Abstract
Apolipoprotein E receptor 2 (Apoer2) is a synaptic receptor in the brain that binds disease-relevant ligand Apolipoprotein E (Apoe) and is highly alternatively spliced. We examined alternative splicing (AS) of conserved Apoer2 exons across vertebrate species and identified gain of exons in mammals encoding functional domains such as the cytoplasmic and furin inserts, and loss of an exon in primates encoding the eighth LDLa repeat, likely altering receptor surface levels and ligand-binding specificity. We utilized single molecule, long-read RNA sequencing to profile full-length Apoer2 isoforms and identified 68 and 48 unique full-length Apoer2 transcripts in the mouse and human cerebral cortex, respectively. Furthermore, we identified two exons encoding protein functional domains, the third EGF-precursor like repeat and glycosylation domain, that are tandemly skipped specifically in mouse. Our study provides new insight into Apoer2 isoform complexity in the vertebrate brain and highlights species-specific differences in splicing decisions that support functional diversity.
Keywords: APOER2, LRP8, alternative splicing
1. Introduction
The human genome contains about 20,000 protein coding genes (Frankish et al., 2021), comparable to much simpler organisms such as worms or flies. However, over 100,000 unique proteins are thought to be synthesized in humans (International Human Genome Sequencing Consortium et al., 2001). One major determinant in proteome diversity is RNA alternative splicing (AS). In AS, different combinations of splice sites are used during pre-mRNA splicing leading to multiple unique RNA isoforms from an individual gene. AS can occur in different forms, including use of alternative 5’ or 3’ splice-sites, cassette-exon inclusion or skipping and intron retention (Nilsen and Graveley, 2010). 92–94% of human genes undergo AS, with 86% of those genes demonstrating a minor isoform frequency of 15% or more (Wang et al., 2008). This greatly expands the proteome and generates complexity from individual genes, as RNA isoforms can possess altered stability, localization, translation competency or coding sequence (Hughes, 2006; Li et al., 2007, p. 22007). Notably, about 15% of disease causing mutations in humans lead to mRNA splicing defects (Cartegni et al., 2002; Krawczak et al., 1992).
AS is more prevalent in higher eukaryotes compared to lower eukaryotes, particularly in vertebrates (Alekseyenko et al., 2007; Keren et al., 2010). The prevalence of cassette exon skipping increases further up the evolutionary tree, suggesting that exon skipping may be the type of AS that contributes most to phenotypic complexity (Keren et al., 2010; Kim et al., 2008). In addition, splicing regulatory protein families have expanded further up the evolutionary tree, including families such as the serine/arginine proteins (SR proteins) and the heterogenous nuclear ribonucleoproteins (hnRNPs) family (Barbosa-Morais, 2005).
In exons themselves, a correlation has been observed between the borders of exons and individual domains at the protein level (Kolkman and Stemmer, 2001) that becomes stronger as organismal complexity increases (Liu and Grigoriev, 2004). This modular design, likely produced through exon shuffling (Dorit and Gilbert, 1991), allows for diversification at the protein level with the addition or subtraction of unique protein domains, provided AS events do not disrupt the protein open reading frame. Interestingly, many modular proteins generated through exon shuffling are involved in mediating cell-cell or cell-extracellular matrix interactions, including members of the Low Density Lipoprotein Receptor (LDLR) family (Patthy, 1999; Sudhof et al., 1985). Conserved alternative exons between mice and humans are also enriched for genes expressed in the brain (Yeo et al., 2005).
Apolipoprotein E receptor 2 (Apoer2), official gene name Lrp8, is a modular type I transmembrane receptor of the LDLR family that is enriched in the vertebrate brain (Kim et al., 1996). Mouse Apoer2 regulates the layering of the neocortex in development (Trommsdorff et al., 1999) and long term potentiation (LTP) in the adult brain (Weeber et al., 2002). Importantly, Apoer2 is an Apolipoprotein E (Apoe) receptor (Kim et al., 1996). Apoe is a secreted glycoprotein that acts as a cholesterol carrier and signaling molecule (Belloy et al., 2019; Huang et al., 2017; Qiu et al., 2004) and serves as an isoform-specific risk factor for sporadic Alzheimer’s disease (Corder et al., 1993; Saunders et al., 1993). Interestingly, human and mouse Apoer2 are highly alternatively spliced in the brain (Clatworthy et al., 1999; Zhang et al., 2014), potentially altering the receptor’s ability to interact with critical ligands like Apoe. Some Apoer2 AS events demonstrate conservation across multiple vertebrate species, including chickens and mice (Brandes et al., 1997; Kim et al., 1996), and exhibit isoform-specific functions, including ligand binding properties (Beffert et al., 2006, 2005; Hibi et al., 2009).
Human APOER2 consists of five functional domains (Kim et al., 1996) that contain distinct modular protein domains. The first is an amino terminal ligand binding domain made up of two types of binding repeats: LDL receptor type A (LDLa) repeats and epidermal growth factor (EGF) precursor-like repeats. The second functional domain is the YWTD β-propeller domain that facilitates ligand release and receptor recycling after endocytosis (Rudenko et al., 2002). The third domain is an O-linked sugar domain critical for receptor glycosylation and surface expression (Wasser et al., 2014). The hydrophobic transmembrane region makes up the fourth functional domain. Lastly, the fifth domain is a short cytoplasmic tail containing an NPXY motif critical for clathrin-mediated endocytosis and binding of adaptor proteins (Dumanis et al., 2011; Minami et al., 2010; Trommsdorff et al., 1998). The cytoplasmic tail of Apoer2 also includes a proline rich 59 amino acid insert that is essential for Reelin-induced enhancement of LTP in mice (Beffert et al., 2005) and is unique to Apoer2 (Kim et al., 1996). These individual APOER2 protein domains are largely encoded by distinct cassette exons that are in phase, allowing for cassette exon skipping events that preserve the open reading frame and generate multiple unique proteins (Kim et al., 1998). Furthermore, conservation of several of these AS events across species, such as chicken, mice and humans (Brandes et al., 2001, 1997; Clatworthy et al., 1999; Kim et al., 1997), suggests these events may have important biological function since the splicing event has avoided negative selection (Keren et al., 2010).
Apoer2 was originally thought to be two distinct receptors of the LDLR family due to the presence of an eighth LDLa repeat that is present in chickens and mice but not in humans (Brandes et al., 1997; Kim et al., 1996; Novak et al., 1996). Further analysis revealed that this exon was lost in monkeys and humans due to mutation of the 3’ splice site (Kim et al., 1998). This indicates that there are evolutionary changes in Apoer2 across vertebrates, suggesting Apoer2 has undergone evolutionary selection in the vertebrate brain. Other exons encoding functional protein domains in Apoer2 have been identified as alternatively spliced, including the cytoplasmic insert in mice and humans (Beffert et al., 2005; Brandes et al., 1997; Hinrich et al., 2016; Kim et al., 1997), the 39-nucleotide exon that introduces a furin cleavage site in mice and humans (Brandes et al., 1997; Koch et al., 2002) and the glycosylation domain in mice and humans (Clatworthy et al., 1999; May et al., 2003; Wasser et al., 2014), among others. However, a systematic tracing of Apoer2 splicing events across the vertebrate lineage is lacking. It is unclear how many distinct Apoer2 isoforms are physiologically produced and how often individual Apoer2 cassette exon splicing events occur in tandem across the entire length of the Apoer2 transcript.
In this study, we have performed a direct comparison of Apoer2 alternatively spliced exons across vertebrates, including zebrafish, chickens, rabbits, mice, monkeys and humans. We demonstrate that AS of Apoer2 increases with evolutionary complexity and show the loss of the eighth LDLa repeat from mice to monkeys. Our analyses indicate the exon encoding a furin cleavage site appears to have evolved between chicken and rabbits, which coincides with the appearance of the alternatively spliced cytoplasmic insert. Furthermore, to investigate coordinated splicing of Apoer2 cassette exons, we have utilized single molecule, long-read sequencing to profile splicing events across Apoer2 in the cerebral cortex of mice and humans. We demonstrate a rich complexity of AS combinations across both humans and mice, some of which are conserved. Conservation of AS events suggests they have significant effects at the functional level, which has already been shown for AS of the cytoplasmic insert in mouse Apoer2 (Beffert et al., 2005). Therefore, it is likely that other AS events or AS combinations in Apoer2 also endow distinct functional properties at the protein level in the brain, such as modulating Apoe binding. We also observed and validated a tandem splicing event of the exons encoding the third EGF precursor-like repeat and the glycosylation domain in mice, but not humans, suggesting differential splicing regulation exists across the two species.
2. Materials and Methods
2.1. Phylogenetic and isoform analysis.
All annotated Lrp8 RNA isoforms were downloaded from each NCBI and Ensembl gene databases for Danio rerio (zebrafish), Gallus gallus (chicken), Oryctolagus cuniculus (rabbit), Mus musculus (mouse), Macaca fascicularis (cynomolgus monkey/macaque) and Homo sapiens (human). Sequences were analyzed and compared to their corresponding reference genome in NCBI and exons annotated manually using SnapGene software. To examine homology between species at the protein level, the longest coding isoform was selected for each species and the corresponding protein sequence was aligned with that of all other species. NCBI’s Constraint-based Multiple Alignment Tool (COBALT) was used to generate protein alignment and phylogenetic tree using the default COBALT parameters (Papadopoulos and Agarwala, 2007). To visualize protein alignment, FASTA output was input into ExPASY’s BoxShade tool with an output of RTF_new (Fig. S1).
2.2. RNA Isolation, cDNA synthesis and RT-PCR.
Total RNA was isolated from the brains of four adult wild-type zebrafish brains using TRIzol reagent (Invitrogen) according to manufacturer instructions. Genomic DNA depletion was performed immediately after using DNase I in Qiagen’s RNeasy Kit according to manufacturer instructions. Total RNA from the whole brain of chicken (CR-201), rabbit (TR-201) and cynomolgus monkey (KR-201) was purchased from Amsbio. Total RNA from the cerebral cortex of mouse (636661) and human (636561) was purchased from Takara Biosciences. RNA was quantified using a NanoDrop spectrophotometer. cDNA synthesis was carried out using Superscript IV First-Strand Synthesis System (Invitrogen). 1.5 μg of RNA was used as input for annealing of oligo-dT primers. Final clean-up was performed with 1 μL of RNase H.
RT-PCR was performed using either PrimeStar Polymerase (Takara Biosciences) or Q5 Hot-Start High-Fidelity Polymerase (NEB) depending on optimization. Optimal annealing temperature was determined using a gradient thermocycler. Primers were designed and screened for specificity using NCBI’s Primer-BLAST (Ye et al., 2012) (Supplemental Dataset S2).
RT-PCR reactions were separated out by agarose gel electrophoresis in 1X TAE Buffer. Ethidium bromide was used for visualization. All DNA bands generated from RT-PCR reactions were individually excised and purified using BioBasic’s Plasmid DNA Isolation Miniprep Kit according to manufacturer protocol for agarose gel DNA purification. Each product was then individually cloned for sequencing using Invitrogen’s Zero Blunt TOPO PCR Cloning Kit. Two to ten clones were picked per individual RT-PCR band and verified by sanger sequencing with vector specific primer M13F. Sequencing analysis was carried out using SnapGene and NCBI BLAST (Altschul et al., 1990).
2.3. Pacific Biosciences library preparation and long read sequencing.
Similar to the methodology used in Treutlein, et al. (Treutlein et al., 2014), gene specific first strand cDNA synthesis was first performed targeting Lrp8, followed by further Lrp8 specific RT-PCR. Briefly, 1 μg of RNA per sample was incubated with 1 μL of 10mM dNTPs and 1 μL Lrp8 reverse primer (Human REV: 5’- TCA GGG TAG TCC ATC ATC TTC AAG GC-3’, Mouse REV: 5’-TCA GGG CAG TCC ATC ATC TTC AAG AC-3’) in a 10 μL reaction for 5 minutes at 65°C. The reaction was then cooled on ice for at least 2 minutes. The reaction was then supplemented to 20 μL with 5X First Strand Buffer (Invitrogen), DTT, Superase In RNAse Inhibitor and SuperScript RTase followed by first strand synthesis: 1 hour at 55°C, 15 minutes at 70°C, hold at 4°C. 2 units (1 μL) of RNase H was then added prior to a 20-minute incubation at 37°C.
