ABSTRACT
The mesenchyme consists of heterogeneous cell populations that support neighboring structures and are integral to intercellular signaling, but are poorly defined morphologically and molecularly. Leveraging single-cell RNA-sequencing, 3D imaging and lineage tracing, we classify the mouse lung mesenchyme into three proximal–distal axes that are associated with the endothelium, epithelium and interstitium, respectively. From proximal to distal: the vascular axis includes vascular smooth muscle cells and pericytes that transition as arterioles and venules ramify into capillaries; the epithelial axis includes airway smooth muscle cells and two populations of myofibroblasts – ductal myofibroblasts, surrounding alveolar ducts and marked by CDH4, HHIP and LGR6, which persist post-alveologenesis, and alveolar myofibroblasts, surrounding alveoli and marked by high expression of PDGFRA, which undergo developmental apoptosis; and the interstitial axis, residing between the epithelial and vascular trees and sharing the marker MEOX2, includes fibroblasts in the bronchovascular bundle and the alveolar interstitium, which are marked by IL33/DNER/PI16 and Wnt2, respectively. Single-cell imaging reveals a distinct morphology of mesenchymal cell populations. This classification provides a conceptual and experimental framework applicable to other organs.
KEY WORDS: Lung development, Mesenchymal cells, Single-cell genomics, 3D imaging, Mouse
Summary: Molecular, anatomical and cellular features allow mouse lung mesenchyme to be classified into three axes (vascular, epithelial and interstitial) and show two populations of myofibroblasts.
INTRODUCTION
Cell types are often named after their characteristic features, as in pyramidal neurons and rod/cone photoreceptors for their shape, basal cells and endothelial cells for their location, and cardiomyocytes and hepatocytes for their tissue origin. In contrast, mesenchymal cells, a heterogeneous group of space-filling fibroblasts present in most tissues, lack distinguishing features and remain poorly defined. Even their major embryonic source, the mesoderm, is simply named for its location between the ectoderm and endoderm. This secondary nature of mesenchymal cells likely reflects the fact that they often do not form a recognizable structure on their own and instead support other cell lineages, such as epithelial and endothelial tubes. This supportive role suggests that mesenchymal cells may be defined relative to their better-understood neighbors, a concept applied in this study.
The typical challenge in defining mesenchymal cells despite their importance is exemplified in the mammalian lung. The lung mesenchyme provides chemical signals and physical constraint necessary for branching and alveolar morphogenesis, forms niches for airway and alveolar stem cells, and goes awry during fibrosis and tumorigenesis (Lambrechts et al., 2018; McCulley et al., 2015; Zepp et al., 2017). However, unlike the epithelial, endothelial and immune lineages in which major cell types have been largely defined (Tabula Muris Consortium et al., 2018; Travaglini et al., 2020), a consensus on the diverse mesenchymal cell types has yet to emerge. Given that the aforementioned cell type-defining features are less forthcoming for mesenchymal cells, the field has relied on components of signaling pathways of known importance in mesenchymal biology, such as the PDGF (Endale et al., 2017; Li et al., 2018; Muhl et al., 2020), FGF (El Agha et al., 2014; Hagan et al., 2020), Shh (Cassandras et al., 2020; Kugler et al., 2017; Li et al., 2015) and Wnt (Lee et al., 2017; Zepp et al., 2017) pathways. Given the dynamic nature of signaling pathways and their deployment in multiple concurrent processes, mesenchymal cell types tagged by candidate signaling molecules might not align with those classically defined by molecular and cellular criteria.
Recent accumulation of single-cell genomic data has allowed unbiased identification of lung mesenchymal cell types (Liu et al., 2021; Riccetti et al., 2020; Tsukui et al., 2020; Xie et al., 2018). However, computationally distinct cell types need to be mapped to the native tissue to integrate the molecular differences with their anatomical and cellular differences. This mapping is challenging in the lung because of its complex, 3D structure in which topologically distal regions may be in the center of a section and the cell body and its ramifying processes may appear discontinuous on a section. These issues can be alleviated by whole-mount immunostaining and 3D imaging, as we have learned from our recent work on the expansive alveolar type 1 cells and identification of Vegfa and NKX2-1 expression in them (Little et al., 2019; Yang et al., 2016). In addition, single-cell genomics now allows cells that have been named after their cellular features, such as lipofibroblasts and matrix fibroblasts (McGowan and Torday, 1997; Xie et al., 2018), to be examined for accuracy based on the corresponding lipid and matrix-related molecules.
In this study, we posit that lung mesenchymal cell types can be mapped relative to the epithelial and endothelial trees they surround and that antibody-based 3D imaging can distinguish intermingled cells and their processes to identify cell type-specific morphology. The resulting molecular, anatomical and cellular features allow us to classify the mouse lung mesenchyme into three axes (vascular, epithelial and interstitial) that are each partitioned proximal-distally with cells of distinct morphology. This classification reveals two populations of myofibroblasts that surround the alveolar ducts or alveoli, respectively, and persist or disappear post-alveologenesis.
RESULTS
Time-course scRNA-seq identifies known and previously unknown mesenchymal cell populations that organize into vascular, epithelial and interstitial axes
Without a consensus surface marker to sort lung mesenchymal cells, we used a negative-gating strategy to separate the other cell lineages, namely epithelial, endothelial and immune lineages that were labeled by CDH1 (also known as E-Cadherin), ICAM2 and CD45 (PTPRC), respectively (Fig. S1A). The resulting four purified cell lineages were remixed in equal proportions for cost-efficiency and sequenced using 10x Genomics. We previously showed that this cell isolation strategy allowed balanced, sufficient sampling of all major cell types in both developing and mature mouse lungs (Cain et al., 2020; Vila Ellis et al., 2020). We collated single-cell RNA-sequencing (scRNA-seq) data of lungs spanning embryonic, neonatal, juvenile and adult stages, and computationally identified the mesenchymal cell clusters as positive for a matrix gene (Col3a1) and negative for other cell lineage markers, including Nkx2-1 (epithelial), Cdh5 (endothelial) and Ptprc (immune) (Fig. S1B).
We used Harmony (Korsunsky et al., 2019) to align comparable cell types across time points without changing the raw gene expression values that we used for subsequent differential analysis. To reduce bias, we performed unsupervised clustering, numerically named the resulting 24 cell clusters and then annotated them based on temporal dynamics and known marker genes (Fig. 1A,B; Table S1). Developmental stage-specific cell clusters were: (1) specific to embryonic day (E) 17, which were thus less-differentiated progenitors and were clustered with their mature counterparts (clusters 19 and 22); (2) proliferative because they were largely limited to before postnatal day (P) 7 and expressed a proliferation marker Mki67 (clusters 11, 14 and 21); or (3) developmentally cleared because they disappeared between P13 and P20 (cluster 6 and the majority of cluster 1).
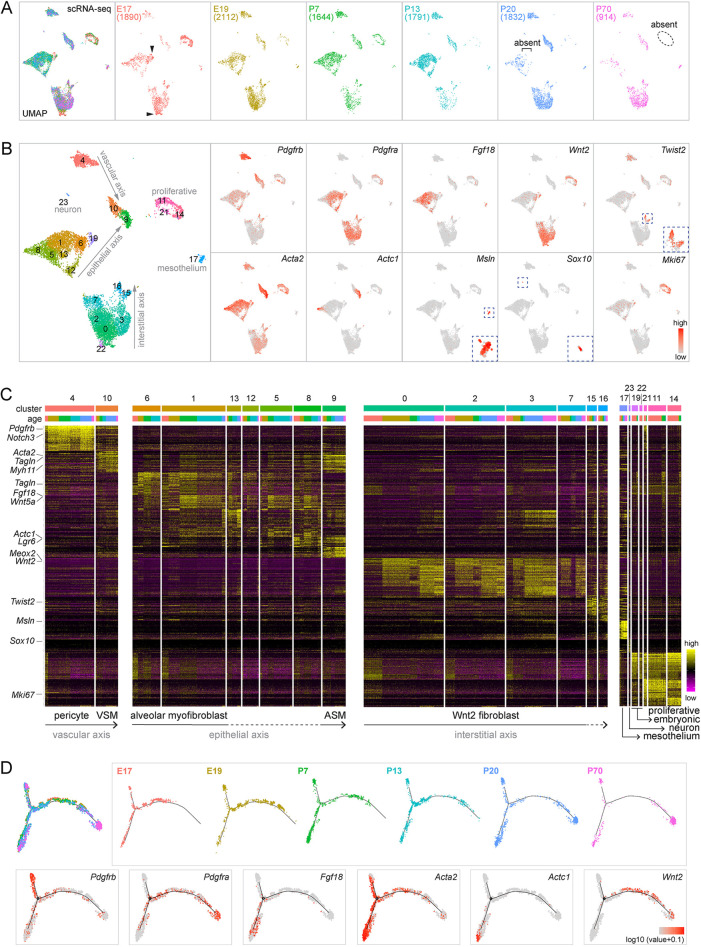
Fig. 1.
Time-course scRNA-seq identifies known and previously unknown lung mesenchymal cell populations that organize into vascular, epithelial and interstitial axes. (A) scRNA-seq UMAPs of lung mesenchymal cells across developmental ages, with cell numbers in parentheses. Embryonic clusters (arrowhead) are absent postnatally. Proliferative cells are absent in the adult lung (P70; dashed oval). The square bracket indicates the absence in mature lungs (P20 and P70) of cells present in younger lungs. (B) Seurat unbiased clustering groups lung mesenchymal cells into 24 clusters. Established markers identify pericytes (Pdgfrb; cluster 4), VSM cells (Acta2/Pdgfrb; cluster 10), ASM cells (Acta2/Actc1; cluster 9), mesothelial cells (Msln; cluster 17), neurons (Sox10; cluster 23) and proliferative cells (Mki67; clusters 11, 14 and 21). Wnt2 and Twist2 mark transcriptionally related populations. Acta2 is high in ASM and VSM cells (clusters 9 and 10, respectively) and in myofibroblasts that are marked by Pdgfra and Fgf18. Embryonic clusters in A are numbered 19 and 22 and express Wnt2. The proposed three axes are labeled. (C) Marker heatmaps with clusters in B grouped by transcriptional similarity and ordered by ages within each cluster. Clusters of unidentified cell types are marked by dashed lines. (D) Monocle coalesces lung mesenchymal cells into three trajectories, the termini of which correspond to ASM cells (Actc1), pericytes (Pdgfrb, Acta2−) and Wnt2 fibroblasts (Wnt2), respectively. Cells from mature lungs (P20 and P70) are largely restricted to the termini.
Known mesenchymal cell types were captured as expected in our dataset, including pericytes marked by Pdgfrb (cluster 4), vascular and airway smooth muscle (VSM and ASM, respectively) cells marked by Acta2 (also known as a-Sma) and distinguishable by Pdgfrb and Actc1 (Ijpma et al., 2020), respectively (clusters 10 and 9), and myofibroblasts marked by Acta2, Pdgfra and Fgf18, albeit with additional heterogeneity to be addressed later in this study (clusters 1, 5, 6, 8, 12 and 13) (Fig. 1C). The remaining major clusters formed a large group (clusters 0, 2, 3 and 7) and a small group (clusters 15 and 16) marked by Wnt2 and Twist2 (also known as Dermo1), respectively. These two groups were recently named Col13a1 and Col14a1 matrix fibroblasts, respectively, after additional markers (Xie et al., 2018); we noted that most mesenchymal cells, as well as endothelial and epithelial cells, produced matrices (Fig. S1C). Via the process of elimination and based on their abundance, the Wnt2- and Twist2-expressing cells should include lipofibroblasts, which were considered to have unique lipid metabolism (McGowan and Torday, 1997); however, the commonly used marker Plin2 (also known as Adrp) was nonspecific (Fig. S1C). In addition, we identified the less-abundant mesothelial cells (cluster 17; marked by Msln) and even rare neurons (cluster 23; marked by Sox10).
