Abstract
We have analyzed genetically three clinical isolates (3180, 3870, and 1244) of Streptococcus pneumoniae with high-level ciprofloxacin resistance. Isolates 3180 and 3870 were atypical because of their insolubility in deoxycholate. However, they hybridized specifically with pneumococcal autolysin and pneumolysin gene probes and have typical pneumococcal atpC and atpA gene sequences. Analysis of the complete sequences of the parC and gyrA genes revealed total variations of 8 and 8.7% (isolate 3180) and 7.4 and 3.6% (isolate 3870), respectively, compared to the wild-type strain R6 sequence. The variations observed between the sequences of R6 and isolate 1244 were less than 0.9%. The structure of the gyrA and parC genes from isolates 3180 and 3870 was organized in sequence blocks that show different levels of divergence, suggesting a pattern of recombination. These results are evidence for recombination at the fluoroquinolone target genes in clinical isolates of S. pneumoniae. The genetically related viridans group streptococci could act as a reservoir for fluoroquinolone resistance genes.
Streptococcus pneumoniae is the most common bacterial cause of community-acquired pneumonia, meningitis, otitis media, and sinusitis. The emergence of resistance to antimicrobial agents commonly used for the treatment of pneumococcal diseases (5, 17, 25, 38) has made very difficult the selection of optimal antimicrobial therapies for the treatment of pneumococcal infections. A parallel increasing resistance to penicillin and macrolide antibiotics has been also observed for the viridans group streptococci (1, 2, 7, 8). These microorganisms are, like S. pneumoniae, commensals of the oropharyngeal tract. Nevertheless, they are causative organisms of infective endocarditis (12, 44, 47) and are also a major cause of bacteremia in neutropenic cancer patients (3, 7, 8, 16).
There is considerable interest in the use of alternative antimicrobial agents, such as the new fluoroquinolones, with good activity against streptococci for the treatment of respiratory tract infections (6). The prevalence of ciprofloxacin resistance in S. pneumoniae has been found to be low in Spain (<3%) (32, 33); similar data have been reported in Canada (9). The prior administration of fluoroquinolones could be an important risk factor for quinolone-resistant strain selection, as has been observed for respiratory tract infections caused by ciprofloxacin-resistant (Cpr) S. pneumoniae (41). Likewise, fluoroquinolone resistance has been reported for blood isolates of viridans group streptococci from neutropenic cancer patients who received quinolone prophylaxis (22, 50). The prevalence of resistance to ciprofloxacin (MIC, ≥4 μg/ml) in viridans group streptococci consecutively isolated from different clinical sources from 1993 to 1998 at Hospital Princeps d'Espanya was as follows: 17.8% (135 of 756 clinical isolates) for Streptococcus mitis, 12.0% (10 of 83 isolates) for Streptococcus salivarius, 2.9% (11 of 378 clinical isolates) for Streptococcus sanguis, and 2.3% (13 of 575 isolates) for Streptococcus anginosus (unpublished data). These data are in accordance with those obtained in Canada, which showed a prevalence of resistance to ciprofloxacin of 11.4% (27 of 236 isolates) for the viridans group streptococci, S. mitis and S. salivarius being the most resistant (10).
The principal targets of the fluoroquinolones are DNA gyrase (gyrase) and DNA topoisomerase IV (topo IV), members of the topoisomerase family of enzymes that control bacterial DNA topology (15). Both enzymes function by passing one DNA double helix through another, using a transient double-strand break (35). Gyrase, an A2B2 complex encoded by the gyrA and gyrB genes, catalyzes ATP-dependent negative supercoiling of DNA and is involved in DNA replication, recombination, and transcription (53); topo IV, a C2E2 complex encoded by the parC and parE genes, is essential in chromosome partitioning (35). The deduced amino acid sequences of ParC and ParE are homologous to those of GyrA and GyrB, respectively (30). Genetic studies from a number of laboratories (23, 28, 36, 39, 49) have shown that topo IV is the primary target for ciprofloxacin in S. pneumoniae and that gyrase is the secondary target. Resistance mutations have been identified in a discrete region of ParC, ParE, GyrA, and GyrB termed the quinolone resistance-determining region (QRDR). We recently reported the same mechanism for viridans group streptococci (22): low-level Cpr strains had mutations altering one of the two subunits of topo IV.
The viridans group streptococci could be a reservoir of fluoroquinolone resistance genes if we assume that resistance in viridans group streptococci and S. pneumoniae arose from horizontal transfer, as has been observed with penicillin resistance (46). A number of observations suggest that this transfer between viridans group streptococci and S. pneumoniae could be a possible mechanism for the spread of fluoroquinolone resistance. The viridans group streptococci and S. pneumoniae share the same mechanism of resistance (22). The nucleotide sequences of their gyrase and topo IV genes show high identity (20, 22), and it is possible to transform S. pneumoniae cells to ciprofloxacin resistance with DNA from Cpr viridans group streptococci (22, 27). Additionally, nucleotide sequence comparisons of the DNA topoisomerase genes of the viridans group streptococci (20, 22) show a high level of intraspecies variation. These observations suggest that the viridans group streptococci could be considered a group of species that interchange genetic material between them and possibly with S. pneumoniae. In this report, we describe the characterization of Cpr S. pneumoniae isolates with a mosaic structure in their parC and gyrA genes, suggesting such interspecies recombination.
MATERIALS AND METHODS
Southern blot identification of S. pneumoniae strains.
