Abstract
Azithromycin (AZT) has widely been used for the treatment of shigellosis in children. Recent studies showed a high rate of decreased susceptibility to azithromycin due to different mechanisms of resistance in Shigella isolates. Accordingly, the purpose of this study was to investigate the role of azithromycin resistance mechanisms of Shigella isolates in Iran during a two-year period. In this study, we investigated the mechanisms of resistance among Shigella spp. that were isolated from children with shigellosis. The minimum inhibitory concentration (MIC) of Shigella isolates to azithromycin was determined by the agar dilution method in the presence and absence of Phe-Arg-β-naphthylamide inhibitor. The presence of 12 macrolide resistance genes was investigated for all isolates by PCR for the first time in Tehran province in Iran. Among the 120 Shigella spp., only the mph(A) gene (49.2%) was detected and other macrolide resistance genes were absent. The phenotypic activity of efflux pump was observed in 1.9% of isolates which were associated with over expression of both omp(A) and omp(W) genes. The high prevalence of the mph(A) gene among DSA isolates may indicate that azithromycin resistance has evolved as a result of antimicrobial selection pressures and inappropriate use of azithromycin.
1. Introduction
Shigella species are Gram-negative and nonmotile rods in the family Enterobacteriaceae that cause shigellosis. Shigellosis was the third leading cause of diarrheal death in children under the age of 5 in 2015 (40,000 deaths per year) [1–3], and it mostly affects children living in developing countries [4–6]. Shigella can be classified into four serogroups or species based on O lipopolysaccharide antigen type: S. dysenteriae (subgroup A), S. flexneri (subgroup B), S. boydii (subgroup C), and S. sonnei (subgroup D) [5, 7]. Based on epidemiological studies, S. flexneri and S. sonnei are the most common species in developing countries, but S. sonnei is the predominant species in developed countries [5, 8–10]. Recent studies have revealed a species shift from S. flexneri to S. sonnei in Iran, and S. sonnei has been the dominant species in most parts of the country [11–14]. Shigella spp. are highly infectious and are transmitted by the fecal-oral route or ingestion of contaminated food or water [5, 8, 15]. While shigellosis is endemic in developing countries with poor water and sanitation conditions, it is usually associated with either returned travelers or men who have sex with men (MSM) in developing countries (3–5). Shigellosis is transmitted in the developing countries as fecal-oral and contaminated food and water, and in developed countries, it is transmitted by traveling to disease-endemic regions and men who having sex with men [8, 10, 16]. Symptoms appear abruptly after an incubation period of 12 hours to approximately 2 days and include high fever, crampy abdominal pain, and diarrhea [17]. The disease is self-limiting, but antibiotic treatment is required in children, elderly, and people with weakened immune systems [18].
Shigella spp. have become resistant to first-line drugs (trimethoprim-sulfamethoxazole and ampicillin) and are no longer prescribed to treat shigellosis due to the emergence of multidrug resistance and has challenged the treatment of the disease in children [4, 5, 19, 20]. These drugs are replaced by ciprofloxacin (CIP), and azithromycin (AZT) for shigellosis treatment in adults and children [4, 5, 21]. Owing to its oral administration and affordability, AZT is recommended by a number of international guidelines for the treatment of shigellosis in children [4]. In Iran, AZT is the most commonly prescribed antibiotic for treatment of children suffering from shigellosis [14, 22]. Reports of Shigella isolates with decreased susceptibility to azithromycin (DSA) are increasing globally, raising concerns about its usefulness as the second-line treatment for children with shigellosis [4, 23]. The most common types of macrolide resistance in Enterobacteriaceae are those encoded in mobile genetic elements, such as target site modification by methylases encoded by erm genes (erm(A), erm(B), erm(C), erm(F), erm(T), erm(X)), inactivation of macrolides, mediated by esterases such as those encoded by ere genes (ereA and ereB), and phosphotransferases encoded by mph genes (mph(A) and mph(B)). Additionally, the macrolide efflux pumps encoded by mef genes (mef(A) or mef(B) and msr(A)) and chromosomal efflux pumps (omp(A) and omp(W)) have been reported to confer resistance to macrolides [24]. To a lesser extent, mutations in the L4 (rplD) and L22 (rplV) ribosomal proteins and in 23S rRNA (rrlH) have been shown to be responsible for macrolide resistance [24, 25].
