Abstract
Introduction
Rheumatoid arthritis (RA) has a huge societal impact due to the high prevalence, irreversible joint damage and systemic complications. Gut microbiota plays an important role in the pathogenesis and progression of RA by regulating the host immune system. Restoring intestinal homeostasis by altering the microbiota could be an attractive strategy for the prevention and treatment of RA. However, the signature features of microbial dysbiosis in RA are still controversial. Therefore, we aim to elucidate the characteristic change in the diversity and composition of gut microbiota in RA.
Methods and analysis
We will systematically search through PubMed, EMBASE, Web of Science and Cochrane Library, as well as dissertations and conference proceedings. The reference lists of all included studies will be also reviewed to retrieve additional relevant studies. The case-control studies that reported either the relative abundance of bacteria at the phylum or genus level or at least one of the alpha-diversity, beta-diversity indexes in both RA and healthy controls will be included. Eligible studies will be screened independently by two reviewers according to the inclusion criteria. The Newcastle-Ottawa Quality Assessment Scale will be used to assess the quality of the included studies. Data extraction, qualitative and quantitative analysis will be performed within the gut microbial dysbiosis in RA. The expected outcomes will be the identification of the specific changes in composition and diversity of the gut microbiota in patients with RA. The quality of evidence will be assessed by the Grading of Recommendations Assessment, Development and Evaluation framework.
Ethics and dissemination
Ethical approval is unnecessary as this review does not address the data and privacy of patients. The results will be published in a peer-reviewed scientific journal and conference presentations.
PROSPERO registration number
CRD42021225229.
Keywords: Rheumatology, Immunology, Microbiology
Strengths and limitations of this study.
This systematic review will identify the characteristic changes in the composition and diversity of gut microbiota in patients with RA, a significant but controversial clinical issue.
The relative abundances of phyla and/or genus levels in the gut microbiota will be used in this meta-analysis.
The Web Plot Digitizer will be used to digitise and extract data from graphs and plots, which may lead to biased results.
This systematic review will only include studies written in English, which may limit available data or result in language bias.
Introduction
Rheumatoid arthritis (RA) is a chronic disease characterised by persistent synovitis, inflammatory and autoantibody changes.1 The prevalence of RA is about 1% globally, and 1.02% in China.2 The prevalence of RA in women is 2–3 times higher than that in men.3 Delays in diagnosis and treatment are associated with worse outcomes, including irreversible joint destruction, disability and disease-related non-articular outcomes such as reduced life span.4 5 In China, 77.6% of RA patients had disabilities, among which moderate and severe disabilities accounted for about 39%, seriously affecting the quality of life of patients.6 The gradual deterioration of RA leads to a sharp increase in the cost of the disease, which imposes a heavy societal and economical burden on individuals and the country.7–9
RA is a lifelong condition and currently no cure for most patients.10 11 European League Against Rheumatism (EULAR) and American College of Rheumatology (ACR) recommend that the purpose of RA treatment should be to enable each patient to achieve the goal of continuous remission or low disease activity.12 Although the prognosis of RA has improved with advances in diagnosis and treatment in recent decades, the exact aetiology and pathogenesis of RA are not fully understood. In order to develop more effective treatment strategies for RA, it is essential to explore its underlying aetiology and pathogenesis.
