ABSTRACT
Glypicans are a family of cell surface heparan sulfate proteoglycans that play critical roles in multiple cell signaling pathways. Glypicans consist of a globular core, an unstructured stalk modified with sulfated glycosaminoglycan chains, and a glycosylphosphatidylinositol anchor. Though these structural features are conserved, their individual contribution to glypican function remains obscure. Here, we investigate how glypican 3 (GPC3), which is mutated in Simpson–Golabi–Behmel tissue overgrowth syndrome, regulates Hedgehog signaling. We find that GPC3 is necessary for the Hedgehog response, surprisingly controlling a downstream signal transduction step. Purified GPC3 ectodomain rescues signaling when artificially recruited to the surface of GPC3-deficient cells but has dominant-negative activity when unattached. Strikingly, the purified stalk, modified with heparan sulfate but not chondroitin sulfate, is necessary and sufficient for activity. Our results demonstrate a novel function for GPC3-associated heparan sulfate and provide a framework for the functional dissection of glycosaminoglycans by in vivo biochemical complementation.
This article has an associated First Person interview with the first author of the paper.
KEY WORDS: Signaling, Glycosaminoglycans, Heparan sulfate, Glypican, Hedgehog
Summary: A novel genetic and biochemical strategy reveals that heparan sulfate modification is necessary and sufficient for the key modulatory function of the GPC3 glypican in the Hedgehog signaling pathway.
INTRODUCTION
Glypicans (GPCs) are a family of cell surface heparan sulfate proteoglycans (HSPGs) deeply implicated in embryogenesis, adult physiology and disease (Selleck, 1999; Filmus et al., 2008; Sarrazin et al., 2011). GPCs were initially biochemically identified as a class of glycosylphosphatidylinositol (GPI)-anchored cell surface HSPGs, comprising a characteristic 64-kDa core protein (de Boeck et al., 1987; David et al., 1990). Subsequent molecular genetic studies revealed that vertebrates contain six GPC paralogs, GPC1 to GPC6 (Filmus et al., 2008). Although the six GPCs diverge considerably in primary sequence, they share a conserved domain architecture, consisting of a globular N-terminal cysteine-rich domain (Kim et al., 2011; Pei and Grishin, 2012; Svensson et al., 2012), followed by a flexible C-terminal stalk, which hosts multiple Ser-Gly attachment sites for sulfated glycosaminoglycan (GAG) chains (Bourdon et al., 1987), and ending with the terminal GPI anchor, which is embedded in the membrane. All vertebrate GPCs are important regulators of cell growth and embryonic development. GPC1 and GPC2 are required for brain development (Stipp et al., 1994; Jen et al., 2009). GPC3, perhaps the best characterized vertebrate GPC, is mutated in Simpson–Golabi–Behmel syndrome, a disorder characterized by tissue overgrowth, and renal medullary dysplasia with cyst formation (Pilia et al., 1996; Shi and Filmus, 2009), phenotypes that are recapitulated by mouse models of GPC3 loss (Cano-Gauci et al., 1999; Grisaru and Rosenblum, 2001; Chiao et al., 2002). GPC4 is involved in cell polarity during embryonic convergent extension (Topczewski et al., 2001) and craniofacial development (LeClair et al., 2009). GPC5 is implicated in cancer (Li et al., 2011) and kidney diseases (Grisaru and Rosenblum, 2001; Roberts and Gleadle, 2008; Okamoto et al., 2011, 2015), and GPC6 is important in bone development (Campos-Xavier et al., 2009; Capurro et al., 2017).
Much of the present thinking about GPC function derives from genetic studies of the two Drosophila GPCs, Division abnormally delayed (Dally) (Nakato et al., 1995) and Dally-like protein (Dlp) (Khare and Baumgartner, 2000), which regulate several of the signaling pathways that orchestrate early metazoan development, including Decapentaplegic (Dpp) (Jackson et al., 1997; Belenkaya et al., 2004), Wingless (Wg)/Wnt (Lin and Perrimon, 1999; Tsuda et al., 1999; Baeg et al., 2001; Franch-Marro et al., 2005; Han et al., 2005), Hedgehog (Hh) (Desbordes and Sanson, 2003; Lum et al., 2003; Han et al., 2004b; Yao et al., 2006; Williams et al., 2010) and fibroblast growth factor (FGF) (Yan and Lin, 2007). Specifically, the Drosophila GPCs regulate both the spread of morphogens through embryonic tissues, as well as signaling responses in target cells, though the two paralogs differ in their mode of regulation within a given pathway, and in their requirement across different pathways and developmental contexts (Häcker et al., 2005).
In addition to the importance of individual GPCs, genetic studies have suggested a critical role for the attached GAG chains. In Drosophila, mutants in GAG biosynthetic enzymes, such as Sugarless (Binari et al., 1997; Häcker et al., 1997; Haerry et al., 1997), Tout-velu (Ttv) (Bellaiche et al., 1998) and its relatives, Botv and Sotv (Han et al., 2004a; Takei et al., 2004), and Sulfateless (Lin and Perrimon, 1999), exhibit developmental phenotypes overlapping with those caused by the loss of GPCs. Similarly, mutations in the heparan sulfate biosynthetic enzyme EXT1 causes hereditary multiple exostoses in humans (Ahn et al., 1995).
In spite of the striking developmental and disease phenotypes caused by impaired GPC function, their molecular basis has remained obscure. In particular, it is not known what accounts for the different activities of GPC paralogs. One possible explanation is that their GAG chains dictate context-specific interactions (Bülow and Hobert, 2006). Heparan sulfate, the primary class of GPC-attached GAGs, consists of a linear chain of repeating disaccharide units, which can be modified by epimerization and by sulfation of four distinct sites, in combinatorial fashion. Moreover, the length of heparan sulfate chains, as well as the extent, type and pattern of their positional modification, can vary (Xu and Esko, 2014). Such structural differences could direct the interaction of a single GPC with multiple partners, an idea supported by the existence of ligand-specific heparan sulfate substructures, such as those involved in binding FGF2 (Turnbull et al., 1992).
Although considerable progress has been made in the structural characterization of GAG chains (Prabhakar et al., 2009), methods for the functional interrogation of specific GAG modifications have been limited. Genetic ablation of GAG-modifying enzymes in whole organisms (Townley and Bülow, 2011) or target cells (Esko et al., 1985) affect multiple proteoglycan classes, each including several family members. Similarly, mutagenesis of GAG attachment sites of a proteoglycan (Williams et al., 2010) addresses the effect of complete GAG loss, rather than that of discrete elements of GAG substructure.
In this study, we develop a genetic and biochemical approach for the precise functional interrogation of GAG substructure, focusing on the role of GPC3 (Pilia et al., 1996; Cano-Gauci et al., 1999; Shi and Filmus, 2009) in cell signaling. GPC3 has been implicated in both Hh and Wnt signaling (De Cat et al., 2003; Capurro et al., 2005; Song et al., 2005; Capurro et al., 2008; Li et al., 2011; Filmus and Capurro, 2014; Capurro et al., 2015; Holtz et al., 2015; Capurro et al., 2017; Kolluri and Ho, 2019), but the mechanism remains unknown. We find that GPC3 is necessary specifically for Hh signaling but not Wnt signaling. Surprisingly, GPC3 does not affect the initial steps involved in vertebrate Hh pathway activation, which occur in primary cilia (Bangs and Anderson, 2017). Instead, the GPC3 requirement occurs downstream of ligand reception, at the level of Gli proteins, the transcriptional effectors of the Hh pathway (Aza-Blanc et al., 2000; Bai and Joyner, 2001). The purified ectodomain of GPC3, when recruited artificially to the cell surface of GPC3-deficient cells, rescues the Hh signaling defect. In contrast, the soluble unattached GPC3 ectodomain exhibits a dominant-negative effect on the Hh pathway. Using protein purified from a panel of cells defective in different GAG biosynthetic enzymes, we demonstrate that the GPC3 ectodomain activity requires heparan sulfate but not chondroitin sulfate. Strikingly, the heparan sulfate-modified unstructured stalk of GPC3 is sufficient to recapitulate the activity of the entire protein, with the rest of the GPC3 molecule dispensable. These results define a novel role for GPC3-derived heparan sulfate in Hh signaling, and outline a systematic biochemical complementation strategy for the functional dissection of GAG substructure.
RESULTS
GPC3 is necessary for Hh signaling
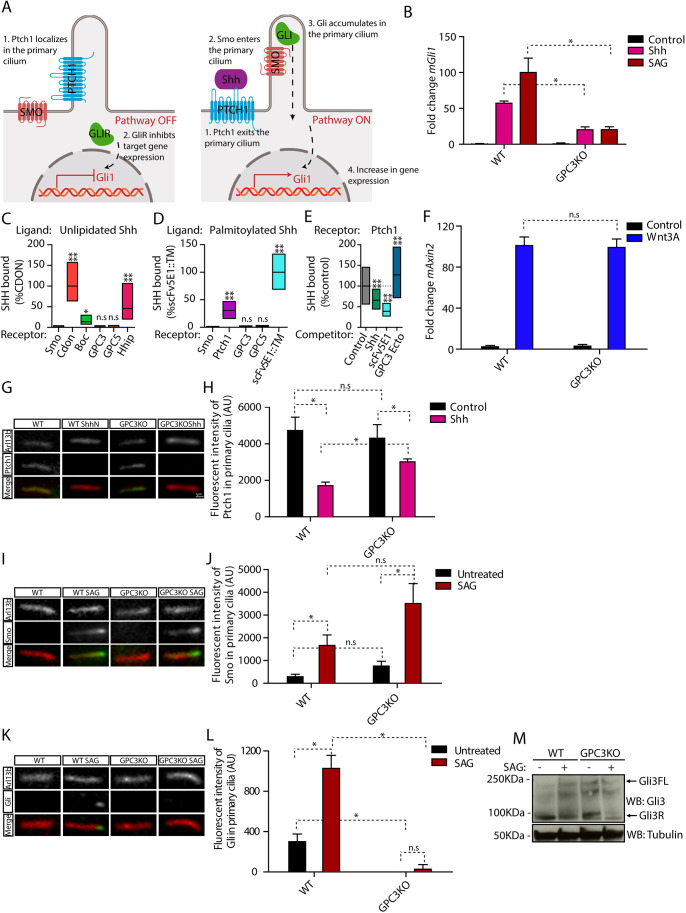
To test whether GPC3 is involved in the Hh pathway (Fig. 1A), we inactivated Gpc3 in mouse embryonic fibroblasts (MEFs) (Fig. S1A), using CRISPR/Cas9 (Table S1) (Ran et al., 2013). We then measured the transcriptional response to Hh pathway stimulation, by assaying induction of the Gli1 target gene (Tukachinsky et al., 2010). As shown in Fig. 1B, when Gpc3KO cells were treated with Shh, which activates Hh signaling by inhibiting the patched1 (Ptch1) receptor, Gli1 induction was significantly lower compared to wild-type cells. As GPC3 has been proposed to compete with Ptch1 for Shh (Capurro et al., 2008), we tested whether GPC3 and Shh interact. Contrary to previous observations (Capurro et al., 2008), we were unable to detect binding of Shh to GPC3 (Fig. 1C,D; Fig. S1B,C), and, furthermore, GPC3 did not compete with Ptch1 for binding to Shh (Fig. 1E; Fig. S1D), suggesting that the effects of GPC3 on Hh signaling are not at the level of the Shh-Ptch1 interaction. Consistent with this idea, Gpc3KO cells showed a blunted response even when the Hh pathway was stimulated downstream of Ptch1 using smoothened agonist (SAG; Chen et al., 2002), a synthetic compound that directly activates Smo (Fig. 1B). These results indicate that GPC3 is necessary for maximal Hh pathway activation, at the level of Smo or downstream of it.
Fig. 1.