The ciprofloxacin-sensitive (Cps) strain of S. pneumoniae used was the wild-type strain R6. The Cpr clinical isolates were obtained from sputum samples at the Hospital Princeps d'Espanya (Barcelona, Spain) in 1992 (strain 1244), 1994 (strain 3180), and 1996 (strain 3870). Plasmid pCE3 (18), containing a 0.65-kb fragment coding for the N terminus of the major pneumococcal autolysin (amidase), was used as a source of the lytA DNA probe. Plasmid pJCP191 (48), containing a 1.6-kb fragment coding for the complete pneumococcal pneumolysin gene, was used as a source of the pnl DNA probe and was kindly provided by S. Taira. The inserts of pCE3 and pJCP191 were isolated after digestion with HindIII-HincII and PvuII, respectively. The resulting DNA inserts were labeled with the Phototope-Star Detection Kit (New England Biolabs). Southern blotting and hybridization were done by following the manufacturer's instructions.
Amplification and analysis of genes.
Genes were amplified from genomic DNA by the PCR as described previously (22). The atpCA region and the parC and gyrA genes were amplified with the following primers, based on published sequences (4, 19, 36, 40): atpCUP (5′-dAAAGGAGAATTTGTTATGAA-3′), corresponding to nucleotides −15 to +5 of atpC, and atpB56 (5′-dGACGGGCTTCTTCAGCTCTGTC-3′), complementary to nucleotides 147 to 169 of atpB; parCUP (5′-dGAACACGCCCTAGATACTGTG-3′), corresponding to nucleotides −103 to −83 of parC, and parCDOWN (5′-dCGTTACTGTCATATTCCACTCC-3′), complementary to nucleotides 120 to 142 downstream of parC; and gyrAUP1 (4) and gyrADOWN (4). DNA fragments were purified with MicroSpin S400 HR columns (Pharmacia) and were sequenced on both strands by use of an Applied Biosystems Prism 377 sequencer with the primers used for PCR amplification and with internal primers. For nucleotide sequence comparisons, in addition to the Cps S. pneumoniae strain R6 (GenBank accession no. AF170996 and AF053121 for parC and gyrA, respectively), two other sequences were used: the sequences of the Cps strain 7785 (accession no. Z67739 and AJ005815 for parC and gyrA, respectively) and of another, unknown isolate, which we call AB (accession no. for gyrA, AB010387).
Nucleotide sequence accession numbers.
The new DNA sequences reported in this paper have been assigned the following GenBank accession no.: AF170996 to AF170999 (parC genes), AF170993 to AF170995 (gyrA genes), and AF171000 to AF171002 (atpCA regions).
RESULTS AND DISCUSSION
Identification of strains.
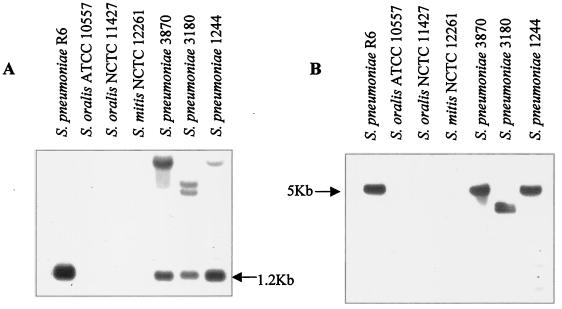
Three clinical isolates, 3180, 3870, and 1244, were analyzed in this work. These isolates were previously described as Streptococcus oralis 3180 (22), S. oralis 3870 (22), and S. pneumoniae 1244 (36). Initial characterization of the three isolates by colony morphology on blood agar and optochin susceptibility identified them as pneumococcal strains. However, strains 3180 and 3870 were insoluble in deoxycholate, while strain 1244 was soluble. Phenotypic characterization of isolates 3180 and 3870 by the API 32 Strep system classified them as S. oralis (22). Given the unreliability of this method for the identification of S. pneumoniae (20, 31), the isolates were studied by hybridization with two pneumococcal probes. One of the probes coded for the N terminus of the major pneumococcal autolysin (lytA), and the other coded for the complete pneumococcal pneumolysin (pnl). The S. pneumoniae strain R6 showed, as expected, hybridization with the lytA probe in a 1.2-kb HindIII chromosomal fragment (21), while S. oralis and S. mitis type strains did not (Fig. 1A). Strains 3180, 3870, and 1244 showed high-molecular-weight hybridization bands with the lytA probe, in addition to the 1.2-kb HindIII fragment (Fig. 1A). These bands could have resulted from hybridization with homologous lytA genes of pneumococcal prophages, which have been described to be very frequent in pneumococcal clinical isolates (43). Hybridization with the pneumolysin probe detected a single 5-kb band in S. pneumoniae R6 ClaI-digested DNA (Fig. 1B), as expected from the physical map of the pnl chromosomal region (48, 52), while no hybridization was observed with the S. oralis and S. mitis type strains. The three Cpr S. pneumoniae clinical isolates (3180, 3870, and 1244) all hybridized with the pnl probe. Because both LytA and pneumolysin proteins have been demonstrated to be species specific (18, 24, 29, 42, 45, 51), these results identified the three isolates as S. pneumoniae.
FIG. 1.
Identification of S. pneumoniae isolates by hybridization with specific DNA probes. Chromosomal DNAs from the S. oralis, S. mitis, and S. pneumoniae strains indicated were cleaved with HindIII (A) or ClaI (B), and the fragments were separated in 1% agarose gels. The gels were blotted, and the blots were probed with biotinylated DNA as follows: A, insert of plasmid pCE3 containing the N terminus of the lytA gene; and B, insert of plasmid pJCP191 containing the pnl gene.
Sequencing of the atpC and atpA genes allowed further characterization of the strains. The atpC gene is responsible for the characteristic optochin susceptibility phenotype of pneumococci (19, 37). The sequences of a region spanning 960 nucleotides, including atpC and atpA, showed high homogeneity (data not shown). The three Cpr S. pneumoniae strains showed less than 0.6% variation, compared with the wild-type strain R6, while the S. oralis NCTC 11427 type strain showed 20% variation. These values are in agreement with the results of comparisons of the amylomaltase gene sequences: ≤0.5% S. pneumoniae intraspecies variation (14) and 4 to 6% divergence between S. pneumoniae and S. oralis (13).
