Abstract
Background/Aims
The composition of the microbiota in the esophagus is only partially understood, especially in patients with achalasia. We aim to investigate the esophageal microbial community and nutritional intakes in patients with achalasia before and after peroral endoscopic myotomies (POEM).
Methods
Twenty-nine patients were prospectively enrolled from 4 referral institutions across Korea. We collected esophageal samples (mucosal biopsies and retention fluid) and conducted dietary surveys for nutritional intake before and 8 weeks after POEM. The esophageal microbiota was analyzed by 16S rRNA gene sequencing targeting the V3-V4 region.
Results
Out of the 105 samples from 29 patients, 99 samples were subjected to microbial bioinformatic analysis after quality control, which excluded samples with no amplification or low-quality sequence data. The overall esophageal microbial compositions of patients with achalasia showed that Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria, and Fusobacteria were the dominant phyla, representing over 95% of the total phyla in all groups. At the genus level, Streptococcus was the most abundant in all groups. The observed operational taxonomic unit number was significantly higher in the retention fluid than in the tissue biopsies. However, the esophageal microbial composition showed no significant changes 8 weeks post POEM. The dietary survey analysis showed that nutritional intake significantly improved post POEM.
Conclusion
This study determined the unique esophageal microbial composition of patients with achalasia, and also found that the microbial composition did not significantly change after POEM in the short-term, despite a significant improvement in the nutritional intake.
Keywords: Esophageal achalasia, Microbiota, Myotomy
Introduction
Although the knowledge regarding the human microbiota composition in the gut is vast, the composition of human microbiota in the esophagus is only partially understood. The microbiota in the esophagus under normal circumstances had been considered as a transitional microbiota originating from the oral cavity and oropharynx.1,2 However, as new techniques such as 16S ribosomal RNA (rRNA) gene sequencing have been introduced, it is now known that the esophagus has a remarkably diverse microbiota that depends on several factors.3,4 The genera Streptococcus, Prevotella, and Veillonella are known to be the major components of the microbiota in a healthy esophagus.5-7 In addition, a previous study reported that dysbiosis, which is associated with the presence of gram-negative taxa, is associated with gastroesophageal reflux disease, Barrett’s esophagus, and eventual adenocarcinoma.5,7-9
Achalasia is a motility disorder that induces aperistalsis and abnormality in lower esophageal sphincter relaxation.10 Therefore, chronic liquid and food stasis causes bacterial fermentation in the esophagus in patients with achalasia.11 Peroral endoscopic myotomy (POEM) is a new option for the treatment of achalasia, and is shown to have excellent efficacy.12 In addition, a previous study reported that POEM may restore the morphologic tortuosity in cases of sigmoid-type achalasia.13 Pajecki et al14 reported that Streptococcus and Veillonella were predominant in patients with achalasia using the culture method, and they also found that the concentration of esophageal microbiota in patients with achalasia consisted mostly of aerobic gram-positive bacteria, and the anaerobic bacteria varied according to the degree of esophageal dilation.14 However, there have been no reports on the esophageal microbiota of patients with achalasia analyzed using 16S rRNA sequencing. Therefore, we aim to evaluate the microbial community in patients with achalasia and investigate the alterations in the microbial composition before and after POEM for treating achalasia.
Materials and Methods
Participant Information
Twenty-nine patients in total were prospectively enrolled from 4 referral institutions across Korea. Written informed consent was obtained from all patients before their enrollment in the study. The inclusion criteria were as follows: (1) patients who were diagnosed with achalasia and planned to undergo POEM, and (2) patients in the age range of 20-80 years. The exclusion criteria were as follows: (1) patients with a history of previous treatment for achalasia except medication; (2) patients with a history of chest and gastrointestinal surgeries, (3) patients with prior treatment with proton pump inhibitors or antibiotics within 8 weeks, (4) patients with the presence of an active infection in the oral cavity, (5) patients with a history of malignancy, (6) patients with a history of bleeding tendency or intake of antithrombotics, and (7) patients with the presence of a systematic disease who were receiving treatment. The study protocol was approved by the institutional review board of each participating institution (IRB No. 3-2018-0246, KCT0005797, KC18TCDI0507, 2018-07-001, 2019-0109).
Demographic Characteristics and Nutrient Intake Status
General information of patients was obtained in terms of their age, sex, height and weight, body mass index, comorbidities (diabetes, hypertension, pulmonary disease, heart disease, and kidney disease), smoking condition (non-smoker, ex-smoker, and current smoker), and alcohol consumption. The dietary survey was conducted as a face-to-face interview, and the eating habits and food intake questionnaire were designed to be an open-ended survey for reporting various foods using the 3 days 24 hours recall method with various measuring aids.15,16 The consumed food was analyzed using the Computer Aided Nutritional Analysis Program for Professionals 5.0 (CAN-Pro 5.0, The Korean Nutrition Society, Seoul, Korea) based on the intake of individual nutrients. Changes in the nutrient intake pre- and post-POEM were analyzed as the amount of daily energy, energy nutrients (carbohydrate, protein, and fat), fiber, vitamins (vitamin A, vitamin D, vitamin E, vitamin K, vitamin C, vitamin B1, vitamin B2, vitamin B6, vitamin B12, niacin, folic acid, and pantothenic acid), and minerals (calcium, phosphorous, sodium, potassium, magnesium, iron, zinc, and selenium).