Endogenous GPC3 is necessary for Hh pathway activation. (A) Schematic of the Hh pathway. Left: in the absence of signaling, Ptch1 in primary cilia inhibits Smo activation and localization to cilia. Right: upon pathway activation by Shh, Ptch1 is inhibited and exits from cilia, whereas Smo becomes active and accumulates in cilia. Smo activates Gli proteins, which accumulate at ciliary tips. Active Gli proteins turn on the transcriptional targets of the pathway, including Gli1. (B) Wild-type (WT) or Gpc3KO MEFs were incubated with Shh, SAG (1 µM) or control medium for 24 h, and Hh signaling was measured by qRT-PCR for Gli1. Data are mean±s.e.m. of three replicates and were normalized from 0% (untreated) to 100% activation of the Hh pathway by saturating SAG. Hh pathway activation by Shh and SAG was significantly decreased in the absence of GPC3. (C) Fluorescently labeled unlipidated Shh was incubated with HEK293T cells transiently transfected with eGFP-tagged GPC3, GPC5 or control receptors, and bound ligand was quantified by fluorescence microscopy. Cdon, Boc and Hhip (left), as well as Ptch1 and a single-chain variant of the anti-Shh monoclonal antibody (scFv5E1) (Wierbowski et al., 2020) (right), were used as positive controls, and Smo was used as negative control. Data are normalized between binding to the negative control (0%) and the highest bound signal (100%). Boxplots represent the median and the first and third quartiles of binding. At least 200 cells were measured per condition. Neither GPC3 nor GPC5 binds unlipidated Shh. (D) As in C but using fluorescently labeled palmitoylated Shh. At least 1000 cells were measured per condition. Neither GPC3 nor GPC5 binds palmitoylated Shh. (E) As in C, except binding of palmitoylated Shh to Ptch1 was measured after preincubation of Shh with purified GPC3 ectodomain (GPC3-Ecto) or control competitors. Data are normalized between binding to Smo (negative control, 0%) and binding to Ptch1 in the presence of control competitor (100%). At least 1000 cells were measured per condition. GPC3-Ecto does not compete with Ptch1 for Shh binding. Shh binding was competed by scFv5E1 or by excess unlipidated Shh. (F) Wild-type or Gpc3KO MEFs were incubated with Wnt3A-conditioned medium or control medium for 24 h, and Wnt signaling was measured by qRT-PCR for Axin2. Wnt pathway activation by Wnt3A was normal in the absence of GPC3. Data are mean±s.e.m. of three replicates. (G) Wild-type or Gpc3KO MEFs stably expressing Ptch1-eGFP were incubated with Shh as in B, and ciliary intensity of Ptch1 was measured by immunofluorescence microscopy. Cilia were detected by staining for endogenous Arl13B. Ptch1 is green, Arl13B is red. In both wild-type and Gpc3KO cells, Shh induces Ptch1 exit from cilia. (H) Quantification of the experiment in G. Data are mean±s.d. (300-400 cilia were measured per condition). (I) As in G but measuring ciliary levels of endogenous Smo following treatment with SAG (1 µM). Smo is green, Arl13B is red. In both wild-type and Gpc3KO cells, SAG causes normal ciliary accumulation of Smo. (J) As in H but quantifying the experiment in I (300-400 cilia were measured per condition). (K) As in G but measuring ciliary intensity of endogenous Gli proteins. The anti-Gli antibody recognizes both full-length Gli2 and Gli3 (Tukachinsky et al., 2010). Gli is green, Arl13B is red. In the absence of GPC3, Gli proteins did not localize to the tips of cilia and their ciliary recruitment in response to SAG was greatly reduced. (L) As in H but quantifying the experiment in K (300-400 cilia were measured per condition). (M) As in B but cells were analyzed by immunoblotting for Gli3. Blotting for tubulin served as loading control. In the absence of GPC3, Gli3R was cleared normally by Hh pathway stimulation. Blots shown are representative of three experiments. *P<0.05; ****P<0.0001; n.s., not significant (two-tailed paired t-test). AU, arbitrary units. Scale bar: 1 μm (shown in G, also applies to I and K).
We next asked whether the GPC3 requirement is specific for the Hh pathway. Previous studies suggested that GPC3 is required for Wnt signaling (De Cat et al., 2003; Capurro et al., 2005; Song et al., 2005). To test this possibility, we treated cells with Wnt3A and measured expression of the Wnt pathway-specific target gene Axin2. We observed no difference in Axin2 induction by Wnt3A between wild-type and Gpc3KO cells (Fig. 1F), indicating that Wnt pathway activation is unaffected by the loss of GPC3; this result is also consistent with a recent report (Dubey et al., 2020). Taken together, these data demonstrate that GPC3 is required for Hh signaling but not Wnt signaling.
GPC3 is required for signaling downstream of Smo
We sought to determine more precisely where GPC3 acts in the Hh pathway, by examining the dynamics of key pathway components in the primary cilium (Fig. 1A), the subcellular site where the initial steps in Hh signal transduction occur in vertebrate cells. In the resting state of the Hh pathway, the Shh receptor Ptch1 is localized to the primary cilium, inhibiting Smo and ensuring its exclusion from the cilium. Upon binding Shh, Ptch1 is inhibited and removed from the cilium, whereas Smo becomes active and accumulates in the cilium (Rohatgi et al., 2007; Tukachinsky et al., 2016; Bangs and Anderson, 2017; Petrov et al., 2017). In Gpc3KO cells, Ptch1 localization to cilia in the absence of stimulation and its Shh-dependent removal from cilia occur normally (Fig. 1G,H), as does activation-dependent accumulation of Smo in cilia (Fig. 1I,J). These results indicate that loss of GPC3 does not impair Ptch1 and Smo ciliary dynamics, suggesting that GPC3 might impinge on a downstream step in Hh signaling.
We next investigated whether loss of GPC3 affects the Gli proteins Gli2 and Gli3, the zinc finger transcriptional factors that function as effectors of the Hh pathway (Dahmane et al., 1997; Aza-Blanc et al., 2000; Bai and Joyner, 2001). In wild-type cells, active Smo leads to Gli2 and Gli3 activation, and accumulation at the tips of cilia (Kim et al., 2009; Tukachinsky et al., 2010; Wen et al., 2010). Strikingly, in Gpc3KO cells, accumulation of Gli proteins at ciliary tips is severely impaired (Fig. 1K,L), suggesting that GPC3 is required for the coupling between active Smo and the downstream Gli proteins.
Finally, we examined the consequence of GPC3 loss on the Gli3 protein. In the absence of Hh stimulation, full-length Gli3 (Gli3FL) is processed into a shorter form (Gli3R), which functions as repressor of the transcriptional output of the Hh pathway. Upon Hh stimulation, Gli3R levels drop and the levels of the transcriptional activator Gli3FL increase, causing increased transcription of Gli1 and accumulation of Gli1 protein. In Gpc3KO cells, we found that Gli3R levels were reduced normally upon SAG stimulation (Fig. 1M), suggesting that the observed inhibition of Hh signaling is not due to repression through Gli3R.
Rescue of GPC3 knockout cells by purified protein complementation
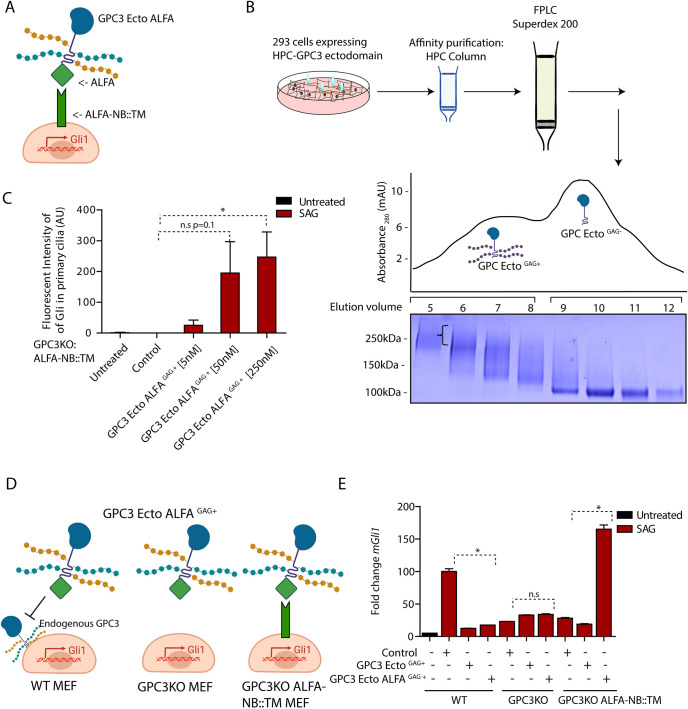
To demonstrate specificity of the Hh signaling defect of Gpc3KO cells, we first tried rescue experiments by expressing full-length GPC3 (Fig. S1E,F). Although we could not establish MEF lines stably expressing full-length GPC3, transient transduction failed to reverse the defect (Fig. S1F); we speculate that this was due to non-physiological levels of GPC3 expression (Fig. S1G). Therefore, we turned to a purified protein complementation strategy for rescuing Gpc3KO cells, which allows precise titration of the amount of added GPC3 (Fig. 2A). Briefly, the entire ectodomain of GPC3 was expressed and purified as a soluble secreted protein (GPC3-Ecto), tagged with the ALFA peptide at the C terminus (GPC3-Ecto-ALFA) (Fig. 2B). The ALFA peptide binds with sub-nanomolar affinity to the nanobody ALFA-NB (Gotzke et al., 2019), an interaction allowing the efficient recruitment of GPC3-Ecto-ALFA to the surface of cells expressing a membrane-anchored ALFA-NB (ALFA-NB::TM) (Fig. S2A); we used this system previously to recruit to the cell surface the ectodomain of GAS1, another GPI-anchored protein, like GPC3 (Wierbowski et al., 2020). When Gpc3KO cells stably expressing ALFA-NB::TM were incubated with purified GAG-modified GPC3-Ecto-ALFA, we observed a dose-dependent rescue of recruitment of Gli proteins to primary cilia in response to Hh pathway stimulation (Fig. 2C); interestingly, too much GPC3-Ecto-ALFA was inhibitory (Fig. S2B). Importantly, GPC3-Ecto-ALFA rescued signal-dependent Gli1 transcription in Gpc3KO cells expressing ALFA-NB::TM (Fig. 2D,E). As expected, rescue was dependent on both ALFA tag and ALFA-NB::TM, as untagged GPC3-Ecto-ALFA did not rescue Gpc3KO cells, and GPC3-Ecto did not rescue Gpc3KO cells expressing ALFA-NB::TM. Together, these data demonstrate that the function of GPC3 in Hh signaling requires its membrane attachment, and, more generally, that surface recruitment of ectodomains is an efficient strategy for reconstituting glypican activity in cells, in a controlled manner.
Fig. 2.
Purified GPC3-GAG (+) recruited to the cell surface rescues loss of endogenous GPC3. (A) Cartoon illustration of Gpc3KO MEFs expressing the membrane-bound ALFA nanobody (NB) construct ALFA-NB::TM, used to recruit GPC3-Ecto-ALFA to the cell surface. (B) Affinity-purified GPC3-Ecto fused to HaloTag was separated by size-exclusion chromatography to isolate GAG-modified and GAG-unmodified fractions. The indicated fractions were analyzed by SDS-PAGE and Coomassie staining. The bracket indicates the high molecular weight imparted by GAG modification of GPC3-Ecto. Gel shown is representative of three experiments. The y axis shows absorbance at 280 nm, expressed in mAU. (C) Gpc3KO MEFs stably expressing ALFA-NB::TM were incubated with SAG (1 µM) for 6 h in the presence of the indicated concentrations of purified GPC3-Ecto-ALFAGAG+. Ciliary intensity of endogenous Gli proteins was measured by immunofluorescence microscopy. Recruitment of GPC3-Ecto-ALFAGAG+ to the cell surface rescued signal-dependent accumulation of Gli at ciliary tips in a dose-dependent manner. Data are mean±s.d. (300-400 cilia were measured per condition). (D) Cartoon illustration of the experiment in E. (E) Wild-type (WT) MEFs, Gpc3KO MEFs or Gpc3KO MEFs stably expressing ALFA-NB::TM were incubated with SAG (1 µM) for 24 h in the presence or absence of 50 nM purified GPC3-Ecto-ALFAGAG+ or GPC3-EctoGAG+. Hh pathway output was measured by qRT-PCR for Gli1. Data are mean±s.e.m. of three replicates. Hh pathway activation is rescued when GPC3-Ecto-ALFAGAG+ is recruited to the cell surface. *P<0.05; n.s., not significant (two-tailed paired t-test). AU, arbitrary units.
