectively, are members of the normal flora of the human pharynx and the human urogenital and intestinal tracts (14, 15). It is thought that they may be responsible for most cases of blood-culture-negative endocarditis and account for 3 to 6% of streptococcal strains isolated from blood in this setting (3). Moreover, these bacteria have been isolated from patients with a variety of other infectious diseases, such as otitis media, endophthalmitis, and brain abscess (2, 4, 10, 14). Cases of infective endocarditis (IE) caused by these microorganisms exhibit much higher rates of relapse, treatment failure, and mortality than those due to other oral streptococci (3, 17). Clinical isolates of Abiotrophia are usually susceptible to antibiotics active against gram-positive bacteria, and most strains reported were susceptible to erythromycin, clindamycin, and tetracycline (6, 8). We report here our studies of the mechanism of erythromycin and tetracycline resistance in a clinical isolate of A. defectiva responsible for sequential episodes of IE in a child.
Case report.
The patient was a 5-year-old boy with a ventricular septal defect and pulmonary stenosis. Because of severe congestive heart failure, a surgical intervention was performed 2 months before the first episode of IE. He remained in a stable condition but was admitted to a hospital because of a 2-week history of intermittent fever. On admission, his temperature was 38°C and the vital signs were normal. A transthoracic echocardiogram revealed normal heart valves without evidence of vegetations. Six blood cultures obtained at the time of the admission were positive within 4 days. Direct Gram stain of these cultures revealed a pleiomorphic gram-positive microorganism with small coccoid forms mixed with long coccobacillar swollen forms. The child was treated with penicillin at 200,000 U/kg of body weight/day administered every 4 h and with gentamicin at 3 mg/kg/day administered every 8 h intravenously for 10 days. All symptoms disappeared on the second day of therapy, and the patient was discharged after a 2-week hospitalization and prescribed oral amoxicillin at 100 mg/kg/day for a further 2 weeks. He was rehospitalized 2 months later because of fever. On admission, the results of his physical examination were the same as those at the previous discharge. Two blood cultures obtained were positive for the same bacteria. The echocardiogram was unchanged, and he was treated for 2 weeks intravenously with amoxicillin (200 mg/kg/day) every 4 h plus gentamicin (3 mg/kg/day) every 8 h, followed by 3 weeks of oral amoxicillin (100 mg/kg/day). Blood cultures obtained during therapy were sterile. However, 1 day after stopping antibiotics, he again became febrile and two blood cultures were positive with the same bacteria. A search for a source of infection was not successful; in particular, a dental examination showed no evidence of disease. Amoxicillin (150 mg/kg/day) associated with rifampin (20 mg/kg/day) was given for 6 months. The patient was monitored for 18 months after this fourth course of antibiotic therapy and has done well.
Identification of A. defectiva NEM1418.
For all positive blood cultures (Organon Tecknica, Fresne, France), small unpigmented colonies with α-hemolysis grew after 48 h on Columbia horse blood agar (Bio-Merieux Ltd., Marcy l'Etoile, France) under 5% CO2 at 37°C. No bacterial growth was observed on chocolate, tryptic soy agar (TSA), or Mueller-Hinton blood agar plates, whereas a positive satellite test was observed around a streak of Staphylococcus aureus on the same media. Gram staining of colonies from Columbia blood agar plates and of bacteria from brain heart infusion broth showed extremely pleiomorphic gram-positive microorganisms with small coccoid forms lying next to elongated forms. The catalase and oxidase reactions were negative. Strains were identified as A. defectiva by conventional biochemical methods using the API 20 and ID32 STREP System (Bio-Merieux Ltd.). The identification at the species level was confirmed by partial sequencing of the gene encoding 16S rRNA and of the gene sodA (13) encoding the manganese-dependent superoxide dismutase (data not shown). Analysis by pulsed-field gel electrophoresis (PFGE) of NotI- and SfiI-digested genomic DNAs of two representative isolates of each of the three infective episodes revealed that the restriction profiles were indistinguishable from each other and were different from those obtained with unrelated strains of A. defectiva from our collection, including the reference strain CIP 103242 T (Fig. 1 and data not shown). This strain was designated NEM1418.
