Abstract
Rheumatoid arthritis is an autoimmune disease that causes significant morbidity. Application of cellular profiling techniques such as single-cell transcriptomics and spatial transcriptomics has uncovered novel pathogenic cell types in RA joint tissues and revealed marked heterogeneity in the cellular composition among RA patients. Together, these insights provide exciting opportunities to translate discoveries into precision medicine in RA. The present review aims to highlight novel insights into RA pathology and discuss key steps needed to translate these discoveries into actionable changes in clinical practice. We review efforts to identify surrogate biomarkers that could be used to predict RA synovial tissue phenotypes and the corresponding responses to therapy. Finally, we discuss the opportunity to develop novel patient-derived organoid systems as a platform for therapeutic target validation.
Keywords: rheumatoid arthritis, precision medicine, single-cell RNAseq, spatial transcriptomics, organoids
I. Introduction
Rheumatoid arthritis (RA) is a systemic autoimmune disease that affects 1% of the population[1][2]. RA causes significant morbidity and carries a substantial societal burden[1]. The treatment of rheumatoid arthritis has advanced significantly over the past decades with the advent of numerous targeted therapies [1]. However, despite these advances, several challenges remain. The first challenge is that we cannot yet predict which therapeutic agent will lead to the optimal response with the least toxicity for each patient. The second challenge is that many RA patients never achieve sustained remission[1], despite rapid advances in RA therapeutics. Third, there is marked disease heterogeneity within RA with regards to synovial tissue phenotypes [3–5] and extraarticular manifestations [6,7]. Together, these unmet needs pose a substantial challenge for precision medicine approaches in RA.
In this review, we start by discussing key technological breakthroughs that have enabled a disease “deconstruction” approach to defining RA pathology at a single-cell level. Next, we summarize recent findings on pathogenic cell states expanded in RA synovia, molecular pathways underlying their expansion, and new insights into RA disease heterogeneity and synovial tissue phenotypes. We describe key steps needed to translate these discoveries into clinical insights. We highlight approaches being taken to identify surrogate biomarkers for RA synovial tissue phenotypes and efforts to reconstruct RA through novel spatial transcriptomics methods. Finally, we discuss the development of novel synovial tissue organoid systems as an experimental platform for precision medicine and therapeutic target validation.
II. Deconstructing rheumatoid arthritis through cellular profiling
The application of transcriptomic and cellular profiling technologies to RA joint tissue previously has focused on preselected cell types, surveyed whole tissues rather than disaggregated cells, or only used a single technology platform. Recently, rapid advances in transcriptomic profiling, cytometry, and spatial transcriptomics have enabled the unprecedented examination of cellular and molecular heterogeneity in RA blood and synovium. We begin the review by discussing key advances in cellular profiling techniques that together, have transformed our ability to examine rheumatoid arthritis at a single-cell resolution.
While fluorescence-based cytometric analysis of cell surface markers has long been the gold standard for immune cell profiling [8], mass cytometry overcomes some of the limitations of conventional fluorescence-based cytometry by utilizing antibodies conjugated to metals[9]. In studies that utilize mass cytometry for cellular profiling, cells are simultaneously labeled with a large panel of antibodies (>40 markers) that encompass key markers that convey information regarding cell lineage, cell type, cytokines, intracellular signaling, and epigenetic status. Moreover, cellular profiling by mass cytometry is scalable, making it feasible to apply mass cytometry to a large number of patient samples.
Transcriptomic profiling through RNA-sequencing (RNA-seq)[10] has enabled a highly detailed examination of gene expression. In particular, the advent of single-cell RNA-seq (scRNAseq)[11] has fundamentally changed the way scientists approach profiling of cell populations by enabling unbiased transcriptomic profiling without a priori knowledge of cell types as defined by known intracellular or surface markers. This unbiased approach is particularly appealing when applied to target tissues of inflammatory disease.. In 2018, Stephenson et al was one of the first groups to apply this technique to the rheumatoid arthritis synovium[12]. Using droplet-based scRNAseq, the authors profiled 20,387 synovial cells to identify 13 putative immune and stromal cell types, including T and NK lymphocyte subsets, B cell populations, and synovial fibroblast subsets[12].
III. Defining inflammatory cell states in RA at the single-cell resolution
The ability to leverage cutting-edge technologies to deconstruct chronic inflammatory diseases became possible as cellular profiling techniques has continued to mature. The overarching goal of the Accelerating Medicines Partnership (AMP) RA/SLE Consortium was to deconstruct inflammatory diseases by applying high-dimensional cellular profiling to organs and tissues affected by chronic inflammation. A major challenge for the network was to implement a standardized protocol for synovia tissue acquisition, processing, data generation, and data analysis across clinical sites. The AMP RA/SLE consortium addressed this issue by first testing different approaches to acquiring and processing synovial biopsy samples across multiple clinical sites[13]. A key advance that enabled synchronization of tissue processing and minimization of technical batch effects was the implementation of a robust fresh tissue synovial biopsy cryopreservation protocol[13]. The AMP RA/SLE consortium demonstrated that cryopreserved synovial tissue maintained high cell viability with intact transcriptomic profiles at a single-cell level[13]. Synovial cells obtained through a cryopreservation pipeline generated largely viable cells and were readily accessible for high-dimensional analyses, including mass cytometry and RNA-sequencing[13]. Next, the ability to recover robust cell yield from synovial tissue biopsies was critical to capture the synovial cellular heterogeneity from individual RA patients. The traditional tissue disaggregation methods utilize collagenase to dissociate cells from synovium which can result in cleavage of surface proteins and altered transcriptomic profiles due to prolonged tissue digestion time. Using a highly purified proteolytic enzyme combined with mechanical trituration, the AMP RA/SLE consortium developed an optimized synovial tissue dissociation protocol that delivers robust cell yield and enables high-quality transcriptomic analyses on a cell-by-cell levell[13].
The AMP RA/SLE consortium applied mass cytometry and scRNAeq to over 40 joint tissues from patients with RA or osteoarthritis (OA). Combining mass cytometry and scRNAseq, the consortium identified 18 unique cell states in RA synovia[14]. Here, we discuss some of the novel cell states in RA synovia uncovered through the cellular profiling approach to deconstructing RA (Figure 1).
Figure 1. Pathogenic cell states in RA.
