Abstract
T follicular helper (Tfh) cells are the conventional drivers of protective, germinal center (GC)-based antiviral antibody responses. However, loss of Tfh cells and GCs has been observed in patients with severe COVID-19. As T cell-B cell interactions and immunoglobulin class switching still occur in these patients, non-canonical pathways of antibody production may be operative during SARS-CoV-2 infection. We found that both Tfh-dependent and -independent antibodies were induced against SARS-CoV-2 infection, SARS-CoV-2 vaccination, and influenza A virus infection. Even though Tfh-independent antibodies to SARS-CoV-2 had evidence of reduced somatic hypermutation, they were still high-affinity, durable, and reactive against diverse spike-derived epitopes and were capable of neutralizing both homologous SARS-CoV-2 and the B.1.351 (beta) variant of concern. Indeed, we found by epitope mapping and BCR sequencing that Tfh cells focused the B cell response and therefore, in the absence of Tfh cells, a more diverse clonal repertoire was maintained. These data support an alternative pathway for the induction of B cell responses during viral infection that enables effective, neutralizing antibody production to complement traditional GC-derived antibodies that might compensate for GCs damaged by viral inflammation.
One-Sentence Summary:
Complementary Tfh cell-dependent and -independent pathways of antibody production mediate neutralizing responses to SARS-CoV-2.
Introduction:
Antibodies mediate protection against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of the coronavirus disease 2019 (COVID-19) pandemic (1). T follicular helper (Tfh) cells are the conventional drivers of protective antibody responses, as they support immunoglobulin class switching, germinal center (GC)-based affinity maturation, and long-lived humoral immunity (2). Indeed, multiple studies have reported a correlation between circulating Tfh (cTfh) cells – in particular, type 1 CXCR3+CCR6− cTfh cells – and neutralizing antibody titers in COVID-19 patients (3). While there have been mixed findings on the association of Tfh cells with disease severity, several groups have observed an absence of Tfh cells and GCs in severely ill patients (4–7). Despite a loss of Tfh cells and GC structures, T cell-B cell interactions and antibody class switching still occur in the secondary lymphoid organs of these patients (6). Findings of enhanced extrafollicular B cell responses associated with severe disease further suggest that non-canonical pathways of antibody production may be operative in these individuals (6, 8). However, a causal relationship between antibody provenance and disease severity cannot be established by existing human studies. Thus, it remains unclear whether antibodies produced through non-canonical pathways without Tfh cell help are also protective against SARS-CoV-2.
No study has evaluated the requirement for Tfh cells in antibody production to SARS-CoV-2. Previous work with a related coronavirus, SARS-CoV-1, has shown that CD4+ T cells are required for neutralizing antibody production (9), but the necessary CD4+ T cell subset has not been identified. For other viruses, as well as bacteria, Tfh cells are thought to be required for class-switched, pathogen-specific antibody production, especially at later timepoints (10–18). In contrast, different studies have found conflicting results on the requirement of Tfh cells during vaccination in mice. Certain vaccine strategies induce robust Tfh-independent antibody responses, though of lower affinity (19, 20), while other strategies fail to induce durable, class-switched, or high-affinity antibodies in mice with Tfh cell deficiency or dysfunction (11, 21–23). In humans though, cTfh cell populations in the blood correlate with the response to influenza vaccination (24, 25). Taken together, while non-Tfh CD4+ T cells can promote effective antibodies in certain contexts, protective anti-pathogen humoral immunity is largely thought to be Tfh cell-dependent.
We therefore directly tested whether non-Tfh CD4+ T cells could compensate for Tfh cell loss in severe COVID-19, and perhaps during acute viral infection in general, by promoting class-switched antibodies. We also sought to compare the Tfh-independent antibody response to SARS-CoV-2 infection with that induced by mRNA vaccination. Based on prior work, we hypothesized that antibodies induced through such non-canonical mechanisms would be lower in quantity and quality. To test this, we characterized the titer, isotype, longevity, affinity, epitope reactivity, and function of antibodies to SARS-CoV-2 infection from Tfh-deficient mice. We also examined antibody titers and isotypes following SARS-CoV-2 vaccination and influenza A virus infection. Both Tfh cells and non-Tfh CD4+ T cells promoted class-switched antibodies to SARS-CoV-2 infection, SARS-CoV-2 vaccination, and influenza A virus. Contrary to our expectations, Tfh-independent antibodies to SARS-CoV-2 were durable and remarkably high-affinity, though B cell receptors from Tfh-deficient mice demonstrated impaired somatic hypermutation and mutation selection. Tfh-independent antibodies were also broadly reactive against diverse SARS-CoV-2 epitopes and effective at neutralizing both homologous and heterologous viruses. Thus, our study suggests a shift in paradigm for T cell-driven antibody responses to viruses in which multiple types of CD4+ T cells in lymphoid organs coordinate the humoral response to viruses. This understanding of T cell-B cell interaction could aid in approaches to develop effective protection against SARS-CoV-2 as well as other viruses.
Results:
SARS-CoV-2 infection and vaccination induce Tfh-dependent and -independent antibodies
To study the cellular pathways that promote antibody production to SARS-CoV-2, we used mice that lack different CD4+ T cell subsets. Bcl6fl/flCd4Cre mice are a well-established model of Tfh cell deficiency due to deletion of the Tfh lineage-defining transcription factor BCL6 (17, 22, 26). Ciita−/− mice lack all CD4+ T cells due to loss of MHC class II expression (27). As SARS-CoV-2 is unable to efficiently interact with mouse ACE2, we utilized two models of human ACE2 (hACE2) overexpression: intratracheal administration of AAV-hACE2, which leads to hACE2 expression in the respiratory tract, and K18-hACE2 transgenic mice, which express hACE2 in epithelial cells (28–30).
We first assessed the cellular responses to SARS-CoV-2 infection in AAV-hACE2 mice. Two weeks after AAV-hACE2 transduction, we infected mice intranasally with SARS-CoV-2 (isolate USA-WA1/2020). At 14 days post infection (dpi), PD-1hiCXCR5hi Tfh cells were efficiently deleted in the mediastinal LN (medLN) of AAV-hACE2 Bcl6fl/flCd4Cre mice (Fig. S1, A to C and Data S1). Consistent with the loss of Tfh cells, GC B cells were severely impaired, and plasmablast formation was reduced (Fig. S1, D to H). Yet, viral burden in the lungs of AAV-hACE2 Bcl6fl/fl and Bcl6fl/flCd4Cre mice was similar at 7 dpi (Fig. S1I). To examine the cellular architecture of the medLN in mice lacking Tfh cells, we performed immunofluorescence on K18-hACE2 Bcl6fl/fl and Bcl6fl/flCd4Cre mice at 14 dpi. K18-hACE2 Bcl6fl/fl medLN exhibited robust GC formation, with large clusters of activated (GL7+) B cells colocalizing with CD35+ follicular dendritic cell (FDC) networks and infiltrated by T cells (Fig. S2A), presumably Tfh cells. In contrast, K18-hACE2 Bcl6fl/flCd4Cre medLN displayed a few small clusters of GL7+ cells that co-localized with sparse FDC networks but lacked T cells (Fig. S2B), congruent with the absence of Tfh cells. As GL7 is a marker of activated lymphocytes, including those outside of GCs, these rare cellular aggregates lacking T cells could represent sites of T-independent antigen-driven B cell stimulation.
We next determined the humoral responses to SARS-CoV-2 infection in AAV-hACE2 mice at 14 dpi (Fig. 1A). Bcl6fl/fl mice produced high levels of spike (S)-specific IgG antibodies (Fig. 1B). While Bcl6fl/flCd4Cre mice had reduced levels of S-specific IgG, they still produced substantially more compared to Ciita−/− mice. Among IgG subclasses, S-specific IgG1 and IgG3 were completely Tfh-dependent, while S-specific IgG2b and IgG2c were promoted by both Tfh and non-Tfh CD4+ T cells (Fig. 1B). Consistent with their divergent requirement for Tfh cell help, S-specific IgG2c was induced earlier than S-specific IgG1 (Fig. S3A). S-specific IgM was unaffected by the absence of Tfh cells (Fig. S3B). Together, these results suggest that both Tfh cells and non-Tfh CD4+ T cells promote the production of class-switched antibodies to SARS-CoV-2, though Tfh cells are specifically able to induce certain subclasses.
Figure 1: SARS-CoV-2 infection and vaccination induce Tfh-dependent and -independent antibodies.
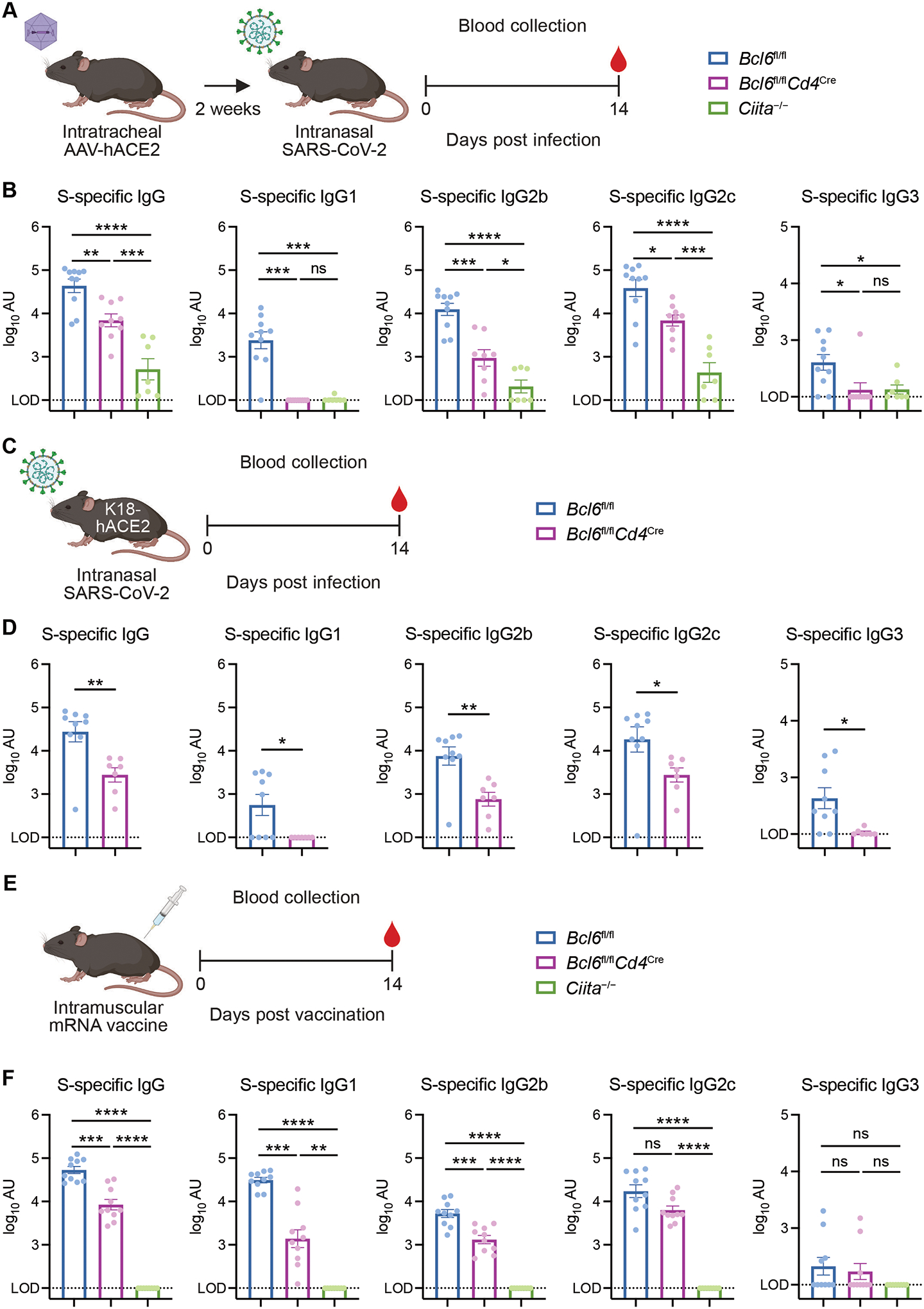
(A) Schematic of experimental design for assessing serum antibody responses against SARS-CoV-2 in AAV-hACE2 mice. Mice were infected intranasally with SARS-CoV-2 (isolate USA-WA1/2020) two weeks following intratracheal administration of AAV-hACE2. Sera were collected 14 days post infection (dpi) for quantification of antibody titers by ELISA.
(B) Spike (S)-specific total IgG, IgG1, IgG2b, IgG2c, and IgG3 antibody titers in sera from control (Bcl6fl/fl; blue symbol), Tfh-deficient (Bcl6fl/flCd4Cre; magenta symbol), or CD4+ T cell-deficient (Ciita−/−; green symbol) mice at 14 dpi with SARS-CoV-2.
(C) Schematic of experimental design for assessing serum antibody responses against SARS-CoV-2 in K18-hACE2 mice. Mice were infected with SARS-CoV-2 (isolate USA-WA1/2020) by intranasal administration. Sera were collected at 14 dpi for quantification of antibody titers by ELISA.
(D) S-specific total IgG, IgG1, IgG2b, IgG2c, and IgG3 antibody titers in sera from K18-hACE2 Bcl6fl/fl (blue) or K18-hACE2 Bcl6fl/flCd4Cre (magenta) mice at 14 dpi with SARS-CoV-2.
(E) Schematic of experimental design for assessing serum antibody responses against SARS-CoV-2 mRNA vaccination. Mice were vaccinated intramuscularly with Moderna mRNA-1273 or Pfizer-BioNTech BNT162b2 mRNA vaccine. Sera were collected 14 days post vaccination (dpv) for quantification of antibody titers by ELISA.
(F) S-specific total IgG, IgG1, IgG2b, IgG2c, and IgG3 antibody titers in sera from Bcl6fl/fl (blue), Bcl6fl/flCd4Cre (magenta), or Ciita−/− (green) mice at 14 dpv with mRNA vaccine.
LOD, limit of detection of the assay. Statistical significance was assessed by one-way ANOVA with Tukey’s test, or Welch’s t-test with Bonferroni multiple hypothesis correction when sample variances were 0 (B and F); two-tailed unpaired t-test or Welch’s t-test, based on the F test for unequal variance (D). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. ns, not significant. Data are expressed as mean ± standard error of mean (SEM) log10 arbitrary units (AU). Each symbol represents an individual mouse. Data are aggregated from at least two independent experiments with a total of seven to ten mice per condition.
To corroborate these findings with a model that does not require AAV pre-transduction, we measured serum antibody titers of K18-hACE2 Bcl6fl/fl and Bcl6fl/flCd4Cre mice at 14 dpi with SARS-CoV-2 (Fig. 1C). K18-hACE2 Bcl6fl/flCd4Cre mice produced substantial levels of S-specific IgG antibodies, though reduced compared to K18-hACE2 Bcl6fl/fl mice (Fig. 1D). K18-hACE2 Bcl6fl/flCd4Cre mice also demonstrated similar patterns of IgG subclass production to AAV-hACE2 Bcl6fl/flCd4Cre mice. While S-specific IgG1 and IgG3 were completely abrogated in the absence of Tfh cells, S-specific IgG2b, IgG2c, and IgM were only partially reduced (Fig. 1D and S3C). This alternative model of SARS-CoV-2 infection therefore confirmed that certain IgG subclasses could be generated through Tfh-independent mechanisms.
Having observed Tfh-independent class-switched antibody production to SARS-CoV-2 infection, we asked whether this could also happen during non-live pathogen-driven immune stimulation such as vaccination. We hypothesized that while the strong and prolonged inflammatory response caused by infection might overcome the requirement for Tfh cell help, antibodies to vaccination would still be fully Tfh-dependent. We therefore vaccinated mice intramuscularly with a single dose of Moderna mRNA-1273 or Pfizer-BioNTech BNT162b2 mRNA vaccine and evaluated antibody responses at 14 days post vaccination (Fig. 1E). As with SARS-CoV-2 infection, mRNA vaccination induced both Tfh- and non-Tfh CD4+ T cell-dependent S-specific IgG antibodies (Fig. 1F). mRNA vaccination promoted higher levels of S-specific IgG1 compared to infection, and some IgG1 could even be made in the absence of Tfh cells, contrasting with the complete Tfh cell dependence of infection-induced IgG1. S-specific IgG2b and IgG2c could similarly be made without Tfh cell help. S-specific IgG3 and IgM were minimally induced by mRNA vaccination (Fig. 1F and S3D). Therefore, class-switched antibodies to SARS-CoV-2 infection and vaccination are generated through both Tfh-dependent and -independent mechanisms.
