Abstract
We developed a simplified assay for estimating efflux by measuring the effect of reserpine on the growth of Streptococcus pneumoniae and Staphylococcus aureus over 7 h. Reserpine enhanced ciprofloxacin and levofloxacin 17 to 68%. The hydrophobic drug trovafloxacin and the drug moxifloxacin, with a bulky C-7 substituent but hydrophilicity similar to that of levofloxacin, showed little (0 to 11%) reserpine-enhancing effect. The ease of resistant mutant strain selection correlated with efflux susceptibility.
Mechanisms of resistance to fluoroquinolones include mutations in DNA gyrase and topoisomerase IV genes (7) and active efflux of agents from the cell (13). Efflux may permit short-term bacterial survival that then leads to adaptive fluoroquinolone resistance via a mutation(s) at key drug target sites. Energy-dependent efflux has been reported in both Staphylococcus aureus and Streptococcus pneumoniae (2, 3, 4, 6, 9, 17–19, 24, 25). Structural differences among fluoroquinolones, notably overall molecular hydrophobicity and bulkiness of the C-7 substituent, are thought to influence the efficiency of efflux (21, 23, 24).
In this work, we developed a simple and sensitive growth inhibition assay to compare the effects of the plant alkaloid reserpine, a specific inhibitor of active efflux (2, 12), on structurally variant fluoroquinolones. We also hypothesized that avoiding active efflux makes a fluoroquinolone more lethal at lower concentrations and thus more difficult for bacteria to develop chromosomal mutations leading to high-level resistance (20). We tested this hypothesis by selecting for S. pneumoniae mutants with different fluoroquinolone agents.
S. aureus strains SA-1199 and SA-1199B (8) and S. pneumoniae CP1000 (22) were used in this study. Todd-Hewitt broth (Difco Laboratories, Detroit, Mich.) supplemented with 0.5% yeast extract (THBY) was used for studies of S. pneumoniae, while S. aureus was grown in Mueller-Hinton broth (MHB; Difco). Casein hydrolysate-yeast extract-tryptone (CAT) agar (Difco) was used for selection of resistant mutants. Fluoroquinolones were provided by their manufacturer: levofloxacin was from Ortho-McNeil Pharmaceuticals (Raritan, N.J.), ciprofloxacin and moxifloxacin were from Bayer Corporation (West Haven, Conn.), sparfloxacin was from Rhône-Poulenc Rorer R-D (Vitry-sur-Seine, France), and trovafloxacin was from Pfizer Pharmaceuticals Group (New York, N.Y.). Susceptibility testing was done according to recommended methods (15). All testing was performed at least in duplicate.
Evaluation of the effect of reserpine.
The accumulation of ciprofloxacin determined by fluorometry was measured by the method of Mortimer and Piddock (14). The growth inhibition assay developed was done as follows. S. aureus and S. pneumoniae were inoculated at 1 × 106 to 2 × 106 CFU/ml into tubes with MHB and THBY medium, respectively, containing each fluoroquinolone at a concentration of one-fourth the determined MIC, either alone or with 10 μg of reserpine per ml (R. Beyer, E. Pestova, V. Stosor, G. A. Noskin, and L. R. Peterson, Abstr. Infect. Dis. Soc. Am. 36th Gen. Meet. 1998, abstr. 50, p. 84, 1998). Growth in ethidium bromide (EtBr) was conducted at 1/4 the MIC for strain SA-1199, 1/8 the MIC for strain SA-1199B, and 1/16 the MIC for strain CP1000. EtBr was included as a known substrate of NorA efflux (10, 16, 17). All strains were grown in reserpine alone, and the data were normalized for any effect on growth. Measurements were determined upon inoculation of each culture and over 6 to 7 h of incubation at 35°C. The extent of growth inhibition by each fluoroquinolone was determined by comparing the optical densities at 550 nm (OD550) of cultures to those of controls at mid-log growth phase (percent decrease in OD) at 6.5 h.
Selection of mutants.
First-step mutants were obtained by exposing S. pneumoniae CP1000 to four doubling fluoroquinolone concentrations, starting at the MIC of each agent. Between 108 and 109 cells from an S. pneumoniae CP1000 culture were plated onto the top layer of CAT agar on a two-layer plate, with a doubled concentration of the corresponding fluoroquinolone in the bottom layer of the agar. A total of 1010 CFU (on multiple plates) was used at each drug concentration, and cells were harvested after 48 h of incubation at 35°C. Second-step mutants of CP1000 were obtained by the same procedure.
Activity of fluoroquinolones against S. pneumoniae and S. aureus.
