Abstract
Anti-HIV broadly neutralizing antibodies (bNAbs) may favor development of antiviral immunity by engaging the immune system during immunotherapy. Targeting integrin α4β7 with an anti-α4β7 monoclonal antibody (Rh-α4β7) affects immune responses in SIV/SHIV-infected macaques. To explore the therapeutic potential of combining bNAbs with α4β7 integrin blockade, SHIVSF162P3-infected, viremic rhesus macaques were treated with bNAbs only (VRC07–523LS and PGT128 anti-HIV antibodies) or a combination of bNAbs and Rh-α4β7 or were left untreated as a control. Treatment with bNAbs alone decreased viremia below 200 copies/ml in all macaques, but seven of eight macaques (87.5%) in the bNAbs-only group rebounded within a median of 3 weeks (95% CI: 2 to 9). In contrast, three of six macaques treated with a combination of Rh-α4β7 and bNAbs (50%) maintained a viremia below 200 copies/ml until the end of the follow-up period; viremia in the other three macaques rebounded within a median of 6 weeks (95% CI: 5 to 11). Thus, there was a modest delay in viral rebound in the macaques treated with the combination antibody therapy compared to bNAbs alone. Our study suggests that α4β7 integrin blockade may prolong virologic control by bNAbs in SHIVSF162P3-infected macaques.
INTRODUCTION
Passive transfer of a number of broadly neutralizing antibodies (bNAbs) targeting the HIV-1 envelope protein (Env) has been shown to reduce viremia in untreated, chronically simian-human immunodeficiency virus (SHIV)–infected macaques (1–3) and HIV-1–infected individuals (4–6). Moreover, bNAbs have been reported to delay viral rebound in HIV-1–infected individuals when administered around the time that combination antiretroviral therapy (cART) is discontinued (7–9), with the combination of two bNAbs exerting a more prolonged effect on viremia (9, 10). In contrast to cART drugs, anti–HIV bNAbs can engage the host immune system but do not prevent viral replication. Very early bNAb administration in SHIV-infected macaques is associated with CD8+ T cell–dependent lasting virologic control in a fraction of treated animals (11). Accumulating evidence indicates that short treatments of SHIV-infected macaques with bNAbs lead to enhanced antiviral immunity (1, 11–13). In HIV-1– infected individuals, bNAb administration was associated with increased potency and breadth of host antibody responses (14) and increased T cell responses after rebound viremia (8). More recently, an increase in HIV-gag–specific CD8+ T cell responses was reported in all study participants after administration of a combination of two anti–HIV bNAbs during the aviremic phase of analytical treatment interruption (ATI) (15). The increased T cell responses were due to both newly detectable reactivity to HIV-1 Gag epitopes and the expansion of preexisting responses (15). These findings indicate that HIV bNAbs therapy during ATI leads to stimulation of host immunity. This could be due, at least in part, to low-grade viral replication and antigen availability in host tissues after ATI. In addition, the formation of bNAb–HIV-1 immune complexes may activate antigen-presenting dendritic cells and enhance their antigen-presenting and cross-presenting capabilities. This enhancement of host immune responses by neutralizing Abs (nAbs), also called the “vaccinal effect,” has been demonstrated for nAbs against other pathogens including the murine leukemia virus (MLV) FrCasE (16–18), respiratory syncytial virus (19), Hendra virus, and Nipah virus (20, 21).
We have recently shown that signaling through integrin α4β7, either by mucosal addressin cell adhesion molecule 1 (MAdCAM-1) or gp120, can promote HIV-1 replication (22, 23). In this regard, we previously demonstrated that a rhesus anti–integrin α4β7 (α4β7) monoclonal antibody (mAb; Rh-α4β7) blocks α4β7 from adopting an active conformation that is critical for MAdCAM-1 or gp120 signaling to occur (24). In addition, we determined that Rh-α4β7 selectively alters trafficking of different T and B cell subsets to mucosal tissues (25) and affects the antibody responses to simian immunodeficiency virus (SIV) infection (26). Administration of vedolizumab, an anti-α4β7 antibody with the same antigen-binding region as Rh-α4β7, in HIV-infected patients with inflammatory bowel disease (IBD) led to an attenuation of lymphoid aggregates in the gut with a decrease in non–plasma B cells particularly in the ileum (27). These alterations in immune cell trafficking and attrition of immune aggregates in mucosal tissues may drive key changes in immune responses and may partially explain the decrease in gut tissue SIV loads when Rh-α4β7 is administered before and throughout the acute phase of SIV infection (28–30).
Here, we investigated whether the immunomodulatory properties of Rh-α4β7 and its ability to affect gut tissue SIV loads may enhance the vaccinal effect of anti–HIV bNAb treatment in SHIVSF162P3-infected macaques. We found that the administration of Rh-α4β7 together with the anti–HIV bNAbs, VRC07–523LS and PGT128, to viremic macaques led to longer virologic control than the administration of VRC07–523LS and PGT128 alone. This effect was associated with reduced SHIV viremia in the colorectal tissue of treated macaques, increased natural killer (NK) cell activation, reduced frequency of B cell follicles and aggregates in the gut, and increased T cell responses against HIV Env.
RESULTS
Rh-α4β7 enhances bNAb-driven virologic control and delays viral rebound
We selected 20 adult rhesus macaques (all but one were negative for the macaque major histocompatibility complex class II alleles Mamu-A*01, Mamu-B*08, and Mamu-B*17; Table 1) that were infected with 300 median tissue culture infectious dose (TCID50) of SHIVSF162P3 intravaginally [controls were from a different study (31)] or intravenously. The plasma viral load (pVL) was between 103 and 105 copies/ml at week 10 post-infection (p.i.). We used macaques infected with this particular SHIVSF162P3 because of its stringent activity. In previous in vivo studies, only 1 of 44 macaques controlled SHIVSF162P3 spontaneously; the median set point pVL at week 10 p.i. (time of bNAb treatment) was 4.5 Log10 (mean, 5.4 Log10). Infection with SHIVSF162P3 consistently led to CD4+ T cell loss during acute infection and to an inversion of the ratio of CD4+ to CD8+ T cells, a marker of immune dysfunction and disease progression (32). All macaques infected with this virus developed high titers of anti-gp120 antibodies; this SHIVSF162P3 had a low passage history and a fully sequenced envelope protein (31). Last, this stock SHIVSF162P3 was sensitive to the combination of bNAbs VRC07–523LS (33) and PGT128 (34, 35) that we intended to use for the study (fig. S1A). These two bNAbs, one specific for the CD4+ T cell binding site and the other directed against the V3/glycan, were chosen because similar anti–HIV bNAbs are currently used together in clinical testing (36).
Table 1.
Animals divided by treatment group.
| Age | Sex | Weight | Infection | Treatment | Week of rebound | Rebound VL (copies/ml) | Tissue collected | |
|---|---|---|---|---|---|---|---|---|
| LE46 | 3.98 | M | 7.80 | IV | None | N/A | N/A | Yes |
| LF12 | 3.91 | M | 10.85 | IV | None | N/A | Yes | |
| JK78 * | 6.74 | F | 9.45 | IVAG | None | N/A | No | |
| FF62 | 13.68 | F | 13.25 | IVAG | None | N/A | No | |
| LA22 | 4.54 | M | 9.50 | IV | None | N/A | Yes | |
|
| ||||||||
| Average | 6.57 | 10.17 | ||||||
|
| ||||||||
| GA95 | 12.6 | F | 8.20 | IVAG | bNAbs | – | <200 | No |
| HM84 | 9.78 | F | 5.90 | IVAG | bNAbs | 15 | 2000 | No |
| IN59 | 8.46 | F | 9.50 | IVAG | bNAbs | 14 | 200 | No |
| JA12 | 7.78 | F | 9.30 | IVAG | bNAbs | 12 | 230 | No |
| KC14 | 6.03 | M | 8.94 | IV | bNAbs | 16 | 230 | Yes |
| LA30 | 4.18 | M | 9.13 | IV | bNAbs | 14 | 3100 | Yes |
| KT51 | 4.88 | M | 9.35 | IV | bNAbs | 19 | 200 | Yes |
| KJ36 | 5.19 | M | 7.60 | IV | bNAbs | 14 | 16,000 | Yes |
|
| ||||||||
| Average | 6.61 | 8.53 | ||||||
|
| ||||||||
| KE78 | 5.96 | M | 10.02 | IV | bNAbs + Rh-α4β7 | – | <200 | Yes |
| JT94 | 6.75 | M | 9.60 | IV | bNAbs + Rh-α4β7 | – | <200 | Yes |
| KL18 | 5.06 | M | 8.20 | IV | bNAbs + Rh-α4β7 | 16 | 630 | Yes |
| LB74 | 4.03 | M | 9.70 | IV | bNAbs + Rh-α4β7 | 15 | 7500 | Yes |
| KB22 | 6.5 | M | 12.10 | IV | bNAbs + Rh-α4β7‡ | – | <200 | Yes |
| KG31 | 6.34 | M | 11.35 | IV | bNAbs + Rh-α4β7‡ | 18 | 320 | Yes |
| KL59 † | 5.59 | M | 9.84 | IV | bNAbs + Rh-α4β7‡ | 15 | 30,000 | Yes |
|
| ||||||||
| Average | 5.75 | 10.12 | ||||||
Mamu-A*01+.
Excluded because of high antidrug antibodies.
Week 11/14.
IVAG, intravaginal; IV, intravenous; N/A, not applicable.
The 20 SHIVSF162P3-infected macaques were assigned to three treatment groups, each with similar pretreatment and peak pVL (fig. S1B). At week 10 p.i., one group of eight macaques was treated with an infusion of the combination of bNAbs: VRC07–523LS (20 mg/kg) (33) and PGT128 (5 mg/kg; bNAbs-only group; Fig. 1A) (34, 35). A second group of seven macaques was treated with a single infusion of anti–HIV bNAbs similar to the first group and with an additional infusion of the Rh-α4β7 mAb (50 mg/kg; bNAbs–Rh-α4β7 group) (37, 38). Rh-α4β7 was given at the same time (n = 4) or 1 week after (n = 3) the bNAb infusion, followed by a second infusion 3 weeks later (Table 1). No major differences were noted between animals infused with Rh-α4β7 1 week apart, and the data were pooled. One animal in the bNAbs–Rh-α4β7 group had to be excluded from analysis because it developed high amounts of antidrug antibodies against Rh-α4β7 and bNAbs within 2 weeks after antibody infusions (fig. S2A). Plasma concentrations of the bNAbs were not significantly different between the groups [VRC07–523LS bNAb–only group: area under the curve (AUC) 244.9, 95% confidence interval (CI): 179.9 to 329.9; bNAbs– Rh-α4β7 group: AUC 361.7, 95% CI: 262.5 to 440.9; PGT128 bNAb–only group: AUC 50.78, 95% CI: 26.98 to 56.57; and bNAbs–Rh-α4β7 group: AUC 80.98, 95% CI: 40 to 110.4 (fig. S2B)].
