Abstract
The enhanced transmissibility and immune evasion associated with emerging SARS-CoV-2 variants demands the development of next-generation vaccines capable of inducing superior protection amid a shifting pandemic landscape. Since a portion of the global population harbors some level of immunity from vaccines based on the original Wuhan-Hu-1 SARS-CoV-2 sequence or natural infection, an important question going forward is whether this immunity can be boosted by next-generation vaccines that target emerging variants while simultaneously maintaining long-term protection against existing strains. Here, we evaluated the immunogenicity of INO-4800, our synthetic DNA vaccine candidate for COVID-19 currently in clinical evaluation, and INO-4802, a next-generation DNA vaccine designed to broadly target emerging SARS-CoV-2 variants, as booster vaccines in nonhuman primates. Rhesus macaques primed over one year prior with the first-generation INO-4800 vaccine were boosted with either INO-4800 or INO-4802 in homologous or heterologous prime-boost regimens. Both boosting schedules led to an expansion of T cells and antibody responses which were characterized by improved neutralizing and ACE2 blocking activity across wild-type SARS-CoV-2 as well as multiple variants of concern. These data illustrate the durability of immunity following vaccination with INO-4800 and additionally support the use of either INO-4800 or INO-4802 in prime-boost regimens.
Keywords: COVID-19, DNA vaccine, SARS-CoV-2, Macaque, Electroporation, Intradermal
1. Introduction
SARS-CoV-2 is a beta-coronavirus belonging to the same family as severe acute respiratory coronavirus (SARS-CoV) and Middle East Respiratory Syndrome coronavirus (MERS-CoV), which share similar structural features including the spike glycoprotein which has been the primary target of vaccine development for each of these viruses [1]. Although the rollout of the EUA vaccines has been underway for several months, global distribution of these vaccines has fallen along entrenched socioeconomic lines, leaving many low- and middle-income countries with inadequate supply [2]. For successful global coverage, many more vaccines will be needed. The rapid expansion of SARS-CoV-2 variants of concern (VOC) has corresponded with a reduction in neutralizing antibody activity in convalescent and vaccinated individuals, suggesting that emerging mutations observed in some lineages are associated with immune escape [3], [4], [5], [6], [7]. Alarmingly, the Beta (B.1.351) variant has demonstrated a reduced sensitivity to neutralizing sera from convalescent and immunized individuals [7]. It has been observed that vaccine effectiveness (either BNT162b2 or ChAdOx1 nCoV-19) was notably lower against the now dominant Delta (B.1.617.2) variant, compared to the Alpha (B.1.1.7) variant [8]. Most recently, the highly mutated Omicron VOC has demonstrated further evasion of the existing humoral immunity to SARS-CoV-2 in the global population. The combination of viral escape mechanisms and waning immunity suggest that heterologous prime-boost strategies may be needed to provide sufficient coverage against novel variants [9].
Synthetic DNA vaccines offer multiple advantages over other vaccine platforms including shortened clinical development timetables for vaccines against emerging infectious diseases, ability to scale up manufacture, and long-term temperature stability that facilitates rapid and efficient deployment in resource-limited settings [10], [11], [12]. We have previously described the design of a synthetic DNA vaccine encoding the wild-type (Wuhan-Hu-1) Spike protein, INO-4800, which is currently in clinical evaluation [10]. In preclinical studies we have shown INO-4800 vaccination induces antigen-specific T cell responses and functional antibodies that neutralize and confer protection against SARS-CoV-2 [10], [13], [14], [15], [16]. In a non-human primate (NHP) challenge model, INO-4800 vaccination was associated with reduced viral loads and protection against respiratory tract disease [13], [14]. Phase 1 and 2 clinical trials of INO-4800 demonstrated a favorable safety and tolerability profile and immunogenicity [17], [18].
In response to the increasing number of SARS-CoV-2 VOCs demonstrating evasion of vaccine- or infection-induced humoral immunity, we have designed INO-4802, a next-generation DNA vaccine expressing a pan-Spike immunogen. INO-4802 was designed using the SynCon strategy with the goal of driving neutralizing coverage against multiple VOCs, as described previously [16]. INO-4802 raises immunity across SARS-CoV-2 VOCs in mice and confers broad protection in hamsters following intranasal challenge with multiple VOCs including Alpha, Beta, Gamma, and Delta [16].
Prime-boost regimens are widely used in the development of vaccines against a variety of infectious diseases [19], [20], including DNA and viral-vector based approaches [21], [22]. DNA vaccines have particular advantages in the prime-boost setting where they have been shown to enhance both humoral and cellular responses without inducing anti-vector immunity [23]. In the boost setting DNA vaccines were found to be superior to the adenovirus platform in expanding responses to simian immunodeficiency virus (SIV) antigens in rhesus macaques [24]. In the clinic, the DNA platform is not limited by the same dose-dependent reactogenicity observed following administration of lipid nanoparticles carrying mRNA vaccines [25], which may be an important consideration in booster acceptance.
In the current study, we investigated the durability and memory recall of antigen-specific SARS-CoV-2 responses in a cohort of non-human primates that were initially primed with the first-generation SARS-CoV-2 vaccine INO-4800. One year following the primary immunization series with INO-4800, the animals were boosted with either INO-4800 or INO-4802 in homologous or heterologous prime-boost regimens, respectively. Boosting with either INO-4800 or INO-4802 led to the induction of antigen-specific T cells and potent neutralizing antibody responses across multiple SARS-CoV-2 VOCs that correlated with ACE2 blocking activity. These data highlight the capability of DNA vaccines to boost SARS-CoV-2 immunity.