FIG. 1.
Analysis of restriction enzyme-digested genomic DNA by PFGE and Southern hybridization. The enzymes NotI (A) and SfiI (B) were used to cleave the DNAs extracted from the strains of A. defectiva isolated during the first, second, and third episodes of a case of IE (lanes 1 to 3, respectively). Southern blot analysis was carried out with digoxigenin-labelled DNA fragments specific for the genes ermB (lanes 3′) and tet(M) (lanes 3"). Bacteriophage λ concatemers were used as molecular-size markers (lanes M). The electrophoretic and hybridization procedures and the DNA probes used were as previously described (12).
Identification of the antibiotic-resistant determinants of A. defectiva NEM1418.
Disk diffusion tests were performed on Columbia agar supplemented with 5% lysed horse blood and 100 μg of pyridoxal hydrochloride per ml, and they were interpreted according to the guidelines of the Comité Français de l'Antibiogramme (1). The MICs were determined on the same medium by a dilution method with an inoculum of 107 CFU/ml. Strain NEM1418 was resistant to erythromycin (MIC, 128 mg/liter) and clindamycin (MIC, 512 mg/liter) and to tetracycline-minocycline (MICs, 64 and 32 mg/liter, respectively). NEM1418 was susceptible to penicillin, amoxicillin, cefotaxime, streptomycin, kanamycin, gentamicin, chloramphenicol, vancomycin, teicoplanin, and rifampin. We tested by Southern blotting under highly stringent conditions for the presence in NEM1418 of nucleotide sequences that are structurally related to ermB and tet(M) and to conjugative transposon Tn916 (12). Sequences homologous to tet(M) and to ermB were detected on 4.8-kb and 800-bp HincII fragments, respectively (data not shown). In addition to these fragments, the Tn916 probe hybridized with 5.5-, 4.5-, and 2.8-kb HincII fragments. These resistant determinants are located on the same 300-kb SfiI and 50-kb NotI DNA fragments (Fig. 1). It is worth mentioning that the tet(M) gene of Tn916 is located on a 4.8-kb HincII fragment and that this element also possesses a 5.5-kb internal HincII fragment (7). We thus interpret our results as indicating that resistance to tetracycline in NEM1418 is due to the presence of one copy of a chromosomally borne element structurally related to Tn916 and that the erythromycin resistance gene ermB resides close to or within this element.
Genetic organization of the Tn916-related element harbored by A. defectiva NEM1418.
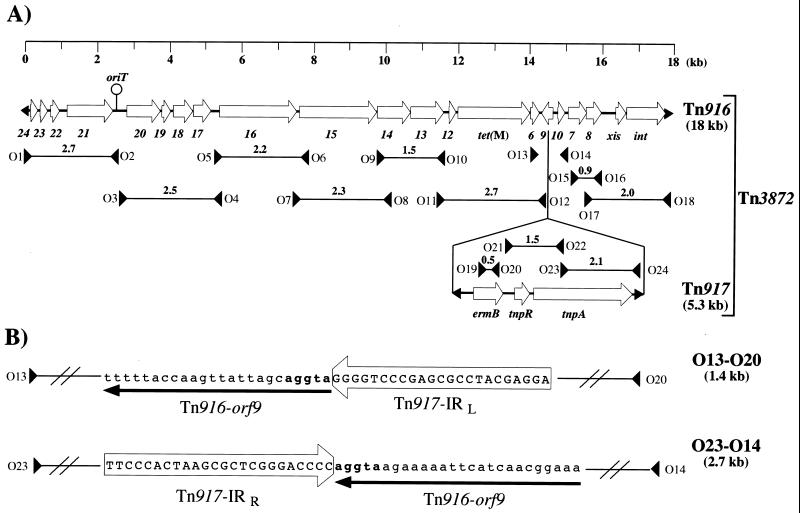
We characterized the genetic organization of the Tn916-like conjugative element harbored by NEM1418 by using pairs of primers (Fig. 2) that enable the amplification of DNA segments spanning all Tn916 (7). PCRs were performed on a Gene Amp System 2400 instrument (Perkin-Elmer Cetus, Roissy, France) in a final volume of 50 μl containing 250 ng of genomic DNA of NEM1418 as the template, 0.5 μM concentrations of each primer, 200 μM concentrations of each deoxynucleoside triphosphate, and 1 U of Taq DNA polymerase in a 1× amplification buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl, 1.5 mM MgCl2). The PCR mixtures were denatured (5 min at 95°C) and then subjected to 35 cycles of amplification (90 s of annealing at 55°C, 2 min of elongation at 72°C, and 30 s of denaturation at 95°C) and 72°C for 10 min for the last elongation cycle. The corresponding PCR products obtained by using DNAs extracted from NEM1418 and NEM420 (Enterococcus faecalis::Tn916) as templates were separated by agarose gel