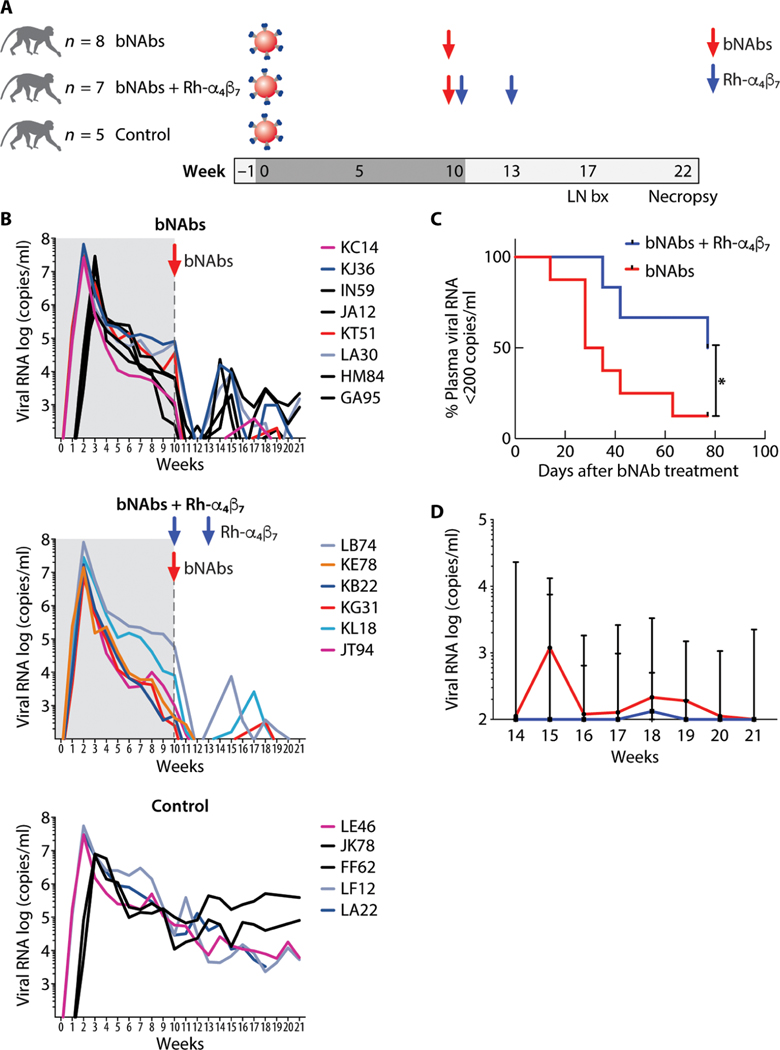
Fig. 1. Anti–HIV bNAbs and Rh-α4β7 mAb combined treatment delays rebound viremia in SHIV-infected macaques.
(A) Shown is a schematic of the study design (LN bx, lymph node biopsy). (B) Log viral RNA (copies/ml) in plasma of macaques from the time of SHIV infection and after antibody treatment is shown (black lines, intravaginal infection; colored lines, intravenous infection). The time before the bNAb infusion is shaded in gray. The top graph shows the pVLs in the bNAbs-only group; the middle graph shows the VLs in the bNAbs–Rh-α4β7 combined treatment group, and the bottom graph shows the VLs in control, untreated animals. (C) Kaplan-Meier curves were generated for time to first detection of plasma viral RNA >200 copies/ml in the bNAbs-only group and bNAbs–Rh-α4β7 group. Curves are compared with the Gehan-Breslow-Wilcoxon test (P = 0.042). (D) Shown is the median ± range of log viral RNA (copies/ml) in plasma after week 14 p.i. (earliest time of pVL rebound) for the animals in the two treatment groups.
Treatment with the VRC07–523LS and PGT128 bNAb combination lowered pVL below 200 copies/ml in all macaques in both bNAb groups. In seven of the eight macaques treated with the bNAb-only combination (87.5%), pVL remained below 200 copies/ml for a median of 3 weeks (95% CI: 2 to 9 weeks) and then rebounded. Only one animal remained below 200 copies/ml until the end of follow-up (~21 weeks p.i.). In contrast, three of six macaques in the bNAbs– Rh-α4β7 group (50%) remained below 200 copies/ml until the end of follow-up (21 weeks p.i.). The remaining three macaques rebounded within a median of 6 weeks after treatment (95% CI: 5 to 11 weeks) (Fig. 1, B and C). Thus, treatment with bNAbs and Rh-α4β7 delayed viral rebound above 200 copies/ml in viremic macaques compared to macaques treated with anti–HIV bNAbs only (P = 0.042; Fig. 1C and data file S1). A subset analysis that included only intravenously infected animals remained significant and is shown in fig. S3A (P = 0.039). Moreover, postrebound pVL was significantly lower in the bNAbs–Rh-α4β7 group compared to the bNAbs-only group [P = 0.020 by two-way analysis of variance (ANOVA) for repeated measures; Fig. 1D and data file S1]. However, the rebound pVL AUCs of the two bNAbs groups did not differ significantly (bNAbs only: 9185, 95% CI: 0 to 20,665; bNAbs–Rh-α4β7: 2226, 95% CI: 0 to 7084).
The plasma concentrations of VRC07–523LS and PGT128 declined rapidly in all bNAb-treated macaques, driven primarily by rising amounts of antidrug antibodies against these human immunoglobulin Gs (IgGs; fig. S2B). However, differential viral rebound in the two treatment groups was not explained by different concentrations of bNAbs in the groups before viral rebound (Fig. 2A). We compared concentrations of VRC07–523LS and PGT128 in each of the animals on the week before viral rebound. Amounts of these bNAbs were modestly lower in the bNAbs–Rh-α4β7 group compared to the bNAbs-only group, although the difference was not statistically significant (Fig. 2A) (an expanded analysis including bNAb concentrations at necropsy in nonrebounding animals is shown in fig. S3B).
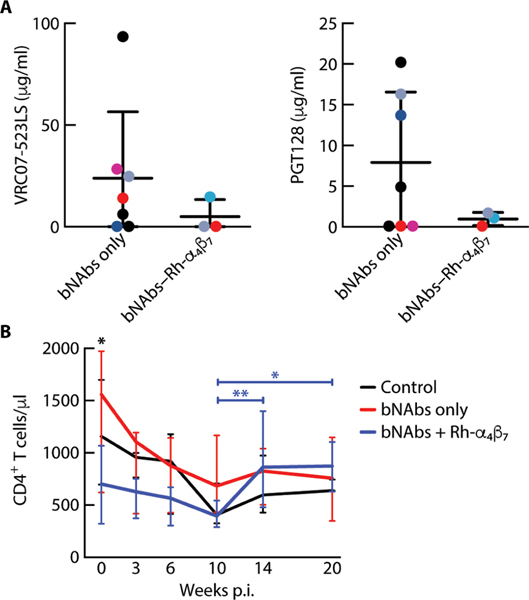
Fig. 2. bNAb plasma concentrations do not differ between treatment groups before viral rebound.
(A) Shown are the plasma concentrations (means ± SD) of anti–HIV bNAbs VRC07–523LS and PGT128 6 days before the first detection of >200 copies/ml of SHIV RNA in the bNAbs-only and bNAbs–Rh-α4β7 treatment groups. (B) Shown are the medians ± range of CD4+ T cell counts in blood for different time points after SHIV infection in the three groups of animals. Differences between groups were tested at each time point by Kruskal-Wallis test, and differences between time points were tested with a mixed-effect analysis using Geisser-Greenhouse correction and Tukey’s multiple comparisons test. *P < 0.05 and **P < 0.01.
Virus isolated at rebound from all the animals treated with bNAbs was sequenced in parallel to virus isolated from the controls at corresponding time points (table S1). The sequences were analyzed for resistance to VRC07–523LS and PGT128 with the bNAb–Resistance Predictor (bNAb-ReP) algorithm, a machine learning algorithm that predicts neutralization resistance to anti–HIV-1 bNAbs given the sequence of viral Env (39). Overall, the analysis suggested that all of the sequences were sensitive to PGT128 (table S2), and most sequences were borderline sensitive to VRC07–523LS. Only two isolates from one macaque, LA30 (bNAbs-only group), were predicted to be resistant to VRC07–523LS (table S3). However, additional isolates from the same time points were sensitive, and a similar resistant isolate was present in one of the control animals. This suggested that, although the bNAbs likely exerted some pressure, viral rebound was not driven by resistance to either of the bNAbs. Two of the three bNAbs–Rh-α4β7–treated macaques that rebounded had viruses with an extensive gp120 deletion (five to six amino acids) at the beginning of the V4 loop (table S2). A similar deletion, but shorter (four amino acids) and further into the V4 loop, was present in a small fraction of viral isolates from one of three control and one of seven bNAbs-only–treated animals.
Regarding CD4+ T cell counts, we noted a significant increase from weeks 10 to 14 (P = 0.002) that was maintained through week 20 p.i. (P = 0.03) in the bNAbs–Rh-α4β7 group, whereas no significant change was observed in either the bNAbs-only group or the control group (Fig. 2B and data file S1). We observed a lower average CD4+ T cell count in the bNAbs–Rh-α4β7 group compared to the other two groups at baseline. However, this did not appear to influence infection.
bNAbs–Rh-α4β7 cotreatment reduces SHIV in colorectal tissue
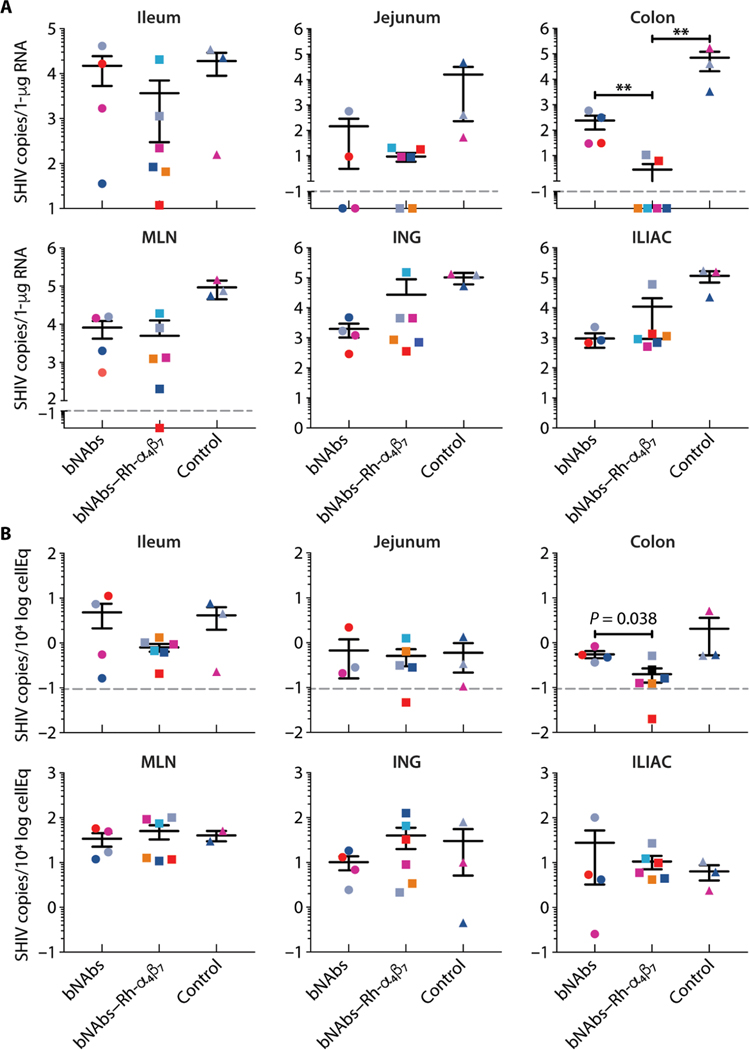
From all intravenously infected animals, lymph nodes and large blood draws were collected at week 17 p.i. (7 weeks after bNAb infusion) and several tissues were collected at necropsy at week 21 p.i. (Table 1). Macaques infected intravaginally (as controls for an SIV acquisition study) (31) were euthanized without tissue collection. Comparison of the SHIV DNA/RNA content in the gut tissues and lymph nodes revealed that the macaques treated with the bNAbs– Rh-α4β7 combination had lower cell-associated SHIV in the colorectal tissue than did the bNAbs-only group or the control group (Fig. 3, A and B). With respect to the bNAbs–Rh-α4β7 group compared to the bNAbs-o nly group, the effect was more pronounced for SHIV RNA (P = 0.009) than for DNA (P = 0.038). Three of the four animals that showed no rebound in pVL also showed undetectable SHIV RNA in colon. Two animals that did have rebound in pVL exhibited detectable SHIV RNA in colorectal tissue. The remaining animal with rebound in pVL had no detectable SHIV RNA in colon tissue (Fig. 3A).