Sample Collection and Preparation
We collected 2 samples, esophageal mucosal biopsies and esophageal retention fluid. We developed collecting methods especially for this research project to avoid potential contamination. First of all, we used gloves and followed aseptic procedures with sterile equipment. A standard video-endoscope was passed through the mouth to the esophagus without touching the oropharynx as carefully as possible and we avoided unnecessary suction. The stasis liquid in the esophageal lumen was aspirated via an endoscopic working channel. We connected tubing (disposable specimen trap) which was used in bronchoscopy (HS-SP-50; Hyupsung Medical Co, LTD, Seoul, Korea) for fluid collection at the entrance of the working channel to avoid contamination. After finishing the fluid collection, biopsies were then taken 2 cm above the squamocolumnar junction using sterile disposable biopsy forceps (EndoJaw; Olympus Co, Ltd, Tokyo, Japan). If there was no retention fluid in esophagus after POEM, we aspirated by washing the esophageal lumen with 10 mL of sterile saline solution. We collected samples twice on the day before or on the day of the POEM and 8 weeks after POEM. The biopsy and fluid samples were placed in a test tube (SNP BiomCare; SNP Genetics Inc, Seoul, Korea) and stored at –70°C. We administered broad-spectrum intravenous antibiotics 30 minutes before the start of the POEM. During the POEM procedure, prophylactic antibiotic solution spraying inside the tunnel was performed before closing the mucosal entry. The use of intravenous antibiotics was discontinued 3 days after the POEM and antibiotics were switched to oral intake for another 7 days.
DNA Extraction and 16S Ribosomal RNA Gene Amplification Sequence Processing
The biopsy and fluid samples (3-10 mL) were centrifuged at 15 000 g for 20 minutes at 4°C to separate the cellular pellet and the cell-free supernatant. DNA was extracted from the pellet using a QIAamp DNA Microbiome Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions.
The V3-V4 region of the 16S rRNA gene was amplified using the 341F (5’ TCG GCA GCG TCA GAT GTG TAT AAG AGA CAG CCT ACG GGN GGC WGC AG 3’) and 805R (5’ GTC TCG TGG GCT CGG AGA TGT GTA TAA GAG ACA GGA CTA CHV GGG TAT CTA ATC C 3’) primers with added Illumina adaptor overhang sequences. The amplicons were purified using a magnetic bead-based clean-up system (Agencourt AMPure XP; Beckman Coulter, Brea, CA, USA). Indexed libraries were prepared by limited-cycle PCR using the Nextera technology, and the samples obtained were further cleaned up and pooled at equimolar concentrations. The final library was denatured with 0.2 N NaOH and diluted to 6 pM with a 20% PhiX control. Sequencing was performed on the Illumina MiSeq platform using a 2 × 300 bp paired-end protocol according to the manufacturer’s instructions.
Bioinformatics and Statistical Methods
The primary analysis of the obtained sequences consisted of their demultiplexation with the MiSeqReporter software (Illumina). The paired-end sequences of each sample were then exported from the MiSeq system for analysis in the FASTQ format. Bioinformatic analysis of the sequences was performed using the QIIME2 package.17 For subsequent data analysis, we used a web-based platform called MicrobiomeAnalyst.18 The sequences were then clustered against the 2013 Greengenes (13_5 release) 97% reference dataset of the ribosomal database.19 The UCLUST algorithm was used to cluster sequences that did not match any entries in this reference into de novo operational taxonomic units (OTUs) at 97% similarity. The OTU table was rarefied to a sequencing depth of 20 000 per sample for subsequent analyses.
Taxonomic analyses were performed after collapsing the OTUs at the genus level. The alpha diversity of each sample was assessed using the Shannon index. Beta diversity was determined based on the Bray-Curtis index distance method using permutational MANOVA, and principal coordinate analysis plots were constructed. The statistical significance of the esophageal microbiota structure between different sampling sites and gut regions was assessed by the non-parametric univariate Mann-Whitney/Kruskal-Wallis test. The linear discriminant analysis effect size (LEfSe) was used to identify significantly different abundances in the bacterial taxa. The false discovery rate correction was used for multiple tests. A P-value of < 0.05 was considered statistically significant.
Results
Characteristics of the Studied Cohort
Between October 2018 and December 2019, we enrolled 29 patients who underwent POEM for achalasia. The baseline characteristics are shown in Table 1. The subtypes of achalasia were 12 (41.4%), 10 (34.5%), and 4 (13.8%) for type I, II, and III, respectively. The subtype of achalasia in the other 3 patients could not be differentiated. The median pre-POEM Eckardt score was 6.0 (range, 1-11) and the median symptom duration was 24 months (range, 3-300 months).
Table 1.
