Abstract
Noroviruses are a leading cause of endemic and epidemic acute gastroenteritis in all age groups. However, in Latin America, there are limited and updated data regarding circulating genotypes. The aim of this study was to assess the prevalence and genetic diversity of norovirus outbreaks in Argentina from 2013 to 2018. Stool samples from 29 acute gastroenteritis (AGE) outbreaks were available for viral testing. Norovirus was detected in samples from 18 (62.1%) outbreaks (2 GI and 16 GII). Both GI outbreaks were typed as GI.6[P11] whereas 10 different GII genotypes were detected, in which GII.4 viruses were the most frequently detected (29.4%, associated with GII.P31 and GII.P16) followed by GII.1[P33] and GII.6[P7] (17.6% each). Like GII.4 viruses, GII.2 viruses were also detected in association with different polymerases (GII.P2 and GII.P16). Our findings underscore the importance of dual RNA-dependent RNA polymerase-VP1 typing since recombinant strains with new polymerase sequences emerge frequently suggesting a possible role in improved fitness of these viruses. This study represents the most recent multi-year assessment of the molecular epidemiology of norovirus strains associated with AGE outbreaks in Argentina. Molecular surveillance of norovirus has to be considered to monitor possible changes in dominant genotypes which may assist to inform the formulation of future vaccines.
Keywords: acute gastroenteritis outbreaks, molecular epidemiology, Norovirus
1 |. INTRODUCTION
Noroviruses are a leading cause of endemic and epidemic acute gastroenteritis (AGE) in people of all age-groups worldwide with the highest disease burden in young children, immunocompromised patients, and the elderly.1,2 They are a genetically and antigenically diverse group of viruses belonging to the genus Norovirus in the family Caliciviridae. The virus is composed of a single-stranded positive-sense RNA genome organized into three open reading frames (ORFs). Noroviruses are classified into at least seven genogroups (G), of which viruses from genogroups GI, GII, and GIV cause disease in humans.3 Based on amino acid diversity of the complete major capsid protein (VP1), GI and GII noroviruses can be further divided into at least 9 GI and 27 GII genotypes.4 Over the last 15 years, GII.4 viruses have been associated with the majority of norovirus outbreaks worldwide.5 However, other genotypes co-circulate and relatively rare genotypes (eg, GII.17 and GII.2) may suddenly emerge and cause the majority of outbreaks in a particular geographic region.6 Most published studies on the prevalence of norovirus in Latin America are from Brazil, Chile, Guatemala, Nicaragua, and Peru7 and the most recent studies on the prevalence of norovirus in Argentina are more than a decade old.8,9 Therefore, the aim of this study was to assess the prevalence and genetic diversity of norovirus outbreaks in Argentina from 2013 to 2018.
2 |. MATERIALS AND METHODS
AGE outbreaks defined as two or more cases of AGE (at least two episodes of watery diarrhea and/or vomiting) linked by time and geographic location, were reported to the Epidemiology Departments in the 24 Argentinian provinces. As molecular diagnosis is not available or is cost-prohibitive in many regional laboratories, norovirus detection is centralized into the National Laboratory of Viral Gastroenteritis in Buenos Aires. For viral testing, whole stool samples were extracted from low-speed centrifuged clarified 10%−20% fecal suspensions with the MagMax-96 Viral RNA Isolation Kit (Ambion) on an automated KingFisher Magnetic Particle Processor (Thermo Fisher Scientific), according to the manufacturer’s protocol. Nucleic acid was screened for norovirus GI and GII, sapovirus, astrovirus and group F adenovirus by real-time quantitative reverse transcription-polymerase chain reaction (RT-qPCR) as described previously.10,11 In addition, samples were also screened for rotavirus by Enzyme-linked immunosorbent assay (ProSpecT, Oxoid, UK). Confirmed norovirus outbreaks (defined as two or more positive RT-qPCR norovirus stool samples within an AGE outbreak) were further genotyped by amplification and sequencing of a partial region of the 3′-end of ORF1 and 5′-end of ORF2 of the genome as previously described.11 Raw sequences were edited using BioEdit version 7.2.5 (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) and genotype assignment was retrieved using the online software Human Calicivirus Typing Tool (https://norovirus.ng.philab.cdc.gov). Norovirus sequences from one representative strain of each outbreak were submitted to GenBank and assigned the following accession numbers MN508755-508756 (GI strains) and MN535185-535201 (GII strains).