2. Results
2.1. Durability of SARS-CoV-2-specific humoral response following immunization with INO-4800
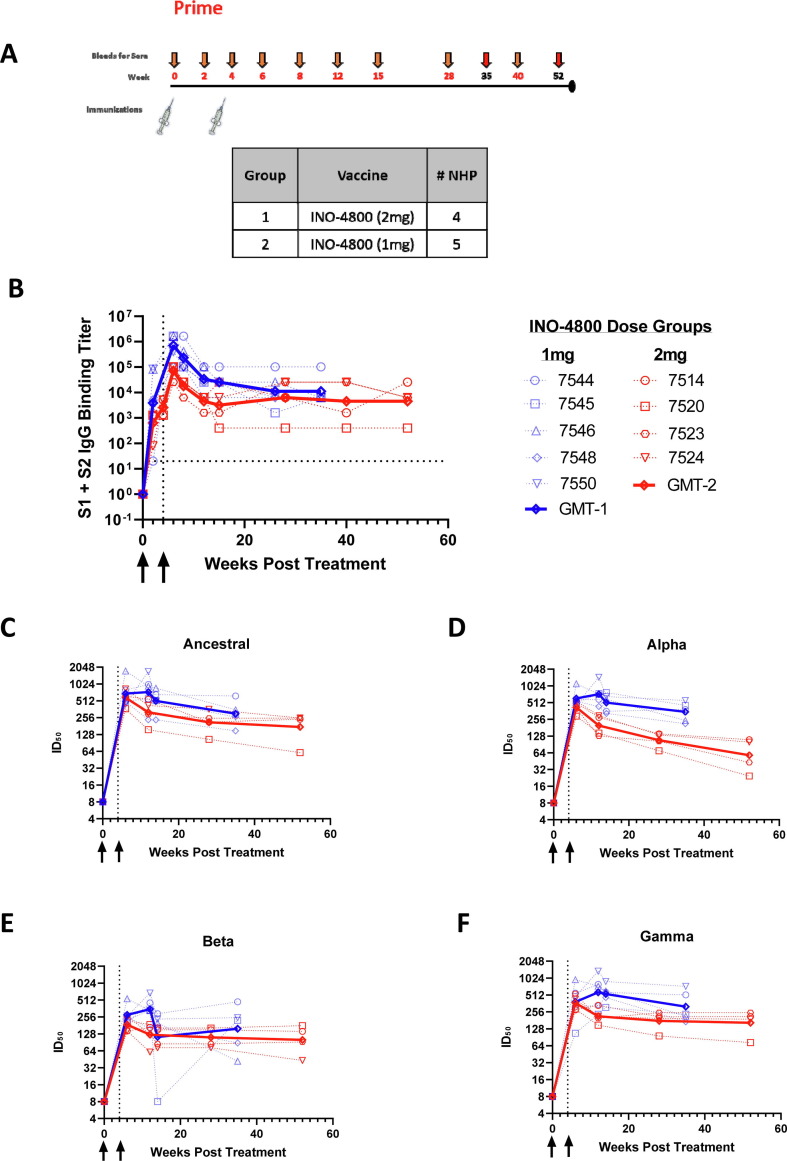
Initial studies investigated the durability of immune responses in non-human primates (NHPs) primed with INO-4800. NHPs were immunized at week 0 and 4 with either a 1 mg or 2 mg dose of INO-4800, and blood was collected over the course of one year (Fig. 1 A). It should be noted that, for Fig. 1 and Supplemental Fig. 1, the NHPs were initially treated on staggered schedules, and therefore the data from the prime immunization portion of the study will show collected data points for NHP IDs #7544, 7545, 7546, 7548, 7550 terminating at Week 35 and for others, IDs #7514, 7520, 7523, 7524, terminating at Week 52. To measure levels of binding antibodies in the sera we used an enzyme-linked immunosorbent assay (ELISA). Peak antibody titers were observed at week 6 with a geometric mean endpoint titer of 258,032, two weeks following the second immunization (Fig. 1 B). Detectable levels of binding antibodies persisted in the serum for the duration of the study, and at the final timepoint prior to boosting, the 1 mg dose group had geometric mean endpoint titers of 11,143 for the S1 + S2 ECD. The 2 mg dose group had geometric mean endpoint titers of 4525 for the S1 + S2 ECD. Similar trends were also observed in the levels of binding antibodies against the SARS-CoV-2 S1, SARS-CoV-2 S2 and RBD proteins (Supplemental Fig. 1).
Fig. 1.
Study design and durability of humoral immune responses in rhesus macaques primed with INO-4800. A) Schematic depicting the prime immunization schedule and sample collection timepoints. Note: The longitudinal collection for the NHPs in the 1 mg dose group ended at Week 35 and for 2 mg dose group at Week 52. B) Longitudinal serum IgG binding titers in rhesus macaques vaccinated with 1 or 2 mg INO-4800 at weeks 0 and 4. Antibody titers in the sera were measured against the wildtype SARS-CoV-2 Spike protein antigen. C-F) Longitudinal serum pseudovirus neutralizing activity in rhesus macaques. Neutralizing activity (ID50) was measured against the ancestral (Wuhan-Hu-1) SARS-CoV-2 (C) as well as the Alpha (D), Beta (E) and Gamma (F) pseudoviruses.
Functional antibody responses were measured in a pseudovirus neutralization assay against the SARS-CoV-2 ancestral, Alpha, Beta and Gamma variants of concern (VOCs) which were in circulation during this time period. Immunization with INO-4800 resulted in the induction of neutralizing antibodies that were increased over baseline for all VOCs (Fig. 1 C-F). SARS-CoV-2 VOC neutralizing antibody responses were durable and remained elevated over baseline at the last collected timepoint, with the 1 mg dose group having a geometric mean titer (GMT) of 301 against ancestral SARS-CoV-2, 349 for Alpha, 158 for Beta, and 317 for Gamma. In Fig. 1E, the authors note that NHP #7545 showed reduced neutralizing activity at Week 14 which was attributed to sampling error during plating. The 2 mg dose group had a GMT of 174.6 for the wild-type variant, 58.2 for Alpha, 100.3 for Beta, and 164.2 for Gamma. Together, these data illustrate that the primary INO-4800 vaccination schedule induced SARS-CoV-2 specific antibodies harboring neutralizing activity that were maintained over the period of 35–52 weeks.
2.2. Humoral responses following delivery of either INO-4800 or INO-4802
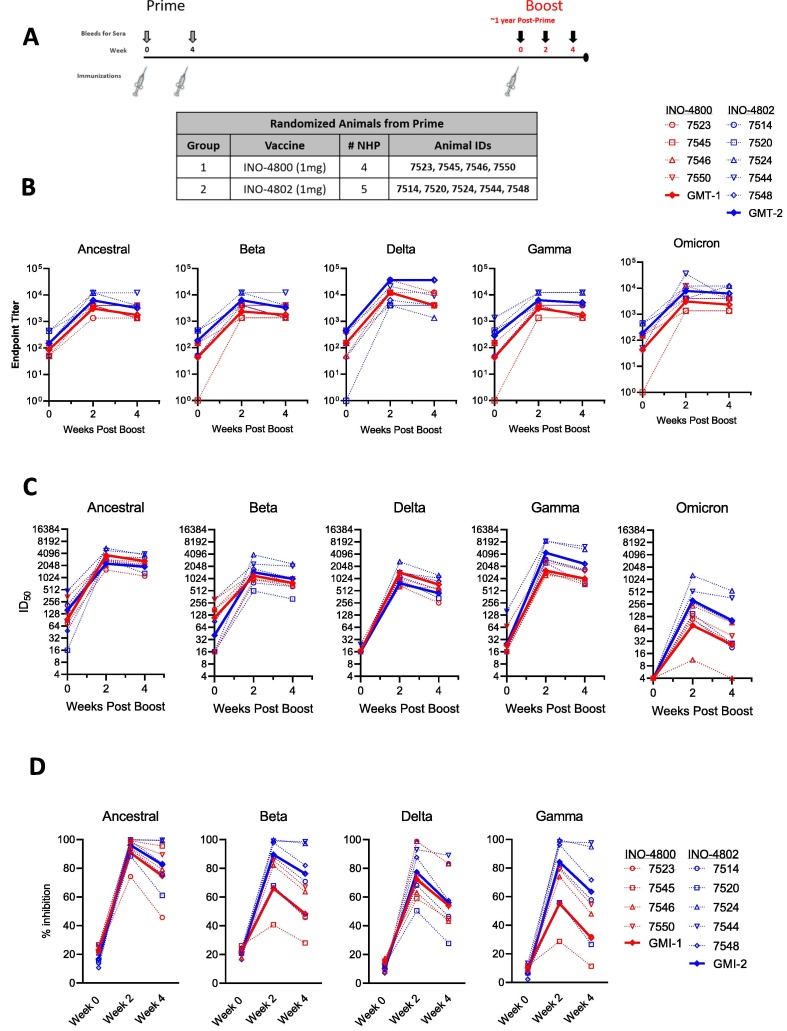
We evaluated INO-4800 and INO-4802 as booster vaccines. The same rhesus macaques that were initially primed with INO-4800 were randomized into two groups and boosted with either INO-4800, homologous to the original vaccine, or INO-4802, an updated pan-SARS-CoV-2 Spike immunogen in a heterologous boost regimen. Rhesus macaques #7544, 7545, 7546, 7548, 7550 were boosted 43 weeks after the initial vaccination while NHPs #7514, 7520, 7523, 7524, were boosted at 64 weeks after the initial vaccination (Fig. 2 A).
Fig. 2.