The results of comparisons of the atpCA region are consistent with the results of Southern blot hybridization and identified the clinical isolates as S. pneumoniae. Since lytA encodes the major pneumococcal autolysin (amidase), responsible for the sodium deoxycholate solubility of pneumococci (34 and references therein), the observed deoxycholate insensitivity of isolates 3870 and 1244 could be due to some alteration in the lytA gene, as has been described for another atypical pneumococcal isolate (11).
Analysis of the sequences of the parC and gyrA genes.
We have previously determined the nucleotide sequences of portions of the parC and gyrA genes that included the QRDRs from strains 3180, 3870, and 1244 (22, 36). Comparison of the sequences of isolate 1244 to those of Cps isolates revealed two sense mutations, one in parC and the other in gyrA. The mutation found in parC produced the amino acid change Ser-79 to Phe (TCT to TTT), while the mutation in gyrA produced an alteration at the equivalent Ser-81 (TCC) residue: a change to Phe (TTC). By means of genetic transformation, it was shown that these amino acid changes were responsible for the Cpr phenotype of isolate 1244 (36). We paid special attention to isolates 3180 and 3870, because they showed an unexpectedly high nucleotide sequence variation in their QRDRs compared to S. pneumoniae strain R6. A comparison of the parC and gyrA QRDR sequences of 13 independent clinical isolates of S. pneumoniae (including 1244) showed variations of ≤1% (unpublished results). However, these variations (excluding mutations involved in Cpr) were 8.6 and 4.3% for the parC QRDRs of isolates 3180 and 3870 and 5% for their gyrA QRDRs. Despite this high nucleotide sequence variation, only two amino acid changes were found in the parC QRDRs of isolates 3180 and 3870: Ser-79 to Phe (TCT to TTT) and Asn-91 to Asp (AAC to GAC) (22). Biological evidence showing that the changes of Ser-79 to Phe were involved in ciprofloxacin resistance has been obtained by genetic transformation. A Cpr recombinant strain was obtained by transformation of competent S. pneumoniae R6 cells with DNA encoding the parC QRDR of isolate 3180. This recombinant strain was shown to carry the Ser-79–to–Phe change but not the Asn-91–to–Asp change (22). On the other hand, the nucleotide changes observed in the gyrA QRDRs of isolates 3180 and 3870 also produced two amino acid changes. One of these changes was Ser-81 to Tyr (TCC to TAC) in isolate 3180 and Ser-81 to Phe (TTC) in isolate 3870. The other change, present in both isolates, was Gly-144 to Ser (AGT to GGT). The analysis of Cpr transformants obtained with DNA from the gyrA QRDRs of isolates 3180 and 3870 showed that the amino acid changes at Ser-81 were indeed involved in resistance (22).
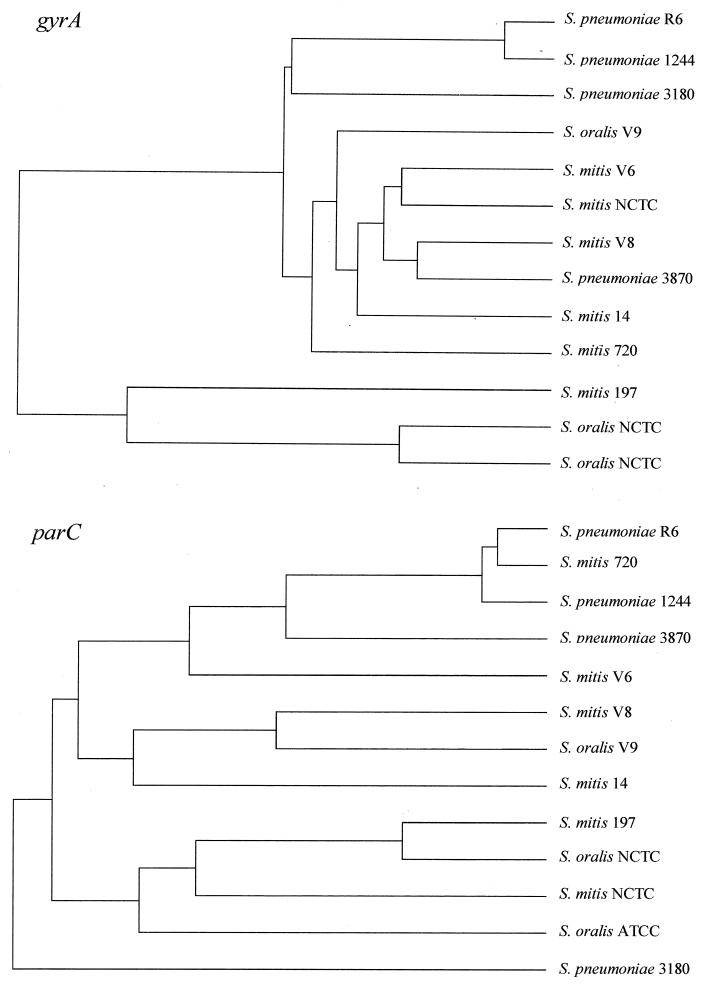
A comparison of the nucleotide sequences of the parC and gyrA QRDRs of isolates 3180, 3870, and 1244, S. pneumoniae R6, and several S. oralis and S. mitis strains is shown in Fig. 2. While isolate 1244 was grouped with S. pneumoniae R6 within the parC or gyrA tree, the location of isolates 3180 and 3870 varied depending on the gene considered. These results suggested a recombinational origin for the genes encoding the fluoroquinolone target proteins of isolates 3180 and 3870. Such a situation would be due to genetic transformation with DNA from other bacterial species, probably the genetically closed related viridans group streptococci. The recombination machinery requires approximately 80% sequence identity between two homologous DNA molecules (19, 26). At least 87% identity was found between the parC and gyrA QRDRs of isolates 3180 and 3870 and those of the viridans group streptococci (Fig. 2). Similarly, horizontal transfer of altered penicillin-binding protein genes between S. pneumoniae and viridans group streptococci (46) has been observed.