Baseline Characteristics
| Variables | |
|---|---|
| Age (yr) | 48.1 ± 15.7 |
| Sex | |
| Male | 15 (51.7) |
| Female | 14 (48.3) |
| Height (cm) | 165.5 ± 7.7 |
| Weight (kg) | 61.6 ± 12.4 |
| BMI (kg/m2) | 22.5 ± 4.3 |
| Comorbidities | |
| Diabetes mellitus | 2 (6.9) |
| Hypertension | 4 (13.8) |
| Pulmonary disease | 0 (0.0) |
| Heart disease | 1 (3.4) |
| Kidney disease | 0 (0.0) |
| Smoking | |
| Non-smoker | 24 (82.8) |
| Ex-smoker | 2 (6.9) |
| Current smoker | 3 (10.3) |
| Alcohol | 11 (37.9) |
| Achalasia type | |
| I | 12 (41.4) |
| II | 10 (34.5) |
| III | 4 (13.8) |
| Not available | 3 (10.3) |
| Eckardt score | 6.0 (1-11) |
| Symptom duration (mo) | 24 (3-300) |
BMI, body mass index.
Data are presented as mean ± SD, n (%), or median (range).
Nutrient Intake in Patients With Achalasia Pre- and Post-peroral Endoscopic Myotomy
The results of the change in nutrient intake status are shown in Table 2. The usual energy intake of the patients increased from 1145.9 ± 358.8 kcal to 1645.6 ± 309.6 kcal after POEM. As the overall food intake increased, the intake status of all nutrients significantly increased too, except the intake status of vitamin E.
Table 2.
Changes of Nutrient Intakes per Daily According to Pre- and Post-peroral Endoscopic Myotomy
| Variables | Pre-POEM | Post-POEM | P-value |
|---|---|---|---|
| Energy (kcal) | 1145.9 ± 358.8 | 1645.6 ± 309.6 | < 0.001 |
| Carbohydrate (g) | 184.0 ± 60.0 | 253.8 ± 46.1 | < 0.001 |
| Protein (g) | 37.2 ± 10.7 | 65.9 ± 14.6 | < 0.001 |
| Fat (g) | 26.1 ± 13.7 | 38.9 ± 14.9 | 0.011 |
| Fiber (g) | 13.3 ± 6.9 | 20.6 ± 6.5 | 0.002 |
| Vitamin A (µg RE) | 186.1 ± 126.3 | 430.7 ± 220.2 | < 0.001 |
| Vitamin D (µg) | 1.6 ± 2.5 | 4.4 ± 3.5 | 0.008 |
| Vitamin E (mg α-TE) | 10.8 ± 7.1 | 14.5 ± 6.3 | 0.112 |
| Vitamin K (µg) | 76.5 ± 78.5 | 237.9 ± 293.4 | 0.031 |
| Vitamin C (mg) | 42.3 ± 45.6 | 84.3 ± 50.1 | 0.013 |
| Vitamin B1 (mg) | 0.9 ± 0.5 | 1.5 ± 0.5 | 0.001 |
| Vitamin B2 (mg) | 0.9 ± 0.4 | 1.4 ± 0.4 | 0.002 |
| Niacin (mg) | 8.0 ± 2.8 | 12.1 ± 5.2 | 0.005 |
| Vitamin B6 (mg) | 1.0 ± 0.4 | 2.5 ± 2.7 | 0.024 |
| Folic acid (µg) | 282.2 ± 156.3 | 498.7 ± 186.0 | 0.001 |
| Vitamin B12 (µg) | 6.8 ± 6.7 | 11.2 ± 8.1 | 0.085 |
| Pantothenic acid (mg) | 2.6 ± 1.2 | 4.8 ± 1.2 | < 0.001 |
| Calcium (mg) | 303.1 ± 165.6 | 500.0 ± 221.7 | 0.005 |
| Phosphorous (mg) | 579.4 ± 246.0 | 1010.6 ± 225.6 | < 0.001 |
| Sodium (mg) | 2399.1 ± 1569.4 | 3702.1 ± 1009.6 | 0.006 |
| Potassium (mg) | 1529.5 ± 808.1 | 2806.4 ± 1059.4 | < 0.001 |
| Magnesium (mg) | 57.5 ± 37.1 | 114.9 ± 56.2 | 0.001 |
| Iron (mg) | 9.4 ± 2.5 | 15.1 ± 4.7 | < 0.001 |
| Zinc (mg) | 6.0 ± 1.7 | 11.0 ± 3.7 | < 0.001 |
| Selenium (µg) | 41.7 ± 21.8 | 64.0 ± 34.1 | 0.026 |
POEM, peroral endoscopic myotomy; RE, retinol equivalents; α-TE, α-tocopherol equivalents.
Data are presented as mean ± SD.