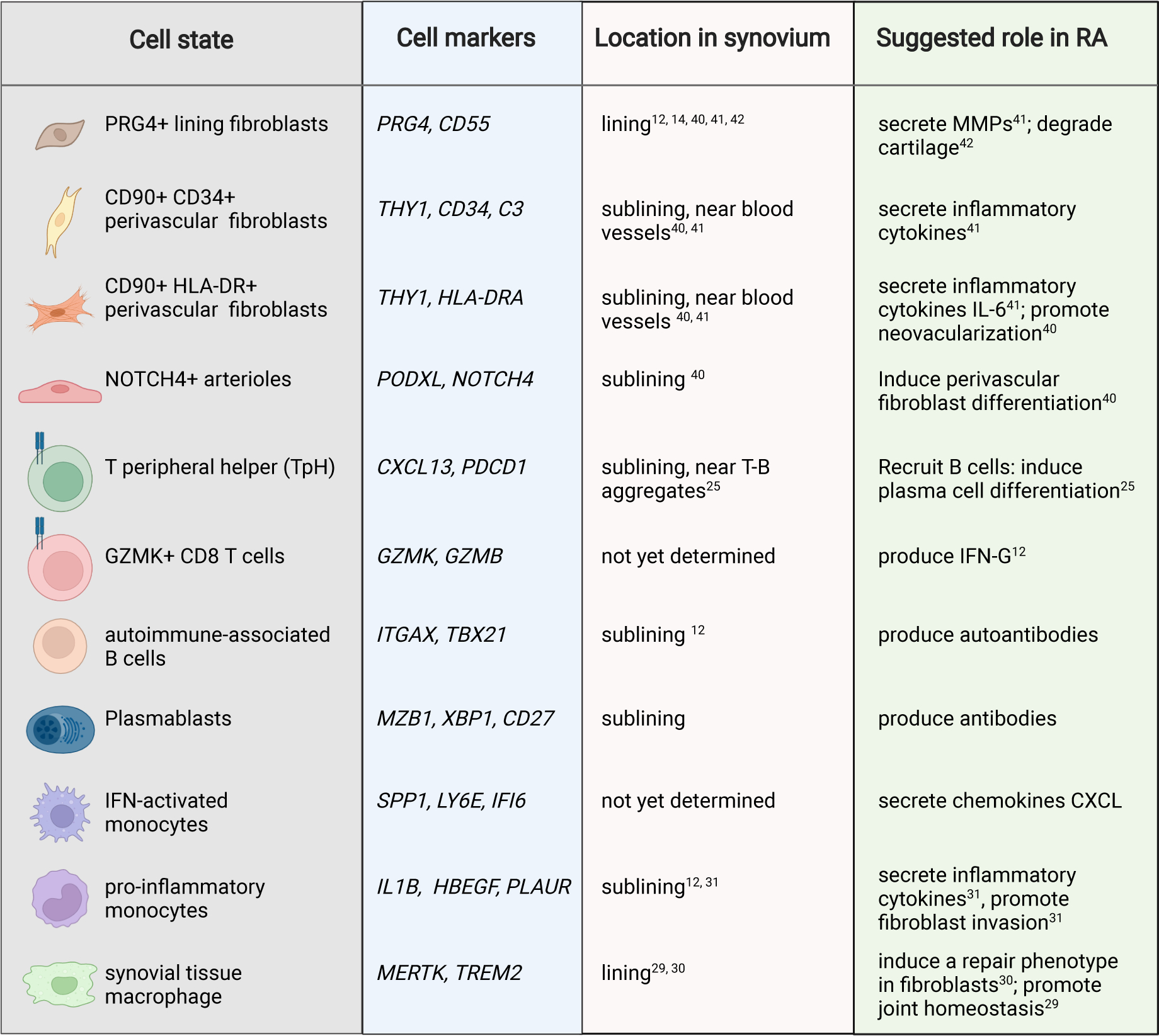
Summary of pathogenic cell states in RA synovia revealed by single-cell profiling. The specific marker genes for each cell state, their known sub-anatomical location within the synovium, and putative pathological functions are highlighted.
A. Pathogenic lymphocyte subsets expanded in RA synovium
The role of B cells in RA pathogenesis has long been appreciated[15] and the efficacy of B-cell depletion strategies strongly supports a role for B cells in driving disease pathogenesis in RA[16][17][18]. In the peripheral blood of RA patients, B cells exhibit abnormal phenotypes, characterized by an expansion of activated memory B cells populations[19][20]. By applying scRNAseq In RA synovia, the AMP RA/SLE consortium identified 4 distinct B cell states: plasmablasts, naive B cells, memory B cells, and autoimmune-associated B cells (ABCs) [14]. ABCs were first reported in aging mice and their presence has been documented in patients with various autoimmune diseases including SLE[21]. In RA, ABCs are highly expanded in patients with active disease and express markers of recently activated B cells [14].
T cell - B cell aggregates can be observed in synovial tissue of patients with seropositive RA[22]. While it has been long appreciated that T cells are important mediators of chronic inflammation in RA[23,24], it was not known until recently which T cell populations drive B cell aggregation and proliferation in RA synovium. In 2017, Rao et al reported a CD4+ PD-1hi CXCR5− T cell population that is highly expanded in synovial tissue, synovial fluid, and peripheral blood of seropositive RA patients[25]. In RA synovia, CD4+ PD-1hi CXCR5− T cells are located adjacent to B cells within lymphoid aggregates[25]. CD4+ PD-1hi CXCR5− T cells express high levels of IL-21 and CXCL13 and induce memory B cell differentiation into plasma cells in vitro[25]. Unlike T follicular helper cells, these cells lack CXCR5 and instead express CCR2 which enables homing of these cells to inflamed peripheral tissue[25]. Due to their ability to shape the B cell responses in peripheral tissues, these cells have been described as T peripheral helper (Tph) cells. Subsequent studies have shown that T cells that resemble Tph are expanded in lupus kidneys[25–27] and celiac disease[28].
In addition to the identification of ABCs and TpH cells, cellular profiling of lymphocytes by scRNAseq also revealed CD8+ T cells as key producers of inflammatory cytokines interferon-gamma and TNF [14]. Interestingly, transcriptomic analysis of CD8+ T cells suggested distinct CD8+ T cell subsets characterized by differential expression of GZMK and GZMB[14]. Further work is warranted to gain insights into the function and origin of GZMK+ CD8+ T cells in RA.
B. Distinct synovial macrophages drive joint inflammation and disease remission
The healthy synovial lining consists of lining fibroblasts and resident macrophages. A recent study in mice demonstrated that synovial lining macrophages are derived from embryonic precursors which populate the synovium during embryogenesis[29]. Synovial lining macrophages renew locally, forming a protective epithelial-like barrier that is disrupted in experimental arthritis[29]. Recent scRNAseq analyses of monocytes and macrophages have shown tremendous heterogeneity among the myeloid compartment in RA synovia [29][30][14,31]. Among the myeloid compartment, two inflammatory monocyte populations were significantly expanded in RA patients with highly inflamed synovium: one myeloid population characterized by high expression of IL1B and HBEGF, and another characterized by an interferon activation signature[14,31]. The IL1B+ HBEGF+ inflammatory macrophages promoted fibroblast invasiveness in vitro[31], suggesting a potential role for IL-1B+ HBEGF+ inflammatory macrophages in driving fibroblast-mediated joint destruction. In a separate study, Alivernini et al examined gene expression profiles of synovial tissue macrophages across patients with early/active RA, treatment-refractory/active RA, and RA in sustained remission[30]. This study highlighted a population of synovial tissue macrophages uniquely expanded during disease remission. These macrophages, characterized by the expression of MERTK and TREM2, produce lipid mediators that suppress inflammation and induce a reparative response in synovial fibroblasts[30]. Interestingly, a reduced frequency of these macrophages in remission was associated with an increased risk of disease flare[30], suggesting a role for local tissue macrophages in sustaining remission. Together, these studies highlight functionally distinct macrophages and monocytes in the synovium.
C. Sublining fibroblast expansion in RA
Fibroblasts are mesenchymal cells that maintain normal organ tissue function[32,33]. Synovial fibroblasts are mesenchymal cells in the joint that form the lining membrane, secret proteoglycans to promote normal joint functions[34][35]. In RA, synovial fibroblasts take on pathological roles that contribute to joint damage by recruiting leukocytes, promoting neoangiogenesis, and degrading cartilage[34][33,35]. Advances in cellular profiling technologies have provided an unprecedented look at fibroblast heterogeneity in chronic inflammatory diseases[35], including inflammatory bowel disease[36–39], RA [40][14,41,42], pulmonary fibrosis[43][44], and cancer[45,46]. To identify pathological fibroblast subsets in RA synovia, Mizoguchi et al[41] applied flow cytometry followed transcriptomic analysis and identified 3 putative synovial fibroblasts subtypes: CD90- CD34- lining fibroblasts, CD90+ CD34+ sublinig fibroblasts, and CD90+ CD34- sublining fibroblasts. Of these three fibroblast subsets, CD90+ subling fibroblasts were highly expanded in RA and correlated with the severity of synovial tissue inflammation[41]. The concept of synovial fibroblast heterogeneity and the expansion of sublining fibroblasts in RA synovia were confirmed in separate studies from the AMP RA/SLE consortium. In the first study, integration of mass cytometry and scRNAseq identified 4 synovial fibroblast subsets: CD90- lining fibroblasts, CD90+CD34+ subliing fibroblast population, CD90+ HLA-DRA+ fibroblasts, and DKK3+ fibroblasts[14]. Consistent with an inflammatory phenotype, CD90+ HLA-DR+ fibroblasts express high levels of IL-6, CCL2, and CXCL12[14]. Reflecting distinct regional functional compartmentalization between the lining and sublining fibroblasts, sublining fibroblasts secrete inflammatory cytokines whereas lining fibroblasts produces matrix-remodeling MMP1 and MMP3[41]. In an animal model of inflammatory arthritis, it was recently shown that CD90+ sublining fibroblasts undergo expansion in joints of arthritis mice[42]. Interestingly, adoptive transfer of CD90+ sublining fibroblasts into arthritic mice worsens joint inflammation, whereas adoptive transfer of CD90- lining fibroblasts worsened cartilage and bone erosion[42]. These results suggest that lining and sublining fibroblasts take on distinct pathological functions in inflammatory arthritis.