Environmental factors are considered to play an important role in RA.13 The gut microbiota is considered an important environmental factor in the development of RA.14 Almost all studies on autoimmune rheumatic diseases show abnormal microbial community structure (ie, dysbiosis).15 Dysbiosis not only affects the proinflammatory and anti-inflammatory process of the intestinal mucosa, but also affects the distal joint through the intestinal-joint axis.16–18 The studies have found dysbiosis in both RA patients and high-risk individuals, indicating that the imbalance of intestinal flora could have occured before the onset of RA.14 19 Dysbiosis has been involved in the pathogenesis of RA in the decade before its diagnosis.20 The intestinal flora imbalance also appeared in the initial peak and relapse stage of RA.21 Dysbiosis is related to the inflammatory response and disease activity of RA, which can be partially recovered by effective treatment.22–24 As a first-line treatment for RA, methotrexate may act in part by modulating the human gut microbiota.24 The results of animal experiments suggest that interventions targeting intestinal microbiota may have the potential to prevent RA in the preclinical stage.25 Probiotics supplementation as adjunctive therapy improves the inflammatory state of RA in human and animal studies.26–29 Therefore, gut microbiota plays an important role in the development of RA, and may be a new therapeutic target.30 31 Gut microbiome studies of RA are essential to elucidate aetiology and pathophysiological mechanisms and to develop potential therapeutic strategies. Regulating the gut microbiota to slow the progression of the disease, especially in the preclinical phase of RA, may be a promising approach for the treatment of RA in the future.32 33
Although numerous studies have shown that dysbiosis of the gut microbiome is a key hallmark of RA, the distinct composition of the gut microbiome in RA patients remains controversial. The abundance of Prevotella increased in patients with early RA, which hurt the development and prognosis of RA.14 34–37 However, it has been reported that the abundance of Prevotella did not significantly change in RA patients.38 Moreover, P. copri and P. histicola of Prevotella have different effects on RA.14 Bacteroidetes were enriched in female patients with RA, while Actinomycetes and Collinsella were enriched in healthy subjects.38 However, the abundance of Bacteroides and Bifidobacterium was found to be reduced in RA patients and animal experiments.23 39 It follows that the results of studies on the gut microbiota of RA patients are contradictory. The identification of specific microbial profiles and patterns that may contribute to the pathogenesis of RA remains a major challenge due to the inconsistent results of studies on the gut microbiota. The conflicting results may stem from inter-study batch effects, such as various biological factors influencing gut microbiome composition, different data processing and analysis methods.40 41 The differences in demographics of the study cohorts (eg, sex, age, ethnicity, geography and diet) also have an important influence on the variability of the results of the gut microbiome study. Through a quantitative review of the existing literature, the changes of RA gut microbiota can be understood more clearly and comprehensively. Recently, several meta-analyses of gut microbiota have identified specific microbial biomarkers associated with disease.42–47 However, there has been no systematic review and meta-analysis focusing on the characteristic dysbiosis of gut microbiota in RA to date. Therefore, we will perform a systematic review and meta-analysis to identify characteristic alterations in the gut microbiota of RA patients.
Objective
The purpose of this protocol is to outline a systematic review and meta-analysis, which evaluates the changes in the diversity of gut microbiota and the relative abundance of bacterial phyla or genera in patients with RA.
Methods
Study design
We plan to conduct a systematic review according to the Cochrane Handbook for Systematic Reviews of Interventions Version 6.1,48 Preferred Reporting Items for Systematic reviews and Meta-Analyses (PRISMA),49 and PRISMA-Protocols (PRISMA-P) 2015,50 as well as the Newcastle-Ottawa Quality Assessment Scale (NOS).51 The PRISMA-P 2015 checklist is shown in table 1. This protocol has been registered at PROSPERO (registration number: CRD42021225229).
Table 1.
PRISMA-P 2015 checklist
| Section and topic | Item no | Checklist item | Reported on page # |
| Administrative information | |||
| Title: | |||
| Identification | 1a | Identify the report as a protocol of a systematic review | 1 |
| Update | 1b | If the protocol is for an update of a previous systematic review, identify as such | N/A |
| Registration | 2 | If registered, provide the name of the registry (such as PROSPERO) and registration number | 1 |
| Authors: | |||
| Contact | 3a | Provide name, institutional affiliation, e-mail address of all protocol authors; provide physical mailing address of corresponding author | 1 |
| Contributions | 3b | Describe contributions of protocol authors and identify the guarantor of the review | 6 |
| Amendments | 4 | If the protocol represents an amendment of a previously completed or published protocol, identify as such and list changes; otherwise, state plan for documenting important protocol amendments | N/A |
| Support: | |||
| Sources | 5a | Indicate sources of financial or other support for the review | 6 |