Fig. 3. bNAbs–Rh-α4β7 treatment reduces viral RNA and DNA in gut tissues of SHIV-infected macaques.
(A and B) Copies of SHIV RNA (A) and SHIV DNA (B) from the indicated tissues at the time of necropsy were quantified by RT-qPCR (normalized to RNA content) and gag-qPCR (normalized to albumin), respectively. Bars represent means ± SD. The dashed line indicates the lower limit of detection of the assay. Data from the treatment groups are compared to data from the control group by Kruskal-Wallis test and the results of the Dunn’s multiple comparisons post hoc test. **P < 0.01. MLN, mesenteric lymph nodes; ING, inguinal lymph nodes; ILIAC, iliac lymph nodes.
bNAbs–Rh-α4β7 cotreatment increases memory T cells in blood and decreases B cells in tissues
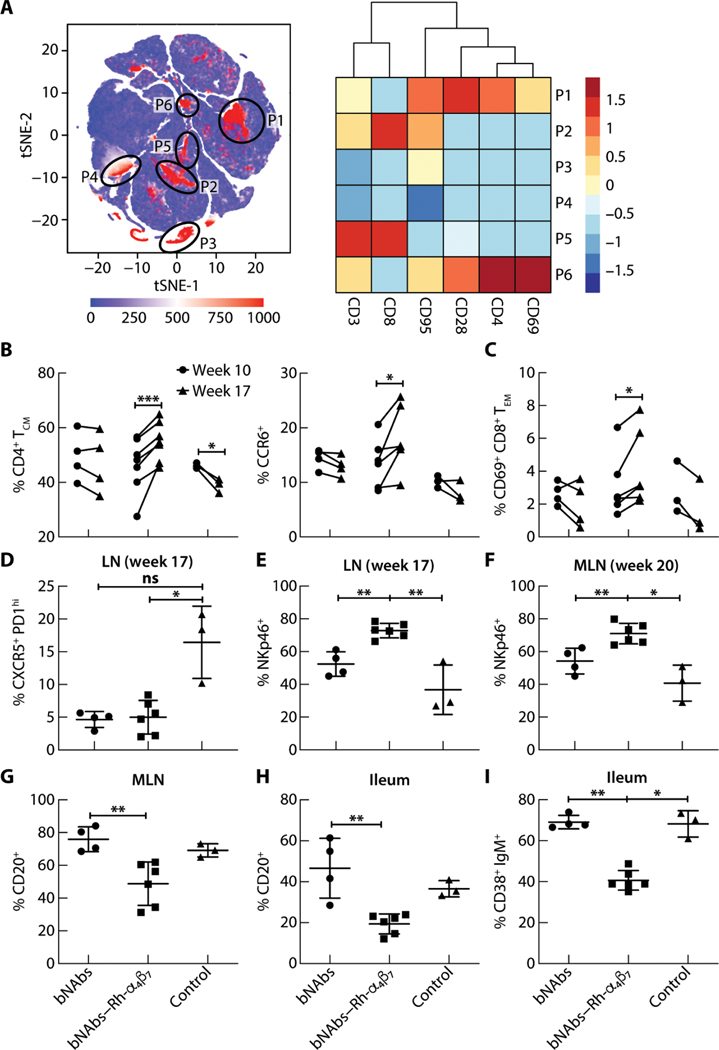
Flow cytometry phenotypic analysis was performed on peripheral blood mononuclear cells (PBMCs) from all macaques in the study at baseline and week 17 p.i. We confirmed that the Rh-α4β7 mAb completely blocked integrin α4β7 on CD4+ T cells at week 17 p.i. for animals receiving Rh-α4β7 (fig. S4A). The impact of targeting α4β7 integrin on different leukocyte subsets was apparent from the t-distributed stochastic neighbor embedding (tSNE) analysis [Fig. 4A (left) and figs. S4B and S5]. In the tSNE analysis of PBMCs before and after treatment, α4β7+ cells were absent from the bNAbs–Rh-α4β7 group plot [Fig. 4A (left) and fig. S4B], specifically in areas [marked P1 to P6 in Fig. 4A (left)] corresponding to CD3+ CD4+ T cells that mostly coexpressed CD28, CD95, and CD69 (shown for areas P1 and P6 in the heatmap; Fig. 4A, right) and in areas corresponding to CD3+ CD8+ T cells (areas P2 and P5 in the heatmap; Fig. 4A, right). Moreover, at least two cell subsets (P3 and P4) absent in the bNAbs– Rh-α4β7 group plot were CD3− T cells (Fig. 4A, right), suggesting that Rh-α4β7 mAb specifically targeted not only T cells but also blood NK cells and myeloid cells (although markers for those cell subsets were not in the panel).
Fig. 4. Effects of bNAbs–Rh-α4β7 treatment of SHIV-infected macaques on T cells, NK cells, and B cells in different tissues.
(A) Left: tSNE plot displaying kinetics of expression of α4β7 on live single-cell populations in blood from SHIV-infected macaques, including four animals from the bNAbs-only group, six from the bNAbs–Rh-α4β7 group, and three controls. Gating of α4β7-positive cell populations on the tSNE plot was based on antigen expression and discrete density clustering. P1 to P6 (red areas in black circles) on the tSNE plot indicate the different immune cell populations expressing α4β7. Right: Heatmap depicting the fold change in expression of the antigens CD3, CD8, CD95, CD28, CD4, and CD69 for each of the immune cell populations in P1 to P6 compared to the channel values for cell populations negative for these antigens. (B and C) Shown are the frequencies of cell subsets within the CD3+ CD4+ T cell population [CD4+ central memory T cells (CM), CCR6+] (B) and the CD3+ CD8+ T cell population [CD69+CD8+ effector memory T cells (EM)] (C) among live cells isolated from PBMCs at weeks 10 and 17 p.i. Data are compared by two-way ANOVA with Bonferroni multiple comparisons correction. (D and E) Shown is the frequency of CXCR5+ PD1high T follicular helper cells within the CD3+ CD4+ T cell population (D) and NKp46+ cells within the NKG2A+ CD3− cell population (E) among live cells isolated from LN bx at weeks 17 or 20 p.i. (F and G) Shown are frequencies of NKp46+ cells (F) within the NKG2A+ CD3− cell population and CD20+ B cells (G) among live cells isolated from MLN at necropsy. (H and I) Shown are frequencies of total CD20+ B cells and CD38+ IgM+ B cells isolated from a small portion of macaque ileum at necropsy. Data from the bNAbs-only and bNAbs–Rh-α4β7 treatment groups were compared directly with a Mann-Whitney nonparametric unpaired test and were compared with the control group using the Kruskal-Wallis test and the results of the Dunn’s multiple comparisons post hoc test. *P < 0.05, **P < 0.01, and ***P < 0.001. Bars represent means ± SD.
A significant increase in the frequency of central memory CD95+ CD28+ T cells (P < 0.001) and CCR6+ CD4+ T cells (P = 0.032) was noted in the bNAbs–Rh-α4β7 group of macaques but not in the other two groups (Fig. 4B). A similar effect of Rh-α4β7 antibody on circulating CD4+ T cells was previously reported by our group in uninfected macaques (25). Our gating strategy was reported elsewhere (25, 40). There was also an increase in the frequency of CD95+ CD28− CD69+ activated effector memory CD8+ T cells in the blood of bNAbs–Rh-α4β7– treated macaques at week 17 p.i. compared to before treatment (Fig. 4C), whereas there was mostly a decrease in this cell population in the other two groups. No other blood cell subsets were modulated by treatment with bNAbs alone or bNAbs–Rh-α4β7 (table S4).
To investigate the impact of bNAbs– Rh-α4β7 cotreatment on macaque tissues, we phenotyped cells isolated from axillary lymph node biopsies collected at week 17 p.i. (week 7 after bNAb infusion) and from mesenteric lymph nodes, ileum, and colorectal tissues collected at necropsy. We found that the bNAbs-only and the bNAbs–Rh-α4β7 groups of macaques had a lower frequency of T follicular helper cells defined as CXCR5+ PD1+ CD4+ T cells in the axillary lymph nodes compared to the control group (Fig. 4D and fig. S4). In contrast, the frequency of NKp46+ cells within the NKG2A+ CD3− NK cell population was higher in the axillary lymph nodes of the macaques treated with bNAbs–Rh-α4β7 compared to bNAbs alone or the control group at week 17 p.i. (Fig. 4E and fig. S6); the increase was still evident in mesenteric lymph nodes a few weeks later at necropsy (P = 0.009; Fig. 4F). This suggested that bNAbs–Rh-α4β7 treatment resulted in an accumulation of activated NK cells in the lymph nodes or elevated expression of NKp46 in lymphoid tissue NK cells. The overall frequency of CD3+ T cells had a tendency to be higher in the mesenteric lymph nodes at necropsy (fig. S7), and the frequency of NKG2A+ NK cells (within the CD3− T cell population) was higher in mesenteric lymph nodes of bNAbs–Rhα4β7–treated animals compared to the bNAbs-only treatment group (P = 0.009; fig. S8A, left). This difference did not appear to be driven by differences in pVL between groups because the frequency of NK cells in the mesenteric lymph nodes did not correlate with pVL (fig. S8A, right).
Another interesting finding from the analysis of tissues at necropsy was a decrease in the frequency of B cells (total CD20+ cells within the CD45+ B cell population) both in the mesenteric lymph nodes and in the ileum of bNAbs–Rhα4β7–treated animals compared to the bNAbs-only treatment group (Fig. 4, G and H). Moreover, among the B cells in the ileum, there was a specific decrease in CD38+ IgM+ cells (Fig. 4I) and a corresponding increase in the proportion of CD38+ IgM− cells (fig. S8B). These cells were all CD27 low or negative. Further characterization of B cell subsets would have required an anti-rhesus IgD antibody, a reagent not available at the time that this work was performed. A similar decrease in the frequency of non–plasma B cells in the ileum has been reported in vedolizumab-treated HIV-positive individuals with IBD (27).