RT-PCR reaction was performed in multiple 50 μL reactions and pooled for subsequent AMPure XP SPRI clean-up. RT-PCR mix utilized: 5X Q5 Buffer (NEB), 5 μg BSA, 1M Betaine, 3% DMSO, 0.2mM dNTP, 0.2 μM FWD primer (Human FWD: 5’-ATG GGC CTC CCC GAG CC −3’, Mouse FWD: 5’- CTA TTA TGG GCC GCC CAG AAC TGG G-3’), 0.2 μM REV primer (Human REV, Mouse REV), Q5 High Fidelity Polymerase, and 2 μL of first strand synthesis reaction per 50μL. Cycle utilized for human RT-PCR is as follows: 98°C 2 minutes; 30 cycles of: 98°C- 10 seconds, 64°C- 30 seconds, 65°C- 1 minute 40 seconds; 72°C- 10 minutes, 4°C hold. Cycle utilized for mouse RT-PCR is as follows: 98°C 2 minutes; 30 cycles of: 98°C- 10 seconds, 68.6°C- 30 seconds, 72°C- 2 minutes; 72°C- 10 minutes, 4°C hold.
RT-PCR reactions were then pooled and subject to 0.5X SPRI clean-up using AMPure XP beads, with a final elution using 20 μL nuclease-free water. To determine concentration and purity, cDNA amplicons were then quantified using both NanoDrop Spectrophotometer and Qubit Fluorometer. 1 μg of DNA amplicon was prepared for PacBio library preparation.
500 ng of each cDNA pool was prepared via the express template preparation kit 2.0 from Pacific Biosciences. Each sample underwent single-strand overhang removal, DNA damage repair, and end repair/a-tailing by standard methods. Barcoded overhang SMRTbell adapters were used for each sample in place of the standard unbarcoded SMRTbell adapters. After ligation, samples were pooled in an equimolar fashion to a total mass of 1000 ng per pool. Pooled libraries were cleaned and concentrated with 0.45X AMPure beads and underwent polymerase annealing and binding by standard methods. Primer v4 was used. Each library was loaded with Sequel loading kit 2.1 at 60pM. A 2-hour pre-extension time and a 24-hour movie was used. CCS and barcode demultiplexing were carried out by SMRTlink v8.0.
2.4. Processing of Pacific Biosciences single molecule, long-read sequencing data.
SMRTlink v8.0 was used to process demultiplexed sequencing files. PacBio primers and Lrp8 specific RT-PCR primers were clipped, primer concatemers were removed and sequences were clustered into polished high-quality isoforms using SMRTlink v8.0 pipeline with standard IsoSeq parameters. High-quality isoforms were collapsed into unique isoforms using cDNA Cupcake (https://github.com/Magdoll/cDNA_Cupcake, v17.0.0) with a maximum 5’ and 3’ difference of 50 base pairs and without merging shorter isoforms. cDNA Cupcake was also used to generate isoform abundance using SMRTLink cluster reports and 5’ fragments were filtered out along with any isoforms with a full-length read count less than two. cDNA Cupcake generated fastq files were then reference corrected against the mouse (mm10) or human (hg38) genome using SQANTI3 v.2.0.0 (Tardaguila et al., 2018).
2.5. Transcript annotation and homology analysis.
Corrected transcript output from SQANTI3 (Tardaguila et al., 2018) was parsed in R (R Core Team, 2020) to include only Apoer2 transcripts. All transcripts were filtered based off the isoform FASTA file so that they contained the final sequence of Apoer2 located in the last exon, just prior to the reverse primer, since the small length of this sequence prevented annotation as an individual exon. The intersection of isoforms after these two filtering steps was moved into further analysis. For exon annotation, the SQANTI3 gff3 output file was parsed for Apoer2 exons, and unique exons were extracted and manually annotated using the Broad Institute’s Integrative Genomics Viewer (IGV) (Robinson et al., 2011). Exons corresponding to individual isoforms were then assembled into a binary splice matrix in R, and exons were labeled using previous manual IGV annotation.
To determine homologous transcripts between mice and humans, homologous exons were matched based off previous Apoer2 protein alignment and exon annotation (Supplementary Dataset S3). Transcripts with exons outside of known homologous exons between the two species were filtered out. Remaining isoforms were matched between the two species in R based on having matching patterns of corresponding exons across the transcript.
2.6. Transcript mapping and exon level analysis.
The binary splice matrix was assembled into a transcript matrix using ggplot2 (Wickham, 2016) and publicly available code deposited in github from Flaherty, et al. (Flaherty et al., 2019). Coincidence was calculated by counting the number of times an exon was spliced in at the same time as each other exon, weighted by transcript abundance and then divided by the total number of transcripts. Heatmaps and barplots were generated using ggplot2. Frequency spliced in was calculated by counting the number of times an exon was spliced in over all transcripts weighted by transcript abundance. Transcript length, cumulative frequency and parts of a whole annotation graphs were generated using GraphPad Prism v 9.2.0. Correlation matrices were generated using the binary splice matrix and R packages Hmsic (Frank E Harrell Jr, with contributions from Charles Dupont and many others. (2020).) and corrplot (Taiyun Wei and Viliam Simko (2017) to analyze the Pearson correlation coefficient. A significance level of 0.01 was used.
3. Results
3.1. Apoer2 cassette exon skipping across vertebrates.
To examine Apoer2 homology across vertebrates, we aligned full-length Apoer2 protein sequences from zebrafish, chicken, rabbit, mouse, macaque and human (Fig. S1). Fig. 1A demonstrates the phylogeny of Apoer2 as phenotypic complexity increases from zebrafish to humans. To understand how Apoer2 protein domains changed over evolution from zebrafish to humans, we mapped the known protein domains of human APOER2 onto the protein alignment to determine whether these domains were conserved in other species. We found that chickens and mice contain an eighth LDLa ligand binding repeat in their extracellular domain, while monkeys and humans do not (Fig. 1B), consistent with previous studies (Brandes et al., 1997; Kim et al., 1997, 1998). We also observed the presence of the eighth LDLa repeat in zebrafish and rabbits. Apoer2 also contains a 39-nucleotide exon at the end of its LDLa repeats in mice and humans that encodes a cleavage site for the furin protease (Brandes et al., 1997; Koch et al., 2002). This 39-nucleotide exon appears to be present in both rabbits and macaques yet lacking in zebrafish and chickens. Lastly, Apoer2 contains a unique 59 amino acid proline rich insert in its cytoplasmic region that has been reported as present in mice but lacking in birds (Brandes et al., 1997; Myant, 2010). Based on our protein alignment, neither zebrafish nor chickens possess the cytoplasmic insert; however, it is apparent in vertebrates higher than the rabbit in this study.
Figure 1. Apoer2 protein and isoform diversity across vertebrates.
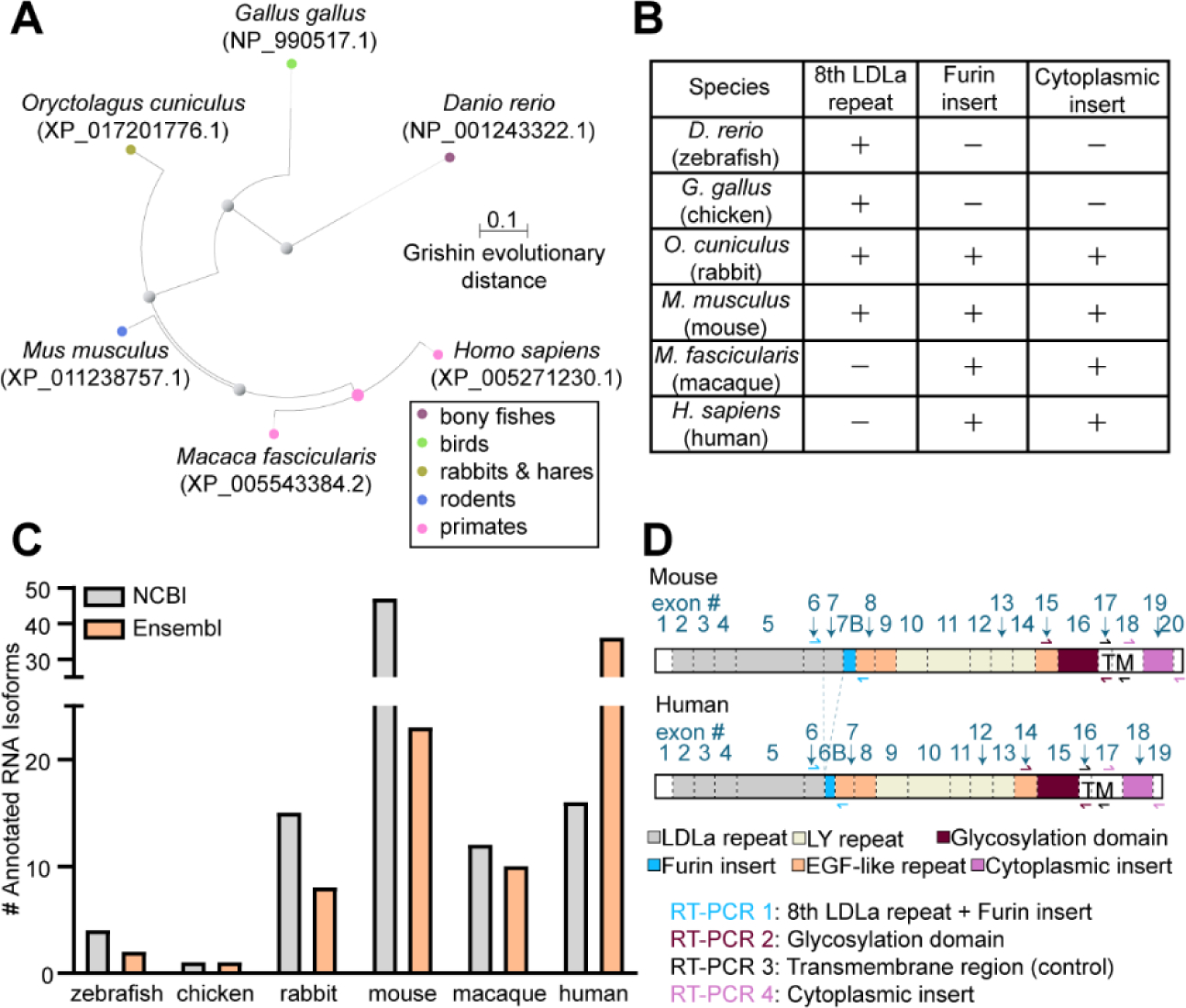
(A) Phylogenetic tree of Apoer2 spanning from Danio rerio (zebrafish) to Homo sapiens (humans) displaying Grishin evolutionary distance. (B) Table indicating presence or absence of Apoer2 protein domains of interest across vertebrates based off protein reference annotations. (C) Bar graph depicting the number of annotated Apoer2 isoforms for each vertebrate species in each the NCBI and Ensembl online databases. (D) Schematic of Apoer2 protein domains and corresponding coding exons in each mice and humans. RT-PCR primer schemes are indicated using arrows.
To understand what is currently known about AS events in Apoer2 across species, we curated all known Apoer2 isoforms deposited in National Center for Biotechnology Information (NCBI) (NCBI Resource Coordinators et al., 2018) and Ensembl (Howe et al., 2021) online databases. There is a general increase in the number of annotated Apoer2 isoforms as evolutionary complexity increases (Fig. 1C). However, published work has demonstrated alternative isoforms of Apoer2 do exist in chickens (Brandes et al., 1997; Novak et al., 1996), which is not currently reflected in these annotation databases. This is perhaps due to the widespread usage of mice or human cells and tissue to study splicing and other biological processes resulting in greater sequencing coverage. To address this discrepancy, we analyzed the splicing patterns of known alternative exons in Apoer2 including the exons encoding the eighth LDLa repeat, the furin cleavage site, the glycosylation domain and the cytoplasmic insert. We mapped the protein sequence of each species used for alignment back onto their corresponding RNA transcript to identify the homologous exons to target (Supplementary Dataset S1). RT-PCR primers were placed as indicated, flanking the alternative exons of interest, as demonstrated for mouse and human Apoer2 in Fig. 1D. RT-PCR reactions will yield one band for exon inclusion and an additional one for exon skipping, if it occurs. The eighth LDLa repeat and the furin cleavage site were captured using the same RT-PCR reaction, generating the possibility of observing four bands indicative of inclusion and exclusion of each the exon encoding the eighth LDLa repeat and the exon encoding the furin cleavage site. Whole brain total RNA from zebrafish, chicken, rabbit and monkey and total RNA from the cerebral cortex of mice and humans were subject to oligo-dT based cDNA synthesis and RT-PCR targeting the Apoer2 regions of interest. RT-PCR reactions were then visualized by gel electrophoresis (Fig. 2A–B) and all bands were confirmed by sequencing.
Figure 2. Apoer2 alternative splicing complexity in the brain increases across vertebrate evolution.