| Sponsor | 5b | Provide name for the review funder and/or sponsor | N/A |
| Role of sponsor or funder | 5c | Describe roles of funder(s), sponsor(s), and/or institution(s), if any, in developing the protocol | N/A |
| Introduction | |||
| Rationale | 6 | Describe the rationale for the review in the context of what is already known | 1-2 |
| Objectives | 7 | Provide an explicit statement of the question(s) the review will address with reference to participants, interventions, comparators and outcomes (PICO) | 2,4 |
| Methods | |||
| Eligibility criteria | 8 | Specify the study characteristics (such as PICO, study design, setting, time frame) and report characteristics (such as years considered, language, publication status) to be used as criteria for eligibility for the review | 4 |
| Information sources | 9 | Describe all intended information sources (such as electronic databases, contact with study authors, trial registers or other grey literature sources) with planned dates of coverage | 4 |
| Search strategy | 10 | Present draft of search strategy to be used for at least one electronic database, including planned limits, such that it could be repeated | 4 |
| Study records: | |||
| Data management | 11a | Describe the mechanism(s) that will be used to manage records and data throughout the review | 4 |
| Selection process | 11b | State the process that will be used for selecting studies (such as two independent reviewers) through each phase of the review (that is, screening, eligibility and inclusion in meta-analysis) | 4 |
| Data collection process | 11c | Describe planned method of extracting data from reports (such as piloting forms, done independently, in duplicate), any processes for obtaining and confirming data from investigators | 4-5 |
| Data items | 12 | List and define all variables for which data will be sought (such as PICO items, funding sources), any preplanned data assumptions and simplifications | 5 |
| Outcomes and prioritisation | 13 | List and define all outcomes for which data will be sought, including prioritisation of main and additional outcomes, with rationale | 5 |
| Risk of bias in individual studies | 14 | Describe anticipated methods for assessing risk of bias of individual studies, including whether this will be done at the outcome or study level, or both; state how this information will be used in data synthesis | 5 |
| Data synthesis | 15a | Describe criteria under which study data will be quantitatively synthesised | 5 |
| 15b | If data are appropriate for quantitative synthesis, describe planned summary measures, methods of handling data and methods of combining data from studies, including any planned exploration of consistency (such as I2, Kendall’s τ) | 5 | |
| 15c | Describe any proposed additional analyses (such as sensitivity or subgroup analyses, meta-regression) | 5 | |
| 15d | If quantitative synthesis is not appropriate, describe the type of summary planned | 5 | |
| Meta-bias(es) | 16 | Specify any planned assessment of meta-bias(es) (such as publication bias across studies, selective reporting within studies) | 5 |
| Confidence in cumulative evidence | 17 | Describe how the strength of the body of evidence will be assessed (such as GRADE) | 6 |
Eligibility criteria
The studies, written in English as eligible, will be selected and screened based on PECOS steps (Population, Exposure, Comparator, Outcomes and Study design).52 53 The data items will be extracted as follows:
Types of participants (P)
The population of interest of the eligible studies should be adults (≥18 years old) diagnosed with RA according to a standardised diagnostic classification system (EULAR/ACR 2010 or ACR 1987 criteria).54 55
Type of exposure (E)
Trials were applied to assess the gut microbiota. Quantitative synthesis of microbiota in faecal samples was performed by using metagenomic shotgun sequencing, 16s rRNA sequencing techniques and/or real-time polymerase chain reaction (rt-PCR).
Comparison (C)
Only healthy adults will be considered eligible for the control group.
Type of outcomes (O)
The primary outcome of the study will be the identification of the composition of the gut microbiome and the relative abundance of bacteria in RA. The secondary outcomes will be considered: changes in the gut microbiota diversity (alpha-diversity, beta-diversity), the effects of different gender and region on the relative abundance of gut microbiota.
Type of studies (S)
We will only include studies with the case-control design, written in English and published in the original peer-reviewed journals. The animal studies, reviews, case reports and the full text unachieved will be excluded from the qualitative and quantitative synthesis.
Data sources and search strategies
We conduct the search using the databases Embase, PubMed, Web of Science and Cochrane Library in the English language published up to September 2020. After reading several documents, a search strategy combining Medical Subject Headings (MeSH) terms and free words was developed: (‘Arthritis, Rheumatoid’ OR ‘Rheumatoid arthritis’ OR ‘RA’) AND (‘Gastrointestinal Microbiome’ OR ‘Gastrointestinal Microbiomes’ OR ‘Microbiome, Gastrointestinal’ OR ‘Gut Microbiome’ OR ‘Gut Microbiomes’ OR ‘Microbiome, Gut’). The search strategy for the Embase database is shown in figure 1. To prevent the omission of the article, two researchers (DWW and XTP) will search the above database independently. Using the snowball method, we manually search for all references contained in the article.
Figure 1.
Embase session results.