To confirm and further explore our findings on a potential Rh-α4β7 antibody– driven decrease in B cells in tissues, we performed histologic analysis of macaque mesenteric lymph nodes and ileum. A decrease in B cell follicles was evident in the mesenteric lymph nodes of macaques treated with bNAbs–Rh-α4β7 when these animals were directly compared to the bNAbs-only treatment group (Mann-Whitney test unadjusted P = 0.019; Kruskal-Wallis and Dunn’s test adjusted P = 0.067; Fig. 5, A and B, and fig. S9) and when compared to the control group (adjusted P = 0.021). Moreover, microscopic analysis of B cell aggregates in random tissue sections (n = 4 to 5 per animal) of the ileum samples collected at necropsy indicated a decrease in the tissue area covered by lymphoid aggregates in the bNAbs–Rh-α4β7–treated macaques compared to the bNAbs-only treatment group (Mann-Whitney test unadjusted P = 0.019; Kruskal-Wallis and Dunn’s adjusted P = 0.024; Fig. 5, C and D, and fig. S10). However, no difference was immediately evident when comparing the bNAbs–Rh-α4β7 group and the control group (Dunn’s adjusted P = 0.539). Last, no significant differences were noted in several other cell subsets analyzed, including CD8+ T cells, NK cells, and other B cell subsets in tissues (table S4).
Fig. 5. bNAbs–Rh-α4β7 treatment reduces B cell follicles and aggregates compared to bNAbs-only treatment.
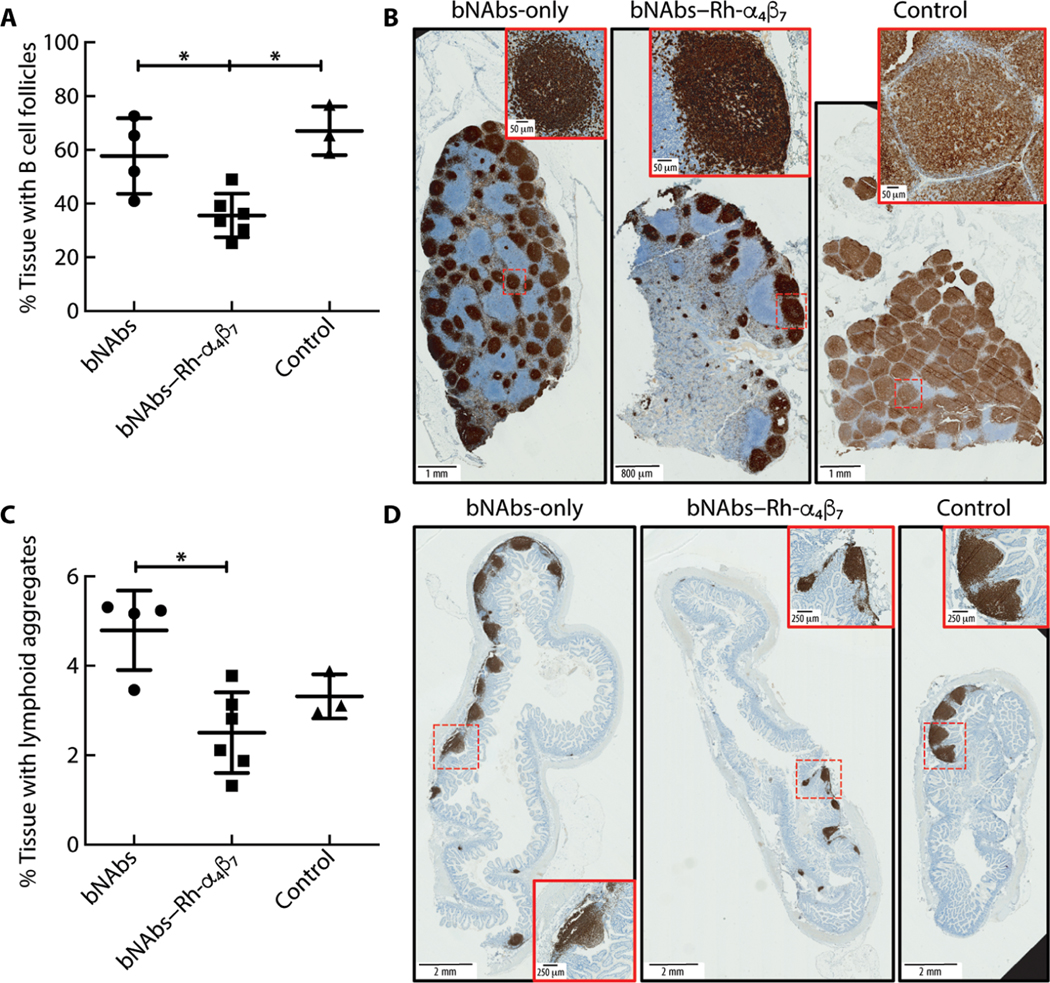
(A to D) Tissue sections (5 μm) were cut from formalin-fixed MLN and ileum tissue blocks (two blocks per animal, one section per block) and were stained with anti-CD20 antibody. Digitally scanned images were analyzed with QuPath version 0.2.3, and results are expressed as a percentage of tissue area covered by B cell follicles (A and B) or lymphoid aggregates (C and D). Representative full-scale images from one tissue per treatment group are shown, with higher magnification of a lymphoid aggregate detailed in the insets (red boxes). Full image scale bars, 2 mm; inset scale bars, 250 μm. Data from the bNAbs-only and bNAbs–Rh-α4β7 treatment groups are compared directly with Mann-Whitney test (nonadjusted P value shown) and with the Kruskal-Wallis test and Dunn’s multiple comparisons post hoc test (shown for comparisons with the control group). *P < 0.05. Bars represent means ± SD.
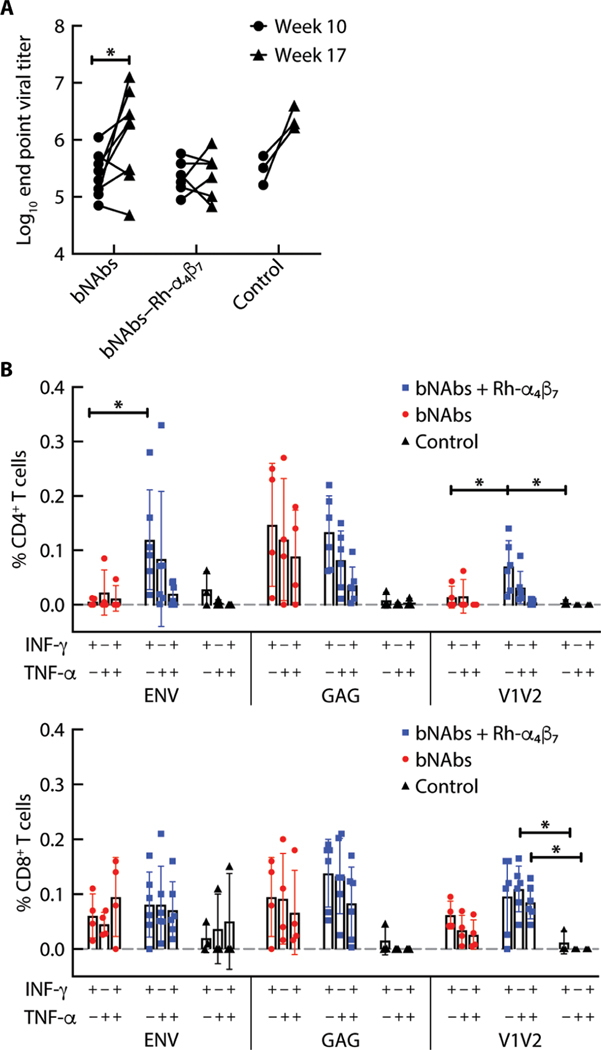
bNAbs–Rh-α4β7 treatment increased T cell responses but not anti-Env antibody responses
Plasma concentrations of total anti-Env antibodies increased from weeks 10 to 17 p.i. in the bNAbs-only and control groups but not in the bNAbs–Rh-α4β7 group (Fig. 6A). T cell responses against viral Env and gag proteins were probed using pools of 15-mer peptides. A higher frequency of interferon-γ–positive (IFN-γ+) CD4+ T cells was noted in the bNAbs–Rh-α4β7 treatment group in response to the Env peptides compared to the bNAbs-only treatment group (Fig. 6B). The IFN-γ+ CD4+ T cell responses of macaques treated with bNAbs– Rh-α4β7 were also higher compared to the other two groups when probed using a pool of seven 15-mer peptides overlapping the V2 loop (Fig. 6B, left). Last, there were more CD8+ T cells expressing tumor necrosis factor–α (TNF-α) and TNF-α+ IFN-γ+ CD8+ T cells in response to V2 loop peptides in the bNAbs–Rh-α4β7 treatment group compared to the control group (Fig. 6B, right). However, the difference compared to the bNAbs-only group did not reach significance. A similar enhanced V2-specific reactivity was previously described in SIV/SHIV-infected macaques treated with the Rh-α4β7 mAb (41). In contrast, no significant differences in response to viral Gag protein were observed among the two treatment groups. However, a tendency toward higher responses was noted in both treatment groups compared to the control group. Last, no additional differences were noted in the frequency of TNF-α or polyreactive T cells [interleukin-2 (IL-2) and IL-17 responses were below the limit of detection] (Fig. 6B).
Fig. 6. bNAbs–Rh-α4β7 treatment increases T cell responses with no change in total anti–HIV Env antibodies.
(A) Antibody titers against the SHIVSF162P3 gp140 trimer were measured before week 10 p.i. and after week 17 p.i. in the bNAbs-only and bNAbs–Rh-α4β7 treatment groups. Results are compared by two-way ANOVA with Sidak’s multiple comparisons test. (B) Shown is the frequency of CD4+ T cells (left) and CD8+ T cells (right) secreting IFN-γ, TNF-α, or both, after PBMC stimulation for 6 hours with CD28/CD49d and a HIV consensus B envelope peptide pool, a gag pool, or a pool of 15-mer overlapping peptides from the V2 loop of the SHIVSF162P3 Env. Data from the two treatment groups are compared by Kruskal-Wallis test and the results of the Dunn’s multiple comparisons post hoc test. *P < 0.05. Bars represent means ± SD.
DISCUSSION
Passive infusion of a combination of anti–HIV bNAbs as immunotherapy, when initiated during early or acute infection in nonhuman primates, has been shown to promote host immunity capable of leading to virologic control several weeks after treatment (11, 42, 43). Human studies with anti–HIV bNAbs also suggest that this therapy may enhance immune responses to HIV and improve virologic control in some individuals (14, 15). However, infusion of bNAbs alone during ATI inevitably leads to viral rebound as bNAbs wane (9). Thus, anti–HIV bNAbs may constitute a promising tool for HIV cure, but they will need to be used in combination with other therapeutic agents and strategies, including immunomodulatory agents. Anti-α4β7 antibody therapy with vedolizumab in individuals with IBD leads to gut-selective immunosuppression (44) and has become a frontline treatment for severe IBD. Vedolizumab and the macaque anti-α4β7 antibody Rh-α4β7 can be considered as immunomodulatory agents. Here, we tested the combination of anti–HIV bNAbs and Rh-α4β7 with the goal of determining whether Rh-α4β7 could affect immune responses against SHIVSF162P3 in a way that enhanced the therapeutic activity of bNAbs. This small proof-of-concept study demonstrated that addition of Rh-α4β7 to an anti-CD4 binding site/anti-V3 loop combination of bNAbs modulated the antiviral immune response and improved virologic control in viremic animals when given in a post-acute or early chronic phase of the infection.