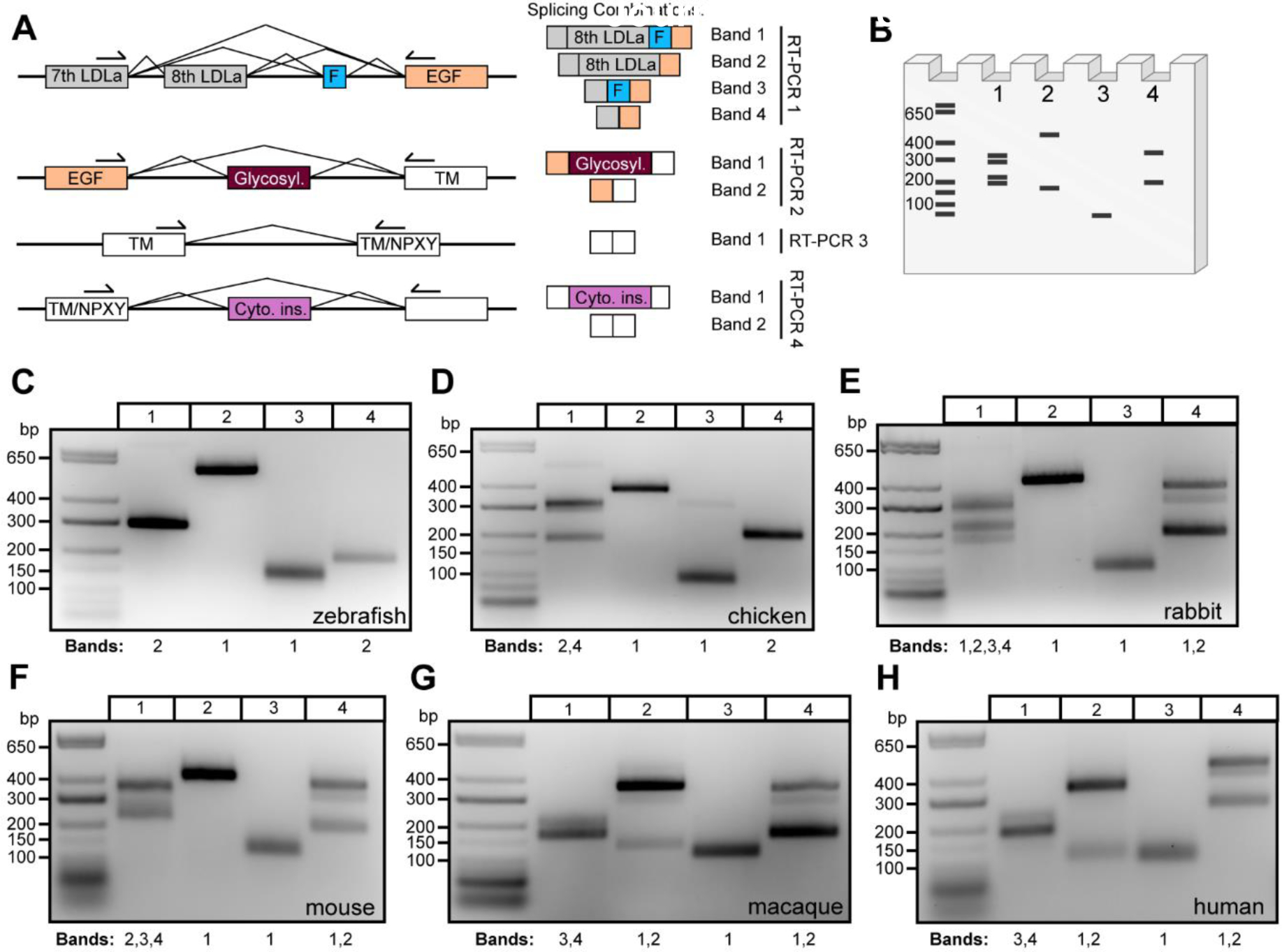
(A) Schematic demonstrating RT-PCR primer design along with the corresponding exon to protein functional domain each primer set amplifies. Expected splicing combinations are depicted on the right side along with a band number corresponding to the cartoon agarose gel (B) (generated using Biorender.com) depicting potential RT-PCR banding patterns if all combinations of cassette exon decisions are used. (C-H) Gels depicting RT-PCR analysis of Apoer2 alternative exons encoding the eighth LDLa repeat and furin insert (Lane 1), glycosylation domain (Lane 2), control transmembrane region (Lane 3) and cytoplasmic insert (Lane 4) across vertebrate species: (C) zebrafish, (D) chicken, (E) rabbit, (F) mouse, (G) macaque and (H) human. Bands in each lane are annotated below the gel according to the numbering displayed in (A).
Zebrafish demonstrate constitutive inclusion of the eighth LDLa ligand binding repeat (Fig. 2C) and do not appear to have the furin insert as indicated by the presence of a strong band in Lane 1 around 300 bp. A faint band just above 700 bp is also visible, which sequencing demonstrated as off-target amplification. In Lanes 2 and 4, there is no evidence of AS of the glycosylation domain or presence of the cytoplasmic insert, respectively. The control reaction in Lane 3 targets the transmembrane region of the receptor, which demonstrates a diffuse band indicative of some nucleotide diversity. At the annotation level, an 18-nucleotide difference is annotated at the boundary of zebrafish exon 18, which appears to be unique to the species. We confirmed the presence of both +/− 18-nucleotide products in this band indicating the use of an alternative 3’ splice site.
In chickens, two bands were observed in Lane 1, just above 300 bp and below 200 bp, indicating the eighth LDLa repeat is alternatively spliced (Fig. 2D). A faint upper band between 400–600 bp is also apparent in Lane 1. Sequencing analysis indicated this band is a mixture of nonspecific amplification and sequence demonstrating inclusion of the eighth LDLa repeat. The band at 300 bp that appears like a doublet revealed only sequence indicating inclusion of the eighth LDLa repeat and no evidence of the 39-nucleotide exon encoding the furin insert, which is consistent with published literature (Brandes et al., 1997; Myant, 2010). To understand whether the exon encoding the furin cleavage site is present in the genome but constitutively spliced out at the RNA level, we examined the chicken Lrp8 locus in comparison to the mouse Lrp8 locus, which does contain this exon. We found no evidence of a conserved region in the chicken genome (Fig. S2), indicating the exon encoding the furin cleavage site was likely introduced into the genome at a point evolutionarily later than chickens. In Lane 2, the glycosylation domain appears to be constitutively included. Interestingly, in Lane 3 we detected an upper band around 300 bp in addition to the expected control product targeting the transmembrane region. Sequencing identified the upper band to be an intron retention event, capturing chicken Apoer2 exon 17, the intervening intron and exon 18. We also saw no evidence of the cytoplasmic insert (Lane 4) in chickens as has been published (Brandes et al., 1997).
Rabbits demonstrated more diversity at the RNA level than both zebrafish and chickens, as shown in Fig. 2E. In Lane 1, at least four distinct bands are present, indicating the AS of the eighth LDLa repeat as well as the furin insert, which are found in all four possible splicing combinations, including skipping of both exons in tandem. Sequencing of clones from the topmost diffuse band also returned a variation of rabbit Apoer2 that has a unique exon following the eighth LDLa repeat, which maps to a region of the genome between the eighth LDLa repeat and the furin insert (Fig. S3). Lane 2 indicates constitutive inclusion of the glycosylation domain. Lane 4 confirms the introduction of the alternatively spliced cytoplasmic insert, which is dynamically spliced in and out. The presence of a third band is intriguing and suggests additional splicing diversity in this region. However, efforts to sequence the third (middle) band have only returned sequence including or excluding the cytoplasmic insert, making it unclear exactly why there is an apparent nucleotide size difference between these bands. One sequenced TOPO clone returned sequence in which 67 bp at the end of rabbit exon 18 were excluded and 61 bp at the beginning of exon 19 were excluded, which would suggest the use of two alternative splice sites. As only one clone returned this sequence, it seems more likely to be a PCR or cloning artifact than AS events, although it is possible these are real AS events present at low frequency.
In the mouse cerebral cortex, multiple bands are apparent in Fig. 2F Lane 1, indicating AS of the eighth LDLa repeat and the furin insert. Sequencing of clones confirmed species containing the following combinations: eighth LDLa repeat but lacking the furin insert, lacking the eighth repeat but containing the furin insert and species lacking both exons. Interestingly, no clones were identified containing both the eighth LDLa repeat and the furin insert. In Lane 2, constitutive inclusion of the glycosylation domain was observed, despite published evidence suggesting this exon is alternatively spliced in mice (May et al., 2003). After extensive optimization of RT-PCR parameters, including testing of multiple annealing temperatures, addition of PCR additives and use of an alternative polymerase, we failed to identify any bands indicating AS of the glycosylation domain in mice. Lane 4 indicates AS of the cytoplasmic insert, as well as the presence of a third band; similar to rabbits (Fig. 2E Lane 4), all sequenced clones returned mouse sequence either containing or lacking the entire cytoplasmic insert.
Loss of the eighth LDLa repeat becomes apparent in primates when examining the expression profile of the macaque. Fig. 2G Lane 1 demonstrates only two bands, neither of which contain the eighth LDLa repeat, but do contain AS of the 39-nucleotide furin insert. In Lane 2, AS of the glycosylation domain is observed, whereas this exon was constitutively included in all other lower vertebrates analyzed (Fig. 2C–F). Consistent with rabbits and mice, monkeys demonstrated AS of the cytoplasmic insert in Lane 4, as well as the presence of a third band shifted in size that returned sequence both including or lacking the entire cytoplasmic insert.
Lastly, we examined the AS pattern of APOER2 in humans (Fig. 2H). As expected, we observed no inclusion of the eighth LDLa repeat but did observe exclusion and inclusion of the 39-nucleotide furin insert in Lane 1. Like macaques, AS of the glycosylation domain was observed in Lane 2. AS of the cytoplasmic insert is also conserved in humans, with bands observed indicating both its presence and absence. As seen in rabbits, mice and monkey, a third band is also present, for which sequencing returned cassette splicing of the cytoplasmic insert.
Overall, the RT-PCR analysis indicates the addition of the furin insert and the cytoplasmic insert sometime between the evolution of chicken and rabbits and the loss of the eighth LDLa ligand binding repeat from mice to monkeys. Interestingly, we only observed AS of the glycosylation domain in primates, including monkeys and humans.
3.2. Full-length Apoer2 isoform mapping in mice and humans identifies homologous isoforms.
In addition to AS of the exons encoding the eighth LDLa repeat, the furin insert, the glycosylation domain and the cytoplasmic insert, other Apoer2 exons have been reported to be alternatively spliced in mouse and human through RT-PCR analyses (Brandes et al., 2001, 1997; Clatworthy et al., 1999; Kim et al., 1997, 1998). Therefore, we asked whether these AS events are often found in different combinations or whether most AS events occur in isolation within the full-length context of the Apoer2 transcript. To understand the full splicing landscape and complexity of Apoer2 isoforms across the brain, we performed Apoer2 specific long-read sequencing on RNA from the mouse and human cerebral cortices. Briefly, we performed Apoer2 specific first strand cDNA synthesis on total RNA from the cerebral cortex of mice and humans as schematized in Fig. 3A. We then performed RT-PCR targeting the entirety of the coding sequence of Apoer2, with a forward primer binding at the ATG site in exon 1 and reverse primer binding at the TGA stop site in exons 19 and 20 for humans and mice, respectively. After size selection, nucleic acid purification and library preparation, Apoer2 cDNA amplicons for mice and humans were subject to Pacific Bioscience’s single-molecule real-time (SMRT) sequencing to generate full-length Apoer2 transcript sequences.
Figure 3. Full-length Apoer2 isoform mapping in the murine cerebral cortex.