Esophageal Microbiota in Patients With Achalasia Pre- and Post-peroral Endoscopic Myotomy
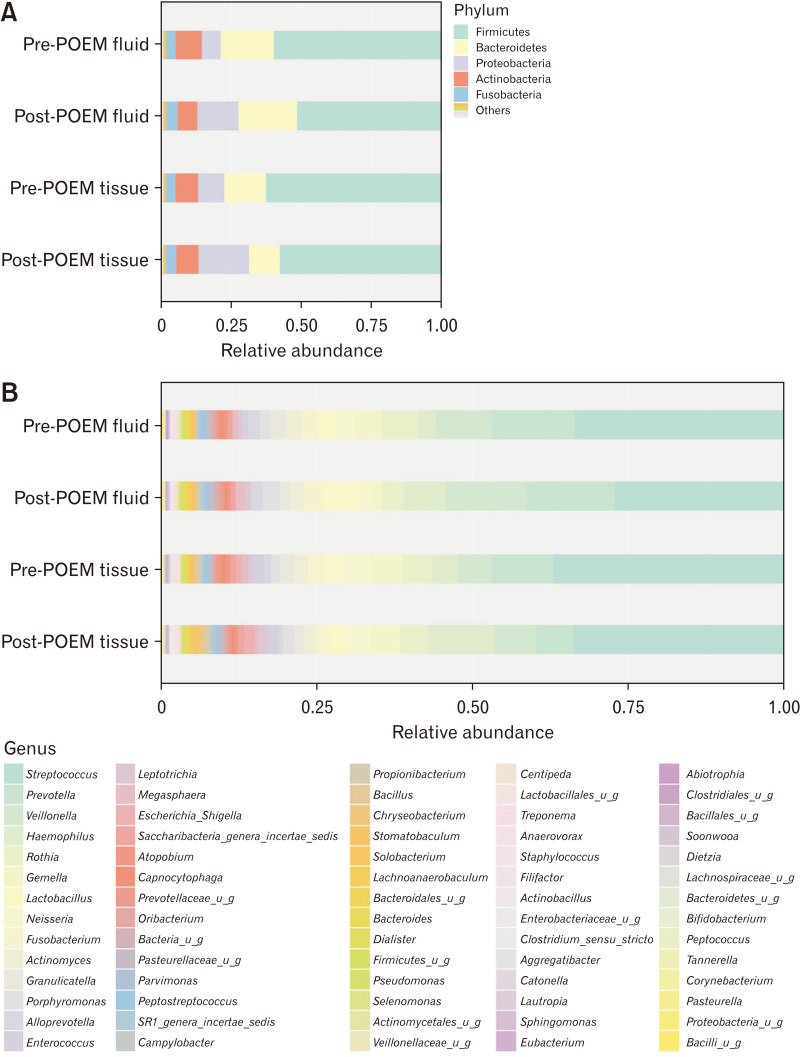
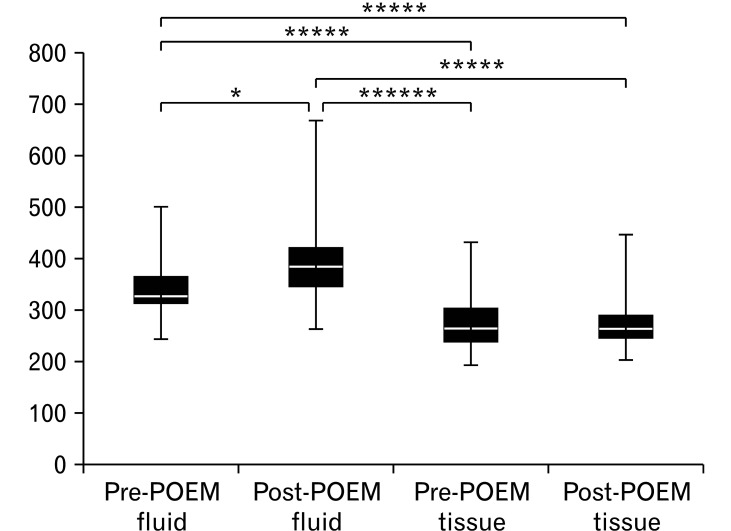
Among the 29 patients, we collected 105 samples (29 mucosal biopsies and 30 retention fluid samples before POEM and 23 mucosal biopsies and fluid retention samples after POEM). After the quality control, which excluded samples with no amplification or low-quality sequence data, the microbiome of 99 samples were subjected to bioinformatic analysis (25 mucosal biopsies and 30 retention fluid samples before POEM and 22 mucosal biopsies and fluid retention samples after POEM). The observation of the overall microbial composition showed that Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria, and Fusobacteria were the dominant phyla, representing over 95% of the total phyla in all groups (Fig. 1A). Firmicutes was the most abundant bacterial phylum in the pre-and post-POEM fluid samples and the pre- and post-POEM tissues, followed by phyla Bacteroidetes and Proteobacteria (Table 3). At the genus level (Fig. 1B), Streptococcus was the most abundant genus in the tissue and fluid samples pre- and post-POEM. Figure 2 shows that the observed OTU number was significantly higher among the fluid samples than in the tissue samples.
Figure 1.
Bar plot of taxonomy profile at (A) phylum level and (B) genus level. POEM, peroral endoscopic myotomy; u_g, unclassified genus.
Table 3.
Most Common Bacterial Phyla According to in the Tissue and Fluid Samples Pre- and Post-peroral Endoscopic Myotomy
| Samples | Firmicutes | Bacteroidetes | Actinobacteria | Proteobacteria | Fusobacteria |
|---|---|---|---|---|---|
| Fluid | |||||
| Pre-POEM | 60% | 19% | 10% | 7% | 3% |
| Post-POEM | 51% | 21% | 7% | 15% | 4% |
| Tissue | |||||
| Pre-POEM | 63% | 15% | 8% | 9% | 3% |
| Post-POEM | 58% | 11% | 8% | 18% | 4% |
Figure 2.
Boxplot of operational taxonomic units in the tissue and fluid samples pre- and post-peroral endoscopic myotomy (POEM) of observed genera. *P < 0.1, *****P < 0.00001, ******P < 0.000001.