Screening procedures of eligible studies
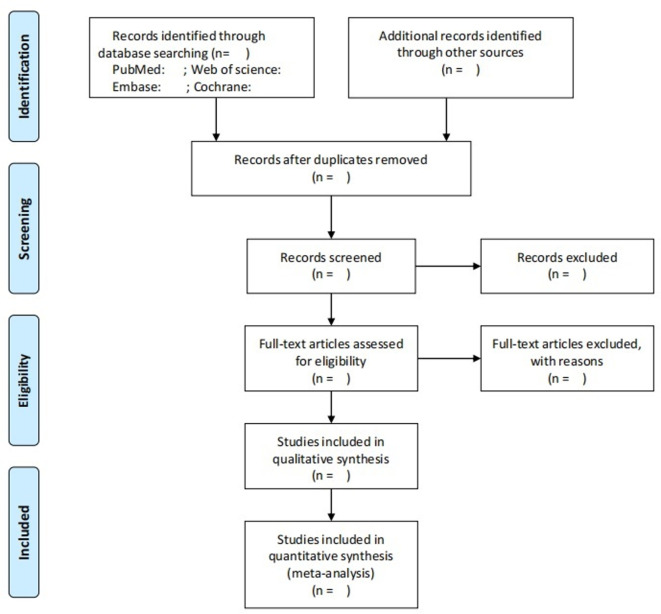
Once the search is complete, the literature will be managed using EndNote X9 (Clarivate Analytics (US) LLC). Duplicates will be identified and deleted according to literature title. Then, the titles and abstracts of the literature will be screened independently by two reviewers (XTP and YFL) according to the inclusion criteria. Retrieval of the full text will be based on the eligible of titles and abstracts, and the literature meeting all the inclusion criteria will be independently assessed. In case of disagreement, a third reviewer (ZLS) will be consulted. To measure inter-rater agreement, the kappa coefficients will be both calculated for the processes of titles/abstract selection and full-text screening. The criteria for judging the scope of the agreement between the evaluators are as follows: 0.00–0.20 = slight agreement, 0.21–0.40 = fair, 0.41–0.60 = moderate, 0.61–0.80 = substantial and 0.81–1.00 = almost perfect agreement.56 The plan of study screening and selection is available in figure 2.
Figure 2.
Plan of study screening and selection process.
Assessment of risk of bias
The quality of the included studies will be assessed using NOS.51 It is a tool mainly used to evaluate the quality of case-control and cohort studies. The parameters considered under each category are ① selection: case definition, representativeness of the cases, selection of controls and definition of controls; ② comparability: comparability of cases and controls based on the basis of the design or analysis; ③ exposure: ascertainment of exposure, the same method of ascertainment for cases and controls, non-response rate. There are 1–2 stars in each category, with a maximum of 9 stars for all. The number of stars is proportional to the mass of the study. The number of stars is directly proportional to the quality of the study. The standard of high quality will be NOS score ≥7 stars.
To ensure consistency in assessments, the two reviewers (HXG and HZ) will independently evaluate the eligible literature according to NOS and will be summarised in a table. When disagreements arise in the review, the third reviewer (ZLS) cooperates with the team to reach a consensus.
Data extraction
Data from each eligible article will be extracted and compiled using a standardised excel sheet. Items required for extraction will be obtained using the PECOS steps. The following data will be extracted for eligible studies: first author’s surname, year of publication, country, classification criteria for RA, number of cases and controls, age and sex, disease duration, antibody positive of RA, 28-joint disease activity score, medication, assessment methods of faecal microbiota, alterations in gut microbial abundance, alpha-diversity indexes (OTUs, Shannon Index and Chao 1 Index) and beta-diversity.
To conduct the meta-analysis, we involve trials that have available and sufficient data to calculate the standardised mean difference (SMD) with 95% CI in RA patients and healthy controls in the analysis of the pooled data set. If additional data or data transformations will be required for analysis, we will download the publicly available raw data from online repositories or links provided in the original publications. If there is no relevant data in the original literature, we will acquire it after personal communication with the authors of the manuscripts. If the authors do not reply, we will use Web Plot Digitizer (v.4.42) to digitise and extract sufficient data from graphs and plots in the articles.45 57
To ensure the accuracy of the extracted data, we will randomly select two eligible pieces of literature to be independently extracted by two reviewers (FQC and RZ). Kappa will be applied to compare the consistency of data extraction from the two literatures by the two reviewers. If there is an almost perfect agreement between the two reviewers (kappa value ≥80%), the remaining literature will extracted by one of the two reviewers.
Data synthesis and analysis
When the number of studies for a single bacterium is five or more, we will conduct the meta-analysis by R language Version 3.4.3 to compare the abundance level of gut microbiota in RA patients with healthy controls. We will adopt SMD with 95% CI of microbiota abundance as summary statistics when gut microbiota was detected by different techniques in the included studies.58–60 The included studies will be analysed at the phylum or genus levels for consitency. The forest plots will be used to visualise the results. We will assess heterogeneity between studies using the Higgin I2 statistic. In relative terms, I2 values are proportional to heterogeneity: I2 values of 25%, 50% and 75% means low, moderate and high heterogeneity, respectively.61 Data analysis will be performed by a random-effect model when there is substantial heterogeneity (I2 >50%); otherwise, a fixed-effects model will be used.47 Additionally, we will conduct subgroup analysis of different genders (man/woman) and regions (east/west) included in the studies.