FIG. 2.
Trees of nucleotide sequences of parC and gyrA QRDRs. The 185-nucleotide parC sequence included positions 213 to 397, and the 280-nucleotide gyrA sequence included positions 175 to 454. Nucleotides are numbered by taking the first gyrA and parC nucleotides as nucleotide 1. The trees were compiled by using the CLUSTAL multiple-alignment program from PCGENE with default parameters. The nucleotide sequences used have been previously reported (20, 22, 36).
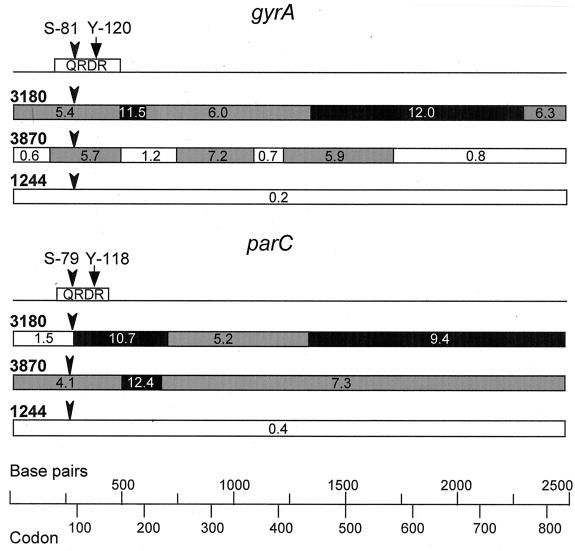
To test this hypothesis, the nucleotide sequences of the complete parC and gyrA genes of the two isolates (3180 and 3870) that showed high divergence in their parC and gyrA QRDRs and one isolate (1244) that did not shown this divergence were determined. The results of sequence comparisons of Cps and Cpr strains (Fig. 3 and 4 and Table 1) clearly showed three groups of strains. One group, with a nucleotide sequence variation of <1%, was formed by the sensitive strains and isolate 1244. Isolates 3180 and 3870 each formed separate groups, since the nucleotide sequence variations between the sequences of the two isolates were 7.6% for their parC sequences and 8% for their gyrA sequences. The average variations in the parC and gyrA sequences between isolate 3180 and the first group of strains (sensitive strains and isolate 1244) were ≥8%, while these values for isolate 3870 were about 7% for parC and about 3% for gyrA (Table 1). The variations found in isolates 3180 and 3870 could be organized in blocks (Fig. 5) with different degrees of relatedness. The limits of the blocks were determined by inspection, with the only limitation being at least a 4% difference in divergence between two contiguous blocks.
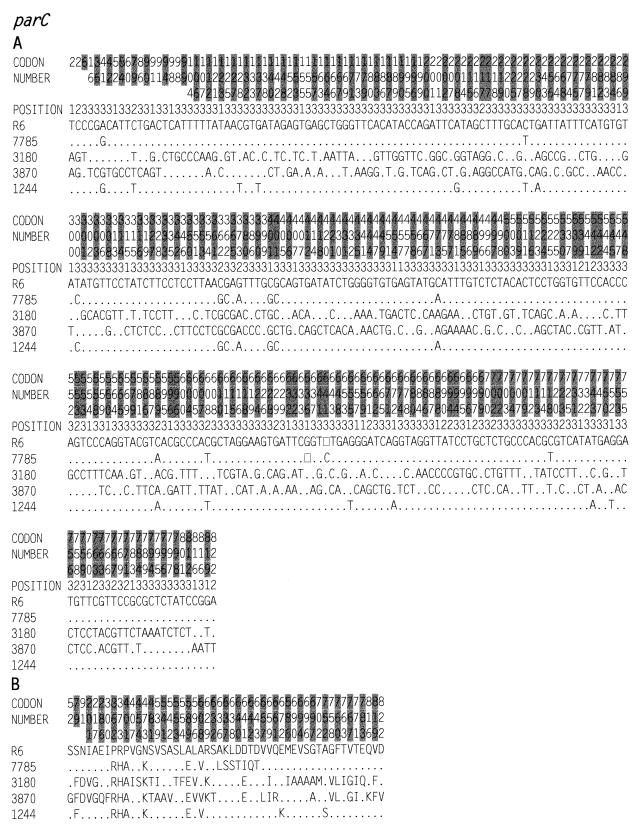
FIG. 3.
Nucleotide (A) and amino acid (B) sequence variations in the parC genes of S. pneumoniae strains. The nucleotides and amino acids present at each polymorphic site are shown in full for strain R6, but only sites that differ are shown for the other strains. Nucleotides and amino acids that are the same as in R6 are shown by dots. The codon numbers are indicated in vertical format above the sequences. The different codons are alternatively shaded in grey for clarity. Positions 1, 2, and 3 in the fourth row refer to the first, second, and third nucleotides in the codon. The sequence in panel A is numbered from the initiation codon of the parC gene. Open squares denote nucleotide deletions. The strains used were R6 (GenBank accession no. AF170996), 7785 (accession no. Z67739), 3180 (accession no. AF170997), 3870 (accession no. AF170998), and 1244 (accession no. AF170999).