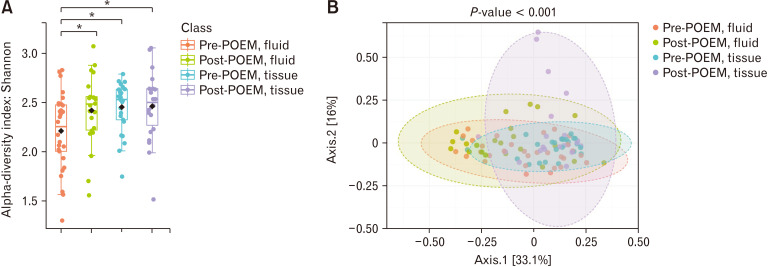
We evaluated the alpha diversity using Shannon analysis (Fig. 3A) and the beta diversity using the Bray-Curtis index (Fig. 3B) among the 4 groups. The results indicated that the structure of the esophageal microbiome in the tissue samples was significantly different from that of the fluid samples.
Figure 3.
Differences in alpha and beta diversity in esophageal microbiome (A) alpha diversity (Shannon index) and (B) beta diversity (Bray-Curtis index) The alpha diversity presented as Shannon index. Each dot represents an individual sample. Pink bars represent pre-peroral endoscopic myotomy (POEM), fluid; green bars represent post-POEM, fluid; blue bars represent pre-POEM, tissue; and purple bars represent post-POEM, tissue. Data are presented as box-and-whisker plots, with whiskers representing the lowest and highest values within 1.5*interquartile range (IQR). Black diamonds indicates the mean of Shannon index. *P < 0.05 as determined by two-way ANOVA followed by Tukey’s post hoc test.
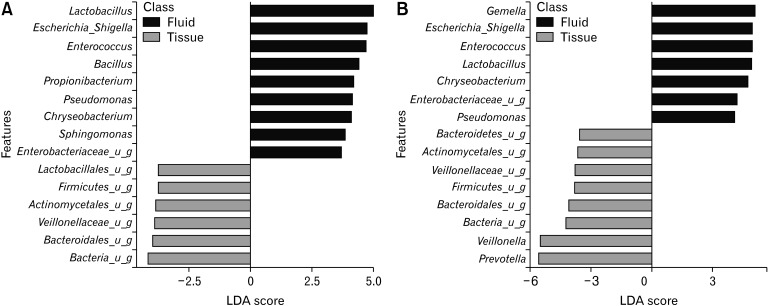
To identify the specific bacterial taxa associated with the tissue and the fluid samples, we compared their esophageal microbiota using LEfSe. Figure 4A shows that 15 genera or species distinguished the esophageal microbial communities between the 2 groups pre-POEM according to the criteria of linear discriminant analysis ≥ 2.5 at P < 0.05. Lactobacillus, Escherichia_Shi, Enterococcus, Bacillus, Propionibacterium, Pseudomonas, Chryseobacterium, Sphingomonas, and Enterobacteriaceae were enriched in the tissue samples. Among the fluid samples, Lactobacillales, Actinomycetales, Veillonellaceae, an unknown genera of Firmicutes, Bacteroidales, and other bacteria were enriched. In the post-POEM group, Gemella, Escherichia_Shi, Enterococcus, Lactobacillus, Chryseobacterium, Enterobacteriaceae, and Pseudomonas were enriched in the tissues. Among the fluid samples, Actinomycetales, Veillonellaceae, Veillonella, Prevotella, an unknown genera of Bacteroidetes, Firmicutes, Bacteroidetes, and other bacteria were enriched after POEM (Fig. 4B).
Figure 4.
The linear discriminant analysis (LDA) effect size of the esophageal microbiome in the tissue and fluid samples pre- and post-peroral endoscopic myotomy (POEM) (genus level). (A) pre-POEM, (B) post-POEM. u_g, unclassified genus.
Discussion
Achalasia causes esophageal stasis and delayed esophageal clearance. The stagnated residues in the esophagus create a propitious environment for the growth of microbiota. In particular, a previous study reported that Veillonella, an anaerobic bacteria, was the predominant organism in the low oxygen environment of achalasia-associated esophagi. In addition, the concentration of microorganisms increased in esophagi on after increased esophageal dilation in patients with achalasia.14 Until now, there is an only one study for esophageal microbiome in achalasia. Although we did not evaluate the esophageal microbiota according to the degree of dilatation of the esophagus in this study, this is the first study to show the esophageal microbiota in patients with achalasia using 16S rRNA sequencing to the best of our knowledge. Therefore, we aimed to provide evidence that support the association between achalasia and microbial composition in the esophagus of patients with achalasia. However, there were no differences in the esophageal microbiota community composition in patients with achalasia after POEM. We performed subgroup analysis according to achalasia type. Among 29 patients, 12 patients are achalasia type I, 10 patients are II, and 4 patients are III, respectively. However, there were no differences in the esophageal microbiota between different types.
