Abstract
The development of human immunodeficiency virus type 1 resistance to delavirdine (DLV) was studied in subjects receiving DLV monotherapy. Phenotypic resistance developed in 28 of 30 subjects within 8 weeks. K103N and Y181C, which confer nonnucleoside reverse transcriptase inhibitor (NNRTI) cross-resistance, were the predominant reverse transcriptase mutations. P236L, which confers DLV resistance but hypersensitivity to other NNRTIs, developed in <10% of isolates.
Nonnucleoside reverse transcriptase inhibitors (NNRTIs) are a structurally diverse group of compounds that specifically inhibit human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) (2), including nevirapine (15), the bis(heteroaryl)piperazine delavirdine (DLV) (4), and efavirenz (19). NNRTIs have a similar mechanism of action, binding to a hydrophobic pocket near the active site of RT (11, 18). Passage of HIV-1 in vitro in the presence of either nevirapine or pyridinones leads to the rapid emergence of resistant virus with mutations at RT codon 181 or 103 (Y181C and K103N) (12, 14). Monotherapy with NNRTIs results in the emergence of resistant variants with Y181C and/or K103N mutations (15, 16).
In vitro passage of HIV-1 in the presence of DLV leads to the emergence of a unique RT mutation, P236L (5). In contrast to Y181C and K103N, P236L confers an increase in HIV-1 susceptibility to other NNRTIs. This observation suggested that cross-resistance to other NNRTIs might not be an inevitable consequence of bis(heteroaryl)piperazine resistance. However, P236L was only rarely detected in patients given DLV in combination with didanosine, and the majority of isolates in these patients had a Y181C and/or K103N mutation (3).
The AIDS Clinical Trials Group (ACTG) conducted a phase I/II trial (ACTG 260) of DLV monotherapy (13). The present report summarizes the occurrence of HIV-1 phenotypic resistance to DLV and the associated RT mutations that developed during the first 8 weeks of the trial.
The study design of ACTG 260, a randomized, multicenter, open-label, dose-ranging phase I/II clinical trial, has been described previously (13). One hundred fifteen HIV-infected subjects with CD4 counts between 200 and 500/mm3 were enrolled from October 1994 to July 1995 (13). Half of the subjects were antiretroviral naive, and the remainder had less than 6 months of prior zidovudine (ZDV) experience. Patients were randomized to receive either nucleoside monotherapy (ZDV for antiretroviral-naive subjects or didanosine for ZDV-experienced subjects) or one of three DLV regimens. For patients randomized to DLV monotherapy, doses were chosen to achieve one of three trough blood DLV concentrations: 3 to 10 μM (low), 11 to 30 μM (middle), or 31 to 50 μM (high). Under the first version of the protocol, volunteers randomized to the low-, mid-, and high-concentration arms initially received 200, 300, or 400 mg of DLV three times a day, respectively. However, because early enrollees achieved unexpectedly low trough DLV concentrations, the protocol was amended to change the initial DLV dose to 400 mg three times a day for all DLV recipients (13).
A subset of 30 patients was chosen for further resistance studies, divided into three subgroups based on plasma HIV-1 RNA responses to DLV. Twenty patients were chosen without regard to their antiviral response (group I). The 16 group I patients essentially represented a convenience sample comprised of patients enrolled early in the trial. These initial 16 subjects were more likely to be ZDV experienced and to have enrolled under the first version of the protocol. To reduce potential bias, four additional subjects who had enrolled under the amended version of the protocol were randomly selected to be added to group I. One patient was selected from the ZDV-experienced subjects, and three were selected from the antiretroviral-naive subjects.
In order to determine whether the frequency of HIV-1 resistance to DLV differed based on virologic response, two additional groups were chosen. Group II included all patients who exhibited at least a 0.5 log10 decline in HIV-1 RNA levels from baseline to week 8 or a similar decline in two consecutive measures after week 8. A total of 10 patients demonstrated this degree of response to DLV, and 9 of these had isolates available for testing. Group III included six subjects who received at least 8 weeks of treatment and had less than a 0.30 log10 decline in HIV-1 RNA levels from baseline to any subsequent week; five of these six patients had isolates available. Because group I patients were chosen without regard to antiviral response, there was some overlap between group I and groups II and III (one patient in group II and three patients in group III were also included in group I).