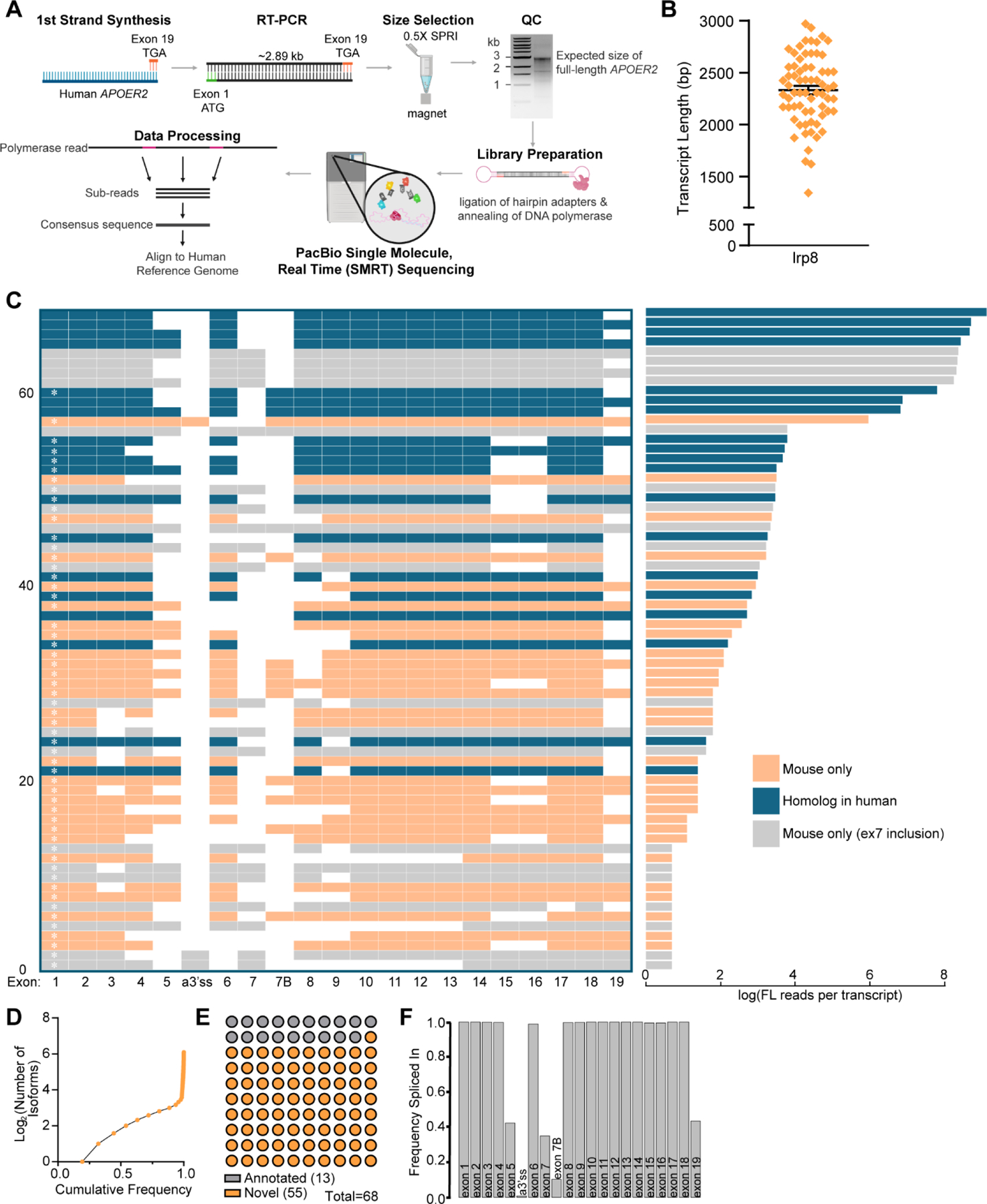
(A) Schematic depicting Apoer2 specific single molecule, long-read sequencing workflow (generated using Biorender.com). (B) Graph indicating mean transcript length ±SEM of unique murine Apoer2 isoforms detected in the cerebral cortex excluding RT-PCR primers. (C) Left: Transcript matrix depicting Apoer2 isoforms identified as individual rows. Transcripts with exons present in less than 10 unique transcripts before filtering were excluded, leaving 68 unique transcripts. Exons spliced in are colored, while skipped exons are white. Numbering at the left-hand margin indicates transcript number. A white asterisk inside of exon 1 indicates a novel or previously unannotated transcript. Right: Bar plot indicating the log of the total number of full-length reads of each corresponding transcript in the adjacent matrix. All transcripts and their corresponding number of reads are colored coded based on whether there is a corresponding homolog isoform in humans (blue), the transcript contains the mouse specific exon encoding the eighth LDLa repeat (grey), or whether the transcript is specific to mouse but does not contain the exon encoding the eighth LDLa repeat (orange). (D) Cumulative frequency of detected Apoer2 isoforms. (E) Graph indicating the proportion of detected transcripts that are either annotated in NCBI or Ensembl or novel. (F) Bar graph demonstrating the frequency at which each Apoer2 exon is spliced in based on the detected transcripts and their number of full-length reads.
Sequencing data was processed into high quality isoforms then further collapsed into unique reference corrected isoforms. After data processing, the mouse and human samples yielded 3,036 and 55,326 high quality full-length reads for analysis, with 87% and 84% respectively of those reads mapping to Apoer2. While the number of full-length, high-quality reads returned for the human was slightly lower than the mouse sample, the proportion of reads mapping to Apoer2 was comparable, giving both RT-PCRs similar precision and enabling transcript mapping and proportional comparisons between the two species. For mouse Apoer2, we identified 68 high quality Apoer2 transcripts that contained exons present in at least ten unique isoforms before filtering. As almost all introns have been spliced out, we are likely capturing mature processed RNA or genuine intron retention splicing events as opposed to pre-mRNA. The mean length of identified murine Apoer2 transcripts clustered just under 2500 bp as shown in Fig. 3B. Full-length murine Apoer2, containing the coding sequence of all 21 exons captured within our primer design, would be about 3 kilobases. The comparatively lower average Apoer2 transcript length suggests there are numerous AS events in the identified transcripts, with some transcripts likely having multiple AS events. When examining the 68 murine Apoer2 transcripts, we see a plethora of cassette exon skipping events occurring individually and in tandem across the length of murine Apoer2 (Fig. 3C). As our reverse primer bound at the nucleotides encoding the stop codon in the last coding exon (human exon 19 and mouse exon 20), we did not annotate this exon as a full exon in the transcript maps (Fig. 3C & 4B) since this full exon is much longer, encoding the 3’UTR which our RT-PCR scheme does not capture. The most abundant murine Apoer2 isoform we identified contained three AS events, with exclusion of exons 5, 7 and 19, which encode 3 LDLa repeats, the eighth LDLa repeat and the cytoplasmic insert, respectively. Of interest, we identified novel AS of exon 3, which would remove an LDLa repeat, yet preserve the open reading frame, as well as transcripts that excluded exons 8 through 13 in tandem, which would remove the entire β-propeller domain and introduce a premature stop codon after the LDLa repeats. We also identified a known alternative 3’ splice site near exon 6, which adds 3 nucleotides of the prior intron, CAG, into the coding sequence of the transcript. Surprisingly, we identified multiple transcripts that contained tandem exon skipping of exons 15 and 16 which code for an EGF precursor-like repeat and the glycosylation domain respectively, while neither exon was identified to be skipped individually.
Figure 4. APOER2 full length isoform mapping in the human cerebral cortex.
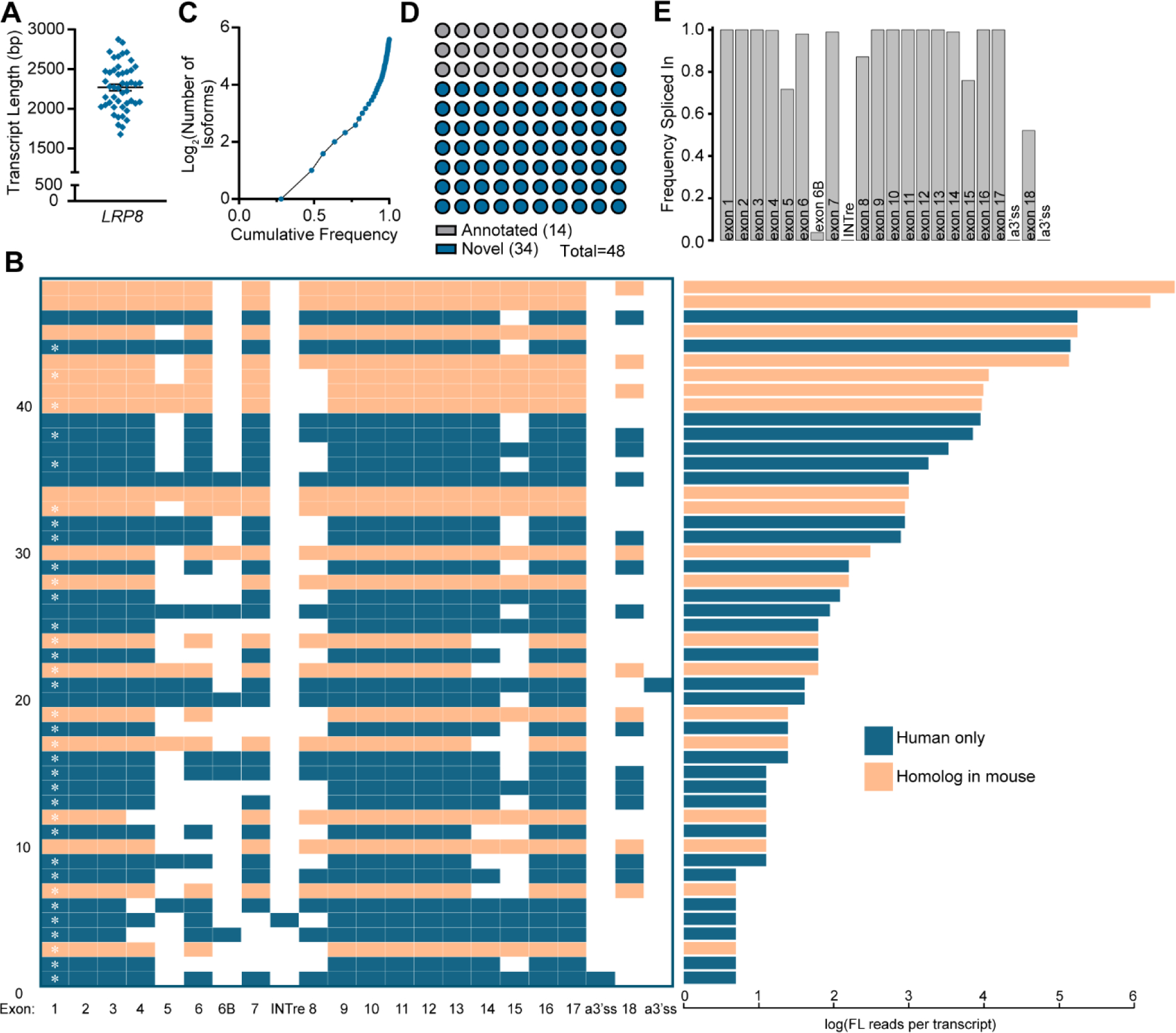
(A) Graph indicating the mean transcript length ±SEM of unique APOER2 isoforms detected in the human cerebral cortex excluding RT-PCR primers. (B) Left: Transcript matrix displaying APOER2 isoforms identified in the human cerebral cortex as individual rows. Exons spliced in are colored, while skipped exons are white. Numbering at the left-hand margin indicates transcript number. A white asterisk inside of exon 1 indicates a novel or previously unannotated transcript. Right: Bar graph displaying the log of the number of full-length reads per isoform in the adjacent transcript matrix. All transcripts and their corresponding number of reads are colored orange if there is a corresponding homolog isoform in mouse or blue if the isoform is specific to humans. (C) Cumulative frequency graph of detected APOER2 isoforms. (D) Parts of a whole graph demonstrating the number of detected transcripts found to be annotated in NCBI or Ensembl databases or novel. (E) Bar graph indicating the frequency at which each APOER2 exon is spliced in across all the detected isoforms weighted by their abundance.
With 68 unique transcripts identified in the mouse cerebral cortex, majority of these AS events result in the inclusion or exclusion of entire protein functional domains at the protein level, suggesting AS may alter or regulate specific Apoer2 protein function in the brain. Based off the number of full-length reads per isoform (Fig. 3C right), we examined the cumulative frequency of individual Apoer2 isoforms to understand how many isoforms make up the majority of Apoer2 expression (Fig. 3D). We observed about 6–7 isoforms make up 75% of Apoer2 expression in the mouse cerebral cortex, with the remaining isoforms found at lower levels. We also compared the 68 identified murine Apoer2 isoforms to the coding region (excluding UTRs which our primer strategy does not capture) of those annotated in the Ensembl gene database (Howe et al., 2021). Using this comparison and a manual confirmation and comparison to the NCBI (NCBI Resource Coordinators et al., 2018) annotated transcripts as well, we identified 13 isoforms that have been previously annotated and 55 novel Apoer2 isoforms (Fig. 3C&E).