Recent studies have reported a relatively high frequency of resistance to azithromycin among Shigella isolates from children with dysentery in Iran [12, 14, 26]. There has been no detailed investigation of the mechanisms of macrolides resistance among the DSA-Shigella isolates in Iran. In this study, we determined the azithromycin MICs for a collection of Shigella isolates recovered from children with shigellosis in Tehran, Iran. Then, we investigated the presence of macrolide resistance genes associated with mobile genetic elements and the expression levels of outer membrane proteins A and W (ompA and ompW) genes related to efflux pump in isolates.
2. Materials and Methods
2.1. Bacterial Isolates and Identification
Shigella isolates were collected between March 2017 and September 2019 from the feces of children under 14 who were suspected to have shigellosis and were referred to Children's Medical Center in Tehran. Initial identification was performed using microbiological and biochemical analysis and Shigella serogroups were determined using latex agglutination serotyping (Figure 1). This study was evaluated by the Local Ethics Committee of Shahid Beheshti University of Medical Sciences (IR.SBMU.MSP.REC.1399.490).
Figure 1.
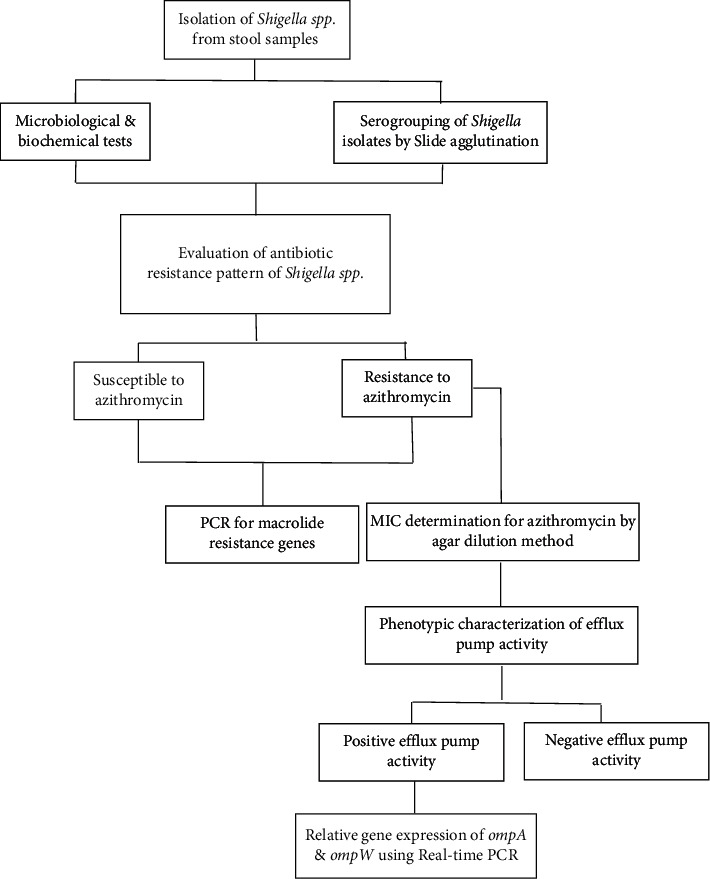
Work flowchart for identification Shigella isolates with decreased susceptibility to azithromycin (DSA) and characterization of the related genetic mechanisms.
2.2. Antibiotic Susceptibility Test and MICs of Azithromycin
The antibiotic susceptibility pattern of all isolates had been previously described [12]. Briefly, antimicrobial susceptibility testing to nine antibiotics was conducted using Kirby–Bauer disk diffusion method. The MICs of DSA isolates were confirmed (ranging from 2 to 512 µg/ml) using the agar dilution method according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (Clinical and Laboratory Standards Institute (CLSI), Performance standards for antimicrobial susceptibility testing, 29th ed., CLSI supplement M100–S29, Wayne, PA; [27]).
2.3. MICs of Azithromycin in the Presence of Efflux Pump Inhibitor
The MICs of DSA isolates were examined by adding efflux pump inhibitor Phe-Arg-β-naphthylamide (PAβN) (20 mg/ml) (Sigma, St. Louis, Mo., USA) to determine the impact of efflux pumps activity on azithromycin resistance. A ≥4-fold reduction in azithromycin MIC in the presence of PAβN suggested the existence of an efflux pump (27, 28).