Peripheral blood mononuclear cell (PBMC) HIV-1 culture was performed according to the ACTG consensus methodology (8). Phenotypic susceptibility testing to DLV was performed on culture supernatants using the ACTG-Department of Defense consensus protocol, with DLV concentrations ranging from 0.01 to 50 μM (3, 10). For sequence analysis, a region of the pol gene was amplified from proviral DNA in cultured PBMCs or plasma RNA by previously described methods (17). The resulting PCR product was directly sequenced by automated methods (Perkin-Elmer Applied Biosystems, Foster City, Calif.). Sequencing primers used included HXB2-88, HXB2-89, and B-reverse. No discordant results were obtained between plasma and PBMC specimens. Plasma HIV-1 RNA levels were measured with the Roche Amplicor Monitor assay, and the syncytium-inducing (SI) phenotype was determined by the ACTG consensus protocol (9). Comparisons of the mean plasma RNA levels among the different patient populations were carried out with Student's t test. Comparisons of the proportion of patients with the SI phenotype at baseline were carried out with Fisher's exact test.
The characteristics of the 84 subjects who were randomized to receive DLV in ACTG 260 and those of the subjects in groups I, II, and III are summarized in Table 1. Of the 84 patients enrolled in ACTG 260, 85% were men, 73% were Caucasian, and 18% had an SI isolate at baseline (Table 1). The mean age was 36 years, and the baseline plasma HIV-1 RNA level was 4.6 log10 copies/ml. The baseline characteristics of group I subjects were comparable to those of the 64 ACTG 260 subjects randomized to receive DLV who were not included in group I (data not shown), with the exception of a higher baseline HIV-1 RNA copy number (4.87 log10 versus 4.52 log10; P = 0.03, Student's t test) and increased frequency of SI phenotype (7 of 20 versus 8 of 61; P = 0.045, Fisher's exact test).
TABLE 1.
Characteristics of patients studied in ACTG 260 and groups I, II, and III
| Characteristic | Value for:
|
|||
|---|---|---|---|---|
| ACTG 260 (n = 84) | Group I (n = 20) | Group II (n = 9) | Group III (n = 5) | |
| Gender [no. (%)] | ||||
| Men | 71 (85) | 19 (95) | 7 (78) | 5 (100) |
| Women | 13 (15) | 1 (5) | 2 (22) | 0 (0) |
| Median age (range) (yr) | 36 (19–61) | 34 (28–50) | 33 (22–48) | 32 (28–44) |
| Race [no. (%)] | ||||
| Caucasian | 61 (73) | 18 (90) | 7 (78) | 3 (60) |
| African-American | 15 (18) | 2 (10) | 1 (11) | 1 (20) |
| Hispanic | 8 (10) | 0 (0) | 1 (11) | 1 (20) |
| ZDV naive [no. (%)] | 41 (49) | 10 (50) | 6 (67) | 1 (20) |
| Enrolled under protocol [no. (%)] | ||||
| Version 1.0 | 42 (50) | 14 (70) | 6 (67) | 5 (100) |
| Version 2.0 | 42 (50) | 6 (30) | 3 (33) | 0 (0) |
| DLV target concn arm [no. (%)] | ||||
| Low dose | 27 (32) | 7 (35) | 3 (33) | 3 (60) |
| Middle dose | 28 (33) | 6 (30) | 4 (44) | 1 (20) |
| High dose | 29 (35) | 7 (35) | 2 (22) | 1 (20) |
| Baseline CD4 count | ||||
| Median | 322 | 356 | 435 | 322 |
| Range | 105–668 | 210–515 | 274–500 | 192–454 |
| Baseline RNA (log10 copies/ml) | ||||
| Mean | 4.60 | 4.87 | 4.32 | 5.11 |
| Range | 2.97–5.95 | 3.74–5.58 | 3.10–5.16 | 4.87–5.58 |
| SI isolate at baseline [no. (%)] | 15 (18) | 7 (35) | 0 (0) | 2 (40) |
| Baseline DLV IC50 (μM) | ||||
| Median | 0.029 | 0.023 | 0.038 | |
| Range | 0.010–0.132 | 0.010–0.120 | 0.017–0.132 | |
| Week 8 DLV IC50 (μM) | ||||
| Median | 9.90 | 1.67 | 3.91 | |
| Range | 0.018–27.53 | 0.010–27.42 | 0.018–27.53 | |
The median baseline DLV 50% inhibitory concentration (IC50) for all samples tested was 0.022 μM (range, 0.01 to 0.132) (Table 1). The median DLV IC50 at week 8 was 5.365 μM (range, 0.01 to 27.53). Overall, only two subjects had a DLV IC50 at week 8 that was unchanged from baseline. The first patient (patient 1) (Table 2) had a <0.30 log10 reduction in plasma HIV-1 RNA and was therefore classified as an RNA nonresponder (this patient was included in both groups I and III). The second patient (patient 27) (Table 2) had an RNA response but developed an isolate with V106A at week 8 (this patient is included in group II only).