Lastly, we wanted to understand the frequency at which individual exons are found to be spliced into the Apoer2 transcript pool. We calculated a frequency spliced in value for each exon across all the isoforms weighted by their abundance as shown in Fig. 3F. Exon 5, encoding three LDLa repeats at the protein level, is spliced in only about 40% of the time, as is exon 7, encoding the eighth LDLa repeat, and exon 19, encoding the cytoplasmic insert. Exon 7B, encoding the furin insert, is included only about 15% of the time. While we observed many isoforms with AS of other exons, when these exons are examined on an individual level across all the transcripts weighted by their abundance, we see that most other exons are included at an almost constitutive level.
We also profiled human APOER2 transcripts using a gene specific RT-PCR coupled with long-read sequencing and compared transcripts across the murine and human cerebral cortex (Fig. 3–4). We identified 48 high quality unique APOER2 isoforms in the human cerebral cortex. These isoforms had a mean length just under 2500 bp (Fig. 4A), similar to the murine sample (Fig. 3B), likely indicating the presence of multiple alternative events since the expected length of the longest canonical APOER2 product would be just over 2.9 kilobases. When we examine the individual human APOER2 transcripts identified (Fig. 4B), the most abundant APOER2 transcript is the canonical full-length APOER2 transcript, closely followed by an APOER2 transcript with exclusion of exon 18, encoding the cytoplasmic insert. Interestingly, in humans the third most abundant APOER2 transcript contains cassette splicing of exon 15, encoding the glycosylation domain, which we did not observe to be alternatively spliced by itself in mice (Fig. 2D & 3C). We did observe tandem exclusion of exons 14 and 15, encoding an EGF precursor-like repeat and the glycosylation domains respectively, as was observed in mice (Fig. 3C). In humans, we observed one intron retention event, with the intron between exons 7 and 8 retained, and likely rendering that isoform a target for nonsense mediated decay. We also observed two alternative 3’ splice sites near exon 18, which encodes the cytoplasmic insert. Of these alternative 3’ sites, one was in the intron before exon 18, resulting in the inclusion of an additional 17 nucleotides, and the other was within exon 18, truncating the exon to only 61 base pairs. In mice, we did observe the presence of alternative 3’ splice sites at exon 19, corresponding to human exon 18; however, these transcripts dropped out of our exon abundance filtering. We analyzed whether those detected alternative sites in mouse were homologous to the two detected in human and did not observe homology at the protein level. However, the use of alternative splice sites at this exon boundary may be conserved.
We examined the cumulative frequency of the identified human APOER2 transcripts (Fig. 4C) and found that 4–5 transcripts make up about 75% of APOER2 expression in the human cerebral cortex, with the remaining 25% of expression made up of numerous unique isoforms. We also compared the identified isoforms to the coding regions of annotated APOER2 transcripts in Ensembl (Howe et al., 2021) and NCBI (NCBI Resource Coordinators et al., 2018) databases. Fourteen of the transcripts we identified are present in existing databases, whereas 34 of the isoforms described here are novel (Fig. 4B&D). Similar to the mouse, we calculated a frequency spliced in value for each individual exon across all the human isoforms (Fig. 4E). Interestingly, we find exon 5 is included at a higher frequency of about 70% in humans compared to mice (Fig. 3F). Exon 8, encoding an EGF precursor-like repeat in humans and corresponding to exon 9 in mice, is also spliced out more in humans, with a frequency spliced in value of about 85% compared to its frequency close to 100% in mice. Exon 15, encoding the glycosylation domain, is also dynamically spliced in humans, with a frequency spliced in of about 75%. Exon 18, encoding the cytoplasmic insert in humans, demonstrates a frequency spliced in value of about 50%, compared to the corresponding exon 19 in mice which is relatively similar around 40%.
3.3. Coordinated alternative splicing in mouse and human Apoer2.
Since we observed that exons 15 and 16 were only spliced out in tandem in the mouse cerebral cortex (Fig. 3C), we wanted to examine the relationship of all individual AS events across Apoer2 to determine whether splicing is coordinated across the length of the transcript. We calculated a weighted exon coincidence value, or the frequency at which each individual exon was spliced in at the same time as each other individual exon, (Fig. 5A, C). Exons such as 1–4 and 8–18, that are included at an almost constitutive level, exhibit high coincidence values as they are almost always spliced in together. Alternative cassette exons including 5, 6, 7, 7B and 19 demonstrate lower coincidence values due to their occasional exclusion. There also appears to be a shift in coincidence value between specific alternative exon pairs, such as exons 5 and 7, 5 and 7B, 7 and 7B, and exon 19 with 5, 7 and 7B. To examine these pairs more closely and determine whether there is any correlation between paired exon inclusion, we generated a correlation matrix (Fig. 5B). We also included exons 15 and 16 due to the tandem exclusion events we observed of these exons in Fig. 3C, as well as exon 9 for comparison with human exon 8 (Fig. 5D). Our results indicate a significant negative correlation between exon 7B and exons 5, 7 and 19, with the strongest correlation observed between exons 7 and 7B, which is supported by our RT-PCR data (Fig. 2F). We also observed significant positive correlations between exons 5 and 7, exons 5 and 19 and exon 7 and 19. Exons 15 and 16 demonstrate a perfect correlation of 1, which reflects the pattern of splicing observed for these exons previously (Fig. 3C).
Figure 5. Mouse Apoer2 exhibits tandem splicing of exons encoding the third EGF precursor-like repeat and glycosylation domain.
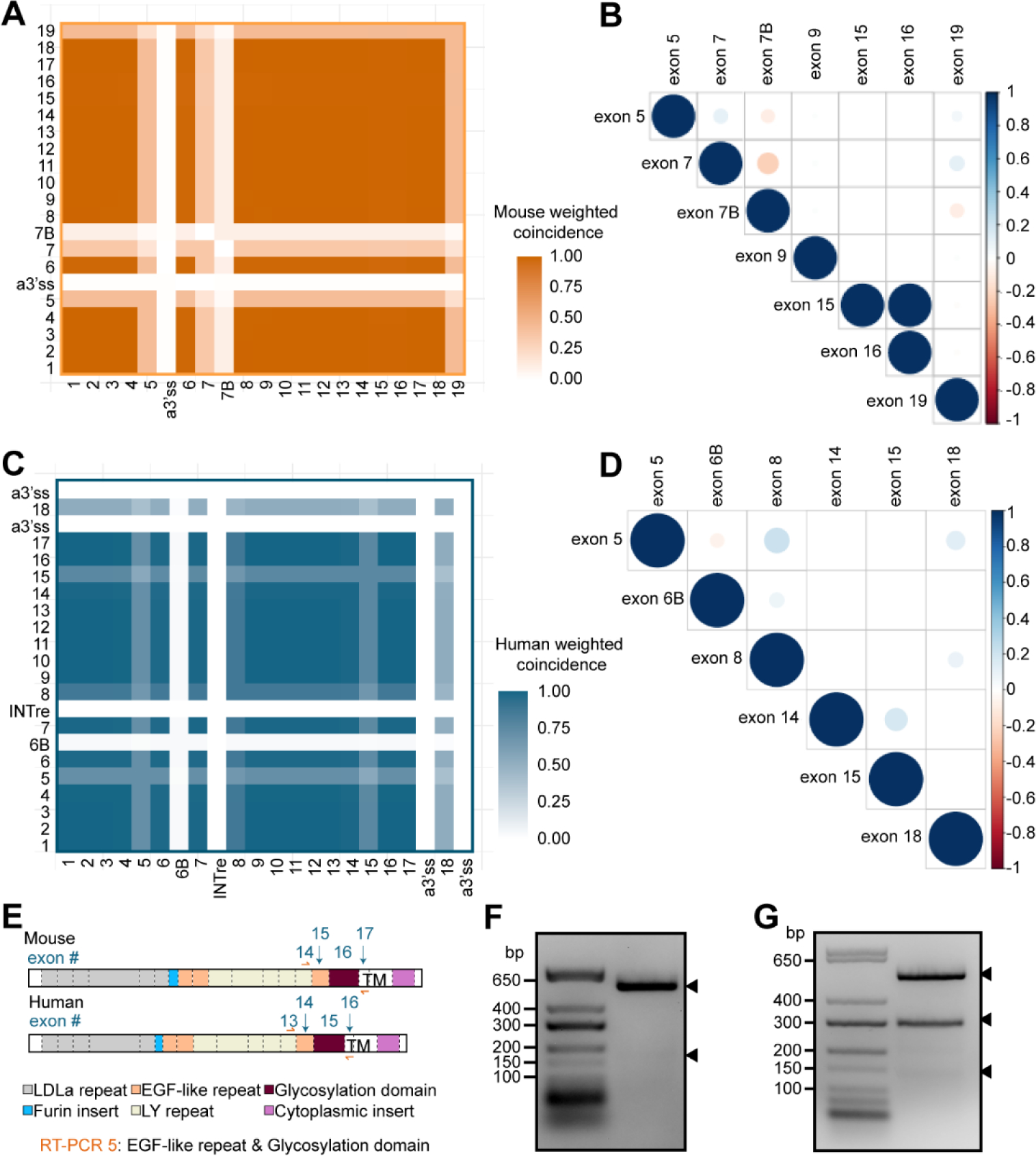
(A) Heatmap examining the coincidence of individual murine Apoer2 exons compared to every other exon. Coincidence was calculated as the number of times the exons in each comparison pair were both spliced into the same transcript divided by the total number of transcripts. (B) Correlation matrix depicting Pearson’s correlation coefficient of alternatively spliced murine Apoer2 exons. Color indicates value of Pearson’s coefficient and size of dot indicates magnitude of significance, with a cutoff of p<0.01. (C) Heatmap displaying coincidence value for human cerebral cortex APOER2 exons. (D) Correlation matrix of Pearson’s correlation coefficient analysis of human alternatively spliced APOER2 exons. Significance is p<0.01. (E) Schematic depicting mouse and human Apoer2 protein domains and corresponding coding exons along with RT-PCR strategy for F and G. (F) Gel depicting RT-PCR of mRNA from the murine cerebral cortex examining coordinated splicing of Apoer2 exons 15 and 16. Arrowheads indicate detected bands that were excised and sequenced. (G) Gel analysis of RT-PCR from human cerebral cortex mRNA examining splicing of exons 14 and 15 in APOER2. Arrowheads indicate detected bands excised for sequencing confirmation.
When we examined coincidence across human APOER2 exons (Fig. 5C), it again highlighted alternatively spliced cassette exons, which have lower coincidence values, including exons 5, 8, 15 and 18. Certain pairs of alternative exons demonstrated an apparent shift in coincidence values which may indicate coordinated splicing, such as the pairs exon 5 and 8, 5 and 18 and 8 and 18. Correlation analysis revealed significant positive correlation between exons 5 and 8, 5 and 18, 6B and 8, and 8 and 18 (Fig. 5D). A small but significant positive correlation is also observed between exons 14 and 15; however, this correlation is smaller when compared to the correlation observed between the corresponding murine exons 15 and 16 (Fig. 5B). We also observed a significant negative correlation between exon 5 and 6B in humans.
To validate the tandem splicing event that we observed in murine Apoer2 of exons 15 and 16 compared to corresponding human APOER2 exons 14 and 15, we designed RT-PCR primers spanning both exons (Fig. 5E) and performed RT-PCR on cDNA generated using oligo-dT primers. In mouse cerebral cortex, we observed two bands present (Fig. 5F), with the top band of stronger intensity. Upon sequencing, we confirmed the top band to contain both murine exons 15 and 16, and the bottom faint band to lack both exons. In humans, we identified the presence of 3 bands (Fig. 5G). Sequencing analysis confirmed the top band to contain exons 14 and 15 and the second band to contain exon 14 but lack exon 15. The smallest band contained tandem exclusion of both exons 14 and 15. We also identified one clone that contained exon 14 as well as a segment of 66 base pairs from the intron between exons 14 and 15, perhaps indicating another alternative cassette exon event (Fig. S4). Together, this confirmed that in mice the last EGF precursor-like repeat and the glycosylation domain are only tandemly excluded, whereas in humans the glycosylation domain can be spliced out individually or in combination with the preceding exon.
4. Discussion
In this study, we utilized a combination of RT-PCR and single molecule, long-read RNA sequencing to map the Apoer2 isoform pool across vertebrates. We found clear evolution of the Apoer2 gene over vertebrate development along with changes in AS patterns, indicating gene diversification at multiple levels. This suggests Apoer2 has been specifically altered over time, likely to introduce new functional consequences at the protein level. Application of SMRT long-read sequencing to Apoer2 has provided us with a new perspective on the complexity of AS events across individual Apoer2 transcripts in the cerebral cortex of both mice and humans. We identified 68 and 48 unique Apoer2 isoforms in the cerebral cortex of each mice and humans, respectively, with only 19 isoforms homologous between the two species. Our findings revealed that Apoer2 AS events occur in a plethora of combinations across the entire transcript and differ between species. Our results parallel recent findings published by the Allen Brain Atlas Team that highlight gene expression divergence in homologous cell types between mouse and human cortex despite overall conserved cell types (Hodge et al., 2019). This diversification in gene expression patterns between mouse and humans emphasizes the importance of species-specific studies, particularly in humans when relating gene function to disease and human functional biology. The identified diversity in Apoer2 isoform expression implicates differential functional effects at the protein level among isoforms and potentially between species and could influence ligand binding, receptor surface expression, receptor proteolysis, and downstream signaling events that affect synaptic function.