Membrane-unattached GPC3 inhibits Hh signaling
In our rescue experiments, we noticed that purified GPC3-Ecto-ALFA inhibited Hh pathway activation in wild-type cells (Fig. 2E). This effect was also observed when wild-type cells were treated with purified GPC3-Ecto (Fig. 2E), or when GPC3-Ecto was expressed by viral transduction (Fig. S1F). Notably, GPC3-Ecto reduced the Hh response of wild-type cells to the levels of Gpc3KO cells but did not further reduce signaling in Gpc3KO cells (Fig. S1F); thus, the inhibitory activity of GPC3-Ecto requires endogenous GPC3. Taken together, these results suggest that GPC3-Ecto, when not membrane anchored, inhibits Hh signaling, perhaps by acting in dominant-negative fashion towards wild-type GPC3.
GAG modification is required for the effect of GPC3-Ecto in Hh signaling
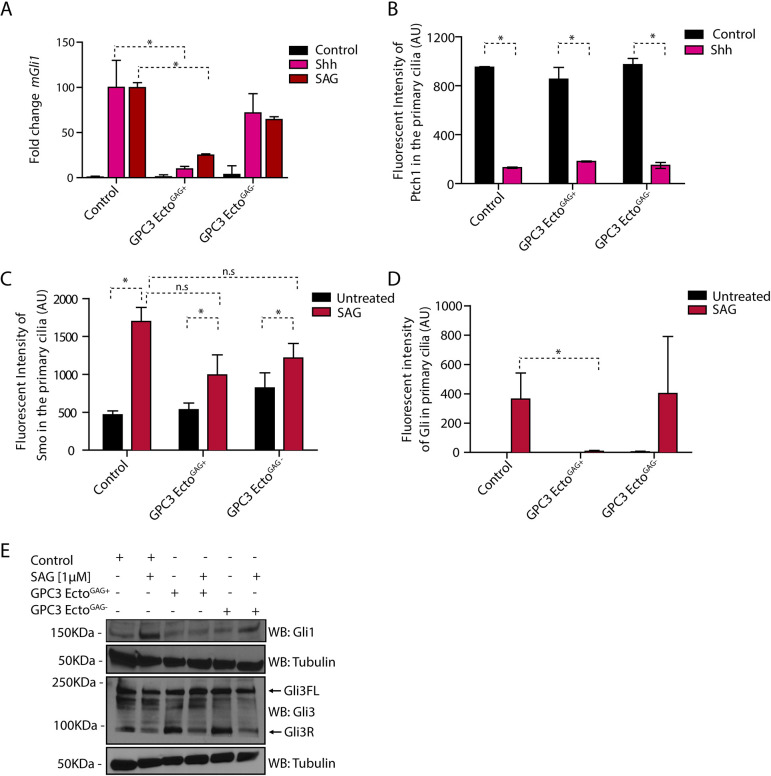
We next investigated the requirements for the inhibitory activity of GPC3-Ecto on the Hh pathway. When affinity-purified GPC3-Ecto was subjected to size-exclusion chromatography, we noticed that it consisted of two pools: a broad high-molecular weight fraction, corresponding to GAG-modified protein (GPC3-EctoGAG+), and a defined monomeric species, corresponding to unmodified protein (GPC3-EctoGAG−) (Fig. 2B). To determine the role of GAG modification, we tested the effect of GPC3-EctoGAG+ and GPC3-EctoGAG− on Hh signaling. Only GPC3-EctoGAG+ inhibited pathway activation by Shh or SAG, with GPC3-EctoGAG− exhibiting inactivity (Fig. 3A). Similarly, constitutive Hh signaling that occurs in Ptch1−/− cells (Dahmane et al., 1997) was reversed by GPC3-EctoGAG+ (Fig. S3A). Together, these results indicate that the GAG modification of GPC3 is critical for its effect on Hh signaling.
Fig. 3.
Purified GPC3 ectodomain inhibits Hh signaling as dominant negative. (A) Wild-type MEFs treated with GPC3-EctoGAG+ or GPC3-EctoGAG− were incubated with SAG (1 µM) or Shh for 24 h, and Hh pathway output was measured by qRT-PCR for Gli1. GPC3-EctoGAG+ inhibits Hh signaling in wild-type MEF cells. Data are mean±s.e.m. of three replicates. (B) Wild-type MEFs stably expressing Ptch1-eGFP were incubated with Shh for 24 h in the presence or absence of 1 µM of purified GPC3-EctoGAG+ or GPC3-EctoGAG−, and ciliary intensity of Ptch1 was measured by immunofluorescence microscopy. Cilia were detected by staining for endogenous Arl13B. GPC3-EctoGAG+ and GPC3-EctoGAG− had no effect on Shh-induced Ptch1 exit from cilia. Data are mean±s.d. (300-400 cilia were measured per condition). (C) As in B but treating wild-type MEFs with SAG (1 µM) and measuring the intensity of endogenous Smo in primary cilia. GPC3-EctoGAG+ and GPC3-EctoGAG− had no effect on SAG-induced ciliary accumulation of Smo. Data are mean±s.d. (300-400 cilia were measured per condition). (D) As in C but measuring the intensity of endogenous Gli proteins in primary cilia. GPC3-EctoGAG+ abolishes Gli recruitment to ciliary tips by Hh pathway activation, whereas GPC3-EctoGAG− has no effect. Data are mean±s.d. (300-400 cilia were measured per condition). (E) As in D but cells were analyzed by immunoblotting for Gli1 and Gli3. Blotting for tubulin served as a loading control. GPC3-EctoGAG+ did not block the reduction in Gli3R levels caused by Hh pathway activation but blocks the accumulation of Gli1. Blots shown are representative of three experiments. *P<0.05; n.s., not significant (two-tailed paired t-test). AU, arbitrary units.
We performed a number of control experiments to assess the specificity of GPC3-EctoGAG+ for the Hh pathway. We first investigated whether the effect is specific to GPC3 or is general to GAG-modified proteins. To address this, we purified the ectodomain of syndecan 1 (SDC1), a member of another class of cell surface HSPGs (Teng et al., 2012), and isolated the SDC1-EctoGAG+ and SDC1-EctoGAG− fractions (Fig. S3B). Neither form of SDC1 had an effect on the Hh pathway response (Fig. S3C), confirming that the antagonistic effect is specific to GAG-modified GPC3. We also tested the effect of the different GPC3-Ecto forms on Wnt signaling. Neither GPC3-EctoGAG+ nor GPC3-EctoGAG− had an effect on Wnt pathway transcriptional output, as measured by Axin2 expression (Fig. S3D), consistent with the specificity of GPC3-EctoGAG+ to the Hh pathway.
We further tested the idea that GPC3-EctoGAG+ acts in a dominant-negative fashion by comparing its effects to those caused by GPC3 loss. Several results indicate that cells treated with purified GPC3-EctoGAG+ behave like Gpc3KO cells. GPC3-EctoGAG+ had no effect on Shh-induced exit of Ptch1 from cilia (Fig. 3B), or Smo accumulation in cilia (Fig. 3C). Importantly, GPC3-EctoGAG+ drastically reduced SAG-induced ciliary accumulation of Gli proteins (Fig. 3D), indicating that GPC3-EctoGAG+ affects a step downstream of Smo, as observed for Gpc3KO cells. Finally, GPC3-EctoGAG+ had no effect on Gli3FL and Gli3R levels but inhibited signal-dependent Gli1 protein accumulation (Fig. 3E), as seen with Gpc3KO cells. In summary, the parallel phenotypes of Gpc3KO cells and wild-type cells treated with GPC3-EctoGAG+ support the idea that the GAG-modified GPC3 ectodomain functions in a dominant-negative fashion to antagonize Hh signaling.
Activity of GPC3-EctoGAG+ in the Hh pathway requires heparan but not chondroitin sulfate
We took advantage of the activity of exogenously added purified GPC3-Ecto to dissect the biochemical requirements for GPC3 function. Specifically, we used this system to test the role of specific GAG modifications, a question impossible to address unambiguously by genetic approaches in cell culture or whole organisms.
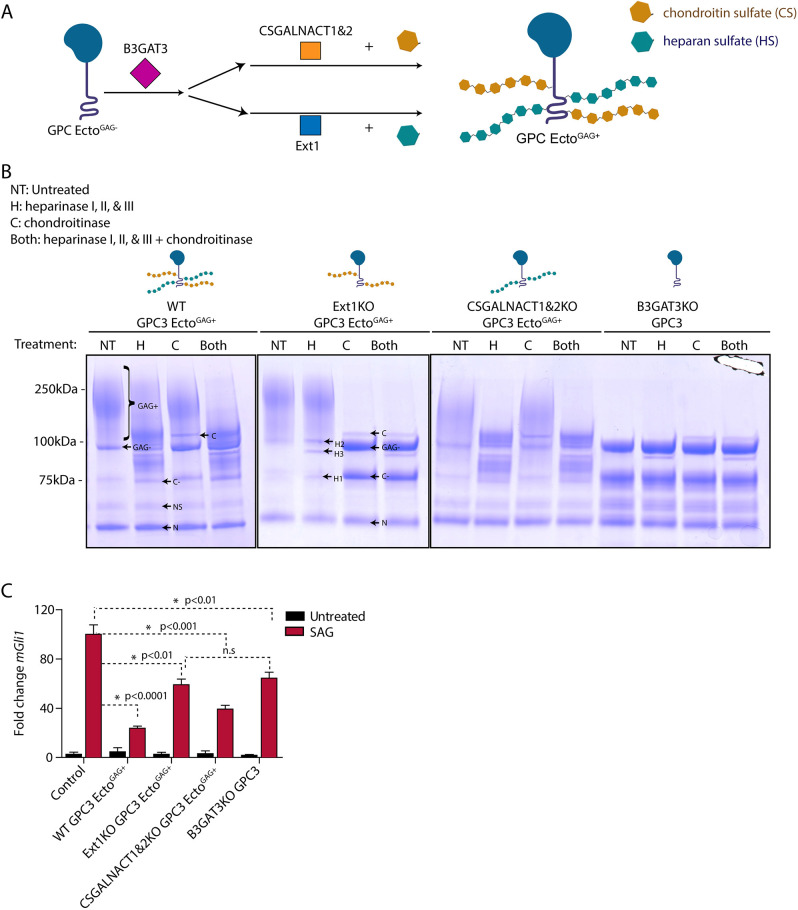
Heparan sulfate and chondroitin sulfate are two broad subfamilies of GAGs attached to proteoglycans (Kusche-Gullberg and Kjellen, 2003; Filmus et al., 2008; Prydz, 2015; Fig. 4A). Although most HSPGs and chondroitin sulfate proteoglycans (CSPGs) carry either heparan sulfate or chondroitin sulfate, others can carry both types of chains (Filmus et al., 2008; Prydz, 2015). Indeed, we observed that purified GPC3-Ecto preparations contained both heparan sulfate and chondroitin sulfate (Fig. 4B). To determine which class of GAG modification is required for GPC3 function in Hh signaling, we generated HEK293T cells lacking enzymes necessary for the attachment of heparan sulfate, chondroitin sulfate or both. Heparan sulfate chains were ablated by knocking out exostosin-1 (EXT1), a glycosyltransferase that catalyzes heparan sulfate elongation (McCormick et al., 2000; Presto et al., 2008). Chondroitin sulfate chains were ablated by knocking out chondroitin sulfate N-acetylgalactosaminyltransferase 1 and 2 (CSGALNACT1 and CSGALNACT2), which are needed for chondroitin sulfate attachment. Heparan sulfate and chondroitin sulfate were removed together by knocking out beta-1,3-glucuronyltransferase 3 (B3GAT3), which is needed for the attachment of both GAGs to proteins (Fig. 4A; Kusche-Gullberg and Kjellen, 2003). We then stably expressed and purified GPC3-Ecto from these knockout cells (Fig. S4A-D). To confirm the identity of attached GAG chains, we treated purified GPC3-Ecto proteins with recombinant heparinase (I, II and III), chondroitinase or both, and assayed the protein by SDS-PAGE. We observed that GPC3-Ecto purified from wild-type cells, when treated with either heparinase or chondroitinase, retained high-molecular weight GAG-modified material (Fig. 4B); however, treatment with both enzymes removed all GAG modifications (Fig. 4B), suggesting that purified GPC3-Ecto contains both heparan sulfate and chondroitin sulfate. As expected, GAG modification of GPC3-Ecto secreted from EXT1KO cells was removed by chondroitinase, whereas GAG modification of GPC3-Ecto secreted from CSGALNACT1,2KO cells was removed by heparinase (Fig. 4B). Finally, GPC3-Ecto secreted from B3GAT3KO cells contained only unmodified core protein (Fig. 4B). Thus, purified GPC3-Ecto is typically modified with a mix of heparan sulfate and chondroitin sulfate but selectively bears either modification when isolated from cells defective for the other branch of GAG biosynthesis.
