SUMMARY
Proteostasis of the endoplasmic reticulum (ER) is controlled by sophisticated signaling pathways that are collectively called the unfolded protein response (UPR) and are initiated by specialized ER membrane-associated sensors. The evidence that complete loss-of-function mutations of the most conserved of the UPR sensors, inositol-requiring enzyme 1 (IRE1), dysregulates tissue growth and development in metazoans and plants raises the fundamental question as to how IRE1 is connected to organismal growth. To address this question, we interrogated the Arabidopsis primary root, an established model for organ development, using the tractable Arabidopsis IRE1 mutant ire1a ire1b, which has marked root development defects in the absence of exogenous stress. We demonstrate that IRE1 is required to reach maximum rates of cell elongation and root growth. We also established that in the actively growing ire1a ire1b mutant root tips the Target of Rapamycin (TOR) kinase, a widely conserved pro-growth regulator, is hyperactive, and that, unlike cell proliferation, the rate of cell differentiation is enhanced in ire1a ire1b in a TOR-dependent manner. By functionally connecting two essential growth regulators, these results underpin a novel and critical role of IRE1 in organ development and indicate that, as cells exit an undifferentiated state, IRE1 is required to monitor TOR activity to balance cell expansion and maturation during organ biogenesis.
Keywords: unfolded protein response, IRE1, TOR, development, differentiation, Arabidopsis thaliana
INTRODUCTION
The endoplasmic reticulum (ER) is responsible for the synthesis of one third of the cellular proteome. Therefore, its biosynthetic capacity is constantly monitored by a set of ER membrane-associated sensors that can upregulate the synthesis of ER protein chaperones and ER membrane while also limiting the rate of protein translation (Han and Kaufman, 2017; Ron and Walter, 2007). The unfolded protein response (UPR) sensors conserved between metazoans and plants include the ER membrane protein kinase and ribonuclease IRE1 and ER membrane-tethered transcription factors (TFs) (metazoan ATF6 and plant bZIP17 and bZIP28) (Angelos et al., 2017; Pastor-Cantizano et al., 2020). Through its ribonuclease domain, IRE1 catalyzes the unconventional cytosolic splicing of mRNA encoding a TF, XBP1 in mammalian cells, bZIP60 in Arabidopsis, and Hac1 in yeast (Calfon et al., 2002; Kawahara et al., 1997; Nagashima et al., 2011). In addition to the unconventional splicing of the mRNA encoding target TFs, IRE1 degrades cytosolic mRNAs through a process known as regulated IRE1-dependent decay (RIDD) to preserve cell proteostasis (Hollien et al., 2009; Mishiba et al., 2013).
Exogenous stress factors, such as hypoxia and metabolic stress in metazoans (Hetz and Papa, 2018) as well as heat stress (Gao et al., 2008), pathogen attack (Guillemette et al., 2014; Zhang et al., 2015), and singlet oxygen generation (Beaugelin et al., 2020) in plants, are known to activate the UPR sensors. Interestingly, these sensors are also activated by endogenous cellular cues during physiological development (Mitra and Ryoo, 2019). For example, the mammalian IRE1 has critical functions in placental and liver development during embryogenesis and during the differentiation of antibody-secreting B-lymphocytes (Reimold et al., 2000, 2001). In Drosophila, IRE1 activity is required for the development of the digestive tract (Huang et al., 2017). In Xenopus and medaka fish, IRE1 is required for proper notochord formation and hatching gland development, respectively (Ishikawa et al., 2017; Tanegashima et al., 2009). In these metazoan models, complete IRE1 loss-of-function mutations are embryo or larval lethal (Mitra and Ryoo, 2019).
In Arabidopsis there are three homologs of the IRE1 gene, IRE1a, IRE1b, and IRE1c, which perform only partially overlapping functions (Mishiba et al., 2019; Pu et al., 2019). IRE1c is expressed primarily in reproductive tissues during gametogenesis, whereas IRE1a is primarily expressed in root tissues, and IRE1b is expressed at a similar level in nearly all tissue types (Pu et al., 2019). Single mutants of the Arabidopsis IRE1 homologs are phenotypically identical to wild-type (WT) plants under physiological conditions of growth; however, high-order mutations cause severe developmental defects. For example, the ire1b ire1c double mutant has a gamete lethal phenotype (Pu et al., 2019), and the ire1a ire1b double mutant, a functional IRE1 knockdown, reproduces normally but has a short root phenotype (Bao et al., 2019; Chen et al., 2014; Chen and Brandizzi, 2012; Deng et al., 2011). Similar to ire1b ire1c, the ire1a ire1b ire1c triple mutant is also gamete lethal (Mishiba et al., 2019), and plants that are heterozygous for ire1c and homozygous for ire1a and ire1b have severe developmental defects in all tissue types (Mishiba et al., 2019). On the whole, these phenotypes support that IRE1 performs critical functions to promote the growth and development of several Arabidopsis tissue types with some degree of specificity likely linked to the expression of the IRE1 isoforms in their respective tissues.
How Arabidopsis IRE1 controls tissue growth and development is completely unknown. In most metazoan model species, IRE1 primarily contributes to development through activation of XBP1 (Ishikawa et al., 2017; Reimold et al., 2000, 2001). Indeed, XBP1 null mutations are also embryo lethal and affect the development of the same tissue types as IRE1 mutations (Mitra and Ryoo, 2019). Surprisingly, loss-of-function mutations in Arabidopsis bZIP60 do not have any growth or reproductive phenotypes (Chen and Brandizzi, 2012; Moreno et al., 2012; Nagashima et al., 2011). Therefore, unlike the metazoan IRE1, the Arabidopsis IRE1 promotes organ growth through mechanisms that are independent of its canonical splicing target.
Similar to IRE1, the Target of Rapamycin (TOR) kinase is highly conserved across eukaryotes (Shi et al., 2018). TOR and its associated protein complexes act as cell regulatory hubs that integrate nutrient availability, energy status, hormone, and stress input signals to coordinate a wide variety of cellular activities ranging from cell proliferation and growth to metabolism and autophagy (Burkart and Brandizzi, 2020; Shi et al., 2018). While several of the key proteins in the TOR complex are conserved between plants and animals (such as LST8 and RAPTOR), the specific inputs and outputs have been evolutionarily adapted to meet organism-specific needs (Burkart and Brandizzi, 2020). Indeed, plant TOR receives activating signals from light availability via photosynthetic production of carbohydrates (photosynthates) and light-dependent synthesis of the plant hormone auxin (Chen et al., 2018; Li et al., 2017). Photosynthate-dependent activation of TOR via glucose is necessary and sufficient to activate root tips and promote cell division in root meristematic zones (MZs) via activation of E2F TFs (Xiong et al., 2013). TOR activity is also required for the polar growth of root hairs (Montané and Menand, 2013), which necessitates substantial synthesis of new cytosolic, membrane, and cell wall components (Ovečka et al., 2005; Retzer and Weckwerth, 2021). Nonetheless, how TOR activities are integrated into other aspects of development in actively growing roots is not well understood.
Despite the evidence that both IRE1 and TOR control growth, a functional connection between these essential regulators in the context of development has yet to be made. Earlier studies of chemically induced ER stress and other stress situations in metazoan cells have demonstrated that TOR activity can lead to an induction of the IRE1-JNK pro-apoptotic kinase signal cascade, (Kato et al., 2012, 2013; Shanware et al., 2014), as well as IRE1 inactivation (Sanchez-Alvarez et al., 2017). Nonetheless, to date, it is yet unknown whether IRE1 controls TOR activity under induced ER stress conditions or physiological conditions of growth in any model organism. To address these fundamental knowledge gaps, we used the tractable Arabidopsis ire1a ire1b model because it avoids the gamete lethality and extreme pleotropic phenotypes of other high-order UPR mutants (Kim et al., 2018; Mishiba et al., 2019). Furthermore, in physiological conditions of growth, the plant phenotype of this mutant is restricted to the root (Chen and Brandizzi, 2012; Ruberti et al., 2018). Due to their invariant cell ontogeny and cell organization the Arabidopsis root is an exquisite development model system (Scheres and Wolkenfelt, 1998), and is therefore suitable to investigate the role of IRE1 in tissue development. Through an in-depth characterization of ire1a ire1b root development in normal conditions of growth, we demonstrate that IRE1 is required to reach maximum rates of root growth afforded by prolonged photoperiods and high carbohydrate availability during the transition from the early seedling stage to the adult vegetative stage. A detailed analysis of this developmental transition carried out in this work indicates that in the root meristem IRE1 is required for the correct timing of cell elongation in a manner that is dependent upon a strict regulation of TOR activity and TOR-dependent cell differentiation. Hence this work brings to light a physiological role of IRE1 in tissue growth by connecting two essential and highly conserved growth-regulating pathways.
RESULTS
IRE1 promotes root growth in an age-dependent manner
The reduced length of the ire1a ire1b root has been documented earlier (Bao et al., 2019; Chen et al., 2014; Chen and Brandizzi, 2012; Deng et al., 2013), but has not been studied in detail during the transition from early seedling development to an adult vegetative state, which corresponds to a highly active growth period. To fill this gap and establish a robust platform for defining the role of IRE1 in organ growth, we set up a time-course analysis of ire1a ire1b growth with measurements of phenotypic traits at 5, 7, 10, and 12 days after germination (D5, D7, D10, D12; Figure 1(a)).
Figure 1.
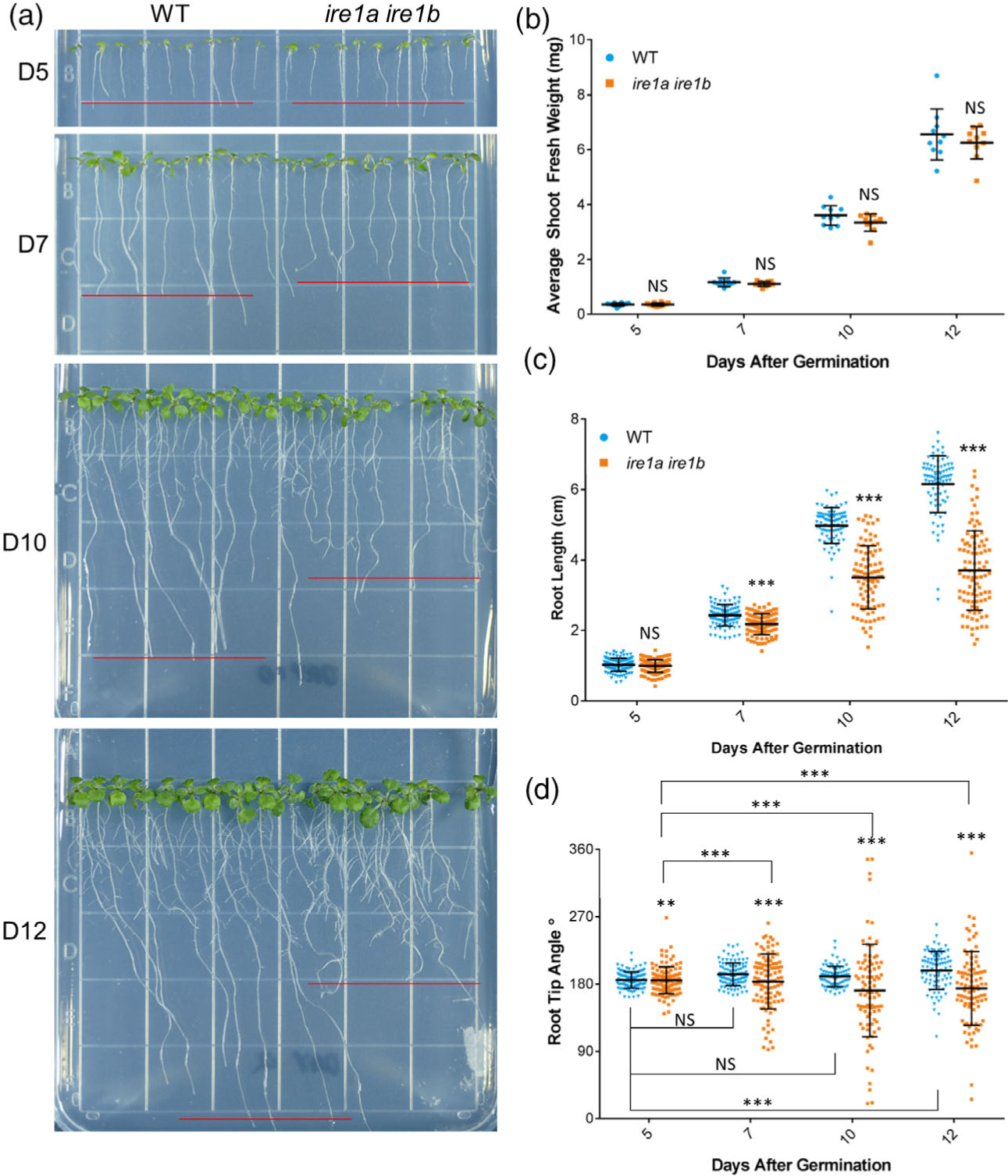
The ire1a ire1b double mutant shows age-dependent primary root growth defects. (a) Representative images of WT and ire1a ire1b mutants grown for 5, 7, 10, or 12 days. Red lines represent approximate average root length of the displayed seedlings. (b) Shoot fresh weight was determined by averaging WT or ire1a ire1b shoots grown in an individual plate for each plate replicate (n = 10; error bars show the standard deviation). (c) The length of individual roots was measured using ImageJ. For Figure 1(b,c): error bars show the standard deviation; significance markers displayed above an ire1a ire1b experimental group are pairwise comparisons to the corresponding WT group for that specific treatment. Brackets denote other specific pairwise comparisons. Significance markers: NS = Padj > 0.01; *0.001 < Padj < 0.01; **0.0001 < Padj < 0.001; ***Padj < 0.0001. (d) The angle of the root tip with the vertical axis (0°) was measured using ImageJ. Significant differences between coefficients of variation were tested using the asymptotic Feltz and Miller test as described in the Experimental Procedures section. Error bars show the standard deviation; significance markers: NS = P > 0.0001; **P < 0.0001 and P > 1.0 × 10−10; ***P < 1.0 × 10−10
We first analyzed shoot development by quantifying the shoot fresh weight (SFW) in WT and ire1a ire1b seedlings at these time points (Figure 1(b)). To test the effect of our experimental variables, i.e., seedling age and genotype, on the SFW, we carried out a two-way between-subjects analysis of variance (hereafter referred to as ANOVA; see the Experimental Procedures section for the statistical analysis pipeline). The analysis indicated that there was not a significant interaction between the effects of seedling age and genotype on SFW (F(3,76) = 0.344, P = 0.793; complete statistics results are presented in Table S1). In both WT and ire1a ire1b genotypes, rapid and similar increases in SFW were found from D0 to D12 (Figure S1(a)). These results demonstrate that the level of IRE1 functional impairment in the ire1a ire1b mutant does not have an effect on shoot development.
We next analyzed primary root development by recording the root length of WT and ire1a ire1b at each time point (Figure 1(c)). We attempted to utilize an ANOVA to test the effect of, and interaction between, seedling age and genotype on root length. However, when we tested the normality and homogeneity of variance assumptions of the ANOVA by the Shapiro–Wilk test and Levene’s test, respectively, we found that these assumptions were violated. Box–Cox and log transformation of the dataset were also attempted; however, the assumptions were still violated. We therefore carried out a two-way between-subjects ANOVA utilizing weighted least squares regression (hereafter referred to as wANOVA; see the Experimental Procedures section and Data S1) to analyze primary root length. The wANOVA showed a highly significant interaction between the effects of seedling age and genotype on root length, although the effect of genotype alone was not significant (F(3,753) = 133, P < 2 × 10−16; Table S1). These results indicate that potential differences between root lengths of WT and ire1a ire1b may be dependent upon seedling age. To determine the specific nature of this interaction, we performed pairwise comparisons between different experimental groups, the results of which are displayed as significance markers in the referenced figures. At D5, we found no significant differences in average root length between WT and ire1a ire1b (Figure 1(c)), indicating that the ire1a ire1b mutation does not affect root growth during early seedling development. Past D5, we found dramatic increases in the rate of root growth in WT from approximately 0.2 cm day−1 between D0 and D5 to approximately 0.9 cm day−1 between D5 and D10 (Figure S1(b)). Noticeably, during the D5–D10 phase of growth, the root growth phenotype of ire1a ire1b became increasingly more severe compared to the earlier phase of growth (i.e., D0–D5). Specifically, the average ire1a ire1b root length was significantly shorter than WT at D7, D10, and D12 (Figure 1(c)). Furthermore, while the rate of root growth in the ire1a ire1b mutant accelerated from approximately 0.2 cm day−1 to approximately 0.7 cm day−1 between D5 and D7, it then declined to approximately 0.5 cm day−1 between D7 and D10 (Figure S1(b)). These results are consistent with the findings that IRE1 is required for root growth (Bao et al., 2019; Chen et al., 2014; Chen and Brandizzi, 2012; Deng et al., 2011), but also expand on these results by demonstrating that IRE1 is required to maintain accelerated rates of primary root growth as seedlings mature.