What drives the functional polarization of synovial fibroblasts? The enrichment of interferon-stimulated genes (ISGs) in CD90+HLA-DR fibroblasts suggests that the inflammatory phenotype is in part driven by interferon signaling[14][47]. Surprisingly, trajectory analysis of synovial fibroblasts suggests that lining and sublining are transcriptionally connected along an anatomical continuum, from synovial lining to the sublining vasculature[40]. We recently identified vascular endothelium-derived Notch signaling as a key driver of CD90+ sublining fibroblast differentiation[40]. Inhibition of Notch receptor signaling in a synovial organoid model abrogated CD90+ fibroblast differentiation, suggesting a critical role for endothelium-derived signals in driving fibroblast positional identity[40]. Together, these observations suggest disease-specific alterations in the fibroblast compartment.
D. Angiogenesis and angiocrine factors in RA.
Endothelial cells that line blood vessels have traditionally been perceived as passive, structural units that provide blood flow to target tissues. However, recent studies suggest that specialized endothelial cells found in niche tissue microenvironments actively orchestrate tissue remodeling through paracrine secretion of morphogenic signals termed angiocrine factors [48]. Across different organs and tissues, specialized capillary endothelial cells secrete angiocrine factors that act as morphogens to determine the shape, architecture, size, and patterning of developing and regenerating tissues[48]. In inflammatory diseases such as RA, the formation of new blood vessels, or angiogenesis, is critical for supplying oxygen and nutrients to immune and stromal cells in inflamed tissue[49]. In inflamed tissues, hypoxia induces the production of proangiogenic factors, including vascular endothelial growth factor (VEGF), placenta growth factor (PGF), fibroblast growth factor (FGF) which in turn induce angiogenesis, neovascularization, and remodeling of the surrounding tissue[49].
The concept of an angiocrine endothelium-mediated synovial tissue remodeling in RA was recently described by our group[40]. Unbiased scRNAseq analysis of synovial endothelial cells from RA patients suggests that synovial endothelial cells exist along the arterial to venous continuum[40]. Arterial endothelial cells are characterized by the expression of PODXL and NOTCH4, whereas venous endothelial cells can be identified by the expression of CLU and ACKR1[40]. Interestingly, Notch ligands delta-like ligand 4 (DLL4) and Jagged-1 (JAG1) were highly expressed by arterial endothelial cells. Endothelial-derived Notch signaling directs the maturation of undifferentiated fibroblasts into mural cells and sublining fibroblasts, leading to sublining expansion in RA. Beyond Notch signaling, synovial endothelial cells express many growth factors, cytokines, and chemokines that together help shape the inflammatory response in RA[40,49]. The identity of additional angiocrine factors in RA and their role in synovial tissue remodeling remains to be defined.
IV. Key steps towards reconstructing rheumatoid arthritis
As discussed above, recent studies utilizing cellular profiling techniques have provided new and exciting insights into key cellular changes relevant to RA pathogenesis. If we consider the overall approach of defining key cell states and molecular pathways in RA as an effort to “deconstruct” RA, what would be the approaches to “reconstruct” RA? As the “disease reconstruction” approach in RA continues to bring forward new insights into RA pathogenesis, it is important to consider the opportunities as well as challenges that are needed to overcome to translate these insights into practical, operational components of clinical therapeutic decision making. The first opportunity is to apply the cellular and molecular data to improve our understanding of RA disease heterogeneity and to guide RA treatment. By designing studies that combine cellular profiling data with deep clinical phenotyping, it may be possible to define distinct RA phenotypes, identify biomarkers that correlate with distinct RA phenotypes, and predict treatment response, leading to a more personalized treatment strategy (Figure 2). The second opportunity is to leverage the rich cellular profiling data to define novel therapies targeting specific cell populations or pathways that are pathologically enriched in RA. Below we discuss ongoing efforts to define RA phenotypes and identify biomarkers that predict treatment response. We also highlight novel experimental approaches that could expedite drug discovery and facilitate target validation in RA. Together, these approaches are critically needed to translate discoveries into precision medicine in RA (Figure 2).
Figure 2. Precision in RA: from disease deconstruction of disease reconstruction.
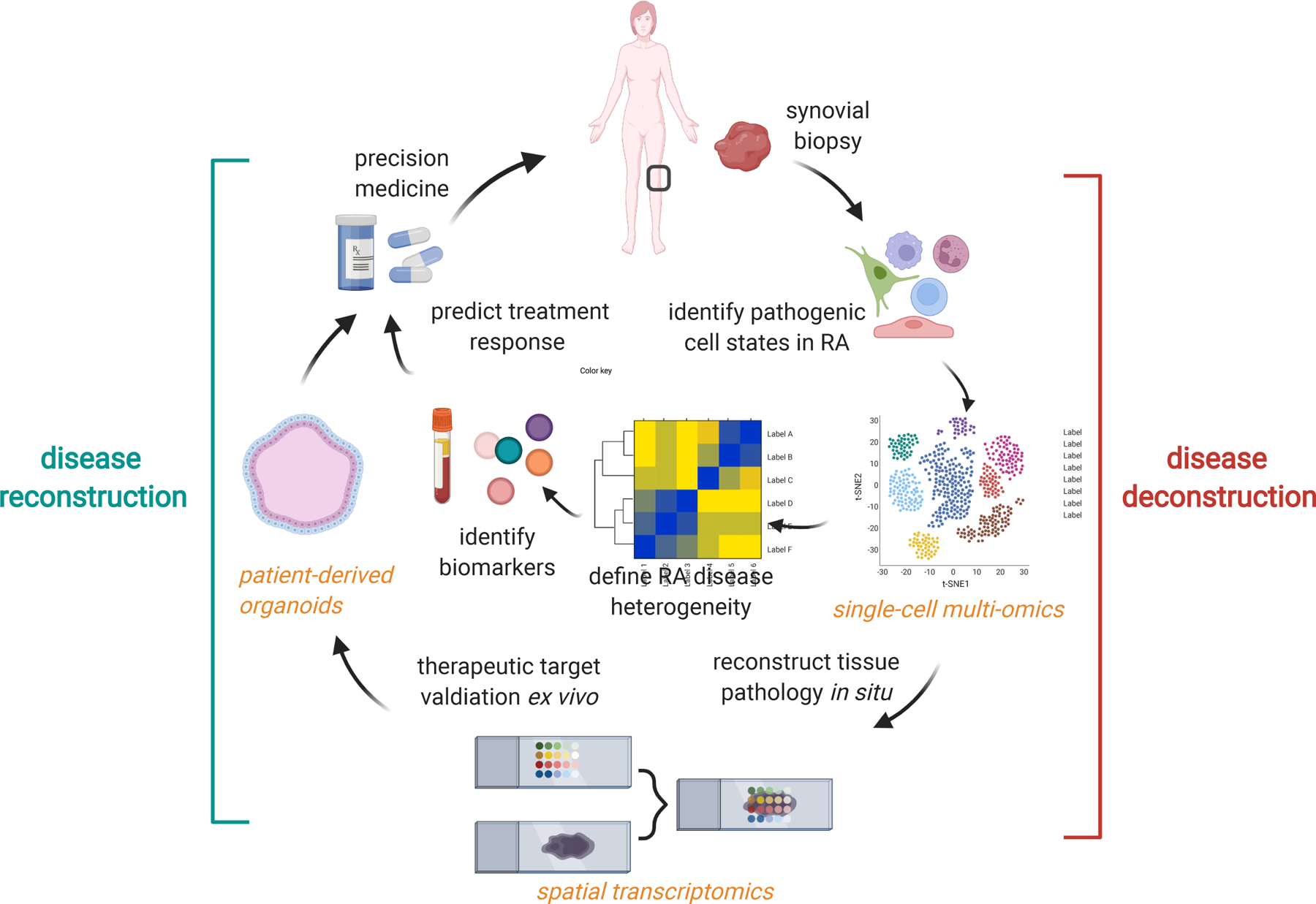
In disease deconstruction (right), synovial biopsy followed by single-cell multi-omics profiling is the first step towards understanding cellular heterogeneity in RA. By integrating synovial cellular profiles with clinical phenotypes, we can begin to define RA phenotypes through molecular taxonomy. Several novel approaches are needed for RA reconstruction (left). First, spatially-resolved transcriptomics could provide new insights into pathological cellular interactions in RA synovia and identify new therapeutic targets. The development of experimental models that retain patient-specific phenotypes (i.e. patient-derived synovial organoids) could provide a platform to test treatment-response and validate new drug targets. Finally, surrogate biomarkers that correlate with distinct RA phenotypes and predict treatment response will be needed to guide clinical decision-making.