As reasoned in the Introduction, mesenchymal cells could be conceptually classified based on their neighboring structures, most notably the vascular and epithelial trees. It was self-evident to assign vascular smooth muscle cells and pericytes to the vascular tree, which we named the vascular axis because the two mesenchymal cell types were situated along a proximal–distal axis. Applying the same concept, we assigned to the epithelial axis the proximal ASM cells (cluster 10) and distal alveolar myofibroblasts (cluster 6 and the majority of cluster 1), both of which constrain and shape the epithelium (Kim and Vu, 2006), and predicted that the other transcriptionally related clusters (5, 8, 12 and 13) were associated with the epithelium in-between, namely the alveolar ducts, for which we provided evidence later in this study. Finally, recognizing that the Twist2-expressing cells were shown in a mouse phenotyping database (Koscielny et al., 2014) to localize between the epithelial and endothelial trees within the proximal bronchovascular bundles and that the ratio of Twist2-expressing and Wnt2-expressing cells was what one would expect for proximal and distal compartments, we predicted that they belonged to a third axis, which we named the interstitial axis to refer to the space between the epithelial and endothelial trees. As a result, the vascular, epithelial and interstitial axes, each arranged proximal distally, accounted for all the major cell clusters with their corresponding markers (Fig. 1B,C). The proliferative clusters (11, 14 and 21) included cells from each of the three axes because cell cycle genes dominated over cell type markers. This dominance of a single biological characteristic in cell clustering also contributed to the proximity of the VSM and ASM clusters.
Supporting this three-axis classification, Monocle trajectory analysis coerced the associated mesenchymal cells into three paths that terminated in Pdgfrb+ pericytes, Actc1+ ASM cells and Wnt2+ fibroblasts, corresponding to the vascular, epithelial and interstitial axes, respectively (Fig. 1D). Reciprocally, cells from the three Monocle trajectories largely mapped to the proposed three axes on the Uniform Manifold Approximation and Projection (UMAP) (Fig. S2). Although the root of Monocle trajectories was arbitrary (Trapnell et al., 2014), cells from the mature lungs (P20 and P70) were largely limited to the three termini, whereas cells from developing lungs were more centrally located, suggesting gradual specialization along the three axes. We then focused on each axis individually to define constituent cell populations, map their proximal–distal distributions in 3D, and categorize the cell morphology.
Within the vascular axis, proximal vascular smooth muscle cells transition to distal pericytes, which mature postnatally
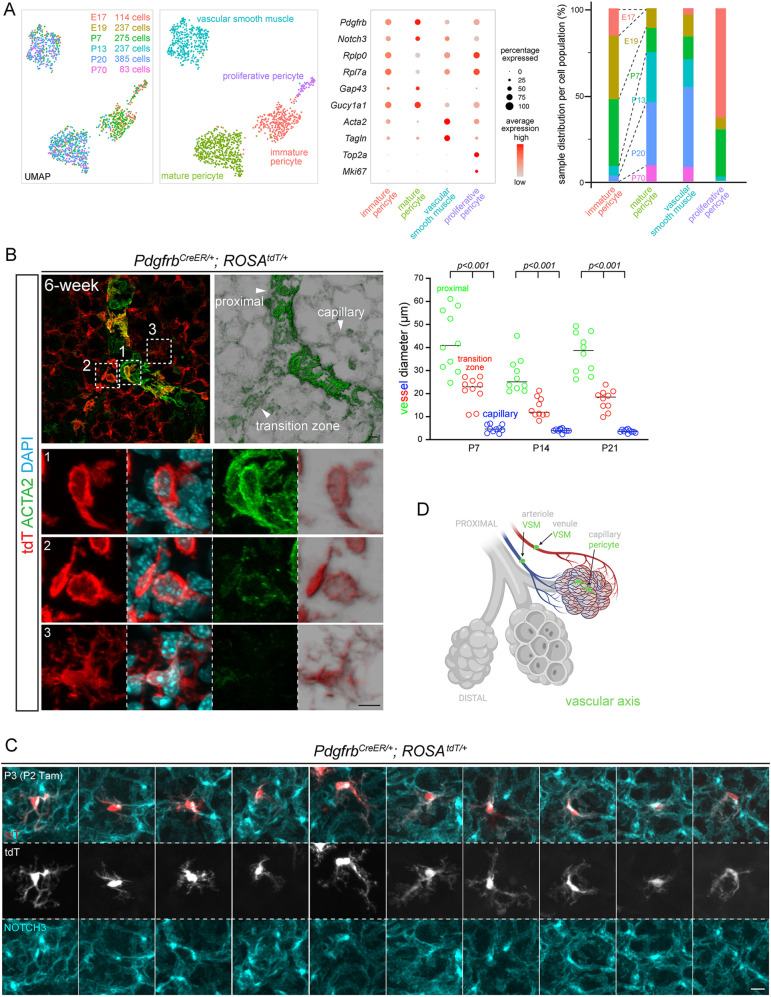
Reclustering of cells of the vascular axis (Fig. 1C) identified one VSM population, marked by contractile genes Acta2 and Tagln (also known as Sm22) and three populations of pericytes: proliferative, immature and mature (Fig. 2A; Table S2). Whereas all cells of the vascular axis expressed Pdgfrb and Notch3, immature pericytes mainly included cells from E17, E19 and P7 lungs and expressed a higher level of ribosomal genes (Rplp0 and Rpl7a), possibly to support cell growth; by contrast, mature pericytes mostly had cells from P13, P20 and P70 and expressed significantly more Gap43 and Gucy1a1 (Fig. 2A; Table S2), suggesting that pericytes, unlike VSM cells, mature after birth.
Fig. 2.
Within the vascular axis, proximal VSM cells transition to distal pericytes, which mature postnatally. (A) scRNA-seq UMAPs of lung mesenchymal cells of the vascular axis, colored for developmental ages (left) or cell types (right), as supported by relevant markers in the dot plot. Immature pericytes are mostly from embryonic (E17 and E19) and neonatal (P7) lungs, as are proliferative pericytes (bar graph). (B) Left: transition zone between VSM cells and pericytes genetically labeled by PdgfrbCreER with 3 mg Tamoxifen administered 48 h before lung harvest. Transition zone cells (box 2) have low ACTA2 but the morphology of VSM cells (box 1) rather than of pericytes (box 3). Imaris normal shading view is shown for ACTA2 and tdT images. Images are representative of at least three biological replicates (same for all subsequent images). Right: decreasing diameters of proximal, transition zone and capillary vessels, as exemplified in the image on the left (ordinary ANOVA with Tukey test; significant for all pair-wise comparisons; all data points and medians are shown). (C) Single-cell morphology of ten pericytes from sparse genetic labeling. (D) Schematic of VSM cells and pericytes of the vascular axis. Created with BioRender.com. Scale bars: 10 µm. Tam, 250 µg Tamoxifen.
To visualize the vascular axis, we used a PdgfrbCreER driver (Cuervo et al., 2017) and whole-mount immunostaining to identify the transition zone from VSM cells to pericytes based on the gradual decrease in vessel diameter, with the transition zone being intermediate between proximal macrovessels and distal capillaries, as well as on ACTA2 staining (Fig. 2B). The ROSAtdT reporter readily defined the nucleus and cell morphology and showed that cells with both high and low ACTA2 expression had nucleus-length, strained cellular projections often at an oblique angle to the vessels, different from the circumferential wrapping of airways by ASM cells (Fig. 2B). In contrast, ACTA2-negative pericytes had long astrocyte-like extensions that were often polarized toward one side of the nucleus and decorated with NOTCH3, a pericyte marker that was also concentrated in a perinuclear vesicular compartment, as was PDGFRB (Huang et al., 2007), a subcellular distribution to be taken into account when enumerating pericytes (Figs S2C, S3A). Therefore, the vascular axis consists of proximal VSM cells and distal pericytes sharing PDGFRB/NOTCH3, albeit with contrasting morphologies (Fig. 2D).
The epithelial axis includes airway smooth muscle cells as well as two populations of myofibroblasts, marked by CDH4/HHIP/LGR6, and high PDGFRA, respectively
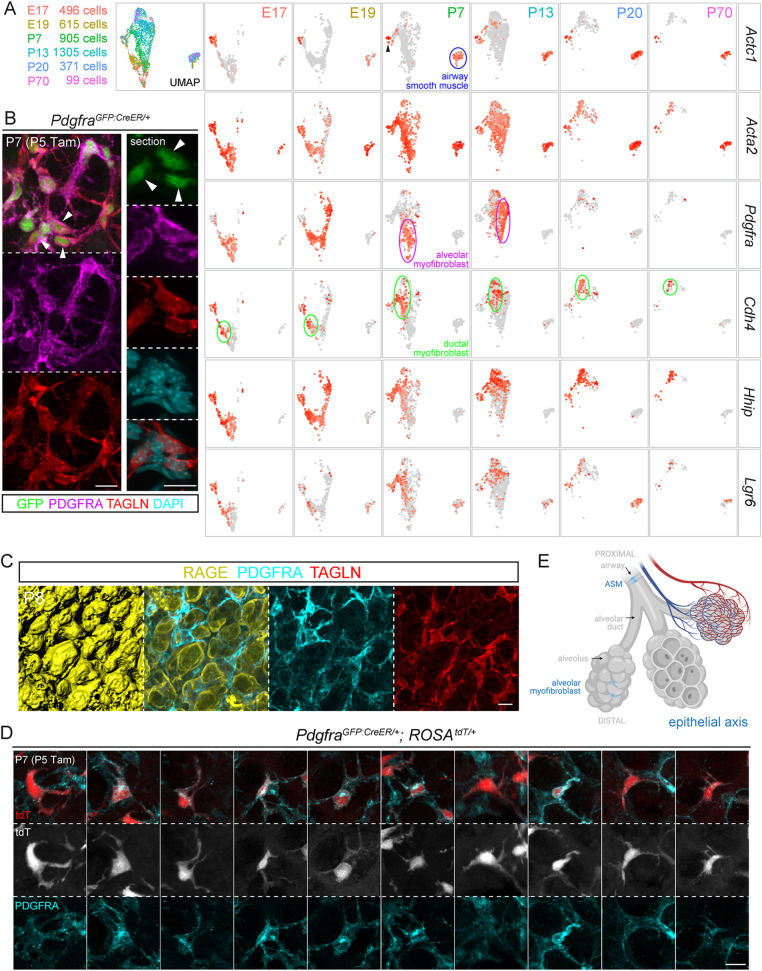
Next, we subsetted and reclustered cells of the epithelial axis, and readily identified an Actc1+ ASM cluster that was largely unchanged on the UMAPs over time, having the highest levels of contractile genes (Acta2/Tagln/Myh11), possibly reflecting their higher mechanical load for airway constriction, as well as an Acta2/Tagln/Myh11+ myofibroblast cluster that shifted on the UMAPs, reflecting changes in gene expression and/or cell composition (Fig. 3A; Fig. S3F, Table S3). Most notably, at P7 and P13, the known PDGFRA-high alveolar myofibroblasts (Endale et al., 2017) only constituted part of the cluster and were adjacent to a second Cdh4/Hhip/Lgr6+ population (Fig. 3A). Intriguingly, Cdh4/Hhip/Lgr6+ cells remained in the same UMAP location at P20 and P70, when the PDGFRA-high alveolar myofibroblasts disappeared (Fig. 3A). These two populations of myofibroblasts (albeit at different UMAP locations, possibly as immature precursors) were also distinguishable at E17 and E19 based on Cdh4 and Lgr6 expression, although Pdgfra and Hhip expression was nondiscriminatory embryonically and only became restricted to their respective myofibroblast populations postnatally (Fig. 3A). Below, we detail the two myofibroblast populations individually.
