Abstract
Background
Altered adipose tissue (AT) metabolism in cancer‐associated weight loss via inflammation, lipolysis, and white adipose tissue (WAT) browning is primarily implicated from rodent models; their contribution to AT wasting in cancer patients is unclear.
Methods
Energy expenditure (EE), plasma, and abdominal subcutaneous WAT were obtained from men (aged 65 ± 8 years) with cancer, with (CWL, n = 27) or without (CWS, n = 47) weight loss, and weight‐stable non‐cancer patients (CON, n = 26). Clinical images were assessed for adipose and muscle area while plasma and WAT were assessed for inflammatory, lipolytic, and browning markers.
Results
CWL displayed smaller subcutaneous AT (SAT; P = 0.05) and visceral AT (VAT; P = 0.034) than CWS, and displayed higher circulating interleukin (IL)‐6 (P = 0.01) and WAT transcript levels of IL‐6 (P = 0.029), IL‐1β (P = 0.042), adipose triglyceride lipase (P = 0.026), and browning markers (Dio2, P = 0.03; PGC‐1a, P = 0.016) than CWS and CON. There was no difference across groups in absolute REE (P = 0.061), %predicted REE (P = 0.18), circulating free fatty acids (FFA, P = 0.13) or parathyroid hormone‐related peptide (PTHrP; P = 0.88), or WAT protein expression of inflammation (IL‐6, P = 0.51; IL‐1β, P = 0.29; monocyte chemoattractant protein‐1, P = 0.23) or WAT protein or gene expression of browning (uncoupling protein‐1, UCP‐1; P = 0.13, UCP‐1, P = 0.14). In patients with cancer, FFA was moderately correlated with WAT hormone‐sensitive lipase transcript (r = 0.38, P = 0.018, n = 39); circulating cytokines were not correlated with expression of WAT inflammatory markers and circulating PTHrP was not correlated with expression of WAT browning markers. In multivariate regression using cancer patients only, body mass index (BMI) directly predicted SAT (N = 25, R 2 = 0.72, P < 0.001), VAT (N = 28, R 2 = 0.64, P < 0.001), and absolute REE (N = 22, R 2 = 0.43, P = 0.001), while BMI and WAT UCP‐1 protein were indirectly associated with %predicted REE (N = 22, R 2 = 0.45, P = 0.02), and FFA was indirectly associated with RQ (N = 22, R 2 = 0.52, P < 0.001).
Conclusions
Cancer‐related weight loss was associated with elevated circulating IL‐6 and elevations in some WAT inflammatory, lipolytic and browning marker transcripts. BMI, not weight loss, was associated with increased energy expenditure. The contribution of inflammation and lipolysis, and lack thereof for WAT browning, will need to be clarified in other tumour types to increase generalizability. Future studies should consider variability in fat mass when exploring the relationship between cancer and adipose metabolism and should observe the trajectory of lipolysis and energy expenditure over time to establish the clinical significance of these associations and to inform more mechanistic interpretation of causation.
Keywords: White adipose tissue, Inflammation, Lipolysis, Adipose browning, Energy expenditure
Introduction
Altered adipose tissue (AT) metabolism leading to low fat mass, particularly abdominal AT, is a common feature of cancer cachexia and is associated with poor prognosis. 1 , 2 , 3 Body mass index (BMI), which is directly associated with fat mass, is a predictor of survival independently from weight loss in cancer. 4 It is therefore difficult to differentiate alterations in whole‐body and AT metabolism in cancer patients due to disease versus variability in AT. Rodent models of cancer cachexia implicate inflammation, lipolysis, and/or browning of white AT (WAT); however, WAT inflammatory genes were unchanged in multiple cancer patient studies. 5 , 6 Increased lipolysis via adipose triglyceride lipase (ATGL) and/or hormone sensitive lipase (HSL) is important for WAT atrophy in rodents, 1 , 5 , 7 but the relative contribution of these to lipolysis in cancer patients is unclear. 8 , 9
Inflammation may also mediate increased lipolysis, resting energy expenditure (REE), and WAT browning in patients with cancer‐associated weight loss. Patients with gastrointestinal (GI) cancer and weight loss displayed elevated circulating interleukin (IL)‐6, tumour necrosis factor (TNF), and free fatty acids (FFAs) and WAT levels of uncoupling protein (UCP)‐1, ATGL, and HSL than weight‐stable patients, and IL‐6 and TNF were directly correlated with FFA in weight‐losing patients. 10 In another GI cohort, circulating IL‐6 and TNF associated directly with REE and weight loss. 11
Animal and in vitro studies demonstrate WAT browning occurs by upregulation of UCP‐1. 12 , 13 Results are conflicting in human cancer studies 14 , 15 and indirect mechanisms like macrophage infiltration may enhance cytokine‐induced WAT browning. This is supported by our recent pre‐clinical findings of cytokine and macrophage co‐localization in WAT of lung cancer‐induced wasting. 16 Circulating parathyroid hormone‐related peptide (PTHrP) was also recently characterized as a tumour factor that may induce WAT browning, AT wasting, and increased REE in rodents. 17 , 18 In cancer patients, circulating PTHrP has been associated with higher relative REE 17 and reduced body fat. 19
The association between inflammation, lipolysis, and WAT browning and their contribution to AT wasting in cancer is poorly understood. The objective of this pilot study was to characterize the relationship between AT atrophy, REE, and systemic and WAT inflammation, browning, and lipolysis in cancer patients with and without weight loss and in non‐cancer controls. We hypothesized that weight‐losing cancer patients would display elevated systemic and WAT markers of inflammation, lipolysis, and browning than weight‐stable cancer patients and non‐cancer controls.
Methods
Participants
This cross‐sectional, observational pilot study was conducted at the Veterans Affairs Puget Sound Health Care System (VAPSHCS) in Seattle, WA, USA. This protocol was approved by the VAPSHCS Institutional Review Board and the Research and Development Committee and was conducted in compliance with the Declarations of Helsinki and its amendments and the International Conference on Harmonization Guideline for Good Clinical Practices.