TABLE 2.
Correlation of phenotypic resistance to DLV with changes in RT genotype
| Group | Patient no. | DLV IC50
|
RT genotypea
|
||||
|---|---|---|---|---|---|---|---|
| Week 0 | Week 8 | Codon 103 (wt = K) | Codon 106 (wt = V) | Codon 181 (wt = Y) | Codon 236 (wt = P) | ||
| I | 1 | 0.020 | 0.018 | K | V | Y | P |
| 2 | 0.010 | 10.060 | N | V | C | P | |
| 3 | 0.013 | 11.518 | N | V | C | P | |
| 4 | 0.014 | 10.823 | N | V | Y | P | |
| 5 | 0.018 | 1.794 | K | V | C | P | |
| 6 | 0.109 | 3.197 | K | V | C | P | |
| 7 | 0.010 | 4.462 | N | V | C | P | |
| 8 | 0.036 | 15.475 | N | V | Y | P | |
| 9 | 0.095 | 9.242 | Q | V | Y | L | |
| 10 | 0.022 | 1.848 | N | V | Y | P | |
| 11 | 0.021 | 25.294 | N | V | C | P | |
| 12 | 0.064 | 6.267 | N | V | C | P | |
| 13 | 0.132 | 9.731 | N | V | Y | P | |
| 14 | 0.126 | 14.142 | N | V | Y | P | |
| 15 | 0.038 | 27.530 | N | V | C | P | |
| 16 | 0.013 | 11.106 | N | V | Y | P | |
| 17 | 0.050 | 10.850 | N | V | Y | P | |
| 18 | 0.074 | 2.915 | N | V | Y | P | |
| 19 | 0.015 | 2.853 | N | V | C | P | |
| 20 | 0.076 | 16.451 | N | V | C | P | |
| II | 8 | 0.036 | 15.475 | N | V | Y | P |
| 21 | 0.020 | 1.250 | N | V | Y | P | |
| 22 | 0.010 | 0.920 | N | V | Y | P | |
| 23 | 0.120 | 8.533 | N | V | Y | P | |
| 24 | 0.050 | 27.420 | N | V | C | P | |
| 25 | 0.011 | 3.141 | N | V | Y | P | |
| 26 | 0.058 | 1.669 | N | V | Y | L | |
| 27 | 0.010 | 0.010 | K | A | Y | P | |
| 28 | 0.023 | 1.025 | N | V | Y | P | |
| III | 1 | 0.020 | 0.018 | K | V | Y | P |
| 13 | 0.132 | 9.731 | N | V | Y | P | |
| 15 | 0.038 | 27.530 | N | V | C | P | |
| 29 | 0.115 | 3.064 | N | V | Y | P | |
| 30 | 0.017 | 3.908 | K | V | C | P | |
A single letter represents the amino acid at that codon (i.e., K = lysine). Mutations known to confer NNRTI resistance are highlighted in boldface. wt, wild type.