The MICs for CP1000 are shown in Table 1. The MIC results for SA-1199 and SA-1199B, respectively, were as follows: EtBr, 4.0 and 32.0 μg/ml; ciprofloxacin, 0.5 and 8.0 μg/ml; levofloxacin, 0.25 and 1.0 μg/ml; moxifloxacin, 0.06 and 0.12 μg/ml; sparfloxacin, 0.12 and 0.25 μg/ml; and trovafloxacin, 0.06 and 0.12 μg/ml.
TABLE 1.
Frequency of resistance selection in S. pneumoniae CP1000
| Selection agent (MIC [μg/ml]) | Drug concn/MIC ratio (fold) | Drug concn (μg/ml) | Mutation frequencya |
|---|---|---|---|
| Levofloxacin (1.0) | 1 | 1.0 | 4.2 × 10−7 |
| 2 | 2.0 | 1.4 × 10−9 | |
| 4 | 4.0 | 1.4 × 10−9 | |
| 8 | 8.0 | 5 × 10−10 | |
| Ciprofloxacin (0.5) | 1 | 0.5 | 2.2 × 10−7 |
| 2 | 1.0 | 1.2 × 10−7 | |
| 4 | 2.0 | 2.2 × 10−9 | |
| 8 | 4.0 | 4.5 × 10−9 | |
| Sparfloxacinb (0.25) | 1 | 0.25 | 2.0 × 10−7 |
| 2 | 0.5 | 3.0 × 10−10 | |
| 4 | 1.0 | <10−10 | |
| 8 | 2.0 | <10−10 | |
| Trovafloxacin (0.12) | 1 | 0.12 | 2.5 × 10−7 |
| 2 | 0.25 | <10−10 | |
| 4 | 0.5 | <10−10 | |
| 8 | 1.0 | <10−10 | |
| Moxifloxacinb (0.12) | 1 | 0.12 | 1.3 × 10−7 |
| 2 | 0.25 | <10−10 | |
| 4 | 0.5 | <10−10 | |
| 8 | 1.0 | <10−10 |
Mutation frequency was calculated as the number of putative mutants divided by the total number of cells in 1 ml of culture determined by the number of colonies on plates containing an appropriate fluoroquinolone compared to the number on plates without drug.
Taken from Pestova et al. (Abstr. 99th Gen. Meet. Am. Soc. Microbiol.).
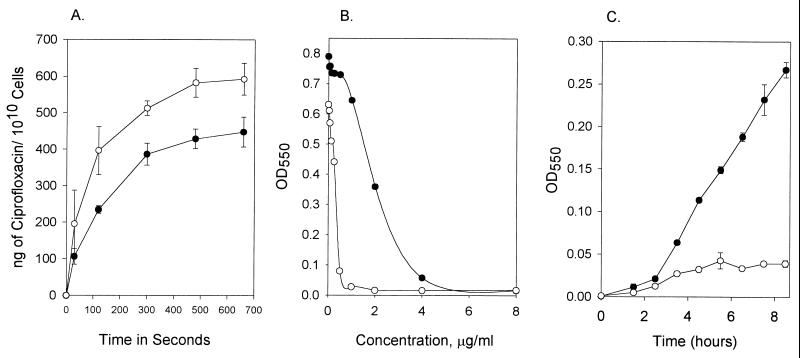
Figure 1A illustrates the effect of 20 μg of reserpine per ml on the accumulation of ciprofloxacin by S. aureus strain SA-1199B as assessed by the fluorometric method and is consistent with the results of previous studies (3, 11, 12). However, this fluorescence method for determining accumulation is effective only for those drugs that fluoresce, such as levofloxacin and ciprofloxacin (with a fluorescence of 34.43 fluorescence units [FU] and 37.42 FU, respectively, at 10 μg/ml). Fluoroquinolones that fluoresce poorly, such as trovafloxacin (with a fluorescence of 0.939 FU), and sparfloxacin, which fluoresces poorly even at 50 μg/ml (21), require a different method for estimating efflux. The growth inhibition assay reproduced the reserpine-mediated effect for both our S. pneumoniae and S. aureus test strains, reflected as a potentiation of ciprofloxacin activity. The dose-response plot in Fig. 1B demonstrates the effect of 10 μg of reserpine per ml on the growth of SA-1199B. Similar patterns were demonstrated for S. aureus SA-1199 and for S. pneumoniae CP1000 (data not shown). This observation reflects the action of reserpine upon the NorA-dependent active efflux mechanism in S. aureus and a homologous system, PmrA, in S. pneumoniae (3, 11, 12). The effect is also demonstrated in our growth inhibition assay (Fig. 1C).