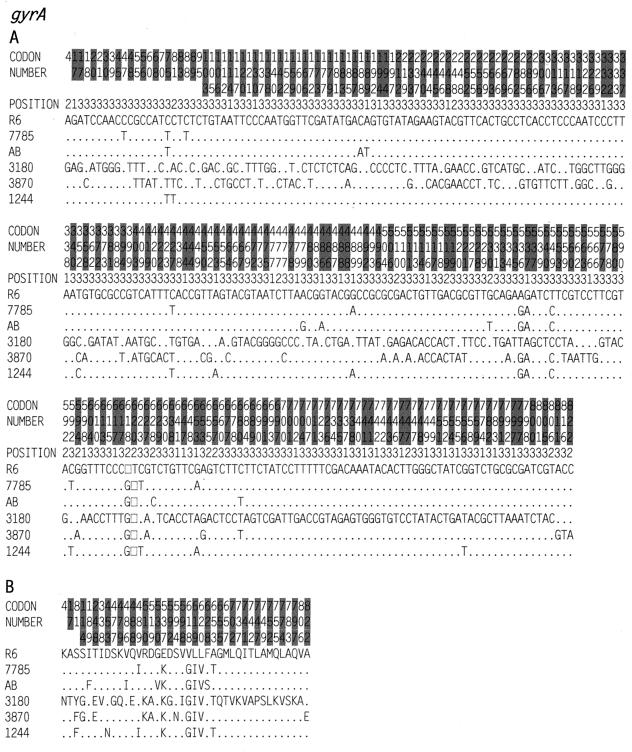
FIG. 4.
Nucleotide (A) and amino acid (B) sequence variations in the gyrA genes of S. pneumoniae strains. See the legend to Fig. 3 for details. Codons are numbered according to the R6 sequence. The strains used were R6 (GenBank accession no. AF053121), 7785 (accession no. AJ005815), AB (accession no. AB010387), 3180 (accession no. AF170993), 3870 (accession no. AF170994), and 1244 (accession no. AF170993).
TABLE 1.
Nucleotide sequence differences for gyrA and parC genesa
| Strain | % (no.) of nucleotide differences in strain:
|
|||||
|---|---|---|---|---|---|---|
| R6 | 7785 | AB | 3180 | 3870 | 1244 | |
| R6 | 0.5 (13) | 0.5 (13) | 8.7 (216) | 3.6 (90) | 0.6 (15) | |
| 7785 | 0.6 (14) | 0.6 (14) | 8.7 (216) | 3.6 (89) | 0.2 (6) | |
| AB | — | — | 8.8 (218) | 3.6 (89) | 0.6 (15) | |
| 3180 | 8.0 (198) | 8.0 (196) | — | 8.1 (200) | 8.6 (213) | |
| 3870 | 7.4 (184) | 7.2 (178) | — | 7.6 (188) | 3.5 (87) | |
| 1244 | 0.8 (20) | 0.5 (12) | — | 8.0 (197) | 7.2 (175) | |
Differences were measured over sequences of 2,469 bp from gyrA (above the diagonal) and 2,472 bp from parC (below the diagonal).
FIG. 5.
Mosaic structure of the gyrA and parC genes of Cpr S. pneumoniae isolates 3180 and 3870. The locations of the QRDRs are represented above the gyrA and parC sequences. The positions of the active Tyr residues (Y-120 in GyrA and Y-118 in ParC) that bind DNA and of the Ser residues that are changed in the Cpr strains (S-81 in GyrA and S-79 in ParC) and are involved in resistance are marked. Blocks showing the percent sequence divergence from the corresponding regions in Cps pneumococci are indicated. White boxes, regions of sequence that differ by ≤1.5%; grey boxes, regions that differ by more than 1.5% but less than 9%; black boxes, regions that differ by >9%.
From these results, we can assume that the first group of strains (sensitive strains and isolate 1244) are nonrecombinant isolates, while isolates 3180 and 3870 show a recombinational pattern probably resulting from gene transfer events. The gyrA sequence of isolate 3870 clearly shows four blocks of low divergence (≤1.2%) and three blocks of high divergence (≥5.7%). This result suggests that the low-divergence blocks represent regions in which interspecies recombination events had occurred. Another block with a 1.5% divergence was observed in parC of isolate 3180, although in this case the second recombination point should be located outside the gene. Likewise, recombination outside the genes would be the origin of the gyrA gene of isolate 3180 and of the parC gene of isolate 3870.
Because both gyrase and topo IV are tetrameric proteins, an interchange of parC would need an accompanying interchange of parE. Since both genes are contiguous in the pneumococcal chromosome, we cannot exclude the possibility of a recombinational event involving both genes. On the other hand, an interchange of both gyrA and gyrB genes would involve two independent recombinations, since the genes are separated by at least 90 kb in the chromosome (36).
Two different processes could lead to the acquisition of fluoroquinolone resistance: spontaneous mutation and transformation. A comparison of the frequencies of these two processes reveals that transformation could be several orders of magnitude more frequent than mutation. The frequency of mutation to Cpr in S. pneumoniae has been shown to be in the range of 10−8 to 10−9 (39). However, the frequencies of transformation to Cpr with chromosomal DNAs from Cpr S. pneumoniae strains were in the range of 10−2 (36, 49) for monogenic transformation (low-level Cpr) (36, 49). The acquisition of low-level Cpr via transformation could then be 106 to 107 times more frequent than that via spontaneous mutation. Likewise, it has been shown that the frequency of transformation of S. pneumoniae competent cells to low-level Cpr with DNA from Cpr S. mitis is in the range of 10−3 (22, 27). Interspecies transformation could thus be 105 to 106 more frequent than spontaneous mutation. These differences are even higher when the acquisition of high-level resistance is considered. The frequencies of transformation with two unlinked markers that gave rise to high-level Cpr were 10−4 when both donor and recipient cells were S. pneumoniae (36, 49) and 10−6 when competent S. pneumoniae cells were transformed with S. mitis DNA (27). However, two spontaneous mutations are necessary to obtain a high level of resistance (i.e., the frequency could be 10−14 to 10−16). Nevertheless, these estimates are not necessarily true since, as pointed out above, an interchange of parC would need an accompanying interchange of parE and an interchange of gyrA would need an accompanying interchange of gyrB. Other factors to be considered for transformation in the natural environment are the availability of DNA and the competence state of the recipient cells.