This study has several limitations. The number of animals is relatively small. Moreover, the treatment groups were not gender balanced, with only male macaques in the bNAbs–Rh-α4β7 group and with a gender mix (~50/50) in the other two groups. However, this most likely did not influence the results of the study because gender differences appear to be less pronounced in SIV/SHIV infection of rhesus macaques compared to humans. Moreover, the lower HIV VLs observed in HIV-infected women would have favored virologic control in the groups with female macaques, instead of which we observed enhanced control in the bNAbs–Rhα4β7 group comprising only male macaques. Another important limitation of the study was that, by design, we chose to test the therapy only in animals with a pVL between 103 and 105 copies/ml and considered suppression of pVL below 200 copies/ml as effective viral control. This study design was based on previous reports that bNAbs decrease pVL below 200 copies/ml only when the pVL is below 105 copies/ml and where the threshold of 200 copies/ml was considered effective suppression (1). In any event, bNAb-based therapeutic strategies are most likely to be used as follow-on therapies to cART where low or undetectable pVLs are already in place. A larger study with more animals in each group and longer follow-up and with bNAbs–Rh-α4β7 treatment administered during cART should be done before the therapeutic potential of combining α4β7 blockade and bNAbs is translated to the clinic.
Our study was robust insofar as the SHIVSF162P3 virus used for the challenge was highly stringent. In contrast to other SHIV strains used for similar studies that frequently lead to spontaneous virologic control, the virus used in this study maintained high peak and set point pVLs and led to CD4+ T cell loss. Unfortunately, the follow-up period was short because all the macaques had to be euthanized relatively early after therapy (~12 weeks). Thus, we could not investigate the long-term effects of the bNAbs–Rh-α4β7 therapy. A longer-term study would have better clarified the nature of the virologic control in the bNAbs-only group and bNAbs–Rh-α4β7 group and would have demonstrated whether the effect of the bNAbs–Rh-α4β7 therapy was transient because of erosion of the gut viral reservoir or whether the dual therapy led to a permanent change in the immune response. It is possible that the animals that controlled viremia in the bNAbs–Rh-α4β7 group may have eventually rebounded. Future studies will need to address how long virologic control lasts in the macaques treated with the bNAbs–Rh-α4β7 combination and underlying mechanisms in greater depth. Our data suggest that the enhanced virologic control may be the result of stronger T cell responses. This is supported by the presence of a higher frequency of central memory CD4+ T cells, activated memory CD8+ T cells, and higher IFN-γ responses to viral Env in the bNAbs–Rh-α4β7 group compared to the bNAbs-only group. Both treatment groups had a tendency toward higher Gag-specific responses both in the CD4+ and CD8+ T cell subsets. This suggests a bNAb-s pecific driven stimulation of host cellular immune responses. However, T cell depletion studies will be needed to determine in a definitive way the role of T cell responses in maintaining virologic control both in bNAb treatment alone and in bNAbs–Rh-α4β7 combination treatment.
As in previous studies with the Rh-α4β7 mAb (28, 30, 41), we noted a decrease in colorectal SHIV load in the macaques treated with bNAbs–Rh-α4β7. However, it is not possible to know whether this was a consequence of the better virologic control in bNAbs–Rh-α4β7–treated macaques or was a direct activity of the Rh-α4β7 antibody, e.g., through reducing reseeding of the gut viral reservoir by blocking trafficking of SHIV-infected α4β7+ CD4+ T cells or by reducing the size of SHIV-loaded B cell aggregates. We noticed that the difference in gut VL was more pronounced with regard to viral RNA than viral DNA. This is likely due to the fact that viral DNA measurements included DNA from cells infected before treatment and also virus in a defective state that was not affected by the treatment.
There are two additional noteworthy findings in this study. First, the combination of bNAbs and Rh-α4β7 mediated an apparent increase in the number of NK cells and their activation in the lymph nodes. However, this observation was limited by the lack of in-depth NK cell phenotyping and functional testing. An increase in the frequency of cytokine-secreting NK cells was previously reported in the blood of Rh-α4β7–treated macaques (26). We previously reported changes in trafficking of NK cells in naïve macaques after Rh-α4β7 infusion (25). Our finding of an increase in NK cells in the present study was limited to lymph nodes. This may be explained by changes in NK cell trafficking to this location and may not necessarily be associated with a change in their activation status or function. Future studies will need to confirm the impact of Rhα4β7 on NK cells in vivo and determine whether NK cells contribute to virologic control in SHIV-infected animals.
A second noteworthy finding is the decrease in B cell follicles in the mesenteric lymph nodes of the bNAbs–Rh-α4β7–treated macaques and the accompanying decrease in B cell aggregates in the ileum. A similar decrease in lymphoid aggregates has been reported in the terminal ileum of HIV-infected individuals with IBD treated with vedolizumab (27). However, whereas, in that study, the decrease was evident after five to six infusions of vedolizumab 22 to 30 weeks after the first treatment, here, we report a difference between groups evident at 9 to 11 weeks after treatment with two Rh-α4β7 infusions. We did not observe a difference in lymphoid aggregates in the ileum between the bNAbs–Rh-α4β7 group and the control group. However, this may have been due to the lack of before and after sampling and the limited scope of the analysis (four to five random sections per animal). The effect of this decrease in gut B cells on SHIV or SIV infection needs to be explored in ad hoc B cell depletion studies. However, disruption of B cell follicles and aggregates may have contributed to the decrease in gut VL. Our finding suggests that specific subsets of B cells may be more affected by Rh-α4β7 than others, and this may have consequences for B cell responses to SHIV, explaining, at least in part, the lack of an increase in antibody responses in the bNAbs– Rh-α4β7–treated animals. Alternatively, this difference may reflect the lower pVL in the bNAbs–Rh-α4β7 treatment group.
In summary, our small study suggests that the Rh-α4β7 antibody may have beneficial immune modulatory activity during SHIV infection of macaques that complements the effect of anti–HIV bNAbs. The exact nature of the activity of both Rh-α4β7 and bNAbs needs further exploration. Nonetheless, the changes that we observed in specific immune cell subsets and in gut VLs after Rh-α4β7 infusion in SHIV-infected macaques, findings that have been recapitulated by several different groups (25, 27, 28, 30, 37, 41, 45), call for further investigation of the therapeutic potential of targeting α4β7 in HIV, even in light of negative results reported in several other studies (46–48).
MATERIALS AND METHODS
Study design
The objective of this study was to determine the effect of treating SHIVSF162P3-infected macaques with a combination of anti–HIV bNAbs and the Rh-α4β7 mAb compared to bNAbs alone or no treatment. Twenty macaques were infected with SHIVSF162P3 at the start of the study either intravaginally or intravenously. They were treated with either anti–HIV bNAbs alone (VRC07–523LS and PGT128; n = 8) or anti–HIV bNAbs and the Rh-α4β7 mAb (n = 7) or were left untreated as controls (n = 7). The study was not blinded, but primary analysis of the data was blinded. One animal was excluded from the bNAbs–Rh-α4β7 group because of the development of antidrug antibodies against Rh-α4β7. There were no outliers, and all of the available data were included in the analysis.
All studies were approved by the Animal Care and Use Committee of the Tulane National Primate Research Center (TNPRC), Office of Laboratory Animal Welfare (assurance no. A4499–01; protocol P0180–3639) and in compliance with the Animal Welfare Act and the Guide for the Care and Use of Laboratory Animals. TNPRC is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC no. 000594).
Animals
A total of 20 adult female and male Indian rhesus macaques [Mamu A*01, B*08, and B*17 negative except for JK78 A*01; average weight, 8.1 kg (range: 4.5, 13.05) and average age, 9.5 years (range: 3.9, 18)] were socially housed (two animals per cage) indoors under climate-controlled conditions with a 12-hour light/12-hour dark cycle until the time of the first viral challenge. After initiation of the viral challenges, all animals were single-housed to avoid cross-infection. All of the macaques in the study were previously assigned to the specific pathogen–free breeding colony for varying lengths of time. Animals were monitored twice daily to ensure their welfare. Any abnormalities including those of appetite, stool, or behavior were recorded and reported to a veterinarian. The animals were fed commercially prepared monkey chow twice daily. Supplemental foods were provided in the form of fruit, vegetables, and foraging treats as part of the TNPRC environmental enrichment program. Water was available at all times through an automatic watering system. The TNPRC environmental enrichment program is reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) semiannually. Veterinarians at the TNPRC Division of Veterinary Medicine have established procedures to minimize pain and distress through several means. Monkeys were anesthetized with ketamine-HCl (10 mg/kg) or tiletamine/zolazepam (6 mg/kg) before all procedures. Preemptive and postprocedural analgesia (buprenorphine; 0.01 mg/kg) was required for procedures that would likely cause more than momentary pain or distress in humans undergoing the same procedures. The above listed anesthetics and analgesics were used to minimize pain or distress associated with this study in accordance with the recommendations of the Weatherall report (49). The animals were euthanized at the end of the study using methods consistent with recommendations of the American Veterinary Medical Association panel on euthanasia and as per the recommendations of the IACUC. Specifically, the animals were anesthetized with tiletamine/zolazepam (8 mg/kg, intramuscularly) and were given buprenorphine (0.01 mg/kg intramuscularly), followed by an overdose of sodium pentobarbital. Death was confirmed by auscultation of the heart and pupillary dilation. None of the animals became severely ill or died before the experimental end point. The TNPRC policy for early euthanasia/humane end point was included in the protocol in case those circumstances arose.
All animals were infected with SHIVSF162P3 at the start of the study either intravaginally or intravenously. Twenty macaques were chosen because their pVL was between 103 and 105 copies/ml at week 10 p.i., and they were divided into three groups of eight, seven, and five animals. At week 10 p.i., the 20 macaques were administered intravenously with the following antibody treatments: (i) VRC07523LS bNAb (20 mg/kg) and PGT128 bNAb (5 mg/kg; n = 8); (ii) VRC07–523LS bNAb (20 mg/kg), PGT128 bNAb (5 mg/kg), and Rh-α4β7 mAb (50 mg/kg) all on the same day (n = 4) or the Rh-α4β7 mAb 1 week after the two bNAbs (n = 3), followed by one infusion of Rh-α4β7 mAb (50 mg/kg) 3 weeks after the first one; and (iii) animals were left untreated as controls (n = 5). Blood was collected weekly to monitor SHIV infection. Lymph node biopsies were collected at week 17 p.i. and at necropsy (~week 22 p.i.). Blood, lymph nodes, and gut tissue were harvested and used for mononuclear cell isolation.
SHIVSF162P3 stock generation, titration, and in vitro sensitivity
SHIVSF162P3 stock used for in vivo challenge was a third-generation growth of virus obtained originally from the National Institutes of Health (NIH) Division of AIDS. The stock was grown in CD8 T cell– depleted allogeneic rhesus macaque PBMCs as described in (31). The harvested virus stock was characterized for p27 content by the enzyme-linked immunosorbent assay (ELISA), SIV gag RNA content by quantitative reverse transcription polymerase chain reaction (qRT-PCR) (24), TCID50 in rhesus macaque PBMCs by Reed and Muench method (50), and focus-forming units in TZM-bl cells by MAGI assay for repeated titration of the same stock over time. The titer used for in vivo infection was TCID50 from macaque PBMCs and the envelope sequenced (31). Macaques were challenged intravaginally (31) or intravenously with 300 TCID50.