Fig. 4.
Heparan sulfate modification is required for the effect of GPC3 on Hh signaling. (A) Schematic of the pathways for glypican modification with heparan sulfate (teal) or chondroitin sulfate (brown). B3GAT3 functions in the synthesis of the tetrasaccharide linker shared between chondroitin sulfate and heparan sulfate. CSGALNACT1 and 2 function in CS biosynthesis, whereas EXT1 functions in HS biosynthesis. (B) GPC3-Ecto was expressed and purified from HEK293T cells of the indicated genetic backgrounds [wild-type (WT), EXT1KO, CSGALNACT1,2KO, or B3GAT3KO]. The proteins were then digested with heparinase I, II and III (H), chondroitinase (C), or both, for 4 h at room temperature. The reactions were analyzed by SDS-PAGE and Coomassie staining. In wild-type cells, GPC3-Ecto was modified with both heparan sulfate and chondroitin sulfate chains. The choice of expression host cells allowed modification of GPC3-Ecto with heparan sulfate, chondroitin sulfate or both. GPC3-Ecto also underwent incomplete Furin-mediated proteolytic cleavage into two fragments (38 and 22 kDa; see Fig. S4F). Band notation is as follows: GAG+, GAG-modified full-length ectodomain and C-terminal cleavage product; GAG−, full-length ectodomain, uncleaved; N, N-terminal Furin cleavage product; C, chondroitinase; H1, heparinase I; H2, heparinase II; H3, heparinase III; C−, GAG− C-terminal Furin cleavage product; NS, nonspecific degradation product. See Fig. S4F for schematic. Gels shown are representative of three experiments. (C) Wild-type MEFs were incubated with SAG (1 µM) or control medium for 24 h in the absence or presence of the indicated differentially GAG-modified purified GPC3-Ecto (1 µM). Hh signaling was measured by qRT-PCR for Gli1. GPC3-Ecto required heparan sulfate chains to inhibit Hh signaling. Data are mean±s.e.m. of three replicates. *P<0.05; n.s., not significant (two-tailed paired t-test).
We used the GPC3-Ecto variants purified above to dissect the role of heparan sulfate and chondroitin sulfate modification. We found that GPC3-EctoGAG+ from CSGALNACT1,2KO cells (i.e. heparan sulfate modified) inhibited Hh pathway activation to a similar level as GPC3-EctoGAG+ from wild-type cells (Fig. 4C), whereas GPC3-EctoGAG+ from EXT1KO cells (i.e. chondroitin sulfate modified) or GPC3-Ecto from B3GAT3KO cells (i.e. unmodified) had a weaker effect than GPC3-Ecto from wild-type or CSGALNACT1,2KO cells. As there was no significant difference between GPC3-Ecto from EXT1KO cells and B3GAT3KO cells, it would appear that heparan sulfate modification is the key contributing modification that causes Hh pathway inhibition. Similar results were obtained for the effect of GPC3-Ecto on constitutive Hh signaling in Ptch1−/− cells (Fig. S3A). Chondroitin sulfate-modified, heparan sulfate-modified and unmodified GPC3-Ecto had no effect on Wnt signaling (Fig. S4E), which is again consistent with the specific role of GPC3 towards the Hh pathway. Together, these data suggest that heparan sulfate modification is necessary for the effect of GPC3-Ecto on Hh signaling.
The GAG-modified unstructured stalk of GPC3 is necessary and sufficient for Hh pathway antagonism
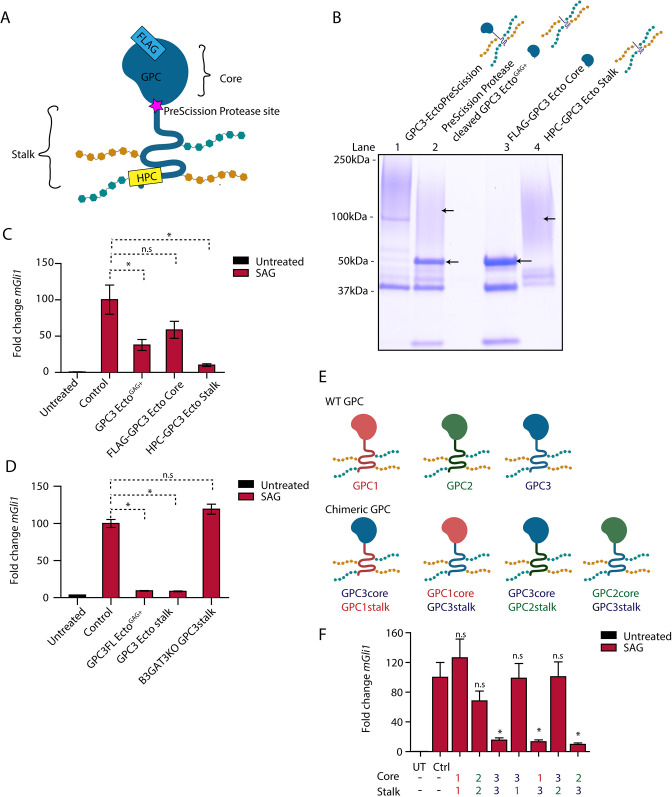
Finally, we investigated which part of GPC3-Ecto is responsible for its activity on Hh signaling. All GPCs contain a large globular core domain and a flexible GAG-modified stalk (Kusche-Gullberg and Kjellen, 2003; Filmus et al., 2008; Prydz, 2015). To assay each portion of GPC3 separately, we purified a version of GPC3-Ecto carrying N- and C-terminal affinity tags, and a PreScission protease site between the globular core and the stalk (GPC3-EctoPreScission; Fig. 5A). This fusion protein was cleaved with PreScission, and the resulting core and stalk fragments were affinity purified (Fig. 5B) and tested separately in signaling assays. We found that the GAG-modified stalk protein potently inhibited Hh signaling in both wild-type (Fig. 5C) and Ptch1−/− cells (Fig. S3A), and the globular core had a slight but not statistically significant effect on signaling (Fig. 5C). Importantly, unmodified stalk purified from B3GAT3KO cells was inactive (Fig. 5D) compared to the GAG-modified GPC3 stalk. These results indicate that the GPC3 stalk region, with its associated heparan sulfate, is necessary and sufficient for activity.
Fig. 5.
The GAG-modified GPC3 stalk is necessary and sufficient for Hh pathway antagonism. (A) Schematic of the GPC3-Ecto variant with an engineered PreScission protease cleavage site between the core and stalk domains (GPC3-EctoPreScission). The fusion is FLAG tagged on the N terminus (to isolate the core domain after cleavage, FLAG-GPC3core) and HPC tagged on the C terminus (to isolate the stalk region, HPC-GPC3 stalk). (B) Affinity-purified GPC3-EctoPreScission (lane 1) was cleaved with PreScission protease (lane 2), followed by purification of the core domain (lane 3) and stalk (lane 4) by FLAG and HPC affinity, respectively. Proteins were analyzed by SDS-PAGE and Coomassie staining. Following protease cleavage, the globular core collapsed to defined bands (lane 3), whereas the stalk region migrated as a high molecular weight smear (lane 4) due to GAG modification. Gel shown is representative of three experiments. (C) Wild-type (WT) MEFs were incubated with SAG (1 µM) in the absence or presence of the indicated purified proteins (1 µM) for 24 h. Hh signaling was measured by qRT-PCR for Gli1. The GAG-modified GPC3 stalk potently inhibited Hh signaling, in contrast to the unmodified globular core. Untreated is no treatment. Cells were kept in DMEM. (D) As in C but with incubation with GPC3 stalk expressed and purified from B3GAT3KO cells. The unmodified GPC3 stalk did not inhibit Hh signaling. (E) Schematic of GPCs GPC1, GPC2 and GPC3, and the chimeric GPCs generated by domain swapping. (F) As in C but with incubation with purified chimeric GPC ectodomains. Hh signaling antagonism required the presence of the GPC3 stalk.; Ctrl, cells treated with the buffer for the purified protein; UT, untreated. Data are mean±s.e.m. of three replicates. *P<0.05; n.s., not significant (two-tailed paired t-test).
The experiments above cannot rule out that the GPC3 globular core might direct GAG modification of the stalk during biosynthesis. To investigate this possibility, we performed domain swaps between GPC3 and GPC1 or GPC2 (Fig. 5E), two GPCs that have no effect on Hh signaling when added as purified ectodomains (Fig. 5F). We found that purified chimeras containing the GPC3 stalk (GPC1core-GPC3stalk-Ecto and GPC2core-GPC3stalk-Ecto) inhibited Hh signaling but the reciprocal chimeras were inactive (Fig. 5F). Together, these results demonstrate that the stalk region of GPC3 is necessary and sufficient for directing its own heparan sulfate modification, which is required for its effect on Hh signaling.
DISCUSSION
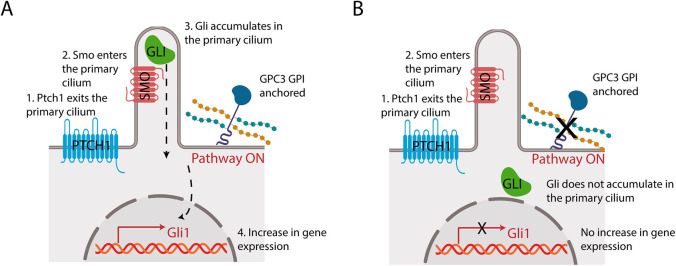
GPCs have long been implicated in cell-cell signaling and tissue homeostasis, yet their mechanisms remain obscure. In this study, we investigated the role of GPC3 in Hh signaling. We found that cells lacking GPC3 exhibit greatly reduced Hh pathway activation, and that the soluble GPC3 ectodomain acts in dominant-negative fashion, phenocopying the effect of GPC3 loss on Hh signaling (Fig. 6B). Interestingly, recruiting the GPC3 ectodomain to the cell surface rescues the full Hh response, indicating that membrane attachment is critical for the role of GPC3 in promoting Hh signaling (Fig. 6A). Surprisingly, epistatic analysis indicated that the upstream steps in Hh pathway activation are unaffected by GPC3, as both Shh-mediated inhibition of Ptch1 and subsequent Smo activation occurred normally. In contrast, activation of downstream Gli proteins was severely affected by GPC3 inhibition, as evidenced by the defective recruitment of full-length Gli to cilia in response to Smo activation and by the reduced Gli-driven transcriptional output of the Hh pathway. Together, these results define a novel glypican-dependent pathway that modulates the strength of Hh signaling in responding cells.
Fig. 6.
Model of GPC3 function in Hh signaling. (A) Endogenous GPI-anchored GPC3 promotes Hh signaling at the level of Gli proteins. (B) Loss of GPC3 impairs Hh signaling.
Our results differ from previous studies of GPCs, which reported both positive (Li et al., 2011; Wilson and Stoeckli, 2013; Capurro et al., 2017) and negative (Capurro et al., 2008) roles in Hh signaling. To our knowledge, all these previous studies posited that GPCs act at the level of Shh reception by responding cells. In studies showing a positive role of GPCs in Hh signaling, GPCs were proposed to enhance Shh recruitment to cells (Witt et al., 2013), or Shh binding to Ptch1 (Li et al., 2011; Wilson and Stoeckli, 2013; Capurro et al., 2017). In instances of Hh antagonism, GPCs were proposed to compete with Ptch1 for binding to Shh (Capurro et al., 2008). We did not detect GPC3 binding to Shh, or an effect of GPC3 on the Shh-Ptch1 interaction. Moreover, we found that GPC3 inhibition affects not only Hh pathway activation by Shh but also downstream activation, at the level of Smo. Finally, we observed a specific defect in the ciliary recruitment of Gli proteins, and Ptch1 and Smo ciliary dynamics were unaffected by GPC3 inhibition. Together, our results suggest that GPC3 is required for Hh signaling at a step subsequent to Smo activation, likely at the level of coupling between active Smo and Gli proteins.