2.4. The Presence of Macrolide Resistance Genes
Genomic DNA was extracted using the High Pure Isolation Kit (Roche, Mannheim, Germany) according to the manufacturer's instructions. Macrolide resistance genes, including, mph(A), mph(B), erm(A), erm(B), erm(C), erm(F), erm(T), erm(X), ere(A), ere(B), mef(A), and msr(A) were amplified by polymerase chain reaction (PCR) using specific primers (Table 1). PCR products were separated using 1.5% agarose gel. Positive PCR products were sequenced (Stab vida, Spain) to confirm the presence of resistance genes (accession number: OL310860).
Table 1.
Primer sequences.
| Target gene | Primer sequence (5′ ⟶ 3′) | Product size (bp) | Annealing temperature (°C) | Reference |
|---|---|---|---|---|
| mph(A) | F: GTGAGGAGGAGCTTCGCGAG | 403 | 59 | Rahman et al. [28] |
| R: TGCCGCAGGACTCGGAGGTC | ||||
| mph(B) | F: GATATTAAACAAGTAATCAGAATAG | 494 | 49 | Rahman et al. [28] |
| R: GCTCTTACTGCATCCATACG | ||||
| erm(A) | F: TCTAAAAAGCATGTAAAAGAAA | 533 | 47 | Rahman et al. [28] |
| R: CGATACTTTTTGTAGTCCTTC | ||||
| erm(B) | F: GAAAAAGTACTCAACCAAATA | 639 | 43 | Rahman et al. [28] |
| R: AATTTAAGTACCGTTACT | ||||
| erm(C) | F: TCAAAACATAATATAGATAAA | 642 | 43 | Rahman et al. [28] |
| R: GCTAATATTGTTTAAATCGTCAAT | ||||
| erm(F) | F: CGACACAGCTTTGGTTGAAC | 302 | 52 | Liu et al. [29] |
| R: GGACCTACCTCATAGACAAG | ||||
| erm(T) | F: CATATAAATGAAATTTTGAG | 369 | 42 | Liu et al. [29] |
| R: ACGATTTGTATTTAGCAACC | ||||
| erm(X) | F: GAGATCGGRCCAGGAAGC | 488 | 54 | Liu et al. [29] |
| R: GTGTGCACCATCGCCTGA | ||||
| ere(A) | F: GCCGGTGCTCATGAACTTGAG | 420 | 55 | Rahman et al. [28] |
| R: CGACTCTATTCGATCAGAGGC | ||||
| ere(B) | F: TTGGAGATACCCAGATTGTAG | 537 | 50 | Rahman et al. [28] |
| R: GAGCCATAGCTTCAACGC | ||||
| mef(A) | F: AGTATCATTAATCACTAGTGC | 345 | 48 | Rahman et al. [28] |
| R: TTCTTCTGGTACTAAAAGTGG | ||||
| msr(A) | F: GCACTTATTGGGGGTAATGG | 384 | 51 | Rahman et al. [28] |
| R: GTCTATAAGTGCTCTATCGTG | ||||
| omp(A) | F: ATGAAAAAGACAGCTATCGCG | 187 | 54 | Gomes et al. [30] |
| R: CACCAAAAGCACCAGCGCCCA | ||||
| omp(W) | F: GTTAACAGTGGCGGGTTTGGC | 154 | 56 | Gomes et al. [30] |
| R: CACGCTGAATCCACCCAGACT |
bp, base pair; F, forward primer; R, reverse primer.
2.5. Quantitative Real-Time PCR (qRT-PCR) for Evaluation of Efflux Pumps Genes Expression
Total RNA was extracted using a BioFACT TM Total RNA Prep Kit (Biofact, South Korea) following the manufacturer's instructions. All extracted RNAs were treated with DNase I (CinnaGen Co., Iran) in order to remove the remaining genomic DNA. Reverse transcription was performed using the Add Script cDNA Synthesis Kit (Add bio, South Korea) with an input of 200 ng/µl of total RNA in a final reaction volume of 20 µL under standard reverse transcription PCR conditions following the manufacturer's instructions.
The expression level of efflux pump genes among phenotypically active isolates was determined by quantitative real-time-PCR (qRT-PCR) using primers targeting omp(A) and omp(W) genes as described previously [30] (Table 1). All reactions were conducted in duplicate, and the 16S rRNA was used as the endogenous control gene. The 2−ΔΔCT method was used to determine the relative expression of the target genes, and a value of ≥4-fold compared to that of S. flexneri ATCC12022 was considered as overexpression [31].