Humoral immune responses following homologous or heterologous boost in INO-4800-primed rhesus macaques. Antibody responses were measured in animals boosted with 1 mg of either the homologous INO-4800 (red symbols) or heterologous INO-4802 (blue symbols) vaccines on the day of the boost (week 0) and at weeks 2 and 4 post-boost. Red lines and blue lines represent geometric mean titers (GMT) or geometric mean inhibition (GMI) for groups 1 and 2, respectively. A) Schematic of the boost schedule showing the vaccine groups with the respective animal IDs. B) Serum IgG binding titers in rhesus macaques boosted with INO-4800 or INO-4802. Binding titers were measured against the ancestral, Beta, Delta, Gamma, and Omicron Spike proteins. C) Serum pseudovirus neutralizing activity in rhesus macaques boosted with INO-4800 or INO-4802. Neutralizing activity was measured against the ancestral, Beta, Delta, Gamma, and Omicron pseudoviruses. D) ACE2 blocking activity in the serum collected from rhesus macaques boosted with INO-4800 or INO-4802. Inhibition of ACE2 binding was measured against the ancestral, Beta, Delta, and Gamma Spike proteins. In panels B-D, comparisons between INO-4800- and INO-4802-boosted animals at Weeks 2 and 4 were performed using a Mann Whitney test. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The homologous boost with INO-4800 resulted in the induction of antibody titers at two weeks post-boost that were increased over pre-boost levels (Fig. 2 B). Increases in binding antibody levels showed similar patterns against the ancestral, Beta, Delta, Gamma, and Omicron Spike proteins, with GMTs of 87, 43, 342, 43, and 43, respectively, pre-boost and 3077, 2338, 21044, 3077, and 3077, respectively, post-boost. Likewise, heterologous boost with INO-4802 also led to increased binding antibodies against all variants tested with GMTs of 150, 187, 44, 290, and 187, respectively, pre-boost and 6285, 6285, 6285, 6285, and 7829, respectively, two weeks post-boost for the wild-type, Beta, Delta, Gamma, and Omicron variants (Fig. 2 B). Binding titers against any of the variants were not significantly different between INO-4800- and INO-4802-boosted animals at either Week 2 or Week 4.
Neutralizing activity against the ancestral, Beta, Delta, Gamma, and Omicron variants was assessed by a pseudovirus neutralization assay, which revealed increased neutralizing antibody responses against all SARS-CoV-2 variants in animals boosted with either INO-4800 or INO-4802 (Fig. 2 C). The GMTs at Week 2 for the NHPs after the homologous INO-4800 boost were 2286.2, 1199.3, 785.6, 1596.1, and 78.3 against the ancestral, Beta, Delta, Gamma, and Omicron pseudoviruses, respectively. The GMTs at Week 2 for the NHPs after the heterologous INO-4802 boost were 3712.0, 1452.1, 1434.8, 4389.6, and 312.9 against the ancestral, Beta, Delta, Gamma, and Omicron pseudoviruses, respectively. At Week 2, INO-4802-boosted NHPs showed significantly greater neutralizing activity against the Gamma and Delta pseudoviruses than animals boosted with INO-4800 (P = 0.0317 and 0.0317, respectively), although by Week 4, there was not a significant difference between the boost groups. INO-4800- and INO-4802-boosted animals did not show a significant difference in neutralization of the ancestral, Beta, and Omicron pseudoviruses at either timepoint. As an additional readout of functional antibody responses, we measured ACE2/SARS-CoV-2 Spike interaction blocking activity of serum antibodies using a Meso Scale Discovery (MSD) assay, by quantifying the level of inhibition of ACE2 binding to a panel of variant SARS-CoV-2 Spike proteins. In line with the pseudovirus neutralization data, all animals showed an increase in the level of functional anti-SARS-CoV-2 antibodies in their serum following the boost immunization (Fig. 2 D). ACE2 blocking activity against any of the variants was not significantly different between INO-4800- and INO-4802-boosted animals at either Week 2 or Week 4 (P = 0.4127, 0.0635, 0.7937, and 0.0635 against the ancestral, Beta, Delta, and Gamma VOCs, respectively, at Week 2 and P = 0.7302, 0.0635, 0.6032, and 0.1111 against the ancestral, Beta, Delta, and Gamma VOCs, respectively, at Week 4). We observed positive correlations between pseudovirus neutralization and inhibition of the ACE2/SARS-CoV-2 Spike interaction (Supplemental Fig. 2 A), supporting the overall functional antibody responses observed in animals receiving either booster vaccine.
We next evaluated T follicular helper cells (Tfh) cells, an important cell type in the generation of high-affinity antibodies [26], [27], [28]. The frequency of circulating Tfh cells positively correlated with ACE2 blocking activity at week 2 in animals boosted with INO-4800 and INO-4802 (Supplemental Fig. 2 B), supporting the generation of functional antibody responses following a boost with SARS-CoV-2 DNA vaccines. Together, these data show an augmentation of humoral responses following a boost with either INO-4800 or INO-4802 in the context of existing SARS-CoV-2 immunity, possibly increasing the breadth of immune response against multiple VOCs.
2.3. Induction of cellular responses by INO-4800 or INO-4802
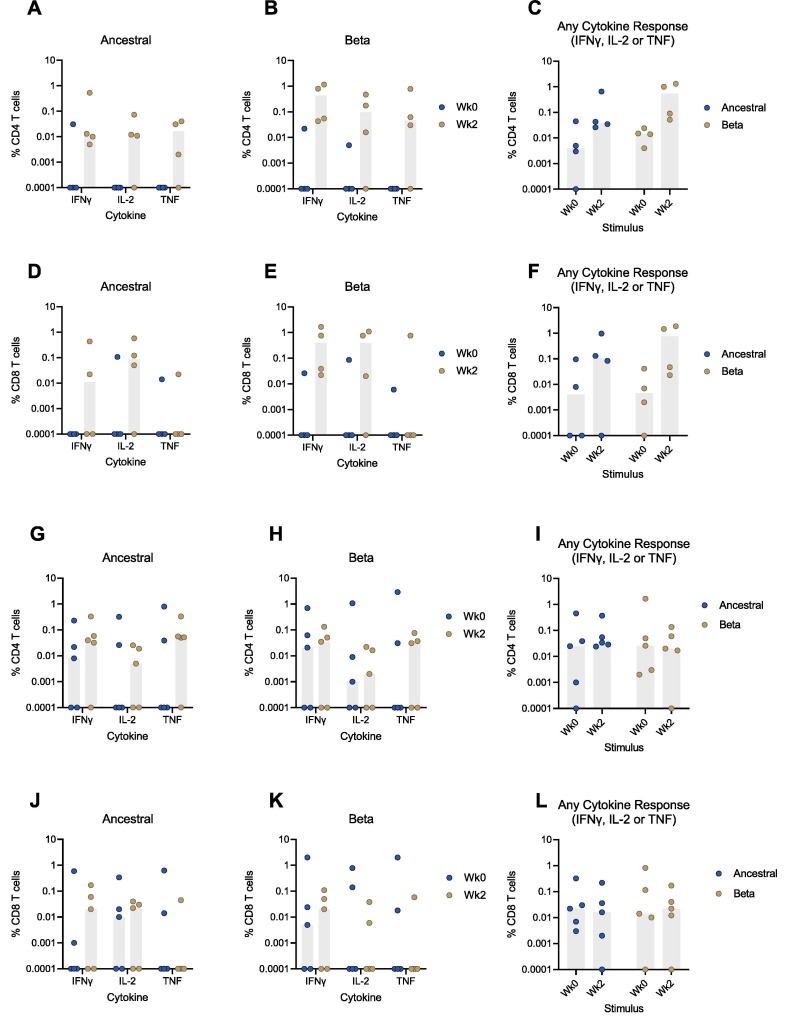
Intracellular cytokine staining (ICS) was performed on peripheral blood mononuclear cells (PBMCs) stimulated with peptides matching the ancestral or Beta SARS-CoV-2 Spike proteins to evaluate cellular responses in rhesus macaques boosted with either INO-4800 or INO-4802. Antigen-specific CD4 and CD8 T cell responses were observed in animals boosted with either vaccine (Fig. 3 ). The magnitude of cellular responses was generally greater at 2 weeks post-boost relative to pre-boost levels and showed that boosting with INO-4800 induced CD4 T cell responses that were maintained across the ancestral and Beta variants (Fig. 3 A-C). Phenotypic analysis of the CD4 T cell responses at Week 2 showed IFNγ secretion in all animals and IL-2 and TNF secretion in 3 of 4 animals (Fig. 3 A-B). Similar responses were observed in the CD8 compartment at Week 2, showing secretion of IFNγ (2 of 4 animals for ancestral and 4 of 4 animals for Beta) and IL-2 (3 of 4 animals for each VOC) (Fig. 3 D-F).