Influenza virus infection induces Tfh-dependent and -independent antibodies
We next determined whether our findings with SARS-CoV-2 infection were generalizable to other models of respiratory viral infection. To this end, we infected mice with mouse-adapted influenza virus A/PR/8/34 H1N1 (PR8) and assessed antibody production at 14 dpi (Fig. 2A). Similar to SARS-CoV-2 infection, PR8 infection induced both Tfh-dependent and non-Tfh CD4+ T cell-dependent IgG antibodies, while IgM was largely CD4+ T cell-independent (Fig. 2, B and C). Again, PR8-specific IgG1 demonstrated a complete dependence on Tfh cell help, while PR8-specific IgG2b and IgG2c were promoted by both Tfh-dependent and -independent pathways (Fig. 2D). PR8-specific IgG3 was only partially dependent on Tfh and CD4+ T cell help (Fig. 2D). Thus, both Tfh and non-Tfh CD4+ T cells contribute to antibody production in two distinct models of respiratory viral infection.
Figure 2: Influenza virus infection induces Tfh-dependent and -independent antibodies.
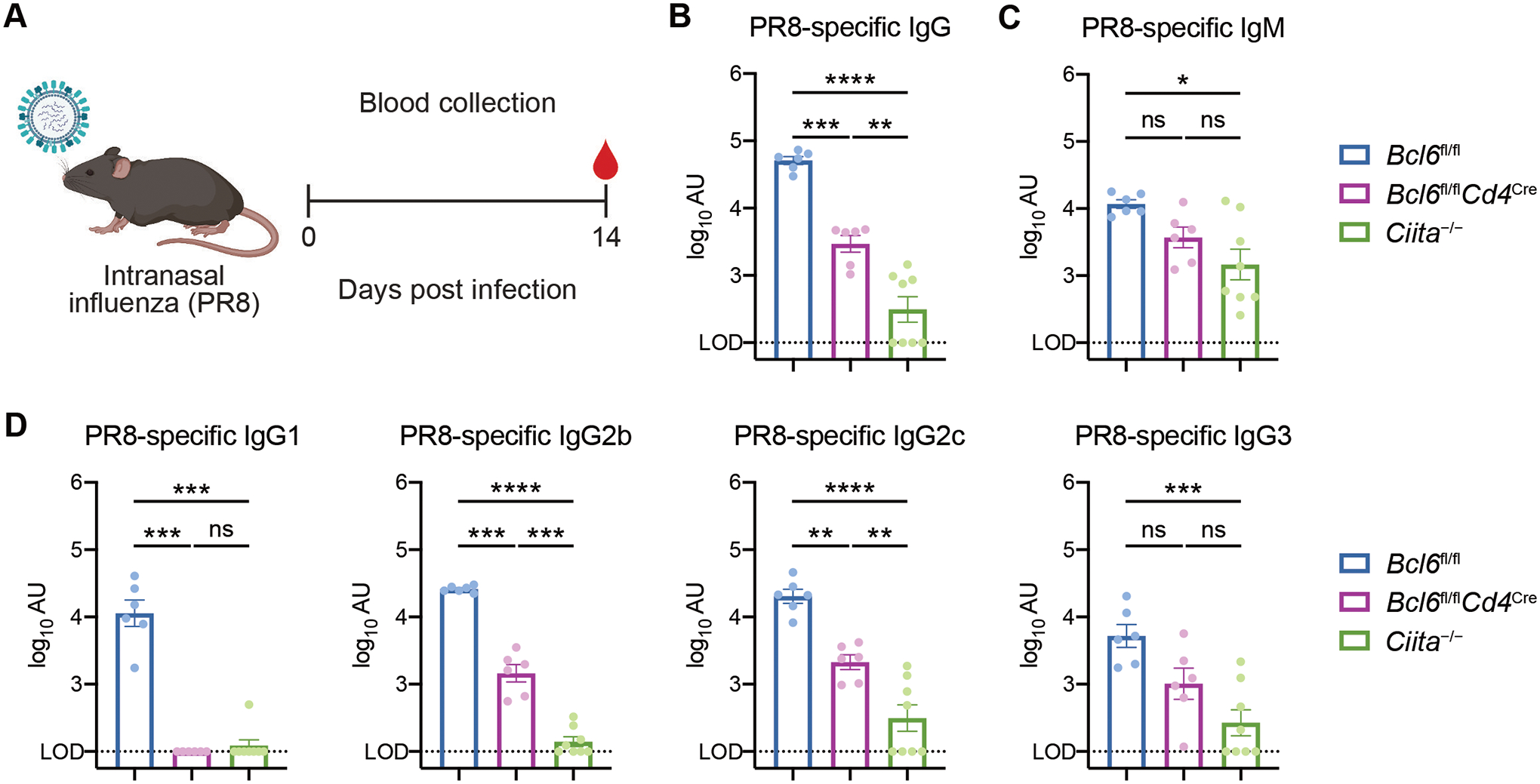
(A) Schematic of experimental design for assessing serum antibody responses against influenza virus. Mice were infected with mouse-adapted influenza virus (PR8) by intranasal administration. Sera were collected at 14 dpi for quantification of antibody titers by ELISA.
(B and C) PR8-specific total IgG (B) and IgM (C) antibody titers in sera from Bcl6fl/fl (blue), Bcl6fl/flCd4Cre (magenta), or Ciita−/− (green) mice at 14 dpi with PR8.
(D) PR8-specific IgG1, IgG2b, IgG2c, and IgG3 antibody titers at 14 dpi with PR8.
LOD, limit of detection of the assay. Statistical significance was assessed by one-way ANOVA with Tukey’s test or Dunnett’s test, or Welch’s t-test with Bonferroni multiple hypothesis correction when sample variances were 0 (B to D). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. ns, not significant. Data are expressed as mean ± SEM log10 arbitrary units (AU). Each symbol represents an individual mouse. Data are aggregated from two independent experiments with a total of six to eight mice per condition.
Durable, high-affinity antibodies to SARS-CoV-2 can be made in the absence of Tfh cells
We then measured the affinity of the antibodies generated during SARS-CoV-2 and PR8 infection. As GCs are the conventional site of somatic hypermutation and affinity maturation (2), we expected that Bcl6fl/fl mice would generate high-affinity antibodies while Bcl6fl/flCd4Cre mice would generate low-affinity antibodies. Using the AAV-hACE2 model, we observed that Bcl6fl/fl mice produced high-affinity IgG antibodies to S as well as the spike receptor-binding domain (RBD) (Fig. 3A), a major target of neutralizing antibodies (31). In contrast, S- and RBD-specific IgG antibodies from Ciita−/− mice displayed minimal affinity compared to Bcl6fl/fl mice (25.7-fold reduction against S; 15.2-fold reduction against RBD). However, we discovered that antibodies from Bcl6fl/flCd4Cre mice still demonstrated substantial affinity toward S and RBD (2.7-fold and 1.9-fold reduction against S and RBD, respectively, relative to Bcl6fl/fl mice), suggesting that non-Tfh CD4+ T cells could also promote high-affinity antibody production.
Figure 3: Durable, high-affinity antibodies to SARS-CoV-2 can be made in the absence of Tfh cells.
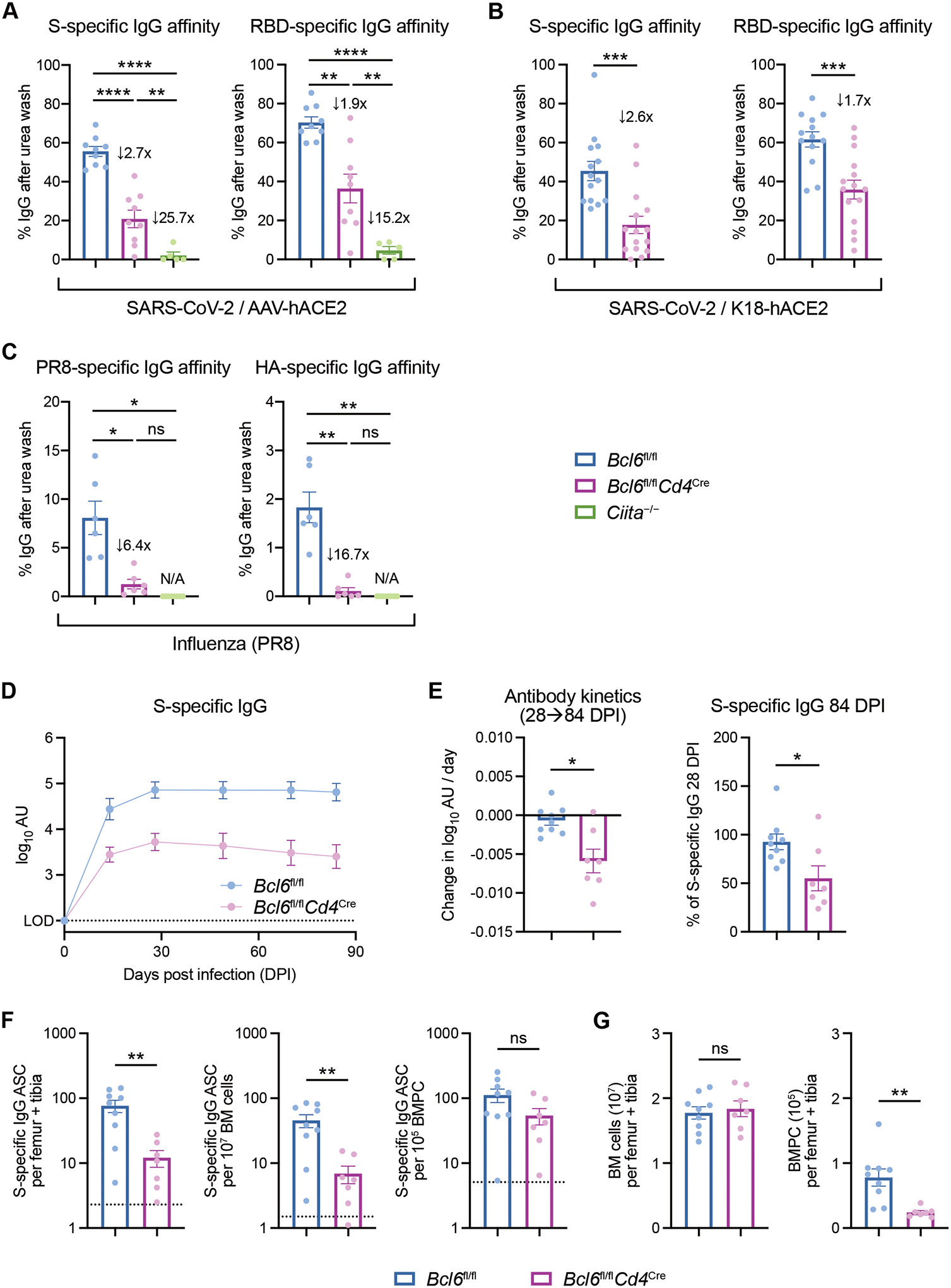
(A) Affinity index of serum IgG antibodies against SARS-CoV-2 S (left) or RBD (right), calculated as relative ELISA signal after urea wash. Sera are from AAV-hACE2 Bcl6fl/fl (blue), Bcl6fl/flCd4Cre (magenta), or Ciita−/− (green) mice infected with SARS-CoV-2 at 14 dpi. Fold changes relative to Bcl6fl/fl mice are annotated.
(B) Affinity index of serum IgG antibodies against SARS-CoV-2 S (left) or RBD (right). Sera are from K18-hACE2 Bcl6fl/fl (blue) or K18-hACE2 Bcl6fl/flCd4Cre (magenta) mice infected with SARS-CoV-2 at 14 dpi. Fold changes relative to K18-hACE2 Bcl6fl/fl mice are annotated.
(C) Affinity index of serum IgG antibodies against PR8 (left) or HA (right), calculated as relative ELISA signal after urea wash. Sera are from Bcl6fl/fl (blue), Bcl6fl/flCd4Cre (magenta), or Ciita−/− (green) mice infected with PR8 at 14 dpi. Fold changes relative to Bcl6fl/fl mice are annotated.
(D) Longitudinal dynamics of S-specific IgG antibodies up to 84 dpi, in sera from K18-hACE2 Bcl6fl/fl or K18-hACE2 Bcl6fl/flCd4Cre mice infected with SARS-CoV-2.
(E) Change in S-specific IgG antibody titer per day between 28 and 84 dpi, as determined by linear regression (left). Relative titers of S-specific IgG at 84 dpi, compared to S-specific IgG at 28 dpi (right).
(F) ELISpot quantification of S-specific IgG antibody-secreting cells (ASC) in the bone marrow of K18-hACE2 Bcl6fl/fl or K18-hACE2 Bcl6fl/flCd4Cre mice at 85 dpi with SARS-CoV-2. S-specific IgG ASC per femur + tibia (left), per 107 bone marrow (BM) cells (middle), or per 105 CD138+ bone marrow plasma cells (BMPC) (right). Dotted line indicates background signal from naïve mice.
(G) Total number of BM cells (left) and CD138+ BMPC (right) in femur + tibia of K18-hACE2 Bcl6fl/fl or K18-hACE2 Bcl6fl/flCd4Cre mice at 85 dpi with SARS-CoV-2.
Statistical significance was assessed by one-way ANOVA with Dunnett’s test, or Welch’s t-test with Bonferroni multiple hypothesis correction when sample variances were 0 (A and C); two-tailed unpaired t-test or Welch’s t-test, based on the F test for unequal variance (B, E, G); two-tailed Mann–Whitney test (F). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. ns, not significant. Data are expressed as mean ± SEM. Each symbol in (A to C and E to G) represents an individual mouse. Each symbol in (D) represents the mean of nine (Bcl6fl/fl) and seven (Bcl6fl/flCd4Cre) mice. Data are aggregated from at least two independent experiments with a total of five to fifteen mice per condition.
Similarly, K18-hACE2 Bcl6fl/flCd4Cre mice produced high-affinity antibodies to S and RBD (Fig. 3B), with a respective 2.6-fold and 1.7-fold reduction in affinity compared to controls. These data indicate that non-Tfh cells can support high-affinity antibody production in two separate models of SARS-CoV-2 infection. However, this was not the case with PR8 infection, as Bcl6fl/flCd4Cre mice produced IgG antibodies of minimal affinity to both PR8 and PR8 surface glycoprotein hemagglutinin (HA) (Fig. 3C). Therefore, the ability of non-Tfh cells to promote high-affinity antibodies may depend on the nature of the viral infection and the antigenic target.
We also evaluated the durability of S-specific IgG antibodies produced by K18-hACE2 Bcl6fl/fl and Bcl6fl/flCd4Cre mice following SARS-CoV-2 infection. Previous studies of viral infection have shown that antibody titers in Tfh-impaired mice are especially reduced at later timepoints (10, 11, 17). In SARS-CoV-2-infected K18-hACE2 Bcl6fl/fl mice, S-specific IgG levels peaked at 28 dpi and were stable through 84 dpi (Fig. 3, D and E). In K18-hACE2 Bcl6fl/flCd4Cre mice, S-specific IgG antibodies slowly declined after 28 dpi, but at 84 dpi still retained 50% of the antibody titer of 28 dpi (Fig. 3E and S4A). AAV-hACE2 Bcl6fl/fl and Bcl6fl/flCd4Cre mice displayed a similar pattern of antibody kinetics (Fig. S4, B and C).
Given that both Bcl6fl/fl and Bcl6fl/flCd4Cre mice demonstrated persistent S-specific IgG antibodies several months after infection, we investigated whether they had developed a virus-specific long-lived plasma cell compartment. We quantified S-specific IgG antibody-secreting cells (ASCs) in the bone marrow of K18-hACE2 Bcl6fl/fl and Bcl6fl/flCd4Cre mice at 85 dpi by ELISpot. S-specific IgG ASCs were detected in both groups of mice, though they were reduced tenfold in Tfh-deficient mice (Fig. 3F). However, Tfh-deficient mice also had fewer total bone marrow plasma cells (BMPCs), so the number of S-specific IgG ASCs per BMPC was similar between Bcl6fl/fl and Bcl6fl/flCd4Cre mice (Fig. 3, F and G). Taken together, these results indicate that Tfh-independent antibodies to SARS-CoV-2 can still be high-affinity and durable – two important qualities usually attributed to Tfh-dependent responses.