electrophoresis, and the results obtained are summarized in Fig. 2A. In both DNA cases, fragments of similar sizes were amplified by using all but one pair of primers, O13-O14. No amplification product was obtained with these primers when NEM1418 DNA was used as the template. These results indicate that the conjugative element harbored by NEM1418 is closely related to Tn916. In dot blot analysis, 32P-labelled oligonucleotides O13 and O14 were found to hybridize to NEM1418 and NEM420 genomic DNAs but not to those extracted from hosts devoid of sequences related to Tn916 (data not shown). We thus interpret the failure to amplify the expected 604-bp O13-O14 amplicon from NEM1418 DNA as indicating that the corresponding segment might contain a large DNA insertion possibly bearing the ermB gene. The primers O13-O14 delineate the gene orf9 of the Tn916 regulatory region, and a survey of the literature revealed that only one Tn916 derivative carrying ermB in this region has been described up to now (9). This element, designated Tn3872, was detected in Streptococcus pneumoniae and was generated by insertion of Tn917 into Tn916 orf9. The presence of sequences related to Tn917 in NEM1418 was demonstrated (Fig. 2A) by amplifying DNA segments corresponding to the regions containing ermB (primers O19 and O20), tnpR and tnpA (primers O21 and O22), and tnpA (primers O23 and O24) genes (16). To further characterize the transposon carried by NEM1418, the Tn916::Tn917 junction fragments present in the O13-O20 and O23-O14 amplicons (Fig. 2B) were sequenced, which revealed their identity with the corresponding junction fragments in Tn3872. We therefore concluded that tetracycline and erythromycin resistances in A. defectiva NEM1418 are due to the presence of a transposon identical or similar to Tn3872.
FIG. 2.
Structural analysis of Tn3872 in A. defectiva NEM1418. (A) The structures of Tn916 (7) and Tn917 (16) are shown, and their main orf genes are labelled as described. The black arrows at the extremities of Tn916 and Tn917 represent their terminal inverted repeats. The location and strand specificity of the PCR primers (O1 to O24) in both transposons are indicated by large black arrows. Positive PCR obtained by using the NEM1418 DNA template are depicted by a horizontal line which joins the primer pairs used, and the size (in kilobases) of the corresponding amplicon is indicated above the line. (B) Partial sequence analysis of the O13-O20 and O23-O14 amplicons. The nucleotide sequence of the 23-bp long left (IRL) and right (IRR) inverted repeat of Tn917 are boxed. The sequence corresponding to Tn916 orf9 is underlined by an arrow oriented according to the direction of transcription. The 5-bp duplications at the Tn917 insertion site are written in bold characters. The sequences (5′ to 3′) of the primers were as follows: O1, GGACTTATCACACTTTATCAAGG; O2, GCCTTGTAATACCAGTCG; O3, CCGTTTAGCTGTTGCGACTGG; O4, CGTAAGCATAACATTCCCCG; O5, GCAAGCTCAAAGCGGTTGCC; O6, CTGTTTACTATTGATGGTTTC; O7, TCAAACTGGACGCTGAAACG; O8, GAGCCAGCACTTCTGCGG; O9, ACAGGTGGAATCGGGCGG; O10, CCCGTCATTCACATAGTAGG; O11, GGTACTTGAAAAGAACGGGAG; O12, TCCATACATAACGGAAAGAGCC; O13, CGGCTCTTTCCGTTATGTATGG; O14, GATGTACTTCATGGCGACG; O15, GTACGTCCACCAATGTGG; O16, GCACGCTTCCACGAAAGGAG; O17, CTCTCCTTTCGTGGAAGCG; O18, GTACTACTAAGCAACAAGACGC; O19, GGTAAAGGGCATTTAACGAC; O20, CGATATTCTCGATTGACCCA; O21, CCAAGGAGCTAAAGAGGTCCC; O22, GTCCCGAGTCCCATGGAAGC; O23, GCTTCCATGGGACTCGGGAC; O24, GCTCCCAATTAATAGGAGA.