Fig. 3.
The epithelial axis includes ASM cells, ductal myofibroblasts and alveolar myofibroblasts, the last of which surround alveoli and disappear after classical alveologenesis. (A) scRNA-seq UMAP and feature plots of lung mesenchymal cells of the epithelial axis, colored for developmental ages. Labeled for the P7 lung, ASM cells form a distinct cluster (outlined in blue), whereas the remaining cluster is also Acta2+ and contains a Pdgfra+ population (also present at P13 but absent at P20 and P70) and a Cdh4/Hhip/Lgr6+ population (present at all time points), corresponding to alveolar (outlined in purple) and ductal (outlined in green) myofibroblasts, respectively. A small Actc1+ group (arrowhead) is adjacent to the myofibroblast cluster, possibly reflecting a transition from the proximal ASM cells to the distal myofibroblasts. (B) PDGFRA-high cells (arrowhead), marked by bright GFP from PdgfraGFP:CreER and perinuclear PDGFRA, express the contractile protein TAGLN. GFP-dim cells are not visible in this imaging setting. (C) Immunostaining images and surface rendering to show PDGFRA-high, TAGLN+ alveolar myofibroblasts situated within grooves of alveolar type 1 cells (stained by RAGE). (D) Single-cell morphology of ten alveolar myofibroblasts from sparse genetic labeling. (E) Schematic of ASM cells and alveolar myofibroblasts of the epithelial axis. Created with BioRender.com. Scale bars: 10 µm. Tam, 300 µg Tamoxifen.
PDGFRA-high myofibroblasts surround alveoli and disappear after classical alveologenesis
Similar to PDGFRB and NOTCH3 (Fig. S3A), PDGFRA staining was diffuse throughout the cell and also concentrated in a perinuclear compartment in the neonatal lung, possibly indicating receptor activation and endocytosis (Pennock et al., 2016) (Fig. 3B); this perinuclear PDGFRA was absent in the mature lung (Fig. S3B), suggesting at least two types of PDGFRA-expressing cell. Consistent with this possibility, a histone-GFP knock-in allele of Pdgfra identified GFP-bright and GFP-dim populations (Endale et al., 2017), which we hypothesized to correspond to those with and without perinuclear PDGFRA staining. Given that the level of histone-GFP would be confounded by the rate of cell division, we resorted to a GFP:CreER fusion knock-in allele of Pdgfra (Miwa and Era, 2015) and found that tamoxifen-induced nuclear GFP:CreER was high in cells with perinuclear PDGFRA, which accumulated TAGLN and, thus, were myofibroblasts (Fig. 3B). Unlike TAGLN, the commonly used marker ACTA2 aligned with PDGFRA cell extensions but did not reliably outline the nucleus for cell counting (Fig. S3C).
These perinuclear-PDGFRA contractile cells occupied grooves over the folding alveolar surface during classical alveologenesis (P3-P14) (Vila Ellis and Chen, 2020) and, thus, corresponded to alveolar myofibroblasts (Fig. 3C). Single-cell imaging using the ROSAtdT reporter showed frequently a strained tri-projection morphology and that each cellular projection was nucleus length, such that multiple cells were expected to assemble a network to constrain alveolar outpocketing (Fig. 3D,E).
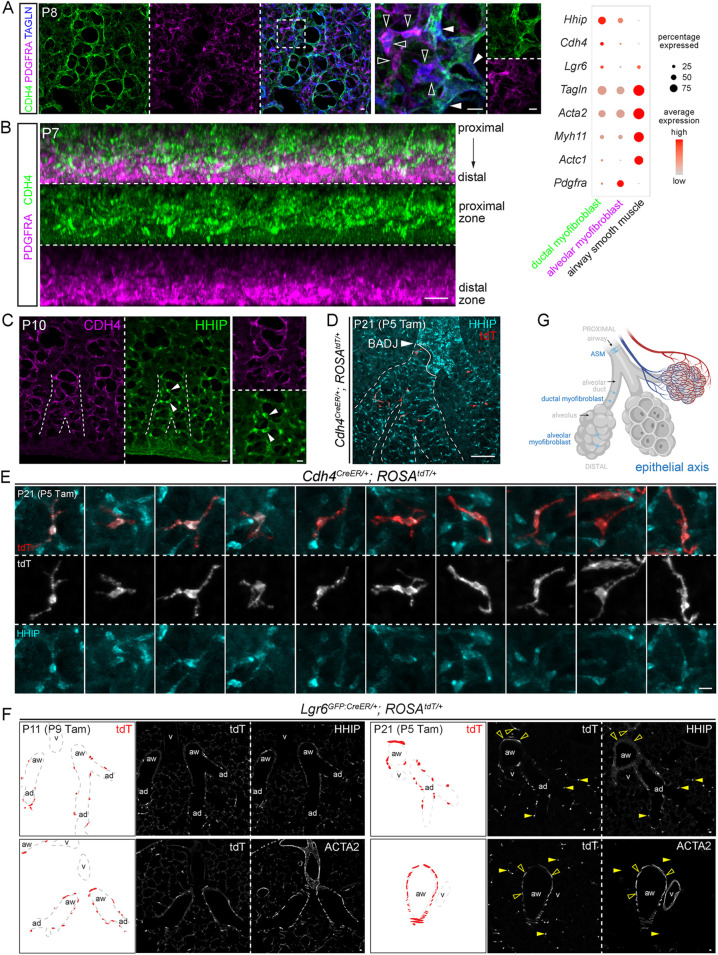
CDH4/HHIP/LGR6 myofibroblasts surround alveolar ducts and persist in the adult lung
As predicted by scRNA-seq (Fig. 3A), CDH4+ myofibroblasts and PDGFRA-high myofibroblasts coexisted in neonatal lungs and constituted the TAGLN+ contractile cells (Fig. 4A). Whole-mount immunostaining revealed in a z-plane view a striking zonation of distal (i.e. surface) PDGFRA and proximal CDH4 (Fig. 4B). The proximal CDH4 zone corresponded to the alveolar ducts, which are tubular extensions of the airways but bordered by alveolar epithelial cells (Fig. S3D). Arising from branch stalks, as the airways do, alveolar ducts had wider airspace than the surrounding alveoli and could be best identified as they extended toward the lateral edge, instead of the lobe surface, in which tissue geometry made tubes less recognizable because they were shorter and interrupted by branching (Vila Ellis and Chen, 2020; Yang and Chen, 2014). We also found an HHIP antibody that marked cellular projections that largely aligned with CDH4 and also perinuclear regions for cell enumeration (Fig. 4C). As predicted from the z-plane view (Fig. 4B), CDH4/HHIP cells specifically wrapped alveolar ducts, reminiscent of ASM cells wrapping the airways, albeit with larger gaps resulting from interruption by alveolar outgrowth, as in an extended accordion (Fig. 4C). This continuation of CDH4/HHIP cells with ASM cells was supported and better visualized by the labeling of both cell populations by CrhCre (Taniguchi et al., 2011), which we found in a screen for new mesenchymal cell drivers (Fig. S4). Notably, CrhCre did not label other mesenchymal cells, such as the VSM cells, supporting our model of three discrete mesenchymal axes.
Fig. 4.
CDH4/HHIP/LGR6 myofibroblasts surround alveolar ducts and persist in the adult lung. (A) TAGLN+ contractile cells include PDGFRA+ (open arrowheads) and CDH4+ (filled arrowheads) populations, as supported by the dot plot, which includes additional markers. CDH4+ cells form ductal structures. (B) Z-plane view of immunostaining images showing that CDH4 and PDGFRA occupy the proximal and distal zones, corresponding to alveolar ducts and alveoli, respectively. (C) HHIP and CDH4 mark the perinuclear region and projections, respectively of cells around alveolar ducts. Arising from branch stalks that form in all directions and lengths, alveolar ducts (dashed lines) are most recognizable when extending toward the lateral edge. (D) Cdh4CreER lineage-labeled cells during the neonatal stage persist in the mature lung and are restricted to HHIP+ ductal myofibroblasts, which surround alveolar ducts (dashed lines) extending beyond the bronchoalveolar duct junction (BADJ, arrowhead). HHIP also labels proximal interstitial cells around the airways. (E) Single-cell morphology of ten ductal myofibroblasts from sparse genetic labeling. (F) Immunostaining images and diagrams to show that Lgr6GFP:CreER labels ASM cells (ACTA2+; open arrowhead) and alveolar duct myofibroblasts (HHIP+; filled arrowhead), but not VSM cells (ACTA2+). Labeled cells in neonatal lungs persist in the mature lung. (G) Schematic of ASM cells, ductal myofibroblasts and alveolar myofibroblasts of the epithelial axis. Created with BioRender.com. Scale bars: 10 µm. ad, alveolar duct; aw, ASM cells; Tam, 500 µg Tamoxifen; v, vessel.
We additionally characterized a Cdh4CreER driver that was generated to label retinal neurons (Rousso et al., 2016) and, despite its inefficiency (∼4%; 734 HHIP+ cells), found it to be specific to CDH4/HHIP+ cells (Fig. S5). Cdh4CreER lineage-labeled cells during the neonatal stage remained in the mature lung and specific to HHIP-marked alveolar ducts (Fig. 4D,E), consistent with persistence of these cells in our scRNA-seq data (Fig. 3A). The resulting sparse cell labeling showed strained cellular projections that were similar to those of PDGFRA-high myofibroblasts, but longer and often bidirectional, as expected for cells constraining a ductal structure wider than a spherical alveolus (Fig. 4E; note the different scale bar from that in Fig. 3D). Similarly, an Lgr6GFP:CreER driver (Snippert et al., 2010), as predicted by scRNA-seq (Fig. 3A), labeled ACTA2+ contractile cells around airways and alveolar ducts (the latter of which were marked by HHIP) but not around vessels; neonatal lineage-labeled cells also persisted in the mature lung (Fig. 4F). This labeling pattern was consistent with a prior report (Lee et al., 2017). Taken together, our data supported two populations of myofibroblasts (CDH4/HHIP/Lgr6 ductal myofibroblasts and PDGFRA-high alveolar myofibroblasts) that are spatially separated, persist or disappear after classical alveologenesis, respectively, and, together with ASM cells, form the proximal–distal epithelial axis, constraining the associated airway, alveolar duct and alveolar epithelium (Fig. 4G). As observed in the gradual transition zone between VSM cells and pericytes, the epithelial axis also has intermediate cells, as evidenced by Actc1 expression in the myofibroblast clusters (Fig. 3A) as well as PDGFRA and CDH4/HHIP double-positive cells (Fig. S3E).