2.6. Statistical Analysis
Pearson's chi-squared test was used to investigate the relationship between antibiotic resistances with regard to the age group of the patients, gender and the species of Shigella isolated from the patients. A p value of <0.05 was considered significant. Data were analyzed using JMP, version16 (SAS Institute Inc., 2021).
3. Results
3.1. Characteristics of the Patients and Isolates
A total of 120 Shigella isolates were collected from the fecal samples of children with shigellosis Sixty percent of patients were male (n = 72), and 40% were female (n = 48) (Table 2). Overall, 55% of patients (n = 66) aged 5 years old or younger, 35% (n = 42) aged 6 to 10, and 10% (n = 12) aged 11 to 14 years old. Among 120 Shigella isolates, S. sonnei was the most common species with 80.8% of the total isolates (n = 97), followed by S. flexneri with 17.5% (n = 21) and S. boydii with 1.7% (n = 2), respectively. The type of Shigella spp. detected in a patient did not vary with respect to age group and gender of the patients (p > 0.05).
Table 2.
Characteristics of the patients with shigellosis.
| Age group | Gender no. | Shigella spp. | ||
|---|---|---|---|---|
| S. sonnei no. | S. flexneri no. | S. boydii no. | ||
| ≥5 | Male: 46 | 37 | 8 | 1 |
| Female: 20 | 15 | 4 | 1 | |
| Total: 66 | 52 | 12 | 2 | |
| 6–10 | Male: 16 | 15 | 1 | 0 |
| Female: 26 | 22 | 4 | 0 | |
| Total: 42 | 37 | 5 | 0 | |
| 11–14 | Male: 10 | 7 | 3 | 0 |
| Female: 2 | 1 | 1 | 0 | |
| Total: 12 | 8 | 4 | 0 | |
| Total | Male: 72 | 59 | 12 | 1 |
| Female: 48 | 38 | 9 | 1 | |
| Total: 120 | 97 | 21 | 2 | |
No.: number.
The azithromycin MICs among the S. sonnei isolates ranged from 32 to 512 µg/ml, and the only S. flexneri isolates had MIC = 32 µg/ml. Of the 54 DSA-Shigella isolates, only one isolate (1.9%) was S. flexneri, and the other 53 isolates (98.1%) were S. sonnei. All DSA isolates were resistant to Trimethoprim/sulfamethoxazole. A high frequency of isolates was resistant to ampicillin (96.2%), nalidixic acid (94.4%), cefotaxime (90.7%), cefixime (90.7%), and minocycline (79.6%). The frequency of resistance to ciprofloxacin and levofloxacin was comparatively low and was 3.7% and 16.6%, respectively. The probability of detecting DSA isolates varied with respect to the age group of the patients (p < 0.05), and children between 11 and 14 years old showed a higher prevalence of DSA isolates. However, the probability of detecting DSA-Shigella isolates did not vary with regard to the gender of the patients (p > 0.05).
3.2. Identification of Efflux Pump-Mediated Resistance
All DSA-Shigella isolates were able to grow in the presence of PAβN. Overall, MIC levels of 8 isolates (14.8%) decreased in the presence of PAβN, irrespective of the initial MIC of azithromycin (MICs ranging from 32 to 256 µg/ml). Only one S. sonnei isolate demonstrated a decrease of ≥4-fold in azithromycin MIC when PAβN was added. The MIC of this isolate dropped from 512 µg/ml to 64 µg/ml in the presence of the inhibitor. The qRT-PCR results also showed overexpression in the mRNA levels of efflux pump genes ompA and ompW (Figure 2). However, mRNA overexpression was not detected among the other isolates that were inhibited by PAβN.
Figure 2.
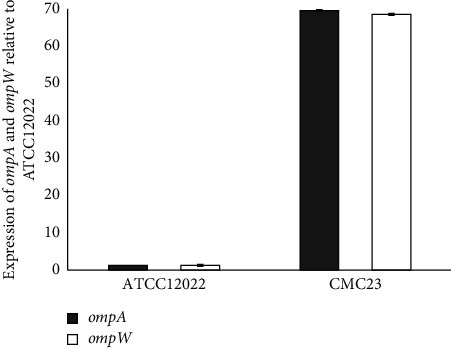
Relative expression of ompA and ompW genes by qRT-PCR. The relative expression level after being normalized to the expression of the reference gene 16S rRNA was compared relative to that in S. flexneri ATCC 12022. Results shown are the mean ± std of two independent experiments.