Male patients with confirmed GI or genitourinary (GU) cancer with or without weight loss (involuntary weight loss >5% or BMI < 20 kg/m2 and weight loss >2% within 6 months) were recruited from surgical oncology or urology clinics. Weight‐stable male patients without cancer history (except non‐melanoma skin cancer) in the last 5 years were recruited as controls from general surgery or urology clinics. All participants were planning elective laparotomy. Participants were excluded for: congestive heart failure; liver disease; renal failure; active/uncontrolled infection; uncontrolled diabetes mellitus (HbA1c ≥ 9%); or actively using an anabolic or investigational agent.
Study visit
Within 2 weeks before laparotomy, participants reported to VAPSHCS after fasting overnight. A blood sample was obtained followed by REE assessment. Absolute REE (kcals/day) and respiratory quotient (RQ, VCO2/VO2 ratio), were assessed by indirect calorimetry in supine position (Vmax Encore, Vyaire Medical, Inc., Mettawa, IL, USA). Per cent predicted REE (%pred) is expressed as the ratio of absolute/predicted; predicted REE was estimated using the Harris–Benedict equation.
Computed tomography analysis
Subcutaneous (SAT), visceral (VAT), and intramuscular (IMAT) AT areas and skeletal muscle (SMA) area (cm2) were quantified from clinically available spiral computed tomography (CT) scans involving the third lumbar level. Control patients had a clinically available CT scan for evaluation of incisional hernia, benign prostatic hyperplasia, unresectable adenomatous colon polyp, gallstones, colonic diverticula, or desmoid‐type fibromatosis. Cross‐sectional area (CSA) was quantified using sliceOmatic software (v5.0, TomoVision, Montreal, Quebec, Canada) with attenuation parameters −190 to −30 (SAT, IMAT) or −150 to −50 (VAT) Hounsfield Units. 20 An intra‐observer coefficient of variation of <1.3% was required for quantification of AT. Slice thickness ranged from 0.6–3.0 mm, tube voltage ranged from 110–120 kilovolts, all images with contrast were obtained from the venous portal phase. Total AT (TAT) area was the summation of SAT, VAT, and IMAT.
Circulating plasma markers
Inflammatory cytokines (IL‐1β, IL‐6, and TNF) were detected by V‐PLEX Human Pro‐inflammatory Panel 1 (Cat# N05049A‐1, Meso Scale Discovery, Rockville, MD, USA); CV < 7.0%. PTHRP was identified by Human PTHLH/PTHRP ELISA kit (Sandwich ELISA, LifeSpan BioSciences, Inc. Cat# LS‐F54974); CV < 10%. FFA were analysed using a commercially available kit (FUJIFILM Wako Diagnostics USA Corporation) adapted to microplates by the Vanderbilt University Lipid Core (DK020593); CV < 1.5%.
Adipose specimen acquisition
A SAT specimen (1.0 g) was excised from the abdominal wall during laparotomy. Each specimen was grossly inspected for vascular or connective tissue, which was quickly cut away. Immediately thereafter, half was flash‐frozen in liquid nitrogen then stored at −80° until analysis. The other half was submerged in RNAlater and stored at 4° no more than 24 h, then any remaining RNAlater was removed and the specimen was stored at −80° until analysis.
Adipose tissue analysis
Total RNA was isolated from 100–200 mg SAT previously submerged in RNAlater by using QiAzol (79306, Qiagen, Valencia, CA, USA) and Qiagen RNeasy mini spin columns (74104, Qiagen). RNA concentration was identified by BioTek Cytation 5, and 250 ng RNA was reverse transcribed to cDNA by using the High Capacity cDNA Reverse Transcription kit with RNase Inhibitor (4374966, Thermo Fisher Scientific, Waltham, MA, USA). Real‐time RT‐PCR was performed on an ABI 7500 instrument (Applied Biosystems) using predesigned Taqman Expression Assays (Thermo Fisher Scientific). Quantification of each gene of interest was normalized to the mean of reference genes CLN3 (Hs00164002_m1) and LRP10 (Hs01047362_m1) and expressed as relative fold‐change of the non‐cancer group by the 2‐ΔCT method. Taqman gene expression assays from Thermo Fisher Scientific (4331182) were Hs00174131_m1 IL‐6 (CV < 2.8%), Hs00174128_m1 TNF (CV < 2.7%), Hs01555410_m1 IL‐1β (CV < 1.1%), Hs00174969_m1 PTHrP (CV < 0.7%), Hs00234140_m1 (monocyte chemoattractant protein‐1; MCP‐1, CV < 1.7%), Hs02836816_g1 CD68 (CV < 5.6%), Hs01084772_m1 UCP‐1 (CV < 9.0%), Hs05050546_s1 Iodothyronine Deiodinase 2 (Dio2; CV < 2.8%), Hs00154455_m1 cell death‐inducing DNA fragmentation factor‐α‐like effector A (Cidea; CV < 2.0%), Hs00173304_m1 peroxisome proliferator‐activated receptor coactivator (PGC)‐1α (CV < 1.0%), Hs00177504_m1 ATGL (CV < 4.3%), and Hs00943410_m1 HSL (CV < 3.8%).
Inflammatory cytokines IL‐1β, IL‐6, and TNF and macrophage marker MCP‐1 proteins in SAT were detected by U‐PLEX Biomarker Group1/Human Assays (K15067L, MSD, Rockville, MD, USA); CV < 5.8%. Proteins were extracted from 100–200 mg SAT by PathScan lysis buffer (Cell Signaling) with protease and phosphatase inhibitors (MSD). Concentration was detected by bicinchoninic acid assay (Thermo Scientific, Rockford, IL, USA) using standard bovine serum albumin and analysed by Discovery Workbench v4.0 (Rockville, MD, USA).
Immunohistochemistry for identifying UCP‐1 protein in SAT was performed using flash‐frozen samples. After mounting with OCT, specimens were sliced at 14 μm by Cryostat (Leica CM3050S, Nussloch, Germany) at −40°C. Slides were incubated in methanol for 15 min at −20°C then 30 min in 3% hydrogen peroxide at room temperature (RT). Slides were then blocked by 2.5% normal horse serum for 1 h RT then incubated in UCP‐1 Polyclonal Antibody (1:200, PA1‐24894, Thermo Fisher Scientific) at 4°C overnight. The following day, signals were visualized using SignalStain® Boost IHC Detection Reagent (8114, Cell Signaling) and SignalStain® DAB Substrate kit (8059, Cell Signaling). Stained slides were dehydrated by 70%, 90%, 100% ethanol, and 100% xylene sequentially and mounted with coverslips using Permount (SP15‐100, Thermo Fisher Scientific). Stained slides were imaged by the Nikon NiE microscope at 10×. Positive areas were quantified and normalized to total area of the section (mm2) using ImageJ.