Measurement of plasma VRC07–523LS, PGT128, and Rh-α4β7 antibodies and antidrug antibodies
Plasma bNAb concentrations were quantified using plates coated with either an anti-idiotypic antibody, 5C9, for specific detection of VRC07–523, or ST0A9 (51), for specific detection of PGT128 as described previously (33). Concentrations of rhesus Rh-α4β7 antibody in macaque plasma were measured using the α4β7-expressing human T cell line HuT 78 (NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: HuT 78 from R. Gallo) in a flow cytometry–based assay as described in (25, 30) using the standard curve method.
Concentrations of anti–VRC07–523LS and PGT128 antibodies were evaluated as follows. Plasma from macaques that had been administered bNAbs was diluted with phosphate-buffered saline (PBS) containing 5% skim milk, 2% bovine serum albumin (BSA), and 0.05% Tween 20. Fivefold serial dilutions ranging from 1:50 to 1:781,250 of these plasmas were then added in duplicate wells to 96-well ELISA plates coated with either of the two bNAbs (2 μg/ml). The plate was incubated for 1 hour at room temperature followed by a PBS-T (PBS with 0.05% Tween 20) wash. Bound monkey IgGs were then probed with a horseradish peroxidase (HRP)–conjugated anti-monkey IgG, Fc-specific (SouthernBiotech) for 30 min at room temperature. The plate was then washed, and SureBlue TMB (Kirkegaard & Perry Laboratories, Gaithersburg, MD) substrate was added. Once color was developed (typically 15 to 20 min), stopping buffer (0.5M) was added and the optical density at 450 nm was read. End point titer was calculated by determining the lowest dilution that had optical density greater than fivefold of that in the background wells.
Concentrations of anti–Rh-α4β7 antibodies were measured via lambda light-chain detection assay. ELISA plates were coated with Rh-α4β7 (10 μg/ml) overnight at 4°C, washed, and blocked with tris-buffered saline (2% BSA and 0.1% Tween 20) for 2 hours at room temperature. Test plasma was serially diluted in dilution buffer (starting at 1:10 then serial 1:4 dilutions), and 100 μl was applied to the plates and incubated for 1 hour at room temperature. Plates were washed and incubated with anti-Ig human lambda light chain–biotin (Miltenyi Biotec), which does not recognize the kappa chain of the Rh-α4β7 for 1 hour at room temperature. Plates were washed and incubated with diluted streptavidin-HRP (Invitrogen) for 1 hour at room temperature. Plates were washed, and enzymatic activity was detected by adding TMB substrate and read on a luminometer at 450 nm. End point was the highest dilution with optical density twofold higher the pretreatment sample.
SHIV VL measurements
Plasma samples were obtained from EDTA-treated whole blood and used for the determination of plasma VL by SIV gag qRT-PCR (quantitative Molecular Diagnostics Core, AIDS and Cancer Virus Program Frederick National Laboratory) (50). DNA and RNA were extracted from snap-frozen tissues using DNeasy/RNeasy blood and tissue kits (QIAGEN) following the manufacturer’s instructions. Tissue viral DNA loads were quantified using the standard curve method and normalized by albumin copy numbers by Gag-qPCR as described in (52). For tissue RNA loads, 1 μg of total RNA was retro-transcribed to DNA using the VILO Kit (Thermo Fisher Scientific) quantified by Gag-qPCR (52). Virus at rebound was isolated from plasma, and single-genome amplification sequencing was performed as previously described (53). Sensitivity to VRC07–523LS and PGT128 was determined using the bNAb-ReP algorithm as previously described (39).
PBMC isolation and flow cytometry
At 17 weeks p.i. and at necropsy, PBMCs were isolated using Ficoll-Hypaque density gradient centrifugation and lymph nodes were cut into small pieces and passed through a 40-μm cell strainer. Cells from iliac and colorectal tissue were isolated by enzymatic digestion in Hanks’ balanced salt solution containing collagenase IV (2 mg/ml; Worthington Biochemical) and human serum albumin (1 mg/ml; Sigma-Aldrich), shaking at 37°C for 50 min. The resulting cell suspension was passed through a 40-μm cell strainer and washed with PBS. Isolated cells were frozen for phenotyping and stimulation experiments at a later time. Cells were phenotyped with the antibody panel listed in table S5. tSNE analyses were performed in R using the “Rtsne” package with default perplexity and theta settings. Graphics were created using the “tidyverse” package in R (54).
For PBMC stimulation experiments, up to 3 × 106 cells per sample were thawed, plated on a plate precoated with goat anti-mouse IgG (2.5 μg/ml), and cross-linked with anti-CD28 and anti-CD49d antibodies (10 μg/ml; Sigma-Aldrich). Cells were stimulated either with SHIVSF162P3 GAG peptide pool (2 μg/ml), with SHIVSF162P3 ENV Peptide Set (2 μg/ml; AIDS Reagents Program, Division of AIDS, NIAID, NIH) or seven peptides from the SHIVSF162P3 Peptide Set corresponding to the V1/V2 loop (2 μg/ml) (41). Pooled cells stimulated with phorbol 12-myristate 13-acetate/ionomycin (1 μg/ml) were used as a positive activation control. One hour later, brefeldin A and monensin (10 μg/ml; GolgiStop, BD Biosciences) were added to each well. After 5 hours, cells were transferred to a fluorescence-activated cell sorting plate and stained with the LIVE/DEAD Aqua viability dye (Thermo Fisher Scientific) and the antibody panel listed in table S5.
Immunohistochemistry analysis of lymphoid aggregates
Sections (5 μm) were cut from formalin-fixed, paraffin-embedded mesenteric lymph nodes and ileum tissue blocks (two blocks per animal, one section per block, and two to three tissues per block) and placed in a 58° to 60°C oven for 1 hour for tissues to adhere. All dewax and retrieval methods were completed using the Leica BOND-MAX Autostainer. Dewax was completed using the Leica BOND Dewax Solution (AR9222), and retrieval was completed using Epitope Retrieval 1 (AR9961, low pH) for 20 min. Using the Leica BOND Polymer Refine Detection Kit (DS9800) with a dispensing of 150 μl of each reagent, first, Peroxide Block was applied for 5 min. Then, CD20 dilution Ready-to-Use (Leica Biosystems, catalog no. PA0200) was applied for 15 min. Then, polymer was dispensed and applied for 8 min. Last, DAB Refine mixes was applied for 10 min, followed by hematoxylin application for 5 min. All slides were rehydrated through alcohol and xylene and mounted, and coverslips were added. Appropriate known control tissue tonsil was used for a positive control, and primary antibodies were omitted in negative controls.
Digital-scanned images were analyzed using open source digital analysis software QuPath version 0.2.3 (55). To improve the staining quality, images were first manually preprocessed with the built-in visual stain editor. The Simple tissue detection tool was then used to create annotation of the tissue regions to be analyzed. Tissue regions with and without B cell aggregates were included for training of an artificial neural network classifier using Pixel Classifier tool. The auto-update tool within QuPath allowed real-time reconfirmation of training efficiency/accuracy. The classifier was applied to all annotated tissue regions, and the detections were also manually confirmed (fig. S7). Detection results were extracted from QuPath and expressed as percentage of tissue area covered by B cell aggregates (>5000 μm2).
Anti-Env antibodies
Ninety-six–well plates were coated with the HIV-1 SF162 gp140 Trimer Recombinant protein (AIDS Reagents Program, Division of AIDS, NIAID, NIH) at 1 μg/ml in coating carbonate buffer for 2 hours at room temperature. After washing the plates four times with washing buffer, the plates were incubated in blocking buffer (2% BSA in PBS) overnight. Plates were washed, and dilutions of test sera or control sera were added to the plates starting at 1:50. Sera were incubated at 37°C for 30 min and plate washed and detected with anti-Rhesus IgG HRP 30 min at 37°C. After washing and substrate incubation at room temperature for 30 min, plates were read at 450/650 nm.
Statistics
To analyze the differences in time to viral rebound, the treatment groups were compared with the control group using the Gehan-Breslow-Wilcoxon test with P value Bonferroni corrected for multiple comparisons. VLs and CD4+ T cell counts were compared using mixed-effect analysis with Geisser-Greenhouse correction and Tukey’s multiple comparisons test. Data from a specific time point were compared between groups by Kruskal-Wallis test and the Dunn’s multiple comparisons post hoc test (Mann-Whitney nonparametric unpaired test was used to compare directly the two treatment groups). Data with repeated time points (baseline and week 17) were compared between groups with two-way ANOVA with Bonferroni multiple comparison correction. All analyses were performed using the GraphPad Prism software V8. Significant P values of α < 0.05, α < 0.01, and α < 0.001 are indicated.
Supplementary Material
Acknowledgments:
We thank D. Burton and D. Sok for providing the PGT128 antibody. We thank D. Magnani, Director of the Nonhuman Primate Reagent Resource of MassBiologics, for providing Rh-α4β7 antibody. We thank J. Lifson, R. Shoemaker, and the AIDS and Cancer Virus Program for measuring pVL. We also thank B. Frederick, B. Shmaltsuyeva, and H. Fan of the Northwestern University Pathology Core Facility Immunohistochemistry laboratory for assistance with immunohistochemical staining. We thank A. Weddle for preparation of the high-resolution figures. We acknowledge the outstanding help and assistance of the professional staff, animal technicians, and caretakers of the Tulane National Primate Research Center and the staff of the Population Council and Rockefeller University resource centers for assistance. Reagents used in these studies were provided by the NIH Nonhuman Primate Reagent Resource (P40 OD028116, U24 AI126683).
Funding: This work was supported by NIAID grant no. R01AI098546-06 to E.M. and was partly supported by NIH grant nos. R37AI094595 to T.J.H. and P51OD011104, U42OD010568, and U42OD024282 to J.L.B. and NCI contract nos. HHSN261200800001E and 75N91019D00024 to B.F.K.