GPC3 is mutated in Simpson–Golabi–Behmel syndrome, with 75% of GPC3 lesions being large deletions, frameshifts and truncating nonsense mutations; most of these mutations likely cause GPC3 loss or misfolding (Vuillaume et al., 2018). Interestingly, among the few characterized GPC3 missense mutations, one occurs in the sequence encoding the GPI anchor, resulting in the production of heparan sulfate-modified GPC3 ectodomain (Vuillaume et al., 2018); this disease mutant is consistent with the dominant-negative effect that we observed with purified GPC3-EctoGAG+.
How might GPC3 regulate the Hh pathway? Although the molecular events that connect GPC3 to Gli protein activation remain to be elucidated, our results permit some speculation on the mechanism of GPC3. Previous work identified GPR161 as a repressor of Hh signaling epistatic to Smo, acting at the level of Gli3R. In the absence of GPR161, Gli3R is completely lost (Mukhopadhyay et al., 2013); in contrast, GPC3 inhibition did not change in Gli3R levels, suggesting that GPC3 might not be involved in the GPR161 branch of the Hh pathway. The phenotype of GPC3 inhibition, characterized by normal Smo entry into cilia but loss of Gli recruitment, resembles that caused by the loss of the KIF7 kinesin (Endoh-Yamagami et al., 2009; He et al., 2014) or one of its upstream regulators, such as DLG5 (Chong et al., 2015) or PPFI1A/PP2A (Liu et al., 2014). One possibility is that GPC3 acts in a signal transduction process upstream of one of these factors; for example, to control KIF7 phosphorylation and thus the regulation of Gli proteins in cilia. Finally, another possibility is that GPC3 might function as a scaffold that brings together other factors on the cell surface to promote a signaling event that impacts Gli recruitment to cilia; such a scaffolding role would explain the inhibitory effect of abnormally high expression of full-length GPC3, or the dominant-negative effect of purified GPC3 ectodomain.
Our results indicate that GPC3 is acutely required for Hh pathway activation. Recruitment of GPC3 ectodomain to the surface of Gpc3KO cells rescues Gli recruitment to cilia tips in less than 6 h. Thus, changes in GPC3 function are translated over a relatively short timescale into corresponding changes in Hh pathway activation, possibly suggesting a transcription-independent mechanism for downstream Hh pathway modulation by GPC3.
Our functional analysis of purified GPC3 constructs afforded several mechanistic insights. First, GPC3 activity depends critically on its modification with GAG chains. Though purified GPC3-Ecto is modified with both heparan sulfate and chondroitin sulfate, we found, using protein purified from cells lacking one or both GAG biosynthetic pathways, that only the heparan sulfate-modified protein is active in Hh signaling. Deletion analysis surprisingly revealed that the GPC3 stalk region is necessary and sufficient for directing its own modification with heparan sulfate, and for its effect on Hh signaling. Although we observed that the GPC3 globular core is dispensable for the dominant-negative effect on the Hh pathway, we cannot rule out the possibility that putative endogenous binding partners might interact not only with stalk-associated heparan sulfate chains but also with the GPC3 core. Interestingly, we found that, in contrast to GPC3-Ecto, the ectodomains belonging to GPCs GPC1 and GPC2 are inactive in Hh signaling. This observation suggests that the discrete molecular structure of glypican-associated heparan sulfate – including the position, pattern and extent of sulfation – may differ between glypican family members. Further characterization of GPC3-associated heparan sulfate, together with functional assays of GPC3 purified from cells deficient in finer-scale heparan sulfate substructure biosynthesis, will be required to define the precise molecular requirements for GPC3 function.
Previous studies of GPCs employed genetic and molecular approaches with limited ability to precisely manipulate the GAG structure of individual GPCs. The present approach, relying on separate systems for GPC biosynthesis and functional interrogation, provides a powerful and general framework for mechanistic dissection. The availability of comprehensive collections of GAG biosynthesis knockout cells (Chen et al., 2018) should greatly facilitate expression and purification of proteoglycans bearing precisely edited GAG chains. Additionally, the heterologous cell-surface recruitment assay we describe can serve as a general platform for biochemical complementation of proteoglycan loss of function. Together, these approaches should catalyze functional studies of proteoglycans with an unprecedented level of mechanistic detail, leading to important basic scientific and therapeutic insights.
MATERIALS AND METHODS
Cell lines
Human embryonic kidney cells (HEK293T; ATCC) and MEFs (gift from Dr Jim R. Woodgett, Lunenfeld-Tanenbaum Research Institute, Toronto, Canada) were grown in Dulbecco's modified Eagle's medium (DMEM), with 10% (v/v) fetal bovine serum (FBS) and penicillin/streptomycin. Stable cell lines were generated by infection with lentiviruses expressing genes of interest. Briefly, genes of interest were subcloned into the third-generation lentiviral vector pHAGE (Mostoslavsky et al., 2006), which was used to produce lentiviruses in HEK293T cells, as described previously (Wierbowski et al., 2020). Lentiviruses were mixed with 1 µg/ml hexadimethrine bromide (Sigma-Aldrich) and were used to infect the desired target cells. 48 h post-infection, stably transduced cells were isolated by selection with blasticidin (InvivoGen) or puromycin (InvivoGen).
CRISPR/Cas9 gene editing
Guide RNA (gRNA) sequences were designed to target murine Gpc3, human EXT1, human CSGALNACT1, human CSGALNACT2 and human B3GAT3 (http://chopchop.cbu.uib.no). Synthetic oligonucleotides (Integrated DNA Technologies) containing gRNA sequences were annealed and cloned into the pX459 vector (Ran et al., 2013). Parental cells were transfected with the gRNA-expressing plasmids, and were transiently selected with puromycin. The cells were then plated and grown clonally. Genomic DNA was extracted from individual clones, and the gRNA target loci were PCR amplified, followed by sequencing using an MiSeq instrument (Illumina). Characterization of CRISPR-induced genomic lesions of the null clones used in this study is shown in Table S1.
Antibodies and chemicals
Primary antibodies for immunoblotting were used at 1 µg/ml in blocking solution, consisting of TBST [10 mM Tris (pH 8), 150 mM NaCl and 0.2% Triton X-100] supplemented with 5% non-fat dry milk. Primary antibodies for immunoblots were as follows: goat anti-Gli1 (R&D Systems, AF3455; 1:500); goat anti-Gli3 (R&D Systems, AF3690; 1:500); and mouse monoclonal anti-tubulin (Sigma-Aldrich, T6199; DM1; 1:1000). Anti-goat IgG and anti-mouse IgG horseradish peroxidase-conjugated secondary antibodies (GE Healthcare) were used (1:5000 dilution) in blocking solution. For immunofluorescence, primary and secondary antibodies were used at 1 µg/ml in TBST supplemented with 5% bovine serum albumin. Primary antibodies for immunofluorescence were as follows: rabbit anti-GFP (1:1000) (Petrov et al., 2020); chicken anti-Arl13B (1:2000) (Petrov et al., 2020); rabbit anti-Gli2/3 (1:500) (Tukachinsky et al., 2016); and goat anti-Smo (1:1000) (Nedelcu et al., 2013). Fluorophore-conjugated secondary antibodies were as follows: donkey anti-chicken IgY-Alexa Fluor 647 (Jackson ImmunoResearch); donkey anti-goat IgG-Alexa Fluor 594 (Jackson ImmunoResearch); and donkey anti-rabbit IgG-Alexa Fluor 488 (Thermo Fisher). SAG (≥98%) was obtained from Axxora.
Hh and Wnt ligands
Shh-conditioned medium was produced as described previously (Nedelcu et al., 2013). Briefly, an expression construct encoding the first 197 amino acids of human Shh cloned in the pCS2 vector was transiently transfected into HEK293T cells, using polyethyleneimine. The next day, the medium was replaced with DMEM, and the cells were incubated for 48 h. The conditioned medium was collected, centrifuged to remove cellular debris and then used in signaling assays. Wnt3A-conditioned medium was produced using L cells stably expressing mouse Wnt3A (American Type Culture Collection). After reaching confluency, the cells were incubated in DMEM supplemented with 1% FBS for 48 h. The medium was harvested, centrifuged and used in Wnt signaling assays.
DNA constructs
Constructs were generated by PCR and were subcloned into the lentiviral pHAGE vector, driven by a human CMV promoter. The pHAGE constructs were used to produce lentiviruses for generating stable cell lines. The construct for expressing mouse Ptch1 tagged with eGFP at the C terminus has been described previously (Tukachinsky et al., 2016). Additional eGFP-tagged receptor constructs for cell-based ligand-receptor binding experiments, including Cdon, Boc and membrane-anchored single chain variable fragment 5E1 antibody (scFv5E1), have been described previously (Wierbowski et al., 2020). The genes used in this study were as follows: human GPC1 (NM_002081.3); human GPC2 (NM_152742.3); mouse GPC3 (NM_016697.3); human GPC4 (NM_001448.3); human GPC5 (NM_004466.6); human GPC6 (NM_005708.5); human SDC1 (NM_001006946.2); and human Hhip (NM_022475.3). The Hhip expression construct consisted of the human calreticulin signal sequence, one copy of eGFP and a flexible (GGGSGGGT)3 linker, followed by the entire Hhip sequence after the portion encoding the N-terminal signal sequence. Full-length GPC expression constructs consisted of the influenza hemagglutinin signal sequence, one copy of eGFP or the FLAG or human protein C (HPC) epitope, followed by the GPC sequence after the portion encoding the N-terminal signal sequence. Constructs for expressing secreted fusion proteins (comprising entire GPC ectodomains or portions thereof) consisted of the following: signal sequence (endogenous or from influenza hemagglutinin), the GPC fragment, a flexible (GGGSGGGT)3 linker, the mutant bacterial dehalogenase HaloTag7 (Ohana et al., 2009) and one copy of the HPC epitope. Some constructs also included PreScission protease cleavage sites for tag removal. Details of the new expression plasmids used in this study are provided in Table S2.
SDS-PAGE and immunoblotting
Protein samples were separated by SDS-PAGE on 4-20% gradient gels (Mini-PROTEAN TGX, Bio-Rad). Gels were soaked in transfer buffer [48 mM Tris (pH 9.2), 39 mM glycine, 1.3 mM SDS and 20% methanol], and were blotted by semi-dry transfer (Trans-Blot SD, Bio-Rad) onto nitrocellulose membranes (Millipore). For immunoblotting, the membranes were incubated for 30 min in blocking solution, followed by overnight incubation at 4°C with primary antibodies. Following three washes with TBST at room temperature, the membranes were incubated with secondary antibodies in blocking solution for 1 h at room temperature. The membranes were then washed three times with TBST and twice with TBS [10 mM Tris (pH 8) and 150 mM NaCl], followed by chemiluminescent detection (ECL, PerkinElmer).
Immunofluorescence microscopy
MEFs were plated on 12-mm diameter gelatin-coated round glass coverslips, in 24-well plates, at a density of 105 cells per well. Following overnight incubation, the complete medium was replaced with serum-free DMEM to induce ciliogenesis. After 24 h, the cells were treated with the indicated factors in serum-free DMEM for another 24 h. The cells were then fixed in PBS with 3.7% formaldehyde for 30 min at room temperature. Following permeabilization with TBST, endogenous Smo, endogenous Gli2/3 or overexpressed Ptch1-eGFP was detected by immunofluorescence microscopy. Cells were co-stained for endogenous Arl13B to detect primary cilia. The stained coverslips were mounted in PBS with 50% glycerol and were imaged using a Nikon TE2000E wide-field epifluorescence microscope equipped with an OrcaER camera (Hamamatsu) and a 40× PlanApo 0.45NA air objective (Nikon), as described previously (Nedelcu et al., 2013). For each condition, MetaMorph software (Molecular Devices) was used to acquire z-series consisting of five focal planes for at least 30 fields of view for fluorescence channels corresponding to Arl13B, and Smo, Gli2/3 or GFP. The z-series were used to generate maximum intensity projections, which were analyzed using custom image analysis scripts written in MATLAB (MathWorks). Briefly, cilia were segmented by local adaptive thresholding of Arl13B images and, for each cilium, background-corrected fluorescence intensity for Smo, Gli2/3 or eGFP fluorescence intensity was calculated. Data are presented as mean±s.e.m. across three biological replicates. P<0.05 was considered significant.
