Abstract
Susceptibility assays by cell culture methods are time-consuming and are particularly difficult to perform with varicella-zoster virus (VZV). To overcome this limitation, we have adapted a functional test of the viral thymidine kinase (TK) in TK-deficient (tdk mutant) bacteria to detect ACV-resistant VZV in clinical samples. After PCR amplification, the complete viral TK open reading frame (ORF) is purified from PCR primers, digested with two restriction enzymes, and ligated in an oriented fashion into a bacterial expression vector. The ligation products are then used to transform tdk mutant bacteria. After transformation, an aliquot of the bacteria is plated onto a plate with minimal medium containing (i) ampicillin to select for plasmids carrying the viral TK ORF and (ii) isopropyl β-d-thiogalactopyranoside (IPTG) to induce its expression. An identical aliquot of bacteria is also plated onto a medium containing, in addition to the components described above, 5-fluorodeoxyuridine (FUdR). Compared to the number of transformants on FUdR-free medium, the number of colonies carrying TK derived from susceptible strains was reduced by 86%, on average, in the presence of FUdR. In contrast, the number of transformants carrying TK from resistant strains with a mutant TK were reduced by only 4%, on average, on FUdR-containing plates. We have assessed the validity of this assay with cell culture isolates and several clinical samples including two cerebrospinal fluid samples from which no virus could be isolated. This colony reduction assay allowed the correct identification of the TK phenotype of each VZV isolate tested and can be completed within 3 days of receipt of the sample.
Herpes simplex virus (HSV) type 1 and 2 and varicella-zoster virus (VZV) belong to the alpha subfamily of the family Herpesviridae. After primary infection, they typically establish a latent infection in spinal or cranial ganglia and from there reactivate to cause mucocutaneous lesions, i.e., recurrent orofacial or genital herpes and zoster, respectively (17, 45). Among immunocompromised hosts, recurrent lesions associated with VZV can progress to ulcerative lesions, to multidermatomal zoster, and to disseminated mucocutaneous or visceral infections (2, 43). Among human immunodeficiency virus (HIV)-infected patients, these clinical presentations are most often observed in patients with advanced disease and profound immune dysfunction. The chronic or recurrent character of HSV- or VZV-associated diseases has led to treatment with protracted or repeated courses of antiviral agents, primarily acyclovir (ACV). Infections which no longer respond to ACV have been observed under these conditions, with the selection of ACV-resistant (ACVr) strains (for a review, see reference 32). ACV resistance mechanisms have been characterized mainly from HSV-infected patients and are very similar for VZV-infected patients. Three kinds of mechanisms can be associated with the generation of ACVr strains (for reviews, see references 6, 8, and 18). Two mechanisms involve the thymidine kinase (TK) gene, the most frequent being the selection of tk mutants which cannot phosphorylate the drug (3, 19, 42). This category of mutants often carries frameshift or nonsense mutations within the TK open reading frame (ORF) resulting in the expression of truncated forms of TK (3, 5, 15, 35, 42). The other, less frequent mechanism involving TK is the selection of viruses with altered TK which are unable to phosphorylate the drug yet which are able to phosphorylate thymidine. Missense mutations are most frequently found in this category of mutants (15, 42). The third mechanism involves the viral DNA polymerase, with the selection of pol mutant viruses which can be propagated even in the presence of high doses of the drug (9, 29, 31). Mutations in tk are found at least 20 times more often than mutations in pol in viruses from patients with ACVr HSV (7, 27). Resistant VZV strains which presumably contain DNA polymerase mutations are also rarely found in patients with suspected clinical resistance (3, 4, 13, 14, 20, 21, 23, 24, 26, 28, 30, 33, 36, 37, 42, 46). Thus, like in HSV infections, most mutations associated with the clinical resistance of VZV to ACV are within tk. Therefore, a phenotypic test that targets the TK protein of VZV could identify the vast majority of resistant strains.