Statistical analysis
SPSS version 26 (SPSS, Inc., Chicago, IL, USA) was used for statistical analysis. One‐way ANOVA with Tukey's post‐hoc pairwise comparison compared continuous variables and Fisher's exact test compared categorical variables across groups. Statistical significance was two‐sided, α ≤ 0.05; data are mean (SD) or N (%). Spearman correlations tested associations between outcomes in cancer patients. Multivariate regression identified significant predictors of AT CSA, RQ, or REE in cancer patients. Effect size was calculated for the pairwise difference between groups using Cohen's d = [(Mean2 − Mean1)/pooled SD] for the primary outcome (WAT IL‐6 gene expression).
Results
Participants
Demographic information for cancer patients with weight loss (CWL, n = 27), weight‐stable cancer patients (CWS, n = 47), and non‐cancer patients (CON, n = 26) is reported in Table 1. Body weight and BMI were lower and recent weight loss was greater in CWL than CWS; BMI and recent weight loss were also different between CWL and CON. A small number of patients in each group were actively taking a diabetes‐related medication: CWL (n = 1), CWS (n = 3), and CON (n = 5).
Table 1.
Descriptive characteristics
| Mean (SD) or N (%) | CON | CWS | CWL | P value |
|---|---|---|---|---|
| N = 26 | N = 47 | N = 27 | ||
| Age (years) | 65.0 (8.2) | 63.6 (9.0) | 66.0 (8.0) | 0.52 |
| Ht (cm) | 176.2 (6.0) | 177.3 (7.5) | 177.1 (9.7) | 0.84 |
| Wt (kg) | 93.6 (16.7) | 97.9 (20.7)* | 82.9 (22.9) | 0.01 |
| BMI (kg/m2) | 30.2 (5.4) | 31.0 (5.5)* | 26.4 (6.0) | 0.003 |
| 6‐mo Wt change (%) | −2.4 (2.2)* | −0.6 (2.3)* | −8.7 (5.9) | <0.001 |
| Ethnicity | 0.81 | |||
| White, non‐Hispanic | 19 (73.1) | 30 (63.8) | 23 (85.2) | |
| White, Hispanic | 0 (0.0) | 3 (6.4) | 1 (3.7) | |
| Black, non‐Hispanic | 5 (19.2) | 6 (12.8) | 2 (7.4) | |
| Asian, Pacific Islander | 1 (2.2) | 3 (6.4) | 1 (3.7) | |
| Native American | 0 (0.0) | 2 (4.3) | 0 (0.0) | |
| Multiple | 0 (0.0) | 2 (4.3) | 0 (0.0) | |
| Unknown/declined | 1 (2.2) | 1 (2.1) | 0 (0.0) | |
| Tumour | 0.47 | |||
| Gastrointestinal | — | 19 (40.4) | 14 (51.9) | |
| Genitourinary | — | 28 (59.6) | 13 (48.1) | |
| Stage | 0.28 | |||
| 1/2 | — | 37 (78.7) | 18 (66.7) | |
| 3/4 | — | 10 (21.3) | 9 (33.3) | |
| Prior 3 month chemo (y) | — | 6 (12.8) | 6 (22.2) | 0.34 |
| Prior 3 month rads (y) | — | 4 (8.5) | 4 (14.8) | 0.45 |
CON, non‐cancer control patients; CWS, weight‐stable cancer patients; CWL, cancer patients with weight loss; BMI, body mass index; chemo, chemotherapy exposure; rads, radiation exposure; y, yes. P values were derived from one‐way ANOVA with Tukey's post‐hoc pairwise comparison for continuous variables or from Fisher's exact test for categorical variables.
vs. CWL, P ≤ 0.05.
Fat mass and energy expenditure
The CWL displayed smaller SAT, VAT, and TAT than CWS and displayed reduced SMA than both CWL and CON (Figure 1A); IMAT was not different across groups (data not shown, P = 0.29). There was a trend for reduced RQ in CWL compared with CON (Figure 1B; ANOVA P = 0.08, CON vs. CWL P = 0.07), with no difference across groups in absolute REE (kcal/day) or %predicted REE (Figure 1C–D). There was a trend for higher REE corrected for skeletal muscle area (REE/SMA) in CWL than CON (Figure 1E; ANOVA P = 0.10, CON vs. CWL P = 0.11).
Figure 1.
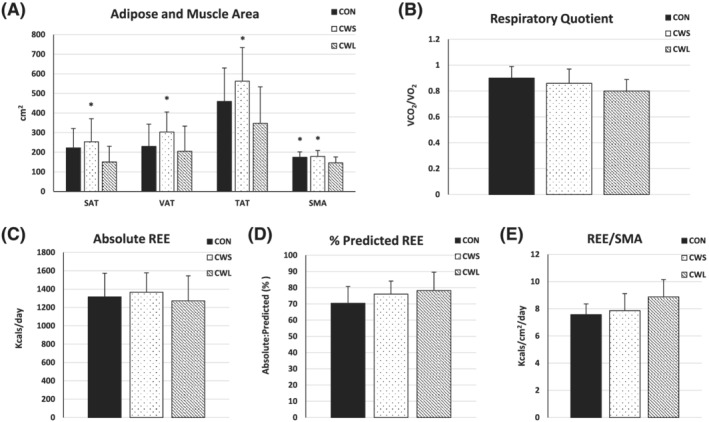
(A–E) Abdominal tissue area and energy expenditure. Means and standard deviations of computed tomography‐derived adipose and muscle cross‐sectional area and energy expenditure. One‐way ANOVA comparison of (A) adipose tissue (AT) area for subcutaneous (SAT), visceral (VAT), and total (TAT) depots and skeletal muscle area (SMA) (CON, N = 17–18; CWS, N = 24–27; CWL, N = 17–22); (B) respiratory quotient (RQ); (C) absolute resting energy expenditure (REE); (D) %predicted REE (B–D): (CON, N = 11; CWS, N = 16; CWL, N = 10); and (E) REE corrected for SMA (CON, N = 7; CWS, N = 10; CWL, N = 7) between cancer patients with weight loss (CWL) or without weight loss (CWS) and non‐cancer patients (CON). *vs CWL, p < 0.05.