Fluorescent Shh ligands
Alexa Fluor594-labeled unlipidated Shh was produced as described previously (Petrov et al., 2021). Briefly, mouse Shh N-terminal signaling domain (amino acids 24-198) was expressed in bacteria as a glutathione S-transferase (GST) fusion and was purified on glutathione agarose (GE Healthcare). Following cleavage of the GST tag with thrombin, Shh was purified by chromatography on heparin agarose (GE Healthcare) using a linear NaCl elution gradient [0.15 M to 1 M NaCl, in 25 mM phosphate (pH 7.2)]. Purified Shh was supplemented with TCEP (10 mM, pH 7.5) and incubated for 3 h at room temperature with a threefold molar excess of Alexa Fluor 594-maleimide (Thermo Fisher Scientific) to label the N-terminal cysteine. Unreacted Alexa Fluor 594-maleimide was removed on a NAP-10 desalting column (GE Healthcare). Tetramethylrhodamine (TMR)-labeled palmitoylated Shh was produced as described previously (Wierbowski et al., 2020). Briefly, human Shh N-terminal signaling domain (amino acids 24-197), tagged C-terminally with HaloTag7 and HPC epitope, was expressed in mammalian cells and purified via anti-HPC affinity chromatography from conditioned medium. The protein was then incubated for 1 h at room temperature with a fivefold molar excess of HaloTag TMR ligand (Promega), followed by removal of the unreacted dye on a NAP-5 desalting column (GE Healthcare).
Cell-based Shh binding assays
HEK293T cells plated in poly-D-lysine-coated wells were transfected with the eGFP-tagged receptor constructs and were grown for 48 h. The cells were incubated in OptiMEM with Alexa Fluor594-labeled unlipidated Shh or TMR-labeled palmitoylated Shh for 1.5 h at 37°C. For competition experiments, the fluorescent Shh ligand and the competitor [purified unlipidated Shh, scFv5E1 anti-Shh single-chain antibody (Wierbowski et al., 2020) or GPC3-Ecto] were pre-incubated before addition to cells for 30 min at room temperature. After incubation, the cells were washed with OptiMEM and were fixed in PBS with 3.7% formaldehyde for 30 min at room temperature. Fixed cells were washed twice with PBS and imaged by fluorescence microscopy using a 10× PlanApo 0.45NA objective (Nikon). For each condition, ligand and receptor images were acquired for four fields of view (MetaMorph software, Molecular Devices). Images were analyzed as described previously (Wierbowski et al., 2020). Briefly, cells were segmented based on the eGFP signal, and the background-subtracted fluorescence intensity of the Shh ligand was calculated. Bound ligand is measured as the ratio of total Shh intensity to area for each segmented cell, and is represented as a boxplot spanning from the first to the third quartile of the distribution. For statistical testing, the median bound ligand was calculated for three random non-overlapping subsets of the cell population measured for each condition. Ordinary one-way ANOVA, with Dunnett's multiple comparisons test, was performed on the sets of three medians to calculate P-values, using the first condition (Smo in Fig. 1C,D, and control competitor for Fig. 1E) as the reference sample.
qRT-PCR
MEF lines were plated in triplicate in six-well plates and, after reaching confluency, were serum starved overnight. Afterwards, the cells were treated with the indicated factors in DMEM for 24 h. Total RNA was isolated using TRIzol (Thermo Fisher Scientific), after which the RNA was treated with DNase I (Promega), followed by a second round of TRIzol purification. The RNA was reverse transcribed using Luna SuperScript (New England Biolabs) and random hexamers. To measure Hh pathway activation, the target gene Gli1 and the control gene cyclophilin were measured with Power SYBR Green (Thermo Fisher Scientific), as described previously (Liu et al., 2014). The primer sequences are listed in Table S3. Control cells were treated with storage buffer [20 mM Na-HEPES (pH 7.5) and 200 mM NaCl] that was used to preserve the purified proteins. The comparative Ct method (Schmittgen and Livak, 2008) was used to compute the expression of Gli1 relative to cyclophilin. Data for Hh pathway activation was normalized from 0% (untreated) to 100% (saturating amounts of the Smo agonist SAG). Data are presented as mean±s.e.m. across three biological replicates. P<0.05 was considered significant.
Protein expression and purification
Secreted proteins (GPC ectodomains and fragments thereof, and scFv5E1) were stably expressed in HEK293T cells and affinity purified from conditioned medium using their C-terminal HPC tags. Briefly, cell lines producing the desired protein fusion were grown in DMEM with 10% FBS, in 10-20 15-cm plates, until they reached confluency. The medium was then changed to DMEM with 1% FBS, and three collections were performed, each over 48 h. Conditioned medium was supplemented with CaCl2 to 2 mM and centrifuged (5000 g) and filtered (0.22 μm) to remove debris. The conditioned medium was loaded onto an anti-HPC affinity column, followed by extensive washing with TBS with 2 mM CaCl2. Bound protein was eluted in elution buffer [20 mM Na-HEPES (pH 7.5), 200 mM NaCl, 5 mM EDTA and 100 µg/ml HPC peptide]. Eluted protein was concentrated using centrifugal filter units (10-kDa cutoff, Millipore) and further purified by size-exclusion chromatography (Superdex 200 10/300 GL column, GE Healthcare). Appropriate fractions were collected, pooled, concentrated using centrifugal filter units, flash-frozen and stored at −80°C until use.
Genes encoding heparinases I, II and III (bt4675, bt4652 and bt4657) (Cartmell et al., 2017), and chondroitinase (bt3350), were cloned from Bacteroides thetaiotaomicron genomic DNA. Sequences encoding the enzymes lacking signal peptides were subcloned into the pGEX bacterial expression vector (GE Healthcare). GST-tagged proteins were expressed in BL21 DE3 pLysS Escherichia coli cells (Novagen) and purified as soluble proteins, according to the manufacturer's instructions. The activity and specificity of GST-tagged recombinant enzymes were confirmed by performing enzyme kinetic assays (measuring A232 absorbance) on porcine intestinal heparin (Sigma-Aldrich) or chondroitin sulfate A (Sigma-Aldrich) substrates. Purified proteins and activity assays are shown in Fig. S5, with specific activity measurements reported in Table S4.
Supplementary Material
Acknowledgements
We thank the Initiative for Gene Editing and Neurodegeneration (Cell Biology, Harvard Medical School) for help with MiSeq experiments; the Harvard Institute for Chemistry and Cell Biology for access to a QuantStudio 7 real-time PCR system; Pengxiang Huang for help with protein purification; Lina Yao (Devlin lab, Harvard Medical School) for the gift of B. thetaiotaomicron genomic DNA; and Marco Catipovic for critical reading of the manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: Y.C.L., B.M.W., A.S.; Methodology: Y.C.L., B.M.W., A.S.; Software: Y.C.L., B.M.W.; Validation: Y.C.L., B.M.W., A.S.; Formal analysis: Y.C.L., B.M.W.; Investigation: Y.C.L., B.M.W., A.S.; Resources: Y.C.L., B.M.W., A.S.; Data curation: Y.C.L., B.M.W., A.S.; Writing - original draft: Y.C.L., B.M.W., A.S.; Writing - review & editing: Y.C.L., B.M.W., A.S.; Visualization: Y.C.L., B.M.W., A.S.; Supervision: A.S.; Project administration: Y.C.L., B.M.W., A.S.; Funding acquisition: Y.C.L., B.M.W., A.S.
Funding
This work was supported by the National Institutes of Health (R01 GM122920 and GM135262 to A.S; T32GM007226 and F31GM120833 to B.M.W.). Y.C.L. was supported by a Kidney Foundation of Canada KRESCENT postdoctoral fellowship. Deposited in PMC for release after 12 months.
Peer review history
The peer review history is available online at https://journals.biologists.com/jcs/article-lookup/doi/10.1242/jcs.259297.
References
- Ahn, J., Lüdecke, H.-J., Lindow, S., Horton, W. A., Lee, B., Wagner, M. J., Horsthemke, B. and Wells, D. E. (1995). Cloning of the putative tumour suppressor gene for hereditary multiple exostoses (EXT1). Nat. Genet. 11, 137-143. 10.1038/ng1095-137 [DOI] [PubMed] [Google Scholar]
- Aza-Blanc, P., Lin, H. Y., Ruiz i Altaba, A. and Kornberg, T. B. (2000). Expression of the vertebrate Gli proteins in Drosophila reveals a distribution of activator and repressor activities. Development 127, 4293-4301. 10.1242/dev.127.19.4293 [DOI] [PubMed] [Google Scholar]
- Baeg, G. H., Lin, X., Khare, N., Baumgartner, S. and Perrimon, N. (2001). Heparan sulfate proteoglycans are critical for the organization of the extracellular distribution of Wingless. Development 128, 87-94. 10.1242/dev.128.1.87 [DOI] [PubMed] [Google Scholar]
- Bai, C. B. and Joyner, A. L. (2001). Gli1 can rescue the in vivo function of Gli2. Development 128, 5161-5172. 10.1242/dev.128.24.5161 [DOI] [PubMed] [Google Scholar]
- Bangs, F. and Anderson, K. V. (2017). Primary cilia and mammalian Hedgehog signaling. Cold Spring Harb. Perspect. Biol. 9, a028175. 10.1101/cshperspect.a028175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belenkaya, T. Y., Han, C., Yan, D., Opoka, R. J., Khodoun, M., Liu, H. and Lin, X. (2004). Drosophila Dpp morphogen movement is independent of dynamin-mediated endocytosis but regulated by the glypican members of heparan sulfate proteoglycans. Cell 119, 231-244. 10.1016/j.cell.2004.09.031 [DOI] [PubMed] [Google Scholar]
- Bellaiche, Y., The, I. and Perrimon, N. (1998). Tout-velu is a Drosophila homologue of the putative tumour suppressor EXT-1 and is needed for Hh diffusion. Nature 394, 85-88. 10.1038/27932 [DOI] [PubMed] [Google Scholar]
- Binari, R. C., Staveley, B. E., Johnson, W. A., Godavarti, R., Sasisekharan, R. and Manoukian, A. S. (1997). Genetic evidence that heparin-like glycosaminoglycans are involved in wingless signaling. Development 124, 2623-2632. 10.1242/dev.124.13.2623 [DOI] [PubMed] [Google Scholar]
- Bourdon, M. A., Krusius, T., Campbell, S., Schwartz, N. B. and Ruoslahti, E. (1987). Identification and synthesis of a recognition signal for the attachment of glycosaminoglycans to proteins. Proc. Natl. Acad. Sci. USA 84, 3194-3198. 10.1073/pnas.84.10.3194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bülow, H. E. and Hobert, O. (2006). The molecular diversity of glycosaminoglycans shapes animal development. Annu. Rev. Cell Dev. Biol. 22, 375-407. 10.1146/annurev.cellbio.22.010605.093433 [DOI] [PubMed] [Google Scholar]
- Campos-Xavier, A. B., Martinet, D., Bateman, J., Belluoccio, D., Rowley, L., Tan, T. Y., Baxová, A., Gustavson, K.-H., Borochowitz, Z. U., Innes, A. M.et al. (2009). Mutations in the heparan-sulfate proteoglycan glypican 6 (GPC6) impair endochondral ossification and cause recessive omodysplasia. Am. J. Hum. Genet. 84, 760-770. 10.1016/j.ajhg.2009.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano-Gauci, D. F., Song, H. H., Yang, H., McKerlie, C., Choo, B., Shi, W., Pullano, R., Piscione, T. D., Grisaru, S., Soon, S.et al. (1999). Glypican-3-deficient mice exhibit developmental overgrowth and some of the abnormalities typical of Simpson-Golabi-Behmel syndrome. J. Cell Biol. 146, 255-264. 10.1083/jcb.146.1.255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capurro, M. I., Xiang, Y.-Y., Lobe, C. and Filmus, J. (2005). Glypican-3 promotes the growth of hepatocellular carcinoma by stimulating canonical Wnt signaling. Cancer Res. 65, 6245-6254. 10.1158/0008-5472.CAN-04-4244 [DOI] [PubMed] [Google Scholar]