We have found that the Apoer2 exon encoding the eighth LDLa repeat is constitutively included in zebrafish and alternatively spliced in chickens, rabbits and mice, yet lost in primates, which is consistent with published literature (Kim et al., 1998; Myant, 2010). The functional consequences of losing the eighth LDLa repeat in primates is unclear, but it is possible that loss of this eighth repeat changes the binding properties of primate Apoer2 compared to that in lower vertebrates, as Apoer2 binds numerous important ligands in addition to Apoe, such as Reelin (D’Arcangelo et al., 1999; Trommsdorff et al., 1999), clusterin (Leeb et al., 2014), selenoprotein P (Burk et al., 2007), and many others (Dlugosz and Nimpf, 2018). AS of the eighth LDLa repeat in combination with AS of the three LDLa repeats encoded by exon 5 has already been shown to decrease Apoer2 affinity for β-VLDL when compared to AS of exon 5 alone (Brandes et al., 2001). Furthermore, exclusion of the eighth LDLa repeat also increases the affinity of Apoer2 for Reelin fragments (Hibi et al., 2009), suggesting perhaps loss of the eighth LDLa repeat occurred in primates to increase the affinity of Apoer2 for its endogenous ligands.
We have also shown that the 39-nucleotide exon encoding a furin cleavage site is present in rabbits, mice and primates, yet absent in zebrafish and chickens. Inclusion of this 39-nucleotide insert introduces a furin cleavage site at the protein level, which has been shown to be cleaved and to generate a soluble Apoer2 ectodomain fragment that acts as an antagonist to Reelin signaling (Koch et al., 2002), which is critical for neuronal migration in the developing brain (Trommsdorff et al., 1999) and ligand induced LTP in the adult brain (Weeber et al., 2002). It is unclear whether this secreted fragment would interfere with Apoer2-Apoe binding, although that seems likely. Furthermore, addition of the furin cleavage site between chickens and rabbits is especially interesting as chickens are not known to synthesize Apoe (Novak et al., 1996), suggesting perhaps diversification of the Apoer2 locus occurred to help modulate Apoe and other ligand binding in higher vertebrates. There is precedence for altered splicing of transmembrane receptors having potent effects on signaling, as ephrin transmembrane receptors have been shown to have either repulsive or adhesive effects on cell migration depending on which isoform is expressed (Holmberg et al., 2000). Furthermore, AS has already been shown to play a role in the Reelin signaling pathway in development, as suppression by the splicing factor Nova2 of a specific isoform of Disabled-1, the adaptor protein that binds to Apoer2, is required for proper neuronal migration (Yano et al., 2010).
Interestingly, we found that AS of the glycosylation domain alone in Apoer2 appears to be restricted to primates. In mice, exon 16, encoding the glycosylation domain, has previously been reported to be alternatively spliced in the brain (May et al., 2003); however, despite thorough efforts to optimize RT-PCR conditions, we were not able to detect a cassette exon skipping event of solely exon 16 in mice. Intriguingly, in mice we did detect coordinated AS of exon 16 with exon 15, which encodes the third EGF precursor-like repeat in Apoer2. As exons 15 and 16 are in the same phase, the open reading frame is likely preserved; however, the functional consequences of this tandem skipping event are unclear. The glycosylation domain has been shown to regulate extracellular processing of Apoer2, as this region contains the extracellular cleavage site for matrix metalloproteases (Wasser et al., 2014). It has also been found that exclusion of exon 16 in combination with AS of the cytoplasmic insert has unique functional consequences in the mouse brain, most notably on Apoer2 cell surface levels, synaptic strength and long term potentiation (Wasser et al., 2014). It is possible that AS of the glycosylation domain, whether in tandem with the EGF precursor-like repeat before it or by itself, emerged in mice and primates as a regulatory mechanism of Apoer2 surface levels, which are critical for maintaining the proper balance of dendritic spines (Dumanis et al., 2011).
The cytoplasmic insert emerged somewhere between chickens and rabbits, likely with the divergence of placental mammals and marsupials, which have been shown to lack the cytoplasmic insert (Myant, 2010). While it is unclear where the cytoplasmic insert originated, as it is unique and not present in other LDLR family members (Kim et al., 1997), it is clear that it adds functional complexity in Apoer2 biology. Inclusion of the cytoplasmic insert is necessary for Reelin-induced LTP (Beffert et al., 2005), neuronal survival (Beffert et al., 2006) and intracellular adaptor protein binding (Gotthardt et al., 2000; He et al., 2007; Stockinger et al., 2000). Inclusion of the cytoplasmic insert in APOER2 has also been positively correlated with human global cognition and shown to be lowered in the temporal cortex of individuals with Alzheimer’s disease. Furthermore, increasing inclusion of the cytoplasmic insert in an amyloid mouse model of Alzheimer’s disease with an antisense oligonucleotide can partially rescue some spatial learning deficits (Hinrich et al., 2016). It is reasonable to posit that incorporation of the cytoplasmic insert into Apoer2 conferred isoform specific functions and helped shape higher level vertebrate regulation of learning and memory. Apoer2 also undergoes sequential cleavage at the membrane, first extracellularly by matrix metalloproteases, and then intramembranously by γ-secretase, releasing an intracellular domain (ICD) that translocates to the nucleus and activates an enhancer profile necessary for the transcription of learning and memory genes (Telese et al., 2015). It remains to be determined which form of the Apoer2 ICD is actually transcriptionally active, as the cytoplasmic insert is located in the ICD and its inclusion or exclusion would naturally change the size and likely structure of the ICD.
From our long-read sequencing data, we observed 68 and 48 unique Apoer2 isoforms in the cerebral cortex of each mice and humans, respectively. The AS events that make up the unique identified isoforms largely occur in regions that encode functional domains at the protein level. This suggests that the different combinations of these AS events will alter the properties of individual Apoer2 isoforms at the protein level. Since Apoer2 AS events occur in many combinations, it highlights the need to understand how these events are regulated and whether they have diverse functional effects. Splicing is often controlled in a spatiotemporal manner (Raj and Blencowe, 2015), and so combinatorial splicing across the Apoer2 transcript provides multiple points at which to fine tune Apoer2 function as needed at different times and places in the brain. Indeed, we observed coordinated splicing in murine Apoer2, where the third EGF precursor-like repeat and the glycosylation domain are only alternatively spliced out together. This suggests some coordinated action of splicing involving these two exons.
Our study utilized bulk tissue from the cerebral cortex for analysis in order to define a clear repertoire of diverse and novel full-length Apoer2 isoforms and highlight species-specific differences in splicing decisions across the human and mouse cerebral cortex. It is likely that individual Apoer2 isoforms are expressed in a spatiotemporal manner or in a cell-type specific manner (Keren et al., 2010), which could give biological impact to some of the lowly expressed transcripts identified. It is also possible that the biological function of having several unique isoforms present at low levels could be to detract from having expression of some of the more abundant isoforms, in a type of splicing regulatory system. Therefore, it will be important for future studies to determine the localization of some of these isoforms within the cerebral cortex as well as determine if they exhibit cell-type specificity or cell to cell heterogeneity. Examining transcript localization is no easy task due to the need to identify the specific combination of exons across the length of the entire transcript. However, new methods are being developed to examine full length transcripts within individual cell types (Hård et al., 2021; Tian et al., 2020), which could prove interesting in the case of Apoer2 biology in the brain. Our finding of 68 unique Apoer2 isoforms in the mouse cerebral cortex is not far off from the identified 50 Apoer2 transcripts identified in the retina and brain by Ray and colleagues (Ray et al., 2020). As such, we are confident our isoform profiling of Apoer2 in mice and humans reflects true isoform diversity present in the brain.
Humans demonstrate substantial heterogeneity at the genomic level, that can affect RNA sequence and splicing decisions. Our study focused on defining the full repertoire of Apoer2 isoforms in one sample per species to understand broadly the potential isoform diversity of Apoer2. We cannot exclude the fact that individual genetic variation will affect Apoer2 isoform biology. Therefore, future studies utilizing biological replicates within species will be necessary to understand if and how human genetic diversity contributes to APOER2 splicing diversity. With genomic sequencing of individual samples done in parallel, the field could also start to understand whether any genomic variation, perhaps in the form of single nucleotide polymorphisms, are associated with certain splicing choices, as has been seen with CD33 (van Bergeijk et al., 2019) and RBM23 (Garrido-Martín et al., 2021).
Overall, we have demonstrated that the Apoer2 gene has evolved over the vertebrate lineage from zebrafish to humans, both at the genomic and AS level. Particularly, Apoer2 shows an extremely high number of AS events that result in the inclusion or exclusion of different functional domains in numerous combinations that implicate differential functions for Apoer2. Therefore, Apoer2 can then be thought of as a multi-functional receptor that can be tightly controlled depending on the AS pattern it possesses. Moving forward, it will be important to understand how the many AS events observed in this study contribute to modulating Apoer2’s roles in brain development, synapse formation (Dumanis et al., 2011) and long-term potentiation (Weeber et al., 2002) based on Apoe and Reelin binding (D’Arcangelo et al., 1999; Kim et al., 1996).
Supplementary Material
Highlights.
Apoer2 splicing complexity in the brain increased over vertebrate evolution.
Apoer2 demonstrates species-specific full-length isoforms in the brain.
Coordinated splicing in Apoer2 varies across species.
5. Acknowledgements
We would like to thank Dr. Shannon Fisher and lab for providing zebrafish brain tissue for this study. We also thank Dr. Rachel Flynn for critical feedback of experimental setup. We also thank Drs. Sara Goodwin and Ying Jin at the Cold Spring Harbor Sequencing Core for their assistance in the long-read sequencing PacBio experiment and initial SMRTLink data analysis.
Funding
This work was supported by the National Institutes of Health [grant numbers R01AG059762, F31AG069498, & 5T32GM008541-22]; and the Harold and Margaret Southerland Alzheimer’s Research Fund.
Footnotes
Sequence data from this article has been deposited with the DDBJ/EMBL/GenBank Data Libraries under Submission ID 2549724.
