The human tryptase locus is a gene-dense region within the sub-telomeric portion of the short (p) arm of chromosome 16 at position 13.3 (16p13.3) [9]. Because the two genes that encode secreted α- and β-tryptases TPSAB1 and TPSB2 are in such close proximity to one another they do not randomly assort and are in near complete linkage disequilibrium. And, individuals inherit tryptase genes as haplotypes from their parents that contain both of these genes. Whereas TPSB2 encodes β-tryptases, TPSAB1 may encode α- or β-tryptases. Thus, the canonical TPSAB1/TPSB2 haplotypes are α/β or β/β - leading to the canonical tryptase genotypes of β/β:β/β, α/β:β/β, or α/β:α/β [9]. Structural homology between tryptase-encoding genes, and other features of the tryptase locus (e.g., GC rich, repetitive sequences) render conventional next generation sequencing incapable of resolving tryptase genotypes. However, a droplet digital PCR assay is available for clinical genotyping of patients [1].
While the three common genotypes β/β:β/β, α/β:β/β, and α/β:α/β account for an estimated >90% of individuals, copy number variation at TPSAB1 and potentially TPSB2 has been reported and may complicate interpretation of tryptase genotype. In many of these cases examination of family pedigrees assists in the clinical interpretation of genotypic findings and Mendelian inheritance of tryptase haplotypes can often clarify seemingly discordant genotype/phenotype observations [2].
Hereditary alpha-tryptasemia (HαT) is an autosomal dominant genetic trait characterized by elevated basal serum tryptase (BST) resulting from increased copy number of the TPSAB1 gene encoding α-tryptase [3]. Several studies in the U.S. and Europe have demonstrated that this is present in 4–6% of the general populations in which it has been studied, affecting an estimated 16 million people in the U.S. Increased TPSAB1 gene copy number and elevated BST can be associated with multisystem complaints, including cutaneous flushing and pruritis [4, 6], anaphylaxis [5, 6], and gastrointestinal (GI) symptoms [1, 3, 6–9]. Herein, we report two symptomatic individuals who presented with elevated BST and were ultimately found to have HαT despite having normal total tryptase gene copy number.
The first patient presented with flushing, itching, hives, joint pain and swelling, and diarrhea. She also had a history of anaphylaxis requiring intramuscular epinephrine administered in an emergency department. Her basal serum tryptase was 18 ng/mL but tryptase genotyping revealed only two copies of TPSAB1 encoding α-tryptase and two copies of TPSB2 encoding β-tryptase (presumed α/β:α/β genotype). Her son was subsequently diagnosed with HαT (β/β:αα/β genotype) after presenting with gastrointestinal food sensitives, abdominal pain, diarrhea, joint pain, and muscle pain. However, his father did not have HαT (β/β:β/β genotype). Testing of the index patient’s parents revealed that her father also had HαT (α/β:αα/β genotype) but her mother had germline loss of a tryptase-encoding sequence at either TPSAB1 or TPSB2 (−/β:α/β genotype) (Fig 1B). The index patient’s father’s symptoms included flushing, joint pain, headaches, food sensitivities, abdominal pain, and diarrhea.
Figure 1. Mendelian inheritance of tryptase haplotypes reveals co-inheritance of TPSAB1 duplications with beta-tryptase sequence loss among individuals with HαT.