A. Biomarkers as key tools towards personalized medicine in RA.
A common theme for personalized medicine is the capability to accurately predict responses to therapies that take into account each patient’s unique characteristics, disease severity, and the likelihood of response to specific targeted therapies [50]. To do this, a critical tool needed to guide personalized therapy in RA is biomarkers that capture each patient’s unique biology, disease phenotype, and response to treatment. Current strategies towards biomarker discovery in RA rely on the utilization of proteomic, genomic, or metabolomic approaches[51]. Despite intense efforts over the past decades to identify markers that predict response to therapy in RA, currently, there are no biomarkers that accurately predict an individual’s response to targeted therapy [50][51–53]. Nevertheless, recent advances in -omic technologies and clinical study design provide exciting opportunities in the space of RA biomarker discovery. Here, we discuss recent studies that utilized -omic approaches to identify novel molecular and cellular biomarkers that correlate with disease phenotypes or predict treatment response.
At the histopathological level, RA synovium exhibit marked heterogeneity at an individual level with regards to the degree of inflammation[54], lining hyperplasia[54], inflammatory cellular composition[55,56], presence of lymphoid aggregates[55,57]. In a recent study of RA phenotype heterogeneity[5], Lewis et al applied RNAseq to synovial tissue and peripheral blood to a large cohort of early, treatment-naive RA patients. Unbiased transcriptomic analysis identified 3 distinct RA phenotypes: fibroblast-enriched macrophage-enriched, and lymphoid-myeloid-enriched phenotypes[5]. Compared to the histologic assessment of RA phenotypes, RNAseq-based stratification of RA phenotypes demonstrated an improvement in predicting therapeutic response[58]. Consistently, several recent studies have similarly shown that synovial tissue phenotype correlates with response to biologics [59][3,5]. To translate the concept of RA synovial tissue phenotype predicting treatment response into routine clinical practice, a surrogate peripheral marker that accurately predicts RA synovial tissue phenotype would be needed since synovial tissue biopsy is not routinely performed as part of the RA management in many countries, including the United States. Transcriptomic profiling of peripheral blood from patients is an attractive approach since peripheral blood is collected routinely clinically for diagnostic and monitoring for drug toxicities. This approach also leverages the relative ease of RNA isolation and preservation from whole blood and rapid advances in deep gene expression profiling techniques [53]. Interestingly, whereas the synovial tissue RNAseq revealed marked transcriptional differences across the 3 RA phenotypes, blood RNAseq exhibited mild differences across distinct RA phenotypes, with type I interferon response genes enriched in the lymphoid-myeloid phenotype[5]. This observation reflects that, compared to the peripheral blood, there is marked variability and heterogeneity in the synovial tissue of patients with RA with regards to the tissue cellular composition. Likely, the local signals from the synovial tissue microenvironment[31,40] are crucial in shaping the tissue inflammatory response across individuals with RA.
In a separate study, Orange et al [60] applied a novel approach utilizing transcriptomic analysis of peripheral blood to identify RNA signatures of flare in patients with established RA. Leveraging weekly home collection of blood in RA patients followed by transcriptomic profiling, Orange et al. established a longitudinal analysis of gene expression changes in the peripheral blood over 364-time points. Using this approach, they identified a gene expression signature consistent with B-cell activation, followed by mesenchymal cell gene signature, in the blood of RA patients before RA disease flare[60]. Interestingly, the mesenchymal gene signature observed from the peripheral blood of these patients is similar to the transcriptomic profiles of synovial fibroblasts in inflamed joints, raising the possibility of circulating mesenchymal cells in peripheral blood of RA patients before disease flares. While additional studies are needed to verify the tissue origin of the mesenchymal cell signature in these patients, this study nevertheless illustrated a novel concept of longitudinal genomics analysis as a strategy to identify surrogate biomarkers in RA patients.
Proteomic technologies enable large-scale profiling of proteins in patient samples[61]. Applying proteomic technologies to identify biomarkers in RA to improve diagnosis and management is an area of intense investigation [62]. The recent discovery of the “pre-RA” stage[63] [64]has led to the identification of autoantibodies against cyclic citrullinated peptides (anti-CCP antibodies) that may predict RA onset in individuals with a high risk for developing RA[65][66] [52] [51]. Identification of precise biomarkers in the pre-RA phase will be crucial for success in clinical trials aimed at preventing RA.
Metabolomics, the comprehensive analysis of metabolites in a biological specimen, is emerging as a powerful tool in precision medicine[67]. Applied to inflammatory arthritis, investigators utilizing metabolomic profiling have identified distinct metabolomic and lipidomic signatures for seronegative RA and psoriatic arthritis[68], RA versus reactive arthritis[69], altered glucose[70–72], and choline[73] metabolism in RA synovial fibroblasts. Utilizing a lipid profiling approach, Gomez et al reported that the concentrations of pro-resolving mediators in the peripheral blood of RA patients predict the responsiveness of DMARD treatment[74]. Furthermore, the levels of baseline lipid mediators are correlated with distinct RA synovial tissue phenotypes[74], indicating lipid profiling of the peripheral blood may serve as a surrogate biomarker for distinct RA phenotypes.
Together, these approaches illustrate that the combination of the cutting-edge omic approach with the deep phenotypic characterization of patients can lead to the novel discovery of peripheral blood markers that may help stratify RA patients, predict response to therapy, and predict disease flares. As single-cell multi-omic techniques continue to improve by incorporating new measurement parameters, including surface proteomics[75], metabolomics[76], and epigenetics[77], our ability to define specific biomarkers in RA will undoubtedly continue to improve. Furthermore, cellular profiling techniques that provide precise identification of cellular sources of relevant biomarkers may provide additional insight into RA disease pathogenesis and disease heterogeneity.
B. Spatial reconstruction of rheumatoid arthritis
Synovitis in RA involves complex cellular interactions between infiltrating immune cells and tissue-resident stromal cells and macrophages that lead to pathology and joint damage. Deconstructing RA synovitis using single-cell technologies has revealed previously underappreciated cellular heterogeneity in RA synovia[14,25,31,40–42]. However, the structural and organizational principles of synovitis in RA remain incompletely understood due to the lack of spatial context and understanding of the architectural design of the microenvironments within inflamed RA synovia. An important next step towards improving our understanding of the tissue context in chronic inflammation is to reconstruct synovitis by determining the spatial localization of key immune and stromal cells within RA synovia.