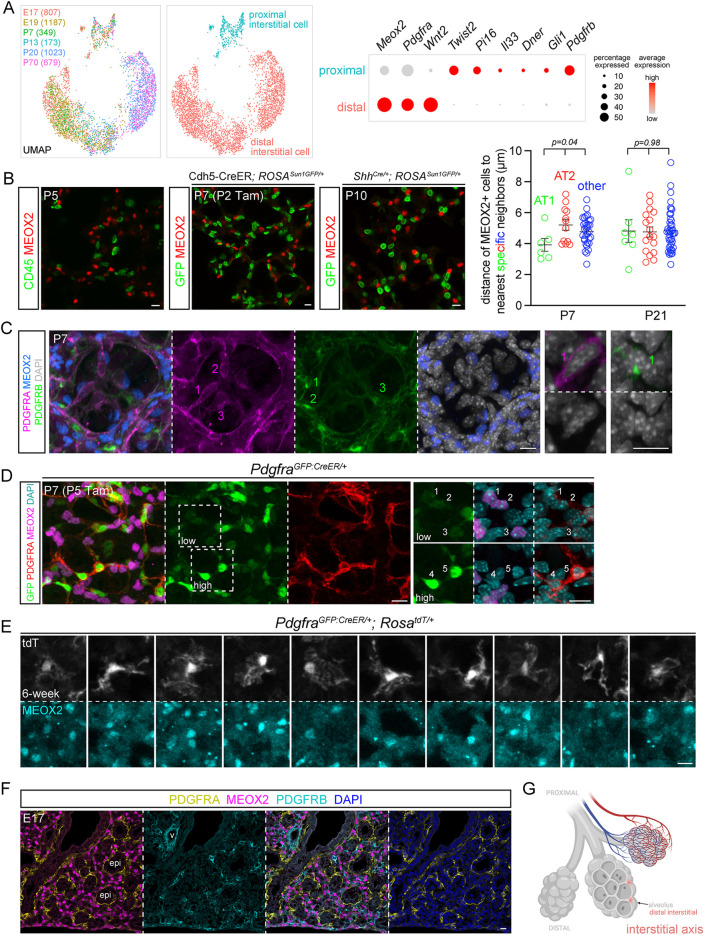
The interstitial axis, visualized by MEOX2, includes distal Wnt2+ PDGFRA-low cells and proximal IL33/DNER/PI16+ cells
Similarly, cells of the interstitial axis were reclustered into the aforementioned proximal Twist2+ (no reliable antibody identified) and distal Wnt2+ populations, the latter of which showed additional heterogeneity between developing and mature lungs, similar to that seen between immature and mature pericytes (Fig. 5A and Fig. 2A; Table S4). Supporting the hypothesis that the interstitial axis could be as discrete an entity as the vascular and epithelial axes, all interstitial mesenchymal cells specifically expressed Meox2, a nuclear protein that would also allow unequivocal cell identification and counting (Fig. S6A). We validated a MEOX2 antibody based on its expected absence in immune, epithelial and endothelial cells, as well as PDGFRA-high alveolar myofibroblasts and PDGFRB pericytes (Fig. 5B,C). Instead, MEOX2 was detected in PDGFRA-low cells in the alveolar region, identifiable by the lack of perinuclear PDGFRA staining and dim GFP from the PdgfraGFP:CreER allele (Fig. 5D), consistent from prior studies using a histone-GFP reporter of Pdgfra (Endale et al., 2017). MEOX2+ cells were labeled by PdgfraGFP:CreER with a high dose of the inducer Tamoxifen, and had long wavy multidirectional protrusions occupying the interstitial space (Fig. 5E). MEOX2+ cells were in the vicinity of alveolar type 2 (AT2) cells, but not significantly closer than alveolar type 1 (AT1) cells or nonepithelial cell nuclei were (Fig. 5B; Fig. S6B). Although a subset of AT2 cells might be particularly juxtaposed with MEOX2+ cells, neither scRNA-seq nor single-cell analysis of accessible chromatin sequencing (scATAC-seq) identified a distinct AT2 cell population in the mouse lung (Little et al., 2021). Oil Red staining, commonly used to detect lipofibroblasts that were expected to correspond to MEOX2+ cells, as discussed above, was not specific to a particular cell type in neonatal or mature lungs (Fig. S6C). Assignment of distal MEOX2+ cells to the interstitial axis was further supported by their discrete localization from PDGFRA+ and PDGFRB+ cells in the embryonic lung before the distal interstitial space became unrecognizable because of postnatal expansion of the alveolar airspace (Fig. 5F,G).
Fig. 5.
The interstitial axis is marked by MEOX2 and includes distal Wnt2-expressing PDGFRA-low cells. (A) scRNA-seq UMAPs of lung mesenchymal cells of the interstitial axis (cell number in parentheses), colored by developmental ages or cell types, as supported by relevant markers shown in the dot plot. Meox2 is lower in a subset of proximal interstitial cells. Distal interstitial cells form subclusters of immature (E17, E19, P7 and P13) and mature (P20 and P70) cells. (B) Left: MEOX2 is not expressed by immune cells (CD45), genetically labeled endothelial (Cdh5-CreER) or epithelial (ShhCre) cells. Genetic labeling is necessary to circumvent co-staining of antibodies from the same species. Right: nearest neighbor analysis of internuclear distance showing that MEOX2+ cells occur at a comparable distance to AT1, AT2 or other nonepithelial cells (ordinary ANOVA with Tukey test; data are mean±s.d.). See Fig. S6B for representative images. (C) MEOX2 is not expressed by PDGFRA-high alveolar myofibroblasts or PDGFRB+ pericytes. Higher magnification images show perinuclear distribution of PDGFRA and PDGFRB. (D) PDGFRA-low cells (1-3) have dim GFP and express MEOX2, whereas PDGFRA-high cells (4,5) have bright GFP and perinuclear PDGFRA. (E) Single-cell morphology of ten distal interstitial cells from sparse genetic labeling with 3 mg Tamoxifen 2 days before lung harvest. (F) Immunostaining of an embryonic lung shows that MEOX2+ distal interstitial cells are distinct from PDGFRA+ and PDGFRB+ cells surrounding the epithelium and vessels, respectively. (G) Schematic of distal interstitial cells. Created with BioRender.com. Scale bars: 10 µm. epi, epithelium; Tam, 250 µg (B) and 300 µg (C) Tamoxifen; v, vessels.
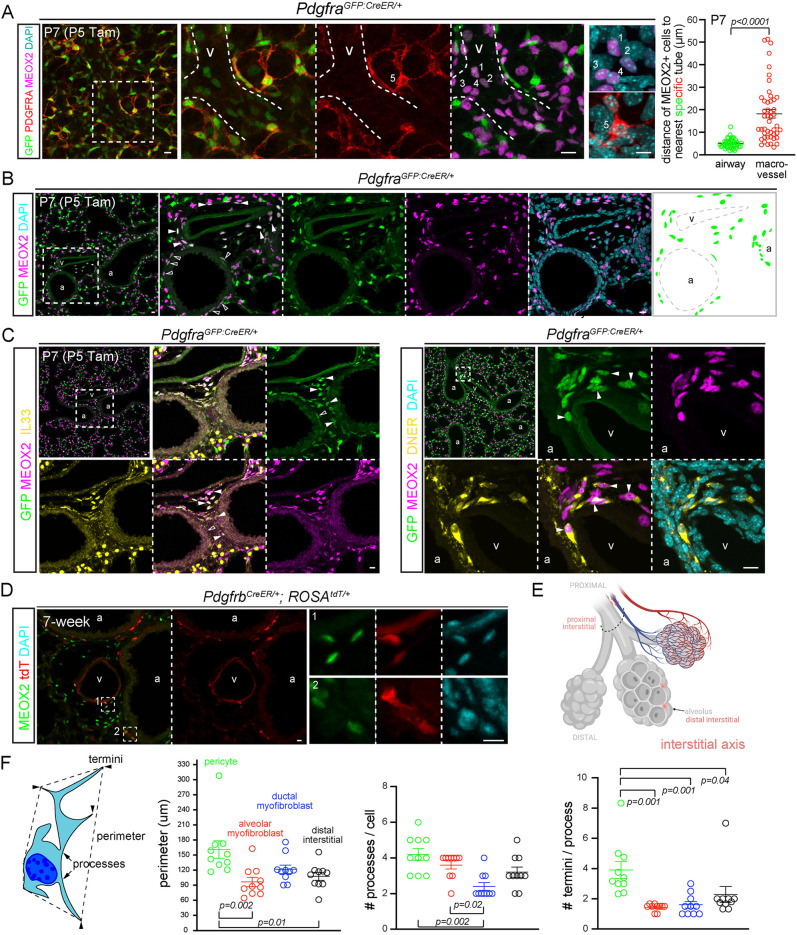
MEOX2 cells were also present more proximally, expressing low PDGFRA (identifiable by dim GFP from PdgfraGFP:CreER) within bronchovascular bundles (Fig. 6A,B). To further characterize these proximal interstitial cells, we resorted to immunostaining for additional markers identified from scRNA-seq, including Il33, Dner and Pi16, because RNA probes would be less reliable for low-abundance genes and their speckly staining uninformative for discerning cell morphology. Consistent with prior studies (Dahlgren et al., 2019; Tsukui et al., 2020), IL33 was nuclear and expressed distally only by AT2 cells and proximally only by MEOX2 cells (Fig. 6C; Fig. S1C and Fig. S7A). Both DNER and PI16 marked the spindly perinuclear portion that tapered into thin processes (Fig. 6C; Fig. S7B). This cell morphology was best visualized using the PdgfrbCreER driver, which scRNA-seq predicted to be active in both pericytes and proximal interstitial cells (Fig. 2A and Fig. 5A; Fig. S6A). Intriguingly, these elongated proximal interstitial cells were wedged between, and basal to, ASM cells (Fig. 6D; Fig. S7B), within 10 μm of the airway basement membrane, whereas those closer to macrovessels were further away in the adventitia (Fig. 6A). Notwithstanding occasional PDGFRA-low cells without MEOX2, resulting from possible additional heterogeneity among the proximal interstitial cells (Fig. 6C), these data supported a distinct interstitial axis marked by MEOX2 and sitting between the vascular and epithelial trees (Fig. 6E).
Fig. 6.
Proximal interstitial cells are in bronchovascular bundles and express MEOX2/IL33/DNER. (A) Left: dim GFP, MEOX2+ cells (1-4) around a vessel (curved dashed lines) and bright GFP, PDGFRA-high cells (5). Dashed box in the first image indicates area shown at higher magnification in the following images in all instances. Tamoxifen (Tam) facilitates nuclear accumulation and, hence, detection of the GFP:CreER fusion protein. Right: nearest neighbor analysis of perpendicular distance of MEOX2+ cells to the basement membrane of the respective tubes. The large spread of the vessel category is from MEOX2+ cells in vascular adventitia (unpaired Student's t-test; data are mean±s.d.). See Fig. 6B,C for representative images. (B) Immunostaining images and diagram showing GFP+, MEOX2+ cells around airways (open arrowhead) and vessels (filled arrowhead) within the bronchovascular bundle. (C) GFP+, MEOX2+ cells (filled arrowhead) are IL33+ (left) and DNER+ (right) within bronchovascular bundles. Occasional GFP+ cells are MEOX2− (open arrowhead). (D) Besides VSM cells, PdgfrbCreER labels MEOX2+ cells within bronchovascular bundles. Tamoxifen 3 mg was administrated 2 days before lung harvest. (E) Proximal interstitial cells within the bronchovascular bundle (dashed semioval). Created with BioRender.com. (F) Schematic and quantification of cell morphology of color-coded cell types in Fig. 2C (P3 pericytes), Fig. 3D (P7 alveolar myofibroblasts), Fig. 4E (P21 ductal myofibroblasts) and Fig. 5E (6-week distal interstitial cells) (ordinary ANOVA with Tukey test; data are mean±s.d.). Each symbol represents a cell; the number of termini per process is averaged over all processes of a given cell. Scale bars: 10 µm. a, airway; Tam, 300 µg Tamoxifen; v, vessel.