The study of esophageal microbiota has advanced from a culture to a culture-independent method. In addition, the methods used to collect esophageal microbiota samples are varied – from biopsies, aspirations, brushes, and esophageal string tests.20 In this study, we collected esophageal samples from patients with achalasia in 2 ways: biopsy and aspiration. Most studies have evaluated the esophageal microbiome using tissue biopsies, which would be the most suitable method, considering the presence of an adherent microbiome at mucosa of esophagus. However, sampling by brushes or aspirates was adopted and this was a less invasive method and used as an alternative to esophageal biopsies. However, esophageal biopsies are always necessary for a direct histological assessment of the diseased tissues in order to avoid misclassification and decrease the risk of contamination by oropharyngeal or gastric secretions. In this study, the bacterial communities in the esophageal tissue and fluid groups pre- and post-POEM were structurally different, with the alpha diversity (OTU number) decreasing from the fluid to tissue samples. Previous studies have reported that reduced microbial diversity, in itself, is a disease status, such as inflammation or cancer.21,22 Therefore, the microbiota of the esophageal tissues may play a pivotal role in patients with achalasia, as the microbiota of the esophageal fluid group is translocated from the oropharynx. Using LEfSe, we identified specific bacterial taxa associated with the esophageal tissue in patients with achalasia, which were the genera Lactobacillus, Escherichia_Shi, Enterococcus, Bacillus, Propionibacterium, Pseudomonas, Chryseobacterium, Sphingomonas, and Enterobacteriaceae. Interestingly, our results showed that at the genus level, Lactobacillus and Enterobacteriaceae were enriched in the esophageal tissue samples, which are abundant in esophageal adenocarcinoma.20 Whereas in the esophageal fluid, the genus Veillonella was abundant, which is seen to be increased in GERD and Barrett’s esophagus.20 Although there is a controversy, regurgitation is frequently observed in patients who have achalasia. Several studies utilizing 24-hour pH monitoring show that untreated achalasia patients experience true reflux. The reasons are that there is a portion of overlap between achalasia and GERD and the refluxed gastric contents in patients with achalasia may be poorly cleared from a dysfunctional esophagus, causing substantial retention. The prevalence of squamous cell carcinoma in patients with achalasia is higher than that in the general population.14 A possible explanation for this phenomenon could be chronic esophagitis due to stasis and bacterial overgrowth.23 The metabolism of nitrates by bacterial species could be involved in carcinogenesis. However, the association between the esophageal microbiota and cancer needs to be studied in future research.
This is the first study to show the esophageal microbiota in patients with achalasia using 16S rRNA sequencing to the best of our knowledge. The most common bacterial phyla in normal esophagus are Firmicutes (70-87%), Bacteroidetes (5-20%), Proteobacteria (2-5%), Actinobacteria (2-4%), Fusobacteria (1-2%), and TM7 (0-1%).24,25 And in other studies, the most common bacterial taxa in a normal esophagus include Streptococcus, Haemophilus, Neisseria, Prevotella, and Veillonella.20 In Barrett’s esophagus, the most common bacterial phyla are Firmicutes (50-55%), Proteobacteria (20-22%), Bacteroidetes (14-19%), Fusobacteria (2-9%), Actinobacteria (2-7%), and TM7 (0-1%).3,25 Streptococcus is the representative bacterial taxa of Firminutes phylum. Pasteurella, Haemophilus, Fusobacterium, Prevotella, and Neisseria were more abundant in the reflux esophagitis group than in the normal group. In esophageal adenocarcinoma, the proportion of Firmicutes phylum decreased and proportion of Lactobacillus, Enterobacteriaceae, and Akkermansia increased in patients with esophageal adenocarcinoma. Until now, there is an only 1 study regarding esophageal microbiome in achalasia. In patients with achalasia, Streptococcus and Veillonella were found to be the most abundant genera using the culture method.14 In this study, the predominant taxa was Firmicutes among all groups, although we did not evaluate the esophageal microbiota of healthy controls. Lactobacillus and Enterobacteriaceae were enriched in the esophageal tissue samples, which are abundant in esophageal adenocarcinoma. Whereas in esophageal fluid, the genus Veillonella was abundant, which is seen to be increased in GERD and Barrett’s esophagus.
Many factors affect the bacterial composition in the esophagus such as aging, dietary fiber intake, proton pump inhibitors, and diseases that cause esophageal dysbiosis.26,27 We conducted a dietary survey of patients with achalasia in this study. It is reported that many patients with achalasia complain of discomfort such as dysphagia, vomiting, decreased appetite, and chest pain, which can lead to weight loss and nutritional deficiencies.28,29 As a result of the survey analysis, nutrient intake pre-POEM was insufficient, and the nutrient intake was significantly improved after POEM in most patients. However, a long-term lack of energy and protein intake can have a direct effect on weight and muscle loss.30 In addition, even after POEM, there were still many nutrients (energy, vitamin A, vitamin C, vitamin B2, niacin, calcium, and magnesium) whose levels in patients did not meet nutritional recommendations.31 Although this study did not analyze the relationship between food intake and the microbiota, the nutrient intake status results show the need for early nutritional intervention and management in patients with achalasia. In addition, decreased acid reflux by PPI administration could affect the esophageal microbiomes. Amir et al32 reported that esophageal microbiome in patients with Barrett’s esophagitis, or a normal distal esophagus, was assessed from distal esophageal biopsies, comparing results before versus after PPIs. After PPIs were administered, Lachnospiraceae, Comamonadaceae, and unclassified Clostridial families significantly increased in distal esophagus.