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alekseyenko AV, Kim N, Lee CJ, 2007. Global analysis of exon creation versus loss and the role of alternative splicing in 17 vertebrate genomes. RNA 13, 661–670. 10.1261/rna.325107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ, 1990. Basic local alignment search tool. Journal of Molecular Biology 215, 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Barbosa-Morais NL, 2005. Systematic genome-wide annotation of spliceosomal proteins reveals differential gene family expansion. Genome Research 16, 66–77. 10.1101/gr.3936206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beffert U, Nematollah Farsian F, Masiulis I, Hammer RE, Yoon SO, Giehl KM, Herz J, 2006. ApoE Receptor 2 Controls Neuronal Survival in the Adult Brain. Current Biology 16, 2446–2452. 10.1016/j.cub.2006.10.029 [DOI] [PubMed] [Google Scholar]
- Beffert U, Weeber EJ, Durudas A, Qiu S, Masiulis I, Sweatt JD, Li W-P, Adelmann G, Frotscher M, Hammer RE, Herz J, 2005. Modulation of synaptic plasticity and memory by Reelin involves differential splicing of the lipoprotein receptor Apoer2. Neuron 47, 567–579. 10.1016/j.neuron.2005.07.007 [DOI] [PubMed] [Google Scholar]
- Belloy ME, Napolioni V, Greicius MD, 2019. A Quarter Century of APOE and Alzheimer’s Disease: Progress to Date and the Path Forward. Neuron 101, 820–838. 10.1016/j.neuron.2019.01.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandes C, Kahr L, Stockinger W, Hiesberger T, Schneider WJ, Nimpf J, 2001. Alternative splicing in the ligand binding domain of mouse ApoE receptor-2 produces receptor variants binding reelin but not alpha 2-macroglobulin. J. Biol. Chem. 276, 22160–22169. 10.1074/jbc.M102662200 [DOI] [PubMed] [Google Scholar]
- Brandes C, Novak S, Stockinger W, Herz J, Schneider WJ, Nimpf J, 1997. Avian and Murine LR8B and Human Apolipoprotein E Receptor 2: Differentially Spliced Products from Corresponding Genes. Genomics 42, 185–191. 10.1006/geno.1997.4702 [DOI] [PubMed] [Google Scholar]
- Burk RF, Hill KE, Olson GE, Weeber EJ, Motley AK, Winfrey VP, Austin LM, 2007. Deletion of Apolipoprotein E Receptor-2 in Mice Lowers Brain Selenium and Causes Severe Neurological Dysfunction and Death When a Low-Selenium Diet Is Fed. Journal of Neuroscience 27, 6207–6211. 10.1523/JNEUROSCI.1153-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L, Chew SL, Krainer AR, 2002. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet 3, 285–298. 10.1038/nrg775 [DOI] [PubMed] [Google Scholar]
- Clatworthy AE, Stockinger W, Christie RH, Schneider WJ, Nimpf J, Hyman BT, Rebeck GW, 1999. Expression and alternate splicing of apolipoprotein E receptor 2 in brain. Neuroscience 90, 903–911. 10.1016/S0306-4522(98)00489-8 [DOI] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA, 1993. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science 261, 921–923. [DOI] [PubMed] [Google Scholar]
- D’Arcangelo G, Homayouni R, Keshvara L, Rice DS, Sheldon M, Curran T, 1999. Reelin is a ligand for lipoprotein receptors. Neuron 24, 471–479. [DOI] [PubMed] [Google Scholar]
- Dlugosz P, Nimpf J, 2018. The Reelin Receptors Apolipoprotein E receptor 2 (ApoER2) and VLDL Receptor. IJMS 19, 3090. 10.3390/ijms19103090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorit RL, Gilbert W, 1991. The limited universe of exons. Current Opinion in Genetics & Development 1, 464–469. 10.1016/S0959-437X(05)80193-5 [DOI] [PubMed] [Google Scholar]
- Dumanis SB, Cha H-J, Song JM, Trotter JH, Spitzer M, Lee J-Y, Weeber EJ, Turner RS, Pak DTS, Rebeck GW, Hoe H-S, 2011. ApoE receptor 2 regulates synapse and dendritic spine formation. PLoS ONE 6, e17203. 10.1371/journal.pone.0017203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty E, Zhu S, Barretto N, Cheng E, Deans PJM, Fernando MB, Schrode N, Francoeur N, Antoine A, Alganem K, Halpern M, Deikus G, Shah H, Fitzgerald M, Ladran I, Gochman P, Rapoport J, Tsankova NM, McCullumsmith R, Hoffman GE, Sebra R, Fang G, Brennand KJ, 2019. Neuronal impact of patient-specific aberrant NRXN1α splicing. Nat Genet 51, 1679–1690. 10.1038/s41588-019-0539-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrell Frank E Jr, with contributions from Charles Dupont and many others. (2020). Hmisc: Harrell Miscellaneous. R package version 4.4–2. https://CRAN.R-project.org/package=Hmisc, n.d. [Google Scholar]
- Frankish A, Diekhans M, Jungreis I, Lagarde J, Loveland JE, Mudge JM, Sisu C, Wright JC, Armstrong J, Barnes I, Berry A, Bignell A, Boix C, Carbonell Sala S, Cunningham F, Di Domenico T, Donaldson S, Fiddes IT, García Girón C, Gonzalez JM, Grego T, Hardy M, Hourlier T, Howe KL, Hunt T, Izuogu OG, Johnson R, Martin FJ, Martínez L, Mohanan S, Muir P, Navarro FCP, Parker A, Pei B, Pozo F, Riera FC, Ruffier M, Schmitt BM, Stapleton E, Suner M-M, Sycheva I, Uszczynska-Ratajczak B, Wolf MY, Xu J, Yang YT, Yates A, Zerbino D, Zhang Y, Choudhary JS, Gerstein M, Guigó R, Hubbard TJP, Kellis M, Paten B, Tress ML, Flicek P, 2021. GENCODE 2021. Nucleic Acids Research 49, D916–D923. 10.1093/nar/gkaa1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido-Martín D, Borsari B, Calvo M, Reverter F, Guigó R, 2021. Identification and analysis of splicing quantitative trait loci across multiple tissues in the human genome. Nat Commun 12, 727. 10.1038/s41467-020-20578-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotthardt M, Trommsdorff M, Nevitt MF, Shelton J, Richardson JA, Stockinger W, Nimpf J, Herz J, 2000. Interactions of the Low Density Lipoprotein Receptor Gene Family with Cytosolic Adaptor and Scaffold Proteins Suggest Diverse Biological Functions in Cellular Communication and Signal Transduction. J. Biol. Chem. 275, 25616–25624. 10.1074/jbc.M000955200 [DOI] [PubMed] [Google Scholar]
- Hård J, Mold JE, Eisfeldt J, Tellgren-Roth C, Häggqvist S, Bunikis I, Contreras-Lopez O, Chin C-S, Rubin C-J, Feuk L, Michaëlsson J, Ameur A, 2021. Long-read whole genome analysis of human single cells (preprint). Genomics. 10.1101/2021.04.13.439527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Cooley K, Chung CHY, Dashti N, Tang J, 2007. Apolipoprotein Receptor 2 and X11 / Mediate Apolipoprotein E-Induced Endocytosis of Amyloid- Precursor Protein and -Secretase, Leading to Amyloid- Production. Journal of Neuroscience 27, 4052–4060. 10.1523/JNEUROSCI.3993-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibi T, Mizutani M, Baba A, Hattori M, 2009. Splicing variations in the ligand-binding domain of ApoER2 results in functional differences in the binding properties to Reelin. Neuroscience Research 63, 251–258. 10.1016/j.neures.2008.12.009 [DOI] [PubMed] [Google Scholar]
- Hinrich AJ, Jodelka FM, Chang JL, Brutman D, Bruno AM, Briggs CA, James BD, Stutzmann GE, Bennett DA, Miller SA, Rigo F, Marr RA, Hastings ML, 2016. Therapeutic correction of ApoER2 splicing in Alzheimer’s disease mice using antisense oligonucleotides. EMBO Molecular Medicine 8, 328–345. 10.15252/emmm.201505846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge RD, Bakken TE, Miller JA, Smith KA, Barkan ER, Graybuck LT, Close JL, Long B, Johansen N, Penn O, Yao Z, Eggermont J, Höllt T, Levi BP, Shehata SI, Aevermann B, Beller A, Bertagnolli D, Brouner K, Casper T, Cobbs C, Dalley R, Dee N, Ding S-L, Ellenbogen RG, Fong O, Garren E, Goldy J, Gwinn RP, Hirschstein D, Keene CD, Keshk M, Ko AL, Lathia K, Mahfouz A, Maltzer Z, McGraw M, Nguyen TN, Nyhus J, Ojemann JG, Oldre A, Parry S, Reynolds S, Rimorin C, Shapovalova NV, Somasundaram S, Szafer A, Thomsen ER, Tieu M, Quon G, Scheuermann RH, Yuste R, Sunkin SM, Lelieveldt B, Feng D, Ng L, Bernard A, Hawrylycz M, Phillips JW, Tasic B, Zeng H, Jones AR, Koch C, Lein ES, 2019. Conserved cell types with divergent features in human versus mouse cortex. Nature 573, 61–68. 10.1038/s41586-019-1506-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg J, Clarke DL, Frisén J, 2000. Regulation of repulsion versus adhesion by different splice forms of an Eph receptor. Nature 408, 203–206. 10.1038/35041577 [DOI] [PubMed] [Google Scholar]
- Howe KL, Achuthan P, Allen James, Allen Jamie, Alvarez-Jarreta J, Amode MR, Armean IM, Azov AG, Bennett R, Bhai J, Billis K, Boddu S, Charkhchi M, Cummins C, Da Rin Fioretto L, Davidson C, Dodiya K, El Houdaigui B, Fatima R, Gall A, Garcia Giron C, Grego T, Guijarro-Clarke C, Haggerty L, Hemrom A, Hourlier T, Izuogu OG, Juettemann T, Kaikala V, Kay M, Lavidas I, Le T, Lemos D, Gonzalez Martinez J, Marugán JC, Maurel T, McMahon AC, Mohanan S, Moore B, Muffato M, Oheh DN, Paraschas D, Parker A, Parton A, Prosovetskaia I, Sakthivel MP, Salam AIA, Schmitt BM, Schuilenburg H, Sheppard D, Steed E, Szpak M, Szuba M, Taylor K, Thormann A, Threadgold G, Walts B, Winterbottom A, Chakiachvili M, Chaubal A, De Silva N, Flint B, Frankish A, Hunt SE, IIsley GR, Langridge N, Loveland JE, Martin FJ, Mudge JM, Morales J, Perry E, Ruffier M, Tate J, Thybert D, Trevanion SJ, Cunningham F, Yates AD, Zerbino DR, Flicek P, 2021. Ensembl 2021. Nucleic Acids Research 49, D884–D891. 10.1093/nar/gkaa942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y-WA, Zhou B, Wernig M, Südhof TC, 2017. ApoE2, ApoE3, and ApoE4 Differentially Stimulate APP Transcription and Aβ Secretion. Cell 168, 427–441.e21. 10.1016/j.cell.2016.12.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TA, 2006. Regulation of gene expression by alternative untranslated regions. Trends in Genetics 22, 119–122. 10.1016/j.tig.2006.01.001 [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium, Whitehead Institute for Biomedical Research, Center for Genome Research:, Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond Christina, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, The Sanger Centre:, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Washington University Genome Sequencing Center, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, US DOE Joint Genome Institute:, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng J-F, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Baylor College of Medicine Human Genome Sequencing Center:, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, RIKEN Genomic Sciences Center:, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Genoscope and CNRS UMR-8030:, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Department of Genome Analysis, Institute of Molecular Biotechnology:, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, GTC Sequencing Center:, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Beijing Genomics Institute/Human Genome Center:, Yang H, Yu J, Wang J, Huang G, Gu J, Multimegabase Sequencing Center, The Institute for Systems Biology:, Hood L, Rowen L, Madan A, Qin S, Stanford Genome Technology Center:, Davis RW, Federspiel NA, Abola AP, Proctor MJ, University of Oklahoma’s Advanced Center for Genome Technology:, Roe BA, Chen F, Pan H, Max Planck Institute for Molecular Genetics:, Ramser J, Lehrach H, Reinhardt R, Cold Spring Harbor Laboratory, Lita Annenberg Hazen Genome Center:, McCombie WR, de la Bastide M, Dedhia N, GBF—German Research Centre for Biotechnology:, Blöcker H, Hornischer K, Nordsiek G, *Genome Analysis Group (listed in alphabetical order, also includes individuals listed under other headings):, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen H-C, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JGR, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AFA, Stupka E, Szustakowki J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang S-P, Yeh R-F, Scientific management: National Human Genome Research Institute, US National Institutes of Health:, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Stanford Human Genome Center:, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, University of Washington Genome Center:, Olson MV, Kaul R, Raymond Christopher, Department of Molecular Biology, Keio University School of Medicine:, Shimizu N, Kawasaki K, Minoshima S, University of Texas Southwestern Medical Center at Dallas:, Evans GA, Athanasiou M, Schultz R, Office of Science, US Department of Energy:, Patrinos A, The Wellcome Trust:, Morgan MJ, 2001. Initial sequencing and analysis of the human genome. Nature 409, 860–921. 10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- Keren H, Lev-Maor G, Ast G, 2010. Alternative splicing and evolution: diversification, exon definition and function. Nature Reviews Genetics 11, 345–355. 10.1038/nrg2776 [DOI] [PubMed] [Google Scholar]