Plasma biomarkers
Circulating IL‐6 was higher in CWL than CWS and CON (Figure 2A), with no difference across groups in circulating TNF (Figure 2A) or proportion of patients with detectable IL‐1β (data not shown, P = 0.62). There was no difference across groups in circulating PTHrP or FFA (Figure 2B,C).
Figure 2.
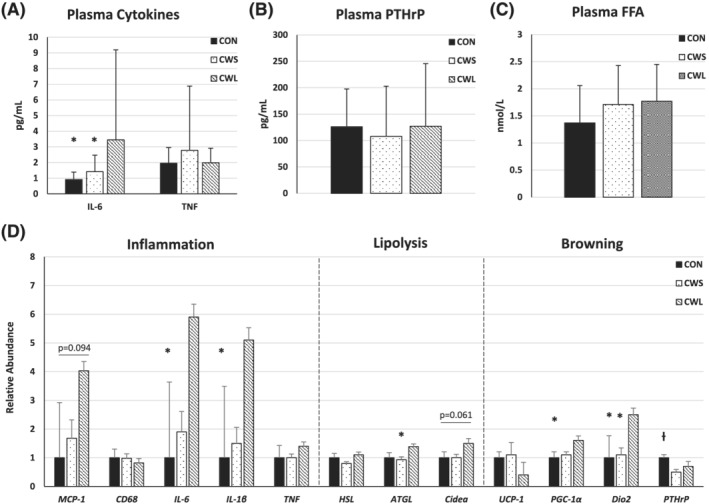
(A–D) Circulating and white adipose tissue biomarkers. Means and standard deviations of plasma and abdominal subcutaneous white adipose tissue biomarkers. One‐way ANOVA comparison of (A) plasma cytokines interleukin (IL)‐6 and tumour necrosis factor (TNF: CON, N = 22; CWS, N = 46; CWL, N = 26), (B) parathyroid hormone related peptide (PTHrP: CON, N = 7; CWS, N = 13; CWL, N = 11), and (C) free fatty acids (FFA: CON, N = 20; CWS, N = 43; CWL, N = 25), (D) mRNA expression of markers of inflammation (MCP‐1, monocyte chemoattractant protein‐1; CD68; macrophage marker; IL‐6, interleukin‐6; IL‐1β, interleukin‐1β; TNF, tumour necrosis factor), lipolysis (HSL, hormone sensitive lipase; ATGL, adipose triglyceride lipase; Cidea, cell death‐inducing DNA fragmentation factor‐α‐like effector (A), and browning (UCP‐1, uncoupling protein‐1; PGC‐1α, peroxisome proliferator‐activated receptor coactivator 1 α; Dio2, iodothyronine deiodinase 2; PTHrP, parathyroid hormone‐related peptide) (CON, N = 25; CWS, N = 27; CWL, N = 14) between cancer patients with weight loss (CWL) or without weight loss (CWS) and non‐cancer patients (CON). *vs. CWL, P ≤ 0.05; ƚ vs. CWS P ≤ 0.05.
Adipose tissue protein and gene expression
Inflammation
There were no differences across groups for protein expression of UCP‐1, MCP‐1, IL‐6, or IL‐1β (Table 2); MCP‐1 and IL‐6 were log‐transformed for analysis, with the raw data reported in the table. TNF protein was undetectable in over 90% of specimens and was therefore not included in the current analysis. CWL displayed higher IL‐6 and IL‐1β than CON (Figure 2D); there were no differences across groups in CD68 or TNF. For the primary outcome (WAT IL‐6), effect size was very large for CWL vs. CWS (day = 6.6) and for CWL vs. CON (day = 2.6).
Table 2.
Adipose protein expression
| Mean (SD) | CON | CWS | CWL | P value |
|---|---|---|---|---|
| N = 20–23 | N = 23–26 | N = 13–14 | ||
| UCP‐1 (% pos stain) | 5.75 (3.74) | 5.96 (4.58) | 9.20 (8.31) | 0.14 |
| MCP‐1 (pg/mg) a | 454.6 (858.6) | 705.7 (1495.9) | 1457.7 (3180.2) | 0.23 |
| IL‐6 (pg/mg) a | 101.9 (241.8) | 185.8 (478.7) | 396.4 (1125.5) | 0.51 |
| IL‐1β (pg/mg) | 4.86 (7.26) | 6.00 (7.40) | 9.23 (9.14) | 0.29 |
CON, non‐cancer control patients; CWS, weight‐stable cancer patients; CWL, cancer patients with weight loss; UCP, uncoupling protein; pos, positive; MCP, monocyte chemoattractant protein; IL, interleukin. P values were derived from one‐way ANOVA with Tukey's post‐hoc pairwise.
Raw data are displayed here; however, P values are from comparison of log‐transformed data.
Lipolysis
The CWL displayed higher ATGL than CWS and (Figure 2D) and there was a trend for Cidea, which is thought to be a lipolysis inhibitor; there was no difference across groups in HSL.
Browning
CWL displayed higher PGC‐1α than CON and higher Dio2 than both CWS and CON while CWS displayed lower PTHrP than CON (Figure 2D); there was no difference in UCP‐1 transcript or protein.
Correlations between plasma and adipose tissue levels in cancer patients
Inflammation
When assessed as a dichotomous variable (above or below the limit of detection), circulating IL‐1β was directly correlated with circulating IL‐6 (r = 0.30, P = 0.012, n = 72) and TNF (r = 0.35, P = 0.002, n = 72), but not with WAT protein or transcript levels of inflammatory markers. Circulating IL‐6 and TNF were directly correlated with each other (r = 0.31, P = 0.008, n = 72), and circulating IL‐6 was directly correlated with IL‐1β protein level in WAT (r = 0.43, P = 0.006, n = 40). There were no other correlations between circulating IL‐6 or TNF and WAT protein or transcript levels of inflammatory markers.
Lipolysis
Circulating FFA were directly correlated with HSL (r = 0.38, P = 0.018, n = 39), with a trend for a direct correlation with ATGL (r = 0.28, P = 0.084, n = 39). HSL (r = 0.56, P < 0.001, n = 40) and ATGL (r = 0.32, P = 0.04, n = 40) were each directly correlated with Cidea and with each other (r = 0.55, P < 0.001, n = 40); neither HSL nor ATGL were correlated with circulating PTHrP or PTHrP.