Isolation of ACVr VZV strains and their characterization are difficult by tissue culture procedures, mainly because VZV is very labile, is highly cell associated, and replicates slowly in cell culture (16). Thus, VZV plaque reduction assay is particularly time-consuming and may take several weeks to complete. In addition, VZV can rarely be isolated from the cerebrospinal fluid (CSF) samples of patients with VZV encephalitis or myelitis (16). This is of particular concern in patients presenting with neurological symptoms and a suspected drug-resistant VZV infection in the absence of skin lesions from which virus could be isolated. To overcome these difficulties and to provide clinicians with a rapid means of diagnosis of ACV-resistant VZV, we have used a phenotypic assay with Escherichia coli that was previously established to select VZV TK mutations generated in vitro (40). This assay depends on the PCR amplification of the entire TK ORF directly from clinical samples, followed by its cloning and expression in TK-deficient (tdk mutant) bacteria. tdk mutants are resistant to the nucleoside analog 5-fluorodeoxyuridine (FUdR) (39). Expression of a functional viral TK restores the sensitivity of these bacteria to the nucleoside analog (40) and results in a significantly reduced colony number on plates containing FUdR compared to that obtained with a nonfunctional viral TK. This colony reduction assay allows identification of ACVr VZV within 3 days of receipt of the clinical sample.
MATERIALS AND METHODS
Bacterial strains.
The TK-deficient strain E. coli B SY211 was obtained from W. Summers (Yale University School of Medicine) (39). It is resistant to 30 to 50 μM FUdR. SY211-pRep4 was generated by stable transformation of SY211 with plasmid pRep4 (Qiagen). It overproduces the Lac repressor, whose gene is carried by pRep4, and is resistant to kanamycin.
Viruses and clinical samples.
ACVr VZV strain LYON-6625, which was isolated from an HIV-infected patient (26), was provided by Françoise Thouvenot (Lyon, France).
Three strains (strains WT1, WT2, and WT3) originated from immunocompetent patients with varicella or zoster. These strains were considered to be wild-type strains owing to the clinical presentation of the patient.
The VZV strains labeled A to K were provided under code to R. Sahli by R. Snoeck. They were subjected to the colony reduction assay without knowledge of their origin or of their susceptibility status in cells in culture.
The two CSF samples (samples 106 and 4.2) were derived from two patients presenting with either zoster (sample 106) or myelitis (sample 4.2), which in this case progressed under ACV treatment (25).
Isolation and propagation of VZV were performed with human embryonic fibroblasts maintained in minimal essential medium (Seromed) supplemented with 10% fetal calf serum (Life Technologies). Determination of the susceptibility of VZV to nucleoside analogs in cells in culture was performed in 96-well dishes by using cytopathic effect as the endpoint as described previously (37). The 50% inhibitory concentration (IC50) was defined as the compound concentration required to reduce the viral cytopathic effect by 50%.
PCR amplification of the VZV TK ORF.
Template DNAs were from cell cultures infected with VZV strains (strains A, B, C, D, E, F, G, H, I, J, K, WT1, WT2, and LYON-6625), from a vesicular fluid sample (strain WT3), or from CSF samples (samples 106 and 4.2). DNAs were purified with the Qiagen blood kit according to the manufacturer's instructions. DNAs were eluted in 200 μl of water.
The sense-strand primer (TKSe) includes an EcoRI restriction site (given below in italics) immediately upstream of the TK initiator codon (given below as the underlined sequence) near position 64806 (indicated below as C*). The antisense primer (TKAh) is downstream of the TK termination codon, near position 65860 (indicated below as C*), and includes a HindIII restriction site (given below in italics). The sequence of primer TKSe is 5′-CTGAATTC*ATGTCAACGGATAAAAC-3′, and the sequence of primer TKAh is 5′-GCGAAGCTTC*TGGTACATACGTAAATAC-3′.
The numbering of the sequences is that of Davison and Scott (10) (Genbank accession nos. X04370, M14891, and M16612).