3 |. RESULTS
A total of 189 stool samples from 29 AGE outbreaks (range 3–6 outbreaks per year) were available for viral testing. The number of samples tested per outbreak ranged from 3 to 18. Norovirus was detected in samples from 18 (62.1%) outbreaks (2 GI and 16 GII), rotavirus in 2 (6.9%), and samples from the remaining 10 outbreaks tested negative for viral enteropathogens (norovirus, rotavirus, sapovirus, group F adenovirus, and astrovirus) (Table 1). In 2015 and 2016, two mixed GII outbreaks were detected, with three and two GII different genotypes, respectively. Both GI outbreaks (#3 and #21; Table 1) were typed as GI.6[P11] whereas 10 different GII genotypes were detected (Figure 1). Of note, GI.6 viruses (associated with GI.P11 polymerase) were detected in the same time-frame as the reported increase of GI.6 outbreaks in the United States.12 GII.4 viruses were detected in 29.4% (5/17) of the GII outbreaks (17.6% associated with GII.P31 and 11.8% with GII.P16) followed by GII.1[P33] and GII.6[P7] each causing 17.6% (3/17) of the outbreaks. Thus, GII was the predominant genogroup and GII.4 represented the most frequently identified genotype and was detected in 5 of the 6 study years. Like GII.4 viruses, GII.2 viruses were also detected in association with different polymerases, GII.P2 and GII.P16.
TABLE 1.
Distribution of AGE outbreaks submitted for viral pathogen detection in Argentina, 2013–2018
| Year | Month | Outbreaka | Province | Number of stool samples tested | Norovirus positive samples (%) | Genotype |
|---|---|---|---|---|---|---|
| 2013 | February | 1 | Neuquén | 6 | 2 (33.3) | GII.6[P7] |
| March | 2 | Neuquén | 14 | 7 (50) | GII.4 Sydney[P31] | |
| May | 3 | San Luis | 6 | 4 (66.7) | GI.6[P11] | |
| 2014 | January | 4 | Tierra del Fuego | 4 | 0 | c |
| February | 5 | Neuquén | 3 | 1 (33.3) | c | |
| December | 6 | Neuquén | 3 | 3 (100) | GII.1[P33] | |
| 2015 | January | 7 | Neuquén | 18 | 7 (38.9) | GII.4 Sydney[P31]; GII.17[PNA6]; GII.6[P7] |
| April | 8 | CABA | 6 | 0 | c | |
| August | 9 | San Luis | 16 | 12 (75) | GII.1[P33] | |
| August | 10 | San Luis | 8 | 6 (75) | GII.1[P33] | |
| October | 11 | Salta | 2 | 0 | c | |
| 2016 | February | 12 | Neuquén | 2 | 0 | c |
| September | 13 | San Luis | 10 | 10 (100) | GII.2[P2]; GII.4 Sydney[P16] | |
| October | 14 | La Pampa | 15 | 0 | c | |
| November | 15 | San Luis | 7 | 0 | c | |
| November | 16 | Chubut | 7 | 5 (71.4) | GII.6[P7] | |
| December | 17 | Santa Fe | 8 | 0 | c | |
| December | 18 | San Luis | 3 | 3 (100) | GII.7[P7] | |
| 2017 | September | 19 | CABA | 2 | 0 | c |
| November | 20 | San Luis | 5 | 4 (80) | GII.4 Sydney[P31] | |
| November | 21 | Santa Cruz | 4 | 3 (75) | GI.6[P11] | |
| November | 22 | Tucumán | 3 | 0 | c | |
| December | 23 | Santa Fe | 3 | 3 (100) | GII.2[P16] | |
| 2018 | February | 24 | San Luis | 3 | 0 | c |
| September | 25 | San Luis | 5 | 4 (80) | GII.17[P17] | |
| September | 26 | Entre Ríos | 5 | 0 | b | |
| September | 27 | Santa Fe | 8 | 1 (12.5) | b | |
| November | 28 | CABA | 7 | 7 (100) | GII.4 Sydney[P16] | |
| December | 29 | Entre Ríos | 6 | 6 (100) | GII.12[P16] |
Abbreviation: AGE, acute gastroenteritis.