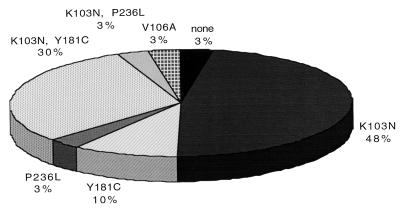
Sequence analysis revealed no NNRTI resistance mutations present at baseline. One baseline isolate (from patient 9) (Table 2) had a polymorphism at codon 103 (K103Q) that has not been reported to be associated with NNRTI resistance; the DLV IC50 of this patient's isolate was 0.095 μM. Only 1 of 30 patients (3%) had no detectable changes in RT sequence during therapy with DLV (Fig. 1); this was the one patient in the study that did not develop phenotypic DLV resistance and had no significant decline in plasma HIV-1 RNA (patient 1) (Table 2). Fourteen patients (48%) developed isolates with the K103N mutation alone, three (10%) developed Y181C alone, one (3%) developed P236L, one (3%) developed V106A, nine (30%) developed K103N in combination with Y181C, and one (3%) developed K103N in combination with P236L (Fig. 1). The viral isolate with the P236L mutation also contained the K103Q polymorphism that had been present before initiation of DLV therapy (patient 9) (Table 2).
FIG. 1.
HIV-1 reverse transcriptase mutations in isolates from patients in ACTG 260. The pie chart represents the frequency of RT mutations in week 8 isolates. Isolates from a total of 30 patients were analyzed. None of the mutations represented here was present in the corresponding baseline isolates.
In summary, we have characterized the frequency and genetic basis of HIV-1 resistance to DLV that occur during DLV monotherapy. Significant increases in DLV IC50, averaging 10 to 1,000 fold, were seen in all but 2 of the 30 patients tested. These rises in DLV IC50 were temporally associated with the return of plasma HIV-1 RNA levels toward baseline. We suspect that one patient did not demonstrate phenotypic or genotypic resistance to DLV because of poor compliance with the medication, since this patient also demonstrated no significant RNA response at any of the time points tested. The second patient had no detectable change in DLV IC50 despite the development of V106A. Codon 106 lines the NNRTI binding pocket, and V106A has been shown to confer resistance to nevirapine (1), although there are no published data on the susceptibility of this mutant to DLV. An X-ray crystal structure of HIV-1 RT bound to DLV has shown that codon 106 is in close proximity to the bound drug, suggesting that a mutation at this site could affect DLV susceptibility (6). We suspect that the selection for the V106A mutation in this patient reflects a low level of DLV resistance not detectable in our phenotypic assay.
Baseline DLV IC50 and the frequency of DLV resistance were similar in the DLV responder and nonresponder groups in this study. This observation is likely due to the fact that plasma RNA reductions in the responder group (and in ACTG 260 as a whole) were modest and not sustained. We did observe that subjects who had better responses to DLV monotherapy tended to have lower baseline plasma HIV-1 RNA levels and were less likely to have an SI isolate at baseline than subjects with minimal plasma HIV-1 RNA responses to DLV.
An unexpected finding was that the P236L mutation, which was seen during in vitro passage of HIV-1 isolates in the presence of DLV, developed in less than 10% of isolates. We hypothesize that the infrequent occurrence of the P236L mutation in patients may be related to the replication defect of this mutant relative to K103N that has been observed in vitro (7).
It is interesting to note that the only isolate in which P236L developed in the absence of other NNRTI resistance mutations contained an unusual polymorphism of the NNRTI binding pocket (K103Q). It may be that the polymorphism at codon 103 in some way compensates for the decreased fitness of the P236L mutant. Because of the infrequent occurrence of the P236L mutant in the isolates we examined, we were unable to determine whether there are specific polymorphisms or factors, such as RNA level or DLV concentration, that predict the development of P236L during DLV monotherapy.
In conclusion, administration of DLV monotherapy led to transient reductions in plasma HIV-1 RNA levels that were accompanied by the almost uniform development of phenotypic DLV resistance. The genetic basis for DLV resistance during monotherapy was predominantly due to the K103N mutation, which confers cross-resistance to other currently available NNRTIs.