Amplification of the complete TK ORF was done with a high-fidelity amplification system (Advantage Klentaq; Clontech) according to the manufacturer's instructions in a 25- to 50-μl reaction mixture with 2.5 or 5 μl of template DNA. As a positive control, VZV DNA was extracted from VZV purified by ultracentrifugation. To estimate its concentration, VZV DNA was digested with HindIII, and the resulting fragments were separated by agarose gel electrophoresis and stained with ethidium bromide, as was a known amount of standard DNA (lambda phage DNA digested with HindIII). Comparison of the fluorescence intensity of a single-copy VZV restriction fragment to that of a fragment of lambda phage DNA digestion with HindIII was used to estimate the amount of VZV DNA.
Cycling was as follows: after an initial denaturation step at 95°C for 5 min, the reactions were subjected to 30 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 90 s, and polymerization at 68 or 72°C for 150 s. After the last polymerization step, the reaction mixtures were further incubated for 6 min at the polymerization temperature and an aliquot was analyzed by electrophoresis through a 1.5% agarose gel. Under these conditions, PCR with vesicular fluid or cell culture samples produced a single DNA fragment corresponding to the 1.1-kb TK ORF. Positive samples were purified with the Qiagen PCR purification kit according to the manufacturer's instructions, and the DNA was eluted in 50 μl of TE (10 mM Tris-HCl, 0.1 mM EDTA [pH 8.0]). CSF samples containing less VZV DNA required up to 40 cycles for the generation of enough material for the colony reduction assay (see below). A low-molecular-weight DNA coamplified with TK in sample 106. This contaminant was not a TK deletion mutant, as determined by DNA sequencing. The specific TK DNA fragment was gel purified with the Qiagen gel purification kit according to the manufacturer's instructions and was eluted in 50 μl of TE. Half of the eluted DNA was subsequently digested with EcoRI and HindIII in a 60-μl reaction volume for 1 h, purified with the Qiagen PCR purification kit, and eluted as indicated above. An aliquot of each TK DNA fragment was analyzed by agarose gel electrophoresis to estimate its quality and its concentration.
Sequencing of TK DNA.
Sequencing of the TK ORFs of VZV strains G, 4.2, and 106 was performed on an ABI310 DNA sequence analyzer with the Big Dye Terminator kit (PE Applied Biosystems), as recommended by the manufacturer. Sequences were determined directly from PCR-amplified DNA instead of cloned DNA to make sure that they were representative of the TK DNA population within the sample and to avoid any artifact due to mutations introduced by PCR. The sequences of the PCR-amplified DNAs were determined before and after gel purification for sample 106 to assess the specificity of the contaminating low-molecular-weight DNA fragment (see above). Reading and editing of the chromatograms were done with Chromas 1.55 software (Technelysium Pty. Ltd., Helensvale, Queensland, Australia), and sequence comparisons were performed with the Vector NTI Suite 1 package (Informax, Inc., Bethesda, Md.).
Expression-cloning of TK in SY211-pRep4 and colony reduction assay.
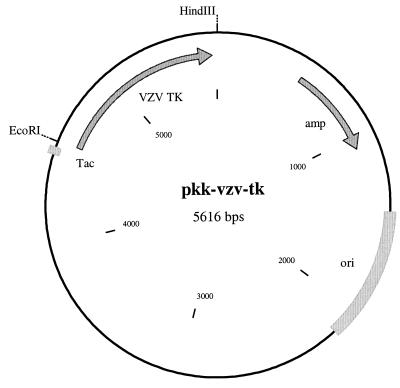
A total of 50 ng of the purified TK product was ligated to 50 ng of gel-purified EcoRI-HindIII-digested pKK223-3 vector (Pharmacia) in a 20-μl reaction mixture containing 2 μl of 10× ligation buffer (Promega) and 0.2 μl (0.2 Weiss units) of T4 DNA ligase (Promega) for at least an hour at room temperature. pKK223-3 allows expression of the inserted gene under the control of the Tac promoter (Fig. 1) in the presence of isopropyl β-d-thiogalactopyranoside (IPTG). Five to 10 μl of the ligation reaction mixture was used to transform 200 μl of CaCl2-competent SY211-pRep4 prepared by standard procedures and kept frozen at −70°C (34). Equal aliquots of the transformation mixture that gave rise to at least 100 colonies (typically, one-fourth of the transformation mixture) were plated onto two minimal medium plates (1.5% agar, M9 salts, 1% glucose, 0.5% Casamino Acids, 1% MgSO4, 40 μg each of cytidine and guanosine per ml, 100 μg each of adenine and uridine per ml, 50 μg of ampicillin per ml, 25 μg of kanamycin per ml, and 25 μM IPTG) containing either 0 or 12.5 μg of FUdR (50 μM) per ml (39). The number of colonies was recorded after an overnight incubation at 37°C and was corrected by subtracting the number of colonies obtained after transformation with a control ligation containing only the vector. Statistical analysis of the results and comparison of the means by the unpaired t test were done with Prism 2 software (GraphPad Software, Inc., San Diego, Calif.).