Norovirus outbreaks are in bold face.
#26 two of the five samples n, #27 four of the eight samples tested positive for rotavirus.
all samples tested negative for rotavirus, astrovirus, sapovirus, and adenovirus.
FIGURE 1.
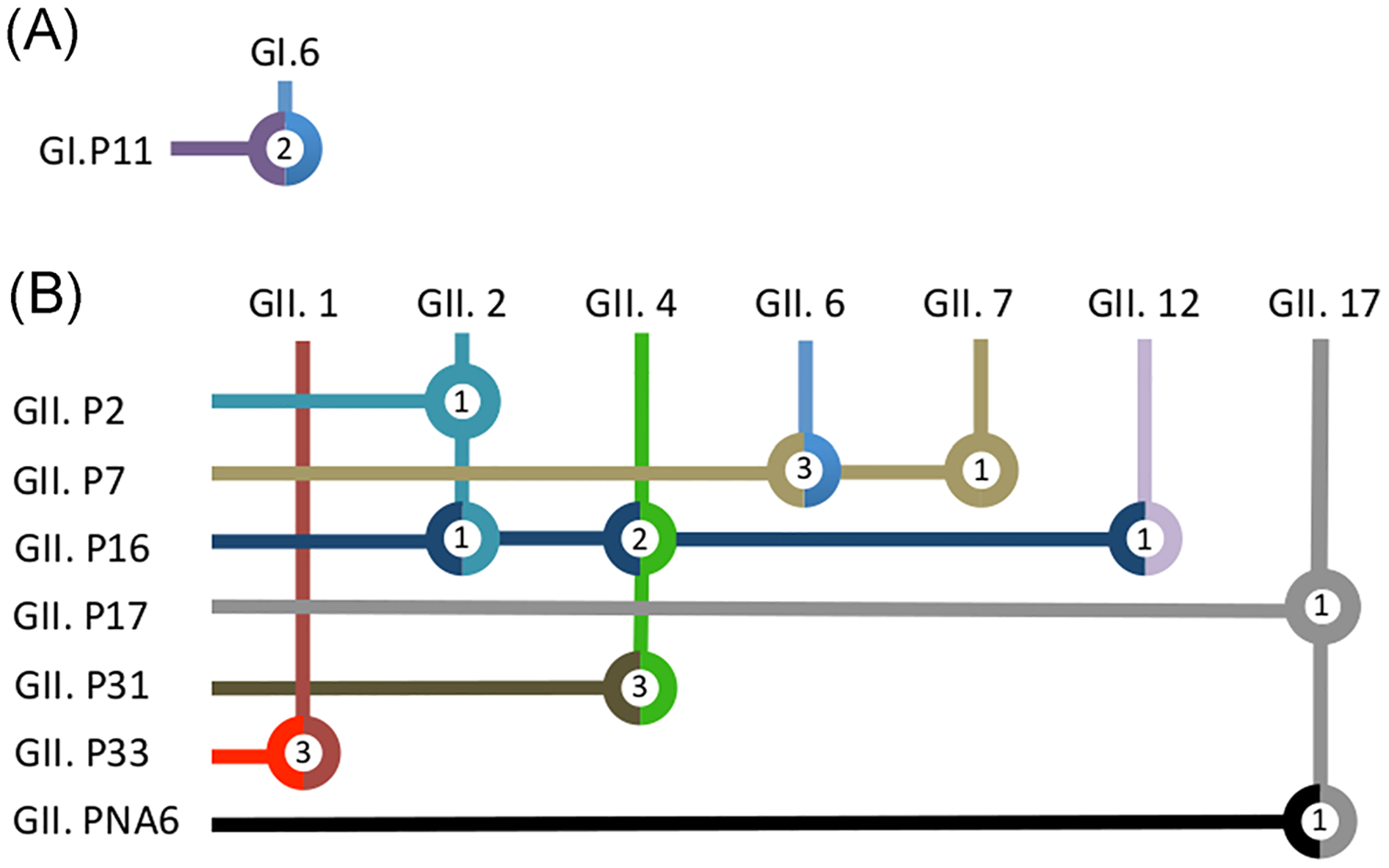
Summarized partial RNA-dependent RNA polymerase (RdRp) and partial VP1 associations in norovirus positive stools from outbreaks detected in Argentina, 2013–2018. Association of the combination of partial RdRp types (in rows) and capsid (VP1) types (in columns) detected in norovirus outbreaks from Argentina. Each genotype, GI viruses (A) and GII viruses (B) is represented with a specific color. Thus, double-colored circles indicate recombinant strains. Inside each circle, the number of outbreaks described in this study is depicted
4 |. DISCUSSION
Our findings underscore the importance of dual typing of norovirus strains since recombinant strains with new polymerase sequences emerge frequently suggesting a possible role in improved fitness of these viruses.13 GII.1[P33] and GII.6[P7] were the most frequently detected recombinant strains although the first one only circulated from December 2014 to August 2015. GII.6[P7] viruses were first reported at the beginning of this decade and are currently one of the most prevalent non-GII.4 circulating strains worldwide.14,15
The new recombinant GII.2[P16] strain, which was associated with the majority of outbreaks in east Asia in 2016, was also detected in Argentina in 2017.16 Noteworthy, this novel polymerase was found to be nearly identical to the GII.P16 polymerase of GII.4 Sydney that predominated in the winter of 2015–2016 in the northern hemisphere.11 GII.P16 polymerases were also detected in association with GII.1, GII.2, GII.3, GII.4, GII.10, and GII.12 viruses.13 Therefore, the rapid spread of viruses with this polymerase may indicate that it provides norovirus strains with an important epidemic potential although in the US no increase in the total number of norovirus outbreaks or the percentage associated with GII.4 viruses has been observed.13 In two outbreaks, more than one norovirus genotype was identified. Outbreaks caused by multiple genotypes are often associated with sewage-contaminated water which when used for growing oysters or for irrigation of produce may lead to foodborne outbreaks.17