Browning
Circulating PTHrP was directly correlated with Cidea (r = 0.43, P = 0.036, n = 24), but not with UCP‐1 protein or other transcript levels of browning markers (UCP‐1, Dio2, PGC‐1α, PTHrP). UCP‐1 protein was directly correlated with Cidea (r = 0.39, P = 0.016, n = 38) and with PTHrP (r = 0.41, P = 0.011, n = 38).
Regression analyses
The following conditional variables were used in multivariate regression models using cancer patient data (N = 74), excluding cases listwise: BMI, relative weight loss over prior 6 months, plasma biomarkers (IL‐6, FFA), WAT protein expression (UCP‐1, IL‐1β), and WAT gene expression (IL‐6, IL‐1β, PGC‐1α, Dio2, ATGL, Cidea). BMI was directly associated with SAT, VAT, and REE (kcals/day) while circulating FFA was indirectly associated with RQ (Table 3). BMI and UCP‐1 protein negatively predicted REE (%pred; Table 3); BMI is indirectly associated with REE (%pred; r = −0.44, P = 0.02) while UCP‐1 protein is not associated with REE (%pred) but displays a trend for an indirect association with BMI (r = −0.27, P = 0.098).
Table 3.
Multivariate regression analyses in cancer patients
| Dependent variable | N | R 2 | Predictors | Unstandardized B (95% CI) | P value |
|---|---|---|---|---|---|
| SAT (cm2) | 25 | 0.72 | BMI | 15.05 (11.01, 19.09) | <0.001 |
| VAT (cm2) | 28 | 0.64 | BMI | 14.61 (10.14, 19.09) | <0.001 |
| TAT (cm2) | 25 | 0.84 | BMI | 29.94 (24.29, 35.59) | <0.001 |
| RQ (VCO2/VO2) | 22 | 0.52 | FFA mmol/L | −0.09 (−0.13, −0.05) | <0.001 |
| REE (kcal/day) | 22 | 0.43 | BMI | 21.94 (10.13, 33.75) | 0.001 |
| REE (%pred) | 22 | 0.45 | BMI | −1.08 (−1.68, −0.48) | 0.001 |
| UCP‐1 (%) | −0.59 (−1.11, −0.08) | 0.027 |
BMI, body mass index; FFA, free fatty acids; REE, resting energy expenditure; RQ, respiratory quotient; SAT, subcutaneous adipose tissue; TAT, total adipose tissue; VAT, visceral adipose tissue; UCP, uncoupling protein. P values are derived from multivariate regression analyses.
Discussion
The specific mechanisms leading to cancer‐associated AT loss rely heavily on animal models. In this pilot study, we looked at tissue and circulating markers of processes thought to be relevant in cancer‐associated AT wasting and looked at physiologically relevant outcomes like AT area, energy expenditure, body weight history, and obesity in cancer patients and non‐cancer controls with a large BMI range. Patients with cancer‐associated weight loss displayed reduced abdominal AT area, increased circulating IL‐6, and higher WAT transcripts of inflammation (cytokines: IL‐6, IL‐1β), lipolysis (lipase: ATGL), and browning (UCP‐1 promoting genes: PGC‐1α, Dio2) than weight‐stable cancer patients and/or controls. We did not observe differences between cancer patients with or without weight loss and non‐cancer controls in energy expenditure, circulating PTHrP or FFA, WAT inflammatory protein expression, or WAT UCP‐1 protein or transcript. Absolute REE was not different across groups despite that CWL experienced significant involuntary weight loss and displayed reduced body mass. In combination with the numerically higher %predicted REE and REE/SMA and numerically lower RQ, these observations may indicate hypermetabolism in CWL. Although, these differences were not significant, we cannot rule out that hypermetabolism was present below the detectable level in this cohort. In cancer patients, BMI itself was a predictor of greater AT area and absolute REE, and circulating FFA was correlated with subcutaneous WAT HSL transcript, but circulating markers of inflammation or browning were not correlated with WAT markers.
Human data suggest that inflammation may mediate increased lipolysis and WAT browning in cancer‐associated weight loss, but WAT inflammatory gene expression has not been consistently reported in patients with cancer. 5 , 6 Here we report elevated circulating IL‐6 and WAT IL‐6 and IL‐1β transcripts in CWL with no difference across groups in circulating TNF or IL‐1β or WAT inflammatory proteins. One possible explanation for this discrepancy is that the variability is greater for the proteins and we were underpowered to detect this difference. An alternative explanation is that these genes are post‐transcriptionally regulated, which has been reported by in‐vitro studies in astrocytes and spleen macrophages. 21 , 22 These observations are partially consistent with a previous report where WAT TNF, IL‐1β, and MCP‐1 transcripts were higher in weight‐losing cancer patients than weight‐stable cancer patients and/or non‐cancer patients. 23 A different report showed that circulating IL‐6 and subcutaneous WAT CD68 transcript were higher in weight‐losing cancer patients than weight‐stable cancer patients and non‐cancer controls. 24 In that study, weight‐stable cancer patients also displayed greater subcutaneous WAT MCP‐1 transcript than weight‐losing cancer patients and weight‐stable non‐cancer patients, 24 whereas we did not observe any difference across groups in WAT MCP‐1 transcript in the current study.