Comparison of cell morphology distinguished various mesenchymal cell types: pericytes were most complex with a larger perimeter, more processes and termini; compared with ductal myofibroblasts, alveolar myofibroblasts were smaller but with more processes, consistent with the geometry of alveolar ducts versus alveoli that they surrounded (Fig. 6F).
Complementary lineage tracing supports apoptotic clearance of PDGFRA-high alveolar myofibroblasts
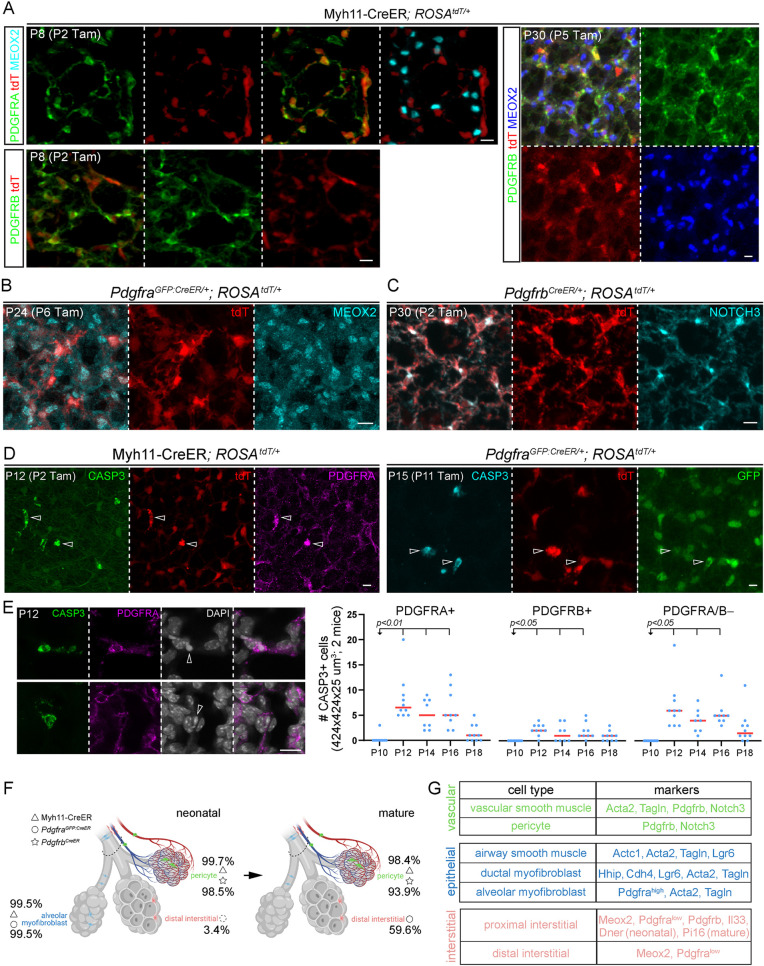
Although developmental maturation was evident for pericytes and interstitial cells within the distal compartment of the three axes (Fig. 2A and Fig. 5A), the most striking change was the disappearance of PDGFRA-high alveolar myofibroblasts but persistence of CDH4/HHIP/LGR6 ductal myofibroblasts, as shown by Lgr6GFP:CreER and Cdh4CreER lineage tracing (Fig. 4E,F). Although myofibroblast clearance was recently reported (Hagan et al., 2020), we sought to clarify the distinct fates of the two myofibroblast populations, as well as genetic driver specificity, using our recently identified molecular markers, including MEOX2.
A contractile-gene driver Myh11-CreER (Wirth et al., 2008) marked alveolar myofibroblasts (99.5% efficiency from 1799 cells in three mice; Table S5), ductal myofibroblasts (98.2% efficiency from 425 cells in three mice), and, unexpectedly, pericytes (99.7% efficiency from 1530 cells in three mice), perhaps reflecting their relatedness to VSM cells, but did not mark MEOX2 interstitial cells in neonatal lungs (Fig. 7A; Fig. S8A). Tracing these labeled neonatal cells to the mature stage when alveolar myofibroblasts disappeared showed labeling in ductal myofibroblasts (98.4% efficiency from 290 cells in three mice) and pericytes (98.4% efficiency from 1501 cells in three mice), but still not in MEOX2 cells, indicating that alveolar myofibroblasts do not become interstitial cells (Fig. 7A; Fig. S8A). To rule out the possibility that alveolar myofibroblasts became pericytes, we lineage traced PdgfraGFP:CreER cells and found that, in the neonatal lung, PdgfraGFP:CreER labeled mostly alveolar myofibroblasts (99.5% efficiency from 660 cells in three mice) and also MEOX2 interstitial cells (initially 3.4% efficiency from 873 cells in three mice, which increased over the tracing time to 59.6% efficiency from 1453 cells in three mice), none of which became pericytes in the mature lung (Fig. 7B; Fig. S8B). Consistent with pericytes being a distinct lineage, PdgfrbCreER-labeled cells remained pericytes in the distal compartment of both neonatal (98.5% efficiency from 416 cells in three mice) and mature (93.9% efficiency from 1037 cells in three mice) lungs (Fig. 7C). Notably, both Myh11-CreER- and PdgfraGFP:CreER-labeled cells, which overlapped for alveolar myofibroblasts, were found to express cleaved Caspase 3 (CASP3), consistent with apoptosis that began after P12, as also supported by chromatin condensation (Fig. 7D,E). We also noted that fewer PDGFRB+ pericytes as well as non-PDGFRA/B cells were positive for cleaved CASP3, implying additional cell trimming in neonatal lungs (Fig. 7E). These data were consistent with the notion that PDGFRA-high alveolar myofibroblasts undergo developmental apoptosis, whereas CDH4/HHIP/LGR6 ductal myofibroblasts persist (Fig. 7F).
Fig. 7.
Complementary lineage tracing supports apoptotic clearance of PDGFRA-high alveolar myofibroblasts. (A) Myh11-CreER labels alveolar myofibroblasts (PDGFRA-high) and pericytes (PDGFRB+), but not distal interstitial cells (MEOX2+) in neonatal (P8) lungs. Pericytes remain labeled after tracing and distal interstitial cells remain unlabeled in mature (P30) lungs. (B) PdgfraGFP:CreER lineage-traced cells are MEOX2+. (C) PdgfrbCreER lineage-traced cells are pericytes (NOTCH3+). (D) Myh11-CreER and PdgfraGFP:CreER-labeled cells are positive for cleaved CASP3 (open arrowhead). (E) Immunostaining images (left) exemplifying PDGFRA+ CASP3+ cells with condensed nuclei (DAPI; open arrowhead), and quantifications (right; ordinary ANOVA with Dunnett test for P10; all data points and medians are shown). Some PDGFRA/B− cells might have downregulated cell type markers during apoptosis. (F) Cells labeled by the three drivers (symbols) and their efficiency (percentages) in neonatal and mature lungs, consistent with developmental apoptosis of alveolar myofibroblasts. Dashed circle indicates inefficient labeling of distal interstitial cells compared with alveolar myofibroblasts by PdgfraGFP:CreER in neonatal lungs. Created with BioRender.com. (G) Summary of the cell types of the three axes and their markers. Scale bars: 10 µm. Tam, 250 µg (C,D) or 300 µg (A,B) Tamoxifen.
DISCUSSION
In this study, we introduced a three-axis classification system for lung mesenchymal cells that integrates single-cell transcriptomic, spatiotemporal, morphological and lineage information (Fig. 7G). Most intuitively, the vascular axis consists of proximal VSM cells and distal pericytes. The epithelial axis includes, from proximal to distal, ASM cells, hitherto unrecognized ductal myofibroblasts, and alveolar myofibroblasts, the latter of which undergo developmental apoptosis. The interstitial axis shares a marker, MEOX2, fills the space between the vascular and epithelial trees, such as that within the bronchovascular bundles, and includes proximal IL33+ cells and distal Wnt2+ cells, the latter of which have been called lipofibroblasts. This classification provides a framework to define heterogeneous lung mesenchymal cells in vivo and in cultured organoids, predicts their functions and signaling interactions with nearby cell lineages and can be extended to other organs. Future studies are needed to probe whether markers such as MEOX2 and CDH4 regulate the transcriptional program and cell sorting of specific mesenchymal cell populations.
Instead of the Cartesian system of XYZ coordinates, our three-axis classification has its root in the cylindrical coordinate system, with an axial height for the proximal–distal location, a radial distance for the layered wrapping around a vascular or epithelial tube, and an azimuth for the cylindrical symmetry. This axial system is a natural way to characterize biology because tubes are fundamental building blocks and radial symmetry dates back to the primitive animal phylum of cnidarians. Locating mesenchyme cells with axial coordinates capitalizes on our better understanding of the vascular and epithelial trees, accounts for their likely supportive roles in each biological tree and is consistent with their distinct developmental origins, as evidenced by the radial versus distal recruitment of VSM and ASM cells, respectively (Greif et al., 2012; Kumar et al., 2014), as well as the possible connection of the epithelial axis and progenitors or the interstitial axis to previously named subepithelial and submesothelial compartments, respectively (White et al., 2006). The resulting indirect demarcation of the proximal and distal compartments of the mesenchyme, possibly mediated by diffusible or mechanical signals, is expected to be more blurred than that for the endothelium or epithelium, leading to intermediate cells, as observed for low-ACTA2 VSM cells, Actc1-expressing myofibroblasts, and PDGFRA/HHIP double-positive cells (Fig. 2B and Fig. 3A; Fig. S3E). Such imprecision might also reflect the intrinsic plasticity or mobility of mesenchymal cells to accommodate changes during tissue homeostasis and injury repair. This tube-centered axial system is conceptually applicable to the mesenchyme in other organs, ranging from smooth muscle cells and pericytes surrounding the omnipresent vascular network, peristaltic muscles along the digestive tract and the hierarchical insulation and organization of axons within nerve bundles by myelination, endoneurium and perineurium.
A notable prediction of the axial system that we have validated experimentally is the presence of ductal myofibroblasts associated with alveolar ducts, an epithelial structure connecting proximal airways with distal alveoli (Fig. 4). Similar to alveolar myofibroblasts, ductal myofibroblasts express contractile genes, have strained cellular projections and cluster nearby on the UMAPs (Figs 3 and 4). Unlike alveolar myofibroblasts, which undergo developmental apoptosis, ductal myofibroblasts persist in the mature lung, perhaps to corroborate or maintain the elastin cable scaffold (Wagner et al., 2015) or to serve as a main source of Wnt5a for nearby Wnt-responsive cells (Nabhan et al., 2018; Zacharias et al., 2018). Intriguingly, genetic polymorphisms in HHIP, a marker of ductal myofibroblasts (Figs 3A and 4), have been linked to chronic obstructive pulmonary disease (COPD) (Zhou et al., 2012). The distinct developmental fates of ductal and alveolar myofibroblasts could explain the remaining Fgf18-lineage cells, which are expected to include both types of myofibroblast (Fig. 1B) (Hagan et al., 2020), and also highlight the heterogeneity within secondary crest myofibroblasts, which include cells around embryonic branch stalks and, thus, future alveolar ducts (Li et al., 2015; Zepp et al., 2021). Future studies are needed to understand the differential regulation, fate and function of the two myofibroblast populations.