Concurrent with altered growth rates in maturing ire1a ire1b primary roots, we observed that directional root growth was increasingly impaired as the seedlings matured. To analyze this, we determined the growth vector of the primary root tip by measuring the root tip angle with the vertical axis (0°) in a counterclockwise orientation, such that roots growing directly downward would have a recorded angle of 180° (Figure 1(d)). We next tested the effect of genotype and seedling age on the average root growth vector and found that there was no significant effect of either variable (Table S1). However, we observed increasingly large differences in standard deviation between WT and ire1a ire1b as seedlings matured (Figure 1(d)). In order to compare the relative distribution of the collected data points, we performed asymptotic Feltz and Miller tests to determine if there were significant differences between coefficients of variation (CoVs) of the different experimental groups (Feltz and Miller, 1996). We found that the CoV was not significantly different between WT root tip angles at D5, D7, and D10, but found a significant difference in WT CoV between D5 and D12 (Figure 1(d)). At all the tested time points, the CoVs of ire1a ire1b root tip angles were significantly different from the corresponding WT CoVs (Figure 1(d)). Furthermore, the ire1a ire1b root tip angle CoV significantly increased with seedling age at every time point compared to the baseline measurements at D5 (Figure 1(d)). Several different types of cell division and organization defects in the root tip can cause a short root phenotype (Lucas et al., 2011; Petricka et al., 2012). However, defective control of the primary root growth vector is more likely due to aberrant cell elongation because tropic growth responses regulate asymmetric cell elongation on one side of the root to direct organ growth in a specific direction (Ishikawa and Evans, 1993; Mullen et al., 1998; Sato et al., 2015). Therefore, the increasingly random root growth vectors displayed by maturing ire1a ire1b support the hypothesis that IRE1 may be required for proper cell elongation.
IRE1 is required for proper cell elongation in the root meristem
Next, we aimed to test the hypothesis that IRE1 is required to control cell elongation and map the role of IRE1 in the different root zones. To do so we performed modified pseudo-Schiff propidium iodide (mPS-PI) staining (Truernit et al., 2008) of fixed WT and ire1a ire1b roots at D5, D7, and D10 followed by confocal microscope imaging and quantitative image analysis (Figure 2; Figures S2 and S3). For each root, we identified the MZ, transition zone (TZ), and elongation zone (EZ) of the root tips according to previously published criteria (Casamitjana-Martinez et al., 2003; Di Mambro et al., 2017). Canonically, cells divide in the MZ and then undergo a transitional stage consisting of genomic endoreduplication and cytoarchitectural changes in the TZ, followed by cell elongation in the EZ (Hayashi et al., 2013; Scheres and Wolkenfelt, 1998). For each root tip zone, we recorded the length, the number of cells, and the average cell length. We then performed a series of two-way wANOVAs (or ANOVAs as indicated) to test the effects of seedling age and genotype on each of these zone metrics in the MZ, TZ, and EZ. As hypothesized, we found significant interactions between the effects of seedling age and genotype on the zone length, cell number, and average cell length of the EZ (wANOVA: F(2,119) = 37.0, P = 3.24 × 10−13, F(2,119) = 15.8, P = 8.48 × 10−7, F(2,119) = 13.2, P = 6.54 × 10−6, respectively). In addition, we also found significant interactions between the effects of seedling age and genotype on the zone length and cell number of the MZ (wANOVA: F(2,119)=9.71, P = 1.23 × 10−04, F(2,119) = 11.4, P = 2.83 × 10−05, respectively). However, for each of these EZ and MZ zone metrics we found that the effect of genotype alone was not significant (complete statistics results are presented in Table S1). Together, these results support that the differences in root tip cell organization between WT and ire1a ire1b map to the MZ and the EZ in a manner that is dependent upon the age of the seedlings. This is further supported by the evidence that at D5, where no significant differences in primary root length were found between WT and ire1a ire1b roots (Figure 1(c)), there were also no significant differences between WT and ire1a ire1b for any of the root tip organization metrics recorded (i.e., zone length, number of cells, and cell length; Figure S2(a–d)). Therefore, the ire1a ire1b mutation does not affect root growth during early seedling development (Figure 1).
Figure 2.
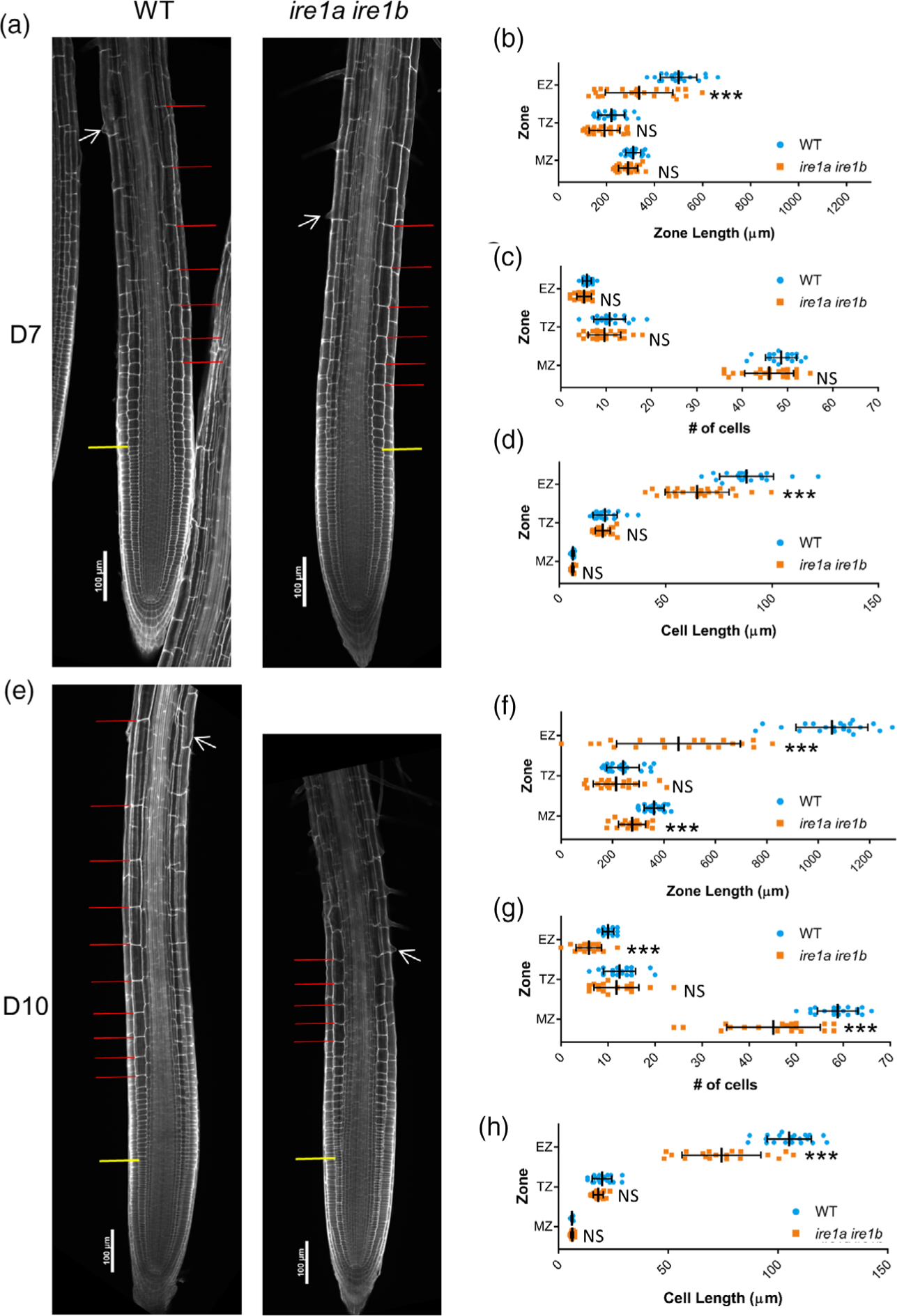
Meristem organization defects in ire1a ire1b are first manifested in cell elongation in the elongation zone. At D7 and D10, root tips were subjected to mPS-PI staining and confocal microscopy to analyze root tip cellular organization. (a) Representative 10× images of mPS-PI-stained WT and ire1a ire1b roots at D7. The yellow line demarks the end of the meristem zone (MZ) and the beginning of the transition zone (TZ), the red lines mark all of the cells in the elongation zone (EZ), and the white arrow marks the first root hair initiation. For all measurements of the MZ secondary 20× images were used to collect data. (b–d) Zone length, number of cells, and cell length at D7. (e) Representative 10× images of mPS-PI-stained WT and ire1a ire1b roots at D10. (f, g) Zone length, number of cells, and cell length at D10. For all graphs error bars show the standard deviation; significance markers displayed above an ire1a ire1b experimental group are pairwise comparisons to the corresponding WT group for that specific treatment. Brackets denote other specific pairwise comparisons. Significance markers: NS = Padj > 0.01; *0.001 < Padj < 0.01; **0.0001 < Padj < 0.001; ***Padj < 0.0001.
We then focused on defining the variables underpinning the dramatic increase in WT primary root growth rates (from approximately 0.2 cm day−1 at D5 to approximately 0.9 cm day−1 at D10; Figure S1(b)). We found that the length of both the MZ and the EZ roughly doubled in WT root tips during this period, while the length of the TZ remained unchanged (Figure S3(a)). Interestingly, we observed a significant 1.7-fold increase in the number of cells in the MZ, while the length of MZ cells remained unchanged, indicating that the increased size of the MZ was due to an increase in the cell number in this zone (Figure S3(b,c)). We also found a 1.25-fold increase in cell length and a 2-fold increase in the number of cells in the EZ at D10 compared to D5, supporting that the observed increased size of the EZ was due to an increase of both cell length and cell number in the EZ (Figure S3(b)).
Next, we compared the root tip organization of WT and ire1a ire1b when the mutant root phenotype is visible (i.e., D7 and D10; Figure 2(a–h)). At D7, we found that the ire1a ire1b MZ was not significantly different from the WT MZ in terms of zone length, number of cells, and cell length (Figure 2(b–d)). However, when we examined the EZ, we found that the ire1a ire1b EZ was significantly shorter than the WT EZ, due to significantly shorter cell length (Figure 2(b,d)). Therefore, at D7, the short root phenotype of ire1a ire1b coincides with the development of a defective EZ. At D10, we found that the ire1a ire1b MZ was significantly shorter than WT and contained a smaller number of cells (Figure 2(f,h)). Contrary to the WT MZ, we did not detect an increase in Ire1a Ire1b MZ size at D10 compared to D7 (Figure S2(b,e)), supporting that the expected increases in MZ size do not take place in the Ire1a Ire1b mutant. At D10, we also observed EZ length reduction in Ire1a Ire1b compared to WT (Figure 2(f)) with a significantly reduced cell size and number (Figure 2(g,h)). In summary, while the EZ length more than doubled between D5 and D10 in WT, it did not significantly change in Ire1a Ire1b over this growth period (Figure S2(a,d)). Together these data indicate that the Ire1a Ire1b roots fail to obtain the rapid rates of root growth achieved by WT plants as they mature, primarily due to defective cell elongation in the EZ. Furthermore, we have successfully established a solid working platform to study the mechanisms underpinning IRE1-dependent control of root growth.
The emergence of the Ire1a Ire1b root growth phenotype depends on high rates of root growth
We then sought to test whether the Ire1a Ire1b root growth phenotype exclusively depends upon seedling age or whether it could be responsive to increased rates of root growth. To do this, we grew seedlings under increasing photoperiod lengths and in the presence of exogenous sucrose supply in the medium. Extending the photoperiod results in an accumulation of photosynthates (i.e., sucrose) (Sulpice et al., 2014) and an increase in the overall rate of rate of root growth (Yazdanbakhsh et al., 2011). Adding sucrose to the growth medium also increases rates of root growth (Yazdanbakhsh et al., 2011). Therefore, we grew WT and Ire1a Ire1b seedlings for 10 days in three different light conditions with increasing photoperiods: 8 h 150 μE light/16 h dark, 16 h 150 μE light/8 h dark, and continuous 150 μE light (hereafter abbreviated 8/16, 16/8, and CL, respectively). In each photoperiod condition, we grew seedlings on plates containing no sucrose or 1% sucrose (Figure 3).
Figure 3.
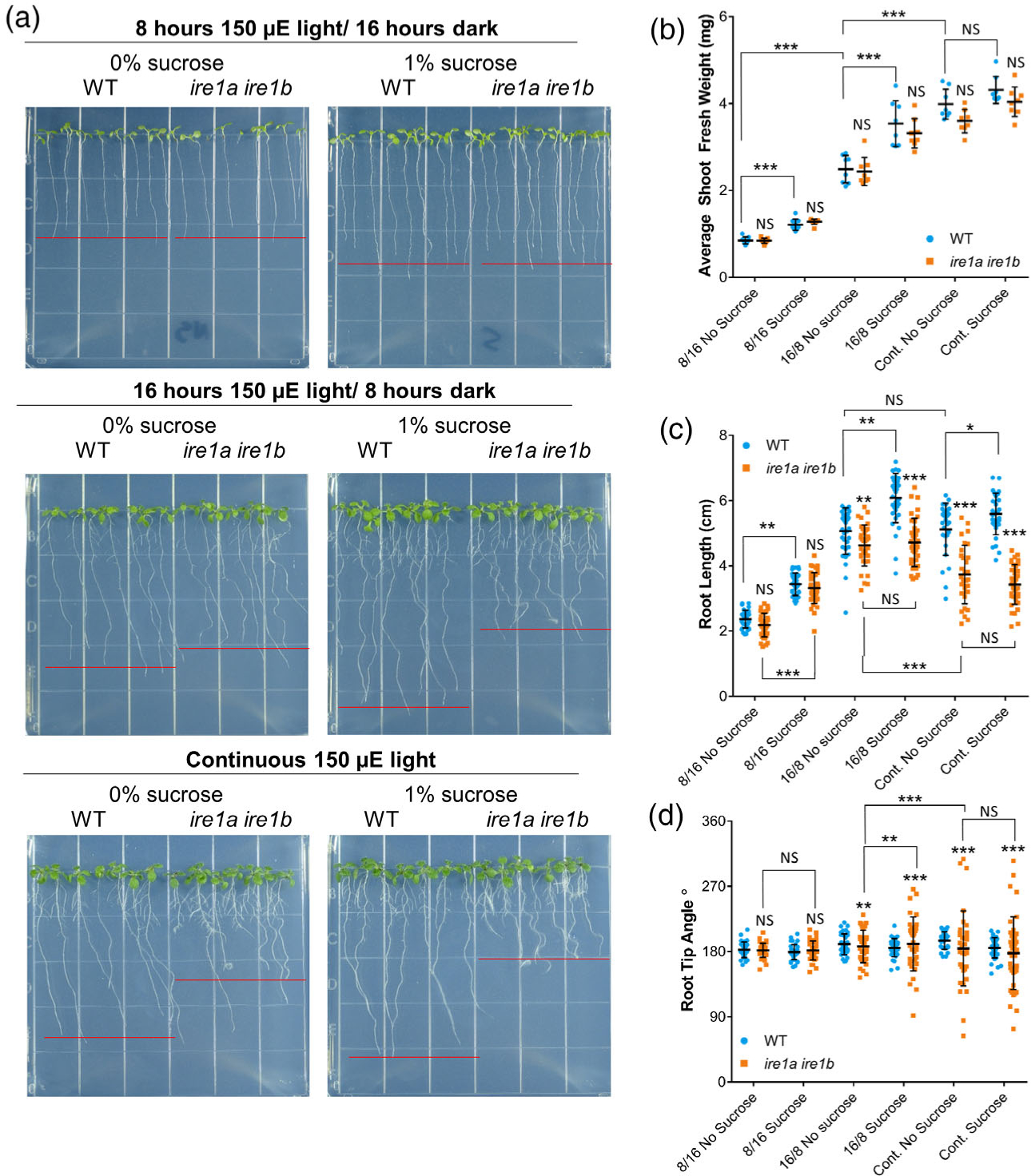
The emergence of the Ire1a Ire1b root phenotype depends on a high rate of root growth. (a) Representative images of WT and Ire1a Ire1b mutants grown in the indicated conditions. Red lines represent approximate average root length of the displayed seedlings. (b) Shoot fresh weight was determined by averaging WT or Ire1a Ire1b shoots grown in an individual plate for each plate replicate (n = 10; error bars show the standard deviation). (c) The length of individual roots was measured using ImageJ. For Figure 1(b,c): error bars show the standard deviation; significance markers displayed above an Ire1a Ire1b experimental group are pairwise comparisons to the corresponding WT group for that specific treatment. Brackets denote other specific pairwise comparisons. Significance markers: NS = Padj > 0.01; *0.001 < Padj < 0.01; **0.0001 < Padj < 0.001; ***Padj < 0.0001. (d) Angle of the root tip with the vertical axis (0°) was measured using ImageJ. Significant differences between coefficients of variation were tested using the asymptotic Feltz and Miller test as described in the Experimental Procedures section. Significance markers: NS = P > 0.05; **P < 0.01 and P > 0.001; ***P < 0.001.