The ability to examine gene expression directly in the context of tissue anatomy is central to understanding disease pathology. Traditional imaging techniques, including immunohistochemistry and in situ hybridization are typically capable of detecting a few genes or protein markers of interest. Recently, the development of advanced multiplexed single-molecule fluorescence in situ hybridization (smFISH)[78] and spatial transcriptomic assays[79] offer exciting possibilities of measuring gene expression directly on tissue sections. Techniques such as spatial transcriptomics[80], MERFISH[81], Slide-seq[82], stereo-seq[83], enable the measurement of hundreds to thousands of genes simultaneously across a tissue section while retaining tissue architecture. Studies applying spatial transcriptomics to RA have begun to emerge over the past two years. In 2019, Carlberg et al. applied spatial transcriptomics to RA and psoriatic arthritis (PsA) synovia and identified an expanded adaptive immune response profile in RA, compared to PsA[84]. As spatial transcriptomic technologies continue to improve concerning spatial resolution and sensitivity, more in-depth studies of RA synovial tissue architecture will no doubt emerge.
C. Patient-derived synovial organoids as an experimental platform for target validation.
As the disease deconstruction approach reveals new disease-associated cell states and pathogenic pathways, there is now an exciting opportunity to identify novel therapeutic targets in RA. A key challenge in target validation is defining experimental systems that faithfully recapitulate synovial tissue pathology in RA. While several animal models of inflammatory arthritis exist and have been crucial in improving our understanding of arthritis pathology[85], there is currently no animal model of inflammatory arthritis that completely recapitulates all aspects of RA pathogenesis or joint pathology. Therefore, experimental systems that utilize patient-derived, organotypic models that recapitulate RA synovitis is highly desirable. In 2010, Kiener et al reported the first in vitro model of synovial lining membrane formation[86]. This “micromass” system leverages the ability of synovial fibroblast to self-assemble into compacted lining membrane when cultured in an extracellular matrix[86]. Since the initial report of the method, the micromass system has been widely used by RA investigators to examine the role of adhesion molecule Cadherin-11 during lining formation and arthritis pathology[87], fibroblast invasion[88], and inflammatory cytokine activation[89].
Recently, the observation that sublining fibroblasts are localized around synovial vascular endothelium led us to hypothesize that endothelial cells are crucial in directing sublining fibroblast differentiation[40]. To reconstruct the synovial sublining, we developed a fibroblast plus endothelial cell organoid system where endothelial cells self-assemble into vascular tubules within the sublining compartment [40]. The formation of vascular tubules in synovial organoids then provides positional cues for fibroblasts, leading to fibroblast differentiation towards a sublining fibroblast phenotype in a NOTCH-dependent manner[40]. This de novo approach relies on the principle of mixing synovial parenchymal cells in a 3-dimensional matrix and then allowing them to self-assemble into their native architecture through homo- and hetero-typic cell-cell contacts. Additional work is needed to determine if this synovial organoid system can be used to determine differentiation cues for other synovial cell types, such as synovial tissue macrophages. Nevertheless, the prospect of using a reconstitution approach to build synovium in a dish is appealing because of its simplicity and the ease of tailoring the organoid system to model specific cell-cell interactions in the synovium.
One limitation of the reconstitution approach to reconstruct the synovium is that these systems often rely on cultured primary cells, often from different donors. The ability to retain patient-specific biology ex vivo is critical for leveraging organoid models for personalized medicine. In oncology, the recent development of patient-derived organoids (PDOs) has accelerated the ability to examine variation in patient response to targeted therapies [90–92]. Tumor PDOs utilize an “en-bloc” approach where tissue can be maintained ex vivo as organoids to assess response to therapeutic modulation[93]. While there has been tremendous technical development of PDOs for cancer therapeutics, little is known if synovial tissue, patient-derived organoids could be leveraged to guide RA therapeutics. The concept for developing synovial tissue PDOs is quite appealing: If a synovial tissue PDO can be developed to preserve the original cellular composition and the native synovial tissue microenvironment unique to each RA patient, synovial tissue PDOs may provide an opportunity to validate therapeutic targets and to determine the therapeutic response to existing therapies. Furthermore, if synovial tissue PDOs can be developed from synovial biopsies, it would provide a powerful platform for which novel therapeutics could be tested directly in RA patient tissues.
IV. Summary
Rheumatoid arthritis is a chronic autoimmune disease that causes significant morbidity. Emerging cellular profiling data from synovial tissue biopsies have revealed novel, pathogenic cell states in RA synovia and that distinct synovial cellular phenotypes exist in RA. There is an urgent unmet need to better understand the clinical implication of RA synovial tissue phenotypes with regards to disease prognosis and therapeutic response to targeted therapies. Defining surrogate biomarkers that could be used to predict RA synovial tissue phenotypes will be critical towards translating these findings into actionable insights in clinical practice. Precision medicine in RA also necessitates the development of novel experimental models that retain each patient’s characteristics. Patient-derived organoid system is a promising approach that may provide a platform to validate therapeutic targets and to determine the therapeutic response to existing therapies.
Practice points
Rheumatoid arthritis (RA) is a heterogeneous disease defined by distinct synovial tissue phenotypes
Emerging evidence for distinct RA phenotypes creates an urgent unmet need to determine how each RA phenotype affects the likelihood of patients’ response to targeted therapies.
Research Agenda
Cellular profiling through single-cell RNAseq enabled RA “disease deconstruction” which has led to the identification of several pathogenic cell states expanded in RA.
Spatial transcriptomic techniques are promising approaches to “reconstruct” RA pathology by defining the anatomical localization of pathogenic cell states in RA synovia
Biomarkers that correlate with RA phenotypes and predict response to targeted therapies are needed for precision medicine in RA
The development of patient-derived synovial tissue organoids will provide a novel platform for drug discovery and target validation in RA.
Acknowledgments
We would like to thank the patients and their families for their invaluable contributions that together help improve our knowledge of rheumatoid arthritis. We would like to thank members of the Accelerating Medicines Partnership (AMP) RA/SLE Consortium for the opportunity to participate in the network. Finally, we would like to thank members of the Wei laboratory, Michael Brenner laboratory, and Soumya Raychaudhuri laboratory for their helpful discussion and insights.
Conflict of interest statement
K.W. serves as a consultant for Mestag, Inc and Gilead Sciences, Inc. K.W. is supported by a sponsored research agreement from Gilead Sciences, Inc.
Funding statement
K.W. is supported by a National Institute of Arthritis and Musculoskeletal and Skin Diseases award (K08AR077037), a Rheumatology Research Foundation Innovative Research award, and a Burroughs Wellcome Fund Career Award for Medical Scientists.
K.B. is supported by a microgrant from the BWH/BCH Joint Biology Consoritum (NIH P30 AR070253)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Kartik Bhamidipati, Department of Medicine, Division of Rheumatology, Inflammation and Immunity, Brigham and Women’s Hospital, Harvard Medical School..