FIG. 1.
Reserpine-mediated inhibition of ciprofloxacin efflux and growth in ciprofloxacin of S. aureus strain SA-1199B. (A) Ciprofloxacin accumulation as measured by a fluorometric method (at 10 μg of ciprofloxacin per ml), both with and without reserpine; (B) mid-log-phase OD values for growth of SA-1199B in various ciprofloxacin concentrations, with and without reserpine; (C) growth inhibition assays for SA-1199B at one-fourth the MIC of ciprofloxacin, with and without reserpine. Symbols: closed circles, experiments done with drug alone; open circles, experiments done with drug and reserpine; vertical bars, range of results obtained for the duplicate experiments.
Reserpine effect on structurally variant fluoroquinolones.
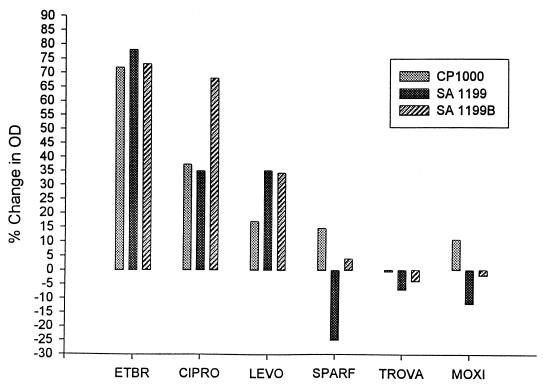
Figure 2 depicts the magnitude of the growth inhibition effect of reserpine, shown as a percent inhibition (decrease) of growth at the 6.5-h time point in mid-log phase. Growth of all three strains in EtBr showed a minimum decrease in OD of 73%, providing evidence of reserpine-mediated inhibition of EtBr efflux. Similarly, there was strong reserpine-mediated growth inhibition for ciprofloxacin and levofloxacin. This decrease in growth with reserpine was most notable in S. aureus strain SA-1199B, which overexpresses the NorA protein.
FIG. 2.
Effect of reserpine on the inhibition of growth by EtBr and structurally variant fluoroquinolone agents (CIPRO, ciprofloxacin; LEVO, levofloxacin; SPARF, sparfloxacin; TROVA, trovafloxacin; MOXI, moxifloxacin). Each column represents the inhibition of growth by reserpine relative to growth without reserpine in the presence of the same fluoroquinolone. All tests were done in duplicate.
Reserpine had little effect on growth in the presence of moxifloxacin, sparfloxacin, and trovafloxacin. In strain SA-1199B, we observed only a 4% decrease in OD for growth for sparfloxacin plus reserpine and increases of 2 and 4% for moxifloxacin and trovafloxacin, respectively. The increase in growth was unexpected, but it must be remembered that export systems have many substrates (1, 16) and that reserpine may compete with quinolones at other sites in the organism which could impede the antibacterial properties of the drug. Thus, it should not be surprising that an increase in growth could occur when bacteria are exposed to multiple compounds.
Emergence of resistant mutants.
The frequencies of first-step mutant selection are shown in Table 1. Those agents with little reserpine-mediated effect demonstrated poor emergence of first-step mutants once their MICs were exceeded. Sequence analysis of mutant clones confirmed the presence of amino acid substitutions in the appropriate fluoroquinolone targets (21; E. Pestova, R. Beyer, G. A. Noskin, and L. R. Peterson, Abstr. 99th Gen. Meet. Am. Soc. Microbiol. 1999, abstr. A-37, p. 8, 1999). We found that once first-step mutants were generated, the emergence of resistance increased with second-step mutants selected on drug concentrations twice the first-step mutant MIC of levofloxacin, ciprofloxacin, trovafloxacin, and moxifloxacin, all occurring with frequencies ranging from 2 × 10−6 to 1.8 × 10−7.
Concluding comments.
We validated a new method for assessing the reserpine-mediated effect on fluoroquinolone efflux that is not dependent upon fluorescence. Such a simplified assay for estimating NorA-type efflux is needed because this resistance mechanism is becoming increasingly important, and an agar screening test may not be reliable since reserpine is poorly soluble and present in agar as a suspension, not a solution.