If meta-analysis is not feasible, we will conduct narrative synthesis to summarise the relevant evidence between RA and gut dysbiosis. The quantitative narrative synthesis will be conducted according to the Synthesis Without Meta-analysis guideline checklist.62 In order to define the characteristics of the gut microbiota in RA, we will perform compositional analysis based on the abundance, diversity, and specific bacterial detection of gut microbiota in RA patients and healthy controls.
Assessment of publication bias
We will apply funnel plot and Egger’s test to assess publication bias.59 If funnel plots present asymmetry, we will use Egger’s test to statistically examination.63 64
Assessment of evidence quality
We will conduct an appraisal of the quality of evidence by applying the Grading of Recommendations Assessment, Development and Evaluation (GRADE) framework.65 Two reviewers (YF and RZ) will assess five domains including limitations of design, inconsistency, indirectness, imprecision and publication bias. The GRADE classifies the quality of evidence as four levels: high, moderate, low and very low. Disagreement on the assessment will be resolved by a third reviewer (ZLS). The GRADE Evidence Profiles will be generated using GRADEpro GDT (https://www.gradepro.org/).
Patient and public involvement
No patient or public involved.
Supplementary Material
Footnotes
Contributors: DWW and XTP drafted the manuscript and contributed equally to this manuscript as joint first authors. YFL provided the materials. HXG and HZ collected and assembled the data. FQC, RZ and YF analysed and interpreted the data. ZLS conceived the study and critically revised the draft. All authors assisted in manuscript editing and approved its contents.
Funding: This work was supported by National Natural Science Foundation of China [81 774 383], Philosophy and Social Science Research of Jiangsu Higher Education Institutions [2020SJA0335], Post-graduate Research & Practice Innovation Programme of Jiangsu Province [KYCX20_1449].
Competing interests: None declared.
Patient and public involvement: Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Provenance and peer review: Not commissioned; externally peer reviewed.
Ethics statements
Patient consent for publication
Not applicable.
References
- 1.Aletaha D, Smolen JS. Diagnosis and management of rheumatoid arthritis: a review. JAMA 2018;320:1360–72. 10.1001/jama.2018.13103 [DOI] [PubMed] [Google Scholar]
- 2.Alamanos Y, Drosos AA. Epidemiology of adult rheumatoid arthritis. Autoimmun Rev 2005;4:130–6. 10.1016/j.autrev.2004.09.002 [DOI] [PubMed] [Google Scholar]
- 3.Gerlag DM, Norris JM, Tak PP. Towards prevention of autoantibody-positive rheumatoid arthritis: from lifestyle modification to preventive treatment. Rheumatology 2016;55:607–14. 10.1093/rheumatology/kev347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taskforce ELAR . Rheuma Map a research roadmap to transform the lives of people with rheumatic and musculoskeletal diseases 2019.