FIG. 1.
pkk-vzv-tk expression vector. The VZV TK ORF (VZV TK) is between the EcoRI and HindIII restriction sites downstream of the Tac promoter (Tac). The ampicillin (amp) resistance gene and the origin of replication (ori) are also indicated.
RESULTS
PCR amplification with primers TKSe and TKAh allowed detection of VZV TK from samples containing as little as 10 copies of VZV DNA (data not shown), indicating that its sensitivity is sufficient enough to warrant analysis of VZV TK from clinical samples such as vesicular fluid after 25 to 30 PCR cycles or even CSF after 35 to 40 PCR cycles. PCR amplification introduced an EcoRI site upstream of the TK initiator codon and a HindIII site downstream of the TK termination codon. Cleavage of the TK fragment by these two restriction enzymes allowed its directional cloning into the bacterial expression vector pKK223-3 (Fig. 1).
To initially test whether VZV TK affected the ability of transformed SY211-pRep4 cells to grow on plates containing FUdR, four control samples were analyzed: three corresponded to wild-type VZV (WT1 to WT3) and one corresponded to a resistant VZV strain (LYON-6625). Following PCR and insertion into pKK223-3, the TK fragments were subjected to the colony reduction assay as described in Materials and Methods. The results presented in Table 1 show that expression of the wild-type TK gave rise to a relative number of colonies of 12% ± 5% (mean ± standard deviation). TK from the resistant strain LYON-6625, analyzed in duplicate, gave rise to a relative number of colonies of 91% ± 6%. The difference between LYON-6625 and the three wild-type strains (P < 0.05) is significant, indicating that the colony reduction assay could discriminate between susceptible and resistant strains of VZV with mutations within tk.
TABLE 1.
Colony reduction assay of control VZV strains
| Strain | Origina | No. of colonies on plates:
|
%b | |
|---|---|---|---|---|
| Without FUdR | With FUdR | |||
| WT1 | Infected cell culture | 3,200 | 360 | 8 |
| WT2 | Infected cell culture | 2,000 | 450 | 17 |
| WT3 | Clinical sample | 1,800 | 330 | 12 |
| LYON-6625 | Infected cell culture | 1,000 | 950 | 95 |
| LYON-6625 | Infected cell culture | 3,000 | 2,600 | 86 |
Template DNA was purified from the indicated source.
Relative number of colonies on FUdR-containing plates expressed as the percentage of the number of colonies on plates not containing FUdR.