Tfh-deficient mice demonstrate similar V gene usage but impaired mutation selection compared to Tfh-sufficient mice
To ascertain how Tfh-deficient mice generate high-affinity antibodies to S and RBD, we considered two non-mutually exclusive hypotheses: 1) unmutated, germline-encoded B cell receptors (BCRs) in the murine V(D)J repertoire may already possess high affinity for S and/or RBD, and 2) S-specific B cells may undergo somatic hypermutation (SHM) even in the absence of Tfh cells and GCs. The former possibility has already been observed for patient-derived SARS-CoV-2-specific antibodies, which demonstrate high potency with minimal SHM (32–36). The latter possibility is supported by prior reports of SHM occurring at extrafollicular sites during chronic autoimmunity and bacterial infection (37–39).
To investigate these two possibilities, we isolated S-specific plasmablasts from K18-hACE2 Bcl6fl/fl and Bcl6fl/flCd4Cre mice at 14 dpi and performed BCR sequencing (Fig. S5, A to B). After read preprocessing and V(D)J gene annotation, we clustered the resulting BCR sequences into clonal families, identifying a range of 131 to 694 distinct clones in each sample (Fig. 4A and S5C). We assessed the relative proportions of the 10 largest clones in each sample, finding that the top-ranked clone in each sample comprised 8% to 56% of total BCR sequences recovered, while the top 10 clones accounted for 40% to 76% of total BCR sequences (Fig. 4A). These findings indicate that clonal expansion had occurred in both Bcl6fl/fl and Bcl6fl/flCd4Cre mice. To further characterize the clonal architecture of the BCR repertoires, we calculated the diversity of each BCR repertoire using the Shannon entropy score and Simpson’s diversity index. The diversity of BCR repertoires in Bcl6fl/fl and Bcl6fl/flCd4Cre mice was not statistically different by either metric, though we noted a trend towards increased diversity in the Bcl6fl/flCd4Cre mice (Fig. 4B).
Figure 4: Tfh-deficient mice demonstrate similar V gene usage but impaired mutation selection compared to Tfh-sufficient mice.
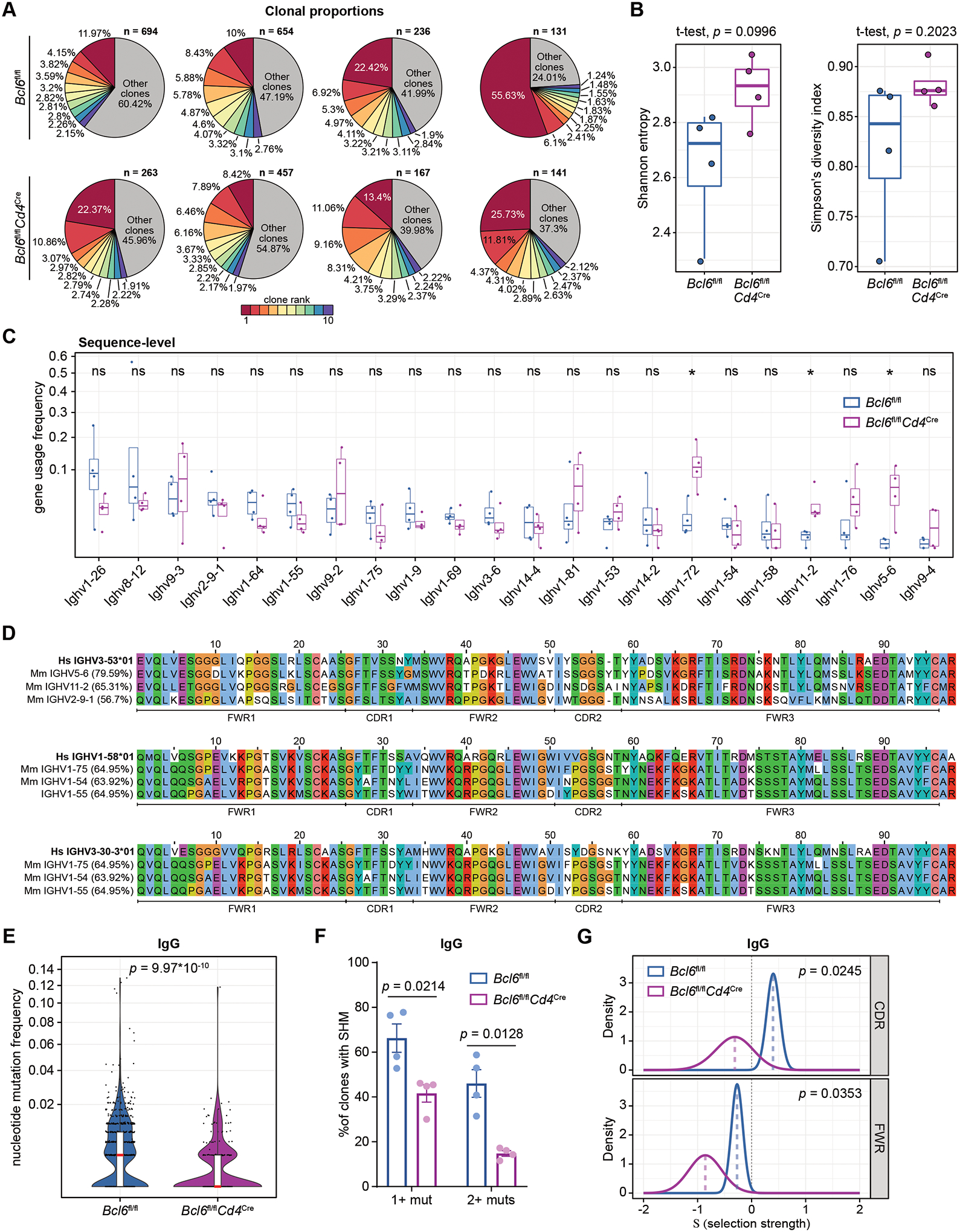
(A) Pie charts detailing the relative proportions of B cell clones identified in K18-hACE2 Bcl6fl/fl (top row) or K18-hACE2 Bcl6fl/flCd4Cre (bottom row) mice infected with SARS-CoV-2 (n = 4 mice per condition from one experiment). The 10 largest clones in each sample are shown individually, with all other clones grouped together. The total number of clones in each sample is annotated above each pie chart.
(B) Tukey boxplots detailing Shannon entropy (left) and Simpson’s diversity index (right) in Bcl6fl/fl or Bcl6fl/flCd4Cre mice. Statistical significance was assessed by two-tailed unpaired Welch’s t-test. Each symbol represents an individual mouse BCR repertoire.
(C) Tukey boxplots detailing V gene usage frequencies, calculated on the level of total BCR sequences in each sample. V genes with ≥ 0.02 usage frequency in ≥ 2 samples are shown here. Statistical significance was assessed by two-tailed unpaired Mann-Whitney test. *P < 0.05. ns, not significant.
(D) Alignment of human IGHV3-53, IGHV1-58, IGHV3-30-3 segments to the 3 murine V segments shown in (C) with the highest homology to each of the human V segments. The percentage sequence identity relative to the corresponding human V gene (bolded) is indicated on the left. The FWR and CDR regions within the V segments are annotated below.
(E) Violin plots of nucleotide mutation frequencies in IgG-predominant B cell clones from Bcl6fl/fl or Bcl6fl/flCd4Cre mice. Red lines indicate median nucleotide mutation frequency. Statistical significance was assessed by two-tailed unpaired Mann-Whitney test.
(F) Quantification of the percentage of IgG-predominant B cell clones with either 1+ or 2+ mutations in Bcl6fl/fl or Bcl6fl/flCd4Cre mice. Statistical significance was assessed by two-tailed unpaired Welch’s t-test. Data are expressed as mean ± SEM.
(G) Probability density function curves of selection strength in IgG-predominant B cell clones from Bcl6fl/fl or Bcl6fl/flCd4Cre mice, as determined by BASELINe analysis. The density curves are separated by region within the V segment: CDRs (top) or FWRs (bottom). Selection was estimated using BASELINe focused test. Statistical differences between the probability density functions were assessed by a two-sample significance test with numerical integration.
Analysis of immunoglobulin heavy-chain variable (V) gene usage frequencies revealed that out of all candidate V genes, 22 were utilized at ≥ 2% frequency in ≥ 2 samples. Usage of most V genes was not significantly different between Bcl6fl/fl and Bcl6fl/flCd4Cre mice (Fig. 4C). However, we observed that Ighv1-72, Ighv11-2, and Ighv5-6 were used more frequently in BCRs from Bcl6fl/flCd4Cre mice compared to Bcl6fl/fl mice. Examining the median usage frequencies, Ighv1-26 was the most frequently used V gene in Bcl6fl/fl mice, while BCRs from Bcl6fl/flCd4Cre mice most frequently used Ighv1-72. Repeating this analysis on the clone-level, rather than the individual BCR sequence-level, similarly revealed that usage of most V genes was not significantly different in the absence of Tfh cells (Fig. S5D). Thus, while Tfh-deficient mice did not exhibit overt changes in V gene usage, there were nevertheless differences that could reflect the distinct nature of the T cell help in these mice.
COVID-19 patient studies have identified potent RBD-specific antibodies with recurrent V gene usage and minimal SHM (32–36). To explore whether there are murine V genes that can similarly contribute to high-affinity S- or RBD-specific antibodies, we identified the murine V genes expressed in our dataset (≥ 2% frequency in ≥ 2 samples) with the greatest homology to frequently used and minimally mutated human V genes (Fig. 4D and S5, E and F) (32–36). Human IGHV3-53 and IGHV3-30 demonstrated the highest homology to murine Ighv5-6 and Ighv11-2, which were utilized more frequently in BCRs from Bcl6fl/flCd4Cre compared to Bcl6fl/fl mice (Fig. 4C). Murine V genes most similar to human IGHV1-58, IGHV3-30-3, and IGHV1-69 were used at comparable frequencies between BCRs from Bcl6fl/fl and Bcl6fl/flCd4Cre mice. Taken together, these data suggest that homologous human and murine V genes may contribute to potent SARS-CoV-2-specific antibodies. Furthermore, preferential usage of these V genes in Tfh-deficient mice may enable the production of high-affinity antibodies with minimal SHM.
To investigate whether SHM could still occur in Tfh-deficient mice, we determined the mutational profiles of each clonal consensus sequence compared to the mouse germline. We found evidence of SHM in S-specific IgG plasmablasts from Bcl6fl/flCd4Cre mice, albeit at significantly lower frequency compared to plasmablasts from Bcl6fl/fl mice (Fig. 4E). Overall, 66.28% ± 6.36% (mean ± s.e.m.) of clones from Bcl6fl/fl mice had somatic mutations, compared to 41.61% ± 3.88% of clones from Bcl6fl/flCd4Cre mice (Fig. 4F). This contrast was especially pronounced when considering the percentage of clones with two or more somatic mutations: 46.04% ± 6.13% of Bcl6fl/fl clones and 14.81% ± 1.25% of Bcl6fl/flCd4Cre clones (Fig. 4F). Similar results were obtained by comparing individual BCR sequences (Fig. S5G).
We further compared mutation selection strength in Bcl6fl/fl and Bcl6fl/flCd4Cre BCR repertoires using BASELINe, which determines the ratio of non-synonymous to synonymous mutations compared to a reference model (40, 41). Validating the BASELINe analytical approach, selection strength was positive in the complementarity determining regions (CDRs) of BCRs from Bcl6fl/fl mice and negative in the framework regions (FWRs) (Fig. 4G). This is consistent with the fact that CDRs are the main determinants of antigen specificity and thus enriched in non-synonymous mutations, while FWRs generally serve as structural scaffolds, making them less tolerant of residue-altering mutations. Across both CDRs and FWRs, we found that the ratio of non-synonymous to synonymous mutations was significantly lower in S-specific IgG plasmablasts from Bcl6fl/flCd4Cre mice compared to Bcl6fl/fl mice (Fig. 4G), indicating that Tfh cells are required for positive selection of mutated B cell clones against SARS-CoV-2. Collectively, our analysis of the BCR repertoire after SARS-CoV-2 infection suggests that both V gene usage patterns and low levels of SHM, though without mutation selection, may contribute to the production of high-affinity Tfh-independent antibodies.
Tfh cells focus the antibody repertoire but are dispensable for broad coverage of SARS-CoV-2 epitopes
We next characterized the antibody epitope repertoire of Tfh-dependent versus -independent responses to SARS-CoV-2. Sera from AAV-hACE2 Bcl6fl/fl and Bcl6fl/flCd4Cre mice at 14 dpi were profiled using a bacterial display library of 2410 linear peptides tiling the entire SARS-CoV-2 proteome (Fig. 5A and Data S2). We first compared the diversity of antibody epitope reactivity, calculating the Shannon entropy, Simpson’s diversity index, and the repertoire focusing index within each sample. We observed that antibody diversity assessed by Shannon entropy and Simpson’s diversity index was significantly decreased in Bcl6fl/fl mice compared to Bcl6fl/flCd4Cre mice, while the degree of repertoire focusing was increased (Fig. 5B and Data S3). These findings were robust to variations in read counts (Fig. S6A). These results suggest that Tfh cells help focus the antibody response to particular SARS-CoV-2 epitopes while non-Tfh cells promote antibodies to a wider array of targets.
Figure 5: Tfh cells focus the antibody repertoire but are dispensable for broad coverage of SARS-CoV-2 epitopes.
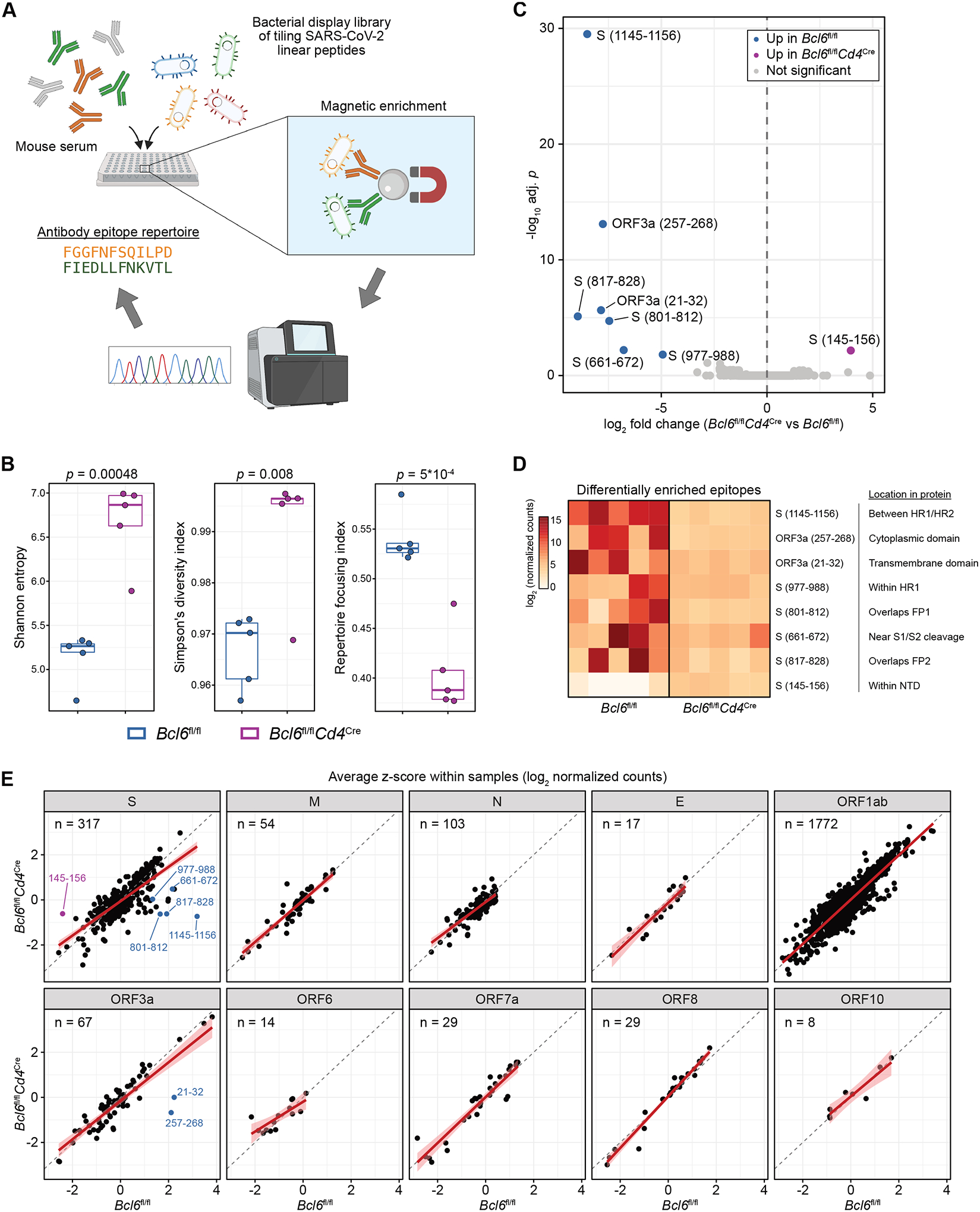
(A) Sera from infected AAV-hACE2 Bcl6fl/fl or Bcl6fl/flCd4Cre mice (n = 5 mice per condition from two independent experiments) were assayed for reactivity against a bacterial display library of overlapping linear epitopes tiling the SARS-CoV-2 proteome.