These data exemplify the complex nature of the relationship between adiposity and inflammation in the cancer setting, which is yet to be fully characterized. This relationship was further complicated here by the general lack of association between circulating and WAT protein or transcript levels of inflammatory markers in the cancer patients. These observations imply that measuring other markers of inflammation in circulation besides IL‐6 may not provide additional information and that assessing inflammation in plasma may not be informative of tissue inflammation. This is clinically relevant as anti‐inflammatory agents are in development for cancer cachexia. 25
We measured the lipolytic markers FFA, HSL, ATGL, and Cidea because lipolysis is thought to play an important role in cancer‐induced weight loss in animal models and humans. 1 We expected CWL to display elevated circulating FFA, a validated marker of lipolysis, 1 and elevated WAT gene expression of the main lipolytic enzymes HSL and/or ATGL as previously reported in other weight‐losing cancer patients 7 , 8 , 10 and weight‐losing non‐cancer patients. 8 Circulating FFA indirectly predicted RQ in multivariate regression in cancer patients in the current study although we did not observe differences across groups. In contrast, circulating FFA was previously reported to increase with obesity due to larger AT mass, increased insulin resistance, and reduced FFA clearance, 26 which were not assessed in the current study, but the large BMI range included here could contribute to these findings. In the current study, FFA levels were moderately, directly correlated with WAT HSL (significantly) and ATGL (trend) transcripts in cancer patients. Also, CWL displayed higher ATGL than CWS with a trend for elevated Cidea. In combination with the inverse relationship between FFA and RQ observed here, these associations suggest a higher reliance on fat as a fuel source in the weight‐losing patients. However, the clinical relevance of these associations cannot be determined from the current cross‐sectional design.
Elevated WAT HSL gene and protein expression were previously reported in weight‐losing cancer patients than weight‐stable cancer patients and/or weight‐losing non‐cancer patients 8 , 9 , 27 with no difference in ATGL transcript. 8 , 9 In another small cohort of cancer patients with various tumour types and weight‐stable non‐cancer patients, visceral WAT from autopsy revealed greater HSL and ATGL activity in cancer patients than non‐cancer patients and in weight‐losing cancer patients than weight‐stable cancer patients. 7 In that study, lipase activity was indirectly correlated with BMI. 7 Cidea, a lipolysis inhibitor, 28 was reportedly elevated after weight loss due to cancer, diet, or bariatric surgery, 29 , 30 , 31 , 32 which may be a compensatory mechanism to balance increased lipolysis during weight loss. It will be important for future studies to determine the association of lipolysis markers like FFA, Cidea, and lipase enzymes with clinical markers like survival, tolerance to therapy, hospitalizations, and future weight loss, to determine their clinical relevance and potential as biomarkers for cancer‐associated weight loss.
To our knowledge, we are the first to report the association between markers of browning in WAT and plasma with REE, body weight history, and fat mass in humans. Cidea may promote WAT browning by inhibiting repression of UCP‐1 enhancer activity. 12 In brown AT from cancer patients, Cidea was associated with PGC‐1α and UCP‐1 transcripts, 33 but this has not been confirmed in WAT from cancer patients. In the current study, Cidea was moderately, directly correlated with WAT UCP‐1 protein expression, but not PGC‐1α or Dio2, despite elevated PGC‐1α and Dio2 in CWL. In support of the premise that WAT browning contributes to AT wasting and hypermetabolism in cancer, UCP‐1 protein was predictive of relative REE in cancer patients. This is likely due to the indirect association between BMI and relative REE and the trend for an indirect association between UCP‐1 protein and BMI. However, these associations were only moderate, and the association between UCP‐1 and WAT browning has not been consistently reported.
In one cohort, UCP‐1 protein was detectable in peritumuoral WAT taken from various fat pads in seven of eight weight‐losing patients with various tumour types but was undetectable in 10 weight‐stable patients with colon cancer. 15 Conversely, a transcriptomics study reported that UCP‐1 was reduced in a small cohort of GI cancer patients compared with non‐cancer patients. 14 Additionally, activation of protein kinase A is thought to activate both thermogenesis and lipolysis, and lipolysis itself can activate thermogenesis in brown AT. 34 It is unknown whether this also occurs in WAT, but this could explain the increase in PGC‐1α and Dio2 without a concomitant effect on UCP‐1. This study may be underpowered to see a difference between groups in UCP‐1 protein, but the fact that it predicts REE suggests it may be a relevant pathway. However, the uncoupling capability of adipocytes has been previously observed independently from UCP‐1 activity, 35 so we cannot determine that WAT browning was not present below the level of our detection.
Circulating PTHrP, a paraneoplastic factor that induces hypercalcaemia, has been shown to indirectly increase HSL phosphorylation, which is required for HSL enzymatic lipase activity, through protein kinase A. 18 , 36 , 37 PTHrP is also reportedly associated with browning via increased UCP‐1 protein 17 in AT in rodent models, making its impact on thermogenesis and lipolysis difficult to differentiate. Other groups have reported that cancer patients with detectable circulating PTHrP levels have higher relative REE 17 and reduced body fat 19 than those with undetectable PTHrP. In the current study, circulating PTHrP was not different across group, but was moderately, directly correlated with Cidea transcript. It was not associated with UCP‐1 protein or other browning markers transcripts, which may suggest that PTHrP is acting as a paracrine lipolytic signal. However, these data suggest that circulating PTHrP may not be a useful marker of cancer‐associated weight loss.
This study has many strengths including fat mass assessments by CT, comparison of age‐matched non‐cancer control patients, simultaneous assessment of physiologic parameters and circulating and WAT biomarkers, homogeneity of the sample consisting of the demographic group with the highest prevalence of cancer (older men), with good functional performance status that made them eligible for surgery, and assessment of both systemic and peripheral pathways. This study may be limited by the amount of tissue procured from surgical biopsies, which precluded a more in‐depth molecular analysis. Because this is a pilot study, we cannot rule out the potential impact of correction for the multiple comparisons presented here. Larger trials powered for extensive pathway analyses will be required to confirm these results; however, any further molecular analysis would require further correction. This study also lacks a comparison between subcutaneous and visceral WAT depots. It was reported that visceral WAT expressed higher inflammatory genes than subcutaneous WAT in cancer and non‐cancer patients; however, browning genes, including UCP‐1, were decreased in visceral WAT from cancer patients in that same cohort. 14 Also, subcutaneous and visceral WAT reportedly decrease to the same extent near the end of life. 3 As a result, there is no consensus as to whether one depot is more impactful in the setting of AT wasting in cancer. This study is also limited by the lack of female patients and its cross‐sectional design that prevents us from establishing causation. Generalizability may be limited to the tumour types included here and/or to patients who are deemed suitable for surgical resection.
To clarify the association between physiologic parameters like BMI, weight loss, energy expenditure, inflammation, lipolysis, and WAT browning markers, we report weight loss‐associated elevations in some WAT inflammatory, lipolytic, and browning marker transcripts. Increased inflammation was confirmed systemically in CWL, but elevated lipolysis as measured by FFA levels was not completely detected at the whole‐body level, and the presence of WAT browning was unlikely due to the absence of alterations in UCP‐1. In cancer patients, energy expenditure was primarily associated with BMI, while RQ was inversely associated with FFA. There was a moderate association between circulating and WAT lipolysis, but this association was not observed between circulating and WAT markers for inflammation or browning, indicating that systemic factors should be interpreted with caution when assessed alone.