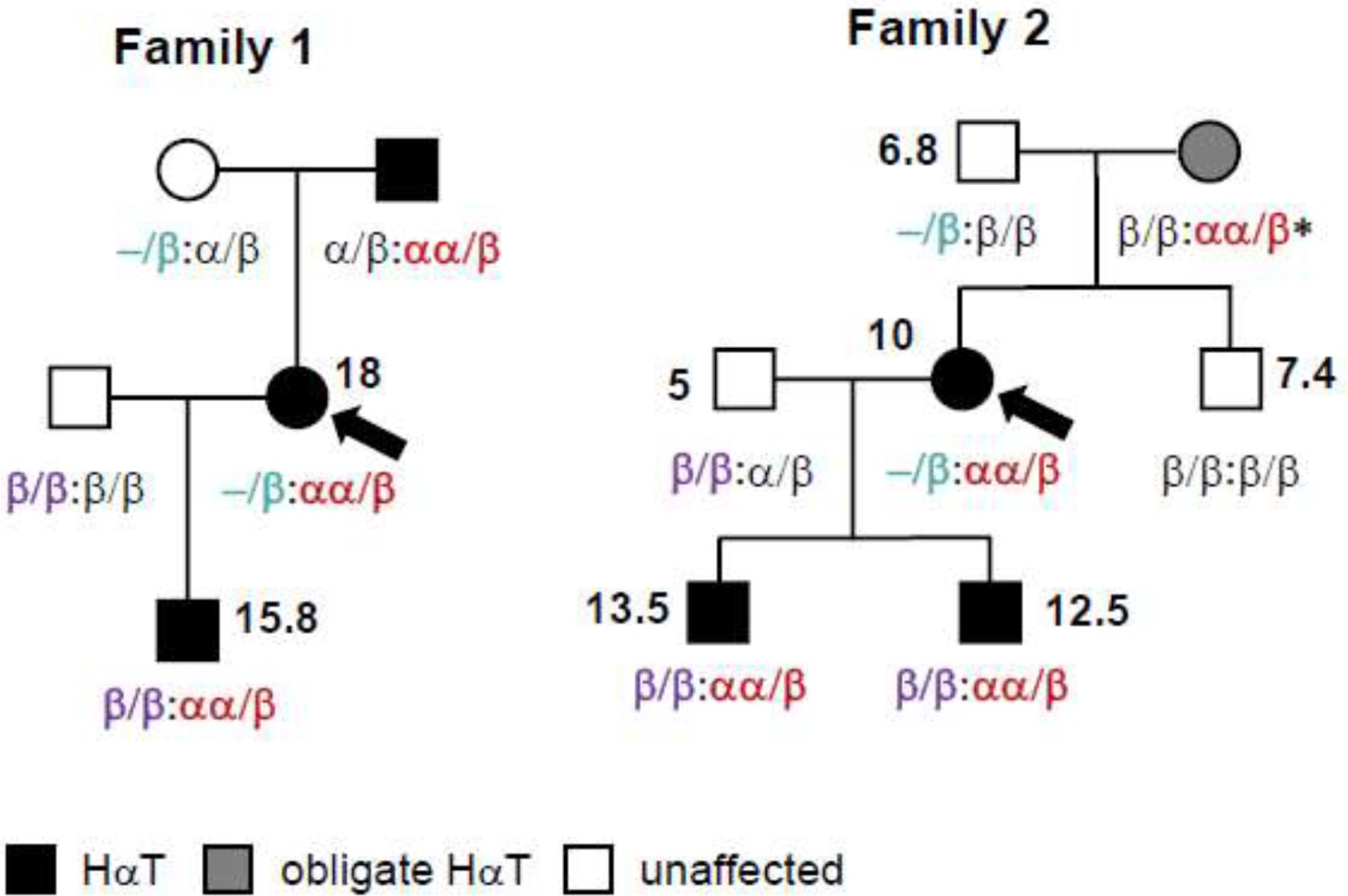
Pedigrees from two families with HαT were identified as individuals (arrows) who co-inherited a haplotype (−/β) with a loss of a tryptase-encoding sequence; inherited haplotypes are in colored fonts; measured BST levels (ng/mL) are shown next to affected individuals in bold. *deduced genotype based upon haplotypes inherited by offspring
The second patient came to attention because both of her sons had been diagnosed with HαT. She reported a history of migraine headaches, asthma, intermittent hives, and joint pain. Her older son (β/β:αα/β genotype) presented with mild pruritus, intermittent hives, joint pain, and a BST of 13.5 ng/mL. Her younger son (also β/β:αα/β genotype) presented with BST of 12.5 ng/mL, multiple food sensitivities, bloody diarrhea, hives, rash, and failure to thrive. When she was tested her BST was 10 ng/mL, but her genotype did not initially appear to be consistent with HαT (presumed α/β:α/β genotype). However, when her sons’ father was tested his BST was only 5 ng/mL, and he had only one copy of TPSAB1 encoding α-tryptase (β/β:α/β genotype). Genotyping of the children’s maternal grandfather found that he did not have any α-tryptase encoding sequences, and had only 3 β-tryptase encoding sequences (−/β:β/β genotype), requiring the maternal grandmother (who was deceased and unavailable for testing) to be an obligate carrier of an extra-allelic copy of TPSAB1 encoding α-tryptase in order for her daughter to have two germline copies, and thus HαT. Her genotype could be fully deduced as an β/β:αα/β genotype since her son - the initial patients’ maternal uncle - also had no α-tryptase-encoding copies (β/β:β/β genotype), thus requiring he received a β/β haplotype from both grandparents (Fig. 1, family 2).