(B) Tukey boxplots detailing Shannon entropy (left), Simpson’s diversity index (middle), and repertoire focusing index (right) in Bcl6fl/fl or Bcl6fl/flCd4Cre mice. Statistical significance was assessed by two-tailed unpaired Welch’s t-test. Each symbol represents an individual mouse.
(C) Volcano plot of differentially enriched linear epitopes in Bcl6fl/fl vs Bcl6fl/flCd4Cre mice. Epitopes that are significantly enriched in Bcl6fl/fl sera are colored in blue, while epitopes that are enriched in Bcl6fl/flCd4Cre sera are colored in magenta. Point labels describe the amino acids within the indicated SARS-CoV-2 protein from which the epitope was derived. Statistical significance was assessed by two-tailed DESeq2 Wald test, with Benjamini-Hochberg multiple hypothesis correction (adjusted p < 0.05).
(D) Heatmap of differentially enriched linear epitopes in Bcl6fl/fl vs Bcl6fl/flCd4Cre mice, from (C). Data are expressed as log2-transformed normalized counts. The position of each epitope in relation to known protein domains is annotated.
(E) Scatter plots of relative enrichment scores for linear epitopes derived from each SARS-CoV-2 protein, comparing Bcl6fl/fl vs Bcl6fl/flCd4Cre sera. Data are expressed as average z-scores (log2 normalized counts were scaled to z-scores within each sample, then averaged across Bcl6fl/fl or Bcl6fl/flCd4Cre mice). The gray dashed line demarcates equivalence between Bcl6fl/fl and Bcl6fl/flCd4Cre, while the red line denotes the linear regression model with 95% confidence intervals shaded in pink. Epitopes that were identified as differentially enriched are annotated as in (C). The number of epitopes derived from each SARS-CoV-2 protein is annotated in the top left.
Given these changes in antibody diversity, we next explored whether there were differences in antibody reactivity at the level of individual linear epitopes. After normalizing for read count variations using the median of ratios method (Fig. S6B), we identified epitopes that were comparatively enriched or depleted in Bcl6fl/fl versus Bcl6fl/flCd4Cre mice (Fig. 5C). We found that seven epitopes were enriched in Bcl6fl/fl mice, while one epitope was depleted (Fig. 5D). Five of the seven enriched epitopes were derived from S: aa661–672 (proximal to S1/S2 cleavage site), aa801–812 (fusion peptide [FP] 1), aa817–828 (FP2), aa977–988 (heptad repeat [HR] 1), and aa1145–1156 (between HR1/HR2). On the other hand, aa145–156 (N-terminal domain) from S was comparatively depleted in Bcl6fl/fl mice.
To further investigate alterations in epitope reactivity, we converted the normalized counts to z-scores on a sample-by-sample basis, such that the z-scores would denote the relative rank of a specific epitope within a particular sample (Fig. S6C). Consistent with our prior analyses, the average z-scores in Bcl6fl/fl versus Bcl6fl/flCd4Cre mice were similar across most SARS-CoV-2 proteins, with the exception of regions within the S and ORF3a proteins (Fig. 5E). In particular, the regression lines for non-S proteins all closely followed the line of identity, indicating that the relative ranks of epitopes from non-S proteins were largely similar in the presence or absence of Tfh cells. These analyses therefore indicate that, while Tfh cells are dispensable for antibody production against most SARS-CoV-2 epitopes, they are required to focus the antibody response against certain S-derived epitopes.
RBD-specific antibodies are generated in the absence of Tfh cells
Analyzing antibody epitope reactivity along the length of S, we observed that the majority of epitopes enriched in Bcl6fl/fl mice (17/21 epitopes with differential average z-score > 1) were found in the S2 domain (aa686–1273; Fisher’s exact test p = 0.0012) (Fig. 6A), which mediates fusion of viral and target cell membranes (42). These included most of the aforementioned S-derived epitopes that were significantly enriched (Fig. 5C), as well as contiguous epitopes whose enrichment did not reach statistical significance in the epitope-level analysis. For example, several epitopes in the fusion peptide were enriched adjacent to the significantly enriched epitopes aa801–812 and aa817–828 (Fig. S6D). Many of these epitopes are highly conserved across human coronaviruses (hCoVs) as well as the emerging variants of concern (Fig. 6A). Interestingly, the enriched epitopes spanning FP1/FP2 and preceding HR2 have also been identified in numerous studies profiling the antibody epitope repertoire of COVID-19 patients (43, 44). Given their immunodominance and conservation across hCoVs, these epitopes have been proposed as targets for a pan-coronavirus vaccine.
Figure 6: RBD-specific antibodies are generated in the absence of Tfh cells.
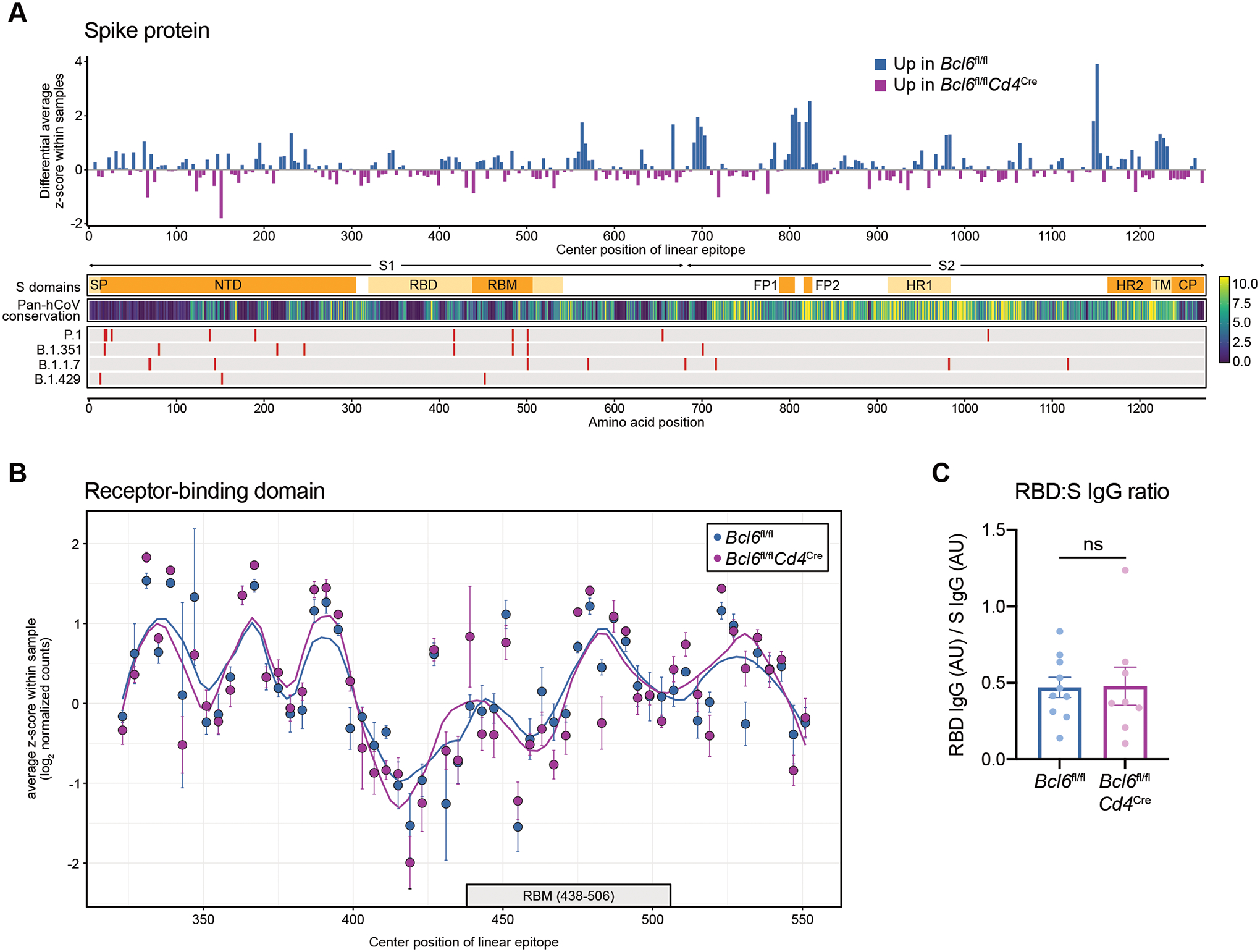
(A) Top: differential enrichment scores for spike (S)-derived linear epitopes, such that epitopes that are comparatively enriched in Bcl6fl/fl mice are represented by positive values (blue), whereas epitopes that are relatively enriched in Bcl6fl/flCd4Cre mice are indicated by negative values (magenta). Linear epitopes are each indicated by their center position within the S protein. Bottom: annotation tracks detailing S protein domains, pan-human coronavirus (hCoV) conservation scores, and mutations found in select SARS-CoV-2 variants of concern. SP, signal peptide. NTD, N-terminal domain. RBD, receptor-binding domain. RBM, receptor-binding motif. FP1, fusion peptide 1. FP2, fusion peptide 2. HR1, heptad repeat 1. HR2, heptad repeat 2. TM, transmembrane domain. CP, cytoplasmic domain.
(B) Relative enrichment scores for linear epitopes derived from RBD, comparing Bcl6fl/fl vs Bcl6fl/flCd4Cre sera. Data are expressed as average z-scores in Bcl6fl/fl or Bcl6fl/flCd4Cre mice, with SEM error bars and loess regression lines. The RBM is annotated below with a gray box.
(C) Ratio of RBD-specific to S-specific IgG antibodies in sera from Bcl6fl/fl or Bcl6fl/flCd4Cre mice at 14 dpi with SARS-CoV-2. Statistical significance was assessed by two-tailed unpaired t-test. ns, not significant. Data are expressed as mean ± SEM. Each symbol represents an individual mouse. Data are aggregated from three independent experiments with a total of eight to ten mice per condition.
In contrast, Bcl6fl/fl and Bcl6fl/flCd4Cre mice demonstrated similar antibody reactivity to most epitopes within the RBD (Fig. 6B), the target of most neutralizing antibodies (31). However, as most antibodies to RBD likely recognize conformational epitopes (43, 44), we also measured RBD-specific antibodies by ELISA using full-length RBD. RBD-specific IgG titers normalized by total S-specific IgG were similar between Bcl6fl/fl and Bcl6fl/flCd4Cre mice (Fig. 6C). Thus, while Tfh cells focus the antibody response against immunodominant S2 epitopes, non-Tfh cells still promote antibodies against the primary target of neutralization, RBD.
Tfh-dependent and -independent antibodies demonstrate similar neutralization potency against homologous SARS-CoV-2 as well as the B.1.351 variant of concern
We next evaluated the function of antibodies from Bcl6fl/fl and Bcl6fl/flCd4Cre mice following SARS-CoV-2 infection. While we had observed that Tfh-independent antibody responses to the virus lacked IgG1/IgG3 subclasses (Fig. 1, B and D) and S2 epitope focusing (Fig. 6A), these antibodies were still high-affinity (Fig. 3, A and B) and could target the RBD (Fig. 6C). We therefore predicted that Tfh-independent antibodies would demonstrate similar neutralizing function against homologous SARS-CoV-2 (USA-WA1/2020) as those generated with Tfh cell help. Using vesicular stomatitis virus (VSV) pseudotyped with USA-WA1/2020 S protein, we measured the neutralization titer (the reciprocal serum dilution achieving 50% neutralization of pseudovirus infection, NT50) of sera from AAV-hACE2 Bcl6fl/fl and Bcl6fl/flCd4Cre mice. Bcl6fl/fl sera exhibited increased NT50 (Fig. 7A), which was expected given their higher levels of S-specific IgG antibodies (Fig. 1B). However, by normalizing NT50 to S-specific IgG levels in each sample, we observed that the neutralization potency indices of Bcl6fl/fl and Bcl6fl/flCd4Cre sera were similar and actually trended higher for Bcl6fl/flCd4Cre sera (Fig. 7A).
Figure 7: Tfh-dependent and -independent antibodies demonstrate similar neutralization potency against homologous SARS-CoV-2 as well as the B.1.351 variant of concern.
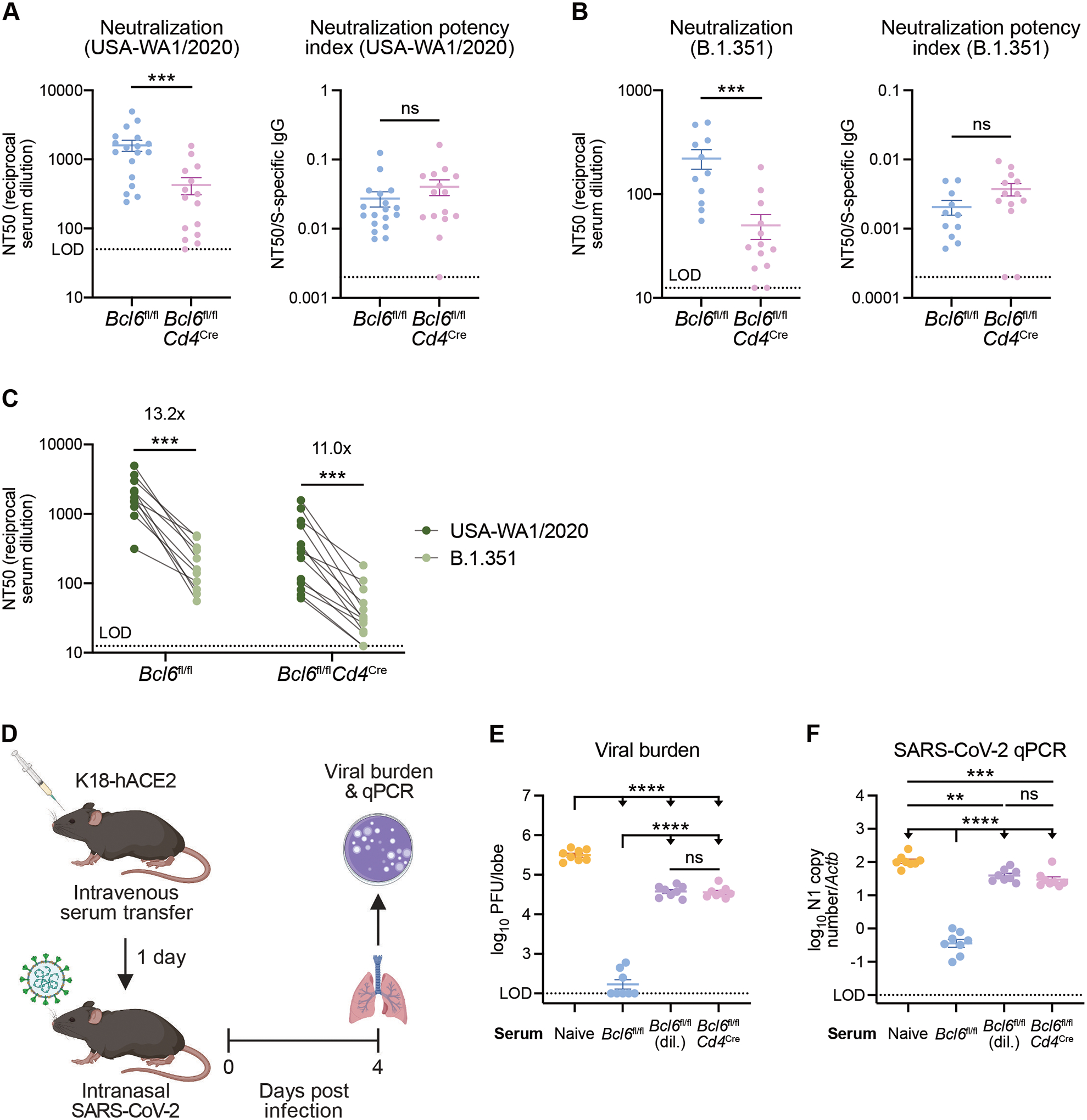
(A) Left: neutralizing titers (inverse of half maximal inhibitory concentration; NT50) of sera from AAV-hACE2 Bcl6fl/fl (blue) or Bcl6fl/flCd4Cre (magenta) mice against VSV particles pseudotyped with homologous S protein (USA-WA1/2020, the same isolate used to infect the mice). Right: neutralization potency indices (NT50 divided by S-specific total IgG) of sera from Bcl6fl/fl or Bcl6fl/flCd4Cre mice against USA-WA1/2020 pseudovirus. Samples with NT50 at the limit of detection (LOD) were assigned a neutralization potency index of 0.002.