Kevin Wei, Department of Medicine, Division of Rheumatology, Inflammation and Immunity, Brigham and Women’s Hospital, Harvard Medical School.; Center for Cellular Profiling - Single Cell Multiomics Core, Brigham and Women’s Hospital Associate Physician, Brigham and Women’s Hospital
References
- 1.Smolen JS, Aletaha D, Barton A, Burmester GR, Emery P, Firestein GS, et al. Rheumatoid arthritis. Nat Rev Dis Primers. 2018. Feb 8;4:18001. [DOI] [PubMed] [Google Scholar]
- 2.McInnes IB, Schett G. The pathogenesis of rheumatoid arthritis. N Engl J Med. 2011. Dec 8;365(23):2205–19. [DOI] [PubMed] [Google Scholar]
- 3.Lliso-Ribera G, Humby F, Lewis M, Nerviani A, Mauro D, Rivellese F, et al. Synovial tissue signatures enhance clinical classification and prognostic/treatment response algorithms in early inflammatory arthritis and predict requirement for subsequent biological therapy: results from the pathobiology of early arthritis cohort (PEAC). Ann Rheum Dis. 2019. Dec;78(12):1642–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Romão VC, Humby F, Kelly S, Di Cicco M, Mahto A, Lazarou I, et al. Treatment-resistant synovitis and radiographic progression are increased in elderly-onset rheumatoid arthritis patients: findings from a prospective observational longitudinal early arthritis cohort study. Semin Arthritis Rheum. 2020. Aug;50(4):735–43. [DOI] [PubMed] [Google Scholar]
- 5.Lewis MJ, Barnes MR, Blighe K, Goldmann K, Rana S, Hackney JA, et al. Molecular Portraits of Early Rheumatoid Arthritis Identify Clinical and Treatment Response Phenotypes. Cell Rep. 2019. Aug 27;28(9):2455–70.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sparks JA, Jin Y, Cho S-K, Vine S, Desai R, Doyle TJ, et al. Prevalence, incidence and cause-specific mortality of rheumatoid arthritis-associated interstitial lung disease among older rheumatoid arthritis patients. Rheumatology. 2021. Aug 2;60(8):3689–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McDermott GC, Doyle TJ, Sparks JA. Interstitial lung disease throughout the rheumatoid arthritis disease course. Curr Opin Rheumatol. 2021. May 1;33(3):284–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perfetto SP, Chattopadhyay PK, Roederer M. Seventeen-colour flow cytometry: unravelling the immune system. Nat Rev Immunol. 2004. Aug;4(8):648–55. [DOI] [PubMed] [Google Scholar]
- 9.Spitzer MH, Nolan GP. Mass Cytometry: Single Cells, Many Features. Cell. 2016. May 5;165(4):780–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009. Jan;10(1):57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015. May 21;161(5):1202–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stephenson W, Donlin LT, Butler A, Rozo C, Bracken B, Rashidfarrokhi A, et al. Single-cell RNA-seq of rheumatoid arthritis synovial tissue using low-cost microfluidic instrumentation. Nat Commun. 2018. Feb 23;9(1):791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donlin LT, Rao DA, Wei K, Slowikowski K, McGeachy MJ, Turner JD, et al. Methods for high-dimensional analysis of cells dissociated from cryopreserved synovial tissue. Arthritis Res Ther. 2018. Jul 11;20(1):139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang F, Wei K, Slowikowski K, Fonseka CY, Rao DA, Kelly S, et al. Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat Immunol. 2019. Jul;20(7):928–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silverman GJ, Carson DA. Roles of B cells in rheumatoid arthritis. Arthritis Res Ther. 2003. Dec 2;5 Suppl 4:S1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anolik JH, Barnard J, Cappione A, Pugh-Bernard AE, Felgar RE, Looney RJ, et al. Rituximab improves peripheral B cell abnormalities in human systemic lupus erythematosus. Arthritis Rheum. 2004. Nov;50(11):3580–90. [DOI] [PubMed] [Google Scholar]
- 17.Edwards JC, Cambridge G. Sustained improvement in rheumatoid arthritis following a protocol designed to deplete B lymphocytes. Rheumatology. 2001. Feb;40(2):205–11. [DOI] [PubMed] [Google Scholar]
- 18.Edwards JCW, Szczepanski L, Szechinski J, Filipowicz-Sosnowska A, Emery P, Close DR, et al. Efficacy of B-cell-targeted therapy with rituximab in patients with rheumatoid arthritis. N Engl J Med. 2004. Jun 17;350(25):2572–81. [DOI] [PubMed] [Google Scholar]
- 19.Adlowitz DG, Barnard J, Biear JN, Cistrone C, Owen T, Wang W, et al. Expansion of Activated Peripheral Blood Memory B Cells in Rheumatoid Arthritis, Impact of B Cell Depletion Therapy, and Biomarkers of Response. PLoS One. 2015. Jun 5;10(6):e0128269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meednu N, Barnard J, Callahan K, Coca A, Marston B, Thiele R, et al. Activated peripheral blood B cells in rheumatoid arthritis and relationship to anti-TNF treatment and response: randomized clinical trial for anti-TNF effects on B cells. Arthritis Rheumatol [Internet]. 2021. Aug 4; Available from: 10.1002/art.41941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang S, Autoimmunity Molecular Medicine Team, Wang J, Kumar V, Karnell JL, Naiman B, et al. IL-21 drives expansion and plasma cell differentiation of autoreactive CD11chiT-bet B cells in SLE [Internet]. Vol. 9, Nature Communications. 2018. Available from: 10.1038/s41467-018-03750-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orr C, Najm A, Biniecka M, McGarry T, Ng C-T, Young F, et al. Synovial Immunophenotype and Anti-Citrullinated Peptide Antibodies in Rheumatoid Arthritis Patients: Relationship to Treatment Response and Radiologic Prognosis. Arthritis Rheumatol. 2017. Nov;69(11):2114–23. [DOI] [PubMed] [Google Scholar]
- 23.Cope AP, Schulze-Koops H, Aringer M. The central role of T cells in rheumatoid arthritis. Clin Exp Rheumatol. 2007. Sep;25(5 Suppl 46):S4–11. [PubMed] [Google Scholar]
- 24.Rao DA. T Cells That Help B Cells in Chronically Inflamed Tissues. Front Immunol. 2018. Aug 23;9:1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rao DA, Gurish MF, Marshall JL, Slowikowski K, Fonseka CY, Liu Y, et al. Pathologically expanded peripheral T helper cell subset drives B cells in rheumatoid arthritis. Nature. 2017. Feb 1;542(7639):110–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bocharnikov AV, Keegan J, Wacleche VS, Cao Y, Fonseka CY, Wang G, et al. PD-1hiCXCR5- T peripheral helper cells promote B cell responses in lupus via MAF and IL-21. JCI Insight [Internet]. 2019. Oct 17;4(20). Available from: 10.1172/jci.insight.130062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arazi A, Rao DA, Berthier CC, Davidson A, Liu Y, Hoover PJ, et al. The immune cell landscape in kidneys of patients with lupus nephritis. Nat Immunol. 2019. Jul;20(7):902–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christophersen A, Lund EG, Snir O, Solà E, Kanduri C, Dahal-Koirala S, et al. Distinct phenotype of CD4 T cells driving celiac disease identified in multiple autoimmune conditions. Nat Med. 2019. May;25(5):734–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Culemann S, Grüneboom A, Nicolás-Ávila JÁ, Weidner D, Lämmle KF, Rothe T, et al. Locally renewing resident synovial macrophages provide a protective barrier for the joint. Nature. 2019. Aug;572(7771):670–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alivernini S, MacDonald L, Elmesmari A, Finlay S, Tolusso B, Gigante MR, et al. Distinct synovial tissue macrophage subsets regulate inflammation and remission in rheumatoid arthritis. Nat Med. 2020. Aug;26(8):1295–306. [DOI] [PubMed] [Google Scholar]