In this study, the median Eckardt score decreased from 6.0 to 0.0, and all patients achieved clinical success (Eckardt score ≤ 3). However, there were no significant alterations of the esophageal microbiota in patients with achalasia pre- and post-POEM. We hypothesize that the small sample size and short follow-up period (8 weeks) in our study may have contributed to the results observed concerning the esophageal microbiota in patients with achalasia. Improved esophageal clearance and acid reflux has been observed in patients after POEM. According to the survey conducted, we know that there is an alteration of food intake in patients post POEM. Therefore, we think that there is a possibility of alterations in the esophageal microbiota in the long term. However, the follow-up period of 8 weeks may not be sufficient to induce esophageal microbiota alteration. In addition, although POEM treatment improves esophageal clearance, the unique esophageal microbiota composition in patients with achalasia may not change because food stasis may still persist to a small extent even after POEM. Therefore, further research is needed to demonstrate whether the esophageal microbiota in patients with achalasia changes in the long term after POEM.
This study has some limitations. First, we did not investigate the microbiome in the oral cavity of the patients and healthy controls. Therefore, we did not determine the origin and role of the oesophageal microbiota in patients with achalasia. Second, the sample size was not sufficient for subgroup analysis according to the type, symptom duration, and severity of disease. Finally, we performed the rarefraction. Rarefying has been criticized as a normalization technique because data can be omitted through the exclusion of either excess sequences or entire samples, depending on the rarefied library size selected.33 However, from another perspective, rarefying enables (1) characterization of the variation introduced to diversity analyses by this random subsampling, and (2) selection of smaller library sizes where necessary to incorporate all samples in the analysis.34 In this study, subsampling via rarefying was also a necessary process to include all possible samples in the analysis.
In conclusion, this study determined the unique oesophageal microbial composition of patients with achalasia by 16S rRNA gene sequencing, and found that this unique esophageal microbial composition did not significantly change in the short-term after POEM despite a significant improvement in the nutritional intake. These esophageal microbial data by 16S rRNA gene sequencing in patients with achalasia will provide novel insights for further research on achalasia.
The sequencing output was uploaded to NCBI SRA archive (Accession No. PRJNA756810).
Footnotes
Financial support: This study was financially supported by a faculty research grant from Yonsei University College of Medicine (6-2017-0154), and by a grant from the Korean Society of Neurogastroenterology and Motility for 2019.
Conflicts of interest: None.
Author contributions: In planning and/or conducting the study, and collecting and/or interpreting data: Da Hyun Jung, Young Hoon Youn, Do Hoon Kim, Chul-Hyun Lim, Hee-Sook Lim, Hee Seok Moon, Ju Yup Lee, Hyojin Park, and Su Jin Hong; and drafting the manuscript: Da Hyun Jung, Young Hoon Youn, and Su Jin Hong.
References
- 1.Corning B, Copland AP, Frye JW. The esophageal microbiome in health and disease. Curr Gastroenterol Rep. 2018;20:39. doi: 10.1007/s11894-018-0642-9. [DOI] [PubMed] [Google Scholar]
- 2.Hunt RH, Yaghoobi M. The esophageal and gastric microbiome in health and disease. Gastroenterol Clin North Am. 2017;46:121–141. doi: 10.1016/j.gtc.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 3.Pei Z, Bini EJ, Yang L, Zhou M, Francois F, Blaser MJ. Bacterial biota in the human distal esophagus. Proc Natl Acad Sci USA. 2004;101:4250–4255. doi: 10.1073/pnas.0306398101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.May M, Abrams JA. Emerging Insights into the esophageal microbiome. Curr Treat Options Gastroenterol. 2018;16:72–85. doi: 10.1007/s11938-018-0171-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Di Pilato V, Freschi G, Ringressi MN, Pallecchi L, Rossolini GM, Bechi P. The esophageal microbiota in health and disease. Ann N Y Acad Sci. 2016;1381:21–33. doi: 10.1111/nyas.13127. [DOI] [PubMed] [Google Scholar]
- 6.Pei Z, Yang L, Peek RM, Levine SM, Jr, Pride DT, Blaser MJ. Bacterial biota in reflux esophagitis and barrett's esophagus. World J Gastroenterol. 2005;11:7277–7283. doi: 10.3748/wjg.v11.i46.7277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fillon SA, Harris JK, Wagner BD, et al. Novel device to sample the esophageal microbiome--the esophageal string test. PLoS One. 2012;7:e42938. doi: 10.1371/journal.pone.0042938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gagliardi D, Makihara S, Corsi PR, et al. Microbial flora of the normal esophagus. Dis Esophagus. 1998;11:248–250. doi: 10.1093/dote/11.4.248. [DOI] [PubMed] [Google Scholar]
- 9.Norder Grusell E, Dahlén G, Ruth M, et al. Bacterial flora of the human oral cavity, and the upper and lower esophagus. Dis Esophagus. 2013;26:84–90. doi: 10.1111/j.1442-2050.2012.01328.x. [DOI] [PubMed] [Google Scholar]