We first measured SFW and found no significant interaction between the effects of exogenous sucrose supply, photoperiod, and genotype via a three-way ANOVA (Figure 3(b); F(2,88) = 0.626, P = 0.537; Table S1). We found that increased photoperiod had a dramatic effect on the development of both WT and Ire1a Ire1b shoots, with an approximately 2-fold SFW increase in 16/8 light compared to 8/16, and a further approximately 2-fold SFW increase in CL compared to 16/8 (Figure 3(b)). Exogenous sucrose had an additional significant growth-promoting effect in both 8/16 and 16/8 light conditions. In both light conditions, SFW was approximately 1.4-fold larger when grown on sucrose-containing media compared to no sucrose controls (Figure 3(b); Table S1). In CL, there was no significant effect of sucrose on SFW accumulation (Figure 3(b); Table S1). In all conditions tested, the Ire1a Ire1b SFW was not significantly different from the respective WT controls (Figure 3(b); Table S1). Therefore, IRE1 is not required for shoot biomass accumulation regardless of the tested growth conditions.
We then measured root length and found a significant interaction between the effects of exogenous sucrose supply, photoperiod, and genotype on primary root length (Figure 3(c); three-way wANOVA: F(2,397) = 9.13, P = 1.32 × 10−4; complete statistics results are presented in Table S1). In WT seedlings, the root length increased significantly in 16/8 light conditions compared to 8/16 light conditions, but no significant differences were noted between 16/8 and CL (Figure 3(c)). We also found that exogenously supplied sucrose significantly increased root length of WT plants grown in 8/16 or 16/8 light photoperiod conditions but not CL, similar to the effect of exogenous sucrose on SFW (Figure 3(b)). In an analysis of root growth vector distribution under these conditions, we also found that the WT CoV was not significantly different from any WT samples across all tested conditions (Figure 3(d)). We then analyzed the Ire1a Ire1b mutant. In 8/16 light conditions, with and without sucrose in the media, the Ire1a Ire1b root length and growth vector distribution were identical to WT (Figure 3(c,d)). These results indicate that the Ire1a Ire1b root growth phenotype is not strictly dependent upon age alone. In 16/8 light, the Ire1a Ire1b roots were slightly but significantly smaller than WT on plates without sucrose (Figure 3(c)). Differently from WT root length, there were no significant differences in overall length between sucrose and no sucrose-treated Ire1a Ire1b roots in 16/8 light (Figure 3(c)). We also observed that 16/8 light led to significant differences in CoV in root growth vector between WT and Ire1a Ire1b (Figure 3(d)). Importantly, we also found that the addition of sucrose to the media significantly increased the root growth vector CoV compared to the Ire1a Ire1b no sucrose control, indicating that exogenous sucrose supply causes aberrant directional root growth in the Ire1a Ire1b mutant (Figure 3(d)). In CL conditions, we found that Ire1a Ire1b roots were significantly shorter than both WT roots grown in CL and Ire1a Ire1b roots grown in 16/8 light conditions. Furthermore, similar to the 16/8 light conditions, the roots of Ire1a Ire1b grown either on sucrose-containing plates or no sucrose controls in CL were similar in length (Figure 3(c)). The Ire1a Ire1b root growth vector CoV was significantly increased by exposure to CL compared to the 16/8 conditions, confirming that the prolonged photoperiod leads to aberrant directional root growth in the Ire1a Ire1b mutant. On the whole, these data indicate that, while a prolonged photoperiod and sucrose availability equally promote shoot growth of WT and the Ire1a Ire1b mutant, IRE1 is absolutely required to reach the maximum rates of root growth afforded by extended photoperiod and increased carbohydrate availability.
TOR inhibition rescues the Ire1a Ire1b primary root growth phenotype
It has been documented that plant TOR integrates light and carbohydrate availability signals to control growth (Li et al., 2017) and that hyperactivation of TOR can lead to a short root phenotype (Cao et al., 2019). Based on the negative effect of pro-growth signals (i.e., prolonged photoperiod and exogenous sucrose supply) on Ire1a Ire1b root growth, we hypothesized that TOR could be hyperactive in Ire1a Ire1b and, therefore, that the IRE1 root growth phenotype might be alleviated by TOR inhibition. As a first step to test this hypothesis, we performed our growth phenotyping assays (Figures 1 and 2), but also supplemented growth media with the TOR inhibitor AZD-8055 (hereafter referred to as AZD; Cao et al., 2019) or DMSO control vehicle. We first conducted a test using concentrations of AZD ranging from 50 to 200 nM in the culture media (Figure S3). At a 150 nM concentration, AZD had a slight inhibitory effect on WT root length in line with previous results (Cao et al., 2019; Montané and Menand, 2013) and significantly altered the Ire1a Ire1b root length phenotype compared to the DMSO control (Figure S3). Therefore, for our analyses we proceeded to use 150 nM AZD in the growth medium to induce a low-level inhibition of TOR compared to the more commonly applied AZD concentration of 1 μM or more (Barrada et al., 2019; Montané and Menand, 2013; Schepetilnikov et al., 2017; Zhuo et al., 2020).
We then performed a time-course analysis of Ire1a Ire1b root growth at D7–D12 on growth media containing either DMSO vehicle or 150 nM AZD (Figure 4(a)). We found a small effect of AZD on WT root growth. We then conducted ANOVAs to test for an interaction between the effects of seedling age, AZD treatment, and genotype on seedling growth phenotypes (i.e., SFW and root length). While we found no significant interactions between the effects of these variables on average SFW (three-way ANOVA: F(2,84) = 0.150, P = 0.861; Figure 4(b)), we did find a highly significant interaction between seedling age, AZD treatment, and genotype on primary root length (three-way wANOVA: F(2,887) = 67.253, P < 2.2 × 1016; Figure 4(c)). Similar to our earlier results (Figure 1), we observed strong, age-dependent root growth defects in the Ire1a Ire1b mutant in the DMSO conditions (Figure 4(c,d)). However, when Ire1a Ire1b was grown in the presence of AZD, at D7 and at D10 we found that the average primary root length was not significantly different from WT (Figure 4(a,c)). At D12, the average primary root length of AZD-treated Ire1a Ire1b was slightly but significantly smaller compared to WT; however, the AZD-treated Ire1a Ire1b roots were nearly 1.5 cm longer than their respective DMSO controls at this time point (Figure 4(c)). In an analysis of directional root growth, we also found that, when Ire1a Ire1b was grown in the presence of AZD, there were no significant differences in the Ire1a Ire1b root CoV compared to AZD-treated WT at D7 and D10 (Figure 4(d)). At D12, while the CoVs of AZD-treated Ire1a Ire1b root tip angles were significantly different from those of AZD-treated WT, we found that the AZD-treated Ire1a Ire1b root tip angles were significantly less variable than DMSO-treated Ire1a Ire1b root tip angles at D12 (Figure 4(d)). These results support the hypothesis that inhibition of TOR activity in Ire1a Ire1b rescues the short root and directional growth phenotypes of this mutant. To confirm these results, we tested the effects of an additional chemical inhibitor of TOR activity, TORIN2 (Cao et al., 2019; Montané and Menand, 2013), on WT and Ire1a Ire1b. Similar to AZD treatment, 200 nM TORIN2 rescued both the short root and root tip angle phenotypes of the Ire1a Ire1b mutant (Figure S5). To confirm that this TOR-responsive root growth phenotype is linked to the IRE1 loss in Ire1a Ire1b, we repeated our AZD growth assay with an Ire1a Ire1b complemented line expressing Ire1a under the control of its own native promoter (Chen and Brandizzi, 2012). We found that IRE1a driven by the native promoter completely rescued the Ire1a Ire1b root growth phenotype and restored the root growth inhibition effects of AZD (Figure S6), supporting that the verified growth phenotypes are specifically due to the loss of IRE1.
Figure 4.
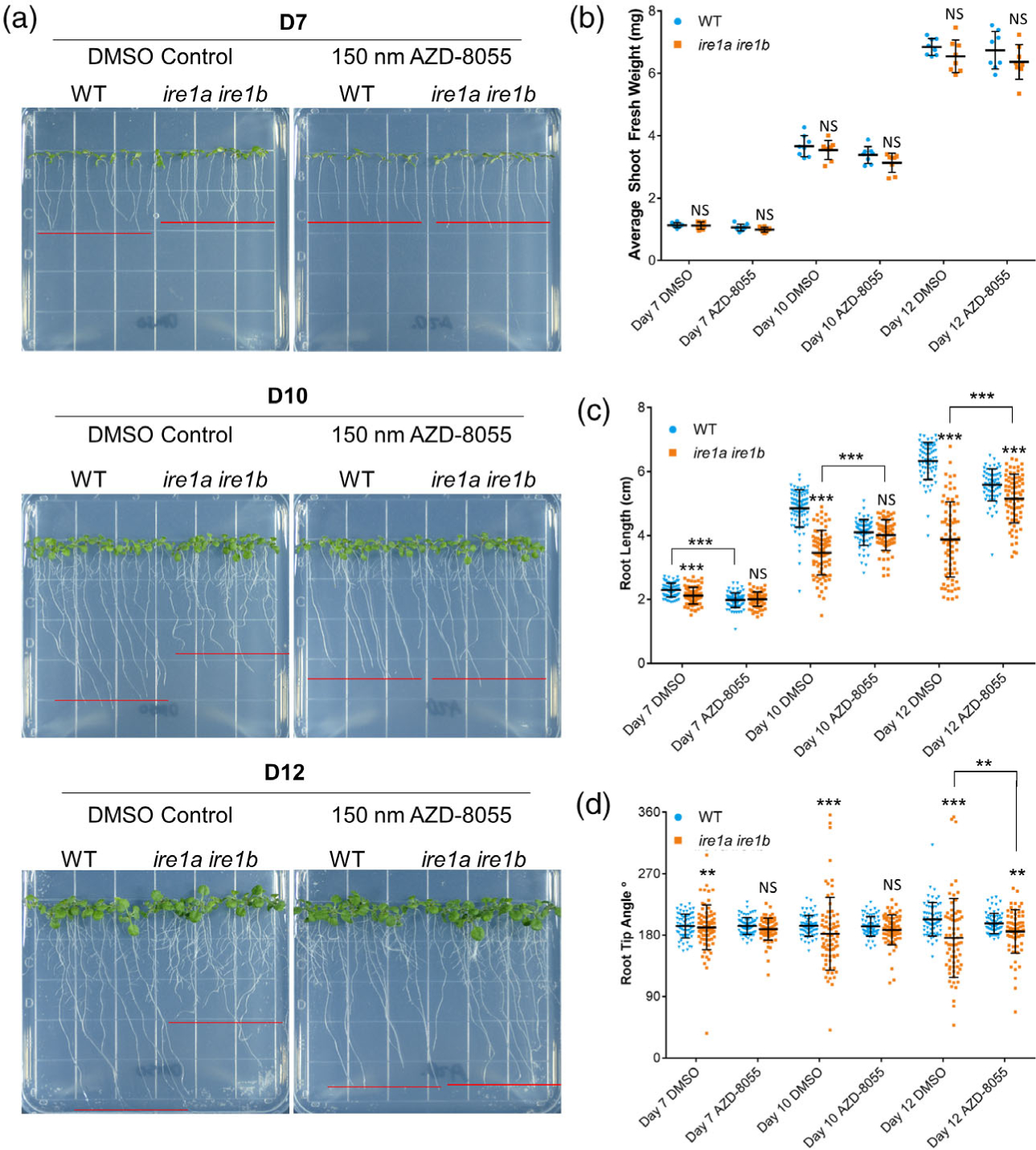
TOR inhibition rescues the Ire1a Ire1b root growth phenotype. (a) Representative images of WT and Ire1a Ire1b grown for 7, 10, or 12 days on plates containing 150 nM AZD-8055 or DMSO control. Red lines represent approximate average root length of the displayed seedlings. (b) Shoot fresh weight was determined by averaging WT or Ire1a Ire1b shoots grown in an individual plate for each plate replicate (n = 10; error bars show the standard deviation). (c) The length of individual roots was measured using ImageJ. For Figure 1(b, c): error bars show the standard deviation; significance markers displayed above an Ire1a Ire1b experimental group are pairwise comparisons to the corresponding WT group for that specific treatment. Brackets denote other specific pairwise comparisons. Significance markers: NS = Padj > 0.01; *0.001 < Padj < 0.01; **0.0001 < Padj < 0.001; ***Padj < 0.0001. (d) The angle of the root tip with the vertical axis (0°) was measured using ImageJ. Significance differences between coefficients of variation were tested using the asymptotic Feltz and Miller test as described in the Experimental Procedures section. Significance markers: NS = P > 0.0001; **P < 0.0001 and P > 1.0 × 10−10; ***P < 1.0 × 10−10.
We then tested whether the rescue of the Ire1a Ire1b root phenotype was specific to TOR inhibition or a general effect of slower root growth rates which could be phenocopied by treatment with other chemical inhibitors or hormones. To do so, we grew WT and Ire1a Ire1b on media containing low concentrations of mitochondrial respiration inhibitors and cytoskeletal inhibitors, which are known to negatively affect the growth of WT roots (Cao et al., 2019; Renna et al., 2018; Van Aken et al., 2016). We found that none of these treatments led to a significant increase in the root length of the Ire1a Ire1b mutant compared to their respective controls and instead led to small but significant decreases in root length, indicating that general inhibitors of root growth are uninfluential to the rescue of the Ire1a Ire1b phenotype (Figures S7 and S8). Additionally, because of the previously established connection between induced ER stress and auxin signaling (Chen et al., 2014) and the previous findings that auxin activates TOR (Retzer and Weckwerth, 2021; Schepetilnikov et al., 2017), we tested whether the Ire1a Ire1b phenotype may be unexpectedly affected in a synergistic manner by concomitant TOR inhibition and auxin treatment. We found that the effects of AZD and the synthetic auxin 1-naphthaleneacetic acid (NAA) were additive such that Ire1a Ire1b seedlings treated with AZD responded to NAA identically to WT plants (Figure S8). Together, these results that TOR inhibition rescues the short root and misdirected primary root growth vector phenotypes of the Ire1a Ire1b mutant indicate that TOR activity strongly and specifically contributes to the root growth phenotypes of the Ire1a Ire1b mutant.
TOR is hyperactive at the growing primary root tips of the Ire1a Ire1b mutant
Next, we sought to map the endogenous alterations to TOR activity in actively growing Ire1a Ire1b roots. To do this we utilized a common immunoblot-based assay of phosphorylated SERINE-KINASE 6 (phospho-S6K), a conserved target of TOR kinase activity (Xiong et al., 2013). We excised approximately 3 mm from 60 root tips and pooled these tips to create an individual biological replicate, and we executed 11 independent biological replicates. The remaining mature root tissue from each root was also excised and pooled. We performed this analysis using 7-day-old seedlings to avoid the possibly confounding effects of the more severe morphological differences between WT and Ire1a Ire1b observed at D10. We compared the relative phospho-S6K signal ratio, which was derived by dividing the phospho-S6K signal by the total S6K signal (αS6K1/2) and subsequently normalizing to the average WT DMSO ratio for each individual blot. We used WT and Ire1a Ire1b seedlings grown in DMSO- or AZD-containing media (150 nM AZD). Via two-way wANOVA, we identified significant effects of both genotype and AZD treatment on the relative phospho-S6K signal ratio (F (1,40) = 9.58, P = 3.59 × 10−3, F(1,40) = 16.7, P = 2.02 × 10−4, respectively). We found that the Ire1a Ire1b root tips had an approximately 2-fold higher phospho-S6K ratio compared to WT in DMSO conditions (Figure 5(a)). As expected, AZD treatment led to a significantly lower phospho-S6K ratio in WT root tips (0.7-fold change) compared to the DMSO control. AZD treatment also significantly reduced the phospho-S6K ratio of Ire1a Ire1b root tips compared to the DMSO control (Figure 5(a)). The phospho-S6K ratio of AZD-treated Ire1a Ire1b root tips was not significantly different from that of AZD-treated WT root tips (Figure 5(a)). When we analyzed the mature root tissues (Figure 5(b)), we found a significant effect of AZD treatment on the relative phospho-S6K signal ratio (two-way ANOVA: F(1,32) = 24.9, P = 2.03 × 10−5) but not genotype (F(1,32) = 1.26, P = 0.269), as there were no significant differences between WT and Ire1a Ire1b phospho-S6K ratios in mature root tissues in either DMSO or AZD treatments (Figure 5(b)). Together these results indicate that the loss of IRE1 leads to increased TOR activity in the root tips but not in mature tissues. Therefore, IRE1 is necessary to maintain proper TOR activity levels specifically in rapidly developing root tips.