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data associated with this study are present in the paper or the Supplementary Materials. VRC07-523LS and PGT128 antibodies were provided by the VRC and TSRI under a material transfer agreement. Rh-α4β7 antibody can be purchased from the Nonhuman Primate Reagent Resource of MassBiologics at www.nhpreagents.org/
REFERENCES AND NOTES
- 1.Barouch DH, Whitney JB, Moldt B, Klein F, Oliveira TY, Liu J, Stephenson KE, Chang HW, Shekhar K, Gupta S, Nkolola JP, Seaman MS, Smith KM, Borducchi EN, Cabral C, Smith JY, Blackmore S, Sanisetty S, Perry JR, Beck M, Lewis MG, Rinaldi W, Chakraborty AK, Poignard P, Nussenzweig MC, Burton DR, Therapeutic efficacy of potent neutralizing HIV-1-specific monoclonal antibodies in SHIV-infected rhesus monkeys. Nature 503, 224–228 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shingai M, Nishimura Y, Klein F, Mouquet H, Donau OK, Plishka R, Buckler-White A, Seaman M, Piatak M Jr., Lifson JD, Dimitrov DS, Nussenzweig MC, Martin MA, Antibody-mediated immunotherapy of macaques chronically infected with SHIV suppresses viraemia. Nature 503, 277–280 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Julg B, Pegu A, Abbink P, Liu J, Brinkman A, Molloy K, Mojta S, Chandrashekar A, Callow K, Wang K, Chen X, Schmidt SD, Huang J, Koup RA, Seaman MS, Keele BF, Mascola JR, Connors M, Barouch DH, Virological control by the CD4-binding site antibody N6 in simian-HIV-infected rhesus monkeys. J. Virol. 91, e00498–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caskey M, Klein F, Lorenzi JC, Seaman MS, West AP Jr., Buckley N, Kremer G, Nogueira L, Braunschweig M, Scheid JF, Horwitz JA, Shimeliovich I, Ben-Avraham S, Witmer-Pack M, Platten M, Lehmann C, Burke LA, Hawthorne T, Gorelick RJ, Walker BD, Keler T, Gulick RM, Fatkenheuer G, Schlesinger SJ, Nussenzweig MC, Viraemia suppressed in HIV-1-infected humans by broadly neutralizing antibody 3BNC117. Nature 522, 487–491 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caskey M, Schoofs T, Gruell H, Settler A, Karagounis T, Kreider EF, Murrell B, Pfeifer N, Nogueira L, Oliveira TY, Learn GH, Cohen YZ, Lehmann C, Gillor D, Shimeliovich I, Unson-O’Brien C, Weiland D, Robles A, Kummerle T, Wyen C, Levin R, Witmer-Pack M, Eren K, Ignacio C, Kiss S, West AP Jr., Mouquet H, Zingman BS, Gulick RM, Keler T, Bjorkman PJ, Seaman MS, Hahn BH, Fatkenheuer G, Schlesinger SJ, Nussenzweig MC, Klein F, Antibody 10–1074 suppresses viremia in HIV-1-infected individuals. Nat. Med. 23, 185–191 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lynch RM, Boritz E, Coates EE, DeZure A, Madden P, Costner P, Enama ME, Plummer S, Holman L, Hendel CS, Gordon I, Casazza J, Conan-Cibotti M, Migueles SA, Tressler R, Bailer RT, McDermott A, Narpala S, O’Dell S, Wolf G, Lifson JD, Freemire BA, Gorelick RJ, Pandey JP, Mohan S, Chomont N, Fromentin R, Chun TW, Fauci AS, Schwartz RM, Koup RA, Douek DC, Hu Z, Capparelli E, Graham BS, Mascola JR, Ledgerwood JE; VRC 601 Study Team, Virologic effects of broadly neutralizing antibody VRC01 administration during chronic HIV-1 infection. Sci. Transl. Med. 7, 319ra206 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bar KJ, Sneller MC, Harrison LJ, Justement JS, Overton ET, Petrone ME, Salantes DB, Seamon CA, Scheinfeld B, Kwan RW, Learn GH, Proschan MA, Kreider EF, Blazkova J, Bardsley M, Refsland EW, Messer M, Clarridge KE, Tustin NB, Madden PJ, Oden K, O’Dell SJ, Jarocki B, Shiakolas AR, Tressler RL, Doria-Rose NA, Bailer RT, Ledgerwood JE, Capparelli EV, Lynch RM, Graham BS, Moir S, Koup RA, Mascola JR, Hoxie JA, Fauci AS, Tebas P, Chun TW, Effect of HIV antibody VRC01 on viral rebound after treatment interruption. N. Engl. J. Med. 375, 2037–2050 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scheid JF, Horwitz JA, Bar-On Y, Kreider EF, Lu CL, Lorenzi JC, Feldmann A, Braunschweig M, Nogueira L, Oliveira T, Shimeliovich I, Patel R, Burke L, Cohen YZ, Hadrigan S, Settler A, Witmer-Pack M, West AP Jr., Juelg B, Keler T, Hawthorne T, Zingman B, Gulick RM, Pfeifer N, Learn GH, Seaman MS, Bjorkman PJ, Klein F, Schlesinger SJ, Walker BD, Hahn BH, Nussenzweig MC, Caskey M, HIV-1 antibody 3BNC117 suppresses viral rebound in humans during treatment interruption. Nature 535, 556–560 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mendoza P, Gruell H, Nogueira L, Pai JA, Butler AL, Millard K, Lehmann C, Suarez I, Oliveira TY, Lorenzi JCC, Cohen YZ, Wyen C, Kummerle T, Karagounis T, Lu CL, Handl L, Unson-O’Brien C, Patel R, Ruping C, Schlotz M, Witmer-Pack M, Shimeliovich I, Kremer G, Thomas E, Seaton KE, Horowitz J, West AP Jr., Bjorkman PJ, Tomaras GD, Gulick RM, Pfeifer N, Fatkenheuer G, Seaman MS, Klein F, Caskey M, Nussenzweig MC, Combination therapy with anti-HIV-1 antibodies maintains viral suppression. Nature 561, 479–484 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bar-On Y, Gruell H, Schoofs T, Pai JA, Nogueira L, Butler AL, Millard K, Lehmann C, Suarez, Oliveira TY, Karagounis T, Cohen YZ, Wyen C, Scholten S, Handl L, Belblidia S, Dizon JP, Vehreschild JJ, Witmer-Pack M, Shimeliovich I, Jain K, Fiddike K, Seaton KE, Yates NL, Horowitz J, Gulick RM, Pfeifer N, Tomaras GD, Seaman MS, Fatkenheuer G, Caskey M, Klein F, Nussenzweig MC, Safety and antiviral activity of combination HIV-1 broadly neutralizing antibodies in viremic individuals. Nat. Med. 24, 1701–1707 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishimura Y, Gautam R, Chun TW, Sadjadpour R, Foulds KE, Shingai M, Klein F, Gazumyan A, Golijanin J, Donaldson M, Donau OK, Plishka RJ, Buckler-White A, Seaman MS, Lifson JD, Koup RA, Fauci AS, Nussenzweig MC, Martin MA, Early antibody therapy can induce long-lasting immunity to SHIV. Nature 543, 559–563 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haigwood NL, Montefiori DC, Sutton WF, McClure J, Watson AJ, Voss G, Hirsch VM, Richardson BA, Letvin NL, Hu SL, Johnson PR, Passive immunotherapy in simian immunodeficiency virus-infected macaques accelerates the development of neutralizing antibodies. J. Virol. 78, 5983–5995 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamamoto H, Kawada M, Takeda A, Igarashi H, Matano T, Post-infection immunodeficiency virus control by neutralizing antibodies. PLOS ONE 2, e540 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schoofs T, Klein F, Braunschweig M, Kreider EF, Feldmann A, Nogueira L, Oliveira T, Lorenzi JC, Parrish EH, Learn GH, West AP Jr., Bjorkman PJ, Schlesinger SJ, Seaman MS, Czartoski J, McElrath MJ, Pfeifer N, Hahn BH, Caskey M, Nussenzweig MC, HIV-1 therapy with monoclonal antibody 3BNC117 elicits host immune responses against HIV-1. Science 352, 997–1001 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niessl J, Baxter AE, Mendoza P, Jankovic M, Cohen YZ, Butler AL, Lu CL, Dube M, Shimeliovich, Gruell H, Klein F, Caskey M, Nussenzweig MC, Kaufmann DE, Combination anti-HIV-1 antibody therapy is associated with increased virus-specific T cell immunity. Nat. Med. 26, 222–227 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gros L, Dreja H, Fiser AL, Plays M, Pelegrin M, Piechaczyk M, Induction of long-term protective antiviral endogenous immune response by short neutralizing monoclonal antibody treatment. J. Virol. 79, 6272–6280 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gros L, Pelegrin M, Michaud HA, Bianco S, Hernandez J, Jacquet C, Piechaczyk M, Endogenous cytotoxic T-cell response contributes to the long-term antiretroviral protection induced by a short period of antibody-based immunotherapy of neonatally infected mice. J. Virol. 82, 1339–1349 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michaud HA, Gomard T, Gros L, Thiolon K, Nasser R, Jacquet C, Hernandez J, Piechaczyk M, Pelegrin M, A crucial role for infected-cell/antibody immune complexes in the enhancement of endogenous antiviral immunity by short passive immunotherapy. PLOS Pathog. 6, e1000948 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kauvar LM, Harcourt JL, Haynes LM, Tripp RA, Therapeutic targeting of respiratory syncytial virus G-protein. Immunotherapy 2, 655–661 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geisbert TW, Mire CE, Geisbert JB, Chan YP, Agans KN, Feldmann F, Fenton KA, Zhu Z, Dimitrov DS, Scott DP, Bossart KN, Feldmann H, Broder CC, Therapeutic treatment of Nipah virus infection in nonhuman primates with a neutralizing human monoclonal antibody. Sci. Transl. Med. 6, 242ra282 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Broder CC, Xu K, Nikolov DB, Zhu Z, Dimitrov DS, Middleton D, Pallister J, Geisbert TW, Bossart KN, Wang LF, A treatment for and vaccine against the deadly Hendra and Nipah viruse-s. Antivir. Res. 100, 8–13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nawaz F, Goes LR, Ray JC, Olowojesiku R, Sajani A, Ansari AA, Perrone I, Hiatt J, Van Ryk D, Wei D, Waliszewski M, Soares MA, Jelicic K, Connors M, Migueles SA, Martinelli E, Villinger F, Cicala C, Fauci AS, Arthos J, MAdCAM costimulation through integrin-α4β7 promotes HIV replication. Mucosal Immunol. 11, 1342–1351 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goes LR, Sajani A, Sivro A, Olowojesiku R, Ray JC, Perrone I, Yolitz J, Girard A, Leyre L, Wibmer CK, Morris L, Gorini G, Franchini G, Mason RD, Roederer M, Mehandru S, Soares MA, Cicala C, Fauci AS, Arthos J, The V2 loop of HIV gp120 delivers costimulatory signals to CD4+T cells through integrin α4β7 and promotes cellular activation and infection. Proc. Natl. Acad. Sci. U.S.A. 117, 32566–32573 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arrode-Bruses G, Goode D, Kleinbeck K, Wilk J, Frank I, Byrareddy S, Arthos J, Grasperge B, Blanchard J, Zydowsky T, Gettie A, Martinelli E, A small molecule, which competes with MAdCAM-1, activates integrin α4β7 and fails to prevent mucosal transmission of SHIV-SF162P3. PLOS Pathog. 12, e1005720 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calenda G, Keawvichit R, Arrode-Bruses G, Pattanapanyasat K, Frank I, Byrareddy SN, Arthos J, Cicala C, Grasperge B, Blanchard JL, Gettie A, Reimann KA, Ansari AA, Martinelli E, Integrin α4β7 blockade preferentially impacts CCR6+ lymphocyte subsets in blood and mucosal tissues of naive rhesus macaques. J. Immunol. 