Fig. 3.
Cellular immune responses following homologous or heterologous boost in INO-4800-primed rhesus macaques. T cell responses were measured in animals boosted with 1 mg of either the homologous INO-4800 (A - F) or heterologous INO-4802 (G - L) vaccines on the day of the boost (week 0) and at week 2 post-boost. A - C) CD4 and (D – F) CD8 T cell responses in INO-4800-boosted animals against ancestral or Beta derived peptide pools. G - I) CD4 and (J – L) CD8 T cell responses in INO-4802-boosted animals against ancestral or Beta derived peptide pools. The sum of IFNγ, IL-2, and TNF responses are represented in C, F, I, and L. Bars represent median.
Alternatively, boost with INO-4802 also induced CD4 T cell responses in most animals (Fig. 3 G-I). Here, CD4 T cell responses against the ancestral and Beta variants at Week 2 were characterized by the secretion of IFNγ (4 of 5 animals and 3 of 5 animals, respectively), IL-2 (3 of 5 animals for both VOCs), and TNF (4 of 5 animals and 3 of 5 animals, respectively) (Fig. 3 G-H). Most INO-4802-boosted animals also showed responses in the CD8 compartment at Week 2 which were predominantly characterized by the secretion of IFNγ (3 of 5 animals for both VOCs) and IL-2 (3 of 5 animals and 2 of 5 animals, for ancestral and Beta respectively) (Fig. 3 J- L).
3. Discussion
Emerging SARS-CoV-2 VOCs and waning immunity will likely lead to recurrent waves of COVID-19 disease [29]. The frequency and severity of future outbreaks will depend on several complex factors that will likely unfold differently across the world [30]. The duration of immunity following SARS-CoV-2 infection and vaccination will be a key determinant of future SARS-CoV-2 transmission dynamics [30]. Accumulating evidence suggests that durable immune responses are maintained in COVID-19 convalescent individuals and vaccinees [31], [32], [33], [34]. However, the emergence of SARS-CoV-2 variants capable of evading humoral immune responses [5], [35] highlights the potential need to update vaccines to mitigate the severity of future SARS-CoV-2 outbreaks. More broadly protective vaccines that can be administered as boosters may be critical in maintaining levels of protection against new outbreak waves with antigenically divergent SARS-CoV-2 lineages. Recent studies have shown that the mutations associated with emerging VOCs have a negative impact on neutralizing antibody responses and on the efficacy of SARS-CoV-2 vaccines [36], [3], [4], [5], [6], [7]. Data show that the adenovirus-vectored ChAdOx1 nCoV-19 vaccine and nanoparticle-based NVX-CoV2373 vaccine have lower efficacy against the Beta variant compared to the overall vaccine efficacy [37], [38].
The development of next-generation vaccines is one approach to broadening immune coverage against emerging SARS-CoV-2 variants, and the immunogenicity of booster vaccines that are heterologous from previous vaccinations will play an important role in informing global immunization strategies. It was found that individuals who received a primary immunization series of mRNA-1273 and subsequently received a booster shot of Moderna’s updated mRNA vaccine encoding the Beta Spike protein, mRNA-1273.351, showed increases in antibody neutralization titers against the Beta and Gamma variants that were superior to those in individuals who received a booster shot of mRNA-1273 [39]. Other studies have assessed the safety and immunogenicity of heterologous prime-boost regimens involving different vaccine platforms. A recent study involving individuals who originally received one of the three EUA vaccines (mRNA-1273, Ad26.COV2.S, or BNT162b2) and were subsequently boosted with a homologous or heterologous vaccine suggests that both boosting schedules increase protective efficacy against symptomatic SARS-CoV-2 infection [40]. In a separate study, favorable increases in humoral responses were observed in individuals previously vaccinated with the ChAdOx1 nCoV-19 vaccine and subsequently boosted with BNT162b2 [41], [42]. Likewise, a heterologous boost of mRNA-1273 also resulted in an increase in humoral responses in individuals primed with ChAdOx1 nCoV-19 [43]. However, homologous boosting strategies have also shown promise in enhancing humoral responses against SARS-CoV-2 VOCs. A third dose of BNT162b2, for instance, was reported to increase neutralizing activity against the Delta variant over 5-fold in 18–55-year-olds and over 11-fold in 65–85-year-olds (Pfizer Second Quarter 2021 Earnings Report). We have observed in Phase 1 trial participants that a third dose of INO-4800, our DNA vaccine candidate for COVID-19, results in higher levels of cellular and humoral immune responses and is well tolerated without increased levels of adverse events [44].
We recently described the design, immunogenicity, and efficacy of INO-4802, a synthetic DNA vaccine expressing a pan-Spike immunogen aimed at inducing broad immunity across SARS-CoV-2 VOCs [16]. In a hamster challenge model, INO-4802 conferred protection following intranasal challenge with the ancestral virus as well as the Alpha, Beta, Gamma, or Delta SARS-CoV-2 variants. Additionally, INO-4802 showed promise as a heterologous booster vaccine by enhancing humoral responses against VOCs in hamsters previously immunized with INO-4800. Here, we addressed the immunogenicity of INO-4800 and INO-4802 as booster regimens in rhesus macaques previously immunized with INO-4800 using clinically relevant dosing parameters.
Rhesus macaques receiving booster immunizations of either INO-4800 or INO-4802 showed a robust induction of humoral responses, supporting the use of either vaccine in a prime/boost regimen. Importantly, boosting of INO-4800-primed animals with INO-4800 or INO-4802 resulted in neutralizing antibody responses that were magnitudes greater compared to pre-boost levels. Both treatment groups induced humoral responses capable of neutralizing wild-type and several VOC pseudoviruses, suggesting broad protection among SARS-CoV-2 variants. Pseudovirus neutralizing activity against the Beta and Gamma variants trended higher in animals boosted with the heterologous INO-4802 vaccine compared to those receiving INO-4800, indicating potential for an enhanced level of protection against some emerging SARS-CoV-2 variants following boosting with the next-generation pan-SARS-CoV-2 vaccine.
Neutralizing antibody responses correlated with inhibition of ACE2 binding activity, further supporting the functional antibody responses following either a homologous or heterologous boost with synthetic DNA vaccine constructs. Levels of antibodies binding variant Spike proteins were also increased following the boost immunization in both treatment groups. Together, these data point to broad functional humoral responses following a boost with both the original INO-4800 and INO-4802 pan-SARS-CoV-2 DNA vaccines. The rapid boost in neutralizing antibody responses can likely be attributed to the maintenance of a memory B cell pool following the priming immunization. Similar increases in humoral responses are observed in COVID-19 convalescent individuals who later received SARS-CoV-2 mRNA vaccines [45]. Longitudinal analyses have also found that SARS-CoV-2-reactive memory B cell clones are stably maintained in convalescent COVID-19 patients for several months following infection [31], [46]. Memory B cell responses persist despite the natural decline of SARS-CoV-2-specific IgG binding titers, suggestive of high-quality and durable memory B cell responses [47].