(B) Left: neutralizing titers of sera from AAV-hACE2 Bcl6fl/fl (blue) or Bcl6fl/flCd4Cre (magenta) mice against VSV particles pseudotyped with variant S protein (B.1.351). Right: neutralization potency indices of sera from Bcl6fl/fl or Bcl6fl/flCd4Cre mice against B.1.351 pseudovirus. Samples with NT50 at the LOD were assigned a neutralization potency index of 0.0002.
(C) Matched comparison of neutralizing titers against homologous (USA-WA1/2020) vs variant (B.1.351) pseudovirus. Fold changes are indicated above each group.
(D) Schematic of experimental design for assessing in vivo neutralization potency of sera.
(E and F) Viral burden in lungs from K18-hACE2 mice infected with SARS-CoV-2 at 4 dpi. Mice were treated with serum from naïve, infected AAV-hACE2 Bcl6fl/fl, or infected AAV-hACE2 Bcl6fl/flCd4Cre mice. Bcl6fl/fl sera was given undiluted or diluted (dil.) 7- to 9-fold to match the S-specific IgG titer of Bcl6fl/flCd4Cre sera in each experiment. Data are expressed as log10 plaque forming units (PFU) per lung lobe by plaque assay (E) or log10 N1 gene copy number by qPCR, normalized to Actb (F).
Statistical significance was assessed by two-tailed Mann–Whitney test (A and B), two-tailed Wilcoxon signed-rank test (C), or one-way ANOVA with Tukey’s test (E and F). **P < 0.01; ***P < 0.001; ****P < 0.0001. ns, not significant. Data are expressed as mean ± SEM. Each symbol represents an individual mouse. Data are aggregated from at least two independent experiments with a total of eight to eighteen mice per condition.
We next tested the same sera against VSV pseudotyped with S protein from the B.1.351 variant of concern. Multiple studies have shown that B.1.351 S mutations, particularly those in the RBD, disrupt binding by neutralizing antibodies and facilitate immune escape (45, 46). We therefore hypothesized that increased focusing of Tfh-dependent antibodies against conserved S2 epitopes would enable Bcl6fl/fl sera to better neutralize B.1.351 pseudovirus than Bcl6fl/flCd4Cre sera. While Bcl6fl/fl sera exhibited greater NT50 than Bcl6fl/flCd4Cre sera, we found that the neutralization potency index of Bcl6fl/flCd4Cre sera again trended higher than that of Bcl6fl/fl sera (Fig. 7B). Nevertheless, both Bcl6fl/fl and Bcl6fl/flCd4Cre sera demonstrated an approximately 10-fold reduction in NT50 against the B.1.351 variant compared to USA-WA1/2020 (Fig. 7C), consistent with previous studies (45, 46). Our findings therefore indicate that Tfh-independent antibodies exhibit similar, if not increased, neutralization potency to Tfh-dependent antibodies in vitro and that S2 epitope focusing does not improve neutralization activity against the B.1.351 variant of concern.
Finally, we assessed the function of Bcl6fl/fl and Bcl6fl/flCd4Cre sera against SARS-CoV-2 in vivo. To this end, we transferred sera from AAV-hACE2 Bcl6fl/fl or Bcl6fl/flCd4Cre mice infected with SARS-CoV-2 (USA-WA1/2020) into naïve K18-hACE2 mice (Fig. 7D). Bcl6fl/fl sera was given undiluted or diluted 7- to 9-fold to match the S-specific IgG titer of Bcl6fl/flCd4Cre sera in a given experiment. One day later, we infected the K18-hACE2 recipients with homologous SARS-CoV-2 and then measured viral burden in the lungs at 4 dpi. Undiluted Bcl6fl/fl sera led to the greatest reduction in viral burden (Fig. 7, E and F), indicating that, as expected, antibody titer is an important determinant of protection against viral challenge. However, once matched for S-specific IgG titer, Bcl6fl/fl and Bcl6fl/flCd4Cre sera provided a similar degree of protection (Fig. 7, E and F), corroborating our in vitro findings that Tfh-dependent and -independent antibodies exhibit similar neutralization potency. Taken together, these results demonstrate that Tfh-independent antibodies efficiently neutralize both homologous SARS-CoV-2 and the B.1.351 variant of concern but that optimal protection in vivo still relies on Tfh cells to generate high antibody titers.
Discussion
While protective antibodies are generally thought to originate from Tfh/GC-dependent pathways, it is unclear what happens to the antibody response when these structures are disrupted, as has been observed in patients with severe SARS-CoV-2 infection (6, 7). We found that certain class-switched antibodies were reduced but still present in Tfh-deficient mice following SARS-CoV-2 or influenza A virus infection, as well as SARS-CoV-2 vaccination. Though BCR analysis demonstrated impairment of SHM and mutation selection, Tfh-independent antibodies to SARS-CoV-2 were still high-affinity. They were also durable and demonstrated more diverse epitope reactivity compared to Tfh-dependent antibodies. Importantly, Tfh-independent antibody responses neutralized both homologous SARS-CoV-2 (USA-WA1/2020) and the B.1.351 variant of concern and were functional in vitro and in vivo (Fig. S7).
Our findings raise the question of which CD4+ T cell subset promotes protective class-switched antibodies in the absence of Tfh cells. It has previously been shown in the setting of influenza vaccination that Th1 cells make IFN-γ along with IL-21 to induce IgG2c antibodies (19). Though less mutated and of lower avidity, these Th1-driven antibodies still neutralize influenza virus in vitro and protect from lethal challenge in vivo (19). Additionally, a recent study proposed a division of labor between Th1 cells and Tfh cells in promoting IgG2c class switching and supporting GC growth, respectively, during influenza virus infection (47). Therefore, it is possible that lymph node-resident Th1 cells are also responsible for the IgG2c antibodies we observed to SARS-CoV-2 infection and vaccination (48). Contrary to the consistent Tfh-independent induction of IgG2c, IgG1 demonstrated a divergent requirement for Tfh cell help between SARS-CoV-2 infection and vaccination. These findings align with prior literature (17, 20, 23), suggesting that there may be different cellular requirements for IgG1 in the setting of diverse immune stimuli. Similarly, CD4+ T cells are dispensable for IgG3 in response to influenza virus infection (49), but we observed that IgG3 to SARS-CoV-2 was completely dependent on Tfh cell help. Thus, the same subclass may be produced by disparate mechanisms even during different viral infections. Finally, it is important to consider whether incomplete deletion of Tfh cells could contribute to antibody responses in Bcl6fl/flCd4Cre mice. This same mouse model demonstrates complete abrogation of IgE and IgG1 to allergens (26), and our characterization of cells and structures in the medLN support a lack of T cell-infiltrated, organized GCs in Bcl6fl/flCd4Cre mice; therefore, we conclude that highly specific and long-lived antibodies can be produced through Tfh-independent mechanisms likely dependent on the nature of infection or vaccination.
A surprising finding from our work was that SARS-CoV-2, but not influenza A virus, induced high-affinity Tfh-independent antibodies. As a possible mechanism for this finding, S-specific BCRs from Tfh-deficient mice used V genes highly homologous to human V genes that generate potent S-specific antibodies with minimal SHM (32–36). Furthermore, S-specific BCRs still experienced low levels of SHM in the absence of Tfh cell help. Though SHM conventionally occurs in GCs (2), it has also been shown to occur at extrafollicular sites during bacterial infection and chronic autoimmunity (37–39). However, in the absence of Tfh cell help, mutated B cell clones did not experience positive selection, differentiating this process from classical affinity maturation. In this non-canonical pathway of high-affinity antibody production, non-Tfh CD4+ T cells may help by selecting naïve B cells that already express potent S-specific BCRs as well as supporting plasmablast differentiation of lowly mutated B cell clones. Our findings also suggest that long-lived plasma cells can emerge from this pathway, providing a durable source of humoral immunity.
Epitope profiling revealed that Tfh cells focus the antibody repertoire against S2-derived epitopes that are highly conserved across human coronaviruses as well as the emerging variants of concern. These same epitopes have been repeatedly identified in studies profiling the antibody repertoire of COVID-19 patients (43, 44), suggesting that the immunodominance of these epitopes in humans is mediated by Tfh cells. It has also been proposed that S2-reactive antibodies in people are primed by prior infections with endemic human coronaviruses (43). Given our findings in mice, which were not exposed to other coronaviruses prior to SARS-CoV-2 infection, it is likely that the intrinsic qualities of these S2-derived epitopes also contribute to their immunodominance. In addition, broadly neutralizing betacoronavirus antibodies with S2 specificity have recently been described, suggesting the use of S2-derived epitopes as targets for a pan-coronavirus vaccine (50). However, S2-specific antibodies are generally less potent neutralizers than RBD-specific antibodies and may even demonstrate little neutralization in vitro despite providing protection in vivo (50, 51). This provides a possible explanation for why we observed slightly lower neutralization potency indices of Tfh-dependent antibodies enriched for S2 epitope reactivity compared to more diverse Tfh-independent antibodies. Alternatively, the less focused antibody repertoire of Tfh-deficient mice may better neutralize SARS-CoV-2 by targeting multiple sites of vulnerability, similar to previous findings of GC inhibition promoting a broader antibody response and enhanced heterosubtypic immunity to influenza virus infection (52).
While long-lived, high-affinity, neutralizing antibody responses conventionally depend on Tfh cells, we found that antibodies of similar quality, though not quantity, could be generated with the help of non-Tfh CD4+ T cells. Therefore, multiple pathways involving different CD4+ T cell subsets likely exist to promote protective antiviral humoral immunity (48). Tfh-independent responses may serve as a parallel mechanism for producing protective antibodies in settings of Tfh/GC impairment, such as COVID-19-induced inflammation and advanced age (6, 53). Understanding this additional axis of antiviral antibody production may therefore inform more effective vaccine design and help broaden our understanding of how T cell-dependent humoral immunity is generated.
Materials and Methods
Study design
The purpose of this study was to define the Tfh-independent antibody response to respiratory viruses, including SARS-CoV-2 and influenza A virus, and SARS-CoV-2 mRNA vaccination. Experiments included intranasal infection or intramuscular vaccination. Analyses included serum antibody profiling, BCR sequencing, T cell and B cell phenotyping from mediastinal lymph nodes and bone marrow, and lung viral burden. Bcl6fl/fl and Bcl6fl/flCd4Cre littermates were used for all experiments. For serum transfer experiments, K18-hACE2 recipients were randomized to age- and sex-matched groups for different treatments. The investigators were not blinded, with the exception of ELISpot counting. The number of experimental replicates is indicated in the figure legends.
Mice
Bcl6fl/fl [B6.129S(FVB)-Bcl6tm1.1Dent/J (22)], Cd4Cre [B6.Cg-Tg(Cd4-cre)1Cwi/BfluJ (54)], Ciita−/− [B6.129S2-Ciitatm1Ccum/J (27)], and K18-hACE2 [B6.Cg-Tg(K18-ACE2)2Prlmn/J (30)] mice were purchased from Jackson Laboratory. Bcl6fl/fl mice were crossed with Cd4Cre mice to generate Bcl6fl/flCd4Cre mice. K18-hACE2 mice were crossed with Bcl6fl/flCd4Cre mice to generate K18-hACE2 Bcl6fl/fl and Bcl6fl/flCd4Cre mice. Mice were bred in-house using mating trios to enable utilization of littermates for experiments. Mice of both sexes between 6 and 10 weeks old were used for this study. Animal use and care was approved in agreement with the Yale Animal Resource Center and Institutional Animal Care and Use Committee according to the standards set by the Animal Welfare Act.
Cell lines
Huh7.5 (CVCL-7927), Vero-E6 (CRL-1586), and 293T (CRL-3216) cells were from ATCC. Cell lines were cultured in Dulbecco’s Modified Eagle Medium (DMEM) with 10% heat-inactivated fetal bovine serum (FBS) and 1% Penicillin/Streptomycin. All cell lines tested negative for Mycoplasma spp.
AAV-hACE2 transduction
Adeno-associated virus 9 expressing hACE2 from a CMV promoter (AAV-hACE2) was purchased from Vector Biolabs (SKU AAV-200183). Mice were anesthetized by intraperitoneal injection of ketamine (50 mg/kg) and xylazine (5 mg/kg). The rostral neck was shaved and disinfected with povidone-iodine. After a 5-mm incision was made, the salivary glands were retracted and the trachea visualized. Using a 31G insulin syringe, 1011 genomic copies of AAV-hACE2 in 50 μl PBS were injected into the trachea. The incision was closed with 3M Vetbond tissue adhesive, and mice were monitored until full recovery.
Viruses
SARS-CoV-2 P1 stock was generated by inoculating Huh7.5 cells with SARS-CoV-2 isolate USA-WA1/2020 (BEI Resources, NR-52281) at a MOI of 0.01 for three days. The P1 stock was then used to inoculate Vero-E6 cells, and the supernatant was harvested after three days at 50% cytopathic effect. The supernatant was clarified by centrifugation (450 × g for 5 min), filtered through a 0.45-micron filter, and stored in aliquots at −80°C. For infection of AAV-hACE2 mice, virus was concentrated by mixing one volume of cold 4X PEG-it Virus Precipitation Solution (40% wt/vol PEG-8000 and 1.2 M NaCl) with three volumes of viral supernatant. The mixture was incubated overnight at 4°C and then centrifuged at 1500 × g for 60 min at 4°C. The pelleted virus was resuspended in PBS and stored in aliquots at −80°C. Virus titer was determined by plaque assay using Vero-E6 cells (55).
Influenza virus A/PR/8/34 H1N1 (PR8) expressing the ovalbumin OT-II peptide was grown for 2.5 days at 37°C in the allantoic cavities of 10-day-old specific-pathogen-free fertilized chicken eggs. Harvested virus was centrifuged at 3000 × g for 20 min at 4°C to remove debris and stored in aliquots at −80°C. Virus titer was determined by plaque assay on Madin-Darby canine kidney cells (56).
For all infections, mice were anesthetized using 30% vol/vol isoflurane diluted in propylene glycol (30% isoflurane) and administered SARS-CoV-2 or PR8 intranasally in 50 μl PBS. AAV-hACE2 Bcl6fl/fl, Bcl6fl/flCd4Cre, and Ciita−/− mice were infected with 1.2×106 PFU of SARS-CoV-2. K18-hACE2 Bcl6fl/fl and Bcl6fl/flCd4Cre mice were infected with 20 PFU of SARS-CoV-2. K18-hACE2 mice in serum transfer experiments were infected with 100 PFU of SARS-CoV-2. Bcl6fl/fl, Bcl6fl/flCd4Cre, and Ciita−/− mice were infected with 70 PFU of PR8. All work with SARS-CoV-2 was performed in a biosafety level 3 (BSL3) facility with approval from the office of Environmental Health and Safety and the Institutional Animal Care and Use Committee at Yale University.
mRNA vaccination
Used vials of Moderna mRNA-1273 and Pfizer-BioNTech BNT162b2 mRNA vaccine were obtained from Yale Health within 6 hours of opening. All vials contained less than one full dose per vial, and no vaccines were diverted for the purpose of this study. Mice were anesthetized using 30% isoflurane and administered 1 μg of either Moderna or Pfizer-BioNTech vaccine intramuscularly in 50 μl PBS. Vaccine was injected into the right hamstring muscles with a 31G insulin syringe.
Enzyme-linked immunosorbent assay (ELISA)
Sera were incubated with a final concentration of 0.5% Triton X-100 and 0.5 mg/ml RNase A for 30 min at room temperature to inactivate potential SARS-CoV-2. SARS-CoV-2 stabilized spike glycoprotein (BEI Resources, NR-53257) (57), SARS-CoV-2 spike glycoprotein receptor-binding domain (RBD) (BEI Resources, NR-52946), and influenza virus A/PR/8/34 H1N1 hemagglutinin (HA) protein (Sino Biological, 11684-V08H) were coated at a concentration of 2 μg/ml in carbonate buffer on 96-well MaxiSorp plates (Thermo Fisher) overnight at 4°C. PR8 was inactivated with 1% Triton X-100 for 1 hr at 37°C and coated at a concentration of 20 μg/ml in carbonate buffer. Plates were blocked with 1% BSA in PBS for 1 hr at room temperature. Serum samples were serially diluted in 1% BSA in PBS and incubated in plates for 2 hr at room temperature. Antibody isotypes were detected with anti-mouse IgG-HRP (1013-05), anti-mouse IgG1-HRP (1073-05), anti-mouse IgG2b-HRP (1093-05), anti-mouse IgG2c-HRP (1077-05), or anti-mouse IgG3-HRP (1103-05) from Southern Biotech or anti-mouse IgM-HRP (550588) from BD Biosciences by incubating for 1 hr at room temperature. Plates were developed with TMB Stabilized Chromogen (Thermo Fisher), stopped with 3 N hydrochloric acid, and read at 450 nm on a microplate reader. Background was determined as twice the average OD value of blank wells. Pooled sera from mice infected with SARS-CoV-2 or PR8 were used as reference standards to calculate arbitrary units. To measure antibody affinity, serial dilutions of serum samples were plated in duplicate. After incubation of serum samples, a 10-min wash with 5.3 M urea was performed on one set of the samples. Percentage of IgG binding after urea wash was calculated by dividing the area under the curve for each sample with urea wash by that without urea wash.