This report reiterates the importance of examining family pedigrees when considering tryptase genotypes [2], in particular when BST seems to be discordant with genotypic findings. We have described two individuals with HαT associated with the previously unreported −/β:αα/β genotype, that would not have been identified without genotyping family members and evaluating inheritance patterns. This analysis prevented the erroneous conclusion that HαT was occurring de novo in the children of these two families, or that elevated BST in the two individuals with HαT and tryptase-encoding gene copy loss was associated with a clonal mast cell disorder inappropriately prompting unnecessary testing. Importantly, while these individuals did not have other indications for bone marrow biopsy, diagnosing HαT should not preclude an evaluation for concomitant clonal mast cell disease where there is a clinical suspicion given that there is an increased prevalence of HαT among individuals with systemic mastocytosis [5, 6, 9].
In addition to loss of ostensibly β-tryptase-encoding sequences, gain of germline β-tryptase-encoding gene copies has been reported [9]. Unlike increased α-tryptase-encoding TPSAB1 copy number, increased β-tryptase-encoding copy number has not been associated with inherited increases in BST levels [2]. Because β-tryptase may be encoded at TPSAB1 or TPSB2, we currently do not know at which locus β-tryptase-encoding gene replications or losses are occurring. However, based upon genotyping performed to date tryptase-encoding gene copy loss appears to occur at TPSAB1 most commonly - or on alleles in which TPSAB1 encodes β-tryptase - since a haplotype containing a single α-tryptase-encoding sequence has not yet been observed despite thousands of individuals having been genotyped [9] (and unpublished data, JJL). Conversely, β-tryptase-encoding gene replications may be occurring at TPSB2, since they have been observed both with β-tryptase sequences at TPSAB1 as well as α-tryptase-encoding gene replications at TPSAB1 [2, 9].
The astounding structural complexity of the tryptase locus will continue to be revealed as new populations are genotyped and additional techniques are applied to resolve the locus in greater detail. Equally complex has been the work to unravel which of the symptoms reported in association with this genetic trait may only be modified, or even unrelated to HαT [9]. Additional studies and insights into the genetic composition of tryptases in different individuals and populations, as well as into associated phenotypes, will continue to refine how and when tryptase gene composition(s) may provide risk, benefit, or modify different clinical presentations as already has been described in some patients [9, 10].
Acknowledgments
This work was supported by extramural funding to Dr. Glover via 1R21TR002639-01A1 and intramural NIH funding to Dr. Lyons via the Division of Intramural Research of the National Institute of Allergy and Infectious Diseases, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have no conflicts of interest to disclose.
References
- 1.Lyons JJ, Hereditary Alpha Tryptasemia: Genotyping and Associated Clinical Features. Immunol Allergy Clin North Am, 2018. 38(3): p. 483–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lyons JJ, On the complexities of tryptase genetics and impact on clinical phenotypes. J Allergy Clin Immunol, 2021. [DOI] [PubMed] [Google Scholar]
- 3.Lyons JJ, et al. , Elevated basal serum tryptase identifies a multisystem disorder associated with increased TPSAB1 copy number. Nat Genet, 2016. 48(12): p. 1564–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Le QT, et al. , Impact of naturally forming human alpha/beta-tryptase heterotetramers in the pathogenesis of hereditary alpha-tryptasemia. J Exp Med, 2019. 216(10): p. 2348–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lyons JJ, et al. , Heritable risk for severe anaphylaxis associated with increased alpha-tryptase-encoding germline copy number at TPSAB1. J Allergy Clin Immunol, 2020. [DOI] [PubMed] [Google Scholar]
- 6.Greiner G, et al. , Hereditary alpha tryptasemia is a valid genetic biomarker for severe mediator-related symptoms in mastocytosis. Blood, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lyons JJ, et al. , Mendelian inheritance of elevated serum tryptase associated with atopy and connective tissue abnormalities. J Allergy Clin Immunol, 2014. 133(5): p. 1471–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robey RC, et al. , Hereditary Alpha-Tryptasemia: UK Prevalence and Variability in Disease Expression. J Allergy Clin Immunol Pract, 2020. [DOI] [PubMed] [Google Scholar]
- 9.Glover SC, et al. , Clinical relevance of inherited genetic differences in human tryptases: Hereditary alpha-tryptasemia and beyond. Ann Allergy Asthma Immunol, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lyons JJ and Yi T, Mast cell tryptases in allergic inflammation and immediate hypersensitivity. Curr Opin Immunol, 2021. 72: p. 94–106. [DOI] [PubMed] [Google Scholar]