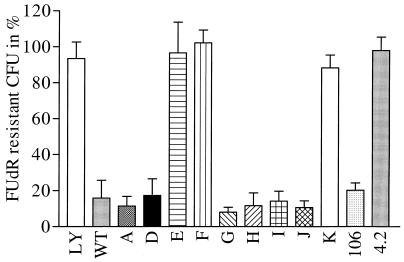
To further evaluate the validity of the colony reduction assay, 11 strains (strains A to K) were tested without prior knowledge of their susceptibility to ACV. We also tested VZV TK from two CSF samples (samples 106 and 4.2) from which VZV could be identified only by PCR with our diagnostic primers (amplification of an internal TK fragment; unpublished data). Amplification of TK from strains B and C resulted only in a short product, suggesting that a large internal deletion occurred in the TKs of both strains. No inhibition of growth of the bacteria was evident with these samples when they were subjected to the colony reduction assay (data not shown). PCR amplification of samples A, samples D to K, and the wild-type and mutant controls resulted in the production of a single TK DNA fragment of the expected size (1.1 kb). These, together with samples 106 and 4.2, were subjected to the colony reduction assay (Fig. 2). Two categories of VZV TK could be distinguished: those (samples A, D, G, H, I, J, and 106) that behaved like the wild-type control in conferring FUdR susceptibility to transformed SY211-pRep4 cells, with median relative colony numbers of between 7% (sample G) and 20% (sample 106) on FUdR-containing plates, and those (samples E, F, K, and 4.2) that behaved like the resistant control LYON-6625, which did not confer FUdR susceptibility to the transformed bacteria, with median relative colony numbers between 91% (sample K) and 99% (sample F). Comparison of the mean relative colony numbers of both categories of samples indicated that they differed significantly (P < 0.05).
FIG. 2.
Relative number of CFU on FUdR-containing plates expressed as the percentage of that on nucleoside analog-free plates. After PCR amplification, the TK ORF was digested with EcoRI and HindIII and was then inserted into pKK223-3 and subjected to the colony reduction assay as described in Materials and Methods. The number of colonies growing in the absence and in the presence of FUdR was recorded from at least three independent experiments. The relative number of CFU on FUdR-containing plates is expressed as the percentage of that on FUdR-free plates. The error bars correspond to the standard deviations. Sample names are indicated below each histogram. LY, LY-6625-resistant control; WT, a wild-type VZV TK control.
Once the results of the colony reduction assay had been obtained, they were compared with those of the susceptibility assay (samples A to K) or to clinical data (samples 106 and 4.2) (Table 2). Strains reported to be ACV resistant by the susceptibility assay (IC50, >3 μg/ml) were also determined to be resistant either by the colony reduction assay (samples E, F, and K) or as indicated by the gross alteration of the size of the PCR-amplified TK DNA (samples B and C). Samples E and F were derived from the OKA reference strain by in vitro selection with ACV and bromovinyldeoxyuridine (BVDU), respectively. Sample K corresponded to a resistant strain VZV O7/1, which carries a missense mutation that results in an aspartic acid-to-asparagine change at position 18 of TK (22). Samples B and C corresponded to two consecutive VZV isolates from an AIDS patient who developed VZV-associated meningoradiculoneuritis, despite ACV treatment (37). The TK genes of viruses in both samples had previously been shown to have large internal deletions, consistent with our PCR data. In contrast, the virus in sample G, which was reported to be susceptible by the colony reduction assay, was resistant to ACV (IC50, 4.7 μg/ml) and to foscarnet (foscavir) (PFAr). It was, however, susceptible to BVDU (IC50, 0.0032 μg/ml), suggesting that it carried a wild-type TK. We confirmed by DNA sequencing that the TK ORF of strain G was the same as that of the wild type, consistent with the result of the colony reduction assay. All other strains for which ACV IC50s were below 3 μg/ml were correctly reported as being susceptible by the colony reduction assay. Those strains included strain A, from the AIDS patient mentioned above for samples B and C, early in her VZV-associated disease; strain D, the OKA reference strain; and strains H, I, and J, which were from immunocompetent patients with varicella. In addition, the identification of a susceptible VZV strain in sample 106, which was derived from an immunocompetent patient with zoster, and of a resistant VZV strain in sample 4.2, which was derived from an HIV-infected patient who presented with myelitis which appeared despite acyclovir treatment, was consistent with their clinical presentations and with DNA sequencing of the TK ORFs of their viruses. The wild-type sequence was found in sample 106, and a mixed population of mutants was found in sample 4.2. The major mutation found in sample 4.2 was a frameshift deletion of the dinucleotide AT at positions 375 and 376 (position 1 refers to the A of the initiator codon), resulting in a predicted TK protein of 129 amino acids devoid of the substrate binding site.