Enzyme-linked immunosorbent spot (ELISpot)
SARS-CoV-2 stabilized spike glycoprotein (BEI Resources, NR-53257) (57) was coated at a concentration of 2 μg/ml in carbonate buffer on 96-well MultiScreen HTS IP Filter plates (Millipore) overnight at 4°C. Plates were blocked with complete RPMI (10% heat-inactivated FBS, 1% Penicillin/Streptomycin, 2 mM L-glutamine, 1 mM sodium pyruvate, 10 mM HEPES, 55 μM 2-mercaptoethanol) for 4 hr at 37°C. Bone marrow cells were isolated from the left femur + tibia of mice infected with SARS-CoV-2. Red blood cells were lysed with RBC Lysis Buffer (BioLegend) for 2 min. Cells were resuspended in complete RPMI and plated in duplicate at three dilutions (1/5, 1/10, and 1/20 of total bone marrow cells) for 20 hr at 37°C. Plates were washed six times with PBS-T (0.01% Tween-20), followed by incubation with anti-mouse IgG-Alkaline Phosphatase (Southern Biotech, 1030-04) in PBS with 0.5% BSA for 2 hr at room temperature. Plates were then washed three times with PBS-T and three times with PBS. Spots were developed with Vector® Blue Substrate Kit (Vector Laboratories, SK-5300) and imaged with an ImmunoSpot analyzer (Cellular Technology Limited). Spots were counted manually by a blinded investigator.
Flow cytometry and cell sorting
Mediastinal lymph nodes (medLN) were homogenized with a syringe plunger and filtered through a 70-μm cell strainer. Red blood cells were lysed with RBC Lysis Buffer (BioLegend) for 1 min. Single-cell preparations were resuspended in PBS with 2% FBS and 1 mM EDTA and pre-incubated with Fc block (2.4G2; BD Biosciences, 553141) for 5 min at room temperature before staining. Cells were stained with the following antibodies or viability dye for 30 min at 4°C: anti-CD4 (RM4-5; BioLegend, 100568), TCRβ (H57-597; BioLegend, 109228), PD-1 (RMP1-30; BioLegend, 109110), CD44 (IM7; BioLegend, 103047), PSGL-1 (2PH1; BD Biosciences, 563448), Ly6C (HK1.4; BioLegend, 128005), B220 (RA3-6B2; BioLegend, 103260), Fas (Jo2; BD Biosciences, 563647), GL7 (GL7; BioLegend, 144610), CD138 (281-2; BioLegend, 142514), and LIVE/DEAD™ Fixable Aqua (Thermo Fisher, L34966). CXCR5 (L138D7; BioLegend, 145510) was stained for 25 min at room temperature. Samples for flow cytometry were fixed with 4% paraformaldehyde for 30 min at room temperature before removal from BSL3 facility. Samples were acquired on a CytoFLEX S (Beckman Coulter) and analyzed using FlowJo software (BD).
For cell sorting, medLN were stained with aforementioned B cell surface markers and viability dye, along with 2 μg/ml FLAG-tagged SARS-CoV-2 spike protein (GenScript, Z03481) for 30 min at 4°C (58). Cells were then stained with both APC- and PE-conjugated anti-FLAG antibodies (BioLegend, 637309 and 637307) to double-stain spike-specific B cells. Spike-specific plasmablasts (live CD138hiSpike+) were sorted into 350 μl Buffer RLT Plus (Qiagen) with 1% β-mercaptoethanol. RLT lysate was vortexed for 1 min, frozen on dry ice, and then stored at −80°C. Cell sorting was performed on a FACSAria II (BD Biosciences) in the BSL3 facility.
BCR library preparation
RNA from spike-specific B cells was isolated using the RNeasy Plus Micro Kit (Qiagen) following manufacturer’s instructions. BCR libraries were prepared using the NEBNext® Single Cell/Low Input cDNA Synthesis & Amplification Module (NEB, E6421) and NEBNext® Immune Sequencing Kit (NEB, E6330), with additional reagents provided by NEB to integrate the two kits. 1–20 ng of high-quality RNA with RIN score ≥ 8 was used as input. Libraries were analyzed by Bioanalyzer High Sensitivity DNA assay, pooled in equal amounts with PhiX spike-in, and sequenced on an Illumina MiSeq using the V3 kit, with 325 bp for read 1 and 275 bp for read 2.
BCR repertoire analysis
UMI-barcoded paired-end sequencing reads were processed and analyzed with the Immcantation suite (Docker image v4.3.0). Read preprocessing was performed with the presto-abseq pipeline interface for pRESTO (v0.7.0) (59) with default settings, using mouse constant region primers, isotype-specific internal constant region sequences, and template switch sequences provided by NEB. The preprocessed reads were aligned to the mouse germline V(D)J genes with the changeo-igblast pipeline interface for Change-O (v1.2.0) (60) and IgBLAST (v1.17.1). Productive sequences aligning to immunoglobulin heavy chains were retained for further analysis. Each sequence was annotated with the corresponding germline V(D)J sequence, masking N/P and D-segment regions. Spectral clustering was performed using SCOPer (v1.2.0) (61) to identify clonally-related sequences, based on the level of junction region homology and the mutation profiles in the V-J segments. Following clonal clustering, consensus germline sequences for each clone were reconstructed as above.
Sequences aligning to constant regions other than IgM, IgA, or IgG were filtered out from further analysis. Relative clonal proportions were calculated by tabulating the number of UMI-barcoded sequences belonging to each clonal family. Clonal proportions were visualized as pie charts, with the top 10 clones each represented individually and all other clones combined into one category. Given the wide variation in the number of clones across samples, in order to characterize the diversity of each BCR repertoire, 100 clones were randomly sampled from each repertoire to calculate Shannon entropy and Simpson’s diversity index. This process was repeated 100 times for each sample, ultimately taking the average value across all repeats.
Downstream analysis was performed using Alakazam (v1.2.0) and SHazaM (v1.1.0) (40, 41). Heavy chain V gene usage frequencies were calculated on either the sequence-level or the clone-level. For the sequence-level analysis, the frequency of individual UMI-barcoded BCR sequences aligned to each V gene was calculated. For the clone-level analysis, the frequency of clones aligned to each V gene was determined, taking the consensus V gene assignment across all constituent UMI-barcoded sequences belonging to each clone. V genes that were present at ≥ 0.02 frequency in ≥ 2 repertoires were retained for visualization purposes. The most frequently used human V genes in SARS-CoV-2 specific antibodies from patients were determined by mining the CoV-AbDab database (62). Out of the mouse V genes that were expressed at ≥ 0.02 clone-level frequency in ≥ 2 repertoires, those that are most similar to the top-ranked human V genes were identified with BLAST. For visualization, alignment of mouse and human V genes was performed using Clustal Omega and visualized using Jalview with the Clustal X color scheme. Lineage trees were constructed with dowser (63), building the maximum parsimony tree for all constituent BCRs in a clone. Branch lengths were scaled by the number of mutations between nodes. To facilitate qualitative comparisons, the pair of Bcl6fl/fl and Bcl6fl/flCd4Cre mice with the greatest similarity in the total number of clones and the size of the largest clone were selected for visualization.
Sequences belonging to the same clone were then collapsed, taking the most common nucleotide at each position along the consensus sequence. Clones were annotated for antibody isotypes by majority vote. IgG isotype clones were retained for further analysis of somatic hypermutation. Nucleotide mutation frequencies of each collapsed clone were calculated across the entire heavy chain sequence (with N/P and D-segment regions masked), normalizing by the length of the input sequence. To evaluate somatic hypermutation propensity, the percentage of clones with 1+ or 2+ mutations was determined in each sample and compared across experimental groups. To estimate selection pressure, the collapsed clone sequences from each experimental group (Bcl6fl/fl or Bcl6fl/flCd4Cre) were analyzed using BASELINe (40, 41), a method that compares the observed vs expected ratios of replacement to synonymous mutations. Specifically, the observed substitution frequencies in each V segment (excluding CDR3), grouped by CDR or FWR, were compared to the expected frequencies of a reference somatic hypermutation targeting model using the “focused” test statistic. As a reference model for somatic hypermutation in murine heavy chains is not currently available, the HH_S5F human heavy chain 5-mer targeting model was used for analysis, since it has been previously been shown that human heavy vs light chains have considerably different mutational patterns (64). Larger BASELINe values (corresponding to an increased ratio of replacement mutations) are consistent with positive selection.
Immunofluorescence
MedLN were fixed with 4% paraformaldehyde in PBS for 4 hr at 4°C, followed by cryopreservation with 20% sucrose in PBS for 2 hr at 4°C. MedLN were snap-frozen in optimal cutting temperature compound and stored at −80°C. Tissues were cut into 7-μm sections and blocked with 10% rat serum in staining solution (PBS with 1% BSA and 0.1% Tween-20) for 1 hr at room temperature. Sections were stained with Fc block, BV421 anti-IgD (11-26c.2a; BioLegend, 405725), AF488 anti-TCRβ (H57-597; BioLegend, 109215), biotinylated anti-CD35 (8C12; BD Biosciences, 553816), and AF647 anti-GL7 (GL7; BioLegend, 144606) overnight at 4°C. Secondary staining was performed with AF594 streptavidin (BioLegend, 405240) for 1 hr at room temperature. Images were acquired on a Nikon TiE Spinning Disk Confocal Microscope with the 20× objective. Tile scan images were taken with the 20× objective with 10% overlap. Image analysis was performed with ImageJ.
Pseudovirus production
Vesicular stomatitis virus (VSV)-based pseudotyped viruses were produced as previously described (55, 65). Vector pCAGGS containing the SARS-CoV-2 Wuhan-Hu-1 spike glycoprotein gene was produced under HHSN272201400008C and obtained through BEI Resources (NR-52310). The sequence of the Wuhan-Hu-1 isolate spike glycoprotein is identical to that of the USA-WA1/2020 isolate. The spike sequence of the B.1.351 variant of concern was generated by introducing the following mutations: L18F, D80A, D215G, R246I, K417N, E484K, N501Y, and A701V. 293T cells were transfected with either spike plasmid, followed by inoculation with replication-deficient VSV expressing Renilla luciferase for 1 hour at 37°C (65). The virus inoculum was then removed, and cells were washed with PBS before adding media with anti-VSV-G (8G5F11) to neutralize residual inoculum. Supernatant containing pseudovirus was collected 24 hours post inoculation, clarified by centrifugation, concentrated with Amicon Ultra Centrifugal Filter Units (100 kDa), and stored in aliquots at −80°C. Pseudoviruses were titrated in Huh7.5 cells to achieve a relative light unit (RLU) signal of 600 times the cell-only control background.
Pseudovirus neutralization assay
Sera for neutralization assay were heat-inactivated for 30 min at 56°C. Sera were tested at a starting dilution of 1:50 for USA-WA1/2020 pseudovirus and 1:12.5 for B.1.351 pseudovirus, with up to eight two-fold serial dilutions. 2×104 Huh7.5 cells were plated in each well of a 96-well plate the day before. Serial dilutions of sera were incubated with pseudovirus for 1 hour at 37°C. Growth media was then aspirated from the cells and replaced with 50 μl of serum/virus mixture. Luciferase activity was measured at 24 hours post infection using the Renilla Luciferase Assay System (Promega). Each well of cells was lysed with 50 μl Lysis Buffer, and 15 μl cell lysate was then mixed with 15 μl Luciferase Assay reagent. Luminescence was measured on a microplate reader (BioTek Synergy). Half maximal inhibitory concentration (IC50) was calculated as previously described (66). Neutralizing titer (NT50) was defined as the inverse of IC50.
Serum transfer
Sera from SARS-CoV-2-infected AAV-hACE2 Bcl6fl/fl or Bcl6fl/flCd4Cre mice at 14 days post infection (dpi) were pooled, and the resulting levels of spike-specific IgG were measured by ELISA. Bcl6fl/fl sera was left undiluted or diluted with naïve sera to match the spike-specific IgG titer of undiluted Bcl6fl/flCd4Cre sera. Serum samples were then mixed 1:1 with PBS, and 200 μl of serum/PBS mixture was transferred intravascularly by retro-orbital injection into K18-hACE2 mice under anesthesia with 30% isoflurane.
Measurement of lung viral burden
The left lobe of the lung was collected and homogenized in 1 ml DMEM supplemented with 2% heat-inactivated FBS and 1% Penicillin/Streptomycin. Lung homogenates were clarified by centrifugation at 3200 × g for 10 min and stored in aliquots at −80°C. Viral burden was measured in lung homogenates by plaque assay on Vero-E6 cells as previously described (55). In addition, 250 μl of lung homogenate was mixed with 750 μl of TRIzol™ LS Reagent (Invitrogen), and RNA was extracted with the RNeasy Mini Kit (Qiagen) following manufacturer’s instructions. cDNA synthesis was performed using random hexamers and ImProm-II™ Reverse Transcriptase (Promega). Quantitative PCR (qPCR) was performed in triplicate for samples and standards using SARS-CoV-2 nucleocapsid (N1)-specific oligonucleotides: Probe: 5’ 6FAM-ACCCCGCATTACGTTTGGTGGACC-BHQ1 3’; Forward primer: 5’ GACCCCAAAATCAGCGAAAT-3’; Reverse primer: 5’ TCTGGTTACTGCCAGTTGAATCTG 3’. The limit of detection was 100 SARS-CoV-2 genome copies/μl. Virus copy numbers were quantified using a control plasmid containing the complete nucleocapsid gene from SARS-CoV-2. SARS-CoV-2 genome copies were normalized to Actb using Actb-specific oligonucleotides: Probe: 5’ 6-JOEN-CACCAGTTC /ZEN/ GCCATGGATGACGA-IABkFQ 3’; Forward primer: 5’ GCTCCTTCGTTGCCGGTCCA 3’; Reverse primer: 5’ TTGCACATGCCGGAGCCGTT 3’. The limit of detection was 100 Actb copies/μl. Samples with undetectable SARS-CoV-2 genome copies were set at 0.01 relative to Actb.
Serum epitope repertoire analysis (SERA) with SARS-CoV-2 linear epitope library
Serum samples for epitope profiling were inactivated with UV light (250 mJ). The SERA platform for next-generation sequencing (NGS)-based analysis of antibody epitope repertoires has been previously described (67). In brief, Escherichia coli were engineered with a surface display vector carrying linear peptides derived from the SARS-CoV-2 proteome (GenBank MN908947.3), designed using oligonucleotides (Twist Bioscience) encoding peptides 12 amino acids in length and tiled with 8 amino acids overlapping. Serum samples (0.5 μl each) were then diluted 1:200 in a suspension of PBS and bacteria carrying the surface display library (109 cells per sample with 3×105 fold library representation), and incubated so that antibodies contained in the serum would bind to the peptides on the surface of the bacteria. After incubating with protein A/G magnetic beads and magnetically isolating bacteria that were bound to antibodies contained in the serum, plasmid DNA was purified and PCR amplified for NGS. Unique molecular identifiers (UMIs) were applied during PCR to minimize amplification bias, designed as an 8 base pair semi-random sequence (NNNNNNHH). After preprocessing and read trimming the raw sequencing data, the resulting reads were filtered by utilizing the UMIs to remove PCR duplicates. The filtered UMI data (hereafter referred to as reads) were then aligned to the original reference of linear epitopes derived from SARS-CoV-2 and quantified.
From the raw mapped read counts for each of the 2410 linear epitopes represented in the library, we first calculated the Shannon entropy and Simpson’s diversity index of each sample using the diversity function in the diverse R package. To calculate the “repertoire focusing index”, we used the formula: 1-(H’/log2(R)), where H’ is Shannon entropy and R is richness, defined here as the number of unique epitopes recognized by a given sample (read count > 0). Statistical differences in these various metrics were assessed by two-tailed unpaired Welch’s t-test, comparing Bcl6fl/fl and Bcl6fl/flCd4Cre conditions.