3.3. Characterization of Macrolide Resistance Genes
The PCR analysis demonstrated that 59 (49.2%) of Shigella isolates carried the mph(A) gene. Five isolates that were susceptible to azithromycin (MICs <16) were found to carry the mph(A) gene. However, other mobile genetic elements associated with the azithromycin resistance (including mph(B), erm(A), erm(B), erm(C), erm(F), erm(T), erm(X), ere(A), ere(B), mef(A), and msr(A) were not detected among these isolates (Figure 3). Of 59 isolates mph(A) positive, 32 (54.2%) exhibited high levels of resistance to azithromycin (MICs ≥64 µg/ml). Demographics and clinical features of pediatric patients are accessible in detail in Table S1.
Figure 3.
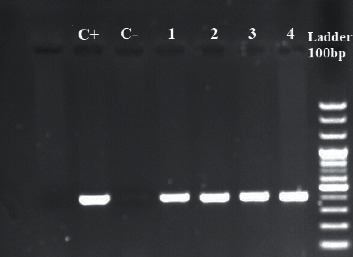
PCR products of mphA gene among Shigella clinical strains. Lane 2, positive control (S. Sonnei accession number: OL310860); lane 3, negative control; and lane 4, 5, 6, and 7 including sample 1 to 4, Shigella isolates harboring mphA gene.
4. Discussion
In the collection of 120 Shigella isolates in this study, 54 isolates (45%) were confirmed to be DSA, indicating more considerations should be taken in prescribing this drug for the treatment of children with shigellosis. Azithromycin MICs of the DSA-Shigella isolates ranged from 32 to 512 µg/ml, and 59.2% of the isolates demonstrated high levels of resistance to azithromycin (MICs ≥64 µg/ml). Previous studies have also reported a relatively high frequency of azithromycin resistance in Shigella isolates. For example, the rate of DSA was 42% in Palestine [32], 20.4% in China [31], 20% in the US [33], 13% in Australia [34], and 5% in Southeast Asia [4]. The lower rate of DSA reported from Southeast Asia has been associated with limited azithromycin usage in the region [4]. A previous study by Ezernitchi et al. [1] found that DSA-Shigella isolates were obtained mainly from children under 9 years of age. The results of our study showed that the age of the patients had a significant impact on the prevalence of DSA isolates among children with shigellosis. The 11- to 14-years-old patients were more likely to harbor DSA-Shigella even though this age group represented only 10% of cases with shigellosis.
Previous studies have established the importance of acquired mobile genetic elements in conferring resistance to macrolides in Shigella and other Enterobacteriaceae [24, 25, 31]. We investigated the presence of 12 mobile genetic elements associated with azithromycin resistance, and 49.2% (59/120) of the isolates were positive for the mph(A) gene. However, other mobile genetic elements associated with the azithromycin resistance were not detected in our Shigella isolates. Five isolates (8.5%) were found to carry the mph(A) gene, but they were susceptible to azithromycin (MICs <16). This heterogeneity could be explained by the differential expression levels in individual cells due to the variation in mph(A) copy numbers, leading to differences in azithromycin resistance levels [24]. Overall, this finding is consistent with that of the previous studies, which reported the role of the mph(A) gene as the principal mechanism for azithromycin resistance in Shigella isolates [4, 29]. For example, Zhang et al. [31] found that 55% of DSA-Shigella isolates were mph(A) positive, and no other resistance gene was detected. Liu et al. [29] reported that 57.8% and 40.7% of S. flexneri and S. sonnei isolates carried the mph(A) gene, but other azithromycin resistance genes were not detected. A very low frequency (0.6%) of DSA-Shigella from Southeast Asia were positive for the erm(B) gene [4]. Likewise, erm(B)-associated azithromycin resistance was detected in 3.4% of the E. coli isolates with DSA in Peru [24].