We found the degree of growth inhibition mediated by reserpine was lessened in the new bulkier and/or more hydrophobic agents moxifloxacin (solubility, 2.4 g/100 ml), sparfloxacin (solubility, 0.11 g/100 ml), and trovafloxacin (solubility, 0.002 g/100 ml). However, the growth of organisms in the smaller, more hydrophilic drugs ciprofloxacin (solubility, 3.5 g/100 ml) and levofloxacin (solubility, 2.5 g/100 ml) was strongly inhibited by the presence of reserpine. Of the fluoroquinolones tested, ciprofloxacin has the least bulky C-7 substituent, a piperasine moiety, with levofloxacin and sparfloxacin each carrying slightly larger piperasine derivatives. Trovafloxacin and moxifloxacin have the bulkiest C-7 substituents, a cycropyl-fused and a piperasine-fused pyrrolidine ring, respectively. It is also of interest that the overall hydrophobicities of sparfloxacin and trovafloxacin are fairly similar, while ciprofloxacin, levofloxacin, and moxifloxacin are much more hydrophilic. Since moxifloxacin is relatively hydrophilic, bulk at C-7 appears to be the key addition for its avoidance of active efflux.
Our study also demonstrated the probability that avoiding efflux results in a significantly decreased potential for emergence of resistance on exposure to a fluoroquinolone. We were unable to select for mutants from S. pneumoniae strain CP1000 at concentrations above the MIC of moxifloxacin and trovafloxacin, while drug concentrations at least 8 times the MIC of ciprofloxacin and levofloxacin yielded resistant mutants. Others have shown that reserpine-mediated inhibition of the efflux mechanism in S. aureus can lower the emergence of resistance (11, 12). The reduced capacity to cause the efflux of fluoroquinolones, assuming that all penetrate the cell with equivalent ease, may allow increased drug accumulation that facilitates faster killing. This appears to eradicate the entire bacterial population before adaptive mutations in DNA gyrase or topoisomerase IV emerge, leading to stable resistance.
The importance of preventing emergence of resistance is pertinent in light of reports showing an increasing prevalence of fluoroquinolone-resistant S. pneumoniae (5). Our results provide an explanation at the molecular level for this clinical observation. We also demonstrated in the second-step mutant selection experiments, where resistant clones appeared more readily with all the fluoroquinolones, that it becomes easier to select for higher-level resistance once bacteria have accumulated an initial resistance mutation. Therefore, to maintain long-term antimicrobial activity, it is imperative to consider the use of bulkier and/or more hydrophobic quinolones in initial fluoroquinolone therapy and to avoid the increasing consumption of less active agents that may enhance fluoroquinolone class resistance development.
Acknowledgments
This work was supported by grants from the Excellence in Academic Medicine program at Northwestern Memorial Hospital, the Pharmaceutical Division of Bayer Corporation, the Pfizer Pharmaceuticals Group, and U.S. Public Health Service grant no. UR8/CCU515081; it was also supported by the Northwestern University Medical School, Chicago, Ill.
We thank G. W. Kaatz, Wayne State University, for generously donating S. aureus strains SA-1199 and SA-1199B.
REFERENCES
- 1.Ahmed M, Borsch C M, Neyfakh A A, Schuldiner S. Mutants of the Bacillus subtilis multidrug transporter Bmr with altered sensitivity to the antihypertensive alkaloid reserpine. J Biol Chem. 1993;268:11086–11089. [PubMed] [Google Scholar]
- 2.Baranova N N, Neyfakh A A. Apparent involvement of a multidrug transporter in the fluoroquinolone resistance of Streptococcus pneumoniae. Antimicrob Agents Chemother. 1997;41:1396–1398. doi: 10.1128/aac.41.6.1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brenwald N P, Gill M J, Wise R. The effect of reserpine, an inhibitor of multidrug efflux pumps, on the in-vitro susceptibilities of fluoroquinolone-resistant strains of Streptococcus pneumoniae to norfloxacin. J Antimicrob Chemother. 1997;40:458–460. doi: 10.1093/jac/40.3.458. [DOI] [PubMed] [Google Scholar]
- 4.Brenwald N P, Gill M J, Wise R. Prevalence of a putative efflux mechanism among fluoroquinolone-resistant clinical isolates of Streptococcus pneumoniae. Antimicrob Agents Chemother. 1998;42:2032–2035. doi: 10.1128/aac.42.8.2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen D K, McGeer A, De Azavedo J C, Low D E. Decreased susceptibility of Streptococcus pneumoniae to fluoroquinolones in Canada. N Engl J Med. 1999;341:233–239. doi: 10.1056/NEJM199907223410403. [DOI] [PubMed] [Google Scholar]