Neutralizing antibody responses are predictive of immune protection against symptomatic SARS-CoV-2 infection [48], and as such, neutralizing antibodies are an important readout in the evaluation of SARS-CoV-2 vaccines [49], [50], [51]. Owing to the critical role of T follicular helper (Tfh) cells in providing help to maturing B cells in germinal centers, Tfh responses serve as a mechanistic indicator of neutralizing antibody responses in infection and vaccination, including for EUA SARS-CoV-2 vaccines [26], [27], [28], [52], [53], [54]. We observed a positive correlation between the frequency of circulating Tfh cells and functional antibody responses, further affirming the immunogenicity of SARS-CoV-2 DNA vaccine boosters in animals with existing vaccine-induced immunity.
Rhesus macaques boosted with either INO-4800 or INO-4802 also showed rapid recall of antigen-specific T cell responses exhibiting a similar magnitude of immune responses against the ancestral and Beta variants. These findings are consistent with clinical evidence showing that T cell responses induced by different vaccine platforms, including INO-4800, are largely maintained across SARS-CoV-2 variants [36], [55], [56]. Together, these data underscore the utility of the synthetic DNA platform for booster vaccination against emerging SARS-CoV-2 variants.
Preliminary data was collected in this study against the SARS-CoV-2 Omicron variant. The Omicron SARS-CoV-2 Spike antigen has an unprecedented number of mutations compared to previous VOCs. There are 15 mutations in the Spike RBD region alone, an important target for neutralizing antibodies. While we observed induction of neutralizing antibodies against the Omicron pseudovirus, these responses were not as robust as levels of neutralizing activity previously observed against other VOCs. Nonetheless, it is encouraging that vaccination can significantly boost the levels of Omicron targeting neutralizing antibodies. Maintenance of T cell responses has been observed against other variants even in the presence of reduced antibody responses [36]. Future studies will provide additional data on vaccine-mediated protection against Omicron and whether T cell immunity is maintained.
Overall, the development of safe and effective booster vaccines will be critical in maintaining control of SARS-CoV-2 in the long term. Ideal treatment regimens should seek to expand immune coverage to emerging variants while maintaining immune responses to existing SARS-CoV-2 variants. Current focus has shifted to evaluating the cross-immunogenicity of booster vaccines against wild-type SARS-CoV-2 antigens and other VOCs and re-designing vaccines to investigate this important question. In this study, we report that the next-generation pan-SARS-CoV-2 vaccine INO-4802 boosts immune responses in animals primed with the wild-type-matched SARS-CoV-2 DNA vaccine INO-4800. These data support the immunogenicity and boosting capability of INO-4800 and INO-4802 in nonhuman primates, which may have broader application in the clinical setting.
4. Methods
4.1. Constructs
The plasmid designs for INO-4800 and INO-4802 have been previous described [10], [16]. For INO-4802, a SynCon® consensus sequence for the SARS-CoV-2 spike harboring focused RBD mutations and 2P mutation was codon-optimized using Inovio’s proprietary optimization algorithm. The final sequence was subcloned into the pGX0001 vector (BamHI/XhoI) and synthesized (Genscript, Piscataway, NJ).
4.2. Animal immunizations, sample collection
All rhesus macaque experiments were approved by the Institutional Animal Care and Use Committee at Bioqual (Rockville, Maryland), an Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) International accredited facility. Nine Chinese rhesus macaques, five males and four females roughly 4 years of age (weights ranging from 4.48 kg to 8.50 kg) were randomized prior to immunization and received one or two injections at 1 mg per dose of INO-4800, at weeks 0 and 4 by intradermal electroporation (ID-EP) administration using the CELLECTRA 2000® Adaptive Constant Current Electroporation Device with a 3P array (Inovio Pharmaceuticals). Approximately one year post prime immunization, the study animals were randomized and received a boost immunization at 1 mg per dose of INO-4800 or INO-4802 by ID-EP. At the indicated time-points, blood was collected to analyse blood chemistry and to isolate peripheral blood mononuclear cells (PBMC) and serum.
4.3. Peripheral blood mononuclear cell isolation and intracellular cytokine staining (ICS)
Blood was collected from each study animal into sodium citrate cell preparation tubes (CPT, BD Biosciences). The tubes were centrifuged to separate plasma and lymphocytes, according to the manufacturer’s protocol. Samples from the prime immunization were transported by same-day shipment on cold-packs from Bioqual to The Wistar Institute, and boost samples were shipped overnight to Inovio Pharmaceuticals for PBMC isolation. PBMCs were washed, and residual red blood cells were removed using ammonium-chloride-potassium (ACK) lysis buffer. Cells were counted using a ViCell counter (Beckman Coulter) and cryopreserved in 90% fetal bovine serum (FBS) / 10% dimethyl sulfoxide (DMSO). For ICS assays, cells were thawed in RPMI 1640 (Corning), supplemented with 10% fetal bovine serum (Seradigm), and 1% penicillin/streptomycin (Gibco).
For ICS, following an overnight rest at 37°C, PBMCs (1x106/sample) were added to each well and stimulated with either 1) SARS-CoV-2-specific peptide pools, 2) R10 with DMSO (negative control), or 3) eBioscience Cell Stimulation Cocktail containing PMA and ionomycin (Invitrogen, 1:1000 dilution) in the presence of GolgiStop and GolgiPlug (Invitrogen) and anti-CD28/CD49d. Plates were incubated for 6 h at 37°C, 5% CO2, washed, and then stained using an antibody cocktail containing anti-CD3 APC-Cy7, anti-CD4 PerCP-Cy5.5, anti-CD8 BV786, and LIVE/DEAD Fixable Aqua Dead Cell Stain (Invitrogen). Cells were then fixed, permeabilized (eBioscience Foxp3/Transcription Factor Fixation/Permeabilization Kit; ThermoFisher), and then stained for intracellular cytokines using an antibody cocktail containing anti-IFNγ BV605, anti-IL-2 BV650, and anti-TNFα APC-R700. Cells were then washed, resuspended and acquired on a BD FACS Celesta. Data were analyzed using FlowJo v10.7 Software (BD Life Sciences).
4.4. Antigen binding ELISA
Serum collected at each time point was evaluated for binding titers as previously described [10]. For prime immunization samples, ninety-six well immunosorbent plates (NUNC) were coated with 1ug/mL recombinant SARS-CoV-2 S1 + S2 ECD protein (Sino Biological 40589-V08B1), S1 protein (Sino Biological 40591-V08H), S2 protein (Sino Biological 40590-V08B), or receptor-binding domain (RBD) protein (Sino Biological 40595-V05H) in PBS overnight at 4°C. For boost samples, ELISA half-area plates were also coated with 1 µg/mL recombinant spike wild-type spike protein, Beta, Gamma, Delta, and Omicron full length spike variant proteins (Acro Biosystems #SPN-C52H8, #SPN-C52Hc, #SPN-C52Hg, #SPN-C52He, and #SPN-C52Hz, respectively). Secondary antibodies included IgG (Bethyl #A140-202P) at 1:50,000. Plates were washed three times with PBS + 0.05% Tween20 (PBS-T) and blocked with 3% FBS in PBS-T for 2 h at room temperature (RT). Sera from vaccinated macaques were serially diluted in PBS-T + 1% FBS, added to the washed ELISA plates, and then incubated for 2 h at RT. Plates were then washed and incubated with an anti-monkey IgG conjugated to horseradish peroxidase (Bethyl A140-202P) 1 h at RT. Within 30 min of development, plates were read at 450 nm using a Biotek Synergy2 plate reader.