- 31.Kuo D, Ding J, Cohn IS, Zhang F, Wei K, Rao DA, et al. HBEGF macrophages in rheumatoid arthritis induce fibroblast invasiveness. Sci Transl Med [Internet]. 2019. May 8;11(491). Available from: 10.1126/scitranslmed.aau8587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koliaraki V, Prados A, Armaka M, Kollias G. The mesenchymal context in inflammation, immunity and cancer. Nat Immunol. 2020. Sep;21(9):974–82. [DOI] [PubMed] [Google Scholar]
- 33.Davidson S, Coles M, Thomas T, Kollias G, Ludewig B, Turley S, et al. Fibroblasts as immune regulators in infection, inflammation and cancer. Nat Rev Immunol. 2021. Nov;21(11):704–17. [DOI] [PubMed] [Google Scholar]
- 34.Nygaard G, Firestein GS. Restoring synovial homeostasis in rheumatoid arthritis by targeting fibroblast-like synoviocytes. Nat Rev Rheumatol. 2020. Jun;16(6):316–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei K, Nguyen HN, Brenner MB. Fibroblast pathology in inflammatory diseases. J Clin Invest [Internet]. 2021. Oct 15;131(20). Available from: 10.1172/JCI149538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parikh K, Antanaviciute A, Fawkner-Corbett D, Jagielowicz M, Aulicino A, Lagerholm C, et al. Colonic epithelial cell diversity in health and inflammatory bowel disease. Nature. 2019. Mar;567(7746):49–55. [DOI] [PubMed] [Google Scholar]
- 37.Kinchen J, Chen HH, Parikh K, Antanaviciute A, Jagielowicz M, Fawkner-Corbett D, et al. Structural Remodeling of the Human Colonic Mesenchyme in Inflammatory Bowel Disease. Cell. 2018. Oct 4;175(2):372–86.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smillie CS, Biton M, Ordovas-Montanes J, Sullivan KM, Burgin G, Graham DB, et al. Intra- and Intercellular Rewiring of the Human Colon during Ulcerative Colitis. Cell. 2019. Jul 25;178(3):714–30.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Friedrich M, Pohin M, Jackson MA, Korsunsky I, Bullers SJ, Rue-Albrecht K, et al. IL-1-driven stromal-neutrophil interactions define a subset of patients with inflammatory bowel disease that does not respond to therapies. Nat Med [Internet]. 2021. Oct 21; Available from: 10.1038/s41591-021-01520-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei K, Korsunsky I, Marshall JL, Gao A, Watts GFM, Major T, et al. Notch signalling drives synovial fibroblast identity and arthritis pathology. Nature. 2020. Jun;582(7811):259–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mizoguchi F, Slowikowski K, Wei K, Marshall JL, Rao DA, Chang SK, et al. Functionally distinct disease-associated fibroblast subsets in rheumatoid arthritis. Nat Commun. 2018. Feb 23;9(1):789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Croft AP, Campos J, Jansen K, Turner JD, Marshall J, Attar M, et al. Distinct fibroblast subsets drive inflammation and damage in arthritis. Nature. 2019. Jun;570(7760):246–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adams TS, Schupp JC, Poli S, Ayaub EA, Neumark N, Ahangari F, et al. Single-cell RNA-seq reveals ectopic and aberrant lung-resident cell populations in idiopathic pulmonary fibrosis. Sci Adv. 2020. Jul;6(28):eaba1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang KY, Petretto E. Cross-species integration of single-cell RNA-seq resolved alveolar-epithelial transitional states in idiopathic pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol. 2021. Sep 1;321(3):L491–506. [DOI] [PubMed] [Google Scholar]
- 45.Buechler MB, Pradhan RN, Krishnamurty AT, Cox C, Calviello AK, Wang AW, et al. Cross-tissue organization of the fibroblast lineage. Nature. 2021. May;593(7860):575–9. [DOI] [PubMed] [Google Scholar]
- 46.Hornburg M, Desbois M, Lu S, Guan Y, Lo AA, Kaufman S, et al. Single-cell dissection of cellular components and interactions shaping the tumor immune phenotypes in ovarian cancer. Cancer Cell. 2021. Jul 12;39(7):928–44.e6. [DOI] [PubMed] [Google Scholar]
- 47.Zhao S, Grieshaber-Bouyer R, Rao DA, Kolb P, Chen H, Andreeva I, et al. JAK inhibition prevents the induction of pro-inflammatory HLA-DR CD90 RA synovial fibroblasts by IFNɣ. Arthritis Rheumatol [Internet]. 2021. Aug 25; Available from: 10.1002/art.41958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rafii S, Butler JM, Ding B-S. Angiocrine functions of organ-specific endothelial cells. Nature. 2016. Jan 21;529(7586):316–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tas SW, Maracle CX, Balogh E, Szekanecz Z. Targeting of proangiogenic signalling pathways in chronic inflammation. Nat Rev Rheumatol. 2016. Feb;12(2):111–22. [DOI] [PubMed] [Google Scholar]
- 50.Robinson WH, Mao R. Biomarkers to guide clinical therapeutics in rheumatology? Curr Opin Rheumatol. 2016. Mar;28(2):168–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lindstrom TM, Robinson WH. Biomarkers for rheumatoid arthritis: making it personal. Scand J Clin Lab Invest Suppl. 2010;242:79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Schaardenburg D, Dijkmans BAC. Clinical approaches to early inflammatory arthritis. Nat Rev Rheumatol. 2009. Nov;5(11):627–33. [DOI] [PubMed] [Google Scholar]
- 53.Ermann J, Rao DA, Teslovich NC, Brenner MB, Raychaudhuri S. Immune cell profiling to guide therapeutic decisions in rheumatic diseases. Nat Rev Rheumatol. 2015. Sep;11(9):541–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krenn V, Morawietz L, Burmester G-R, Kinne RW, Mueller-Ladner U, Muller B, et al. Synovitis score: discrimination between chronic low-grade and high-grade synovitis. Histopathology. 2006. Oct;49(4):358–64. [DOI] [PubMed] [Google Scholar]
- 55.Pitzalis C, Kelly S, Humby F. New learnings on the pathophysiology of RA from synovial biopsies. Curr Opin Rheumatol. 2013. May;25(3):334–44. [DOI] [PubMed] [Google Scholar]
- 56.van Baarsen LGM, Wijbrandts CA, Timmer TCG, van der Pouw Kraan TCTM, Tak PP, Verweij CL. Synovial tissue heterogeneity in rheumatoid arthritis in relation to disease activity and biomarkers in peripheral blood. Arthritis Rheum. 2010. Jun;62(6):1602–7. [DOI] [PubMed] [Google Scholar]
- 57.Bugatti S, Manzo A, Bombardieri M, Vitolo B, Humby F, Kelly S, et al. Synovial tissue heterogeneity and peripheral blood biomarkers. Curr Rheumatol Rep. 2011. Oct;13(5):440–8. [DOI] [PubMed] [Google Scholar]
- 58.Humby F, Durez P, Buch MH, Lewis MJ, Rizvi H, Rivellese F, et al. Rituximab versus tocilizumab in anti-TNF inadequate responder patients with rheumatoid arthritis (R4RA): 16-week outcomes of a stratified, biopsy-driven, multicentre, open-label, phase 4 randomised controlled trial. Lancet. 2021. Jan 23;397(10271):305–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dennis G Jr, Holweg CTJ, Kummerfeld SK, Choy DF, Setiadi AF, Hackney JA, et al. Synovial phenotypes in rheumatoid arthritis correlate with response to biologic therapeutics. Arthritis Res Ther. 2014. Apr 30;16(2):R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Orange DE, Yao V, Sawicka K, Fak J, Frank MO, Parveen S, et al. RNA Identification of PRIME Cells Predicting Rheumatoid Arthritis Flares. N Engl J Med. 2020. Jul 16;383(3):218–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat Biotechnol. 2006. Aug;24(8):971–83. [DOI] [PubMed] [Google Scholar]
- 62.Hueber W, Robinson WH. Proteomic biomarkers for autoimmune disease. Proteomics. 2006. Jul;6(14):4100–5. [DOI] [PubMed] [Google Scholar]