PAβN is an efflux pump inhibitor, which competes with macrolides for its specific binding point. The role of PAβN-inhibitable efflux pumps in azithromycin resistance has been demonstrated in Shigella spp. and E. coli [24, 30]. In this study, one S. sonnei isolate (1.9%) demonstrated azithromycin resistance associated with the efflux pump activity. This isolate contained omp(A), omp(W) and mph(A) genes. Several studies have reported that mutations in the ribosomal proteins L4 (rplD) and ribosomal proteins L22 (rplV) and in 23S rRNA (rrlH) can confer macrolide resistance [24, 35]. Unfortunately, we did not determine the nucleotide sequence changes of the specific regions of the three genes, and we could not determine the azithromycin resistance mechanism in one S. sonnei isolate. Further studies are required to understand the possible additional mechanisms responsible for the DSA in Shigella spp. The present study demonstrated that the plasmid-mediated mph(A) gene is the most common macrolide resistance gene in Shigella isolates collected from children with shigellosis in Tehran, Iran. The high prevalence of the mph(A) gene among DSA isolates may indicate that azithromycin resistance has evolved as a result of antimicrobial selection pressures and inappropriate use of azithromycin. The plasmid-mediated mph(A) gene can spread quickly among different members of the Enterobacteriaceae and yield either the same or different strains with DSA [36].
5. Conclusion
Contrary to most studies, which have shown that efflux pump has no role in azithromycin resistance, our study showed that one of our DSA isolates increased omp(A) and omp(W) expression levels and consequently, efflux pump can play a role in resistance.
Acknowledgments
This work was financially supported by the Shahid Beheshti University of Medical Sciences, Tehran, Iran (grant no. 25845), and this article has been extracted from the thesis written by Parisa Behruznia in the School of Medicine, Shahid Beheshti University of Medical Sciences (registration no: 522).
Data Availability
All the data generated or analyzed during this study were included in this article.
Conflicts of Interest
The authors declare that there are no conflicts of interest.
Authors' Contributions
P. Behruznia conducted the molecular tests and designed the study. M. Sadredinamin performed the data collection and phenotypic tests. A. Hashemi conducted the microbiological experiments. B. Hajikhani conceptualized the study and designed the methodology. N. Yousefi performed the bacterial isolation. M. Behruznia conducted the statistical analysis and drafted the manuscript. Z. Ghalavand critically revised the manuscript and supervised and administered the project. All authors approved the final version of the manuscript and authorship list.
Supplementary Materials
Table S1: clinical data of pediatric patients, drug resistance, and distribution of azithromycin resistance genes in DSA-Shigella spp.
References
- 1.Ezernitchi A. V., Sirotkin E., Dana D., Agmon1 V., Valinsky L., Rokney A. Azithromycin non-susceptible Shigella circulating in Israel, 2014–2016. PLoS One . 2019;14(10) doi: 10.1371/journal.pone.0221458.e0221458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lima I. F. N., Havt A., Lima A. A. M. Update on molecular epidemiology of Shigella infection. Current Opinion in Gastroenterology . 2015;31(1):30–37. doi: 10.1097/mog.0000000000000136. [DOI] [PubMed] [Google Scholar]
- 3.Williams P. C. M., Berkley J. A. Guidelines for the treatment of dysentery (shigellosis): a systematic review of the evidence. Paediatrics and International Child Health . 2018;38(1):S50–S65. doi: 10.1080/20469047.2017.1409454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Darton T. C., Tuyen H. T., Chung The H., et al. Azithromycin resistance in Shigella spp. in southeast Asia. Antimicrobial Agents and Chemotherapy . 2018;62(4):e01748–17. doi: 10.1128/AAC.01748-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kotloff K. L., Riddle M. S., Platts-Mills J. A., Pavlinac P., Zaidi A. K. M. Shigellosis. The Lancet . 2018;391(10122):801–812. doi: 10.1016/s0140-6736(17)33296-8. [DOI] [PubMed] [Google Scholar]