4.5. Meso Scale Discovery ACE2 inhibition assay
Meso Scale Discovery (MSD) V-PLEX SARS-CoV-2 ACE2 Neutralization Kit, Panels 5 and 14 were used to evaluate sera collected from immunized study animals according to the manufacturer’s instructions with the MSD Sector S 600 instrument. Briefly, MSD plates containing SARS-CoV-2 Spike proteins (wildtype, Beta, Gamma and Delta) were blocked, washed, and incubated with sera from vaccinated animals at a 1:27 dilution. Plates were then washed and incubated with SULFO-TAG ACE2 and developed according to the manufacturer’s protocol. Functional antibody activity was measured as percent inhibition of binding of SULFO-TAG ACE2 to Spike protein.
4.6. Pseudovirus neutralization assay
SARS-CoV-2 pseudovirus stocks encoding for the wildtype, Alpha, Gamma, Beta, Delta, or Omicron Spike proteins were produced as previously described [10], [36]. To assess the extent of which neutralizing antibodies are present in the sera, CHO cells stably expressing ACE2 (ACE2-CHOs – Creative Biolabs) were used as target cells at 7,000 cells/well. Sera was heat inactivated and serially diluted prior to incubation with the different SARS-CoV-2 variant pseudoviruses. After a 90-minute incubation, sera-pseudovirus mixture was added to ACE2-CHOs, then 72 h later, cells were lysed using Britelite plus Reporter Gene Assay System (PerkinElmer) and RLU was measured using an automated luminometer. Neutralization titers (ID50) were calculated using GraphPad Prism 9 and defined as the reciprocal serum dilution that is reduced by 50% compared to the signal in the infected control wells.
4.7. Flow cytometry
Thawed, cryopreserved PBMCs were assessed to determine the frequency of circulating T follicular helper (Tfh) using a panel which included the following antibodies: CD3 (BD Biosciences; clone SP34-2), CD4 (BD Biosciences; clone L200), CXCR5 (eBioscience; clone MU5UBEE), and PD-1 (BioLegend; clone EH12.2H7). Tfh cells were identified as CD3+/CD4+/CXCR5+/PD-1 +. Samples were acquired on a BD FACS Celesta flow cytometer and analysed using FlowJo v10.7 Software version (BD Life Sciences).
CRediT authorship contribution statement
Jewell N. Walters: Conceptualization, Methodology, Investigation, Writing – original draft, Writing – review & editing, Supervision, Project administration. Blake Schouest: Writing – review & editing, Methodology, Investigation, Writing – original draft, Supervision, Project administration, Project administration. Ami Patel: Supervision, Writing – review & editing, Conceptualization, Methodology, Investigation, Project administration. Emma L. Reuschel: Methodology, Investigation. Katherine Schultheis: Methodology, Investigation. Elizabeth Parzych: Methodology, Investigation. Igor Maricic: Investigation. Ebony N. Gary: Investigation. Mansi Purwar: Investigation. Viviane M. Andrade: Investigation. Arthur Doan: Investigation. Dustin Elwood: Investigation. Zeena Eblimit: Investigation. Brian Nguyen: Investigation. Drew Frase: Investigation. Faraz I. Zaidi: Investigation. Abhijeet Kulkarni: Investigation. Alison Generotti: Investigation. J Joseph Kim: Writing – review & editing, Project administration, Funding acquisition. Laurent M. Humeau: Writing – review & editing, Project administration, Funding acquisition. Stephanie J. Ramos: Conceptualization, Supervision, Project administration. Trevor R.F. Smith: Supervision, Writing – review & editing, Conceptualization, Methodology, Project administration. David B. Weiner: Writing – review & editing, Conceptualization, Methodology, Supervision, Project administration, Funding acquisition. Kate E. Broderick: Conceptualization, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Kate Broderick reports financial support was provided by Coalition for Epidemic Preparedness Innovations. David B. Weiner reports a relationship with Inovio Pharmaceuticals Inc that includes: board membership, consulting or advisory, and equity or stocks. A.P., E.L.R., E.P., E.N.G., M.P., D.F., F.I.Z, A.K., declare no competing interests. J.N.W., B.S., K.S., I.M., Z.E., A.D., D.E., A.G., V.M.A., J.J.K., L.M.H., S.J.R., T.R.F.S., K.E.B. are employees of Inovio Pharmaceuticals and as such receive salary and benefits, including ownership of stock and stock options, from the company. D.B.W. discloses the following paid associations with commercial partners: GeneOne (Consultant), Geneos (Advisory Board), AstraZeneca (Advisory Board, Speaker), Inovio (BOD, SRA, Stock), Pfizer (Speaker), Merck (Speaker), Sanofi (Advisory Board), BBI (Advisory Board).
Acknowledgements
The studies described in this manuscript were funded by a grant from the Coalition for Epidemic Preparedness Innovations (CEPI). The authors would like to additional thank Maria Yang, Roi Ferrer, Joseph Fader, Francisco Vega Vega and Jon Schantz at Inovio Pharmaceuticals for their assistance, and John Harrison and Fabian Paz at Bioqual for their expert assistance.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.vaccine.2022.03.060.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Fig. S1. Humoral Responses in INO-4800 vaccinated rhesus macaques. IgG binding was measured in sera from INO-4800 vaccinated rhesus macaques to SARS-CoV-2 RBD (A), S1 (B) and S2 (C) protein antigens. Fig. S2. Functional antibody responses following homologous or heterologous boost in INO-4800-primed rhesus macaques. A) Spearman correlation of ACE2 blocking activity and neutralizing activity among animals boosted with either INO-4800 or INO-4802. Correlations relating to functional antibody responses against the wildtype (left) Beta SARS-CoV-2 (center), and Delta (right) variants at weeks 2 and 4 post-boost are shown. B) Spearman correlation of the frequency of circulating T follicular helper cells with ACE-2 binding inhibition at week 2 post-boost.
References
- 1.Tse L.V., Meganck R.M., Graham R.L., Baric R.S. The Current and Future State of Vaccines, Antivirals and Gene Therapies Against Emerging Coronaviruses. Front Microbiol. 2020;11:658. doi: 10.3389/fmicb.2020.00658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mullard A. How COVID vaccines are being divvied up around the world. Nature. 2020 doi: 10.1038/d41586-020-03370-6. [DOI] [PubMed] [Google Scholar]
- 3.Wibmer C.K., Ayres F., Hermanus T., Madzivhandila M., Kgagudi P., Oosthuysen B., et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat Med. 2021;27(4):622–625. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- 4.Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593(7857):130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 5.Garcia-Beltran W.F., Lam E.C., St. Denis K., Nitido A.D., Garcia Z.H., Hauser B.M., et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021;184(9):2372–2383.e9. doi: 10.1016/j.cell.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu K, Werner AP, Moliva JI, Koch M, Choi A, Stewart-Jones GBE, et al. mRNA-1273 vaccine induces neutralizing antibodies against spike mutants from global SARS-CoV-2 variants. bioRxiv. 2021:2021.01.25.427948. doi: 10.1101/2021.01.25.427948.