- Kim DH, Iijima H, Goto K, Sakai J, Ishii H, Kim HJ, Suzuki H, Kondo H, Saeki S, Yamamoto T, 1996. Human apolipoprotein E receptor 2. A novel lipoprotein receptor of the low density lipoprotein receptor family predominantly expressed in brain. J. Biol. Chem. 271, 8373–8380. 10.1074/jbc.271.14.8373 [DOI] [PubMed] [Google Scholar]
- Kim D-H, Magoori K, Inoue TR, Mao CC, Kim H-J, Suzuki H, Fujita T, Endo Y, Saeki S, Yamamoto TT, 1997. Exon/Intron Organization, Chromosome Localization, Alternative Splicing, and Transcription Units of the Human Apolipoprotein E Receptor 2 Gene. J. Biol. Chem. 272, 8498–8504. 10.1074/jbc.272.13.8498 [DOI] [PubMed] [Google Scholar]
- Kim E, Goren A, Ast G, 2008. Alternative splicing: current perspectives. Bioessays 30, 38–47. 10.1002/bies.20692 [DOI] [PubMed] [Google Scholar]
- Kim H-J, Kim D-H, Magoori K, Saeki S, Yamamoto TT, 1998. Evolution of the Apolipoprotein E Receptor 2 Gene by Exon Loss. Journal of Biochemistry 124, 451–456. 10.1093/oxfordjournals.jbchem.a022134 [DOI] [PubMed] [Google Scholar]
- Koch S, Strasser V, Hauser C, Fasching D, Brandes C, Bajari TM, Schneider WJ, Nimpf J, 2002. A secreted soluble form of ApoE receptor 2 acts as a dominant-negative receptor and inhibits Reelin signaling. EMBO J 21, 5996–6004. 10.1093/emboj/cdf599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolkman JA, Stemmer WPC, 2001. Directed evolution of proteins by exon shuffling. Nat Biotechnol 19, 423–428. 10.1038/88084 [DOI] [PubMed] [Google Scholar]
- Krawczak M, Reiss J, Cooper DavidN., 1992. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: Causes and consequences. Hum Genet 90. 10.1007/BF00210743 [DOI] [PubMed] [Google Scholar]
- Leeb C, Eresheim C, Nimpf J, 2014. Clusterin Is a Ligand for Apolipoprotein E Receptor 2 (ApoER2) and Very Low Density Lipoprotein Receptor (VLDLR) and Signals via the Reelin-signaling Pathway. Journal of Biological Chemistry 289, 4161–4172. 10.1074/jbc.M113.529271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Lee J-A, Black DL, 2007. Neuronal regulation of alternative pre-mRNA splicing. Nature Reviews Neuroscience 8, 819–831. 10.1038/nrn2237 [DOI] [PubMed] [Google Scholar]
- Liu M, Grigoriev A, 2004. Protein domains correlate strongly with exons in multiple eukaryotic genomes – evidence of exon shuffling? Trends in Genetics 20, 399–403. 10.1016/j.tig.2004.06.013 [DOI] [PubMed] [Google Scholar]
- May P, Bock HH, Nimpf J, Herz J, 2003. Differential Glycosylation Regulates Processing of Lipoprotein Receptors by γ-Secretase. J. Biol. Chem. 278, 37386–37392. 10.1074/jbc.M305858200 [DOI] [PubMed] [Google Scholar]
- Minami SS, Sung YM, Dumanis SB, Chi SH, Burns MP, Ann E-J, Suzuki T, Turner RS, Park H-S, Pak DTS, Rebeck GW, Hoe H-S, 2010. The cytoplasmic adaptor protein X11α and extracellular matrix protein Reelin regulate ApoE receptor 2 trafficking and cell movement. The FASEB Journal 24, 58–69. 10.1096/fj.09-138123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myant NB, 2010. Reelin and apolipoprotein E receptor 2 in the embryonic and mature brain: effects of an evolutionary change in the apoER2 gene. Proc. R. Soc. B 277, 345–351. 10.1098/rspb.2009.1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI Resource Coordinators, Agarwala R, Barrett T, Beck J, Benson DA, Bollin C, Bolton E, Bourexis D, Brister JR, Bryant SH, Canese K, Cavanaugh M, Charowhas C, Clark K, Dondoshansky I, Feolo M, Fitzpatrick L, Funk K, Geer LY, Gorelenkov V, Graeff A, Hlavina W, Holmes B, Johnson M, Kattman B, Khotomlianski V, Kimchi A, Kimelman M, Kimura M, Kitts P, Klimke W, Kotliarov A, Krasnov S, Kuznetsov A, Landrum MJ, Landsman D, Lathrop S, Lee JM, Leubsdorf C, Lu Z, Madden TL, Marchler-Bauer A, Malheiro A, Meric P, Karsch-Mizrachi I, Mnev A, Murphy T, Orris R, Ostell J, O’Sullivan C, Palanigobu V, Panchenko AR, Phan L, Pierov B, Pruitt KD, Rodarmer K, Sayers EW, Schneider V, Schoch CL, Schuler GD, Sherry ST, Siyan K, Soboleva A, Soussov V, Starchenko G, Tatusova TA, Thibaud-Nissen F, Todorov K, Trawick BW, Vakatov D, Ward M, Yaschenko E, Zasypkin A, Zbicz K, 2018. Database resources of the National Center for Biotechnology Information. Nucleic Acids Research 46, D8–D13. 10.1093/nar/gkx1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR, 2010. Expansion of the eukaryotic proteome by alternative splicing. Nature 463, 457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak S, Hiesberger T, Schneider WJ, Nimpf J, 1996. A New Low Density Lipoprotein Receptor Homologue with 8 Ligand Binding Repeats in Brain of Chicken and Mouse. J. Biol. Chem. 271, 11732–11736. 10.1074/jbc.271.20.11732 [DOI] [PubMed] [Google Scholar]
- Papadopoulos JS, Agarwala R, 2007. COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics 23, 1073–1079. 10.1093/bioinformatics/btm076 [DOI] [PubMed] [Google Scholar]
- Patthy L, 1999. Genome evolution and the evolution of exon-shuffling — a review. Gene 238, 103–114. 10.1016/S0378-1119(99)00228-0 [DOI] [PubMed] [Google Scholar]
- Qiu Z, Hyman BT, Rebeck GW, 2004. Apolipoprotein E Receptors Mediate Neurite Outgrowth through Activation of p44/42 Mitogen-activated Protein Kinase in Primary Neurons. J. Biol. Chem. 279, 34948–34956. 10.1074/jbc.M401055200 [DOI] [PubMed] [Google Scholar]
- R Core Team, 2020. R: A language and environment for statistical computing.
- Raj B, Blencowe BJ, 2015. Alternative Splicing in the Mammalian Nervous System: Recent Insights into Mechanisms and Functional Roles. Neuron 87, 14–27. 10.1016/j.neuron.2015.05.004 [DOI] [PubMed] [Google Scholar]
- Ray TA, Cochran K, Kozlowski C, Wang J, Alexander G, Cady MA, Spencer WJ, Ruzycki PA, Clark BS, Laeremans A, He M-X, Wang X, Park E, Hao Y, Iannaccone A, Hu G, Fedrigo O, Skiba NP, Arshavsky VY, Kay JN, 2020. Comprehensive identification of mRNA isoforms reveals the diversity of neural cell-surface molecules with roles in retinal development and disease. Nat Commun 11, 3328. 10.1038/s41467-020-17009-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP, 2011. Integrative genomics viewer. Nat Biotechnol 29, 24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudenko G, Henry L, Henderson K, Ichtchenko K, Brown MS, Goldstein JL, Deisenhofer J, 2002. Structure of the LDL Receptor Extracellular Domain at Endosomal pH. Science 298, 2353. 10.1126/science.1078124 [DOI] [PubMed] [Google Scholar]
- Saunders AM, Strittmatter WJ, Schmechel D, St. George-Hyslop PH, Pericak-Vance MA, Joo SH, Rosi BL, Gusella JF, Crapper-MacLachlan DR, Alberts MJ, Hulette C, Crain B, Goldgaber D, Roses AD, 1993. Association of apolipoprotein E allele 4 with late-onset familial and sporadic Alzheimer’s disease. Neurology 43, 1467–1467. 10.1212/WNL.43.8.1467 [DOI] [PubMed] [Google Scholar]
- Stockinger W, Brandes C, Fasching D, Hermann M, Gotthardt M, Herz J, Schneider WJ, Nimpf J, 2000. The Reelin Receptor ApoER2 Recruits JNK-interacting Proteins-1 and −2. J. Biol. Chem. 275, 25625–25632. 10.1074/jbc.M004119200 [DOI] [PubMed] [Google Scholar]
- Sudhof T, Russell D, Goldstein J, Brown M, Sanchez-Pescador R, Bell G, 1985. Cassette of eight exons shared by genes for LDL receptor and EGF precursor. Science 228, 893–895. 10.1126/science.3873704 [DOI] [PubMed] [Google Scholar]
- Wei Taiyun and Simko Viliam (2017). R package “corrplot”: Visualization of a Correlation Matrix (Version 0.84). Available from https://github.com/taiyun/corrplot, n.d.
- Tardaguila M, de la Fuente L, Marti C, Pereira C, Pardo-Palacios FJ, del Risco H, Ferrell M, Mellado M, Macchietto M, Verheggen K, Edelmann M, Ezkurdia I, Vazquez J, Tress M, Mortazavi A, Martens L, Rodriguez-Navarro S, Moreno-Manzano V, Conesa A, 2018. SQANTI: extensive characterization of long-read transcript sequences for quality control in full-length transcriptome identification and quantification. Genome Res. 28, 396–411. 10.1101/gr.222976.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telese F, Ma Q, Perez PM, Notani D, Oh S, Li W, Comoletti D, Ohgi KA, Taylor H, Rosenfeld MG, 2015. LRP8-Reelin-Regulated Neuronal Enhancer Signature Underlying Learning and Memory Formation. Neuron 86, 696–710. 10.1016/j.neuron.2015.03.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Jabbari JS, Thijssen R, Gouil Q, Amarasinghe SL, Kariyawasam H, Su S, Dong X, Law CW, Lucattini A, Chung JD, Naim T, Chan A, Ly CH, Lynch GS, Ryall JG, Anttila CJA, Peng H, Anderson MA, Roberts AW, Huang DCS, Clark MB, Ritchie ME, 2020. Comprehensive characterization of single cell full-length isoforms in human and mouse with long-read sequencing (preprint). Genomics. 10.1101/2020.08.10.243543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein B, Gokce O, Quake SR, Südhof TC, 2014. Cartography of neurexin alternative splicing mapped by single-molecule long-read mRNA sequencing. Proc Natl Acad Sci USA 111, E1291–E1299. 10.1073/pnas.1403244111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trommsdorff M, Borg J-P, Margolis B, Herz J, 1998. Interaction of Cytosolic Adaptor Proteins with Neuronal Apolipoprotein E Receptors and the Amyloid Precursor Protein. J. Biol. Chem. 273, 33556–33560. 10.1074/jbc.273.50.33556 [DOI] [PubMed] [Google Scholar]
- Trommsdorff M, Gotthardt M, Hiesberger T, Shelton J, Stockinger W, Nimpf J, Hammer RE, Richardson JA, Herz J, 1999. Reeler/Disabled-like Disruption of Neuronal Migration in Knockout Mice Lacking the VLDL Receptor and ApoE Receptor 2. Cell 97, 689–701. 10.1016/S0092-8674(00)80782-5 [DOI] [PubMed] [Google Scholar]
- van Bergeijk P, Seneviratne U, Aparicio-Prat E, Stanton R, Hasson SA, 2019. SRSF1 and PTBP1 Are trans -Acting Factors That Suppress the Formation of a CD33 Splicing Isoform Linked to Alzheimer’s Disease Risk. Mol Cell Biol 39, e00568–18, /mcb/39/18/MCB.00568-18.atom. 10.1128/MCB.00568-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB, 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456, 470–476. 10.1038/nature07509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasser CR, Masiulis I, Durakoglugil MS, Lane-Donovan C, Xian X, Beffert U, Agarwala A, Hammer RE, Herz J, 2014. Differential splicing and glycosylation of Apoer2 alters synaptic plasticity and fear learning. Sci Signal 7, ra113. 10.1126/scisignal.2005438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeber EJ, Beffert U, Jones C, Christian JM, Förster E, Sweatt JD, Herz J, 2002. Reelin and ApoE Receptors Cooperate to Enhance Hippocampal Synaptic Plasticity and Learning. J. Biol. Chem. 277, 39944–39952. 10.1074/jbc.M205147200 [DOI] [PubMed] [Google Scholar]
- Wickham H, 2016. ggplot2: Elegant Graphics for Data Analysis, 2nd ed. 2016. ed, Use R! Springer International Publishing : Imprint: Springer, Cham. 10.1007/978-3-319-24277-4 [DOI] [Google Scholar]
- Yano M, Hayakawa-Yano Y, Mele A, Darnell RB, 2010. Nova2 Regulates Neuronal Migration through an RNA Switch in Disabled-1 Signaling. Neuron 66, 848–858. 10.1016/j.neuron.2010.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL, 2012. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13, 134. 10.1186/1471-2105-13-134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo GW, Van Nostrand E, Holste D, Poggio T, Burge CB, 2005. Identification and analysis of alternative splicing events conserved in human and mouse. Proceedings of the National Academy of Sciences 102, 2850–2855. 10.1073/pnas.0409742102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, Deng S, Liddelow SA, Zhang C, Daneman R, Maniatis T, Barres BA, Wu JQ, 2014. An RNA-Sequencing Transcriptome and Splicing Database of Glia, Neurons, and Vascular Cells of the Cerebral Cortex. Journal of Neuroscience 34, 11929–11947. 10.1523/JNEUROSCI.1860-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