- 63.Deane KD, Holers VM. Rheumatoid Arthritis Pathogenesis, Prediction, and Prevention: An Emerging Paradigm Shift. Arthritis Rheumatol. 2021. Feb;73(2):181–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Deane KD, El-Gabalawy H. Pathogenesis and prevention of rheumatic disease: focus on preclinical RA and SLE. Nat Rev Rheumatol. 2014. Apr;10(4):212–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nielen MMJ, van Schaardenburg D, Reesink HW, van de Stadt RJ, van der Horst-Bruinsma IE, de Koning MHMT, et al. Specific autoantibodies precede the symptoms of rheumatoid arthritis: a study of serial measurements in blood donors. Arthritis Rheum. 2004. Feb;50(2):380–6. [DOI] [PubMed] [Google Scholar]
- 66.Rantapää-Dahlqvist S, de Jong BAW, Berglin E, Hallmans G, Wadell G, Stenlund H, et al. Antibodies against cyclic citrullinated peptide and IgA rheumatoid factor predict the development of rheumatoid arthritis. Arthritis Rheum. 2003. Oct;48(10):2741–9. [DOI] [PubMed] [Google Scholar]
- 67.Clish CB. Metabolomics: an emerging but powerful tool for precision medicine. Cold Spring Harb Mol Case Stud. 2015. Oct;1(1):a000588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Souto-Carneiro M, Tóth L, Behnisch R, Urbach K, Klika KD, Carvalho RA, et al. Differences in the serum metabolome and lipidome identify potential biomarkers for seronegative rheumatoid arthritis versus psoriatic arthritis. Ann Rheum Dis. 2020. Apr;79(4):499–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dubey D, Kumar S, Chaurasia S, Guleria A, Ahmed S, Singh R, et al. NMR-Based Serum Metabolomics Revealed Distinctive Metabolic Patterns in Reactive Arthritis Compared with Rheumatoid Arthritis. J Proteome Res. 2019. Jan 4;18(1):130–46. [DOI] [PubMed] [Google Scholar]
- 70.de Oliveira PG, Farinon M, Sanchez-Lopez E, Miyamoto S, Guma M. Fibroblast-Like Synoviocytes Glucose Metabolism as a Therapeutic Target in Rheumatoid Arthritis. Front Immunol. 2019. Aug 2;10:1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bustamante MF, Garcia-Carbonell R, Whisenant KD, Guma M. Fibroblast-like synoviocyte metabolism in the pathogenesis of rheumatoid arthritis. Arthritis Res Ther. 2017. May 31;19(1):110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Garcia-Carbonell R, Divakaruni AS, Lodi A, Vicente-Suarez I, Saha A, Cheroutre H, et al. Critical Role of Glucose Metabolism in Rheumatoid Arthritis Fibroblast-like Synoviocytes. Arthritis Rheumatol. 2016. Jul;68(7):1614–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guma M, Sanchez-Lopez E, Lodi A, Garcia-Carbonell R, Tiziani S, Karin M, et al. Choline kinase inhibition in rheumatoid arthritis. Ann Rheum Dis. 2015. Jul;74(7):1399–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gomez EA, Colas RA, Souza PR, Hands R, Lewis MJ, Bessant C, et al. Blood pro-resolving mediators are linked with synovial pathology and are predictive of DMARD responsiveness in rheumatoid arthritis. Nat Commun. 2020. Oct 27;11(1):5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H, et al. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods. 2017. Sep;14(9):865–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guo S, Zhang C, Le A. The limitless applications of single-cell metabolomics. Curr Opin Biotechnol. 2021. Oct;71:115–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015. Jul 23;523(7561):486–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pichon X, Lagha M, Mueller F, Bertrand E. A Growing Toolbox to Image Gene Expression in Single Cells: Sensitive Approaches for Demanding Challenges [Internet]. Vol. 71, Molecular Cell. 2018. p. 468–80. Available from: 10.1016/j.molcel.2018.07.022 [DOI] [PubMed] [Google Scholar]
- 79.Rao A, Barkley D, França GS, Yanai I. Exploring tissue architecture using spatial transcriptomics. Nature. 2021. Aug;596(7871):211–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ståhl PL, Salmén F, Vickovic S, Lundmark A, Navarro JF, Magnusson J, et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016. Jul 1;353(6294):78–82. [DOI] [PubMed] [Google Scholar]
- 81.Moffitt JR, Hao J, Wang G, Chen KH, Babcock HP, Zhuang X. High-throughput single-cell gene-expression profiling with multiplexed error-robust fluorescence in situ hybridization. Proc Natl Acad Sci U S A. 2016. Sep 27;113(39):11046–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rodriques SG, Stickels RR, Goeva A, Martin CA, Murray E, Vanderburg CR, et al. Slide-seq: A scalable technology for measuring genome-wide expression at high spatial resolution. Science. 2019. Mar 29;363(6434):1463–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen A, Liao S, Cheng M, Ma K, Wu L, Lai Y, et al. Spatiotemporal transcriptomic atlas of mouse organogenesis using DNA nanoball patterned arrays [Internet]. bioRxiv. 2021. [cited 2021 Nov 21]. p. 2021.01.17.427004. Available from: 10.1101/2021.01.17.427004v3.abstract [DOI] [PubMed] [Google Scholar]
- 84.Carlberg K, Korotkova M, Larsson L, Catrina AI, Ståhl PL, Malmström V. Exploring inflammatory signatures in arthritic joint biopsies with Spatial Transcriptomics. Sci Rep. 2019. Dec 12;9(1):18975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chang MH, Nigrovic PA. Antibody-dependent and -independent mechanisms of inflammatory arthritis [Internet]. Vol. 4, JCI Insight. 2019. Available from: 10.1172/jci.insight.125278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kiener HP, Watts GFM, Cui Y, Wright J, Thornhill TS, Sköld M, et al. Synovial fibroblasts self-direct multicellular lining architecture and synthetic function in three-dimensional organ culture. Arthritis Rheum. 2010. Mar;62(3):742–52. [DOI] [PubMed] [Google Scholar]
- 87.Lee DM, Kiener HP, Agarwal SK, Noss EH, Watts GFM, Chisaka O, et al. Cadherin-11 in synovial lining formation and pathology in arthritis. Science. 2007. Feb 16;315(5814):1006–10. [DOI] [PubMed] [Google Scholar]
- 88.Karonitsch T, Beckmann D, Dalwigk K, Niederreiter B, Studenic P, Byrne RA, et al. Targeted inhibition of Janus kinases abates interfon gamma-induced invasive behaviour of fibroblast-like synoviocytes. Rheumatology. 2018. Mar 1;57(3):572–7. [DOI] [PubMed] [Google Scholar]
- 89.Friščić J, Böttcher M, Reinwald C, Bruns H, Wirth B, Popp S-J, et al. The complement system drives local inflammatory tissue priming by metabolic reprogramming of synovial fibroblasts. Immunity. 2021. May 11;54(5):1002–21.e10. [DOI] [PubMed] [Google Scholar]
- 90.Neal JT, Li X, Zhu J, Giangarra V, Grzeskowiak CL, Ju J, et al. Organoid Modeling of the Tumor Immune Microenvironment. Cell. 2018. Dec 13;175(7):1972–88.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ganesh K, Wu C, O’Rourke KP, Szeglin BC, Zheng Y, Sauvé C-EG, et al. A rectal cancer organoid platform to study individual responses to chemoradiation. Nat Med. 2019. Oct;25(10):1607–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yao Y, Xu X, Yang L, Zhu J, Wan J, Shen L, et al. Patient-Derived Organoids Predict Chemoradiation Responses of Locally Advanced Rectal Cancer. Cell Stem Cell. 2020. Jan 2;26(1):17–26.e6. [DOI] [PubMed] [Google Scholar]
- 93.Nagle PW, Plukker JTM, Muijs CT, van Luijk P, Coppes RP. Patient-derived tumor organoids for prediction of cancer treatment response. Semin Cancer Biol. 2018. Dec;53:258–64. [DOI] [PubMed] [Google Scholar]