As whole-genome sequencing has reversed the classical flow from protein biochemistry to gene discovery, single-cell genomics has cataloged transcriptionally and epigenetically distinct cell states in search of spatial, morphological and functional features that traditionally define a cell type. Compared with in situ hybridization, immunostaining validates the protein products of transcriptional markers from single-cell genomics and reveals cell morphology in 3D tissues, but could be challenging to interpret as a result of the distinct subcellular localization of protein markers, especially for intertwined lung cells. Notably, colocalization of image pixels does not always result from co-expression within the same cell, as exemplified by under-the-diffraction-limit juxtaposition and, thus, apparent colocalization of alveolar type 1 cell membrane markers, such as AQP5 or RAGE (AGER), with endothelial cell membrane markers, such as ICAM2 or EMCN. By contrast, co-expressed markers often do not colocalize, as exemplified by the nuclear localized NKX2-1 and junctional CDH1, which are expectedly present in every lung epithelial cell. Given the complex morphology of lung mesenchymal cells (Fig. 2C, Fig. 3D, Fig. 4E, Fig. 5E and Fig. 6F), it is paramount to pinpoint the nucleus to localize and enumerate the cell populations identified in single-cell genomics. Accordingly, we identified a previously unknown marker, MEOX2, for interstitial cells, complemented ACTA2 with TALGN and CDH4 with HHIP and characterized the perinuclear accumulation of PDGFRA and PDGFRB, with the added benefit of using perinuclear PDGFRA to distinguish PDGFRA-high versus -low cells.
This battery of molecular tools shed light on lineage-tracing experiments. For example, PdgfraGFP:CreER labels alveolar myofibroblasts and, less efficiently because of lower Pdgfra expression, interstitial MEOX2 cells (Figs 5, 6 and 7), whereas PdgfrbCreER labels pericytes and proximal interstitial cells (Figs 2 and 6). The latter are also referred to as adventitial fibroblasts (Tsukui et al., 2020), which might give rise to pathological myofibroblasts (Rock et al., 2011; Tsukui et al., 2020) and might also be Gli1CreER-lineage cells promoting airway epithelium repair (Cassandras et al., 2020; Moiseenko et al., 2020). Although the reported Axin2+ myofibrogenic progenitors (AMPs) appear to correspond to pericytes and the mesenchymal alveolar niche cells (MANCs) to proximal MEOX2 cells based on their distributions on the UMAPs, future work is needed to clarify the additional heterogeneity as a result of Wnt signaling, as reflected by the apparent stochastic expression of Axin2 (Fig. S1C), as well as enriched expression of Wnt co-receptors Lgr5 and Lgr6 in cells of the epithelial axis (Figs 3 and 4). These molecular analyses, integrated with cell type-specific morphology [analogous to Golgi staining of neuronal cell morphology (Vints et al., 2019)] and functional studies, will lead to a multifaceted definition of lung mesenchymal cell types.
MATERIALS AND METHODS
Mus musculus
The mouse protocols used for this research complied with the regulations of MD Anderson Cancer Center's Institutional Animal Care and Use Committee. Wild-type C57BL/J mice were used for validation of gene-based molecular markers with immunostaining. Both male and female mice were used. The mouse strains for lineage labeling and tracing experiments were: CrhCre (Taniguchi et al., 2011), PdgfraGFP:CreER (Miwa and Era, 2015), Cdh4CreER (Rousso et al., 2016), Myh11-CreER (Wirth et al., 2008), PdgfrbCreER (Cuervo et al., 2017), Lgr6GFP:CreER (Snippert et al., 2010), ShhCre (Harfe et al., 2004), Cdh5-CreER (Sorensen et al., 2009), ROSASun1GFP (Mo et al., 2015), and ROSAtdT (Madisen et al., 2010). Intraperitoneal injections of Tamoxifen (T5648; Sigma) dissolved in corn oil (C8267; Sigma) were administered to induce Cre recombination; specific dosages are detailed in the figure legends. P0-P12 was considered the neonatal stage.
Antibodies
The following antibodies were used for immunofluorescence: goat anti-PDGFRA (1:1000, AF1062, R&D Systems), rat anti-PDGFRA (1:1000, 14-1401-82, eBioscience), rat anti-cadherin-4 (CDH4, 1:20, MRCD5, Developmental Studies Hybridoma Bank), rat anti-PDGFRB (1:1000, 14-1402-82, eBioscience), goat anti-PDGFRB (1:1000, AF1042, R&D Systems), goat anti-NOTCH3 (1:500, AF1308, R&D Systems), rabbit anti-MEOX2 (1:1000, NBP2-30647, Novus Biological), rabbit anti-cleaved CASP3 (1:500, 9661, Cell Signaling), goat anti-PI16 (1:1000, AF4929, R&D Systems), rat anti-CD45 (1:2000, 14-0451-81, eBioscience), Alexa488-conjugated mouse anti-ACTA2 (1:1000, sc-32251 AF488, Santa Cruz Biotechnology), Cy3-conjugated mouse anti-ACTA2 (1:1000, C6198, Sigma), Alexa647-conjugated mouse anti-ACTA2 (1:1000, sc-32251 AF647, Santa Cruz Biotechnology), rat anti-RAGE (1:1000, MAB1179, R&D Systems), rabbit anti-TAGLN (1:1000, ab14106, Abcam), chicken anti-GFP (1:5000, ab13970, Abcam), goat anti-HHIP (1:1000, AF1568, R&D Systems), guinea pig anti-LAMP3 (1:500, 391005, Synaptic Systems), mouse anti-NKX2-1 (1:500, TTF-1-L-CE, Leica Biosystems), goat anti-IL33 (1:500, AF3626, R&D Systems), rabbit anti-CNN1 (1:1000, ab46794, Abcam), rabbit anti-AQP5 (1:2500, ab78486, Abcam) and goat anti-DNER (1:1000, AF2264, R&D Systems). Alexa 488-, Cy3- or Alexa 647-conjugated donkey anti-rabbit, anti-guinea pig, anti-rat, anti-goat, anti-chicken and anti-mouse secondary antibodies were used (all from Jackson ImmunoResearch; 711-545-152, 711-165-152, 711-605-152, 706-545-148, 706-165-148, 706-605-148, 712-545-153, 712-165-153, 712-605-153, 705-545-147, 705-165-147, 705-605-147, 703-545-155, 703-165-155, 703-605-155, 715-545-151, 715-165-151 and 715-605-151; 1:1000).
Cell dissociation and FACS
The experiment protocol for cell dissociation and fluorescence-activating cell sorting (FACS) was performed as described (Vila Ellis et al., 2020). After perfusion through the right heart ventricle with PBS, the whole lung was removed, separated into lobes in PBS and collected in ice-cold Eppendorf tubes with RPMI (11875093, Thermo Fisher Scientific) or Lebovitz's Media (21083027, Gibco). Lung lobes were minced with forceps and digested with 2 mg/ml Collagenase Type I (CLS-1, LS004197, Worthington), 2 mg/ml Elastase (ESL, LS002294, Worthington) and 0.5 mg/ml DNase I (D, LS002007, Worthington) for 30 min at 37°C using a dry block incubator (Accublock, D1100, Labnet). The tissue was resuspended after 15 min of incubation and enzymatic digestion was stopped at 30 min by adding fetal bovine serum (FBS; 10082-139, Invitrogen) to a 20% concentration.
Samples were placed on ice and the following steps were performed in the cold room. The samples were resuspended until homogeneous, filtered with a 70 µm Falcon cell strainer (352350, Falcon) and centrifuged in 2 ml tubes for 1 min at 2300 g. After supernatant removal, 1 ml red blood cell lysis buffer (15 mM NH4Cl, 12 mM NaHCO3, 0.1 mM EDTA, pH 8.0) was added gently to the sample, followed by a wait time of 3 min on ice and centrifugation. After performing this step twice, cells were washed and resuspended with 1 ml of ice-cold Lebovitz's Media supplemented with 10% FBS. Cells were filtered into a 5 ml glass tube with a cell strainer cap (352235, Falcon) and transferred to a 2 ml Eppendorf tube for immunostaining for 30 min on ice for immune cells (CD45-PE/Cy7, 1:250, 103114, BioLegend), endothelial cells (ICAM2-A647, 1:250, A15452, Life Technologies) and epithelial cells (ECAD-488, 1:250, 53-3249-80, eBioscience). Cells were centrifuged, washed with Lebovitz/10%FBS and filtered into a 5 ml glass tube with a cell strainer cap. After incubating cells with SYTOX Blue (S34857, Invitrogen), cells were analyzed using the BD FACSAria Fusion Cell Sorter to evaluate cell viability and gating strategy for epithelial, endothelial, immune and mesenchymal cells. CD45-negative cells were selected, from which ICAM2-negative cells were then selected and, from those, ECAD negative cells were finally selected, resulting in the triple-negative mesenchymal cell population. An equal proportion of immune, endothelial, epithelial and mesenchymal lineage cells were collected into a single tube and concentrated through centrifugation at 4°C for 5 min at 2300 g. Supernatant that contained ambient RNA was removed, resuspended in 250 µl Lebovitz/10% FBS and used for 10x Genomics library preparation.
scRNA-seq
Sequencing and analysis were performed as described (Vila Ellis et al., 2020). Sorted samples were prepared for sequencing using the Chromium Single Cell Gene Expression Solution Platform (10x Genomics) along with the Chromium Single Cell 3′ Library and Gel Bead Kit (V2, revD; 10x Genomics). Samples were sequenced with an Illumina NextSeq500 using a 26X124 sequencing run format with 8 bp index (Read1). Sequencing output files underwent pipeline analysis with ‘cellranger count’ and ‘cellranger aggregate’, which resulted in files for easy analysis with Loupe Cell Browser (10x Genomics) and Seurat 3.1, an R-package for quality control, normalization and data exploration (https://satijalab.org/seurat/). Quality control metrics were analyzed per sample. Cells were filtered using the following criteria: no fewer than 200 genes, no more than 6000 transcripts and a mitochondrial percentage up to 15% to preserve embryonic cells enriched in mitochondria. Additional normalization using the R package, Harmony, reduced batch effects across samples (https://portals.broadinstitute.org/harmony/index.html). Clusters were analyzed with the FindAllMarkers function to identify differentially expressed genes and markers for known and previously unknown lung mesenchymal populations. Clusters positive for Col3a1 but negative for Nkx2-1, Cdh5 and Ptprc were subsetted as mesenchymal cells. Trajectory analysis was performed with Monocle 2.8 using the top 1500 genes (Qiu et al., 2017). The DoMultiBarHeatmap function is from (GitHub http://github.com/elliefewings/DoMultiBarHeatmap). R script is available in Supplementary File 1. Raw data were deposited in the Gene Expression Omnibus under accession number GSE180822.
Lung harvest
Lungs were processed as described in our previous publications (Little et al., 2019; Vila Ellis et al., 2020; Yang et al., 2016). Briefly, mice were anesthetized with Avertin (2,2,2-tribromoethanol; T48402, Sigma). After opening the abdominal cavity, the aorta was cut for exsanguination, followed by opening of the rib cage and perfusion of the right ventricle of the heart with PBS. The trachea was separated from posterior structures using blunt dissection and incised for canula insertion. Thread and a surgical knot were used to stabilize the canula to allow gravity drip lung inflation at 25 cm H2O pressure with 0.5% paraformaldehyde (PFA; P6148, Sigma) in PBS for 5 min. Lungs were collected in 0.5% PFA/PBS and incubated for 3 h at room temperature on a rocker. After fixation, samples were washed again in PBS overnight at 4C for further processing.