TABLE 2.
Correlation of clinical resistance, colony reduction assay with bacteria, and susceptibility assay with cells in culturea
| VZV strain (origin) | Sourceb | Clinical resistance | Phenotype (CRA) | IC50 (μg/ml)c
|
Size of the TK PCR fragmentd | TK genotypee | |
|---|---|---|---|---|---|---|---|
| ACV | BVDU | ||||||
| A (NCLU II, CSF) | CC | No | S | 0.28 ± 0.28 (S) | 0.0018 ± 0.0013 (S) | WT | WT |
| B (NCLU III, CSF) | CC | Yes | R | 8.8 ± 2.0 (R) | >50 (R) | Del | ND |
| C (NCLU IV, CSF) | CC | Yes | R | 12.5 ± 7.2 (R) | >50 (R) | Del | ND |
| D (OKA) | CC | NA | S | 0.22 ± 0.14 (S) | 0.0008 ± 0.0003 (S) | WT | ND |
| E (OKA, ACVr in vitro) | CC | NA | R | 20.7 ± 4.7 (R) | >20* (R) | WT | ND |
| F (OKA, BVDUr in vitro) | CC | NA | R | 27.4 ± 6.5 (R) | >20* (R) | WT | ND |
| G (OKA, PFAr in vitro) | CC | NA | S | 4.7 ± 3.7 (R) | 0.0032 ± 0.0014 (S) | WT | WT |
| H (CI-22) | CC | No | S | 0.33 ± 0.27 (S) | 0.0009 ± 0.0001 (S) | WT | ND |
| I (CI-13) | CC | No | S | 0.24 ± 0.35 (S) | 0.0014 ± 0.0012 (S) | WT | ND |
| J (CI-16) | CC | No | S | 0.32 ± 0.36 (S) | 0.0008 ± 0.0006 (S) | WT | ND |
| K (O7-1) | CC | NA | R | 13.5 ± 4.4 (R) | 24.8 ± 19.4 (R) | WT | ND |
| 106 | CSF | No | S | ND | ND | WT | WT |
| 4.2 | CSF | Yes | R | ND | ND | WT | Mut |
Abbreviations and symbols: CRA, colony reduction assay; CC, cell culture; S, susceptible; R, resistant; NA, not applicable; ND, not determined; WT, wild type; Del, deletion; Mut, mutant; ∗, only one determination.
DNA was purified from the indicated source of material, either cell culture or CSF.
Drug concentration (mean ± standard deviation of at least two independent experiments) required to reduce the viral cytopathic effect by 50% in the susceptibility assay.
The size of the PCR product either was that of the wild type or was shorter owing to an internal deletion, as determined by gel electrophoresis.
TK was either that of the wild type or that of the mutant as determined from the sequence of its corresponding PCR product.