This study has several limitations. First, the number of outbreaks from which stool samples were available for testing was small and therefore the relative frequency of the circulating genotypes may not be reliable. Second, a large proportion of outbreaks tested negative for a virus, but since no information was available on testing for bacterial or parasitic pathogens or the appropriate time of specimen collection after the onset of symptoms, the precise contribution of norovirus as the cause of AGE outbreaks in Argentina is not known. Because most outbreaks occurred in the central and southern provinces of Argentina, the genetic strain diversity might not be representative for the entire country although neighboring countries have described similar strain distribution trends.15,18
In summary, this study represents the most recent multi-year assessment of the molecular epidemiology of norovirus strains associated with AGE outbreaks in Argentina. Norovirus is not a notifiable disease even when food handlers are involved and self-limited diarrheal episodes in immunocompetent adults are underestimated because in most cases they do not seek medical assistance. Also, when a norovirus outbreak in a semi-closed setting is suspected, Kaplan criteria are often used to indicate a viral cause over etiological confirmation.19 In 2015, rotavirus vaccination was introduced in Argentina and with an expected decrease of the number of rotavirus infections, norovirus may become the leading cause of pediatric AGE as has occurred in other countries that have introduced rotavirus vaccines.20
With several norovirus vaccines in clinical trials,21 molecular surveillance of norovirus in hospitalized children with AGE can be considered to monitor possible changes in dominant genotypes which may assist to inform the formulation of these vaccines to reduce the burden of norovirus disease.