Figure 5.
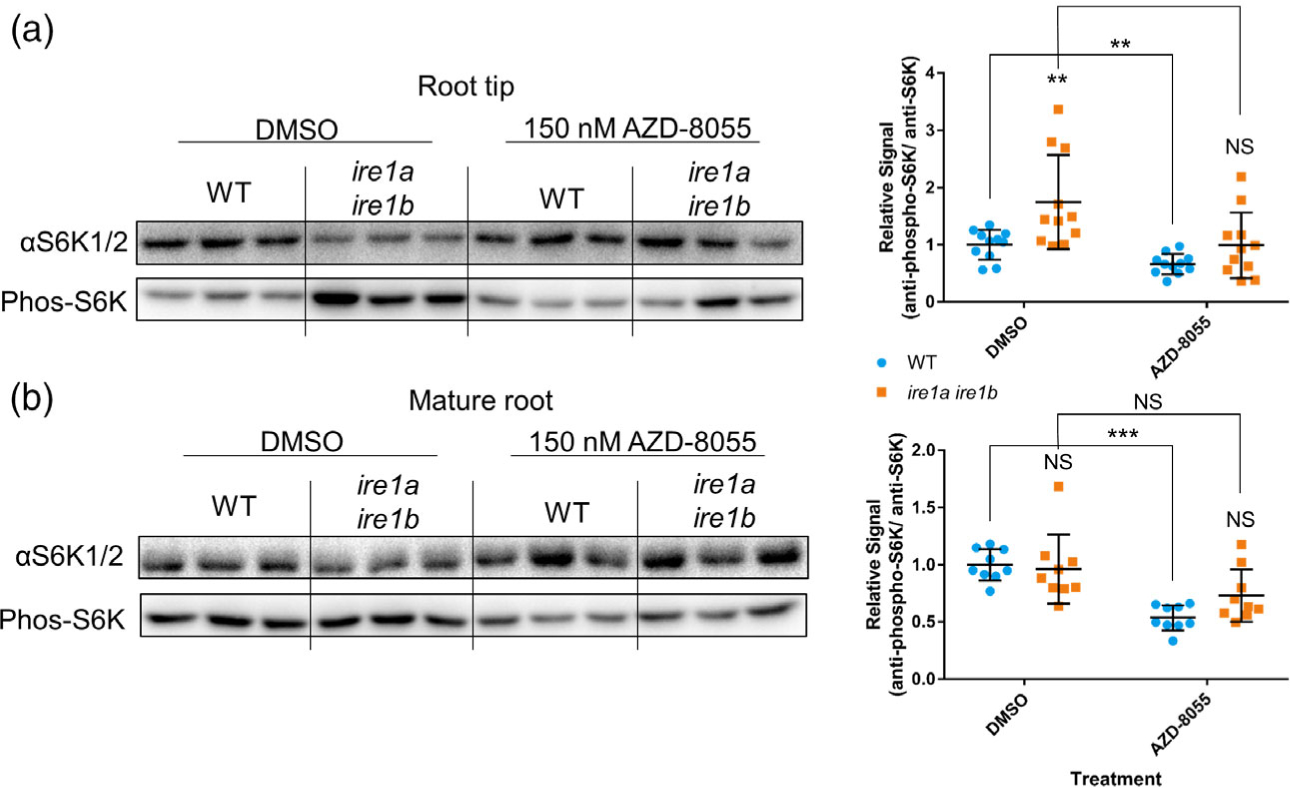
TOR is hyperactive in Ire1a Ire1b mutant root tips but not in the mature root. (a) Representative immunoblot assay to determine relative S6K phosphorylation levels. Antibodies against total S6K (αS6K1/2) or phosphorylated S6K (Phos-S6K) were used against total soluble protein extracted from excised WT or Ire1a Ire1b root tips grown for 7 days on media containing AZD-8055 or DMSO control (see Experimental Procedures). Relative signals (Phos-S6K/αS6K1/2) in each experimental group were normalized to WT DMSO control (n = 11). (b) Same immunoblot method used in (a) but used against total soluble protein extracted from excised mature WT or Ire1a Ire1b root tissues grown for 7 days on media containing AZD-8055 or DMSO control (n = 9). For all graphs error bars show the standard deviation; significance markers displayed above an Ire1a Ire1b experimental group are pairwise comparisons to the corresponding WT group for that specific treatment. Brackets denote other specific pairwise comparisons. Significance markers: NS = Padj > 0.05; *0.005 < Padj < 0.05; **0.0005 < Padj < 0.005; ***Padj < 0.0005. See Figure S9 for full blot images and Ponceau stain loading controls.
TOR inhibition rescues the Ire1a Ire1b cell elongation phenotype at the root meristem
The spatial specificity of TOR hyperactivation verified in growing root tips but not in mature tissues of the Ire1a Ire1b roots (Figure 5) prompted us to establish the cellular consequences of the TOR hyperactivity in the Ire1a Ire1b mutant. To do so, we performed mPS-PI analysis of root tips from WT and Ire1a Ire1b plants grown on DMSO- or AZD-containing media. We performed our analysis at D10 in order to test the effects of TOR inhibition on the strong defects in cell elongation in the EZ as well as the moderate defects in the MZ, which only were found at D10 (Figure 2 (e–h)). In WT plants grown on AZD-containing media, we observed a small but significant decrease in the number of cells in the MZ, and a small but significant increase in the number of cells in the TZ (Figure 6(c)), consistent with previously published results with respect to the effects of TOR inhibition on root tip meristem organization (Montané and Menand, 2013). We then performed a series of two-way wANOVAs (or ANOVAs as indicated) to test the effects of AZD treatment and genotype on each of these zone metrics in the MZ, TZ, and EZ. We found significant interactions between the effects of AZD treatment and genotype on zone length, cell number, and average cell length of the EZ (wANOVA: F(1,76) = 62.8, P = 1.51 × 10−11, F(1,76) = 21.4, P = 1.51 × 10−5, F(1,76) = 34.1, P = 1.21 × 10−7, respectively). We also found significant interactions between the effects of AZD treatment and genotype on the zone length and cell number of the MZ (wANOVA: F(1,76) = 24.2, P = 4.80 × 10−6, F(1,76) = 43.9, P = 4.39 × 10−9, respectively) and a marginally significant interaction with the effects on MZ cell length (F(1,76) = 6.44, P = 0.0131). On the whole, these results demonstrate that the zone metrics exhibiting significant interactions between seedling age and genotype (Figure 2) also showed significant interactions between AZD treatment and genotype.
Figure 6.
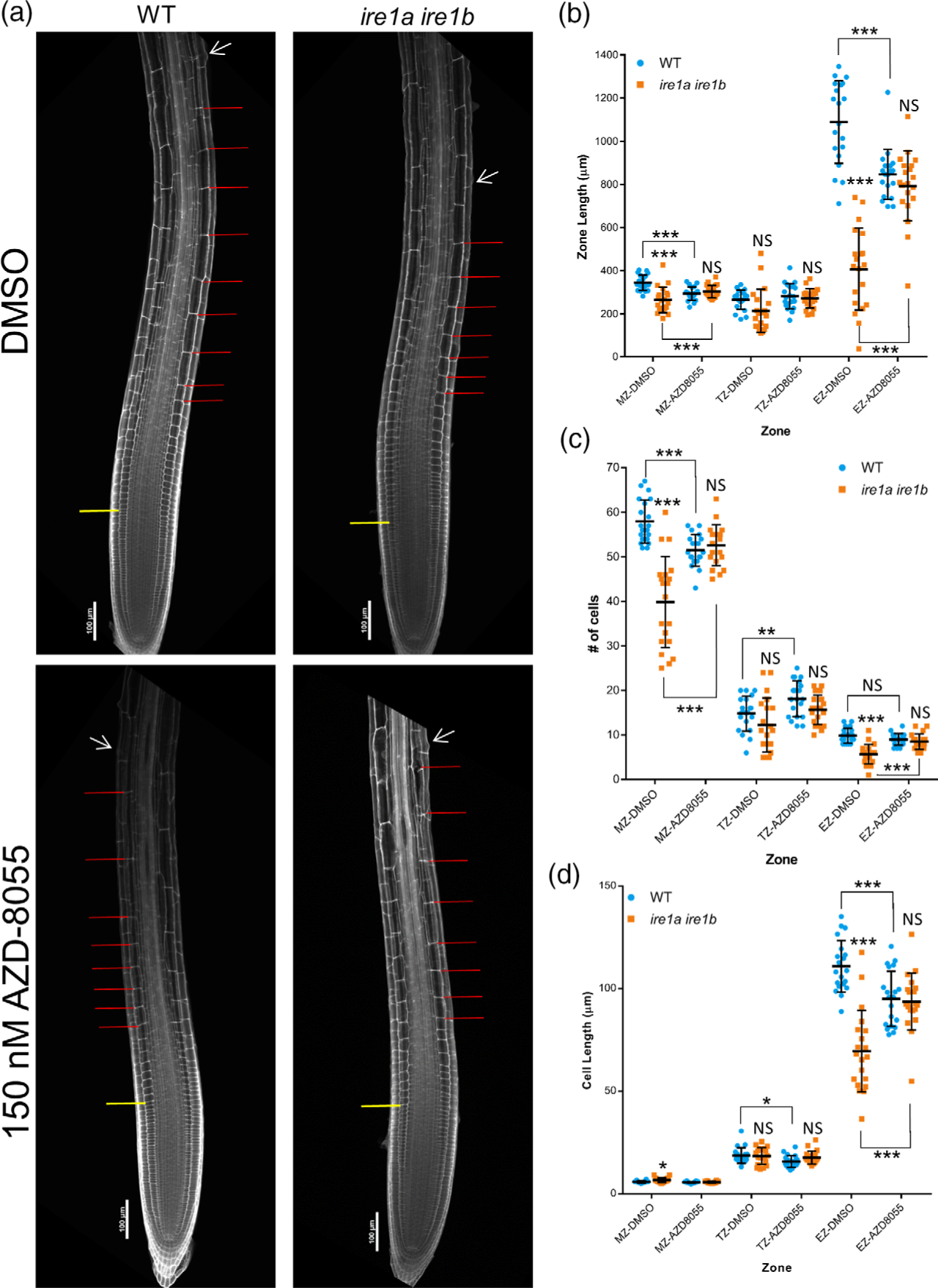
Meristem organization defects in Ire1a Ire1b are rescued by TOR inhibition. At D10, root tips grown on DMSO- or AZD-8055-containing media were subjected to mPS-PI staining and confocal microscopy to analyze root tip cellular organization. (a) Representative 10 images of mPS-PI-stained WT and ire1a ire1b roots. The yellow line demarks the end of the meristem zone (MZ) and the beginning of the transition zone (TZ), the red lines mark all of the cells in the elongation zone (EZ), and the white arrow marks the first root hair initiation. For all measurements of the MZ secondary 20 images were used to collect data. (b–d) Zone length, number of cells, and cell length at D10 in roots gown on DMSO- or AZD-8055-containing media. For all graphs error bars show the standard deviation; significance markers displayed above an Ire1a Ire1b experimental group are pairwise comparisons to the corresponding WT group for that specific treatment. Brackets denote other specific pairwise comparisons. Significance markers: NS = Padj>0.01; *0.001 < Padj<0.01; **0.0001 <Padj<0.001; ***Padj<0.0001.
We then narrowed our analysis to determine the nature of the interaction between AZD treatment and genotype in the MZ and EZ by performing pairwise comparisons. In addition to the small but significant decrease in MZ cell number and the increase in TZ cell number of the WT root tips treated with AZD, we also found that the EZ length was significantly shorter in AZD-treated WT compared to DMSO controls (Figure 6(b)). Interestingly, we observed that while the number of EZ cells remained unchanged, the average cell length in the EZ of the AZD-treated WT root tips was significantly smaller than that of DMSO controls, indicating that the decreased size of the EZ was specifically due to a decrease in cell length in this zone (Figure 6(c,d)). This suggests that TOR activity is required to increase rates of cell elongation, consistent with previous reports (Yuan et al., 2020). However, in net contrast, we found that AZD inhibition of TOR in the Ire1a Ire1b mutant led to an increase in the zone length and the number of cells in both the MZ and EZ compared to DMSO-treated Ire1a Ire1b controls (Figure 6(b,c)). Additionally, we also verified an increase in cell size in the EZ of AZD-treated Ire1a Ire1b (Figure 6(d)). In every measured zone metric of the root tip organization, we found no significant differences between AZD-treated WT and AZD-treated Ire1a Ire1b root tips (Figure 6), consistent with an AZD-mediated rescue of the Ire1a Ire1b root growth phenotype verified at D10 (Figure 4). These results support that, while basal TOR activity is needed to increase rates of cell elongation, TOR hyperactivity in the Ire1a Ire1b mutant is detrimental to elongation processes.
TOR hyperactivity in the Ire1a Ire1b mutant promotes cell differentiation rather than cell proliferation
Based on the results that TOR hyperactivity in the Ire1a Ire1b mutant has a detrimental effect on cellular elongation (Figure 6) and on previously published reports that after cell elongation TOR activity is necessary to actuate root hair growth in the differentiation zone (also referred to as the maturation zone; Retzer and Weckwerth, 2021), we hypothesized that a hyperactive TOR may lead to increased rates of differentiation, which would halt cell elongation. Therefore, we sought to test whether TOR hyperactivity in the Ire1a Ire1b mutant affects rates of cell proliferation, which have already been associated with TOR activity (Xiong et al., 2013), or leads to increased rates of cell differentiation. We utilized 5-ethynyl-2′-deoxyuridine (EdU; a thymidine analog that marks cell cycle entry into the S-phase; Hayashi et al., 2013) to perform a pulse-chase experiment of labeled nuclei in intact roots. This would allow us to determine the rate of DNA synthesis at the root tip as a measure of cell proliferation and would also allow us to track labeled meristematic cells over time to determine relative rates of cell differentiation, which is marked by root hair initiation (Dolan and Davies, 2004). At D7, whole seedlings grown on DMSO- or AZD-containing media were treated in liquid half-strength Linsmaier and Skoog (½ LS) media containing EdU for 20 min. Subsets of seedlings were then immediately fixed (0 h; Figure 7) while the rest of the seedlings were returned to their original ½ LS plates for an additional 6 h allowing for further root growth before fixation (6 h; Figure 7). Z-series of consecutive images for each root tip were then collected by confocal microscopy and assembled into max intensity projections (Figure 7(a)). For each root dataset, the sum EdU intensity from the entire image (Figure 7(b)) was quantified after background subtraction, which was identical for each image. With this experimental setup, we then sought to determine if there were differences in cell proliferation between WT and Ire1a Ire1b under conditions of TOR inhibition. We therefore tested the effects of EdU-chase time, genotype, and AZD treatment on sum EdU intensity via three-way wANOVA. We did not find a significant interaction between these variables, nor did we find a significant effect of genotype or AZD treatment alone on sum EdU intensity (Table S1). Pairwise comparisons of these values did not indicate any significant differences between WT and Ire1a ire1 upon DMSO or AZD treatment, suggesting that the IRE1 mutation and low-level TOR inhibition do not have a significant effect on the total rates of EdU incorporation over the entire root and, therefore, cell proliferation (Figure 7(b)). However, from visual observations (Figure 7(a)) it seemed that the AZD treatment altered the size of the root region marked by the EdU signal, which is inconsistent with our results in Figure 7(b). We hypothesized that this discrepancy (between visual observations in Figure 7(a) and data in Figure 7(b)) could be due to altered spatial distribution of the EdU incorporation, which may not be immediately evident to the naked eye. To address this, we measured the sum Edu intensity as a function of distance away from the quiescent center (QC; Figure 7(c), Figure S10(a,b)). Consistent with the visual observations of Figure 7(a), we did find a significantly lower level of EdU incorporation in AZD-treated WT and Ire1a Ire1b between 350 and 450 μm in 0 h samples and between 400 and 600 μm in 6 h samples (see Region 2; Figure S10(c,d); Table S1). In contrast we found that AZD treatment of WT and Ire1a Ire1b samples leads to increased levels of EdU incorporation between 100 and 250 μm in both 0 and 6 h samples (see Region 1; Figure S10(c,d); Table S1). Together these results indicate that both the loss of IRE1 and low-level treatment of roots with AZD do not significantly compromise the rate of cell proliferation as measured by total EdU incorporation; however, chronic, low-level AZD treatment does lead to alterations in the spatial distribution of cell proliferation at the root meristem.