For further analysis, we normalized the raw count data using the median ratio approach implemented in DESeq2 (68). Differential enrichment analysis was performed using the Wald test in DESeq2, comparing Bcl6fl/fl vs Bcl6fl/flCd4Cre samples. Multiple hypothesis correction was performed by the Benjamini-Hochberg method, setting a statistical significance threshold of adjusted p < 0.05. After identifying differentially enriched epitopes, the normalized counts were log2 transformed (hereafter referred to as log2 normalized counts) for downstream visualization and analysis.
For converting the log2 normalized counts into relative enrichment scores on a sample-by-sample basis, we scaled the log2 normalized counts within each sample to z-scores. In this manner, a z-score of 0 would correspond to epitopes that exhibited an average level of enrichment in a given sample; a positive z-score would indicate that an epitope is relatively enriched in a sample, while a negative z-score would denote a low-scoring epitope. Where applicable, statistical significance was assessed on the within-sample z-scores by two-tailed unpaired Welch’s t-test. For visualization purposes, we also calculated the average z-scores in Bcl6fl/fl or Bcl6fl/flCd4Cre groups.
Pan-human coronavirus (hCoV) conservation scores were calculated through multiple alignment of several hCoV spike sequences using Clustal Omega. The following hCoV spike sequences were used: HKU1 (UniProt: Q0ZME7), OC43 (P36334), 229E (P15423), NL63 (Q6Q1S2), SARS-CoV (P59594), SARS-CoV-2 (P0DTC2), and MERS-CoV (K9N5Q8). Amino-acid level conservation scores for SARS-CoV-2 spike were extracted through JalView.
Statistical analysis
Data analysis was performed using GraphPad Prism 9 unless otherwise indicated. Data were analyzed using one-way ANOVA with Tukey’s test or Dunnett’s test; Welch’s t-test with Bonferroni multiple hypothesis correction; Student’s two-tailed, unpaired t-test; Welch’s two-tailed, unpaired t-test; two-tailed Mann–Whitney test; or two-tailed Wilcoxon signed-rank test, as indicated. P < 0.05 was considered statistically significant.
Supplementary Material
Data S1. Raw data file
Data S2. Design and composition of the SARS-CoV-2 linear epitope library
Data S3. Raw count matrix of the antibody epitope repertoire
Figure S1. Cellular response to SARS-CoV-2 in Tfh-sufficient and -deficient mice
Figure S2. Lymph node structures of Tfh-sufficient and -deficient mice following SARS-CoV-2 infection
Figure S3. Humoral response to SARS-CoV-2 in Tfh-sufficient and -deficient mice
Figure S4. Long-term antibody production in Tfh-sufficient and -deficient mice infected with SARS-CoV-2
Figure S5. Analysis of SARS-CoV-2-reactive BCRs from mice and humans
Figure S6. Analysis of SARS-CoV-2 linear epitopes recognized by antibodies from Tfh-sufficient and -deficient mice
Figure S7. Complementary Tfh cell-dependent and -independent pathways of antibody production mediate neutralizing responses to SARS-CoV-2
Acknowledgements:
We thank all members of the Eisenbarth and Wilen labs for helpful discussions. We would like to acknowledge Benhur Lee, Ann Haberman, Joao Pereira, Jonathan Klein, Steven Kleinstein, Kenneth Hoehn, Ruoyi Jiang, Minh Pham, Joerg Nikolaus, Yale West Campus Imaging Core, Yale Center for Genome Analysis, and BEI Resources for providing critical reagents, resources, and expertise. We thank Yale Environmental Health and Safety for providing necessary training and support for SARS-CoV-2 research. Illustrations were created with BioRender.com.
Funding:
This study was supported by National Institutes of Health grants T32GM136651 (JSC, RDC, ES), F30HL149151 (JSC), F30CA250249 (RDC), F30CA239444 (ES), T32AI007019 (TM), T32AI007517 (BI), R37AR040072 and R01AR074545 (JC), K08AI128043 and R01AI148467 (CBW), and R01AI136942 (SCE); Edward L. Tatum Fellowship from the Graduate School of Arts & Sciences and the Gershon-Trudeau Fellowship from the Department of Immunobiology of Yale University (WS); Bill & Melinda Gates Foundation (LC); DoD grant IIAR W81XWH-21-1-0019 (SC); Burroughs Wellcome Fund Career Award for Medical Scientists (CBW); Mathers Charitable Foundation (CBW); Ludwig Family Foundation (CBW); and Emergent Ventures Fast Grants (CBW, SCE).
Footnotes
Competing interests:
KK, JB, WAH, and JCS declare the following competing interests: ownership of stocks or shares at Serimmune, paid employment at Serimmune, and patent applications on behalf of Serimmune. Yale University (CBW) has a patent pending entitled “Compounds and Compositions for Treating, Ameliorating, and/or Preventing SARS-CoV-2 Infection and/or Complications Thereof.”
Data and materials availability:
BCR sequencing data are available via the Gene Expression Omnibus (GSE188792). All other data needed to evaluate the conclusions of the paper are available in the paper or the Supplementary Materials. SARS-CoV-2 stabilized spike glycoprotein was made available from University of Washington through a material transfer agreement. Reagents required for integrating the NEBNext® Single Cell/Low Input cDNA Synthesis & Amplification Module and NEBNext® Immune Sequencing Kit were made available under a material transfer agreement from New England Biolabs.
References:
- 1.McMahan K, Yu J, Mercado NB, Loos C, Tostanoski LH, Chandrashekar A, Liu J, Peter L, Atyeo C, Zhu A, Bondzie EA, Dagotto G, Gebre MS, Jacob-Dolan C, Li Z, Nampanya F, Patel S, Pessaint L, Van Ry A, Blade K, Yalley-Ogunro J, Cabus M, Brown R, Cook A, Teow E, Andersen H, Lewis MG, Lauffenburger DA, Alter G, Barouch DH, Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature. 590, 630–634 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crotty S, T Follicular Helper Cell Biology: A Decade of Discovery and Diseases. Immunity. 50, 1132–1148 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumjohann D, Fazilleau N, Antigen-dependent multistep differentiation of T follicular helper cells and its role in SARS-CoV-2 infection and vaccination. Eur J Immunol (2021), doi: 10.1002/eji.202049148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang J, Wu Q, Liu Z, Wang Q, Wu J, Hu Y, Bai T, Xie T, Huang M, Wu T, Peng D, Huang W, Jin K, Niu L, Guo W, Luo D, Lei D, Wu Z, Li G, Huang R, Lin Y, Xie X, He S, Deng Y, Liu J, Li W, Lu Z, Chen H, Zeng T, Luo Q, Li Y-P, Wang Y, Liu W, Qu X, Spike-specific circulating T follicular helper cell and cross-neutralizing antibody responses in COVID-19-convalescent individuals. Nature Microbiology. 6, 51–58 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Moderbacher CR, Ramirez SI, Dan JM, Grifoni A, Hastie KM, Weiskopf D, Belanger S, Abbott RK, Kim C, Choi J, Kato Y, Crotty EG, Kim C, Rawlings SA, Mateus J, Tse LPV, Frazier A, Baric R, Peters B, Greenbaum J, Saphire EO, Smith DM, Sette A, Crotty S, Antigen-Specific Adaptive Immunity to SARS-CoV-2 in Acute COVID-19 and Associations with Age and Disease Severity. Cell. 183, 996–1012.e19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaneko N, Kuo H-H, Boucau J, Farmer JR, Allard-Chamard H, Mahajan VS, Piechocka-Trocha A, Lefteri K, Osborn M, Bals J, Bartsch YC, Bonheur N, Caradonna TM, Chevalier J, Chowdhury F, Diefenbach TJ, Einkauf K, Fallon J, Feldman J, Finn KK, Garcia-Broncano P, Hartana CA, Hauser BM, Jiang C, Kaplonek P, Karpell M, Koscher EC, Lian X, Liu H, Liu J, Ly NL, Michell AR, Rassadkina Y, Seiger K, Sessa L, Shin S, Singh N, Sun W, Sun X, Ticheli HJ, Waring MT, Zhu AL, Alter G, Li JZ, Lingwood D, Schmidt AG, Lichterfeld M, Walker BD, Yu XG, Padera RF, Pillai S, Loss of Bcl-6-Expressing T Follicular Helper Cells and Germinal Centers in COVID-19. Cell, S0092867420310679 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duan Y, Xia M, Ren L, Zhang Y, Ao Q, Xu S, Kuang D, Liu Q, Yan B, Zhou Y, Chu Q, Liu L, Yang X-P, Wang G, Deficiency of Tfh Cells and Germinal Center in Deceased COVID-19 Patients. CURR MED SCI. 40, 618–624 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woodruff MC, Ramonell RP, Nguyen DC, Cashman KS, Saini AS, Haddad NS, Ley AM, Kyu S, Howell JC, Ozturk T, Lee S, Suryadevara N, Case JB, Bugrovsky R, Chen W, Estrada J, Morrison-Porter A, Derrico A, Anam FA, Sharma M, Wu HM, Le SN, Jenks SA, Tipton CM, Staitieh B, Daiss JL, Ghosn E, Diamond MS, Carnahan RH, Crowe JE, Hu WT, Lee FE-H, Sanz I, Extrafollicular B cell responses correlate with neutralizing antibodies and morbidity in COVID-19. Nature Immunology. 21, 1506–1516 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J, Lau YF, Lamirande EW, Paddock CD, Bartlett JH, Zaki SR, Subbarao K, Cellular Immune Responses to Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) Infection in Senescent BALB/c Mice: CD4+ T Cells Are Important in Control of SARS-CoV Infection. Journal of Virology. 84, 1289–1301 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crotty S, Kersh EN, Cannons J, Schwartzberg PL, Ahmed R, SAP is required for generating long-term humoral immunity. Nature. 421, 282–287 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Kamperschroer C, Dibble JP, Meents DL, Schwartzberg PL, Swain SL, SAP Is Required for Th Cell Function and for Immunity to Influenza. The Journal of Immunology. 177, 5317–5327 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Fahey LM, Wilson EB, Elsaesser H, Fistonich CD, McGavern DB, Brooks DG, Viral persistence redirects CD4 T cell differentiation toward T follicular helper cells. Journal of Experimental Medicine. 208, 987–999 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu X, Chen X, Zhong B, Wang A, Wang X, Chu F, Nurieva RI, Yan X, Chen P, van der Flier LG, Nakatsukasa H, Neelapu SS, Chen W, Clevers H, Tian Q, Qi H, Wei L, Dong C, Transcription factor achaete-scute homologue 2 initiates follicular T-helper-cell development. Nature. 507, 513–518 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ray JP, Marshall HD, Laidlaw BJ, Staron MM, Kaech SM, Craft J, Transcription factor STAT3 and type I interferons are corepressive insulators for differentiation of follicular helper and T helper 1 cells. Immunity. 40, 367–377 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greczmiel U, Kräutler NJ, Pedrioli A, Bartsch I, Agnellini P, Bedenikovic G, Harker J, Richter K, Oxenius A, Sustained T follicular helper cell response is essential for control of chronic viral infection. Science Immunology. 2, eaam8686 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Xin G, Zander R, Schauder DM, Chen Y, Weinstein JS, Drobyski WR, Tarakanova V, Craft J, Cui W, Single-cell RNA sequencing unveils an IL-10-producing helper subset that sustains humoral immunity during persistent infection. Nature Communications. 9, 5037 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang H, Tang J, Liu Z, Liu Y, Huang Y, Xu Y, Hao P, Yin Z, Zhong J, Ye L, Jin X, Wang H, ZIKV infection induces robust Th1-like Tfh cell and long-term protective antibody responses in immunocompetent mice. Nature Communications. 10, 3859 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee SK, Rigby RJ, Zotos D, Tsai LM, Kawamoto S, Marshall JL, Ramiscal RR, Chan TD, Gatto D, Brink R, Yu D, Fagarasan S, Tarlinton DM, Cunningham AF, Vinuesa CG, B cell priming for extrafollicular antibody responses requires Bcl-6 expression by T cells. Journal of Experimental Medicine. 208, 1377–1388 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyauchi K, Sugimoto-Ishige A, Harada Y, Adachi Y, Usami Y, Kaji T, Inoue K, Hasegawa H, Watanabe T, Hijikata A, Fukuyama S, Maemura T, Okada-Hatakeyama M, Ohara O, Kawaoka Y, Takahashi Y, Takemori T, Kubo M, Protective neutralizing influenza antibody response in the absence of T follicular helper cells. Nature Immunology. 17, 1447–1458 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Kotov JA, Jenkins MK, Cutting Edge: T Cell–Dependent Plasmablasts Form in the Absence of Single Differentiated CD4 + T Cell Subsets. J.I 202, 401–405 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Junt T, Fink K, Förster R, Senn B, Lipp M, Muramatsu M, Zinkernagel RM, Ludewig B, Hengartner H, CXCR5-Dependent Seeding of Follicular Niches by B and Th Cells Augments Antiviral B Cell Responses. The Journal of Immunology. 175, 7109–7116 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Hollister K, Kusam S, Wu H, Clegg N, Mondal A, Sawant DV, Dent AL, Insights into the role of Bcl6 in follicular Th cells using a new conditional mutant mouse model. J. Immunol 191, 3705–3711 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toyama H, Okada S, Hatano M, Takahashi Y, Takeda N, Ichii H, Takemori T, Kuroda Y, Tokuhisa T, Memory B Cells without Somatic Hypermutation Are Generated from Bcl6-Deficient B Cells. Immunity. 17, 329–339 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Bentebibel S-E, Lopez S, Obermoser G, Schmitt N, Mueller C, Harrod C, Flano E, Mejias A, Albrecht RA, Blankenship D, Xu H, Pascual V, Banchereau J, Garcia-Sastre A, Palucka AK, Ramilo O, Ueno H, Induction of ICOS+CXCR3+CXCR5+ TH Cells Correlates with Antibody Responses to Influenza Vaccination. Science Translational Medicine. 5, 176ra32–176ra32 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spensieri F, Borgogni E, Zedda L, Bardelli M, Buricchi F, Volpini G, Fragapane E, Tavarini S, Finco O, Rappuoli R, Del Giudice G, Galli G, Castellino F, Human circulating influenza-CD4+ ICOS1+IL-21+ T cells expand after vaccination, exert helper function, and predict antibody responses. Proc Natl Acad Sci U S A. 110, 14330–14335 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kobayashi T, Iijima K, Dent AL, Kita H, Follicular helper T cells mediate IgE antibody response to airborne allergens. Journal of Allergy and Clinical Immunology. 139, 300–313.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang C-H, Guerder S, Hong S-C, van Ewijk W, Flavell RA, Mice Lacking the MHC Class II Transactivator (CIITA) Show Tissue-Specific Impairment of MHC Class II Expression. Immunity. 4, 167–178 (1996). [DOI] [PubMed] [Google Scholar]