- 5.Veale DJ, Orr C, Fearon U. Cellular and molecular perspectives in rheumatoid arthritis. Semin Immunopathol 2017;39:343–54. 10.1007/s00281-017-0633-1 [DOI] [PubMed] [Google Scholar]
- 6.Zhou Y, Wang X, An Y, et al. Disability and health-related quality of life in Chinese patients with rheumatoid arthritis: a cross-sectional study. Int J Rheum Dis 2018;21:1709–15. 10.1111/1756-185X.13345 [DOI] [PubMed] [Google Scholar]
- 7.Hu H, Luan L, Yang K, et al. Burden of rheumatoid arthritis from a societal perspective: a prevalence-based study on cost of this illness for patients in China. Int J Rheum Dis 2018;21:1572–80. 10.1111/1756-185X.13028 [DOI] [PubMed] [Google Scholar]
- 8.Furneri G, Mantovani LG, Belisari A, et al. Systematic literature review on economic implications and pharmacoeconomic issues of rheumatoid arthritis. Clin Exp Rheumatol 2012;30:S72–84. [PubMed] [Google Scholar]
- 9.Shafrin J, Tebeka MG, Price K, et al. The economic burden of ACPA-positive status among patients with rheumatoid arthritis. J Manag Care Spec Pharm 2018;24:4–11. 10.18553/jmcp.2017.17129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smolen JS, Landewé RBM, Bijlsma JWJ, et al. EULAR recommendations for the management of rheumatoid arthritis with synthetic and biological disease-modifying antirheumatic drugs: 2019 update. Ann Rheum Dis 2020;79:685–99. 10.1136/annrheumdis-2019-216655 [DOI] [PubMed] [Google Scholar]
- 11.Alam J, Jantan I, Bukhari SNA. Rheumatoid arthritis: recent advances on its etiology, role of cytokines and pharmacotherapy. Biomed Pharmacother 2017;92:615–33. 10.1016/j.biopha.2017.05.055 [DOI] [PubMed] [Google Scholar]
- 12.Smolen JS, Landewé RBM, Bijlsma JWJ, et al. EULAR recommendations for the management of rheumatoid arthritis with synthetic and biological disease-modifying antirheumatic drugs: 2019 update. Ann Rheum Dis 2020;79:685–99. 10.1136/annrheumdis-2019-216655 [DOI] [PubMed] [Google Scholar]
- 13.Karami J, Aslani S, Jamshidi A, et al. Genetic implications in the pathogenesis of rheumatoid arthritis; an updated review. Gene 2019;702:8–16. 10.1016/j.gene.2019.03.033 [DOI] [PubMed] [Google Scholar]
- 14.Maeda Y, Takeda K. Host-microbiota interactions in rheumatoid arthritis. Exp Mol Med 2019;51:1–6. 10.1038/s12276-019-0283-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Konig MF. The microbiome in autoimmune rheumatic disease. Best Pract Res Clin Rheumatol 2020;34:101473. 10.1016/j.berh.2019.101473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Häger J, Bang H, Hagen M, et al. The role of dietary fiber in rheumatoid arthritis patients: a feasibility study. Nutrients 2019;11. 10.3390/nu11102392. [Epub ahead of print: 07 Oct 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van de Wiele T, Van Praet JT, Marzorati M, et al. How the microbiota shapes rheumatic diseases. Nat Rev Rheumatol 2016;12:398–411. 10.1038/nrrheum.2016.85 [DOI] [PubMed] [Google Scholar]
- 18.Sun Y, Chen Q, Lin P, et al. Characteristics of gut microbiota in patients with rheumatoid arthritis in Shanghai, China. Front Cell Infect Microbiol 2019;9:369. 10.3389/fcimb.2019.00369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alpizar-Rodriguez D, Lesker TR, Gronow A, et al. Prevotella copri in individuals at risk for rheumatoid arthritis. Ann Rheum Dis 2019;78:590–3. 10.1136/annrheumdis-2018-214514 [DOI] [PubMed] [Google Scholar]
- 20.Nguyen Y, Mariette X, Salliot C, et al. Chronic diarrhoea and risk of rheumatoid arthritis: findings from the French E3N-EPIC cohort study. Rheumatology 2020;59:3767–75. 10.1093/rheumatology/keaa133 [DOI] [PubMed] [Google Scholar]
- 21.Nemoto N, Takeda Y, Nara H, et al. Analysis of intestinal immunity and flora in a collagen-induced mouse arthritis model: differences during arthritis progression. Int Immunol 2020;32:49–56. 10.1093/intimm/dxz058 [DOI] [PubMed] [Google Scholar]
- 22.Zhang X, Zhang D, Jia H, et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med 2015;21:895–905. 10.1038/nm.3914 [DOI] [PubMed] [Google Scholar]
- 23.Chiang H-I, Li J-R, Liu C-C, et al. An association of gut microbiota with different phenotypes in Chinese patients with rheumatoid arthritis. J Clin Med 2019;8. 10.3390/jcm8111770. [Epub ahead of print: 24 10 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nayak RR, Alexander M, Stapleton-Grey K. Perturbation of the human gut microbiome by a non-antibiotic drug contributes to the resolution of autoimmune disease. bioRxiv 2019. [Google Scholar]