- Capurro, M. I., Xu, P., Shi, W., Li, F., Jia, A. and Filmus, J. (2008). Glypican-3 inhibits Hedgehog signaling during development by competing with patched for Hedgehog binding. Dev. Cell 14, 700-711. 10.1016/j.devcel.2008.03.006 [DOI] [PubMed] [Google Scholar]
- Capurro, M., Shi, W., Izumikawa, T., Kitagawa, H. and Filmus, J. (2015). Processing by convertases is required for glypican-3-induced inhibition of Hedgehog signaling. J. Biol. Chem. 290, 7576-7585. 10.1074/jbc.M114.612705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capurro, M., Izumikawa, T., Suarez, P., Shi, W., Cydzik, M., Kaneiwa, T., Gariepy, J., Bonafe, L. and Filmus, J. (2017). Glypican-6 promotes the growth of developing long bones by stimulating Hedgehog signaling. J. Cell Biol. 216, 2911-2926. 10.1083/jcb.201605119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartmell, A., Lowe, E. C., Baslé, A., Firbank, S. J., Ndeh, D. A., Murray, H., Terrapon, N., Lombard, V., Henrissat, B., Turnbull, J. E.et al. (2017). How members of the human gut microbiota overcome the sulfation problem posed by glycosaminoglycans. Proc. Natl. Acad. Sci. USA 114, 7037-7042. 10.1073/pnas.1704367114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J. K., Taipale, J., Young, K. E., Maiti, T. and Beachy, P. A. (2002). Small molecule modulation of Smoothened activity. Proc. Natl. Acad. Sci. USA 99, 14071-14076. 10.1073/pnas.182542899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y.-H., Narimatsu, Y., Clausen, T. M., Gomes, C., Karlsson, R., Steentoft, C., Spliid, C. B., Gustavsson, T., Salanti, A., Persson, A.et al. (2018). The GAGOme: a cell-based library of displayed glycosaminoglycans. Nat. Methods 15, 881-888. 10.1038/s41592-018-0086-z [DOI] [PubMed] [Google Scholar]
- Chiao, E., Fisher, P., Crisponi, L., Deiana, M., Dragatsis, I., Schlessinger, D., Pilia, G. and Efstratiadis, A. (2002). Overgrowth of a mouse model of the Simpson-Golabi-Behmel syndrome is independent of IGF signaling. Dev. Biol. 243, 185-206. 10.1006/dbio.2001.0554 [DOI] [PubMed] [Google Scholar]
- Chong, Y. C., Mann, R. K., Zhao, C., Kato, M. and Beachy, P. A. (2015). Bifurcating action of Smoothened in Hedgehog signaling is mediated by Dlg5. Genes Dev. 29, 262-276. 10.1101/gad.252676.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahmane, N., Lee, J., Robins, P., Heller, P. and Ruiz i Altaba, A. (1997). Activation of the transcription factor Gli1 and the Sonic hedgehog signalling pathway in skin tumours. Nature 389, 876-881. 10.1038/39918 [DOI] [PubMed] [Google Scholar]
- David, G., Lories, V., Decock, B., Marynen, P., Cassiman, J. J. and Van den Berghe, H. (1990). Molecular cloning of a phosphatidylinositol-anchored membrane heparan sulfate proteoglycan from human lung fibroblasts. J. Cell Biol. 111, 3165-3176. 10.1083/jcb.111.6.3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boeck, H., Lories, V., David, G., Cassiman, J. J. and van den Berghe, H. (1987). Identification of a 64 kDa heparan sulphate proteoglycan core protein from human lung fibroblast plasma membranes with a monoclonal antibody. Biochem. J. 247, 765-771. 10.1042/bj2470765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cat, B., Muyldermans, S.-Y., Coomans, C., Degeest, G., Vanderschueren, B., Creemers, J., Biemar, F., Peers, B. and David, G. (2003). Processing by proprotein convertases is required for glypican-3 modulation of cell survival, Wnt signaling, and gastrulation movements. J. Cell Biol. 163, 625-635. 10.1083/jcb.200302152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbordes, S. C. and Sanson, B. (2003). The glypican Dally-like is required for Hedgehog signalling in the embryonic epidermis of Drosophila. Development 130, 6245-6255. 10.1242/dev.00874 [DOI] [PubMed] [Google Scholar]
- Dubey, R., van Kerkhof, P., Jordens, I., Malinauskas, T., Pusapati, G. V., McKenna, J. K., Li, D., Carette, J. E., Ho, M., Siebold, C.et al. (2020). R-spondins engage heparan sulfate proteoglycans to potentiate WNT signaling. eLife 9, e54469. 10.7554/eLife.54469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endoh-Yamagami, S., Evangelista, M., Wilson, D., Wen, X., Theunissen, J.-W., Phamluong, K., Davis, M., Scales, S. J., Solloway, M. J., de Sauvage, F. J.et al. (2009). The mammalian Cos2 homolog Kif7 plays an essential role in modulating Hh signal transduction during development. Curr. Biol. 19, 1320-1326. 10.1016/j.cub.2009.06.046 [DOI] [PubMed] [Google Scholar]
- Esko, J. D., Stewart, T. E. and Taylor, W. H. (1985). Animal cell mutants defective in glycosaminoglycan biosynthesis. Proc. Natl. Acad. Sci. USA 82, 3197-3201. 10.1073/pnas.82.10.3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filmus, J. and Capurro, M. (2014). The role of glypicans in Hedgehog signaling. Matrix Biol. 35, 248-252. 10.1016/j.matbio.2013.12.007 [DOI] [PubMed] [Google Scholar]
- Filmus, J., Capurro, M. and Rast, J. (2008). Glypicans. Genome Biol. 9, 224. 10.1186/gb-2008-9-5-224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franch-Marro, X., Marchand, O., Piddini, E., Ricardo, S., Alexandre, C. and Vincent, J.-P. (2005). Glypicans shunt the Wingless signal between local signalling and further transport. Development 132, 659-666. 10.1242/dev.01639 [DOI] [PubMed] [Google Scholar]
- Gotzke, H., Kilisch, M., Martinez-Carranza, M., Sogratte-Idrissi, S., Rajavel, A., Schlichthaerle, T., Engels, N., Jungmann, R., Stenmark, P., Opazo, F.et al. (2019). The ALFA-tag is a highly versatile tool for nanobody-based bioscience applications. Nat. Commun. 10, 4403. 10.1038/s41467-019-12301-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisaru, S. and Rosenblum, N. D. (2001). Glypicans and the biology of renal malformations. Pediatr. Nephrol. 16, 302-306. 10.1007/s004670000530 [DOI] [PubMed] [Google Scholar]
- Häcker, U., Lin, X. and Perrimon, N. (1997). The Drosophila sugarless gene modulates Wingless signaling and encodes an enzyme involved in polysaccharide biosynthesis. Development 124, 3565-3573. 10.1242/dev.124.18.3565 [DOI] [PubMed] [Google Scholar]
- Häcker, U., Nybakken, K. and Perrimon, N. (2005). Heparan sulphate proteoglycans: the sweet side of development. Nat. Rev. Mol. Cell Biol. 6, 530-541. 10.1038/nrm1681 [DOI] [PubMed] [Google Scholar]
- Haerry, T. E., Heslip, T. R., Marsh, J. L. and O'Connor, M. B. (1997). Defects in glucuronate biosynthesis disrupt Wingless signaling in Drosophila. Development 124, 3055-3064. 10.1242/dev.124.16.3055 [DOI] [PubMed] [Google Scholar]
- Han, C., Belenkaya, T. Y., Khodoun, M., Tauchi, M., Lin, X. and Lin, X. (2004a). Distinct and collaborative roles of Drosophila EXT family proteins in morphogen signalling and gradient formation. Development 131, 1563-1575. 10.1242/dev.01051 [DOI] [PubMed] [Google Scholar]
- Han, C., Belenkaya, T. Y., Wang, B. and Lin, X. (2004b). Drosophila glypicans control the cell-to-cell movement of Hedgehog by a dynamin-independent process. Development 131, 601-611. 10.1242/dev.00958 [DOI] [PubMed] [Google Scholar]
- Han, C., Yan, D., Belenkaya, T. Y. and Lin, X. (2005). Drosophila glypicans Dally and Dally-like shape the extracellular Wingless morphogen gradient in the wing disc. Development 132, 667-679. 10.1242/dev.01636 [DOI] [PubMed] [Google Scholar]
- He, M., Subramanian, R., Bangs, F., Omelchenko, T., Liem, K. F., Jr, Kapoor, T. M. and Anderson, K. V. (2014). The kinesin-4 protein Kif7 regulates mammalian Hedgehog signalling by organizing the cilium tip compartment. Nat. Cell Biol. 16, 663-672. 10.1038/ncb2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtz, A. M., Griffiths, S. C., Davis, S. J., Bishop, B., Siebold, C. and Allen, B. L. (2015). Secreted HHIP1 interacts with heparan sulfate and regulates Hedgehog ligand localization and function. J. Cell Biol. 209, 739-758. 10.1083/jcb.201411024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, S. M., Nakato, H., Sugiura, M., Jannuzi, A., Oakes, R., Kaluza, V., Golden, C. and Selleck, S. B. (1997). dally, a Drosophila glypican, controls cellular responses to the TGF-beta-related morphogen, Dpp. Development 124, 4113-4120. 10.1242/dev.124.20.4113 [DOI] [PubMed] [Google Scholar]
- Jen, Y.-H. L., Musacchio, M. and Lander, A. D. (2009). Glypican-1 controls brain size through regulation of fibroblast growth factor signaling in early neurogenesis. Neural Dev. 4, 33. 10.1186/1749-8104-4-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare, N. and Baumgartner, S. (2000). Dally-like protein, a new Drosophila glypican with expression overlapping with wingless. Mech. Dev. 99, 199-202. 10.1016/S0925-4773(00)00502-5 [DOI] [PubMed] [Google Scholar]
- Kim, J., Kato, M. and Beachy, P. A. (2009). Gli2 trafficking links Hedgehog-dependent activation of Smoothened in the primary cilium to transcriptional activation in the nucleus. Proc. Natl. Acad. Sci. USA 106, 21666-21671. 10.1073/pnas.0912180106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M.-S., Saunders, A. M., Hamaoka, B. Y., Beachy, P. A. and Leahy, D. J. (2011). Structure of the protein core of the glypican Dally-like and localization of a region important for hedgehog signaling. Proc. Natl. Acad. Sci. USA 108, 13112-13117. 10.1073/pnas.1109877108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolluri, A. and Ho, M. (2019). The role of Glypican-3 in regulating Wnt, YAP, and Hedgehog in liver cancer. Front. Oncol. 9, 708. 10.3389/fonc.2019.00708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusche-Gullberg, M. and Kjellen, L. (2003). Sulfotransferases in glycosaminoglycan biosynthesis. Curr. Opin. Struct. Biol. 13, 605-611. 10.1016/j.sbi.2003.08.002 [DOI] [PubMed] [Google Scholar]
- LeClair, E. E., Mui, S. R., Huang, A., Topczewska, J. M. and Topczewski, J. (2009). Craniofacial skeletal defects of adult zebrafish Glypican 4 (knypek) mutants. Dev. Dyn. 238, 2550-2563. 10.1002/dvdy.22086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, F., Shi, W., Capurro, M. and Filmus, J. (2011). Glypican-5 stimulates rhabdomyosarcoma cell proliferation by activating Hedgehog signaling. J. Cell Biol. 192, 691-704. 10.1083/jcb.201008087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, X. and Perrimon, N. (1999). Dally cooperates with Drosophila Frizzled 2 to transduce Wingless signalling. Nature 400, 281-284. 10.1038/22343 [DOI] [PubMed] [Google Scholar]
- Liu, Y. C., Couzens, A. L., Deshwar, A. R., McBroom-Cerajewski, L. D. B., Zhang, X., Puviindran, V., Scott, I. C., Gingras, A. C., Hui, C.-C. and Angers, S. (2014). The PPFIA1-PP2A protein complex promotes trafficking of Kif7 to the ciliary tip and Hedgehog signaling. Sci. Signal. 7, ra117. 10.1126/scisignal.2005608 [DOI] [PubMed] [Google Scholar]
- Lum, L., Yao, S., Mozer, B., Rovescalli, A., Von Kessler, D., Nirenberg, M. and Beachy, P. A. (2003). Identification of Hedgehog pathway components by RNAi in Drosophila cultured cells. Science 299, 2039-2045. 10.1126/science.1081403 [DOI] [PubMed] [Google Scholar]