Section immunostaining
The immunostaining protocol was performed as described in our previous publications (Little et al., 2019; Vila Ellis et al., 2020; Yang et al., 2016). Fixed lungs were separated into lobes. The middle and caudal lobes were cryoprotected in 20% sucrose in PBS containing 10% optimal cutting temperature (OCT) embedding compound (4583, Tissue-Tek) overnight at 4°C for rapid dry-ice freezing and −80°C storage. Cryosections of 20 µm were cut and dried for 1 h at room temperature. After hydrophobic rectangles were drawn on the sections with a PAP pen, the samples were placed in a humid chamber and washed with PBS three times for 5 min each. Tissues were blocked with diluted 5% donkey serum (017-000-121, Jackson ImmunoResearch) in PBS with 0.3% Triton X-100 (PBST) for 1 h at room temperature, followed with primary antibody incubation overnight at 4°C. Next, lung tissue sections were washed in a Coplin jar with PBS for 30 min at room temperature and PBS excess was removed with a vacuum. Tissues were incubated with secondary antibody mixture for 1-2 h, followed with a 30 min PBS wash in a Coplin jar at room temperature. Sections were mounted with Aquamount (18606, Polysciences) and covered with 24×50 mm coverslip (No. 1, VWR). Samples were imaged using Nikon A1 Plus and Olympus FV1000 confocal microscopes.
For Oil Red staining, a stock solution of 0.25 g Oil Red O Sigma (O0625-25G) was dissolved in 50 ml of isopropanol. Lung tissue sections of 20 µm thick were immunostained as above without normal donkey serum and post-fixed with 2% PFA for 1 h. A working Oil Red solution was freshly prepared by diluting the stock with distilled deionized water (ddH2O) in a 6:4 ratio, which was left to stand for 10 min before being filtered first with a 70 µm Falcon cell strainer and then with a 0.22 µm pore-size filter membrane to remove Oil Red precipitate. Immunostained lung sections were washed with PBS three times for 5 min each and incubated in working Oil Red solution for 30 min, followed by three 30-s washes in ddH2O and then washing under running tap water for 10 min.
Whole-mount immunostaining
The outer edges of the cranial and left lung lobes were cut into strips and blocked with donkey serum for 1 h. The strips were incubated with primary antibody mixture at 4°C in a rocker overnight, followed by three 1-h washes with PBS with 1% Triton X-100 and 1% Tween-20 (PBSTT). Lung strips were incubated with a secondary antibody mixture overnight at 4°C, followed by PBSTT washes, as previously done for primary antibody incubation. Lung strips were then fixed using 2% PFA for 2-4 h and washed with PBS three times for 5 min each. Samples were placed on plain microscope slides (8201, Premier) with Aquamount and covered with 22×22 mm coverslips (No 1, 48366 067, VWR). Immunostained whole lung lobes were imaged by using an optical projection tomography scanner (Bioptonics, 3001 M).
Confocal imaging and processing
Samples were imaged with Nikon A1 Plus and Olympus FV1000 confocal microscopes using four-color imaging (DAPI, GFP, TXR and Cy5) and 1-µm-step-size 20- to 50-µm stacks. Images were analyzed using Imaris 7.7.2 (Bitplane, http://www.bitplane.com/imaris), ImageJ (US National Institutes of Health, https://imagej.nih.gov/ij/) software. Nearest neighbor analyses of MEOX2+ cells measured the distance between the center of the MEOX2+ nucleus to the center of NKX2-1± nuclei, which were categorized as AT1 and AT2 cells based on LAMP3 staining, or the perpendicular distance to the basement membrane to airway and macrovascular tubes. Cell morphology was quantified as shown in Fig. 6F. Cell perimeter was measured with cells projected to show their largest profile. Cell processes were defined as projections originating from the perinuclear region. Cell termini were defined as >1 µm protrusions from a cell process. For CASP3+ quantification, ten random areas from two mice at various developmental stages were imaged without knowledge of CASP3 staining. Biological replicates and mouse number are specified within the figures.
Data analysis
All statistical analyses were carried out using Prism software as detailed in figure legends. Analyses of images and genomic data are described in their respective sections.
Supplementary Material
Acknowledgements
We thank Dr Margo P. Cain and Belinda J. Hernandez in the Chen lab for generating scRNA-seq data for embryonic and P70 lungs. We thank Dalia Hassan, Celine S. L. Kong and Andrés Gutiérrez for assistance with quantification of genetic drivers. The University of Texas MD Anderson Cancer Center DNA Analysis Facility and Flow Cytometry and Cellular Imaging Core Facility are supported by the Cancer Center Support Grant (CA #16672).
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: O.N.P., J.C.; Investigation: O.N.P., M.J.G.G., J.C.; Writing - original draft: O.N.P., J.C.; Writing - review & editing: O.N.P., M.J.G.G., J.C.; Supervision: J.C.; Project administration: J.C.; Funding acquisition: O.N.P., J.C.
Funding
This work was supported by the University of Texas MD Anderson Cancer Center Start-up and Retention Fund, and the National Institutes of Health (R01HL130129 and R01HL153511 to J.C.; F31HL149232 to O.N.P.). Deposited in PMC for release after 12 months.
Data availability
Raw data have been deposited in GEO under accession number GSE180822.
Peer review history
The peer review history is available online at https://journals.biologists.com/dev/article-lookup/doi/10.1242/dev.200081.
References
- Cain, M. P., Hernandez, B. J. and Chen, J. (2020). Quantitative single-cell interactomes in normal and virus-infected mouse lungs. Dis. Model. Mech. 13, dmm044404. 10.1242/dmm.044404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassandras, M., Wang, C., Kathiriya, J., Tsukui, T., Matatia, P., Matthay, M., Wolters, P., Molofsky, A., Sheppard, D., Chapman, H.et al. (2020). Gli1(+) mesenchymal stromal cells form a pathological niche to promote airway progenitor metaplasia in the fibrotic lung. Nat. Cell Biol. 22, 1295-1306. 10.1038/s41556-020-00591-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo, H., Pereira, B., Nadeem, T., Lin, M., Lee, F., Kitajewski, J. and Lin, C. S. (2017). PDGFRbeta-P2A-CreER(T2) mice: a genetic tool to target pericytes in angiogenesis. Angiogenesis 20, 655-662. 10.1007/s10456-017-9570-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlgren, M. W., Jones, S. W., Cautivo, K. M., Dubinin, A., Ortiz-Carpena, J. F., Farhat, S., Yu, K. S., Lee, K., Wang, C., Molofsky, A. V.et al. (2019). Adventitial stromal cells define Group 2 innate lymphoid cell tissue niches. Immunity 50, 707-722. 10.1016/j.immuni.2019.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Agha, E., Herold, S., Al Alam, D., Quantius, J., MacKenzie, B., Carraro, G., Moiseenko, A., Chao, C. M., Minoo, P., Seeger, W.et al. (2014). Fgf10-positive cells represent a progenitor cell population during lung development and postnatally. Development 141, 296-306. 10.1242/dev.099747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endale, M., Ahlfeld, S., Bao, E., Chen, X., Green, J., Bess, Z., Weirauch, M. T., Xu, Y. and Perl, A. K. (2017). Temporal, spatial, and phenotypical changes of PDGFRalpha expressing fibroblasts during late lung development. Dev. Biol. 425, 161-175. 10.1016/j.ydbio.2017.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greif, D. M., Kumar, M., Lighthouse, J. K., Hum, J., An, A., Ding, L., Red-Horse, K., Espinoza, F. H., Olson, L., Offermanns, S.et al. (2012). Radial construction of an arterial wall. Dev. Cell 23, 482-493. 10.1016/j.devcel.2012.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan, A. S., Zhang, B. and Ornitz, D. M. (2020). Identification of a FGF18-expressing alveolar myofibroblast that is developmentally cleared during alveologenesis. Development 147, dev181032. 10.1242/dev.181032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe, B. D., Scherz, P. J., Nissim, S., Tian, H., McMahon, A. P. and Tabin, C. J. (2004). Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell 118, 517-528. 10.1016/j.cell.2004.07.024 [DOI] [PubMed] [Google Scholar]
- Huang, M., Duhadaway, J. B., Prendergast, G. C. and Laury-Kleintop, L. D. (2007). RhoB regulates PDGFR-beta trafficking and signaling in vascular smooth muscle cells. Arterioscler. Thromb. Vasc. Biol. 27, 2597-2605. 10.1161/ATVBAHA.107.154211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ijpma, G., Kachmar, L., Panariti, A., Matusovsky, O. S., Torgerson, D., Benedetti, A. and Lauzon, A. M. (2020). Intrapulmonary airway smooth muscle is hyperreactive with a distinct proteome in asthma. Eur. Respir. J. 56, 1902178. 10.1183/13993003.02178-2019 [DOI] [PubMed] [Google Scholar]
- Kim, N. and Vu, T. H. (2006). Parabronchial smooth muscle cells and alveolar myofibroblasts in lung development. Birth Defects Res. C Embryo Today 78, 80-89. 10.1002/bdrc.20062 [DOI] [PubMed] [Google Scholar]
- Korsunsky, I., Millard, N., Fan, J., Slowikowski, K., Zhang, F., Wei, K., Baglaenko, Y., Brenner, M., Loh, P. R. and Raychaudhuri, S. (2019). Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289-1296. 10.1038/s41592-019-0619-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koscielny, G., Yaikhom, G., Iyer, V., Meehan, T. F., Morgan, H., Atienza-Herrero, J., Blake, A., Chen, C. K., Easty, R., Di Fenza, A.et al. (2014). The International Mouse Phenotyping Consortium Web Portal, a unified point of access for knockout mice and related phenotyping data. Nucleic Acids Res. 42, D802-D809. 10.1093/nar/gkt977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugler, M. C., Loomis, C. A., Zhao, Z., Cushman, J. C., Liu, L. and Munger, J. S. (2017). Sonic hedgehog signaling regulates myofibroblast function during alveolar septum formation in murine postnatal lung. Am. J. Respir. Cell Mol. Biol. 57, 280-293. 10.1165/rcmb.2016-0268OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, M. E., Bogard, P. E., Espinoza, F. H., Menke, D. B., Kingsley, D. M. and Krasnow, M. A. (2014). Mesenchymal cells. Defining a mesenchymal progenitor niche at single-cell resolution. Science 346, 1258810. 10.1126/science.1258810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrechts, D., Wauters, E., Boeckx, B., Aibar, S., Nittner, D., Burton, O., Bassez, A., Decaluwe, H., Pircher, A., Van den Eynde, K.et al. (2018). Phenotype molding of stromal cells in the lung tumor microenvironment. Nat. Med. 24, 1277-1289. 10.1038/s41591-018-0096-5 [DOI] [PubMed] [Google Scholar]