- 28.Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L, Chen H-D, Chen J, Luo Y, Guo H, Jiang R-D, Liu M-Q, Chen Y, Shen X-R, Wang X, Zheng X-S, Zhao K, Chen Q-J, Deng F, Liu L-L, Yan B, Zhan F-X, Wang Y-Y, Xiao G-F, Shi Z-L, A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 579, 270–273 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Israelow B, Song E, Mao T, Lu P, Meir A, Liu F, Alfajaro MM, Wei J, Dong H, Homer RJ, Ring A, Wilen CB, Iwasaki A, Mouse model of SARS-CoV-2 reveals inflammatory role of type I interferon signaling. J Exp Med. 217 (2020), doi: 10.1084/jem.20201241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCray PB, Pewe L, Wohlford-Lenane C, Hickey M, Manzel L, Shi L, Netland J, Jia HP, Halabi C, Sigmund CD, Meyerholz DK, Kirby P, Look DC, Perlman S, Lethal Infection of K18-hACE2 Mice Infected with Severe Acute Respiratory Syndrome Coronavirus. Journal of Virology. 81, 813–821 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dejnirattisai W, Zhou D, Ginn HM, Duyvesteyn HME, Supasa P, Case JB, Zhao Y, Walter TS, Mentzer AJ, Liu C, Wang B, Paesen GC, Slon-Campos J, López-Camacho C, Kafai NM, Bailey AL, Chen RE, Ying B, Thompson C, Bolton J, Fyfe A, Gupta S, Tan TK, Gilbert-Jaramillo J, James W, Knight M, Carroll MW, Skelly D, Dold C, Peng Y, Levin R, Dong T, Pollard AJ, Knight JC, Klenerman P, Temperton N, Hall DR, Williams MA, Paterson NG, Bertram FKR, Siebert CA, Clare DK, Howe A, Radecke J, Song Y, Townsend AR, Huang K-YA, Fry EE, Mongkolsapaya J, Diamond MS, Ren J, Stuart DI, Screaton GR, The antigenic anatomy of SARS-CoV-2 receptor binding domain. Cell. 184, 2183–2200.e22 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brouwer PJM, Caniels TG, van der Straten K, Snitselaar JL, Aldon Y, Bangaru S, Torres JL, Okba NMA, Claireaux M, Kerster G, Bentlage AEH, van Haaren MM, Guerra D, Burger JA, Schermer EE, Verheul KD, van der Velde N, van der Kooi A, van Schooten J, van Breemen MJ, Bijl TPL, Sliepen K, Aartse A, Derking R, Bontjer I, Kootstra NA, Wiersinga WJ, Vidarsson G, Haagmans BL, Ward AB, de Bree GJ, Sanders RW, van Gils MJ, Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 369, 643–650 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robbiani DF, Gaebler C, Muecksch F, Lorenzi JCC, Wang Z, Cho A, Agudelo M, Barnes CO, Gazumyan A, Finkin S, Hägglöf T, Oliveira TY, Viant C, Hurley A, Hoffmann H-H, Millard KG, Kost RG, Cipolla M, Gordon K, Bianchini F, Chen ST, Ramos V, Patel R, Dizon J, Shimeliovich I, Mendoza P, Hartweger H, Nogueira L, Pack M, Horowitz J, Schmidt F, Weisblum Y, Michailidis E, Ashbrook AW, Waltari E, Pak JE, Huey-Tubman KE, Koranda N, Hoffman PR, West AP, Rice CM, Hatziioannou T, Bjorkman PJ, Bieniasz PD, Caskey M, Nussenzweig MC, Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 584, 437–442 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuan M, Liu H, Wu NC, Lee C-CD, Zhu X, Zhao F, Huang D, Yu W, Hua Y, Tien H, Rogers TF, Landais E, Sok D, Jardine JG, Burton DR, Wilson IA, Structural basis of a shared antibody response to SARS-CoV-2. Science. 369, 1119–1123 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang L, Zhou T, Zhang Y, Yang ES, Schramm CA, Shi W, Pegu A, Oloniniyi OK, Henry AR, Darko S, Narpala SR, Hatcher C, Martinez DR, Tsybovsky Y, Phung E, Abiona OM, Antia A, Cale EM, Chang LA, Choe M, Corbett KS, Davis RL, DiPiazza AT, Gordon IJ, Hait SH, Hermanus T, Kgagudi P, Laboune F, Leung K, Liu T, Mason RD, Nazzari AF, Novik L, O’Connell S, O’Dell S, Olia AS, Schmidt SD, Stephens T, Stringham CD, Talana CA, Teng I-T, Wagner DA, Widge AT, Zhang B, Roederer M, Ledgerwood JE, Ruckwardt TJ, Gaudinski MR, Moore PL, Doria-Rose NA, Baric RS, Graham BS, McDermott AB, Douek DC, Kwong PD, Mascola JR, Sullivan NJ, Misasi J, Ultrapotent antibodies against diverse and highly transmissible SARS-CoV-2 variants. Science. 373, eabh1766 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feldman J, Bals J, Altomare CG, St Denis K, Lam EC, Hauser BM, Ronsard L, Sangesland M, Bracamonte Moreno T, Okonkwo V, Hartojo N, Balazs AB, Bajic G, Lingwood D, Schmidt AG, Naive human B cells engage the receptor binding domain of SARS-CoV-2, variants of concern, and related sarbecoviruses. Sci Immunol, eabl5842 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Di Niro R, Lee S-J, Vander Heiden JA, Elsner RA, Trivedi N, Bannock JM, Gupta NT, Kleinstein SH, Vigneault F, Gilbert TJ, Meffre E, McSorley SJ, Shlomchik MJ, Salmonella Infection Drives Promiscuous B Cell Activation Followed by Extrafollicular Affinity Maturation. Immunity. 43, 120–131 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trivedi N, Weisel F, Smita S, Joachim S, Kader M, Radhakrishnan A, Clouser C, Rosenfeld AM, Chikina M, Vigneault F, Hershberg U, Ismail N, Shlomchik MJ, Liver Is a Generative Site for the B Cell Response to Ehrlichia muris. Immunity. 51, 1088–1101.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.William J, Euler C, Christensen S, Shlomchik MJ, Evolution of Autoantibody Responses via Somatic Hypermutation Outside of Germinal Centers. Science. 297, 2066–2070 (2002). [DOI] [PubMed] [Google Scholar]
- 40.Yaari G, Uduman M, Kleinstein SH, Quantifying selection in high-throughput Immunoglobulin sequencing data sets. Nucleic Acids Res. 40, e134 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yaari G, Vander Heiden JA, Uduman M, Gadala-Maria D, Gupta N, Stern JNH, O’Connor KC, Hafler DA, Laserson U, Vigneault F, Kleinstein SH, Models of somatic hypermutation targeting and substitution based on synonymous mutations from high-throughput immunoglobulin sequencing data. Front Immunol. 4, 358 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tortorici MA, Veesler D, in Advances in Virus Research, Rey FA, Ed. (Academic Press, 2019; https://www.sciencedirect.com/science/article/pii/S0065352719300284), vol. 105 of Complementary Strategies to Understand Virus Structure and Function, pp. 93–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ladner JT, Henson SN, Boyle AS, Engelbrektson AL, Fink ZW, Rahee F, D’ambrozio J, Schaecher KE, Stone M, Dong W, Dadwal S, Yu J, Caligiuri MA, Cieplak P, Bjørås M, Fenstad MH, Nordbø SA, Kainov DE, Muranaka N, Chee MS, Shiryaev SA, Altin JA, Epitope-resolved profiling of the SARS-CoV-2 antibody response identifies cross-reactivity with endemic human coronaviruses. CR Med. 2 (2021), doi: 10.1016/j.xcrm.2020.100189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y, Ma M, Lei Q, Wang F, Hong W, Lai D, Hou H, Xu Z, Zhang B, Chen H, Yu C, Xue J, Zheng Y, Wang X, Jiang H, Zhang H, Qi H, Guo S, Zhang Y, Lin X, Yao Z, Wu J, Sheng H, Zhang Y, Wei H, Sun Z, Fan X, Tao S, Linear epitope landscape of the SARS-CoV-2 Spike protein constructed from 1,051 COVID-19 patients. Cell Reports. 34 (2021), doi: 10.1016/j.celrep.2021.108915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoffmann M, Arora P, Groß R, Seidel A, Hörnich BF, Hahn AS, Krüger N, Graichen L, Hofmann-Winkler H, Kempf A, Winkler MS, Schulz S, Jäck H-M, Jahrsdörfer B, Schrezenmeier H, Müller M, Kleger A, Münch J, Pöhlmann S, SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell. 184, 2384–2393.e12 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou D, Dejnirattisai W, Supasa P, Liu C, Mentzer AJ, Ginn HM, Zhao Y, Duyvesteyn HME, Tuekprakhon A, Nutalai R, Wang B, Paesen GC, Lopez-Camacho C, Slon-Campos J, Hallis B, Coombes N, Bewley K, Charlton S, Walter TS, Skelly D, Lumley SF, Dold C, Levin R, Dong T, Pollard AJ, Knight JC, Crook D, Lambe T, Clutterbuck E, Bibi S, Flaxman A, Bittaye M, Belij-Rammerstorfer S, Gilbert S, James W, Carroll MW, Klenerman P, Barnes E, Dunachie SJ, Fry EE, Mongkolsapaya J, Ren J, Stuart DI, Screaton GR, Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell. 184, 2348–2361.e6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mendoza A, Yewdell WT, Hoyos B, Schizas M, Bou-Puerto R, Michaels AJ, Brown CC, Chaudhuri J, Rudensky AY, Assembly of a spatial circuit of T-bet–expressing T and B lymphocytes is required for antiviral humoral immunity. Science Immunology. 6 (2021), doi: 10.1126/sciimmunol.abi4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eisenbarth SC, Baumjohann D, Craft J, Fazilleau N, Ma CS, Tangye SG, Vinuesa CG, Linterman MA, CD4+ T cells that help B cells – a proposal for uniform nomenclature. Trends in Immunology. 42, 658–669 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee BO, Rangel-Moreno J, Moyron-Quiroz JE, Hartson L, Makris M, Sprague F, Lund FE, Randall TD, CD4 T Cell-Independent Antibody Response Promotes Resolution of Primary Influenza Infection and Helps to Prevent Reinfection. The Journal of Immunology. 175, 5827–5838 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Pinto D, Sauer MM, Czudnochowski N, Low JS, Tortorici MA, Housley MP, Noack J, Walls AC, Bowen JE, Guarino B, Rosen LE, di Iulio J, Jerak J, Kaiser H, Islam S, Jaconi S, Sprugasci N, Culap K, Abdelnabi R, Foo C, Coelmont L, Bartha I, Bianchi S, Silacci-Fregni C, Bassi J, Marzi R, Vetti E, Cassotta A, Ceschi A, Ferrari P, Cippà PE, Giannini O, Ceruti S, Garzoni C, Riva A, Benigni F, Cameroni E, Piccoli L, Pizzuto MS, Smithey M, Hong D, Telenti A, Lempp FA, Neyts J, Havenar-Daughton C, Lanzavecchia A, Sallusto F, Snell G, Virgin HW, Beltramello M, Corti D, Veesler D, Broad betacoronavirus neutralization by a stem helix–specific human antibody. Science. 373, 1109–1116 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hsieh C-L, Werner AP, Leist SR, Stevens LJ, Falconer E, Goldsmith JA, Chou C-W, Abiona OM, West A, Westendorf K, Muthuraman K, Fritch EJ, Dinnon KH, Schäfer A, Denison MR, Chappell JD, Baric RS, Graham BS, Corbett KS, McLellan JS, Stabilized Coronavirus Spike Stem Elicits a Broadly Protective Antibody. Cell Reports, 109929 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Keating R, Hertz T, Wehenkel M, Harris TL, Edwards BA, McClaren JL, Brown SA, Surman S, Wilson ZS, Bradley P, Hurwitz J, Chi H, Doherty PC, Thomas PG, McGargill MA, The kinase mTOR modulates the antibody response to provide cross-protective immunity to lethal infection with influenza virus. Nat Immunol. 14, 1266–1276 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Linterman MA, How T follicular helper cells and the germinal centre response change with age. Immunology & Cell Biology. 92, 72–79 (2014). [DOI] [PubMed] [Google Scholar]
- 54.Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, Pérez-Melgosa M, Sweetser MT, Schlissel MS, Nguyen S, Cherry SR, Tsai JH, Tucker SM, Weaver WM, Kelso A, Jaenisch R, Wilson CB, A Critical Role for Dnmt1 and DNA Methylation in T Cell Development, Function, and Survival. Immunity. 15, 763–774 (2001). [DOI] [PubMed] [Google Scholar]
- 55.Wei J, Alfajaro MM, DeWeirdt PC, Hanna RE, Lu-Culligan WJ, Cai WL, Strine MS, Zhang S-M, Graziano VR, Schmitz CO, Chen JS, Mankowski MC, Filler RB, Ravindra NG, Gasque V, de Miguel FJ, Patil A, Chen H, Oguntuyo KY, Abriola L, Surovtseva YV, Orchard RC, Lee B, Lindenbach BD, Politi K, van Dijk D, Kadoch C, Simon MD, Yan Q, Doench JG, Wilen CB, Genome-wide CRISPR Screens Reveal Host Factors Critical for SARS-CoV-2 Infection. Cell (2020), doi: 10.1016/j.cell.2020.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pillai PS, Molony RD, Martinod K, Dong H, Pang IK, Tal MC, Solis AG, Bielecki P, Mohanty S, Trentalange M, Homer RJ, Flavell RA, Wagner DD, Montgomery RR, Shaw AC, Staeheli P, Iwasaki A, Mx1 reveals innate pathways to antiviral resistance and lethal influenza disease. Science. 352, 463–466 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walls AC, Park Y-J, Tortorici MA, Wall A, McGuire AT, Veesler D, Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 181, 281–292.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen Y, Zuiani A, Fischinger S, Mullur J, Atyeo C, Travers M, Lelis FJN, Pullen KM, Martin H, Tong P, Gautam A, Habibi S, Bensko J, Gakpo D, Feldman J, Hauser BM, Caradonna TM, Cai Y, Burke JS, Lin J, Lederer JA, Lam EC, Lavine CL, Seaman MS, Chen B, Schmidt AG, Balazs AB, Lauffenburger DA, Alter G, Wesemann DR, Quick COVID-19 Healers Sustain Anti-SARS-CoV-2 Antibody Production. Cell. 183, 1496–1507.e16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vander Heiden JA, Yaari G, Uduman M, Stern JNH, O’Connor KC, Hafler DA, Vigneault F, Kleinstein SH, pRESTO: a toolkit for processing high-throughput sequencing raw reads of lymphocyte receptor repertoires. Bioinformatics. 30, 1930–1932 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gupta NT, Vander Heiden JA, Uduman M, Gadala-Maria D, Yaari G, Kleinstein SH, Change-O: a toolkit for analyzing large-scale B cell immunoglobulin repertoire sequencing data. Bioinformatics. 31, 3356–3358 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nouri N, Kleinstein SH, Somatic hypermutation analysis for improved identification of B cell clonal families from next-generation sequencing data. PLOS Computational Biology. 16, e1007977 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Raybould MIJ, Kovaltsuk A, Marks C, Deane CM, CoV-AbDab: the coronavirus antibody database. Bioinformatics. 37, 734–735 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hoehn KB, Pybus OG, Kleinstein SH, “Phylogenetic analysis of migration, differentiation, and class switching in B cells” (2020), p. 2020.05.30.124446, doi: 10.1101/2020.05.30.124446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cui A, Niro RD, Heiden JAV, Briggs AW, Adams K, Gilbert T, O’Connor KC, Vigneault F, Shlomchik MJ, Kleinstein SH, A Model of Somatic Hypermutation Targeting in Mice Based on High-Throughput Ig Sequencing Data. The Journal of Immunology. 197, 3566–3574 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Avanzato VA, Oguntuyo KY, Escalera-Zamudio M, Gutierrez B, Golden M, Pond SLK, Pryce R, Walter TS, Seow J, Doores KJ, Pybus OG, Munster VJ, Lee B, Bowden TA, A structural basis for antibody-mediated neutralization of Nipah virus reveals a site of vulnerability at the fusion glycoprotein apex. PNAS. 116, 25057–25067 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Decker JM, Bibollet-Ruche F, Wei X, Wang S, Levy DN, Wang W, Delaporte E, Peeters M, Derdeyn CA, Allen S, Hunter E, Saag MS, Hoxie JA, Hahn BH, Kwong PD, Robinson JE, Shaw GM, Antigenic conservation and immunogenicity of the HIV coreceptor binding site. Journal of Experimental Medicine. 201, 1407–1419 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kamath K, Reifert J, Johnston T, Gable C, Pantazes RJ, Rivera HN, McAuliffe I, Handali S, Daugherty PS, Antibody epitope repertoire analysis enables rapid antigen discovery and multiplex serology. Sci Rep. 10 (2020), doi: 10.1038/s41598-020-62256-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Raw data file
Data S2. Design and composition of the SARS-CoV-2 linear epitope library
Data S3. Raw count matrix of the antibody epitope repertoire
Figure S1. Cellular response to SARS-CoV-2 in Tfh-sufficient and -deficient mice
Figure S2. Lymph node structures of Tfh-sufficient and -deficient mice following SARS-CoV-2 infection
Figure S3. Humoral response to SARS-CoV-2 in Tfh-sufficient and -deficient mice
Figure S4. Long-term antibody production in Tfh-sufficient and -deficient mice infected with SARS-CoV-2
Figure S5. Analysis of SARS-CoV-2-reactive BCRs from mice and humans
Figure S6. Analysis of SARS-CoV-2 linear epitopes recognized by antibodies from Tfh-sufficient and -deficient mice
Figure S7. Complementary Tfh cell-dependent and -independent pathways of antibody production mediate neutralizing responses to SARS-CoV-2
Data Availability Statement
BCR sequencing data are available via the Gene Expression Omnibus (GSE188792). All other data needed to evaluate the conclusions of the paper are available in the paper or the Supplementary Materials. SARS-CoV-2 stabilized spike glycoprotein was made available from University of Washington through a material transfer agreement. Reagents required for integrating the NEBNext® Single Cell/Low Input cDNA Synthesis & Amplification Module and NEBNext® Immune Sequencing Kit were made available under a material transfer agreement from New England Biolabs.