Figure 7.
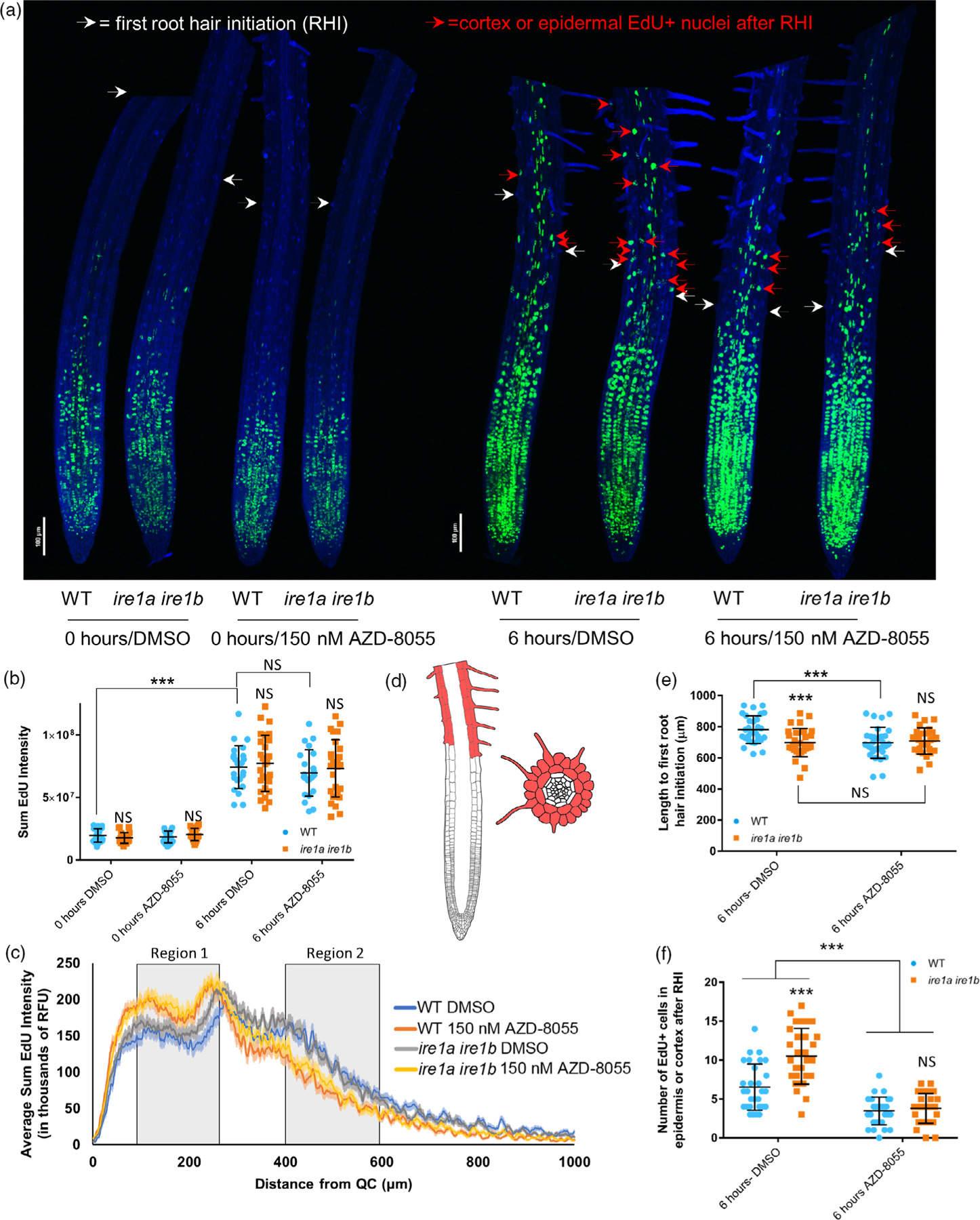
TOR hyperactivity in the Ire1a Ire1b mutant promotes differentiation rather than cellular proliferation. Seedlings grown for 7 days on media containing DMSO or AZD-8055 were treated briefly with EdU and immediately fixed (0 h) or returned to plates and allowed to grow for a further 6 h (6 h). See the Experimental Procedures section for full analysis methods. (a) Composite image compiled from representative 10× max projection images of root tips from all treatments. (b) Sum EdU intensity and (c) average EdU sum intensity along the root tip (6 h) was determined from max projection images of individual roots. (d) Diagrams of vertical and horizontal cross-sections of an Arabidopsis root tip highlighting the epidermal and cortex cell layers above the first RHI where EdU-positive nuclei were counted. (e) Length to the first RHI and (f) the number of differentiated EdU+ nuclei were determined for each root by manual assessment of Z-series images. For all graphs error bars show the standard deviation; significance markers displayed above an Ire1a Ire1b experimental group are pairwise comparisons to the corresponding WT group for that specific treatment. Brackets denote other specific pairwise comparisons. Significance markers: NS = Padj > 0.01; *0.001 < Padj < 0.01; **0.0001 < Padj < 0.001; ***Padj < 0.0001.
We next aimed to test the effect of IRE1 on rate of cell differentiation. To do this, we manually analyzed the Z-series images for each root in the 6-h chase sample pool to identify the first cells bearing root hairs along the root tip axis (i.e., first root hair initials [RHI]), as markers for tissue differentiation (Figure 7(d,e); Dolan and Davies, 2004). We then counted the number of EdU-positive (EdU+) nuclei in the cortex or epidermal cells past the first RHI (hereafter referred to as differentiated EdU+ cells; Figure 7(d,f)). Nuclei that were uniformly labeled (corresponding to labeling during the early S-phase) and nuclei that displayed a speckled pattern (corresponding to labeling during late S-phase) were both counted as EdU+ (Hayashi et al., 2013). We specifically counted EdU+ nuclei in the cortex and epidermal cell layers because they are unambiguously identifiable based on their size and because they only undergo cell division in the MZ (Dolan and Costa, 2001). We found that there was a significant interaction between the effects of AZD treatment and genotype on the length to the first RHI (two-way ANOVA: F(1,113) = 7.92, P = 5.76 × 10−3; Figure 7(e)) as well as significant effects of AZD treatment and genotype alone on the length to the first RHI (F(1,113) = 12.3, P = 6.38 × 10−4; F(1,113) = 12.6, P = 5.48 × 10−4, respectively). We also established that the root tip length to first RHI was significantly shorter in Ire1a Ire1b than WT in DMSO conditions. AZD treatment significantly shortened the length to first RHI in WT but not in Ire1a Ire1b, which remained unchanged (Figure 7(a,f)). This pattern matches the D7 root growth phenotype where AZD-treated WT and Ire1a Ire1b roots have primary root lengths that are identical to the Ire1a Ire1b roots grown in DMSO (Figure 4(a,c)).
While we found that the outward morphological characteristics and rates of cell proliferation were similar between DMSO- and AZD-treated Ire1a Ire1b roots, the number of differentiated EdU+ cells was markedly different (Figure 7(e)). Specifically, we found a significant interaction between the effects of AZD treatment and genotype on the number of differentiated EdU+ cells (two-way wANOVA: F(1,113) = 13.4, P = 3.81 × 10−4), as well as significant effects of AZD treatment and genotype alone on the number of differentiated EdU+ cells (F(1,113) = 22.0, P = 7.84 × 10−6; F(1,113) = 20.5, P = 1.51 × 10−5, respectively). In the Ire1a Ire1b root tips, which had higher levels of TOR activity in DMSO conditions (Figure 5), we found that the number of differentiated EdU+ cells was nearly 2-fold higher than WT. We also established that AZD treatment led to a significant reduction in the number of differentiated EdU+ cells in both WT (approximately 2-fold) and Ire1a Ire1b (approximately 4-fold), such that there was not a significant difference between WT and Ire1a Ire1b in AZD conditions. Together these results support that an IRE1-dependent limitation of TOR activity is required to prevent uncontrolled increases in the rate of cell differentiation from the meristem in the shootward direction.
DISCUSSION
Loss-of-function mutations of IRE1, the most conserved master regulator of the UPR across eukaryotes, cause a wide variety of defects in growth and development in plants and metazoans (Bao et al., 2019; Chen et al., 2014; Kim et al., 2018; Mishiba et al., 2019; Mitra and Ryoo, 2019). In metazoans, some of the causative relationships between the loss of IRE1 activity and developmental defects have been defined (Mitra and Ryoo, 2019). In marked contrast in plants, prior to this work, a functional connection between the loss of IRE1 and developmental defects had yet to be made. To address this significant gap, we performed a detailed analysis of the tractable Ire1a Ire1b model, which exhibits a distinctive defect in primary root growth. We found that the Ire1a Ire1b root growth defects are specifically brought about by age-related increases in rates of organ growth, which are most likely tied to increased availability of carbohydrates as the plants mature. We established that such defects primarily manifest through ineffective actuation of cellular elongation at the root tip, leading to shorter roots that do not maintain gravity-driven growth vectors. We found that in actively growing root tips of the Ire1a Ire1b mutant, TOR activity is significantly elevated compared to WT, and that low-level inhibition of TOR restores the Ire1a Ire1b root growth phenotype to WT levels. We further demonstrated that such TOR hyperactivation drives increased rates of cell differentiation at the root tips. Therefore, our work demonstrates that IRE1 controls TOR activity in specific developmental stages in physiological conditions of growth. In addition to supporting the canonical role for TOR as a driver of cell proliferation in the root tip, our results also reveal a new role of TOR in cell differentiation whose functional homeostasis depends on IRE1 availability.
IRE1 regulates TOR activity in a multicellular eukaryote
In mammalian models, some connections between mammalian TOR (mTOR) activity and IRE1 regulation have been found previously (Kato et al., 2012, 2013; Li et al., 2014; Pfaffenbach et al., 2010; Sanchez-Alvarez et al., 2017; Shanware et al., 2014; Young et al., 2013). In cases of light-induced retinal injury, hepatic lipotoxicity, chemically induced ER stress, cadmium toxicity, and lipid-starved solid tumor microenvironments, mTOR activity induces cell apoptosis either through aggravation of general ER stress (Li et al., 2014; Pfaffenbach et al., 2010) or through specific activation of the IRE1-induced apoptosis via the IRE1-JNK kinase signal cascade (Kato et al., 2012, 2013; Young et al., 2013). Significantly, in all of these studies modulation of IRE1 activity is a downstream effect of mTOR activation. In this work, we show that in actively growing root tips of the Ire1a Ire1b mutant TOR is hyperactive (Figure 5). These results, in conjunction with the observations that low-level TOR inhibition completely rescues all aspects of the Ire1a Ire1b root growth phenotype (Figures 4, 6, and 7), lead us to conclude not only that IRE1 controls TOR activity, but also that this is the primary pathway by which IRE1 promotes proper organ biogenesis. To our knowledge, the results presented in this work are the first to demonstrate that IRE1 regulates TOR activity in any model organism and that such activity occurs in the absence of induced ER stress.
IRE1 is necessary to control TOR activity at tissue- and development-specific levels
Previous reports indicated that IRE1 contributes to Arabidopsis growth and development on a broad level and have repeatedly demonstrated that the Ire1a Ire1b mutant has a short root phenotype (Chen and Brandizzi, 2012; Deng et al., 2013; Mishiba et al., 2019; Pu et al., 2019). However, a detailed dissection of events leading to this phenotype at the cell and tissue levels and an accounting of how variation in standard growth conditions could alter it were lacking. In our work, we have demonstrated that the increased rates of root growth maintained in WT plants as they mature cannot be actuated in the Ire1a Ire1b mutant (Figure 1). We established that the observed defects are primarily restricted to the EZ and secondarily to the MZ as a function of seedling age (Figure 2), and are concurrent with hyperactivation of TOR in the Ire1a Ire1b mutant specifically at the root tips (Figure 5). At the initial phase of rapid root growth (day 7), we found that the Ire1a Ire1b EZ exhibits fewer and smaller cells compared to WT (Figure 2), but we did not observe any significant differences in cell size and proliferation rate in the Ire1a Ire1b MZ compared to WT (Figures 2 and 7). Therefore, our results argue that, in early developmental stages in WT, IRE1 is necessary to maintain homeostatic levels of TOR activity in the EZ but not in the MZ. At the later stages of the Ire1a Ire1b phenotype development (i.e., D10), we found that the Ire1a Ire1b MZ is shorter, and has not increased in size like the WT MZ (Figure 2). Fascinatingly, we also found that the reduction in the number of cells in the Ire1a Ire1b MZ is reverted to WT levels by chemical inhibition of TOR (Figure 6). Based on these results, we conclude that TOR activity may have opposite effects in the MZ during the rapid growth of the root tip: basal TOR activity promotes cell proliferation in the MZ, as reported earlier (Li et al., 2017; Xiong et al., 2013), but TOR hyperactivity may also dampen it, as demonstrated in this work. In connection with our Ire1a Ire1b root phenotypic results, we infer that IRE1 activity is required to control TOR activity especially at stages of development requiring increased cell proliferation and elongation. Although the underlying mechanisms on the MZ size control exerted by IRE1 through TOR are yet unknown, our data support that IRE1 is absolutely required to antagonize TOR hyperactivation to maintain proper organ development.
IRE1-dependent repression of TOR is independent of the unconventional splicing of bZIP60
In conditions of induced ER stress, IRE1 splices the mRNA of its effector TF, bZIP60 (Nagashima et al., 2011). It is well established that in physiological conditions of growth, a bzip60 complete loss-of-function mutant does not exhibit a short root phenotype (Chen and Brandizzi, 2012; Moreno et al., 2012; Nagashima et al., 2011). Based on our results that Ire1a Ire1b has a marked root length phenotype (Figure 1) and the notion that an Ire1a Ire1b bzip60 triple mutant is phenotypically indistinguishable from Ire1a Ire1b under normal growth conditions (Ruberti et al., 2018), we deduce that the molecular mechanisms by which IRE1 controls root development and TOR activity are independent of a functional interaction with bZIP60. The loss of the mammalian homolog of bZIP60, XBP1, compromises tissue development in a similar way to IRE1 loss-of-function mutations; for example, both mutations lead to defective embryonic liver development (Mitra and Ryoo, 2019). Hence, the results that in physiological conditions of growth a bzip60 mutant does not exhibit a visible phenotype highlight a functional divergence between the IRE1-dependent TFs XBP1 and bZIP60 in organ development across kingdoms.
Metazoan and plant IRE1 proteins are known to cleave transcripts other than XBP1 or bZIP60 through RIDD (Hollien et al., 2009; Mishiba et al., 2013). Therefore, we hypothesize that under high growth pressure due to a prolonged photoperiod and abundant carbohydrate supply, conditions that we selectively applied in our work (Figure 3), IRE1-mediated RIDD of a single or multiple RNA targets could lead to a strong limitation or cap on TOR activity in certain tissue types. Due to the fact that known Arabidopsis IRE1-RIDD targets have thus far been identified exclusively in ER stress conditions using RNA derived from whole seedlings (Mishiba et al., 2013), the identification of the intermediate targets between IRE1 and TOR under physiological conditions in actively growing root tissues remains an exciting topic for future study.