- 7.Planas D., Bruel T., Grzelak L., Guivel-Benhassine F., Staropoli I., Porrot F., et al. Sensitivity of infectious SARS-CoV-2 B.1.1.7 and B.1.351 variants to neutralizing antibodies. Nat Med. 2021;27(5):917–924. doi: 10.1038/s41591-021-01318-5. [DOI] [PubMed] [Google Scholar]
- 8.Lopez Bernal J., Andrews N., Gower C., Gallagher E., Simmons R., Thelwall S., et al. Effectiveness of Covid-19 Vaccines against the B.1.617.2 (Delta) Variant N Engl J Med. 2021;385(7):585–594. doi: 10.1056/NEJMoa2108891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barros-Martins J., Hammerschmidt S.I., Cossmann A., Odak I., Stankov M.V., Morillas Ramos G., et al. Immune responses against SARS-CoV-2 variants after heterologous and homologous ChAdOx1 nCoV-19/BNT162b2 vaccination. Nat Med. 2021 doi: 10.1038/s41591-021-01449-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith T.R.F., Patel A., Ramos S., Elwood D., Zhu X., Yan J., et al. Immunogenicity of a DNA vaccine candidate for COVID-19. Nat Commun. 2020;11(1) doi: 10.1038/s41467-020-16505-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maslow J.N. Vaccine development for emerging virulent infectious diseases. Vaccine. 2017;35(41):5437–5443. doi: 10.1016/j.vaccine.2017.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tebas P., Kraynyak K.A., Patel A., Maslow J.N., Morrow M.P., Sylvester A.J., et al. Intradermal SynCon(R) Ebola GP DNA Vaccine Is Temperature Stable and Safely Demonstrates Cellular and Humoral Immunogenicity Advantages in Healthy Volunteers. J Infect Dis. 2019;220(3):400–410. doi: 10.1093/infdis/jiz132. [DOI] [PubMed] [Google Scholar]
- 13.Gooch K.E., Smith T.R.F., Salguero F.J., Fotheringham S.A., Watson R.J., Dennis M.J., et al. One or two dose regimen of the SARS-CoV-2 synthetic DNA vaccine INO-4800 protects against respiratory tract disease burden in nonhuman primate challenge model. Vaccine. 2021;39(34):4885–4894. doi: 10.1016/j.vaccine.2021.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patel A., Walters J.N., Reuschel E.L., Schultheis K., Parzych E., Gary E.N., et al. Intradermal-delivered DNA vaccine induces durable immunity mediating a reduction in viral load in a rhesus macaque SARS-CoV-2 challenge model. Cell Rep Med. 2021;2(10):100420. doi: 10.1016/j.xcrm.2021.100420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gary E.N., Warner B.M., Parzych E.M., Griffin B.D., Zhu X., Tailor N., et al. A novel mouse AAV6 hACE2 transduction model of wild-type SARS-CoV-2 infection studied using synDNA immunogens. iScience. 2021;24(7):102699. doi: 10.1016/j.isci.2021.102699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reed CC, Schultheis K, Andrade VM, Kalia R, Tur J, Schouest B, et al. Design, immunogenicity and efficacy of a Pan-SARS-CoV-2 synthetic DNA vaccine. bioRxiv. 2021:2021.05.11.443592. doi: 10.1101/2021.05.11.443592.
- 17.Tebas P., Yang ShuPing, Boyer J.D., Reuschel E.L., Patel A., Christensen-Quick A., et al. Safety and immunogenicity of INO-4800 DNA vaccine against SARS-CoV-2: A preliminary report of an open-label, Phase 1 clinical trial. EClinicalMedicine. 2021;31:100689. doi: 10.1016/j.eclinm.2020.100689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mammen MP, Tebas P, Agnes J, Giffear M, Kraynyak KA, Blackwood E, et al. Safety and immunogenicity of INO-4800 DNA vaccine against SARS-CoV-2: a preliminary report of a randomized, blinded, placebo-controlled, Phase 2 clinical trial in adults at high risk of viral exposure. medRxiv. 2021:2021.05.07.21256652. doi: 10.1101/2021.05.07.21256652.
- 19.Kardani K., Bolhassani A., Shahbazi S. Prime-boost vaccine strategy against viral infections: Mechanisms and benefits. Vaccine. 2016;34(4):413–423. doi: 10.1016/j.vaccine.2015.11.062. [DOI] [PubMed] [Google Scholar]
- 20.Lu S. Heterologous prime-boost vaccination. Curr Opin Immunol. 2009;21(3):346–351. doi: 10.1016/j.coi.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bakari M., Aboud S., Nilsson C., Francis J., Buma D., Moshiro C., et al. Broad and potent immune responses to a low dose intradermal HIV-1 DNA boosted with HIV-1 recombinant MVA among healthy adults in Tanzania. Vaccine. 2011;29(46):8417–8428. doi: 10.1016/j.vaccine.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koblin B.A., Casapia M., Morgan C., Qin L.i., Wang Z.M., Defawe O.D., et al. Safety and immunogenicity of an HIV adenoviral vector boost after DNA plasmid vaccine prime by route of administration: a randomized clinical trial. PLoS One. 2011;6(9):e24517. doi: 10.1371/journal.pone.0024517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sardesai N.Y., Weiner D.B. Electroporation delivery of DNA vaccines: prospects for success. Curr Opin Immunol. 2011;23(3):421–429. doi: 10.1016/j.coi.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hirao L.A., Wu L., Satishchandran A., Khan A.S., Draghia-Akli R., Finnefrock A.C., et al. Comparative analysis of immune responses induced by vaccination with SIV antigens by recombinant Ad5 vector or plasmid DNA in rhesus macaques. Mol Ther. 2010;18(8):1568–1576. doi: 10.1038/mt.2010.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sahin U., Muik A., Derhovanessian E., Vogler I., Kranz L.M., Vormehr M., et al. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature. 2020;586(7830):594–599. doi: 10.1038/s41586-020-2814-7. [DOI] [PubMed] [Google Scholar]
- 26.Crotty S. T Follicular Helper Cell Biology: A Decade of Discovery and Diseases. Immunity. 2019;50(5):1132–1148. doi: 10.1016/j.immuni.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pardi N., Hogan M.J., Naradikian M.S., Parkhouse K., Cain D.W., Jones L., et al. Nucleoside-modified mRNA vaccines induce potent T follicular helper and germinal center B cell responses. J Exp Med. 2018;215(6):1571–1588. doi: 10.1084/jem.20171450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tauzin A., Nayrac M., Benlarbi M., Gong S.Y., Gasser R., Beaudoin-Bussieres G., et al. A single dose of the SARS-CoV-2 vaccine BNT162b2 elicits Fc-mediated antibody effector functions and T cell responses. Cell Host Microbe. 2021;29(7):1137–50 e6. doi: 10.1016/j.chom.2021.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kissler S.M., Tedijanto C., Goldstein E., Grad Y.H., Lipsitch M. Projecting the transmission dynamics of SARS-CoV-2 through the postpandemic period. Science. 2020;368(6493):860–868. doi: 10.1126/science.abb5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scudellari M. How the pandemic might play out in 2021 and beyond. Nature. 2020;584(7819):22–25. doi: 10.1038/d41586-020-02278-5. [DOI] [PubMed] [Google Scholar]
- 31.Dan J.M., Mateus J., Kato Y.u., Hastie K.M., Yu E.D., Faliti C.E., et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;371(6529) doi: 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hartley G.E., Edwards E.S.J., Aui P.M., Varese N., Stojanovic S., McMahon J., et al. Rapid generation of durable B cell memory to SARS-CoV-2 spike and nucleocapsid proteins in COVID-19 and convalescence. Sci Immunol. 2020;5(54) doi: 10.1126/sciimmunol.abf8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Widge A.T., Rouphael N.G., Jackson L.A., Anderson E.J., Roberts P.C., Makhene M., et al. Durability of Responses after SARS-CoV-2 mRNA-1273 Vaccination. N Engl J Med. 2021;384(1):80–82. doi: 10.1056/NEJMc2032195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garrido C., Curtis A.D., Dennis M., Pathak S.H., Gao H., Montefiori D., et al. SARS-CoV-2 vaccines elicit durable immune responses in infant rhesus macaques. Sci Immunol. 2021;6(60) doi: 10.1126/sciimmunol.abj3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geers D., Shamier M.C., Bogers S., den Hartog G., Gommers L., Nieuwkoop N.N., et al. SARS-CoV-2 variants of concern partially escape humoral but not T-cell responses in COVID-19 convalescent donors and vaccinees. Sci Immunol. 2021;6(59) doi: 10.1126/sciimmunol.abj1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andrade VM, Christensen-Quick A, Agnes J, Tur J, Reed C, Kalia R, et al. INO-4800 DNA Vaccine Induces Neutralizing Antibodies and T cell Activity Against Global SARS-CoV-2 Variants. bioRxiv. 2021:2021.04.14.439719. doi: 10.1101/2021.04.14.439719. [DOI] [PMC free article] [PubMed]
- 37.Madhi S.A., Baillie V., Cutland C.L., Voysey M., Koen A.L., Fairlie L., et al. Efficacy of the ChAdOx1 nCoV-19 Covid-19 Vaccine against the B.1.351 Variant. N Engl J Med. 2021;384(20):1885–1898. doi: 10.1056/NEJMoa2102214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shinde V., Bhikha S., Hoosain Z., Archary M., Bhorat Q., Fairlie L., et al. Efficacy of NVX-CoV2373 Covid-19 Vaccine against the B.1.351 Variant. N Engl J Med. 2021;384(20):1899–1909. doi: 10.1056/NEJMoa2103055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu K, Choi A, Koch M, Ma L, Hill A, Nunna N, et al. Preliminary Analysis of Safety and Immunogenicity of a SARS-CoV-2 Variant Vaccine Booster. medRxiv. 2021:2021.05.05.21256716. doi: 10.1101/2021.05.05.21256716.