- Lee, J. H., Tammela, T., Hofree, M., Choi, J., Marjanovic, N. D., Han, S., Canner, D., Wu, K., Paschini, M., Bhang, D. H.et al. (2017). Anatomically and functionally distinct lung mesenchymal populations marked by Lgr5 and Lgr6. Cell 170, 1149-1163. 10.1016/j.cell.2017.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C., Li, M., Li, S., Xing, Y., Yang, C. Y., Li, A., Borok, Z., De Langhe, S. and Minoo, P. (2015). Progenitors of secondary crest myofibroblasts are developmentally committed in early lung mesoderm. Stem Cells 33, 999-1012. 10.1002/stem.1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, R., Bernau, K., Sandbo, N., Gu, J., Preissl, S. and Sun, X. (2018). Pdgfra marks a cellular lineage with distinct contributions to myofibroblasts in lung maturation and injury response. eLife 7, e36865. 10.7554/eLife.36865.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little, D. R., Gerner-Mauro, K. N., Flodby, P., Crandall, E. D., Borok, Z., Akiyama, H., Kimura, S., Ostrin, E. J. and Chen, J. (2019). Transcriptional control of lung alveolar type 1 cell development and maintenance by NK homeo box 2-1. Proc. Natl. Acad. Sci. USA 116, 20545-20555. 10.1073/pnas.1906663116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little, D. R., Lynch, A. M., Yan, Y., Akiyama, H., Kimura, S. and Chen, J. (2021). Differential chromatin binding of the lung lineage transcription factor NKX2-1 resolves opposing murine alveolar cell fates in vivo. Nat. Commun. 12, 2509. 10.1038/s41467-021-22817-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X., Rowan, S. C., Liang, J., Yao, C., Huang, G., Deng, N., Xie, T., Wu, D., Wang, Y., Burman, A.et al. (2021). Categorization of lung mesenchymal cells in development and fibrosis. iScience 24, 102551. 10.1016/j.isci.2021.102551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen, L., Zwingman, T. A., Sunkin, S. M., Oh, S. W., Zariwala, H. A., Gu, H., Ng, L. L., Palmiter, R. D., Hawrylycz, M. J., Jones, A. R.et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133-140. doi:10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulley, D., Wienhold, M. and Sun, X. (2015). The pulmonary mesenchyme directs lung development. Curr. Opin. Genet. Dev. 32, 98-105. 10.1016/j.gde.2015.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan, S. E. and Torday, J. S. (1997). The pulmonary lipofibroblast (lipid interstitial cell) and its contributions to alveolar development. Annu. Rev. Physiol. 59, 43-62. 10.1146/annurev.physiol.59.1.43 [DOI] [PubMed] [Google Scholar]
- Miwa, H. and Era, T. (2015). Generation and characterization of PDGFRalpha-GFPCreERT2 knock-In mouse line. Genesis 53, 329-336. 10.1002/dvg.22853 [DOI] [PubMed] [Google Scholar]
- Mo, A., Mukamel, E. A., Davis, F. P., Luo, C., Henry, G. L., Picard, S., Urich, M. A., Nery, J. R., Sejnowski, T. J., Lister, R.et al. (2015). Epigenomic signatures of neuronal diversity in the mammalian brain. Neuron 86, 1369-1384. 10.1016/j.neuron.2015.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moiseenko, A., Vazquez-Armendariz, A. I., Kheirollahi, V., Chu, X., Tata, A., Rivetti, S., Gunther, S., Lebrigand, K., Herold, S., Braun, T.et al. (2020). Identification of a repair-supportive mesenchymal cell population during airway epithelial regeneration. Cell Rep 33, 108549. 10.1016/j.celrep.2020.108549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhl, L., Genove, G., Leptidis, S., Liu, J., He, L., Mocci, G., Sun, Y., Gustafsson, S., Buyandelger, B., Chivukula, I. V.et al. (2020). Single-cell analysis uncovers fibroblast heterogeneity and criteria for fibroblast and mural cell identification and discrimination. Nat. Commun. 11, 3953. 10.1038/s41467-020-17740-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabhan, A. N., Brownfield, D. G., Harbury, P. B., Krasnow, M. A. and Desai, T. J. (2018). Single-cell Wnt signaling niches maintain stemness of alveolar type 2 cells. Science 359, 1118-1123. 10.1126/science.aam6603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennock, S., Kim, L. A. and Kazlauskas, A. (2016). Vascular endothelial cell growth factor a acts via platelet-derived growth factor receptor alpha to promote viability of cells enduring hypoxia. Mol. Cell. Biol. 36, 2314-2327. 10.1128/MCB.01019-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, X., Mao, Q., Tang, Y., Wang, L., Chawla, R., Pliner, H. A. and Trapnell, C. (2017). Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979-982. https://doi.org/10.1038/nmeth.4402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccetti, M., Gokey, J. J., Aronow, B. and Perl, A. T. (2020). The elephant in the lung: Integrating lineage-tracing, molecular markers, and single cell sequencing data to identify distinct fibroblast populations during lung development and regeneration. Matrix Biol. 91-92, 51-74. 10.1016/j.matbio.2020.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock, J. R., Barkauskas, C. E., Cronce, M. J., Xue, Y., Harris, J. R., Liang, J., Noble, P. W. and Hogan, B. L. (2011). Multiple stromal populations contribute to pulmonary fibrosis without evidence for epithelial to mesenchymal transition. Proc. Natl. Acad. Sci. USA 108, E1475-E1483. 10.1073/pnas.1117988108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousso, D. L., Qiao, M., Kagan, R. D., Yamagata, M., Palmiter, R. D. and Sanes, J. R. (2016). Two pairs of ON and OFF retinal ganglion cells are defined by intersectional patterns of transcription factor expression. Cell Rep 15, 1930-1944. 10.1016/j.celrep.2016.04.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snippert, H. J., Haegebarth, A., Kasper, M., Jaks, V., van Es, J. H., Barker, N., van de Wetering, M., van den Born, M., Begthel, H., Vries, R. G.et al. (2010). Lgr6 marks stem cells in the hair follicle that generate all cell lineages of the skin. Science 327, 1385-1389. 10.1126/science.1184733 [DOI] [PubMed] [Google Scholar]
- Sorensen, I., Adams, R. H. and Gossler, A. (2009). DLL1-mediated Notch activation regulates endothelial identity in mouse fetal arteries. Blood 113, 5680-5688. 10.1182/blood-2008-08-174508 [DOI] [PubMed] [Google Scholar]
- Tabula Muris Consortium, Overall coordination, Logistical coordination, Organ collection and processing, Library preparation and sequencing, Computational data analysis, Cell type annotation, Writing group, Supplemental text writing group, Principal investigators (2018). Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562, 367-372. 10.1038/s41586-018-0590-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi, H., He, M., Wu, P., Kim, S., Paik, R., Sugino, K., Kvitsiani, D., Fu, Y., Lu, J., Lin, Y.et al. (2011). A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron 71, 995-1013. 10.1016/j.neuron.2011.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell, C., Cacchiarelli, D., Grimsby, J., Pokharel, P., Li, S., Morse, M., Lennon, N. J., Livak, K. J., Mikkelsen, T. S. and Rinn, J. L. (2014). The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381-386. 10.1038/nbt.2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travaglini, K. J., Nabhan, A. N., Penland, L., Sinha, R., Gillich, A., Sit, R. V., Chang, S., Conley, S. D., Mori, Y., Seita, J.et al. (2020). A molecular cell atlas of the human lung from single-cell RNA sequencing. Nature 587, 619-625. 10.1038/s41586-020-2922-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukui, T., Sun, K. H., Wetter, J. B., Wilson-Kanamori, J. R., Hazelwood, L. A., Henderson, N. C., Adams, T. S., Schupp, J. C., Poli, S. D., Rosas, I. O.et al. (2020). Collagen-producing lung cell atlas identifies multiple subsets with distinct localization and relevance to fibrosis. Nat. Commun. 11, 1920. 10.1038/s41467-020-15647-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila Ellis, L. and Chen, J. (2020). A cell-centric view of lung alveologenesis. Dev. Dyn. 250, 482-496. 10.1002/dvdy.271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila Ellis, L., Cain, M. P., Hutchison, V., Flodby, P., Crandall, E. D., Borok, Z., Zhou, B., Ostrin, E. J., Wythe, J. D. and Chen, J. (2020). Epithelial vegfa specifies a distinct endothelial population in the mouse lung. Dev. Cell 52, 617-630. 10.1016/j.devcel.2020.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vints, K., Vandael, D., Baatsen, P., Pavie, B., Vernaillen, F., Corthout, N., Rybakin, V., Munck, S. and Gounko, N. V. (2019). Modernization of Golgi staining techniques for high-resolution, 3-dimensional imaging of individual neurons. Sci. Rep. 9, 130. 10.1038/s41598-018-37377-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, W., Bennett, R. D., Ackermann, M., Ysasi, A. B., Belle, J., Valenzuela, C. D., Pabst, A., Tsuda, A., Konerding, M. A. and Mentzer, S. J. (2015). Elastin cables define the axial connective tissue system in the murine lung. Anat. Rec. 298, 1960-1968. 10.1002/ar.23259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, A. C., Xu, J., Yin, Y., Smith, C., Schmid, G. and Ornitz, D. M. (2006). FGF9 and SHH signaling coordinate lung growth and development through regulation of distinct mesenchymal domains. Development 133, 1507-1517. 10.1242/dev.02313 [DOI] [PubMed] [Google Scholar]
- Wirth, A., Benyo, Z., Lukasova, M., Leutgeb, B., Wettschureck, N., Gorbey, S., Orsy, P., Horvath, B., Maser-Gluth, C., Greiner, E.et al. (2008). G12-G13-LARG-mediated signaling in vascular smooth muscle is required for salt-induced hypertension. Nat. Med. 14, 64-68. 10.1038/nm1666 [DOI] [PubMed] [Google Scholar]
- Xie, T., Wang, Y., Deng, N., Huang, G., Taghavifar, F., Geng, Y., Liu, N., Kulur, V., Yao, C., Chen, P.et al. (2018). Single-cell deconvolution of fibroblast heterogeneity in mouse pulmonary fibrosis. Cell Rep 22, 3625-3640. 10.1016/j.celrep.2018.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J. and Chen, J. (2014). Developmental programs of lung epithelial progenitors: a balanced progenitor model. Wiley Interdiscip. Rev. Dev. Biol. 3, 331-347. 10.1002/wdev.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J., Hernandez, B. J., Martinez Alanis, D., Narvaez del Pilar, O., Vila-Ellis, L., Akiyama, H., Evans, S. E., Ostrin, E. J. and Chen, J. (2016). The development and plasticity of alveolar type 1 cells. Development 143, 54-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias, W. J., Frank, D. B., Zepp, J. A., Morley, M. P., Alkhaleel, F. A., Kong, J., Zhou, S., Cantu, E. and Morrisey, E. E. (2018). Regeneration of the lung alveolus by an evolutionarily conserved epithelial progenitor. Nature 555, 251-255. 10.1038/nature25786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zepp, J. A., Zacharias, W. J., Frank, D. B., Cavanaugh, C. A., Zhou, S., Morley, M. P. and Morrisey, E. E. (2017). Distinct mesenchymal lineages and niches promote epithelial self-renewal and myofibrogenesis in the lung. Cell 170, 1134-1148. 10.1016/j.cell.2017.07.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zepp, J. A., Morley, M. P., Loebel, C., Kremp, M. M., Chaudhry, F. N., Basil, M. C., Leach, J. P., Liberti, D. C., Niethamer, T. K., Ying, Y.et al. (2021). Genomic, epigenomic, and biophysical cues controlling the emergence of the lung alveolus. Science 371, eabc3172. 10.1126/science.abc3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, X., Baron, R. M., Hardin, M., Cho, M. H., Zielinski, J., Hawrylkiewicz, I., Sliwinski, P., Hersh, C. P., Mancini, J. D., Lu, K.et al. (2012). Identification of a chronic obstructive pulmonary disease genetic determinant that regulates HHIP. Hum. Mol. Genet. 21, 1325-1335. 10.1093/hmg/ddr569 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.