- 6.Gill M J, Brenwald N P, Wise R. Identification of an efflux pump gene, pmrA, associated with fluoroquinolone resistance in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1999;43:187–189. doi: 10.1128/aac.43.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hooper D C. Bacterial topoisomerases, anti-topoisomerases, and anti-topoisomerase resistance. Clin Infect Dis. 1998;27(Suppl. 1):S54–S63. doi: 10.1086/514923. [DOI] [PubMed] [Google Scholar]
- 8.Kaatz G W, Seo S M, Ruble C A. Mechanisms of fluoroquinolone resistance in Staphylococcus aureus. J Infect Dis. 1991;163:1080–1086. doi: 10.1093/infdis/163.5.1080. [DOI] [PubMed] [Google Scholar]
- 9.Kaatz G W, Seo S M, Ruble C A. Efflux-mediated fluoroquinolone resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1993;37:1086–1094. doi: 10.1128/aac.37.5.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaatz G W, Seo S M. Inducible NorA-mediated multidrug resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1995;39:2650–2655. doi: 10.1128/aac.39.12.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Markham P N, Neyfakh A A. Inhibition of the multidrug transporter NorA prevents emergence of norfloxacin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1996;40:2252–2257. doi: 10.1128/aac.40.11.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Markham P N. Inhibition of the emergence of ciprofloxacin resistance in Streptococcus pneumoniae by the multidrug efflux inhibitor reserpine. Antimicrob Agents Chemother. 1999;43:988–989. doi: 10.1128/aac.43.4.988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez J L, Alonso A, Gomez-Gomez J M, Baquero F. Quinolone resistance by mutations in chromosomal gyrase genes. Just the tip of the iceberg? J Antimicrob Chemother. 1998;42:683–688. doi: 10.1093/jac/42.6.683. [DOI] [PubMed] [Google Scholar]
- 14.Mortimer P G S, Piddock L J V. A comparison of methods used for measuring the accumulation of quinolones by Enterobacteriaceae, Pseudomonas aeruginosa and Staphylococcus aureus. J Antimicrob Chemother. 1991;28:639–653. doi: 10.1093/jac/28.5.639. [DOI] [PubMed] [Google Scholar]
- 15.National Committee for Clinical Laboratory Standards. Method for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. M7-A4. Villanova, Pa: National Committee for Laboratory Standards; 1997. [Google Scholar]
- 16.Neyfakh A A. Natural functions of bacterial multidrug transporters. Trends Microbiol. 1997;5:309–313. doi: 10.1016/S0966-842X(97)01064-0. [DOI] [PubMed] [Google Scholar]
- 17.Neyfakh A A, Borsch C M, Kaatz G W. Fluoroquinolone resistance protein NorA of Staphylococcus aureus is a multidrug efflux transporter. Antimicrob Agents Chemother. 1993;37:128–129. doi: 10.1128/aac.37.1.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ng E Y, Trucksis M, Hooper D C. Quinolone resistance mediated by norA: physiologic characterization and relationship to flqB, a quinolone resistance locus on the Staphylococcus aureus chromosome. Antimicrob Agents Chemother. 1994;38:1345–1355. doi: 10.1128/aac.38.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paulsen I T, Brown M H, Skurray R A. Proton-dependent multidrug efflux systems. Microbiol Rev. 1996;60:575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pestova E, Beyer R, Cianciotto N P, Noskin G A, Peterson L R. Contribution of topoisomerase IV and DNA gyrase mutations in Streptococcus pneumoniae to resistance to novel fluoroquinolones. Antimicrob Agents Chemother. 1999;43:2000–2004. doi: 10.1128/aac.43.8.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Piddock L J V, Zhu M. Mechanism of sparfloxacin against and mechanism of resistance in gram-negative and gram-positive bacteria. Antimicrob Agents Chemother. 1991;35:2423–2427. doi: 10.1128/aac.35.11.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shoemaker N B, Guild W R. Destruction of low efficiency markers is a slow process occurring at a heteroduplex stage of transformation. Mol Gen Genet. 1974;128:283–290. doi: 10.1007/BF00268516. [DOI] [PubMed] [Google Scholar]
- 23.Takenouchi T, Tabata F, Iwata Y, Hanzawa H, Sugawara M, Ohya S. Hydrophilicity of quinolones is not an exclusive factor for decreased activity in efflux-mediated resistant mutants of Staphylococcus aureus. Antimicrob Agents Chemother. 1996;40:1835–1842. doi: 10.1128/aac.40.8.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshida H, Bogaki M, Nakamura S, Ubukata K, Konno M. Nucleotide sequence and characterization of the Staphylococcus aureus norA gene, which confers resistance to quinolones. J Bacteriol. 1990;172:6942–6949. doi: 10.1128/jb.172.12.6942-6949.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeller V, Janoir C, Kitzis M-D, Gutmann L, Moreau N J. Active efflux as a mechanism of resistance to ciprofloxacin in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1997;41:1973–1978. doi: 10.1128/aac.41.9.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]