- 10.Jung DH, Park H. Is gastroesophageal reflux disease and achalasia coincident or not? J Neurogastroenterol Motil. 2017;23:5–8. doi: 10.5056/jnm16121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Husebye E. Gastrointestinal motility disorders and bacterial overgrowth. J Intern Med. 1995;237:419–427. doi: 10.1111/j.1365-2796.1995.tb01196.x. [DOI] [PubMed] [Google Scholar]
- 12.Park CH, Jung DH, Kim DH, et al. Comparative efficacy of per-oral endoscopic myotomy and heller myotomy in patients with achalasia: a meta-analysis. Gastrointest Endosc. 2019;90:546–558. e3. doi: 10.1016/j.gie.2019.05.046. [DOI] [PubMed] [Google Scholar]
- 13.Yoon HJ, Lee JE, Jung DH, Park JC, Youn YH, Park H. Morphologic restoration after peroral endoscopic myotomy in sigmoid-type achalasia. J Neurogastroenterol Motil. 2020;26:67–73. doi: 10.5056/jnm19144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pajecki D, Zilberstein B, dos Santos MA, et al. Megaesophagus microbiota: a qualitative and quantitative analysis. J Gastrointest Surg. 2002;6:723–729. doi: 10.1016/S1091-255X(02)00028-8. [DOI] [PubMed] [Google Scholar]
- 15.Asakuma Y, Matsui S, Kawasaki M, Sakurai T, Kashida H, Kudo M. Prevention of delayed bleeding after endoscopic submucosal dissection (ESD) for gastric tumors. 2011. [DOI] [Google Scholar]
- 16.Salvador Castell G, Serra-Majem L, Ribas-Barba L. What and how much do we eat? 24-hour dietary recall method. Nutr Hosp. 2015;31(suppl 3):46–48. doi: 10.3305/nh.2015.31.sup3.8750. [DOI] [PubMed] [Google Scholar]
- 17.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dhariwal A, Chong J, Habib S, King IL, Agellon LB, Xia J. MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 2017;45:W180–W188. doi: 10.1093/nar/gkx295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeSantis TZ, Hugenholtz P, Larsen N, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park CH, Lee SK. Exploring esophageal microbiomes in esophageal diseases: a systematic review. J Neurogastroenterol Motil. 2020;26:171–179. doi: 10.5056/jnm19240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ren Z, Jiang J, Xie H, et al. Gut microbial profile analysis by MiSeq sequencing of pancreatic carcinoma patients in China. Oncotarget. 2017;8:95176–95191. doi: 10.18632/oncotarget.18820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gevers D, Kugathasan S, Denson LA, et al. The treatment-naive microbiome in new-onset crohn's disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loviscek LF, Cenoz MC, Badaloni AE, Agarinakazato O. Early cancer in achalasia. Dis Esophagus. 1998;11:239–247. doi: 10.1093/dote/11.4.239. [DOI] [PubMed] [Google Scholar]
- 24.Arthur JC, Perez-Chanona E, Mühlbauer M, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338:120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mannell A, Plant M, Frolich J. The microflora of the oesophagus. Ann R Coll Surg Engl. 1983;65:152–154. [PMC free article] [PubMed] [Google Scholar]
- 26.Deshpande NP, Riordan SM, Castaño-Rodríguez N, Wilkins MR, Kaakoush NO. Signatures within the esophageal microbiome are associated with host genetics, age, and disease. Microbiome. 2018;6:227. doi: 10.1186/s40168-018-0611-4.bb788bf818b64abe9859936a3dbc92ff [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nobel YR, Snider EJ, Compres G, et al. Increasing dietary fiber intake is associated with a distinct esophageal microbiome. Clin Transl Gastroenterol. 2018;9:199. doi: 10.1038/s41424-018-0067-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Milito P, Aquilino K, Lazzari V, et al. The malnutrition universal screening tool can predict malnutrition in patients with esophageal achalasia. Eur J Gastroenterol Hepatol. 2020;32:1135–1140. doi: 10.1097/MEG.0000000000001798. [DOI] [PubMed] [Google Scholar]
- 29.Cappell MS, Stavropoulos SN, Friedel D. Updated systematic review of achalasia, with a Focus on POEM therapy. Dig Dis Sci. 2020;65:38–65. doi: 10.1007/s10620-019-05784-3. [DOI] [PubMed] [Google Scholar]
- 30.Newberry C, Vajravelu RK, Pickett-Blakely O, Falk G, Yang YX, Lynch KL. Achalasia patients are at nutritional risk regardless of presenting weight category. Dig Dis Sci. 2018;63:1243–1249. doi: 10.1007/s10620-018-4985-8. [DOI] [PubMed] [Google Scholar]
- 31.Ministry of health and welfare, author. Dietary reference intakes for Koreans 2015. Korean Nutrition Society; Seoul: 2015. [DOI] [Google Scholar]
- 32.Amir I, Konikoff FM, Oppenheim M, Gophna U, Half EE. Gastric microbiota is altered in oesophagitis and Barrett's oesophagus and further modified by proton pump inhibitors. Environ Microbiol. 2014;16:2905–2914. doi: 10.1111/1462-2920.12285. [DOI] [PubMed] [Google Scholar]
- 33.McMurdie PJ, Holmes S. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol. 2014;10:e1003531. doi: 10.1371/journal.pcbi.1003531.23df78e998f94d62ab52c12b001f49e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cameron ES, Schmidt PJ, Tremblay BJM, Emelko MB, Müller KM. Enhancing diversity analysis by repeatedly rarefying next generation sequencing data describing microbial communities. Scientific Reports. 2021;11:22302. doi: 10.1038/s41598-021-01636-1.ac2f8ee28b1a482b837a75acc69d8a76 [DOI] [PMC free article] [PubMed] [Google Scholar]