DISCUSSION
The aim of this work was to evaluate a rapid method for the detection of ACVr VZV in clinical samples. This method involves PCR amplification of the VZV TK ORF followed by the phenotypic analysis of its encoded protein in bacteria which are otherwise deficient in endogenous TK activity (tdk mutant). Expression of the wild-type VZV TK ORF in tdk mutant bacteria resulted, on average, in an 86% reduction in the bacterial colony number in the presence of FUdR. In contrast, expression of the TK ORF derived from resistant VZV strains only slightly affected colony formation of the transformed bacteria, with a 4% reduction in colony number, on average. This marked difference between susceptible and resistant VZV strains has allowed the correct prediction of the phenotypes of all strains tested except strain G. This resistant strain was obtained after selection of the reference OKA strain with foscavir in vitro. Such selection is expected to result in a strain with a mutation in the polymerase gene (8). This hypothesis is supported by the susceptibility of strain G to BVDU, which must be activated by TK to exert its antiviral effect, and by the wild-type genotype of its encoded TK, as determined by DNA sequencing. The incorrect attribution of a phenotype of ACV susceptibility to strain G by the colony reduction assay must be considered a false-negative result, which underlines the fact that other methods that target pol (plaque reduction assay, DNA sequencing) must be performed to identify all drug-resistant VZV isolates in clinical samples. This false negativity would be of particular concern if the frequency of mutations that affect tk was similar to or lower than the frequency of mutations that affect pol. However, this is not the case. Indeed, one can expect that most strains will be correctly identified by the colony reduction assay, for only a few strains with mutations in pol (4, 14, 28) have been reported, and these represent no more than 5% of all ACVr VZV strains (3, 4, 13, 14, 20, 21, 23, 24, 26, 28, 30, 33, 36, 37, 42, 46). In addition, VZV TK mutations selected in tdk mutant bacteria can confer either deficient or altered TK phenotypes (40). Therefore, the colony reduction assay is expected to allow detection of the two kinds of TK mutations known to be associated with ACV resistance of VZV in vivo. While the colony reduction assay does not distinguish between mutations that confer phenotypes of TK deficiency or TK alteration, one could analyze VZV TK biochemically in vitro using bacterial clone extracts as described by Suzutani et al. (40) to get this information. In this case, it will be necessary to ascertain by DNA sequencing that the TK sequence of the clone being analyzed corresponds to the sequence of the parental TK PCR fragment.
We did not encounter difficulties in amplifying the VZV TK ORF from vesicular fluid samples expected to contain high-copy-number VZV DNA. In addition, we could also amplify TK from CSF, as illustrated by samples 106 and 4.2, which contained small amounts of VZV DNA. No virus could be isolated by cell culture from the patients who submitted these CSF samples; cell culture is known to be particularly insensitive for detection of VZV from CSF samples (16). Thus, for sample 4.2, the colony reduction assay was the only way by which detection of resistant VZV could be achieved. The ability to analyze the VZV TK ORF from CSF will thus be useful for the detection of ACVr VZV in patients who develop VZV-associated central nervous system disease that is refractory to ACV treatment but that shows no signs of cutaneous involvement, as was the case for the patient from whom sample 4.2 was obtained (25).
Rapid assays for the detection of resistant VZV or HSV have been reported (1, 11, 13, 38, 41, 44), and all these assays depend on cell culture methods to obtain the initial virus sample for analysis. This may be limiting and time-consuming, in particular, for VZV. As an alternative, direct sequence analysis of PCR-amplified tk or pol DNA could be very rapid (22). Sequencing, however, suffers from the limitation that prior knowledge of the effects of mutations, especially of the missense type, on the activity of the protein is necessary to identify resistant viruses unambiguously. While hot-spot mutations that target the ATP and nucleoside binding sites have been identified in the tk genes from HSV as well as from VZV, mutations at other positions throughout tk are also associated with resistance (3, 15, 22, 40, 42). It may therefore not be possible to associate as yet undescribed mutations with either wild-type or mutant TK phenotypes. The colony reduction assay will thus be useful as a complement to DNA sequencing in uncovering mutations in tk associated with clinical resistance in VZV-infected patients, while DNA sequencing by itself will be necessary to validate the findings of VZV resistance by the colony reduction assay.
The colony reduction assay can also provide an assessment of the homogeneity of the virus population in a sample. Thus, a reduction in colony number of more than 20% in the presence of FUdR may be a hint that there is a mixed population of resistant and susceptible VZV in the sample being tested. The background rate of occurrence of 10 to 20% of mutant TK in the colony reduction assay performed with samples considered to contain wild-type VZV may be contributed by mutations introduced during the PCR process (22), despite the use of a high-fidelity amplification system, or may represent a background of strains with mutant TK present in the clinical or infected cell culture sample itself. The latter possibility is suggested by the heterogeneity of the TK phenotypes of HSV strains within clinical samples (12, 27, 31).