Mobility of the Tn916-like transposon Tn3872 element of A. defectiva NEM1418.
Amplification of the Tn916 attachment site by PCR, as previously described (18), revealed that Tn3872 excised in A. defectiva NEM1418 (data not shown). When NEM1418 was filter mated (12) with E. faecalis JH2-2, transfer of the erythromycin and tetracycline resistance determinant was detected at a very low frequency (0.2 × 10−8 per donor). The presence of Tn3872 in these transconjugants was confirmed by a PCR analysis carried out by using the pairs of primers described in Fig. 2A (data not shown). Six JH2-2::Tn3872 transconjugants independently obtained were used as donors in ensuing matings with E. faecalis BM4110 recipients. Conjugative transposition of Tn3872 was then detected at frequencies ranging from 4 × 10−6 to 2.8 × 10−8, depending upon the donor strains. These frequencies of conjugative transposition are similar to those obtained with Tn916 when transferred between E. faecalis strains (5). On the other hand, conjugal transfer of Tn916-Km from E. faecalis EFCP2 (11) to A. defectiva NEM1418 was repeatedly detected at a low frequency (0.4 × 10−8 per donor). Taken together, these results indicate that members of the genus Abiotrophia can contribute to the dissemination of Tn916-related elements by acting both as donors and recipients. They also demonstrate that conjugative transposition of this type of element does not require expression of orf9.
Conclusions.
There are rare reports of strains of Abiotrophia spp. resistant to antibiotics (6, 8, 10), but the genetic basis of the resistance phenotypes has never been characterized. A. defectiva NEM1418, responsible for an IE in a child, was resistant to erythromycin and tetracycline, and we have shown that this resistance phenotype was due to the presence of a Tn916-related element similar to the pneumococcal element Tn3872 (9). To the best of our knowledge, these results constitute the first demonstration of the involvement of conjugative transposons in the dissemination of antibiotic resistance in the genus Abiotrophia.
Acknowledgments
We thank C. Tinsley for critical reading of the manuscript.
This work was supported by a grant from PRFMMIP-98.
REFERENCES
- 1.Acar J, Carret G, Cavallo J D, Chardon H, Choutet P, Courvalin P, Dabernat H, Drugeon H, Dubreuil L, Goldstein F, Jarlier V, Leclercq R, Nicolas-Chanoine M H, Philippon A, Rouveix B, Sirot J, Soussy C J, Thabault A. Le communiqué 1998 du comité de l'antibiogramme de la société française de microbiologie. Pathol Biol. 1998;46:1–16. [PubMed] [Google Scholar]
- 2.Biermann C, Fries G, Jehnichen P, Bhakdi S, Husmann M. Isolation of Abiotrophia adiacens from a brain abscess which developed in a patient after neurosurgery. J Clin Microbiol. 1999;37:769–771. doi: 10.1128/jcm.37.3.769-771.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bouvet A. Human endocarditis due to nutitionally variant streptococci: Streptococcus adjacens and Streptococcus defectivus. Eur Heart J. 1995;16(Suppl. B):24–27. doi: 10.1093/eurheartj/16.suppl_b.24. [DOI] [PubMed] [Google Scholar]
- 4.Bouvet A, Grimont F, Grimont P A D. Streptococcus defectivus sp. nov. and Streptococcus adjacens sp. nov., nutritionally variant streptococci from human clinical specimens. Int J Syst Bacteriol. 1989;39:290–294. doi: 10.1099/00207713-41-4-483. [DOI] [PubMed] [Google Scholar]
- 5.Clewell D B, Gawron-Burke C. Conjugative transposons and the dissemination of antibiotic resistance. Annu Rev Microbiol. 1986;40:635–659. doi: 10.1146/annurev.mi.40.100186.003223. [DOI] [PubMed] [Google Scholar]