While BMI range may introduce variability to future findings, including those observed here, it is representative of the current US cancer population; therefore, future studies should consider variability in fat mass when exploring the relationship between cancer and AT metabolism. Assessment of the trajectory of lipolysis and energy expenditure over time may provide more mechanistic interpretation of causation than the single measurement assessed here. The contribution of inflammation and lipolysis, and lack thereof for WAT browning, will need to be clarified in other tumour types to increase the generalizability of these results. Additionally, larger longitudinal studies will be needed to establish the clinical significance of these associations and whether they are causal.
Funding
J. G. receives research support from the US Department of Veterans Affairs (VA; BX002807), the Congressionally Directed Medical Research Programs (PC170059), and from the NIH (R01CA239208, R01AG061558, R01AR067319). LJA receives research support from the University of Washington (DK007247) and US Department of Veterans Affairs (RX03245). This project was also supported by the NIH National Institute of Diabetes and Digestive and Kidney Diseases funded Nutrition Obesity Research Center, University of Washington (DK035816) and Diabetes Research Center (P30 DK017047) at the University of Washington.
Conflict of interest
These authors have no conflict of interest to declare.
Acknowledgements
We extend our great thanks to Dr Dianne Lattemann for her insight during revision of this manuscript. The authors of this manuscript certify that they comply with the ethical guidelines for authorship and publishing in the Journal of Cachexia, Sarcopenia and Muscle. 38
Anderson L. J., Lee J., Anderson B., Lee B., Migula D., Sauer A., Chong N., Liu H., Wu P. C., Dash A., Li Y.‐P., and Garcia J. M. (2022) Whole‐body and adipose tissue metabolic phenotype in cancer patients, Journal of Cachexia, Sarcopenia and Muscle, 13, 1124–1133, 10.1002/jcsm.12918
References
- 1. Tsoli M, Swarbrick MM, Robertson GR. Lipolytic and thermogenic depletion of adipose tissue in cancer cachexia. Semin Cell Dev Biol 2016;54:68–81. [DOI] [PubMed] [Google Scholar]
- 2. Fouladiun M, Korner U, Bosaeus I, Daneryd P, Hyltander A, Lundholm KG. Body composition and time course changes in regional distribution of fat and lean tissue in unselected cancer patients on palliative care—correlations with food intake, metabolism, exercise capacity, and hormones. Cancer 2005;103:2189–2198. [DOI] [PubMed] [Google Scholar]
- 3. Murphy RA, Wilke MS, Perrine M, Pawlowicz M, Mourtzakis M, Lieffers JR, et al. Loss of adipose tissue and plasma phospholipids: relationship to survival in advanced cancer patients. Clin Nutr 2010;29:482–487. [DOI] [PubMed] [Google Scholar]
- 4. Martin L, Senesse P, Gioulbasanis I, Antoun S, Bozzetti F, Deans C, et al. Diagnostic criteria for the classification of cancer‐associated weight loss. J Clin Oncol 2015;33:90–99. [DOI] [PubMed] [Google Scholar]
- 5. Dahlman I, Mejhert N, Linder K, Agustsson T, Mutch DM, Kulyte A, et al. Adipose tissue pathways involved in weight loss of cancer cachexia. Br J Cancer 2010;102:1541–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ryden M, Agustsson T, Laurencikiene J, Britton T, Sjolin E, Isaksson B, et al. Lipolysis—not inflammation, cell death, or lipogenesis—is involved in adipose tissue loss in cancer cachexia. Cancer 2008;113:1695–1704. [DOI] [PubMed] [Google Scholar]
- 7. Das SK, Eder S, Schauer S, Diwoky C, Temmel H, Guertl B, et al. Adipose triglyceride lipase contributes to cancer‐associated cachexia. Science 2011;333:233–238. [DOI] [PubMed] [Google Scholar]
- 8. Agustsson T, Ryden M, Hoffstedt J, van Harmelen V, Dicker A, Laurencikiene J, et al. Mechanism of increased lipolysis in cancer cachexia. Cancer Res 2007;67:5531–5537. [DOI] [PubMed] [Google Scholar]
- 9. Silverio R, Lira FS, Oyama LM, Oller do Nascimento CM, Otoch JP, Alcantara PSM, et al. Lipases and lipid droplet‐associated protein expression in subcutaneous white adipose tissue of cachectic patients with cancer. Lipids Health Dis 2017;16:159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Han J, Meng Q, Shen L, Wu G. Interleukin‐6 induces fat loss in cancer cachexia by promoting white adipose tissue lipolysis and browning. Lipids Health Dis 2018;17:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ravasco P, Monteiro‐Grillo I, Camilo M. How relevant are cytokines in colorectal cancer wasting? Cancer J 2007;13:392–398. [DOI] [PubMed] [Google Scholar]
- 12. Jash S, Banerjee S, Lee MJ, Farmer SR, Puri V. CIDEA transcriptionally regulates UCP1 for britening and thermogenesis in human fat cells. iScience 2019;20:73–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tsoli M, Moore M, Burg D, Painter A, Taylor R, Lockie SH, et al. Activation of thermogenesis in brown adipose tissue and dysregulated lipid metabolism associated with cancer cachexia in mice. Cancer Res 2012;72:4372–4382. [DOI] [PubMed] [Google Scholar]
- 14. Miller J, Dreczkowski G, Ramage MI, Wigmore SJ, Gallagher IJ, Skipworth RJE. Adipose depot gene expression and intelectin‐1 in the metabolic response to cancer and cachexia. J Cachexia Sarcopenia Muscle 2020;11:1141–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Petruzzelli M, Schweiger M, Schreiber R, Campos‐Olivas R, Tsoli M, Allen J, et al. A switch from white to brown fat increases energy expenditure in cancer‐associated cachexia. Cell Metab 2014;20:433–447. [DOI] [PubMed] [Google Scholar]