Nucleotide sequence accession numbers.
All nucleotide sequences have been submitted to GenBank (accession no. AF090466 to AF090532).
Acknowledgments
This research was supported in part by the National Institutes of Health (AI-38858, AI-27658, RR-00044-34S2, AI-041387, AI-27666, AI-25924, and AI-27675) and Pharmacia and Upjohn.
We thank Peter Gerondelis and Angela Dexter for the performance of resistance assays and Michael Chiulli and Luis Berrios for the performance of p24 antigen assays.
REFERENCES
- 1.Byrnes V W, Sardana V V, Schleif W A, Condra J H, Waterbury J A, Wolfgang J A, Long W J, Schneider C L, Schlabach A J, Wolanski B S, et al. Comprehensive mutant enzyme and viral variant assessment of human immunodeficiency virus type 1 reverse transcriptase resistance to nonnucleoside inhibitors. Antimicrob Agents Chemother. 1993;37:1576–1579. doi: 10.1128/aac.37.8.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Clercq E. Non-nucleoside reverse transcriptase inhibitors (NNRTIs) for the treatment of human immunodeficiency virus type 1 (HIV-1) infections: strategies to overcome drug resistance development. Med Res Rev. 1996;16:125–157. doi: 10.1002/(SICI)1098-1128(199603)16:2<125::AID-MED1>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 3.Demeter L M, Meehan P M, Morse G, Gerondelis P, Dexter A, Berrios L, Cox S, Freimuth W, Reichman R C. HIV-1 drug susceptibilities and reverse transcriptase mutations in patients receiving combination therapy with didanosine and delavirdine. J Acquir Immune Defic Syndr Hum Retrovirol. 1997;14:136–144. doi: 10.1097/00042560-199702010-00006. [DOI] [PubMed] [Google Scholar]
- 4.Dueweke T J, Poppe S M, Romero D L, Swaney S M, So A G, Downey K M, Althaus I W, Reusser F, Busso M, Resnick L, et al. U-90152, a potent inhibitor of human immunodeficiency virus type 1 replication. Antimicrob Agents Chemother. 1993;37:1127–1131. doi: 10.1128/aac.37.5.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dueweke T J, Pushkarskaya T, Poppe S M, Swaney S M, Zhao J Q, Chen I S, Stevenson M, Tarpley W G. A mutation in reverse transcriptase of bis(heteroaryl)piperazine-resistant human immunodeficiency virus type 1 that confers increased sensitivity to other nonnucleoside inhibitors. Proc Natl Acad Sci USA. 1993;90:4713–4717. doi: 10.1073/pnas.90.10.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esnouf R M, Ren J, Hopkins A L, Ross C K, Jones E Y, Stammers D K, Stuart D I. Unique features in the structure of the complex between HIV-1 reverse transcriptase and the bis(heteroaryl)piperazine (BHAP) U-90152 explain resistance mutations for this nonnucleoside inhibitor. Proc Natl Acad Sci USA. 1997;94:3984–3989. doi: 10.1073/pnas.94.8.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerondelis P, Archer R H, Palaniappan C, Reichman R C, Fay P J, Bambara R A, Demeter L M. The P236L delavirdine-resistant human immunodeficiency virus type 1 mutant is replication defective and demonstrates alterations in both RNA 5′-end- and DNA 3′-end-directed RNase H activities. J Virol. 1999;73:5803–5813. doi: 10.1128/jvi.73.7.5803-5813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hollinger F B, Bremer J W, Myers L E, Gold J W M, McQuay L the NIH/NIAID/DAIDS/ACTG Virology Laboratories. Standardization of sensitive human immunodeficiency virus coculture procedures and establishment of a multicenter quality assurance program for the AIDS Clinical Trials Group. J Clin Microbiol. 1992;30:1787–1794. doi: 10.1128/jcm.30.7.1787-1794.