- 25.Jubair WK, Hendrickson JD, Severs EL, et al. Modulation of inflammatory arthritis in mice by gut microbiota through mucosal inflammation and autoantibody generation. Arthritis Rheumatol 2018;70:1220–33. 10.1002/art.40490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mandel DR, Eichas K, Holmes J. Bacillus coagulans: a viable adjunct therapy for relieving symptoms of rheumatoid arthritis according to a randomized, controlled trial. BMC Complement Altern Med 2010;10:1. 10.1186/1472-6882-10-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vaghef-Mehrabany E, Alipour B, Homayouni-Rad A, et al. Probiotic supplementation improves inflammatory status in patients with rheumatoid arthritis. Nutrition 2014;30:430–5. 10.1016/j.nut.2013.09.007 [DOI] [PubMed] [Google Scholar]
- 28.So J-S, Kwon H-K, Lee C-G, et al. Lactobacillus casei suppresses experimental arthritis by down-regulating T helper 1 effector functions. Mol Immunol 2008;45:2690–9. 10.1016/j.molimm.2007.12.010 [DOI] [PubMed] [Google Scholar]
- 29.Fan Z, Yang B, Ross RP, et al. Protective effects of Bifidobacterium adolescentis on collagen-induced arthritis in rats depend on timing of administration. Food Funct 2020;11:4499–511. 10.1039/D0FO00077A [DOI] [PubMed] [Google Scholar]
- 30.Lee YH. Causal association of gut microbiome on the risk of rheumatoid arthritis: a Mendelian randomisation study. Ann Rheum Dis 2022;81:e3. 10.1136/annrheumdis-2019-216747 [DOI] [PubMed] [Google Scholar]
- 31.Uchiyama K, Naito Y, Takagi T. Intestinal microbiome as a novel therapeutic target for local and systemic inflammation. Pharmacol Ther 2019;199:164–72. 10.1016/j.pharmthera.2019.03.006 [DOI] [PubMed] [Google Scholar]
- 32.Horta-Baas G, Sandoval-Cabrera A, Romero-Figueroa MdelS, MJCrr R-F. Modification of gut microbiota in inflammatory arthritis: highlights and future challenges. Curr Rheumatol Rep 2021;23:67. 10.1007/s11926-021-01031-9 [DOI] [PubMed] [Google Scholar]
- 33.Gupta VK, Cunningham KY, Hur B, et al. Gut microbial determinants of clinically important improvement in patients with rheumatoid arthritis. Genome Med 2021;13:149. 10.1186/s13073-021-00957-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Drago L. Prevotella Copri and Microbiota in Rheumatoid Arthritis: Fully Convincing Evidence? J Clin Med 2019;8. 10.3390/jcm8111837. [Epub ahead of print: 01 11 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pianta A, Arvikar S, Strle K, et al. Evidence of the immune relevance of Prevotella copri, a gut microbe, in patients with rheumatoid arthritis. Arthritis Rheumatol 2017;69:964–75. 10.1002/art.40003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lorenzo D, GianVincenzo Z, Carlo Luca R, et al. Oral-Gut microbiota and arthritis: is there an evidence-based axis? J Clin Med 2019;8. 10.3390/jcm8101753. [Epub ahead of print: 22 10 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maeda Y, Takeda K. Role of gut microbiota in rheumatoid arthritis. J Clin Med 2017;6:60. 10.3390/jcm6060060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jeong Y, Kim J-W, You HJ, et al. Gut microbial composition and function are altered in patients with early rheumatoid arthritis. J Clin Med 2019;8. 10.3390/jcm8050693. [Epub ahead of print: 16 05 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li X, Lu C, Fan D, et al. Human umbilical mesenchymal stem cells display therapeutic potential in rheumatoid arthritis by regulating interactions between immunity and gut microbiota via the aryl hydrocarbon Receptor. Front Cell Dev Biol 2020;8:131. 10.3389/fcell.2020.00131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu Y, Jiao N, Zhu R, et al. Identification of microbial markers across populations in early detection of colorectal cancer. Nat Commun 2021;12:3063. 10.1038/s41467-021-23265-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Najafi S, Abedini F, Azimzadeh Jamalkandi S, et al. The composition of lung microbiome in lung cancer: a systematic review and meta-analysis. BMC Microbiol 2021;21:315. 10.1186/s12866-021-02375-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ho NT, Li F, Lee-Sarwar KA, et al. Meta-analysis of effects of exclusive breastfeeding on infant gut microbiota across populations. Nat Commun 2018;9:4169. 10.1038/s41467-018-06473-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shen T, Yue Y, He T, et al. The Association Between the Gut Microbiota and Parkinson’s Disease, a Meta-Analysis. Front Aging Neurosci 2021;13:636545. 10.3389/fnagi.2021.636545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wirbel J, Pyl PT, Kartal E, et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat Med 2019;25:679–89. 10.1038/s41591-019-0406-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nikolova VL, Hall MRB, Hall LJ, et al. Perturbations in gut microbiota composition in psychiatric disorders. JAMA Psychiatry 2021;78:1343. 