- 16. Liu H, Luo J, Guillory B, Chen JA, Zang P, Yoeli JK, et al. Ghrelin ameliorates tumor‐induced adipose tissue atrophy and inflammation via Ghrelin receptor‐dependent and ‐independent pathways. Oncotarget 2020;11:3286–3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kir S, White JP, Kleiner S, Kazak L, Cohen P, Baracos VE, et al. Tumour‐derived PTH‐related protein triggers adipose tissue browning and cancer cachexia. Nature 2014;513:100–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Larsson S, Jones HA, Goransson O, Degerman E, Holm C. Parathyroid hormone induces adipocyte lipolysis via PKA‐mediated phosphorylation of hormone‐sensitive lipase. Cell Signal 2016;28:204–213. [DOI] [PubMed] [Google Scholar]
- 19. Hong N, Yoon HJ, Lee YH, Kim HR, Lee BW, Rhee Y, et al. Serum PTHrP predicts weight loss in cancer patients independent of hypercalcemia, inflammation, and tumor burden. J Clin Endocrinol Metab 2016;101:1207–1214. [DOI] [PubMed] [Google Scholar]
- 20. Mourtzakis M, Prado CM, Lieffers JR, Reiman T, McCargar LJ, Baracos VE. A practical and precise approach to quantification of body composition in cancer patients using computed tomography images acquired during routine care. Appl Physiol Nutr Metab 2008;33:997–1006. [DOI] [PubMed] [Google Scholar]
- 21. Neininger A, Kontoyiannis D, Kotlyarov A, Winzen R, Eckert R, Volk HD, et al. MK2 targets AU‐rich elements and regulates biosynthesis of tumor necrosis factor and interleukin‐6 independently at different post‐transcriptional levels. J Biol Chem 2002;277:3065–3068. [DOI] [PubMed] [Google Scholar]
- 22. Spooren A, Mestdagh P, Rondou P, Kolmus K, Haegeman G, Gerlo S. IL‐1β potently stabilizes IL‐6 mRNA in human astrocytes. Biochem Pharmacol 2011;81:1004–1015. [DOI] [PubMed] [Google Scholar]
- 23. Camargo RG, Riccardi DM, Ribeiro HQ, Carnevali LC Jr, de Matos‐Neto EM, Enjiu L, et al. NF‐κBp65 and expression of its pro‐inflammatory target genes are upregulated in the subcutaneous adipose tissue of cachectic cancer patients. Nutrients 2015;7:4465–4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Batista ML Jr, Henriques FS, Neves RX, Olivan MR, Matos‐Neto EM, Alcantara PS, et al. Cachexia‐associated adipose tissue morphological rearrangement in gastrointestinal cancer patients. J Cachexia Sarcopenia Muscle 2016;7:37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hong DS, Janku F, Naing A, Falchook GS, Piha‐Paul S, Wheler JJ, et al. Xilonix, a novel true human antibody targeting the inflammatory cytokine interleukin‐1 alpha, in non‐small cell lung cancer. Invest New Drugs 2015;33:621–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Frohnert BI, Jacobs DR Jr, Steinberger J, Moran A, Steffen LM, Sinaiko AR. Relation between serum free fatty acids and adiposity, insulin resistance, and cardiovascular risk factors from adolescence to adulthood. Diabetes 2013;62:3163–3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cao DX, Wu GH, Yang ZA, Zhang B, Jiang Y, Han YS, et al. Role of β1‐adrenoceptor in increased lipolysis in cancer cachexia. Cancer Sci 2010;101:1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Puri V, Ranjit S, Konda S, Nicoloro SMC, Straubhaar J, Chawla A, et al. Cidea is associated with lipid droplets and insulin sensitivity in humans. Proc Natl Acad Sci U S A 2008;105:7833–7838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dahlman I, Kaaman M, Jiao H, Kere J, Laakso M, Arner P. The CIDEA gene V115F polymorphism is associated with obesity in Swedish subjects. Diabetes 2005;54:3032–3034. [DOI] [PubMed] [Google Scholar]
- 30. Karczewska‐Kupczewska M, Nikolajuk A, Majewski R, Filarski R, Stefanowicz M, Matulewicz N, et al. Changes in adipose tissue lipolysis gene expression and insulin sensitivity after weight loss. Endocr Connect 2020;9:90–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Laurencikiene J, Stenson BM, Arvidsson Nordstrom E, Agustsson T, Langin D, Isaksson B, et al. Evidence for an important role of CIDEA in human cancer cachexia. Cancer Res 2008;68:9247–9254. [DOI] [PubMed] [Google Scholar]
- 32. Nordstrom EA, Ryden M, Backlund EC, Dahlman I, Kaaman M, Blomqvist L, et al. A human‐specific role of cell death‐inducing DFFA (DNA fragmentation factor‐alpha)‐like effector A (CIDEA) in adipocyte lipolysis and obesity. Diabetes 2005;54:1726–1734. [DOI] [PubMed] [Google Scholar]
- 33. Jespersen NZ, Larsen TJ, Peijs L, Daugaard S, Homoe P, Loft A, et al. A classical brown adipose tissue mRNA signature partly overlaps with brite in the supraclavicular region of adult humans. Cell Metab 2013;17:798–805. [DOI] [PubMed] [Google Scholar]
- 34. Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev 2004;84:277–359. [DOI] [PubMed] [Google Scholar]
- 35. Nyman E, Bartesaghi S, Melin Rydfalk R, Eng S, Pollard C, Gennemark P, et al. Systems biology reveals uncoupling beyond UCP1 in human white fat‐derived beige adipocytes. NPJ Syst Biol Appl 2017;3:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hu W, Xiong H, Ru Z, Zhao Y, Zhou Y, Xie K, et al. Extracellular vesicles‐released parathyroid hormone‐related protein from Lewis lung carcinoma induces lipolysis and adipose tissue browning in cancer cachexia. Cell Death Dis 2021;12:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sagar G, Sah RP, Javeed N, Dutta SK, Smyrk TC, Lau JS, et al. Pathogenesis of pancreatic cancer exosome‐induced lipolysis in adipose tissue. Gut 2016;65:1165–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. von Haehling S, Morley JE, Coats AJS, Anker SD. Ethical guidelines for publishing in the Journal of Cachexia, Sarcopenia and Muscle: update 2019. J Cachexia Sarcopenia Muscle 2019;10:1143–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]