- 6.Sousa M. Â. B., Mendes E. N., Collares G. B., Péret-Filho L. A., Penna F. J., Magalhães P. P. Shigella in Brazilian children with acute diarrhoea: prevalence, antimicrobial resistance and virulence genes. Memórias Do Instituto Oswaldo Cruz . 2013;108(1):30–35. doi: 10.1590/s0074-02762013000100005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taneja N., Mewara A. Shigellosis: epidemiology in India. The Indian Journal of Medical Research . 2016;143(5):p. 565. doi: 10.4103/0971-5916.187104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baker K. S., Dallman T. J., Ashton P. M., et al. Intercontinental dissemination of azithromycin-resistant shigellosis through sexual transmission: a cross-sectional study. The Lancet Infectious Diseases . 2015;15(8):913–921. doi: 10.1016/s1473-3099(15)00002-x. [DOI] [PubMed] [Google Scholar]
- 9.Kotloff K. L., Winickoff J. P., Bernard I., et al. Global burden of Shigella infections: implications for vaccine development and implementation of control strategies. Bulletin of the World Health Organization . 1999;77(8):p. 651. [PMC free article] [PubMed] [Google Scholar]
- 10.Muthuirulandi Sethuvel D. P., Ragupathi N. K. D., Anandan S., Veeraraghavan B. Update on: Shigella new serogroups/serotypes and their antimicrobial resistance. Letters in Applied Microbiology . 2017;64(1):8–18. doi: 10.1111/lam.12690. [DOI] [PubMed] [Google Scholar]
- 11.Abbasi E., Hamid A., van Belkum A., Ghaznavi-Rad E. Multidrug-resistant Shigella infection in pediatric patients with diarrhea from central Iran. Infection and Drug Resistance . 2019;12:p. 1535. doi: 10.2147/idr.s203654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karimi-Yazdi M., Ghalavand Z., Shabani M., et al. High rates of antimicrobial resistance and virulence gene distribution among Shigella spp. isolated from pediatric patients in Tehran, Iran. Infection and Drug Resistance . 2020;13:p. 485. doi: 10.2147/idr.s238559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ranjbar R., Bolandian M., Behzadi P. Virulotyping of Shigella spp. isolated from pediatric patients in Tehran, Iran. Acta Microbiologica et Immunologica Hungarica . 2017;64(1):71–80. doi: 10.1556/030.64.2017.007. [DOI] [PubMed] [Google Scholar]
- 14.rizi S., Kobra H. F., Saeed Sasan M. High rate of resistance to ceftriaxone and azithromycin among Shigella spp. isolates at three children’s referral hospitals in northeast Iran. Journal of Infection and Chemotherapy . 2020;26(9):955–958. doi: 10.1016/j.jiac.2020.04.022. [DOI] [PubMed] [Google Scholar]
- 15.Chang Z., Lu S., Chen L., Jin Qi, Yang J. Causative species and serotypes of shigellosis in mainland China: systematic review and meta-analysis. PLoS One . 2012;7(12) doi: 10.1371/journal.pone.0052515.e52515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ingle D. J., Andersson P., Valcanis M., et al. Prolonged outbreak of multidrug-resistant Shigella sonnei harboring bla CTX-M-27 in victoria, Australia. Antimicrobial Agents and Chemotherapy . 2020;64(12):e01518–e01520. doi: 10.1128/AAC.01518-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ashkenazi S. Shigella infections in children: new insights. Seminars in Pediatric Infectious Diseases . 2004;15 doi: 10.1053/j.spid.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 18.Schroeder G. N., Hubert H. Molecular pathogenesis of Shigella spp.: controlling host cell signaling, invasion, and death by type III secretion. Clinical Microbiology Reviews . 2008;21(1):134–156. doi: 10.1128/cmr.00032-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jain S. K., Gupta A., Glanz B., Dick J., Siberry G. K. Antimicrobial-resistant Shigella sonnei: limited antimicrobial treatment options for children and challenges of interpreting in vitro azithromycin susceptibility. The Pediatric Infectious Disease Journal . 2005;24(6):494–497. doi: 10.1097/01.inf.0000164707.13624.a7. [DOI] [PubMed] [Google Scholar]
- 20.Niyogi S. K. Shigellosis. Journal of Microbiology . 2005;43(2):133–143. [PubMed] [Google Scholar]
- 21.Bowen A., Grass J., Bicknese A., Campbell D., Hurd J., Kirkcaldy R. D. Elevated risk for antimicrobial drug–resistant Shigella infection among men who have sex with men, United States, 2011–2015. Emerging Infectious Diseases . 2016;22(9):p. 1613. doi: 10.3201/eid2209.160624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Behzadi P., Gajdács M. . Writing a strong scientific paper in medicine and the biomedical sciences: a checklist and recommendations for early career researchers. Biologia Futura . 2021;72(4):395–407. doi: 10.1007/s42977-021-00095-z. [DOI] [PubMed] [Google Scholar]