- 40.Atmar R.L., Lyke K.E., Deming M.E., Jackson L.A., Branche A.R., El Sahly H.M., et al. Heterologous SARS-CoV-2 Booster Vaccinations – Preliminary Report. medRxiv. 2021 doi: 10.1101/2021.10.10.21264827. [DOI] [Google Scholar]
- 41.Liu X., Shaw R.H., Stuart A.S.V., Greenland M., Aley P.K., Andrews N.J., et al. Safety and immunogenicity of heterologous versus homologous prime-boost schedules with an adenoviral vectored and mRNA COVID-19 vaccine (Com-COV): a single-blind, randomised, non-inferiority trial. Lancet. 2021 doi: 10.1016/S0140-6736(21)01694-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tenbusch M., Schumacher S., Vogel E., Priller A., Held J., Steininger P., et al. Heterologous prime-boost vaccination with ChAdOx1 nCoV-19 and BNT162b2. Lancet Infect Dis. 2021 doi: 10.1016/S1473-3099(21)00420-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Normark J., Vikström L., Gwon Y.-D., Persson I.-L., Edin A., Björsell T., et al. Heterologous ChAdOx1 nCoV-19 and mRNA-1273 Vaccination. N Engl J Med. 2021;385(11):1049–1051. doi: 10.1056/NEJMc2110716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kraynyak KA, Blackwood E, Agnes J, Tebas P, Giffear M, Amante D, et al. SARS-CoV-2 DNA Vaccine INO-4800 Induces Durable Immune Responses Capable of Being Boosted in a Phase 1 Open-Label Trial. medRxiv. 2021:2021.10.06.21264584. doi: 10.1101/2021.10.06.21264584. [DOI] [PMC free article] [PubMed]
- 45.Wang Z., Muecksch F., Schaefer-Babajew D., Finkin S., Viant C., Gaebler C., et al. Naturally enhanced neutralizing breadth against SARS-CoV-2 one year after infection. Nature. 2021;595(7867):426–431. doi: 10.1038/s41586-021-03696-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sokal A., Chappert P., Barba-Spaeth G., Roeser A., Fourati S., Azzaoui I., et al. Maturation and persistence of the anti-SARS-CoV-2 memory B cell response. Cell. 2021;184(5):1201–1213.e14. doi: 10.1016/j.cell.2021.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Winklmeier S, Eisenhut K, Taskin D, Rübsamen H, Schneider C, Eichhorn P, et al. Persistence of functional memory B cells recognizing SARS-CoV-2 variants despite loss of specific IgG. medRxiv. 2021:2021.05.15.21257210. doi: 10.1101/2021.05.15.21257210. [DOI] [PMC free article] [PubMed]
- 48.Khoury D.S., Cromer D., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A., et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat Med. 2021;27(7):1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- 49.Wu K., Werner A.P., Koch M., Choi A., Narayanan E., Stewart-Jones G.B.E., et al. Serum Neutralizing Activity Elicited by mRNA-1273 Vaccine. N Engl J Med. 2021;384(15):1468–1470. doi: 10.1056/NEJMc2102179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sahin U., Muik A., Vogler I., Derhovanessian E., Kranz L.M., Vormehr M., et al. BNT162b2 vaccine induces neutralizing antibodies and poly-specific T cells in humans. Nature. 2021;595(7868):572–577. doi: 10.1038/s41586-021-03653-6. [DOI] [PubMed] [Google Scholar]
- 51.Folegatti P.M., Ewer K.J., Aley P.K., Angus B., Becker S., Belij-Rammerstorfer S., et al. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet. 2020;396(10249):467–478. doi: 10.1016/S0140-6736(20)31604-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang J., Wu Q., Liu Z., Wang Q., Wu J., Hu Y., et al. Spike-specific circulating T follicular helper cell and cross-neutralizing antibody responses in COVID-19-convalescent individuals. Nat Microbiol. 2021;6(1):51–58. doi: 10.1038/s41564-020-00824-5. [DOI] [PubMed] [Google Scholar]
- 53.Juno J.A., Tan H.-X., Lee W.S., Reynaldi A., Kelly H.G., Wragg K., et al. Humoral and circulating follicular helper T cell responses in recovered patients with COVID-19. Nat Med. 2020;26(9):1428–1434. doi: 10.1038/s41591-020-0995-0. [DOI] [PubMed] [Google Scholar]
- 54.Corbett K.S., Flynn B., Foulds K.E., Francica J.R., Boyoglu-Barnum S., Werner A.P., et al. Evaluation of the mRNA-1273 Vaccine against SARS-CoV-2 in Nonhuman Primates. N Engl J Med. 2020;383(16):1544–1555. doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tarke A., Sidney J., Methot N., Yu E.D., Zhang Y., Dan J.M., et al. Impact of SARS-CoV-2 variants on the total CD4(+) and CD8(+) T cell reactivity in infected or vaccinated individuals. Cell Rep Med. 2021;2(7) doi: 10.1016/j.xcrm.2021.100355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tarke A., Coelho C.H., Zhang Z., Dan J.M., Yu E.D., Methot N., et al. SARS-CoV-2 vaccination induces immunological T cell memory able to cross-recognize variants from Alpha to Omicron. Cell. 2022;185(5):847–859.e11. doi: 10.1016/j.cell.2022.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Humoral Responses in INO-4800 vaccinated rhesus macaques. IgG binding was measured in sera from INO-4800 vaccinated rhesus macaques to SARS-CoV-2 RBD (A), S1 (B) and S2 (C) protein antigens. Fig. S2. Functional antibody responses following homologous or heterologous boost in INO-4800-primed rhesus macaques. A) Spearman correlation of ACE2 blocking activity and neutralizing activity among animals boosted with either INO-4800 or INO-4802. Correlations relating to functional antibody responses against the wildtype (left) Beta SARS-CoV-2 (center), and Delta (right) variants at weeks 2 and 4 post-boost are shown. B) Spearman correlation of the frequency of circulating T follicular helper cells with ACE-2 binding inhibition at week 2 post-boost.