- McCormick, C., Duncan, G., Goutsos, K. T. and Tufaro, F. (2000). The putative tumor suppressors EXT1 and EXT2 form a stable complex that accumulates in the Golgi apparatus and catalyzes the synthesis of heparan sulfate. Proc. Natl. Acad. Sci. USA 97, 668-673. 10.1073/pnas.97.2.668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostoslavsky, G., Fabian, A. J., Rooney, S., Alt, F. W. and Mulligan, R. C. (2006). Complete correction of murine Artemis immunodeficiency by lentiviral vector-mediated gene transfer. Proc. Natl. Acad. Sci. USA 103, 16406-16411. 10.1073/pnas.0608130103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay, S., Wen, X., Ratti, N., Loktev, A., Rangell, L., Scales, S. J. and Jackson, P. K. (2013). The ciliary G-protein-coupled receptor Gpr161 negatively regulates the Sonic hedgehog pathway via cAMP signaling. Cell 152, 210-223. 10.1016/j.cell.2012.12.026 [DOI] [PubMed] [Google Scholar]
- Nakato, H., Futch, T. A. and Selleck, S. B. (1995). The division abnormally delayed (dally) gene: a putative integral membrane proteoglycan required for cell division patterning during postembryonic development of the nervous system in Drosophila. Development 121, 3687-3702. 10.1242/dev.121.11.3687 [DOI] [PubMed] [Google Scholar]
- Nedelcu, D., Liu, J., Xu, Y., Jao, C. and Salic, A. (2013). Oxysterol binding to the extracellular domain of Smoothened in Hedgehog signaling. Nat. Chem. Biol. 9, 557-564. 10.1038/nchembio.1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohana, R. F., Encell, L. P., Zhao, K., Simpson, D., Slater, M. R., Urh, M. and Wood, K. V. (2009). HaloTag7: a genetically engineered tag that enhances bacterial expression of soluble proteins and improves protein purification. Protein Expr. Purif. 68, 110-120. 10.1016/j.pep.2009.05.010 [DOI] [PubMed] [Google Scholar]
- Okamoto, K., Tokunaga, K., Doi, K., Fujita, T., Suzuki, H., Katoh, T., Watanabe, T., Nishida, N., Mabuchi, A., Takahashi, A.et al. (2011). Common variation in GPC5 is associated with acquired nephrotic syndrome. Nat. Genet. 43, 459-463. 10.1038/ng.792 [DOI] [PubMed] [Google Scholar]
- Okamoto, K., Honda, K., Doi, K., Ishizu, T., Katagiri, D., Wada, T., Tomita, K., Ohtake, T., Kaneko, T., Kobayashi, S.et al. (2015). Glypican-5 increases susceptibility to nephrotic damage in diabetic kidney. Am. J. Pathol. 185, 1889-1898. 10.1016/j.ajpath.2015.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei, J. and Grishin, N. V. (2012). Cysteine-rich domains related to Frizzled receptors and Hedgehog-interacting proteins. Protein Sci. 21, 1172-1184. 10.1002/pro.2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov, K., Wierbowski, B. M. and Salic, A. (2017). Sending and Receiving Hedgehog Signals. Annu. Rev. Cell Dev. Biol. 33, 145-168. 10.1146/annurev-cellbio-100616-060847 [DOI] [PubMed] [Google Scholar]
- Petrov, K., Wierbowski, B. M., Liu, J. and Salic, A. (2020). Distinct cation gradients power cholesterol transport at different key points in the Hedgehog signaling pathway. Dev. Cell 55, 314-327.e17. 10.1016/j.devcel.2020.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov, K., de Almeida Magalhaes, T. and Salic, A. (2021). Mechanism and ultrasensitivity in Hedgehog signaling revealed by Patched1 disease mutations. Proc. Natl Acad. Sci. USA 118, e2006800118. 10.1073/pnas.2006800118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilia, G., Hughes-Benzie, R. M., MacKenzie, A., Baybayan, P., Chen, E. Y., Huber, R., Neri, G., Cao, A., Forabosco, A. and Schlessinger, D. (1996). Mutations in GPC3, a glypican gene, cause the Simpson-Golabi-Behmel overgrowth syndrome. Nat. Genet. 12, 241-247. 10.1038/ng0396-241 [DOI] [PubMed] [Google Scholar]
- Prabhakar, V., Capila, I. and Sasisekharan, R. (2009). The structural elucidation of glycosaminoglycans. Methods Mol. Biol. 534, 147-156. 10.1007/978-1-59745-022-5_11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presto, J., Thuveson, M., Carlsson, P., Busse, M., Wilen, M., Eriksson, I., Kusche-Gullberg, M. and Kjellen, L. (2008). Heparan sulfate biosynthesis enzymes EXT1 and EXT2 affect NDST1 expression and heparan sulfate sulfation. Proc. Natl. Acad. Sci. USA 105, 4751-4756. 10.1073/pnas.0705807105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prydz, K. (2015). Determinants of Glycosaminoglycan (GAG) Structure. Biomolecules 5, 2003-2022. 10.3390/biom5032003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran, F. A., Hsu, P. D., Wright, J., Agarwala, V., Scott, D. A. and Zhang, F. (2013). Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281-2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, I. S. D. and Gleadle, J. M. (2008). Familial nephropathy and multiple exostoses with exostosin-1 (EXT1) gene mutation. J. Am. Soc. Nephrol. 19, 450-453. 10.1681/ASN.2007080842 [DOI] [PubMed] [Google Scholar]
- Rohatgi, R., Milenkovic, L. and Scott, M. P. (2007). Patched1 regulates hedgehog signaling at the primary cilium. Science 317, 372-376. 10.1126/science.1139740 [DOI] [PubMed] [Google Scholar]
- Sarrazin, S., Lamanna, W. C. and Esko, J. D. (2011). Heparan sulfate proteoglycans. Cold Spring Harb. Perspect. Biol. 3, a004952. 10.1101/cshperspect.a004952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen, T. D. and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101-1108. 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- Selleck, S. B. (1999). Overgrowth syndromes and the regulation of signaling complexes by proteoglycans. Am. J. Hum. Genet. 64, 372-377. 10.1086/302266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, W. and Filmus, J. (2009). A patient with the Simpson-Golabi-Behmel syndrome displays a loss-of-function point mutation in GPC3 that inhibits the attachment of this proteoglycan to the cell surface. Am. J. Med. Genet. A 149A, 552-554. 10.1002/ajmg.a.32669 [DOI] [PubMed] [Google Scholar]
- Song, H. H., Shi, W., Xiang, Y.-Y. and Filmus, J. (2005). The loss of glypican-3 induces alterations in Wnt signaling. J. Biol. Chem. 280, 2116-2125. 10.1074/jbc.M410090200 [DOI] [PubMed] [Google Scholar]
- Stipp, C. S., Litwack, E. D. and Lander, A. D. (1994). Cerebroglycan: an integral membrane heparan sulfate proteoglycan that is unique to the developing nervous system and expressed specifically during neuronal differentiation. J. Cell Biol. 124, 149-160. 10.1083/jcb.124.1.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson, G., Awad, W., Håkansson, M., Mani, K. and Logan, D. T. (2012). Crystal structure of N-glycosylated human glypican-1 core protein: structure of two loops evolutionarily conserved in vertebrate glypican-1. J. Biol. Chem. 287, 14040-14051. 10.1074/jbc.M111.322487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei, Y., Ozawa, Y., Sato, M., Watanabe, A. and Tabata, T. (2004). Three Drosophila EXT genes shape morphogen gradients through synthesis of heparan sulfate proteoglycans. Development 131, 73-82. 10.1242/dev.00913 [DOI] [PubMed] [Google Scholar]
- Teng, Y. H.-F., Aquino, R. S. and Park, P. W. (2012). Molecular functions of syndecan-1 in disease. Matrix Biol. 31, 3-16. 10.1016/j.matbio.2011.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topczewski, J., Sepich, D. S., Myers, D. C., Walker, C., Amores, A., Lele, Z., Hammerschmidt, M., Postlethwait, J. and Solnica-Krezel, L. (2001). The zebrafish glypican knypek controls cell polarity during gastrulation movements of convergent extension. Dev. Cell 1, 251-264. 10.1016/S1534-5807(01)00005-3 [DOI] [PubMed] [Google Scholar]
- Townley, R. A. and Bülow, H. E. (2011). Genetic analysis of the heparan modification network in Caenorhabditis elegans. J. Biol. Chem. 286, 16824-16831. 10.1074/jbc.M111.227926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda, M., Kamimura, K., Nakato, H., Archer, M., Staatz, W., Fox, B., Humphrey, M., Olson, S., Futch, T., Kaluza, V.et al. (1999). The cell-surface proteoglycan Dally regulates Wingless signalling in Drosophila. Nature 400, 276-280. 10.1038/22336 [DOI] [PubMed] [Google Scholar]
- Tukachinsky, H., Lopez, L. V. and Salic, A. (2010). A mechanism for vertebrate Hedgehog signaling: recruitment to cilia and dissociation of SuFu-Gli protein complexes. J. Cell Biol. 191, 415-428. 10.1083/jcb.201004108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukachinsky, H., Petrov, K., Watanabe, M. and Salic, A. (2016). Mechanism of inhibition of the tumor suppressor Patched by Sonic Hedgehog. Proc. Natl. Acad. Sci. USA 113, E5866-E5875. 10.1073/pnas.1606719113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull, J. E., Fernig, D. G., Ke, Y., Wilkinson, M. C. and Gallagher, J. T. (1992). Identification of the basic fibroblast growth factor binding sequence in fibroblast heparan sulfate. J. Biol. Chem. 267, 10337-10341. 10.1016/S0021-9258(19)50023-0 [DOI] [PubMed] [Google Scholar]
- Vuillaume, M.-L., Moizard, M.-P., Rossignol, S., Cottereau, E., Vonwill, S., Alessandri, J.-L., Busa, T., Colin, E., Gérard, M., Giuliano, F.et al. (2018). Mutation update for the GPC3 gene involved in Simpson-Golabi-Behmel syndrome and review of the literature. Hum. Mutat. 39, 790-805. 10.1002/humu.23428 [DOI] [PubMed] [Google Scholar]
- Wen, X., Lai, C. K., Evangelista, M., Hongo, J.-A., de Sauvage, F. J. and Scales, S. J. (2010). Kinetics of hedgehog-dependent full-length Gli3 accumulation in primary cilia and subsequent degradation. Mol. Cell. Biol. 30, 1910-1922. 10.1128/MCB.01089-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierbowski, B. M., Petrov, K., Aravena, L., Gu, G., Xu, Y. and Salic, A. (2020). Hedgehog pathway activation requires coreceptor-catalyzed, lipid-dependent relay of the Sonic Hedgehog ligand. Dev. Cell 55, 450-467.e58. 10.1016/j.devcel.2020.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, E. H., Pappano, W. N., Saunders, A. M., Kim, M.-S., Leahy, D. J. and Beachy, P. A. (2010). Dally-like core protein and its mammalian homologues mediate stimulatory and inhibitory effects on Hedgehog signal response. Proc. Natl. Acad. Sci. USA 107, 5869-5874. 10.1073/pnas.1001777107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, N. H. and Stoeckli, E. T. (2013). Sonic hedgehog regulates its own receptor on postcrossing commissural axons in a glypican1-dependent manner. Neuron 79, 478-491. 10.1016/j.neuron.2013.05.025 [DOI] [PubMed] [Google Scholar]
- Witt, R. M., Hecht, M.-L., Pazyra-Murphy, M. F., Cohen, S. M., Noti, C., van Kuppevelt, T. H., Fuller, M., Chan, J. A., Hopwood, J. J., Seeberger, P. H.et al. (2013). Heparan sulfate proteoglycans containing a glypican 5 core and 2-O-sulfo-iduronic acid function as Sonic Hedgehog co-receptors to promote proliferation. J. Biol. Chem. 288, 26275-26288. 10.1074/jbc.M112.438937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, D. and Esko, J. D. (2014). Demystifying heparan sulfate-protein interactions. Annu. Rev. Biochem. 83, 129-157. 10.1146/annurev-biochem-060713-035314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, D. and Lin, X. (2007). Drosophila glypican Dally-like acts in FGF-receiving cells to modulate FGF signaling during tracheal morphogenesis. Dev. Biol. 312, 203-216. 10.1016/j.ydbio.2007.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, S., Lum, L. and Beachy, P. (2006). The ihog cell-surface proteins bind Hedgehog and mediate pathway activation. Cell 125, 343-357. 10.1016/j.cell.2006.02.040 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.