- 6.Cooksey R C, Swenson J M. In vitro antimicrobial inhibition patterns of nutritionally variant streptococci. Antimicrob Agents Chemother. 1979;16:514–518. doi: 10.1128/aac.16.4.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flannagan S E, Zitzow L A, Su Y A, Clewell D B. Nucleotide sequence of the 18-kb conjugative transposon Tn916 from Enterococcus faecalis. Plasmid. 1994;32:350–354. doi: 10.1006/plas.1994.1077. [DOI] [PubMed] [Google Scholar]
- 8.Gephart J F, Washington J A., II Antimicrobial susceptibilities of nutritionally variant streptococci. J Infect Dis. 1982;146:536–539. doi: 10.1093/infdis/146.4.536. [DOI] [PubMed] [Google Scholar]
- 9.McDougal L K, Tenover F, Lee L N, Rasheed J K, Patterson J E, Jorgensen J H, Leblanc D J. Detection of Tn917-like sequences within a Tn916-like conjugative transposon (Tn3872) in erythromycin-resistant isolates of Streptococcus pneumoniae. Antimicrob Agents Chemother. 1998;42:2312–2318. doi: 10.1128/aac.42.9.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Namdari H, Kintner K, Jackson B A, Namdari S, Hughes J L, Peairs R R, Savage D J. Abiotrophia species as a cause of endophthalmitis following cataract extraction. J Clin Microbiol. 1999;37:1564–1566. doi: 10.1128/jcm.37.5.1564-1566.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poyart C, Celli J, Trieu-Cuot P. Conjugative transposition of Tn916-related elements from Enterococcus faecalis to Escherichia coli and Pseudomonas fluorescens. Antimicrob Agents Chemother. 1995;39:500–506. doi: 10.1128/aac.39.2.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poyart C, Pierre C, Quesne G, Pron B, Berche P, Trieu-Cuot P. Emergence of vancomycin resistance in the genus Streptococcus: characterization of a vanB transferable determinant in Streptococcus bovis. Antimicrob Agents Chemother. 1997;41:24–29. doi: 10.1128/aac.41.1.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poyart C, Quesne G, Coulon S, Berche P, Trieu-Cuot P. Identification of streptococci to species level by sequencing the gene encoding the manganese-dependent superoxide dismutase. J Clin Microbiol. 1998;36:41–47. doi: 10.1128/jcm.36.1.41-47.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roberts R B, Krieger A G, Schiller N L, Gross K C. Viridans streptococcal endocarditis: the role of various species, including pyridoxal-dependent streptococci. Rev Infect Dis. 1979;1:955–966. doi: 10.1093/clinids/1.6.955. [DOI] [PubMed] [Google Scholar]
- 15.Ruoff K L. Nutritionally variant streptococci. Clin Microbiol Rev. 1991;4:184–190. doi: 10.1128/cmr.4.2.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shaw J H, Clewell D B. Complete nucleotide sequence of macrolide-lincosamide-streptogramin B-resistance transposon Tn917 in Streptococcus faecalis. J Bacteriol. 1985;164:782–796. doi: 10.1128/jb.164.2.782-796.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stein D S, Nelson K E. Endocarditis due to nutritionally deficient streptococci: therapeutic dilemma. Rev Infect Dis. 1987;5:908–916. doi: 10.1093/clinids/9.5.908. [DOI] [PubMed] [Google Scholar]
- 18.Storrs M J, Poyart-Salmeron C, Trieu-Cuot P, Courvalin P. Conjugative transposition of Tn916 requires the excisive and integrative activities of the transposon-encoded integrase Int-Tn. J Bacteriol. 1991;173:4347–4352. doi: 10.1128/jb.173.14.4347-4352.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]