ACKNOWLEDGMENTS
We thank P. A. Lazo for allowing us to use the PCGENE program and A. Rodriguez-Bernabé for excellent technical assistance.
M.J.F. has a fellowship from Comunidad Autónoma de Madrid. This work was supported by grant 97/2026 from Fondo de Investigación Sanitaria.
REFERENCES
- 1.Alcaide F, Carratala J, Liñares J, Gudiol F, Martín R. In vitro activities of eight macrolide antibiotics and RP-59500 (quinupristin-dalfopristin) against viridans group streptococci isolated from blood of neutropenic cancer patients. Antimicrob Agents Chemother. 1996;40:2117–2120. doi: 10.1128/aac.40.9.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alcaide F, Liñares J, Pallarés R, Carratala J, Benítez M A, Gudiol F, Martín R. In vitro activity of 22 β-lactam antibiotics against penicillin-resistant and penicillin-susceptible viridans group streptococci isolated from blood. Antimicrob Agents Chemother. 1995;39:2243–2247. doi: 10.1128/aac.39.10.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Awada A P, Van der Auwera P, Meunier P, Daneau D, Klastersky J. Streptococcal and enterococcal bacteremia in patients with cancer. Clin Infect Dis. 1992;15:33–48. doi: 10.1093/clinids/15.1.33. [DOI] [PubMed] [Google Scholar]
- 4.Balas D, Fernández-Moreira E, de la Campa A G. Molecular characterization of the gene encoding the DNA gyrase A subunit of Streptococcus pneumoniae. J Bacteriol. 1998;180:2854–2861. doi: 10.1128/jb.180.11.2854-2861.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baquero F. Epidemiology and management of penicillin-resistant pneumococci. Curr Opin Infect Dis. 1996;9:372–379. [Google Scholar]
- 6.Bartlett J G, Breiman R F, Mandell L, File T M. Community-acquired pneumonia in adults: guidelines for management. Clin Infect Dis. 1998;26:811–838. doi: 10.1086/513953. [DOI] [PubMed] [Google Scholar]
- 7.Bochud P Y, Eggiman P H, Calandra T, Van Melle G, Saghafi L, Francioli P. Bacteremia due to viridans streptococcus in neutropenic patients with cancer: clinical spectrum and risk factors. Clin Infect Dis. 1994;18:25–31. doi: 10.1093/clinids/18.1.25. [DOI] [PubMed] [Google Scholar]
- 8.Carratalá J, Alcaide F, Fernández-Sevilla A, Corbell X, Liñares J, Gudiol F. Bacteremia due to viridans streptococci that are highly resistant to penicillin: increase among neutropenic patients with cancer. Clin Infect Dis. 1995;20:1169–1173. doi: 10.1093/clinids/20.5.1169. [DOI] [PubMed] [Google Scholar]
- 9.Chen D K, McGeer A, De Acevedo J C, Low D E. Decreased susceptibility of Streptococcus pneumoniae to fluoroquinolones in Canada. N Engl J Med. 1999;341:233–239. doi: 10.1056/NEJM199907223410403. [DOI] [PubMed] [Google Scholar]
- 10.de Azevedo J C S, Trpeski L, Pong-Porter S, Matsumura S, Network T C B S, Low D E. In vitro activity of fluoroquinolones against antibiotic-resistant blood culture isolates of viridans group streptococci across Canada. Antimicrob Agents Chemother. 1999;43:2299–2301. doi: 10.1128/aac.43.9.2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Díaz E, López R, García J L. Role of the major pneumococcal autolysin in the atypical response of a clinical isolate of Streptococcus pneumoniae. J Bacteriol. 1992;174:5508–5515. doi: 10.1128/jb.174.17.5508-5515.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Douglas C W I, Heath J, Hampton K K, Preston F E. Identity of viridans streptococci isolated from cases of infective endocarditis. J Med Microbiol. 1993;39:179–182. doi: 10.1099/00222615-39-3-179. [DOI] [PubMed] [Google Scholar]
- 13.Dowson C G, Hutchinson A, Woodford N, Johnson A P, George R C, Spratt B G. Penicillin-resistant viridans streptococci have obtained altered penicillin-binding protein genes from penicillin-resistant strains of Streptococcus pneumoniae. Proc Natl Acad Sci USA. 1990;87:5858–5862. doi: 10.1073/pnas.87.15.5858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dowson C G, Hutchison A, Brannigan J A, George R C, Hansman D, Liñares J, Tomasz A, Maynard Smith J, Spratt B G. Horizontal transfer of penicillin-binding protein genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Proc Natl Acad Sci USA. 1989;86:8842–8846. doi: 10.1073/pnas.86.22.8842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drlica K, Zhao X. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol Mol Biol Rev. 1997;61:377–392. doi: 10.1128/mmbr.61.3.377-392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elting L S, Bodey G P, Keefe B H. Septicemia and shock syndrome due to viridans streptococci: a case-control study predisposing factors. Clin Infect Dis. 1992;14:1201–1207. doi: 10.1093/clinids/14.6.1201. [DOI] [PubMed] [Google Scholar]
- 17.Fenoll A, Jado I, Vicioso D, Pérez A, Casal J. Evolution of Streptococcus pneumoniae serotypes and antibiotic resistance in Spain: update (1990–1996) J Clin Microbiol. 1998;36:3447–3454. doi: 10.1128/jcm.36.12.3447-3454.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fenoll A, Martínez-Suárez J, Muñoz R, Casal J, García J L. Identification of atypical strains of Streptococcus pneumoniae by a specific DNA probe. Eur J Clin Microbiol Infect Dis. 1990;9:396–401. doi: 10.1007/BF01979468. [DOI] [PubMed] [Google Scholar]