In Arabidopsis, TOR activity depends on a variety of cues (e.g., mitochondrial respiration, auxin, amino acids; Burkart and Brandizzi, 2020; Li et al., 2017; Schepetilnikov et al., 2017; Shi et al., 2018). Hence potential RIDD targets may affect one or several of these pathways. Nonetheless, based on our observations that link the Ire1a Ire1b phenotype to photoperiod- and carbohydrate-related increases in rates of root growth (Figure 3), we speculate that the most probable target may be associated with IRE1 and regulation of mitochondrial respiration. This is predicated by the evidence that the assembly of the mTOR complex 1 depends on the TTT-RUVBL1/2 complex, which leads to activation of mTOR through formation of mTOR obligate dimers (Kim et al., 2013). The activity of the TTT-RUVBL1/2 complex is in turn strongly dependent on mitochondrial ATP generation through respiration. ER functions and mitochondrial metabolism are closely linked in metazoan models primarily through calcium delivery to mitochondria via the ER (Gutiérrez et al., 2020; Hirabayashi et al., 2017; Kaufman and Malhotra, 2014; Rieusset, 2018; Szabadkai et al., 2006). In Arabidopsis, IRE1 activity has been tied to a regulation of mitochondrial stress responses, albeit under induced ER stress situations (Ng et al., 2013). Therefore, it is possible that in Arabidopsis, IRE1 may regulate mitochondrial respiration in rapidly growing tissues under physiological conditions.
TOR activity regulates cellular differentiation and elongation in actively growing Arabidopsis root tips
Previous work supports that TOR activity can promote cell elongation. Specifically, TOR was shown to promote accumulation of the auxin efflux transporter PIN2 at the root meristem through a direct protein–protein interaction, leading to increased size of cells in the EZ without affecting cell proliferation (Yuan et al., 2020). In our work, we have demonstrated that low-level TOR inhibition of rapidly growing WT roots leads to a small reduction in root length (Figure 4, D7) and is sufficient to significantly impact TOR activity at the root tip (Figure 5). However, this minimal TOR inhibition does not strongly affect overall rates of cell proliferation and instead leads to smaller cells in the EZ and a reduced rate of cell differentiation (Figures 6 and 7). In the Ire1a Ire1b root tips, we found that TOR hyperactivity coincided with increased rates of cell differentiation compared to WT, which were reduced to WT levels with low-level TOR inhibition (Figure 7).
A plausible model to illustrate the effect of Ire1a Ire1b mutation and TOR inhibition on cell differentiation is presented in Figure S11. In this model, TOR activity is needed in the later stages of cell maturation to actuate cell differentiation programs in addition to the known roles of TOR in promoting cell proliferation. In contrast to the effect on cell differentiation, which responds in a linear manner to TOR activity, it seems that cell elongation has a biphasic response to TOR activation (Figure S11). Similar to cell differentiation, our data support that a basal level of TOR activity is also required to promote cell elongation. However, at high levels of TOR activity cell elongation is negatively impacted and is therefore rescued by TOR inhibition. We speculate that under TOR hyperactivity the increased rate of cellular differentiation negatively impacts the time that cells have to undergo cell elongation processes prior to root hair initiation, leading to a smaller maximum cell size in the EZ cells. Together these results provide further evidence that TOR has significant effects in determining cell fate outside of cell proliferation in the MZ and demonstrate that IRE1 is an upstream regulatory factor of TOR in these contexts. Therefore, this study provides an important foundation for future work by uncovering a novel link between two ancient eukaryotic signaling pathways. With this tractable Ire1a Ire1b model further investigation could yield important information related to UPR- and TOR-dependent control over multicellular organism development.
EXPERIMENTAL PROCEDURES
Plant material and high-quality seed production
Seeds of the Arabidopsis thaliana Ire1a (WISCDSLOX420D09) and Ire1b (SAIL_238_F07) mutants were obtained from the Arabidopsis Biological Resource Center (Columbus, OH, USA), and the Ire1a Ire1b double mutant was published previously (Chen and Brandizzi, 2012). Special care was taken to produce high-quality seeds to enable vigorous and reproducible growth of seedlings on sterile culture plates without any exogenously supplied carbohydrates. WT and Ire1a Ire1b seeds used for the same experiments were always produced simultaneously. Plate-grown seedlings were transplanted to potting soil and grown in standard Arabidopsis growth chamber conditions (16 h 150 μE light/8 h dark; 50% humidity, 23°C). Plants were watered exclusively with Hoagland’s nutrient solution every approximately 7 days prior to bolting and then as needed after bolting (usually every 4 days). Prior to any silique senescence, inflorescences were staked and tied after they could not support their own weight to prevent seed loss. Two to four weeks after bolting, prior to any significant rosette senescence, one quarter to one half of all formed siliques had senesced. Seeds were harvested by tapping or gentle gripping of the tied inflorescence, and seeds that fell off with minimal physical disturbance were harvested and the remaining plant was discarded. Large numbers of plants were grown simultaneously to compensate for smaller seed yield per plant. Plants were not dried prior to seed harvest. In our experience seeds produced under these conditions had initial germination rates near 100% on 0% sucrose-containing plates and had germination rates of >98% on 0% sucrose after a year of storage in ideal conditions (seed envelope in a humidity-controlled, dark environment).
Plant phenotyping
Unless otherwise stated, all sterile culture plates were made with ½ LS basal salts-containing buffer (LSP03, Caisson Labs, Smithfield, UT, USA) with 1.0% w/v agar (NCM0236A, Neogen, Lansing, MI, USA) added and without exogenous sucrose (or other carbohydrates). Medium was adjusted to pH 5.7 and autoclaved for 20 min under standard conditions. Medium was cooled on a stir plate until the external temperature of the bottle was 40°C, dispensed into plates, and allowed to cool in a single layer (as opposed to stacked plates) to allow for even cooling rates. Plates were always made on the same day the seeds were sterilized and plated. Seeds were sterilized with 1× wash of 100% ethanol for 30 sec, 1× wash of 50% bleach with 0.1% Tween 20 for 1 min, and 6× washes with sterile double distilled water (ddH2O). Seeds were sterilized and wet plated using a 1-ml pipette immediately after plates had cooled. We suspect that extended incubation in ddH2O in the microcentrifuge tube after sterilization may affect germination and initial growth rates on 0% sucrose medium. After seed plating, plates were left open in the sterile hood for 3–5 min or until water from the plating method evaporated. Plates were wrapped with 1-inch surgical tape (70200534694, 3 M, MN, USA) with overlapping sections on the bottom of the plate and then wrapped in aluminum foil. Seeds were stratified at 4°C for 48 h. Plates were incubated vertically in Percival growth chambers in continuous 150 μE light (verified with an external PAR light meter) for the indicated growth periods. A minimum of five plate replicates per experimental group were used for each phenotyping experiment.
TORIN2 (MedChemExpress, Monmouth Junction, NJ, USA), AZD (MedChemExpress), Oryzalin (Sigma-Aldrich, St. Louis, MO, USA), Latrunculin b (Sigma-Aldrich), NAA (Sigma-Aldrich), and Oligomycin were dissolved in DMSO and Antimycin A in 100% ethanol to 10 mM and stored at −80°C. These chemicals were taken out of the freezer after the external temperature of the media bottle reached 40°C and added simultaneously to all bottles in that experiment at the required concentrations.
After the indicated incubation period, plates were removed from the Percival growth chamber as needed, imaged, used for downstream analysis, and then discarded. No repeated measures were performed on individual plates (i.e., different time points within the same experiment were recorded from two separate populations of seedlings). This was done to prevent possible confounding effects of altered gravity vectors from plate movement on the Ire1a Ire1b root phenotype during critical growth stages. Average SFW was determined by excising shoots from all plants on one plate (usually 8–10 shoots), weighing the total, and dividing by the number of seedlings. Root length was measured using ImageJ. Root tip angles were also measured using ImageJ by placing an approximately 3-mm line over the root tip (beginning at the apex) and using the Feret diameter measurement function which was then converted into a 360° scale.
mPS-PI staining and meristem cellular organization analysis
The mPS-PI staining protocol was adapted from Truernit et al. (2008). After the indicated growth period, whole seedlings (for 5- or 7-day-old seedlings) or 2-cm excised roots sections (for 10-day-old seedlings) were fixed in a 5:4:1 methanol:ddH2O:acetic acid solution and stored at 4°C for a minimum of 12 h and a maximum of 2 weeks in capped 2-dram vials. For the staining procedure seedlings/roots were treated in six-well plates to avoid mechanical damage. After removal of fixative, tissue was washed with 10 ml of ddH2O and then treated with 5 ml of a 1% periodic acid (PA) solution for 40 min. After removing the PA solution, tissue was washed with 10 ml of ddH2O and then treated with 5 ml of freshly made mPS-PI working reagent (100 mM sodium metabisulfite, 0.15 m HCl, 100 μg ml−1 propidium iodide) for 40 min in the dark. After removing the mPS-PI solution samples were was washed with another 10 ml of ddH2O and then submerged in 3 ml of the chloral hydrate alternative Visikol to clear the tissue. Samples were sealed with parafilm and incubated for 2–3 days prior to confocal microscopy analysis.
Samples were imaged using a Nikon A1 confocal microscope with excitation at 488 nm and detection at 520–720 nm. Transmitted light detection images were also collected in parallel to assist in root tip zone identification. For each root, one 20× image was collected for accurate identification and quantification of MZ and TZ metrics and a 10× image was collected for identification of TZ and EZ metrics. The beginning of the TZ was defined as the first cell in the cortical cell layer which was >2 times as long as the previous one (Casamitjana-Martinez et al., 2003; Di Mambro et al., 2017). The first cortical cell which was twice as long as it was wide was identified as the beginning of the EZ. The end of the EZ was defined as the last cortical cell before the first visible root hair initiation, which was identified using the transmitted light image if no root hair initiations were visible in the same focal plane as the cortex cells. In some instances (particularly for WT D10 samples), two images were required to fully measure the length of the EZ.
TOR activity assays
Tissue for TOR activity assays was collected from 7-day-old seedlings. One biological replicate consisted of 60 seedlings grown on two plates. The plates were removed from the Percival growth chamber and approximately 3 mm of the root tip from all 60 seedlings was quickly but carefully cut in situ using surgical scissors. A 3-mm reference object was used to ensure accuracy. After cutting, forceps were used to gently collect root tips into a prepared microcentrifuge tube with two glass beads and placed in liquid nitrogen. Then a second cut was used to excise the mature root tissues, which were collected into a second microcentrifuge tube with two glass beads. The excision and collection of both tissue types took less than 5 min total for each biological replicate. This process was repeated, alternating experimental groups until all biological replicates were collected. Continuous light was used to eliminate potential sample variation caused by circadian-dependent processes over the multiple hours required to harvest these samples by an individual researcher. Biological replicates in Figure 5 were collected over four separate experiments.
Frozen samples were ground to a powder using a Retch Mixer Mill (Retch; Haan, Germany) in 2× 10-sec bursts and then 4× 20-sec bursts with refreezing in liquid nitrogen between bursts and after the final grinding. To the frozen root tip tissue, 100 μl of extraction buffer (EB) containing phosphate-buffered saline (PBS) pH 7.4, plant protease inhibitor cocktail (P9599, Sigma-Aldrich), and PhosSTOP phosphatase inhibitor (4906845001, Roche) was added to the root tip samples and 200 μl of EB to the mature root samples. Tubes were shaken and vortexed until the sample/buffer was melted and homogenized and then kept on ice (30 sec). The entire sample/supernatant was transferred into a new tube without the beads and sequentially spun down at 21 000 ×g for 5, 10, and 15 min in a 4°C cooled centrifuge, transferring the supernatant to a new tube between each spin. Protein content of the root tip extracts was sufficient to load 2.5 μg of total protein on two 12% SDS-PAGE gels. Gels were transferred to PVDF membranes (1620177, Bio-Rad, Hercules, CA, USA), which were blocked with 3% bovine serum albumin (BSA) in Tris-buffered saline plus Tween 20 (TBST) for 1 h at room temperature and then incubated overnight at 4°C with primary antibodies in TBST to detect total S6K (αS6K1/2, AS12 1855; Agrisera, Vännäs, Sweden) or phospho-S6K (ab207399, Abcam, Cambridge, UK). Blots were washed three times with TBST for 20 min each and then incubated with secondary HRP-conjugated goat anti-rabbit antibody (A0545, Sigma-Aldrich) for 1 h at room temperature. Blots were developed using the SuperSignal West Femto Kit (34096, ThermoFisher Scientific, Waltham, MA, USA).
EdU pulse-chase experiments
EdU pulse-chase was performed on 7-day-old seedlings grown on plates containing DMSO or 150 nM AZD using the Click-iT EdU Alexa Fluor 488 Imaging Kit (C10337, ThermoFisher Scientific). Seedlings were transferred from Petri dishes to six-well plates containing 10 μM EdU in ½ LS medium (no sucrose) and then placed back in the Percival growth chamber for 20 min. At the end of the incubation period, the ½ LS medium with EdU was removed and the seedlings were gently washed 3× with 5 ml of ½ LS media (without EdU). Half of the seedlings were then transferred to fixation buffer (4% paraformaldehyde, 0.1% Triton X-100, 1 × PBS pH 7.4) while the other half were returned to their growth plates. Plates were rewrapped in surgical tape and placed back in the Percival growth chamber for 6 h. Chase samples were transferred to fixation buffer after the 6-h chase period. Samples were then stored for between 12 h and 1 week at 4°C. Samples were stained and imaged in small batches (with some samples from each experimental group) on each day over that week and no degradation of sample was found.
The Click-iT reaction was performed according to the manufacturer’s protocol with some modifications. Seedlings were removed from the fixation buffer and washed three times with 3% BSA in PBS. Click-iT reaction cocktail was prepared without modification and samples were incubated with the cocktail for 1 h in the dark. Shorter incubation periods lead to incomplete tissue penetration of the reaction cocktail into the MZ. After incubation, samples were washed 1× with 3% BSA in PBS and then 3× with PBS. Samples were then incubated with Hoechst 33342 provided with the Click-iT kit (working solution created by diluting 1 μl Hoechst 33342 in 1 ml of PBS) for 40 min in the dark. Samples were then washed with two washes of PBS. Shoot tissue was removed and roots were mounted in PBS on slides using polypropylene packing tape to create a watertight imaging chamber of uniform depth (approximately 70 μm). Z-series images were collected over the entire depth of the root tips using a Nikon A1 confocal microscope equipped with a 10× objective. Sum EdU intensity was quantified in each root tip using ImageJ after transformation of Z-series images into max intensity projects and background subtraction (which was equally applied to all images) in the Nikon NIS-Elements Advanced Research software. Length to the first RHI and the number of EdU+ nuclei found after the first RHI were manually quantified using the Z-series images.
Data reporting and statistical analysis
No statistical methods were used to determine the sample size for these experiments. Sample sizes were determined by the maximum number of replicates which could be grown simultaneously in a uniform incubation environment (i.e., on the same shelf of a Percival growth chamber) or by the maximum number of samples that could be collected by a single researcher in a reasonable time frame.
All statistical analysis was performed using R. Two-way or three-way between-subjects ANOVA was conducted as needed on each dataset to determine the effects of experimental variables on test outcomes. Type III ANOVAs were used for unbalanced datasets. For each dataset, residual analysis was performed in order to test for the assumptions of the ANOVAs. The Shapiro–Wilk test was used to check normality and homogeneity of variance was assessed via the studentized Breusch–Pagan test and Levene’s test. In most of the datasets the homogeneity of variance assumption was violated due to the increased variance in the Ire1a Ire1b short root phenotype (see changes in standard deviation of the Ire1a Ire1b root length over time in Figure 1(c) and Figure S3(d, e)). If the assumption tests were violated (P < 0.05), Box–Cox or log transformations were applied to the dataset and the ANOVA was rerun using transformed data. If the assumption checks were still violated after Box–Cox or log transformations, then weighted least squares regression was applied to the ANOVA model (referred to in the text as wANOVA). All datasets which were subjected to weighted least squares regression passed the residual analysis. From the appropriate ANOVA model for each dataset (standard, transformed, or weighted least squares) pairwise comparisons (CRAN R package emmeans) were then run with Bonferroni adjustment applied. Significance markers in figure graphs were based on the results of these pairwise comparison tests. Code of the R packages used for the analysis pipeline is found in Data S1.
To determine if there were significant differences in the CoV between root tip angles of two different experimental groups, we utilized the asymptotic Feltz and Miller test (Feltz and Miller, 1996) as applied by Rodriguez et al. (2020) using the CRAN R package cvequality.
Supplementary Material
Figure S1. Rate of shoot fresh weight accumulation and rate of primary root growth for data displayed in Figure 1. (a) Average shoot fresh weight values from Figure 1(b) were reprocessed to show the rate of shoot fresh weight accumulation for a specific growth period (value = average shoot fresh weight accumulated during a growth period divided by the number of days in that growth period). (b) Average root length values from Figure 1(c) were reprocessed to show the rate of primary root length increases for a specific growth period (value = average root length increases accumulated during a growth period divided by the number of days in that growth period).