REFERENCES
- 1.Ahmed SM, Hall AJ, Robinson AE, et al. Global prevalence of norovirus in cases of gastroenteritis: a systematic review and meta-analysis. Lancet Infect Dis. 2014;14(8):725–730. 10.1016/S1473-3099(14)70767-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Green KY. Norovirus infection in immunocompromised hosts. Clin Microbiol Infect. 2014;20(8):717–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vinjé J. Advances in laboratory methods for detection and typing of norovirus. J Clin Microbiol. 2015;53(2):373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chhabra P, de Graaf M, Parra GI, et al. Updated classification of norovirus genogroups and genotypes. J Gen Virol. 2019;100:1393–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siebenga JJ, Vennema H, Zheng D-P, et al. Norovirus illness is a global problem: emergence and spread of norovirus GII.4 variants, 2001–2007. J Infect Dis. 2009;200(5):802–812. [DOI] [PubMed] [Google Scholar]
- 6.de Graaf M, van Beek J, Vennema H, et al. Emergence of a novel GII.17 norovirus—end of the GII.4 era? Euro Surveill. 2015;20(26):21178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Ryan M, Riera-Montes M, Lopman B. Norovirus in Latin America: systematic review and meta-analysis. Pediatr Infect Dis J. 2017;36(2): 127–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gomes KA, Stupka JA, Diana A, Parra GI. Molecular characterization of calicivirus strains detected in outbreaks of gastroenteritis occurring in Argentina during 2005 and 2006. Rev Argent Microbiol. 2008; 40(4):222–228. [PubMed] [Google Scholar]
- 9.Gomes KA, Stupka JA, Gómez J, Parra GI. Molecular characterization of calicivirus strains detected in outbreaks of gastroenteritis in Argentina. J Med Virol. 2007;79(11):1703–1709. [DOI] [PubMed] [Google Scholar]
- 10.Grant L, Vinjé J, Parashar U, et al. Epidemiologic and clinical features of other enteric viruses associated with acute gastroenteritis in American Indian infants. J Pediatr. 2012;161(1):110–115. [DOI] [PubMed] [Google Scholar]
- 11.Cannon JL, Barclay L, Collins NR, et al. Genetic and epidemiologic trends of norovirus outbreaks in the United States from 2013 to 2016 demonstrated emergence of Novel GII.4 recombinant viruses. J Clin Microbiol. 2017;55(7):2208–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leshem E, Barclay L, Wikswo M, et al. Genotype GI.6 norovirus, United States, 2010–2012. Emerging Infect Dis. 2013;19(8): 1317–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barclay L, Cannon JL, Wikswo ME, et al. Emerging Novel GII.P16 noroviruses associated with multiple capsid genotypes. Viruses. 2019; 11(6):535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fajardo Á, Tort FL, Victoria M, et al. Phylogenetic analyses of Norovirus strains detected in Uruguay reveal the circulation of the novel GII.P7/GII.6 recombinant variant. Infect Genet Evol. 2014;28:328–332. [DOI] [PubMed] [Google Scholar]
- 15.Cantelli CP, da Silva MFM, Fumian TM, et al. High genetic diversity of noroviruses in children from a community-based study in Rio de Janeiro, Brazil, 2014–2018. Arch Virol. 2019;164(5): 1427–1432. [DOI] [PubMed] [Google Scholar]
- 16.Niendorf S, Jacobsen S, Faber M, et al. Steep rise in norovirus cases and emergence of a new recombinant strain GII.P16-GII.2, Germany, winter 2016. Euro Surveill. 2017;22(4):30447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Zhang J, Shen Z. The impact of calicivirus mixed infection in an oyster-associated outbreak during a food festival. J Clin Virol. 2015; 73:55–63. [DOI] [PubMed] [Google Scholar]
- 18.Galeano ME, Martinez M, Amarilla AA, et al. Molecular epidemiology of norovirus strains in Paraguayan children during 2004–2005: description of a possible new GII.4 cluster. J Clin Virol. 2013;58(2):378–384. [DOI] [PubMed] [Google Scholar]
- 19.Kaplan JE. Epidemiology of Norwalk gastroenteritis and the role of Norwalk virus in outbreaks of acute nonbacterial gastroenteritis. Ann Intern Med. 1982;96(6 Pt 1):756–761. [DOI] [PubMed] [Google Scholar]
- 20.Payne DC, Vinjé J, Szilagyi PG, et al. Norovirus and medically attended gastroenteritis in US children. N Engl J Med. 2013;368(12): 1121–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mattison CP, Cardemil CV, Hall AJ. Progress on norovirus vaccine research: public health considerations and future directions. Expert Rev Vaccines. 2018;17(9):773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]