- 19.Fenoll A, Muñoz R, García E, de la Campa A G. Molecular basis of the optochin-sensitive phenotype of pneumococcus: characterization of the genes encoding the F0 complex of the Streptococcus pneumoniae and Streptococcus oralis H+-ATPases. Mol Microbiol. 1994;12:587–598. doi: 10.1111/j.1365-2958.1994.tb01045.x. [DOI] [PubMed] [Google Scholar]
- 20.Ferrándiz M J, Oteo J, Aracil B, Gómez-Garcés J L, de la Campa A G. Drug efflux and parC mutations are involved in fluoroquinolone resistance in viridans group streptococci. Antimicrob Agents Chemother. 1999;43:2520–2523. doi: 10.1128/aac.43.10.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.García P, García J L, García E, López R. Nucleotide sequence and expression of the pneumococcal autolysin gene from its own promoter in Escherichia coli. Gene. 1986;43:265–272. doi: 10.1016/0378-1119(86)90215-5. [DOI] [PubMed] [Google Scholar]
- 22.González I, Georgiou M, Alcaide F, Balas D, Liñares J, de la Campa A G. Fluoroquinolone resistance mutations in the parC, parE, and gyrA genes of clinical isolates of viridans group streptococci. Antimicrob Agents Chemother. 1998;42:2792–2798. doi: 10.1128/aac.42.11.2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gootz T D, Zaniewski R, Haskell S, Schmieder B, Tankovic J, Girard D, Courvalin P, Polzer R J. Activity of the new fluoroquinolone trovafloxacin (cp-99,219) against DNA gyrase and topoisomerase IV mutants of Streptococcus pneumoniae selected in vitro. Antimicrob Agents Chemother. 1996;40:2691–2697. doi: 10.1128/aac.40.12.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guillespie S, Ullman C, Smith M D, Emery V. Detection of Streptococcus pneumoniae in sputum samples by PCR. J Clin Microbiol. 1994;32:1308–1311. doi: 10.1128/jcm.32.5.1308-1311.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoffman J, Cetron M S, Farley M M, Baughman W S, Facklam R R, Elliot J A, Deaver K A, Breiman R F. The prevalence of drug-resistant Streptococcus pneumoniae in Atlanta. N Engl J Med. 1995;333:481–486. doi: 10.1056/NEJM199508243330803. [DOI] [PubMed] [Google Scholar]
- 26.Humbert O, Prudhomme M, Hakenbeck R, Dowson C G, Claverys J-P. Homologous recombination and mismatch repair during transformation in Streptococcus pneumoniae: saturation of the Hex mismatch repair system. Proc Natl Acad Sci USA. 1995;92:9052–9056. doi: 10.1073/pnas.92.20.9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janoir C, Podglajen I, Kitzis M D, Poyart C, Gutmann L. In vitro exchange of fluoroquinolone resistance determinants between Streptococcus pneumoniae and viridans streptococci and genomic organization of the parE-parC region in S. mitis. J Infect Dis. 1999;180:555–558. doi: 10.1086/314888. [DOI] [PubMed] [Google Scholar]
- 28.Janoir C, Zeller V, Kitzis M-D, Moreau N J, Gutmann L. High-level fluoroquinolone resistance in Streptococcus pneumoniae requires mutations in parC and gyrA. Antimicrob Agents Chemother. 1996;40:2760–2764. doi: 10.1128/aac.40.12.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kalin M, Klanclerski K, Granstrom M, Móllby R. Diagnosis of pneumococcal pneumonia by enzyme-linked immunosorbent assay of antibodies to pneumococcal hemolysin (pneumolysin) J Clin Microbiol. 1987;25:226–229. doi: 10.1128/jcm.25.2.226-229.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kato J, Nishimura Y, Imamura R, Niki H, Higara S, Suzuki H. New topoisomerase essential for chromosome segregation in E. coli. Cell. 1990;63:393–404. doi: 10.1016/0092-8674(90)90172-b. [DOI] [PubMed] [Google Scholar]
- 31.Kikuchi K, Enari T, Totsuka K, Shimizu K. Comparison of phenotypic characteristics, DNA-DNA hybridization results, and results with a commercial rapid biochemical and enzymatic reaction system for identification of viridans group streptococci. J Clin Microbiol. 1995;33:1215–1222. doi: 10.1128/jcm.33.5.1215-1222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liñares J, de la Campa A G, Pallarés R. Fluoroquinolone resistance in Streptococcus pneumoniae. N Engl J Med. 1999;341:1546–1548. doi: 10.1056/nejm199911113412013. [DOI] [PubMed] [Google Scholar]
- 33.Liñares J, Tubau F, Domínguez M A. Antibiotic resistance in Streptococcus pneumoniae in Spain: an overview in the 1990s. In: Tomasz A, editor. Streptococcus pneumoniae. Molecular biology and mechanisms of disease–update for the 1990s. New York, N.Y: Mary Ann Liebert Inc.; 1999. pp. 399–407. [Google Scholar]
- 34.López R, García E, García P, García J L. The pneumococcal cell wall degrading enzymes: a modular design to create new lysins? Microb Drug Resist. 1997;3:199–211. doi: 10.1089/mdr.1997.3.199. [DOI] [PubMed] [Google Scholar]
- 35.Luttinger A. The twisted life of DNA in the cell: bacterial DNA topoisomerases. Mol Microbiol. 1995;15:601–606. doi: 10.1111/j.1365-2958.1995.tb02369.x. [DOI] [PubMed] [Google Scholar]