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Japour A J, Fiscus S A, Arduino J M, Mayers D L, Reichelderfer P S, Kuritzkes D R. Standardized microtiter assay for determination of syncytium-inducing phenotypes of clinical human immunodeficiency virus type 1 isolates. J Clin Microbiol. 1994;32:2291–2294. doi: 10.1128/jcm.32.9.2291-2294.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Japour A J, Mayers D L, Johnson V A, Kuritzkes D R, Beckett L A, Arduino J-M, Lane J, Black R J, Reichelderfer P S, D'Aquila R T, Crumpacker C S the RV-43 Study Group; the AIDS Clinical Trials Group Virology Committee Resistance Working Group. Standardized peripheral blood mononuclear cell culture assay for determination of drug susceptibilities of clinical human immunodeficiency virus type 1 isolates. Antimicrob Agents Chemother. 1993;37:1095–1101. doi: 10.1128/aac.37.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kohlstaedt L A, Wang J, Friedman J M, Rice P A, Steitz T A. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 12.Nunberg J H, Schlief W A, Boots E J, O'Brien J A, Quintero J C, Hoffman J M, Emini E, Goldman M E. Viral resistance to human immunodeficiency virus type 1-specific pyridinone reverse transcriptase inhibitors. J Virol. 1991;65:4887–4892. doi: 10.1128/jvi.65.9.4887-4892.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Para M F, Meehan P, Holden-Wiltse J, Fischl M, Morse G, Shafer R, Demeter L M, Wood K, Nevin T, Virani-Ketter N, Freimuth W W for the AIDS Clinical Group Protocol 260 Team. ACTG 260: a randomized, phase I-II, dose-ranging trial of the anti-human immunodeficiency virus activity of delavirdine monotherapy. Antimicrob Agents Chemother. 1999;43:1373–1378. doi: 10.1128/aac.43.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richman D, Shih C-K, Lowy I, Rose J, Prodanovich P, Goff S, Griffin J. Human immunodeficiency virus type 1 mutants resistant to nonnucleoside inhibitors of reverse transcriptase arise in tissue culture. Proc Natl Acad Sci USA. 1991;88:11241–11245. doi: 10.1073/pnas.88.24.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richman D D, Havlir D, Corbeil J, Looney D, Ignacio C, Spector S A, Sullivan J, Cheeseman S, Barringer K, Pauletti D, et al. Nevirapine resistance mutations of human immunodeficiency virus type 1 selected during therapy. J Virol. 1994;68:1660–1666. doi: 10.1128/jvi.68.3.1660-1666.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saag M S, Emini E A, Laskin O L, Douglas J, Lapidus W I, Shlief W A, Whitley R J, Hildebrand C, Byrnes V W, Kappes J C, Anderson K W, Massari F E, Shaw G M the L697,661 Working Group. A short-term clinical evaluation of L-697,661, a non-nucleoside inhibitor of HIV-1 reverse transcriptase. N Engl J Med. 1993;329:1065–1072. doi: 10.1056/NEJM199310073291502. [DOI] [PubMed] [Google Scholar]
- 17.Shafer R W, Eisen J A, Merigan T C, Katzenstein D A. Sequence and drug susceptibility of subtype C reverse transcriptase from human immunodeficiency virus type 1 seroconverters in Zimbabwe. J Virol. 1997;71:5441–5448. doi: 10.1128/jvi.71.7.5441-5448.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spence R A, Kati W M, Anderson K S, Johnson K A. Mechanism of inhibition of HIV-1 reverse transcriptase by nonnucleoside inhibitors. Science. 1995;267:988–993. doi: 10.1126/science.7532321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Young S D, Britcher S F, Tran L O, Payne L S, Lumma W C, Lyle T A, Huff J R, Anderson P S, Olsen D B, Carroll S S, et al. L-743,726 (DMP-266): a novel, highly potent nonnucleoside inhibitor of the human immunodeficiency virus type 1 reverse transcriptase. Antimicrob Agents Chemother. 1995;39:2602–2605. doi: 10.1128/aac.39.12.2602. [DOI] [PMC free article] [PubMed] [Google Scholar]