Figure S2. At D5 there are no significant defects in Ire1a Ire1b meristem organization. At D5, root tips were subjected to mPS-PI staining and confocal microscopy to analyze root tip cellular organization. (a) Representative 10× images of mPS-PI stained WT and Ire1a Ire1b roots. The yellow line demarks the end of the meristem zone (MZ) and the beginning of the transition zone (TZ), the red lines marks all of the cells in the elongation zone (EZ), and the white arrow marks the first root hair initiation. For all measurements of the MZ secondary 20× images were used to collect data. (b) The average length of each zone. (c) The number of cells in each zone. (d) Average cell length in each zone. For all graphs error bars show the standard deviation; significance markers displayed above an Ire1a Ire1b experimental group are pairwise comparisons to the corresponding WT group for that specific treatment. Brackets denote other specific pairwise comparisons. Significance markers: NS = Padj> 0.01; *0.001 < Padj < 0.01; **0.0001 < Padj < 0.001; ***Padj < 0.0001.
Figure S3. Average root tip cellular organization metrics displayed over time in WT and Ire1a Ire1b. Average values from the data presented in Figure S2 and Figure 2 are plotted over time to compare differences over time in each genotype separately. (a–c) Zone length, number of cells, and cell length in the WT root tips. (d–f) Zone length, number of cells, and cell length in the Ire1a Ire1b root tips. For all graphs: error bars are standard deviation; significance markers displayed over an experimental group at D7 or D10 are comparisons to the corresponding D5 value for that group. Brackets denote other specific pairwise comparisons. Significance markers: NS = Padj > 0.01; *0.001 < Padj < 0.01; **0.0001 < Padj < 0.001; ***Padj < 0.0001. P-value indicator color matches the color of the corresponding root tip zone.
Figure S4. TOR inhibitor AZD-8055 concentration response analysis. (a) Representative images from DMSO, 100 nM, and 200 nM AZD-8055 conditions. Red lines represent approximate average root length of the displayed seedlings. (b) Root length of individual roots was measured using ImageJ. For all graphs, error bars show the standard deviation; significance markers displayed above an Ire1a Ire1b experimental group are pairwise comparisons to the corresponding WT group for that specific treatment. Brackets denote other specific pairwise comparisons. Significance markers: NS = Padj > 0.01; *0.001 < Padj < 0.01; **0.0001 < Padj < 0.001; ***Padj < 0.0001. For this experiment the growth chamber used had 200 μE continuous light which caused proportionally shorter roots in both WT and Ire1a Ire1b compared to 150 μE light; however, the general conclusions from the DMSO versus AZD treatments are the same in both conditions.
Figure S5. The TOR inhibitor TORIN2 rescues the Ire1a Ire1b root growth phenotypes. (a) Representative images from DMSO and TORIN2 conditions. Red lines represent approximate average root length of the displayed seedlings. (b) Root length of individual roots was measured using ImageJ. Error bars show the standard deviation; significance markers displayed above an Ire1a Ire1b experimental group are pairwise comparisons to the corresponding WT group for that specific treatment. Brackets denote other specific pairwise comparisons. Significance markers: NS = Padj > 0.01; *0.001 < Padj < 0.01; **0.0001 < Padj < 0.001; ***Padj < 0.0001. (c) The angle of the root tip with the vertical axis (0°) was measured using ImageJ. Significant differences between coefficients of variation were tested using the asymptotic Feltz and Miller test as described in the Experimental Procedures section. Significance markers: NS = P > 0.05; **0.001 < P < 0.01; ***P < 0.001.
Figure S6. Complementation of the Ire1a Ire1b mutant rescues root growth and AZD-8055 sensitivity phenotypes. (a) Representative images from DMSO and AZD-8055 treatment conditions. Red lines represent approximate average root length of the displayed seedlings. (b) Root length of individual roots was measured using ImageJ. Error bars show the standard deviation; significance markers displayed above an Ire1a Ire1b experimental group are pairwise comparisons to the corresponding WT group for that specific treatment. Brackets denote other specific pairwise comparisons. Significance markers: NS = Padj > 0.01; *0.001 < Padj < 0.01; **0.0001 < Padj < 0.001; ***Padj < 0.0001. (c) The angle of the root tip with the vertical axis (0°) was measured using ImageJ. Significant differences between coefficients of variation were tested using the asymptotic Feltz and Miller test as described in the Experimental Procedures section. Significance markers: NS = P > 0.05; **0.001 < P < 0.01; ***P < 0.001.
Figure S7. Other chemical inhibitors of root growth do not rescue the Ire1a Ire1b root length phenotype. (a) Representative images from DMSO, Oryzalin, Latrunculin b, and Oligomycin growth conditions. Red lines represent approximate average root length of the displayed seedlings. This experiment and TORIN2 experiments (Figure S5) were performed simultaneously and share the DMSO control data. (b) Root length of individual roots was measured using ImageJ. (c) Representative images from Antimycin A and ethanol control growth conditions. Red lines represent approximate average root length of the displayed seedlings. (c) Root lengths of individual roots for ethanol and Antimycin A growth conditions. For all graphs, error bars show the standard deviation; significance markers displayed above an Ire1a Ire1b experimental group are pairwise comparisons to the corresponding WT group for that specific treatment. Brackets denote other specific pairwise comparisons. Significance markers: NS = Padj > 0.01; *0.001 < Padj < 0.01; **0.0001 < Padj < 0.001; ***Padj < 0.0001.
Figure S8. Auxin inhibition of root growth and TOR inhibition of root growth are additive effects in WT plants. (a) Representative images of seedlings grown on plates containing various concentrations of NAA, with or without addition of AZD-8055. Red lines represent approximate average root length of the displayed seedlings. (b) Primary root length after 10 days of growth. For graphs error bars show the standard deviation; significance markers displayed above an Ire1a Ire1b experimental group are pairwise comparisons to the corresponding WT group for that specific treatment. Brackets denote other specific pairwise comparisons. Significance markers: NS = Padj > 0.01; *0.001 < Padj < 0.01; **0.0001 < Padj < 0.001; ***Padj < 0.0001.
Figure S9. Full blot images (from Figure 6) and the Ponceau stain images to demonstrate equal protein loading.
Figure S10. Chronic, low-level TOR inhibition leads to a spatial redistribution of EdU incorporation at the root tip. For each image EdU sum intensity along the root tip was measured in ImageJ for (a) 0 h and (b) 6 h samples (standard error displayed as lighter shaded area above and below each line). Then, for each root, the mean sum EdU intensity value was determined for two specific regions of interest for (c) 0 h and (d) 6 h samples. AZD treatment, but not genotype significantly affected the average sum EdU intensity in all regions of interest (see Table S1 for ANOVA results) For all graphs error bars show the standard deviation; significance markers displayed above an Ire1a Ire1b experimental group are pairwise comparisons to the corresponding WT group for that specific treatment. Brackets denote other specific pairwise comparisons. Significance markers: NS = Padj > 0.01; *0.001 < Padj < 0.01; **0.0001 < Padj < 0.001; ***Padj < 0.0001.
Figure S11. Simplified model based on our results showing that TOR activity must be balanced by IRE1 to maintain optimal cell elongation. Basal TOR activity is needed for cell elongation, illustrated by the difference between WT DMSO and WT AZD-8055. However, TOR hyperactivity promotes faster and unbalanced rates of differentiation, which favors premature root hair initiation and cessation of cell elongation. This in turn leads to shorter length of mature cells and an overall shorter root length in the Ire1a Ire1b mutant.
Table S1. Full results for all ANOVAs performed.
Data S1. R code used for the ANOVA statistical analysis pipeline.
ACKNOWLEDGMENTS
We acknowledge support by the Chemical Sciences, Geosciences and Biosciences Division, Office of Basic Energy Sciences, Office of Science, U.S. Department of Energy (award number DE-FG02-91ER20021) for infrastructure, NASA (award 80NSSC19K0707), NIH R35-GM136637, and a fellowship from Michigan State University under the Training Program in Plant Biotechnology for Health and Sustainability (T32-GM110523). We would also like to thank the Center for Statistical Training and Consulting at Michigan State University for their kind assistance and for providing the necessary code to perform the ANOVA tests utilizing weighted least squares regression.
Footnotes
CONFLICT OF INTEREST
The authors of this work declare that they have no conflicts of interest associated with this work.
SUPPORTING INFORMATION
Additional Supporting Information may be found in the online version of this article.
DATA AVAILABILITY STATEMENT
Data supporting the findings of this work can be found within the manuscript and its supporting materials. All data and materials are available from the corresponding author.
REFERENCES
- Angelos E, Ruberti C, Kim S-J & Brandizzi F (2017) Maintaining the factory: the roles of the unfolded protein response in cellular homeostasis in plants. The Plant Journal, 90(4), 671–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y, Bassham DC & Howell SH (2019) A Functional unfolded protein response is required for normal vegetative development. Plant Physiology, 179(4), 1834–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrada A, Djendli M, Desnos T, Mercier R, Robaglia C, Montané MH et al. (2019) A TOR-YAK1 signaling axis controls cell cycle, meristem activity and plant growth in Arabidopsis. Development, 146(3). [DOI] [PubMed] [Google Scholar]
- Beaugelin I, Chevalier A, d’Alessandro S, Ksas B & Havaux M (2020) Endoplasmic reticulum-mediated unfolded protein response is an integral part of singlet oxygen signalling in plants. The Plant Journal, 102(6), 1266–1280. [DOI] [PubMed] [Google Scholar]
- Burkart GM & Brandizzi F (2020) A tour of TOR complex signaling in plants. Trends in Biochemical Sciences, 46(5), 417–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP et al. (2002) IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature, 415(6867), 92–96. [DOI] [PubMed] [Google Scholar]
- Cao P, Kim S-J, Xing A, Schenck CA, Liu L, Jiang N et al. (2019) Homeostasis of branched-chain amino acids is critical for the activity of TOR signaling in Arabidopsis, eLife 8, e50747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casamitjana-Martinez E, Hofhuis HF, Xu J, Liu CM, Heidstra R & Scheres B (2003) Root-specific CLE19 overexpression and the sol1/2 suppressors implicate a CLV-like pathway in the control of Arabidopsis root meristem maintenance. Current Biology, 13(16), 1435–1441. [DOI] [PubMed] [Google Scholar]
- Chen G-H, Liu M-J, Xiong Y, Sheen J & Wu S-H (2018) TOR and RPS6 transmit light signals to enhance protein translation in deetiolating Arabidopsis seedlings. Proceedings of the National Academy of Sciences, 115(50), 12823–12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Aung K, Rolčík J, Walicki K, Friml J & Brandizzi F (2014) Inter-regulation of the unfolded protein response and auxin signaling. The Plant Journal, 77(1), 97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y & Brandizzi F (2012) AtIRE1A/AtIRE1B and AGB1 independently control two essential unfolded protein response pathways in Arabidopsis. The Plant Journal, 69(2), 266–277. [DOI] [PubMed] [Google Scholar]
- Deng Y, Humbert S, Liu J-X, Srivastava R, Rothstein SJ & Howell SH (2011) Heat induces the splicing by IRE1 of a mRNA encoding a transcription factor involved in the unfolded protein response in Arabidopsis. Proceedings of the National Academy of Sciences, 108(17), 7247–7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Srivastava R & Howell SH (2013) Protein kinase and ribonuclease domains of IRE1 confer stress tolerance, vegetative growth, and reproductive development in Arabidopsis. Proceedings of the National Academy of Sciences, 110(48), 19633–19638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Mambro R, De Ruvo M, Pacifici E, Salvi E, Sozzani R, Benfey PN et al. (2017) Auxin minimum triggers the developmental switch from cell division to cell differentiation in the Arabidopsis root. Proceedings of the National Academy of Sciences, 114(36), E7641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan L & Costa S (2001) Evolution and genetics of root hair stripes in the root epidermis. Journal of Experimental Botany, 52(suppl_1), 413–417. [DOI] [PubMed] [Google Scholar]
- Dolan L & Davies J (2004) Cell expansion in roots. Current Opinion in Plant Biology, 7(1), 33–39. [DOI] [PubMed] [Google Scholar]
- Feltz CJ & Miller GE (1996) An asymptotic test for the equality of coefficients of variation from k populations. Statistics in Medicine, 15(6), 646–658. [DOI] [PubMed] [Google Scholar]
- Gao H, Brandizzi F, Benning C & Larkin RM (2008) A membrane-tethered transcription factor defines a branch of the heat stress response in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, 105(42), 16398–16403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemette T, Calmes B & Simoneau P (2014) Impact of the UPR on the virulence of the plant fungal pathogen A. brassicicola. Virulence, 5(2), 357–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez T, Qi H, Yap MC, Tahbaz N, Milburn LA, Lucchinetti E et al. (2020) The ER chaperone calnexin controls mitochondrial positioning and respiration. Science Signaling, 13(638), eaax6660. [DOI] [PubMed] [Google Scholar]
- Han J & Kaufman RJ (2017) Physiological/pathological ramifications of transcription factors in the unfolded protein response. Genes & Development, 31(14), 1417–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Hasegawa J & Matsunaga S (2013) The boundary of the meristematic and elongation zones in roots: endoreduplication precedes rapid cell expansion. Scientific Reports, 3(1), 2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C & Papa FR (2018) The unfolded protein response and cell fate control. MolecularCell,69(2),169–181. [DOI] [PubMed] [Google Scholar]
- Hirabayashi Y, Kwon S-K, Paek H, Pernice WM, Paul MA, Lee J et al. (2017) ER-mitochondria tethering by PDZD8 regulates Ca2+ dynamics in mammalian neurons. Science, 358(6363), 623–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollien J, Lin JH, Li H, Stevens N, Walter P & Weissman JS (2009) Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. Journal of Cell Biology, 186(3), 323–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H-W, Zeng X, Rhim T, Ron D & Ryoo HD (2017) The requirement of IRE1 and XBP1 in resolving physiological stress during Drosophila development. Journal of Cell Science, 130(18), 3040–3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H & Evans ML (1993) The role of the distal elongation zone in the response of maize roots to auxin and gravity. Plant Physiology, 102 (4), 1203–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T, Kashima M, Nagano AJ, Ishikawa-Fujiwara T, Kamei Y, Todo T et al. (2017) Unfolded protein response transducer IRE1-mediated signaling independent of XBP1 mRNA splicing is not required for growth and development of medaka fish, eLife, 6, e26845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Katoh R & Kitamura M (2013) Dual regulation of cadmium-induced apoptosis by mTORC1 through selective induction of IRE1 branches in unfolded protein response. PLoS One, 8(5), e64344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Nakajima S, Saito Y, Takahashi S, Katoh R & Krtamura M (2012) mTORC1 serves ER stress-triggered apoptosis via selective activation of the RE1–JNK pathway.Cell Death & Differentiation,19(2),310–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman RJ & Malhotra JD (2014) Calcium trafficking integrates endoplasmic reticulum function with mitochondrial bioenergetics. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, 1843(10), 2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara T, Yanagi H, Yura T & Mori K (1997) Endoplasmic reticulum stress-induced mRNA splicing permits synthesis of transcription factor Hac1p/Ern4p that activates the unfolded protein response. Molecular Biology of the Cell, 8(10), 1845–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J-S, Yamaguchi-Shinozaki K & Shinozaki K (2018) ER-anchored transcription factors bZIP17 and bZIP28 regulate root elongation. Plant Physiology, 176(3), 2221–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SG, Hoffman GR, Poulogiannis G, Buel GR, Jang YJ, Lee KW et al. (2013) Metabolic stress controls mTORC1 lysosomal localization and dimerization by regulating the TTT-RUVBL1/2 complex. Molecular Cell, 49(1), 172–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li GY, Fan B & Jiao YY (2014) Rapamycin attenuates visible light-induced injury in retinal photoreceptor cells via inhibiting endoplasmic reticulum stress. Brain Research, 1563, 1–12. [DOI] [PubMed] [Google Scholar]