200, 810–820 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Byrareddy SN, Arthos J, Cicala C, Villinger F, Ortiz KT, Little D, Sidell N, Kane MA, Yu J, Jones JW, Santangelo PJ, Zurla C, McKinnon LR, Arnold KB, Woody CE, Walter L, Roos C, Noll A, Van Ryk D, Jelicic K, Cimbro R, Gumber S, Reid MD, Adsay V, Amancha PK, Mayne AE, Parslow TG, Fauci AS, Ansari AA, Sustained virologic control in SIV+ macaques after antiretroviral and α4β7 antibody therapy. Science 354, 197–202 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uzzan M, Tokuyama M, Rosenstein AK, Tomescu C, SahBandar IN, Ko HM, Leyre L, Chokola A, Kaplan-Lewis E, Rodriguez G, Seki A, Corley MJ, Aberg J, La Porte A, Park EY, Ueno H, Oikonomou I, Doron I, Iliev ID, Chen BK, Lui J, Schacker TW, Furtado GC, Lira SA, Colombel JF, Horowitz A, Lim JK, Chomont N, Rahman AH, Montaner LJ, Ndhlovu LC, Mehandru S, Anti-α4β7 therapy targets lymphoid aggregates in the gastrointestinal tract of HIV-1-infected individuals. Sci. Transl. Med. 10, eaau4711 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santangelo PJ, Cicala C, Byrareddy SN, Ortiz KT, Little D, Lindsay KE, Gumber S, Hong JJ, Jelicic K, Rogers KA, Zurla C, Villinger F, Ansari AA, Fauci AS, Arthos J, Early treatment of SIV+ macaques with an α4β7 mAb alters virus distribution and preserves CD4(+) T cells in later stages of infection. Mucosal Immunol. 11, 932–946 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santangelo PJ, Rogers KA, Zurla C, Blanchard EL, Gumber S, Strait K, Connor-Stroud F, Schuster DM, Amancha PK, Hong JJ, Byrareddy SN, Hoxie JA, Vidakovic B, Ansari AA, Hunter E, Villinger F, Whole-body immunoPET reveals active SIV dynamics in viremic and antiretroviral therapy-treated macaques. Nat. Methods 12, 427–432 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Byrareddy SN, Kallam B, Arthos J, Cicala C, Nawaz F, Hiatt J, Kersh EN, McNicholl JM, Hanson D, Reimann KA, Brameier M, Walter L, Rogers K, Mayne AE, Dunbar P, Villinger T, Little D, Parslow TG, Santangelo PJ, Villinger F, Fauci AS, Ansari AA, Targeting α4β7 integrin reduces mucosal transmission of simian immunodeficiency virus and protects gut-associated lymphoid tissue from infection. Nat. Med. 20, 1397–1400 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Derby N, Lal M, Aravantinou M, Kizima L, Barnable P, Rodriguez A, Lai M, Wesenberg A, Ugaonkar S, Levendosky K, Mizenina O, Kleinbeck K, Lifson JD, Peet MM, Lloyd Z, Benson M, Heneine W, O’Keefe BR, Robbiani M, Martinelli E, Grasperge B, Blanchard J, Gettie A, Teleshova N, Fernandez-Romero JA, Zydowsky TM, Griffithsin carrageenan fast dissolving inserts prevent SHIV HSV-2 and HPV infections in vivo. Nat. Commun. 9, 3881 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McBride JA, Striker R, Imbalance in the game of T cells: What can the CD4/CD8 T-cell ratio tell us about HIV and health? PLOS Pathog. 13, e1006624 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rudicell RS, Kwon YD, Ko SY, Pegu A, Louder MK, Georgiev IS, Wu X, Zhu J, Boyington C, Chen X, Shi W, Yang ZY, Doria-Rose NA, McKee K, O’Dell S, Schmidt SD, Chuang GY, Druz A, Soto C, Yang Y, Zhang B, Zhou T, Todd JP, Lloyd E, Eudailey J, Roberts KE, Donald BR, Bailer RT, Ledgerwood J, Program NCS, Mullikin JC, Shapiro L, Koup RA, Graham BS, Nason MC, Connors M, Haynes BF, Rao SS, Roederer M, Kwong PD, Mascola JR, Nabel GJ, Enhanced potency of a broadly neutralizing HIV-1 antibody in vitro improves protection against lentiviral infection in vivo. J. Virol. 88, 12669–12682 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong CH, Phogat S, Wrin T, Simek MD, Protocol GPI, Koff WC, Wilson IA, Burton DR, Poignard P, Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477, 466–470 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pejchal R, Doores KJ, Walker LM, Khayat R, Huang PS, Wang SK, Stanfield RL, Julien P, Ramos A, Crispin M, Depetris R, Katpally U, Marozsan A, Cupo A, Maloveste S, Liu Y, McBride R, Ito Y, Sanders RW, Ogohara C, Paulson JC, Feizi T, Scanlan CN, Wong CH, Moore JP, Olson WC, Ward AB, Poignard P, Schief WR, Burton DR, Wilson IA, A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science 334, 1097–1103 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caskey M, Klein F, Nussenzweig MC, Broadly neutralizing anti-HIV-1 monoclonal antibodies in the clinic. Nat. Med. 25, 547–553 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ansari AA, Reimann KA, Mayne AE, Takahashi Y, Stephenson ST, Wang R, Wang X, Li J, Price AA, Little DM, Zaidi M, Lyles R, Villinger F, Blocking of α4β7 gut-homing integrin during acute infection leads to decreased plasma and gastrointestinal tissue viral loads in simian immunodeficiency virus-infected rhesus macaques. J. Immunol. 186, 1044–1059 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pereira LE, Onlamoon N, Wang X, Wang R, Li J, Reimann KA, Villinger F, Pattanapanyasat K, Mori K, Ansari AA, Preliminary in vivo efficacy studies of a recombinant rhesus anti-α(4)β(7) monoclonal antibody. Cell. Immunol. 259, 165–176 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rawi R, Mall R, Shen CH, Farney SK, Shiakolas A, Zhou J, Bensmail H, Chun TW, Doria-Rose NA, Lynch RM, Mascola JR, Kwong PD, Chuang GY, Accurate prediction for antibody resistance of clinical HIV-1 isolates. Sci. Rep. 9, 14696 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martinelli E, Tharinger H, Frank I, Arthos J, Piatak M Jr., Lifson JD, Blanchard J, Gettie A, Robbiani M, HSV-2 infection of dendritic cells amplifies a highly susceptible HIV-1 cell target. PLOS Pathog. 7, e1002109 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Calenda G, Frank I, Arrode-Bruses G, Pegu A, Wang K, Arthos J, Cicala C, Rogers KA, Shirreff L, Grasperge B, Blanchard JL, Maldonado S, Roberts K, Gettie A, Villinger F, Fauci AS, Mascola JR, Martinelli E, Delayed vaginal SHIV infection in VRC01 and anti-α4β7 treated rhesus macaques. PLOS Pathog. 15, e1007776 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nishimura Y, Donau OK, Dias J, Ferrando-Martinez S, Jesteadt E, Sadjadpour R, Gautam R, Buckler-White A, Geleziunas R, Koup RA, Nussenzweig MC, Martin MA, Immunotherapy during the acute SHIV infection of macaques confers long-term suppression of viremia. J. Exp. Med. 218, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Borducchi EN, Liu J, Nkolola JP, Cadena AM, Yu WH, Fischinger S, Broge T, Abbink P, Mercado NB, Chandrashekar A, Jetton D, Peter L, McMahan K, Moseley ET, Bekerman E, Hesselgesser J, Li W, Lewis MG, Alter G, Geleziunas R, Barouch DH, Antibody and TLR7 agonist delay viral rebound in SHIV-infected monkeys. Nature 563, 360–364 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sheridan J, Doherty GA, Editorial: Gut selective immunosuppression-is it a double edged sword? Aliment. Pharmacol. Ther. 46, 373 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Sivro A, Schuetz A, Sheward D, Joag V, Yegorov S, Liebenberg LJ, Yende-Zuma N, Stalker RS Mwatelah, Selhorst P, Garrett N, Samsunder N, Balgobin A, Nawaz F, Cicala C, Arthos J, Fauci AS, Anzala AO, Kimani J, Bagaya BS, Kiwanuka N, Williamson C, Kaul R, Passmore JS, Phanuphak N, Ananworanich J, Ansari A, Karim QA, Karim SSA, McKinnon LR; CAPRISA004 and RV254 study groups, Integrin α4β7 expression on peripheral blood CD4(+) T cells predicts HIV acquisition and disease progression outcomes. Sci. Transl. Med. 10, eaam6354 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Di Mascio M, Lifson JD, Srinivasula S, Kim I, DeGrange P, Keele BF, Belli AJ, Reimann KA, Wang Y, Proschan M, Lane HC, Fauci AS, Evaluation of an antibody to α4β7 in the control of SIVmac239-nef-stop infection. Science 365, 1025–1029 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abbink P, Mercado NB, Nkolola JP, Peterson RL, Tuyishime H, McMahan K, Moseley ET, Borducchi EN, Chandrashekar A, Bondzie EA, Agarwal A, Belli AJ, Reimann KA, Keele BF, Geleziunas R, Lewis MG, Barouch DH, Lack of therapeutic efficacy of an antibody to α4β7 in SIVmac251-infected rhesus macaques. Science 365, 1029–1033 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iwamoto N, Mason RD, Song K, Gorman J, Welles HC, Arthos J, Cicala C, Min S, King HAD, Belli AJ, Reimann KA, Foulds KE, Kwong PD, Lifson JD, Keele BF, Roederer M, Blocking α4β7 integrin binding to SIV does not improve virologic control. Science 365, 1033–1036 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arnason G, The ethical justification for the use of non-human primates in research: The Weatherall report revisited. J. Med. Ethics 44, 328–331 (2018). [DOI] [PubMed] [Google Scholar]
- 50.Aravantinou M, Frank I, Hallor M, Singer R, Tharinger H, Kenney J, Gettie A, Grasperge B, Blanchard J, Salazar A, Piatak M Jr., Lifson JD, Robbiani M, Derby N, PolyICLC exerts pro-and anti-HIV effects on the DC-T cell milieu in vitro and in vivo. PLOS ONE 11, e0161730 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou T, Zhu J, Yang Y, Gorman J, Ofek G, Srivatsan S, Druz A, Lees CR, Lu G, Soto C, Stuckey J, Burton DR, Koff WC, Connors M, Kwong PD, Transplanting supersites of HIV-1 vulnerability. PLOS ONE 9, e99881 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guerra-Perez N, Aravantinou M, Veglia F, Goode D, Truong R, Derby N, Blanchard J, Grasperge B, Gettie A, Robbiani M, Martinelli E, Rectal HSV-2 infection may increase rectal SIV acquisition even in the context of SIVΔnef vaccination. PLOS ONE 11, e0149491 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Francica JR, Sheng Z, Zhang Z, Nishimura Y, Shingai M, Ramesh A, Keele BF, Schmidt SD, Flynn BJ, Darko S, Lynch RM, Yamamoto T, Matus-Nicodemos R, Wolinsky D, Program NCS, Nason M, Valiante NM, Malyala P, De Gregorio E, Barnett SW, Singh M, O’Hagan DT, Koup RA, Mascola JR, Martin MA, Kepler TB, Douek DC, Shapiro L, Seder RA, Analysis of immunoglobulin transcripts and hypermutation following SHIV(AD8) infection and protein-plus-adjuvant immunization. Nat. Commun. 6, 6565 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wickham H, Averick M, Bryan J, Chang W, McGowan LD, François R, Grolemund G, Hayes, Henry L, Hester J, Kuhn M, Pedersen TL, Miller E, Bache SM, Müller K, Ooms J, Robinson D, Seidel DP, Spinu V, Takahashi K, Vaughan D, Wilke C, Woo K, Yutani H, Welcome to the tidyverse. J. Open. Source. Softw. 4, 1686 (2019). [Google Scholar]
- 55.Bankhead P, Loughrey MB, Fernandez JA, Dombrowski Y, McArt DG, Dunne PD, McQuaid S, Gray RT, Murray LJ, Coleman HG, James JA, Salto-Tellez M, Hamilton PW, QuPath: Open source software for digital pathology image analysis. Sci. Rep. 7, 16878 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.