- 23.Houpt E. R., Ferdous T., Ara R., et al. Clinical outcomes of drug-resistant shigellosis treated with azithromycin in Bangladesh. Clinical Infectious Diseases . 2021;72(10):1793–1798. doi: 10.1093/cid/ciaa363. [DOI] [PubMed] [Google Scholar]
- 24.Gomes C., Ruiz-Roldán L., Mateu J., Ochoa T. J., Ruiz J. Azithromycin resistance levels and mechanisms in Escherichia coli. Scientific Reports . 2019;9(1):p. 6089. doi: 10.1038/s41598-019-42423-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gomes C., Martínez-Puchol S., Palma N., et al. Macrolide resistance mechanisms in enterobacteriaceae: focus on azithromycin. Critical Reviews in Microbiology . 2017;43(1):1–30. doi: 10.3109/1040841x.2015.1136261. [DOI] [PubMed] [Google Scholar]
- 26.Aminshahidi M., Arastehfar A., Pouladfar G., Arman E., Fani F. Diarrheagenic Escherichia coli and Shigella with high rate of extended-spectrum beta-lactamase production: two predominant etiological agents of acute diarrhea in Shiraz, Iran. Microbial Drug Resistance . 2017;23(8):1037–1044. doi: 10.1089/mdr.2017.0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clinical and Laboratory Standards Institute (CLSI) CLSI Supplement M100–S29 . 29th. Wayne, PA, USA: Clinical and Laboratory Standards Institute; 2019. Performance standards for antimicrobial susceptibility testing. [Google Scholar]
- 28.Rahman M., Haque A. F., Mallik Deeba I., et al. Emergence of extensively drug-resistant Shigella sonnei in Bangladesh. Immunology of Infectious Diseases . 2017;5:1–9. doi: 10.13189/iid.2017.050101. [DOI] [Google Scholar]
- 29.Liu Y., Li H., Lv N., et al. Prevalence of plasmid-mediated determinants with decreased susceptibility to azithromycin among Shigella isolates in anhui, China. Frontiers in Microbiology . 2020;11(1181) doi: 10.3389/fmicb.2020.01181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gomes C., Pons M. J., Magallon-Tejada A., et al. In vitro development and analysis of Escherichia coli and Shigella boydii azithromycin–resistant mutants. Microbial Drug Resistance . 2013;19(2):88–93. doi: 10.1089/mdr.2012.0036. [DOI] [PubMed] [Google Scholar]
- 31.Zhang C., Zhang R., Qi Y., Chu X., Sun J., Liu Q. Decreased susceptibility to azithromycin among clinical Shigella isolates from China. Microbial Drug Resistance . 2017;23(5):596–601. doi: 10.1089/mdr.2016.0134. [DOI] [PubMed] [Google Scholar]
- 32.Salah M., Shtayeh I., Ghneim R., et al. Evaluation of Shigella species azithromycin CLSI epidemiological cutoff values and macrolide resistance genes. Journal of Clinical Microbiology . 2019;57(4):e01422–e01518. doi: 10.1128/JCM.01422-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murray K., Reddy V., Kornblum J., et al. Increasing antibiotic resistance in Shigella spp. from infected New York city residents, New York, USA. Emerging Infectious Disease Journal . 2017;23(2):p. 332. doi: 10.3201/eid2302.161203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown J. D., Willcox S. J., Franklin N., et al. Shigella species epidemiology and antimicrobial susceptibility: the implications of emerging azithromycin resistance for guiding treatment, guidelines and breakpoints. Journal of Antimicrobial Chemotherapy . 2017;72(11):3181–3186. doi: 10.1093/jac/dkx268. [DOI] [PubMed] [Google Scholar]
- 35.Diner E. J., Hayes C. S. Recombineering reveals a diverse collection of ribosomal proteins L4 and L22 that confer resistance to macrolide antibiotics. Journal of Molecular Biology . 2009;386(2):300–315. doi: 10.1016/j.jmb.2008.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phuc N. M. C., Woerther P.-L., Bouvet M., Antoine A., Leclercq R., Canu A. Escherichia coli as reservoir for macrolide resistance genes. Emerging Infectious Disease Journal . 2009;15(10):p. 1648. doi: 10.3201/eid1510.090696. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: clinical data of pediatric patients, drug resistance, and distribution of azithromycin resistance genes in DSA-Shigella spp.
Data Availability Statement
All the data generated or analyzed during this study were included in this article.