- Li X, Cai W, Liu Y, Li H, Fu L, Liu Z et al. (2017) Differential TOR activation and cell proliferation in Arabidopsis root and shoot apexes. Proceedings of the National Academy of Sciences, 114(10), 2765–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas M, Swarup R, Paponov IA, Swarup K, Casimiro I, Lake D et al. (2011) SHORT-ROOT regulates primary, lateral, and adventitious root development in Arabidopsis. Plant Physiology, 155(1), 384–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishiba K-I, Iwata Y, Mochizuki T, Matsumura A, Nishioka N, Hirata R et al. (2019) Unfolded protein-independent IRE1 activation contributes to multifaceted developmental processes in Arabidopsis. Life Science Alliance, 2(5), e201900459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishiba K-I, Nagashima Y, Suzuki E, Hayashi N, Ogata Y, Shimada Y et al. (2013) Defects in IRE1 enhance cell death and fail to degrade mRNAs encoding secretory pathway proteins in the Arabidopsis unfolded protein response. Proceedings ofthe National Academy of Sciences, 110(14), 5713–5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra S & Ryoo HD (2019) The unfolded protein response in metazoan development. Journal of Cell Science, 132(5), jcs217216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montané M-H & Menand B (2013) ATP-competitive mTOR kinase inhibitors delay plant growth by triggering early differentiation of meristematic cells but no developmental patterning change. Journal of Experimental Botany, 64(14), 4361–4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno AA, Mukhtar MS, Blanco F, Boatwright JL, Moreno I, Jordan MR et al. (2012) IRE1/bZIP60-mediated unfolded protein response plays distinct roles in plant immunity and abiotic stress responses. PLoS One, 7(2), e31944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen JL, Ishikawa H & Evans ML (1998) Analysis of changes in relative elemental growth rate patterns in the elongation zone of Arabidopsis roots upon gravistimulation. Plants, 206(4), 598–603. [DOI] [PubMed] [Google Scholar]
- Nagashima Y, Mishiba K-I, Suzuki E, Shimada Y, Iwata Y & Koizumi N (2011) Arabidopsis IRE1 catalyses unconventional splicing of bZIP60 mRNA to produce the active transcription factor. Scientific Reports, 1, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S, Ivanova A, Duncan O, Law SR, Van Aken O, De Clercq I et al. (2013) A membrane-bound NAC transcription factor, ANAC017, mediates mitochondrial retrograde signaling in Arabidopsis. The Plant Cell, 25(9), 3450–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovečka M, Lang I, Baluška F, Ismail A, Illeš P & Lichtscheidl I (2005) Endocytosis and vesicle trafficking during tip growth of root hairs. Protoplasma, 226(1–2), 39–54. [DOI] [PubMed] [Google Scholar]
- Pastor-Cantizano N, Ko DK, Angelos E, Pu Y & Brandizzi F (2020) Functional diversification of ER stress responses in Arabidopsis. Trends in Biochemical Sciences, 45(2), 123–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petricka JJ, Winter CM & Benfey PN (2012) Control of Arabidopsis root development. Annual Review of Plant Biology, 63(1), 563–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffenbach KT, Nivala AM, Reese L, Ellis F, Wang D, Wei Y et al. (2010) Rapamycin inhibits postprandial-mediated X-Box-binding protein-1 splicing in rat liver. The Journal of Nutrition, 140(5), 879–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu Y, Ruberti C, Angelos ER & Brandizzi F (2019) AtIRE1C, an unconventional isoform of the UPR master regulator AtIRE1, is functionally associated with AtIRE1B in Arabidopsis gametogenesis. Plant Direct, 3 (11),e00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimold AM, Etkin A, Clauss I, Perkins A, Friend DS, Zhang J et al. (2000) An essential role in liver development for transcription factor XBP-1. Genes & Development, 14(2), 152–157. [PMC free article] [PubMed] [Google Scholar]
- Reimold AM, Iwakoshi NN, Manis J, Vallabhajosyula P, Szomolanyi-Tsuda E, Gravallese EM et al. (2001) Plasma cell differentiation requires the transcription factor XBP-1. Nature, 412(6844), 300–307. [DOI] [PubMed] [Google Scholar]
- Renna L, Stefano G, Slabaugh E, Wormsbaecher C, Sulpizio A, Zienkiewicz K et al. (2018) TGNap1 is required for microtubule-dependent homeostasis of a subpopulation of the plant trans-Golgi network. Nature Communications, 9(1), 5313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retzer K & Weckwerth W (2021) The TOR–auxin connection upstream of root hair growth. Plants, 10(1), 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieusset J (2018) The role of endoplasmic reticulum-mitochondria contact sites in the control of glucose homeostasis: an update. Cell Death & Disease, 9(3), 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez E, Chevalier J, Olsen J, Ansbøl J, Kapousidou V, Zuo Z et al. (2020) Autophagy mediates temporary reprogramming and dedifferentiation in plant somatic cells. The EMBO Journal, 39(4). 10.15252/embj.2019103315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D & Walter P (2007) Signal integration in the endoplasmic reticulum unfolded protein response. Nature Reviews Molecular Cell Biology, 8(7), 519–529. [DOI] [PubMed] [Google Scholar]
- Ruberti C, Lai Y & Brandizzi F (2018) Recovery from temporary endoplasmic reticulum stress in plants relies on the tissue-specific and largely independent roles of bZIP 28 and bZIP 60, as well as an antagonizing function of BAX-Inhibitor 1 upon the pro-adaptive signaling mediated by bZIP 28. The Plant Journal, 93(1), 155–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Alvarez M, del Pozo MA & Bakal C (2017) AKT-mTOR signaling modulates the dynamics of IRE1 RNAse activity by regulating ER-mitochondria contacts. Scientific Reports, 7(1), 16497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato EM, Hijazi H, Bennett MJ, Vissenberg K & Swarup R (2015) New insights into root gravitropic signalling. Journal of Experimental Botany, 66(8), 2155–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepetilnikov M, Makarian J, Srour O, Geldreich A, Yang Z, Chicher Jet a/. (2017) GTPase ROP2 binds and promotes activation of target of rapamycin, TOR, in response to auxin. The EMBO Journal, 36(7), 886–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B & Wolkenfelt H (1998) The Arabidopsis root as a model to study plant development. Plant Physiology and Biochemistry, 36(1), 21–32. [Google Scholar]
- Shanware NP, Bray K, Eng CH, Wang F, Follettie M, Myers J et al. (2014) Glutamine deprivation stimulates mTOR-JNK-dependent chemokine secretion. Nature Communications, 5, 4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Wu Y & Sheen J (2018) TOR signaling in plants: conservation and innovation. Development, 145(13), dev160887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulpice R, Flis A, Ivakov AA, Apelt F, Krohn N, Encke B et al. (2014) Arabidopsis coordinates the diurnal regulation of carbon allocation and growth across a wide range of photoperiods. Molecular Plant, 7(1), 137–155. [DOI] [PubMed] [Google Scholar]
- Szabadkai G, Bianchi K, Varnai P, De Stefani D, Wieckowski MR, Cavagna D et a/. (2006) Chaperone-mediated coupling of endoplasmic reticulum and mitochondrial Ca2+ channels. The Journal of Cell Biology, 175(6),901–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanegashima K, Zhao H, Rebbert ML & Dawid IB (2009) Coordinated activation of the secretory pathway during notochord formation in the Xenopusembryo. Development, 136(21), 3543–3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truernit E, Bauby H, Dubreucq B, Grandjean O, Runions J, Barthélémy J et al. (2008) High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of Phloem development and structure in Arabidopsis. The Plant Cell, 20(6), 1494–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aken O, De Clercq I, Ivanova A, Law SR, Van Breusegem F, Millar AH et al. (2016) Mitochondrial and chloroplast stress responses are modulated in distinct touch and chemical inhibition phases. Plant Physiology, 171(3), 2150–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, McCormack M, Li L, Hall Q, Xiang C & Sheen J (2013) Glucose–TOR signalling reprograms the transcriptome and activates meristems. Nature, 496(7444), 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdanbakhsh N, Sulpice R, Graf A, Stitt M & Fisahn J (2011) Circadian control of root elongation and C partitioning in Arabidopsis thaliana. Plant, Cell & Environment, 34(6), 877–894. [DOI] [PubMed] [Google Scholar]
- Young RM, Ackerman D, Quinn ZL, Mancuso A, Gruber M, Liu L et al. (2013) Dysregulated mTORC1 renders cells critically dependent on desaturated lipids for survival under tumor-like stress. Genes & Development, 27(10), 1115–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X, Xu P, Yu Y & Xiong Y (2020) Glucose-TOR signaling regulates PIN2 stability to orchestrate auxin gradient and cell expansion in Arabidopsis root. Proceedings of the National Academy of Sciences USA, 117(51), 32223–32225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L,Chen H, Brandizzi F,Verchot J & Wang A (2015) The UPR branch IRE1-bZIP60 in plants plays an essential role in viral infection and is complementary to the only UPR pathway in yeast. PLoS Genetics, 11(4), e1005164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo F, Xiong F, Deng K, Li Z & Ren M (2020) Target of Rapamycin (TOR) negatively regulates ethylene signals in Arabidopsis. International Journal of Molecular Sciences, 21(8), 2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Rate of shoot fresh weight accumulation and rate of primary root growth for data displayed in Figure 1. (a) Average shoot fresh weight values from Figure 1(b) were reprocessed to show the rate of shoot fresh weight accumulation for a specific growth period (value = average shoot fresh weight accumulated during a growth period divided by the number of days in that growth period). (b) Average root length values from Figure 1(c) were reprocessed to show the rate of primary root length increases for a specific growth period (value = average root length increases accumulated during a growth period divided by the number of days in that growth period).
Figure S2. At D5 there are no significant defects in Ire1a Ire1b meristem organization. At D5, root tips were subjected to mPS-PI staining and confocal microscopy to analyze root tip cellular organization. (a) Representative 10× images of mPS-PI stained WT and Ire1a Ire1b roots. The yellow line demarks the end of the meristem zone (MZ) and the beginning of the transition zone (TZ), the red lines marks all of the cells in the elongation zone (EZ), and the white arrow marks the first root hair initiation. For all measurements of the MZ secondary 20× images were used to collect data. (b) The average length of each zone. (c) The number of cells in each zone. (d) Average cell length in each zone. For all graphs error bars show the standard deviation; significance markers displayed above an Ire1a Ire1b experimental group are pairwise comparisons to the corresponding WT group for that specific treatment. Brackets denote other specific pairwise comparisons. Significance markers: NS = Padj> 0.01; *0.001 < Padj < 0.01; **0.0001 < Padj < 0.001; ***Padj < 0.0001.
Figure S3. Average root tip cellular organization metrics displayed over time in WT and Ire1a Ire1b. Average values from the data presented in Figure S2 and Figure 2 are plotted over time to compare differences over time in each genotype separately. (a–c) Zone length, number of cells, and cell length in the WT root tips. (d–f) Zone length, number of cells, and cell length in the Ire1a Ire1b root tips. For all graphs: error bars are standard deviation; significance markers displayed over an experimental group at D7 or D10 are comparisons to the corresponding D5 value for that group. Brackets denote other specific pairwise comparisons. Significance markers: NS = Padj > 0.01; *0.001 < Padj < 0.01; **0.0001 < Padj < 0.001; ***Padj < 0.0001. P-value indicator color matches the color of the corresponding root tip zone.
Figure S4. TOR inhibitor AZD-8055 concentration response analysis. (a) Representative images from DMSO, 100 nM, and 200 nM AZD-8055 conditions. Red lines represent approximate average root length of the displayed seedlings. (b) Root length of individual roots was measured using ImageJ. For all graphs, error bars show the standard deviation; significance markers displayed above an Ire1a Ire1b experimental group are pairwise comparisons to the corresponding WT group for that specific treatment. Brackets denote other specific pairwise comparisons. Significance markers: NS = Padj > 0.01; *0.001 < Padj < 0.01; **0.0001 < Padj < 0.001; ***Padj < 0.0001. For this experiment the growth chamber used had 200 μE continuous light which caused proportionally shorter roots in both WT and Ire1a Ire1b compared to 150 μE light; however, the general conclusions from the DMSO versus AZD treatments are the same in both conditions.
Figure S5. The TOR inhibitor TORIN2 rescues the Ire1a Ire1b root growth phenotypes. (a) Representative images from DMSO and TORIN2 conditions. Red lines represent approximate average root length of the displayed seedlings. (b) Root length of individual roots was measured using ImageJ. Error bars show the standard deviation; significance markers displayed above an Ire1a Ire1b experimental group are pairwise comparisons to the corresponding WT group for that specific treatment. Brackets denote other specific pairwise comparisons. Significance markers: NS = Padj > 0.01; *0.001 < Padj < 0.01; **0.0001 < Padj < 0.001; ***Padj < 0.0001. (c) The angle of the root tip with the vertical axis (0°) was measured using ImageJ. Significant differences between coefficients of variation were tested using the asymptotic Feltz and Miller test as described in the Experimental Procedures section. Significance markers: NS = P > 0.05; **0.001 < P < 0.01; ***P < 0.001.
Figure S6. Complementation of the Ire1a Ire1b mutant rescues root growth and AZD-8055 sensitivity phenotypes. (a) Representative images from DMSO and AZD-8055 treatment conditions. Red lines represent approximate average root length of the displayed seedlings. (b) Root length of individual roots was measured using ImageJ. Error bars show the standard deviation; significance markers displayed above an Ire1a Ire1b experimental group are pairwise comparisons to the corresponding WT group for that specific treatment. Brackets denote other specific pairwise comparisons. Significance markers: NS = Padj > 0.01; *0.001 < Padj < 0.01; **0.0001 < Padj < 0.001; ***Padj < 0.0001. (c) The angle of the root tip with the vertical axis (0°) was measured using ImageJ. Significant differences between coefficients of variation were tested using the asymptotic Feltz and Miller test as described in the Experimental Procedures section. Significance markers: NS = P > 0.05; **0.001 < P < 0.01; ***P < 0.001.
Figure S7. Other chemical inhibitors of root growth do not rescue the Ire1a Ire1b root length phenotype. (a) Representative images from DMSO, Oryzalin, Latrunculin b, and Oligomycin growth conditions. Red lines represent approximate average root length of the displayed seedlings. This experiment and TORIN2 experiments (Figure S5) were performed simultaneously and share the DMSO control data. (b) Root length of individual roots was measured using ImageJ. (c) Representative images from Antimycin A and ethanol control growth conditions. Red lines represent approximate average root length of the displayed seedlings. (c) Root lengths of individual roots for ethanol and Antimycin A growth conditions. For all graphs, error bars show the standard deviation; significance markers displayed above an Ire1a Ire1b experimental group are pairwise comparisons to the corresponding WT group for that specific treatment. Brackets denote other specific pairwise comparisons. Significance markers: NS = Padj > 0.01; *0.001 < Padj < 0.01; **0.0001 < Padj < 0.001; ***Padj < 0.0001.
Figure S8. Auxin inhibition of root growth and TOR inhibition of root growth are additive effects in WT plants. (a) Representative images of seedlings grown on plates containing various concentrations of NAA, with or without addition of AZD-8055. Red lines represent approximate average root length of the displayed seedlings. (b) Primary root length after 10 days of growth. For graphs error bars show the standard deviation; significance markers displayed above an Ire1a Ire1b experimental group are pairwise comparisons to the corresponding WT group for that specific treatment. Brackets denote other specific pairwise comparisons. Significance markers: NS = Padj > 0.01; *0.001 < Padj < 0.01; **0.0001 < Padj < 0.001; ***Padj < 0.0001.
Figure S9. Full blot images (from Figure 6) and the Ponceau stain images to demonstrate equal protein loading.
Figure S10. Chronic, low-level TOR inhibition leads to a spatial redistribution of EdU incorporation at the root tip. For each image EdU sum intensity along the root tip was measured in ImageJ for (a) 0 h and (b) 6 h samples (standard error displayed as lighter shaded area above and below each line). Then, for each root, the mean sum EdU intensity value was determined for two specific regions of interest for (c) 0 h and (d) 6 h samples. AZD treatment, but not genotype significantly affected the average sum EdU intensity in all regions of interest (see Table S1 for ANOVA results) For all graphs error bars show the standard deviation; significance markers displayed above an Ire1a Ire1b experimental group are pairwise comparisons to the corresponding WT group for that specific treatment. Brackets denote other specific pairwise comparisons. Significance markers: NS = Padj > 0.01; *0.001 < Padj < 0.01; **0.0001 < Padj < 0.001; ***Padj < 0.0001.
Figure S11. Simplified model based on our results showing that TOR activity must be balanced by IRE1 to maintain optimal cell elongation. Basal TOR activity is needed for cell elongation, illustrated by the difference between WT DMSO and WT AZD-8055. However, TOR hyperactivity promotes faster and unbalanced rates of differentiation, which favors premature root hair initiation and cessation of cell elongation. This in turn leads to shorter length of mature cells and an overall shorter root length in the Ire1a Ire1b mutant.
Table S1. Full results for all ANOVAs performed.
Data S1. R code used for the ANOVA statistical analysis pipeline.
Data Availability Statement
Data supporting the findings of this work can be found within the manuscript and its supporting materials. All data and materials are available from the corresponding author.