- 36.Muñoz R, de la Campa A G. ParC subunit of DNA topoisomerase IV of Streptococcus pneumoniae is a primary target of fluoroquinolones and cooperates with DNA gyrase A subunit in forming resistance phenotype. Antimicrob Agents Chemother. 1996;40:2252–2257. doi: 10.1128/aac.40.10.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muñoz R, García E, de la Campa A G. Quinine specifically inhibits the proteolipid subunit of the F0F1 H+-ATPase of Streptococcus pneumoniae. J Bacteriol. 1996;178:2455–2458. doi: 10.1128/jb.178.8.2455-2458.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pallarés R, Liñares J, Vadillo M, Cabellos C, Manresa F, Viladrich P F, Martín R, Gudiol F. Resistance to penicillin and cephalosporin and mortality from severe pneumococcal pneumonia in Barcelona, Spain. N Engl J Med. 1995;333:474–480. doi: 10.1056/NEJM199508243330802. [DOI] [PubMed] [Google Scholar]
- 39.Pan X-S, Ambler J, Mehtar S, Fisher L M. Involvement of topoisomerase IV and DNA gyrase as ciprofloxacin targets in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1996;40:2321–2326. doi: 10.1128/aac.40.10.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pan X-S, Fisher L M. Cloning and characterization of the parC and parE genes of Streptococcus pneumoniae encoding DNA topoisomerase IV: role in fluoroquinolone resistance. J Bacteriol. 1996;178:4060–4069. doi: 10.1128/jb.178.14.4060-4069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pérez-Trallero E, García-Arenzana J M, Jiménez J A, Peris A. Therapeutic failure and selection of resistance to quinolones in a case of pneumococcal pneumonia treated with ciprofloxacin. Eur J Clin Microbiol Infect Dis. 1990;9:905–906. doi: 10.1007/BF01967510. [DOI] [PubMed] [Google Scholar]
- 42.Pozzi G, Oggioni M R, Tomasz A. DNA probe for the identification of Streptococcus pneumoniae. J Clin Microbiol. 1989;27:370–372. doi: 10.1128/jcm.27.2.370-372.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramirez M, Severina E, Tomasz A. A high incidence of prophage carriage among natural isolates of Streptococcus pneumoniae. J Bacteriol. 1999;181:3618–3625. doi: 10.1128/jb.181.12.3618-3625.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roberts R B, Krieger A G, Schiller N L, Gross K C. Viridans streptococcal endocarditis: the role of various species, including pyridoxal-dependent streptococci. Rev Infect Dis. 1979;1:955–965. doi: 10.1093/clinids/1.6.955. [DOI] [PubMed] [Google Scholar]
- 45.Rudolph K M, Parkison A J, Black C M, Mayer L W. Evaluation of polymerase chain reaction for diagnosis of pneumococcal pneumonia. J Clin Microbiol. 1993;31:2661–2666. doi: 10.1128/jcm.31.10.2661-2666.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spratt B G. Resistance to antibiotics mediated by target alterations. Science. 1994;264:388–393. doi: 10.1126/science.8153626. [DOI] [PubMed] [Google Scholar]
- 47.Sussman J I, Baron J, Tenenbaum M J, Kaplan M H, Greenspan R R, Facklam R R, Tyburski M B, Goldman M A, Kanzer B F, Pizzarello R A. Viridans streptococcal endocarditis: clinical, microbiological, and echocardiographic correlations. J Infect Dis. 1986;154:597–603. doi: 10.1093/infdis/154.4.597. [DOI] [PubMed] [Google Scholar]
- 48.Taira S, Jalonen E, Paton J C, Sarvas M, Runeberg-Nyman K. Production of pneumolysin, a pneumococcal toxin, in Bacillus subtilis. Gene. 1989;77:211–218. doi: 10.1016/0378-1119(89)90069-3. [DOI] [PubMed] [Google Scholar]
- 49.Tankovic J, Perichon B, Duval J, Courvalin P. Contribution of mutations in gyrA and parC genes to fluoroquinolone resistance of mutants of Streptococcus pneumoniae obtained in vivo and in vitro. Antimicrob Agents Chemother. 1996;40:2505–2510. doi: 10.1128/aac.40.11.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Venditti M, Baiocchi P, Barandimarte C, Serra P, Gentile G, Girmenia C, Martino P. Antimicrobial susceptibilities of Streptococcus species that cause septicemia in neutropenic patients. Antimicrob Agents Chemother. 1989;33:580–582. doi: 10.1128/aac.33.4.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Virolainen A, Salo P, Jero J, Karma P, Eskola J, Leinonen M. Comparison of PCR assay with bacterial culture for detecting Streptococcus pneumoniae in middle ear fluid of children with acute otitis media. J Clin Microbiol. 1994;32:2667–2670. doi: 10.1128/jcm.32.11.2667-2670.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walker J A, Allen R L, Falgame P, Johnson M K, Boulnois G J. Molecular cloning, characterization, and complete nucleotide sequence of the gene for pneumolysin, the sulfhydryl-activated toxin of Streptococcus pneumoniae. Infect Immun. 1987;55:1184–1189. doi: 10.1128/iai.55.5.1184-1189.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang J C. DNA topoisomerases. Annu Rev Biochem. 1985;54:665–697. doi: 10.1146/annurev.bi.54.070185.003313. [DOI] [PubMed] [Google Scholar]