10.1001/jamapsychiatry.2021.2573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iglesias-Vázquez L, Van Ginkel Riba G, Arija V, et al. Composition of gut microbiota in children with autism spectrum disorder: a systematic review and meta-analysis. Nutrients 2020;12:792. 10.3390/nu12030792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu M, Xu X, Li J, et al. Association between gut microbiota and autism spectrum disorder: a systematic review and meta-analysis. Front Psychiatry 2019;10:473. 10.3389/fpsyt.2019.00473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.et alHiggins JPT TJ, Chandler J, Cumpston M. Cochrane Handbook for systematic reviews of interventions version 6.1, 2020. Available: www.training.cochrane.org/handbook
- 49.Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med 2009;151:264–9. 10.7326/0003-4819-151-4-200908180-00135 [DOI] [PubMed] [Google Scholar]
- 50.Shamseer L, Moher D, Clarke M, et al. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015: elaboration and explanation. BMJ 2015;349:g7647. 10.1136/bmj.g7647 [DOI] [PubMed] [Google Scholar]
- 51.Wells BS, D GA, O'Connell JP, Welch V, et al. The Newcastle-Ottawa scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses. Available: www.ohri.ca/programs/clinical_epidemiology/oxford.asp
- 52.Morgan RL, Thayer KA, Bero L, et al. GRADE: assessing the quality of evidence in environmental and occupational health. Environ Int 2016;92-93:611–6. 10.1016/j.envint.2016.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morgan RL, Whaley P, Thayer KA, et al. Identifying the PECO: a framework for formulating good questions to explore the association of environmental and other exposures with health outcomes. Environ Int 2018;121:1027–31. 10.1016/j.envint.2018.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aletaha D, Neogi T, Silman AJ, et al. 2010 rheumatoid arthritis classification criteria: an American College of Rheumatology/European League against rheumatism collaborative initiative. Arthritis Rheum 2010;62:2569–81. 10.1002/art.27584 [DOI] [PubMed] [Google Scholar]
- 55.Arnett FC, Edworthy SM, Bloch DA, et al. The American rheumatism association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum 1988;31:315–24. 10.1002/art.1780310302 [DOI] [PubMed] [Google Scholar]
- 56.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics 1977;33:159–74. 10.2307/2529310 [DOI] [PubMed] [Google Scholar]
- 57.Safadi JM, Quinton AMG, Lennox BR, et al. Gut dysbiosis in severe mental illness and chronic fatigue: a novel trans-diagnostic construct? A systematic review and meta-analysis. Mol Psychiatry 2021;42. 10.1038/s41380-021-01032-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li F, Ye J, Shao C, et al. Compositional alterations of gut microbiota in nonalcoholic fatty liver disease patients: a systematic review and meta-analysis. Lipids Health Dis 2021;20:22. 10.1186/s12944-021-01440-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim KN, Yao Y, Ju SY. Short chain fatty acids and fecal microbiota abundance in humans with obesity: a systematic review and meta-analysis. Nutrients 2019;11:2512. 10.3390/nu11102512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Creedon AC, Hung ES, Berry SE, et al. Nuts and their effect on gut microbiota, gut function and symptoms in adults: a systematic review and meta-analysis of randomised controlled trials. Nutrients 2020;12:2347. 10.3390/nu12082347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Higgins JPT, et al. Measuring inconsistency in meta-analyses. BMJ 2003;327:557–60. 10.1136/bmj.327.7414.557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Campbell M, McKenzie JE, Sowden A, et al. Synthesis without meta-analysis (SWiM) in systematic reviews: reporting guideline. BMJ 2020;72:l6890. 10.1136/bmj.l6890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang L, Alammar N, Singh R, et al. Gut microbial dysbiosis in the irritable bowel syndrome: a systematic review and meta-analysis of case-control studies. J Acad Nutr Diet 2020;120:565–86. 10.1016/j.jand.2019.05.015 [DOI] [PubMed] [Google Scholar]
- 64.Ji R, Zhao X, Cao X, et al. Changes in gastric mucosal microbiota in gastric carcinogenesis: a systematic review protocol. BMJ Open 2021;11:e045810. 10.1136/bmjopen-2020-045810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bobbio E, Lingbrant M, Nwaru BI, et al. Inflammatory cardiomyopathies: short- and long-term outcomes after heart transplantation—a protocol for a systematic review and meta-analysis. Heart Fail Rev 2020;25:481–5. 10.1007/s10741-020-09919-x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.