The colony reduction assay gives results within 3 days of receipt of the clinical sample, which is by far faster than the VZV plaque reduction assay or susceptibility assays, whose completion may take several weeks. This will help clinicians to rapidly adjust therapy in the presence of an ACVr VZV strain and will necessitate the use of antiviral agents like cidofovir or foscarnet, whose use is fraught with serious adverse effects and will require careful monitoring of patients (18). We are extending the colony reduction assay to HSV types 1 and 2. Our first analyses indicate that it can also be adapted to these viruses.
ACKNOWLEDGMENTS
This work has been supported by grant 24 of the Association pour la Collaboration Vaud/Genève and by the Fonds for Geneeskundig Wetenschappelijk Onderzoek (Krediet 3-0180-95).
We thank Françoise Thouvenot and William Summers for gifts of material and the staff of the viral diagnostic laboratory of the Institute of Microbiology for technical help.
REFERENCES
- 1.Agut H, Aubin J T, Ingrand D, Blanc S, Clayton A L, Chantler S M, Huraux J M. Simplified test for detecting the resistance of herpes simplex virus to acyclovir. J Med Virol. 1990;31:209–214. doi: 10.1002/jmv.1890310307. [DOI] [PubMed] [Google Scholar]
- 2.Arvin A M. Varicella-zoster virus. Clin Microbiol Rev. 1996;9:361–381. doi: 10.1128/cmr.9.3.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boivin G, Edelman C K, Pedneault L, Talarico C L, Biron K K, Balfour H H., Jr Phenotypic and genotypic characterization of acyclovir-resistant varicella-zoster viruses isolated from persons with AIDS. J Infect Dis. 1994;170:68–75. doi: 10.1093/infdis/170.1.68. [DOI] [PubMed] [Google Scholar]
- 4.Breton G, Fillet A M, Katlama C, Bricaire F, Caumes E. Acyclovir-resistant herpes zoster in human immunodeficiency virus-infected patients: results of foscarnet therapy. Clin Infect Dis. 1998;27:1525–1527. doi: 10.1086/515045. [DOI] [PubMed] [Google Scholar]
- 5.Chatis P A, Crumpacker C S. Analysis of the thymidine kinase gene from clinically isolated acyclovir-resistant herpes simplex viruses. Virology. 1991;180:793–797. doi: 10.1016/0042-6822(91)90093-q. [DOI] [PubMed] [Google Scholar]
- 6.Chatis P A, Crumpacker C S. Resistance of herpesviruses to antiviral drugs. Antimicrob Agents Chemother. 1992;36:1589–1595. doi: 10.1128/aac.36.8.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christophers J, Clayton J, Craske J, Ward R, Collins P, Trowbridge M, Darby G. Survey of resistance of herpes simplex virus to acyclovir in northwest England. Antimicrob Agents Chemother. 1998;42:868–872. doi: 10.1128/aac.42.4.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coen D M. Nucleosides and foscarnet—mechanism. In: Richman D D, editor. Antiviral drug resistance. Chichester, England: John Wiley & Sons Ltd.; 1996. pp. 81–102. [Google Scholar]
- 9.Collins P, Larder B A, Oliver N M, Kemp S, Smith I W, Darby G. Characterization of a DNA polymerase mutant of herpes simplex virus from a severely immunocompromised patient receiving acyclovir. J Gen Virol. 1989;70(Pt. 2):375–382. doi: 10.1099/0022-1317-70-2-375. [DOI] [PubMed] [Google Scholar]
- 10.Davison A J, Scott J E. The complete DNA sequence of varicella-zoster virus. J Gen Virol. 1986;67(Pt. 9):1759–1816. doi: 10.1099/0022-1317-67-9-1759. [DOI] [PubMed] [Google Scholar]
- 11.de la Iglesia P, Melon S, Lopez B, Rodriguez M, Blanco M I, Mellado P, de Ona M. Rapid screening tests for determining in vitro susceptibility of herpes simplex virus clinical isolates. J Clin Microbiol. 1998;36:2389–2391. doi: 10.1128/jcm.36.8.2389-2391.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellis M N, Keller P M, Fyfe J A, Martin J L, Rooney J F, Straus S E, Lehrman S N, Barry D W. Clinical isolate of herpes simplex virus type 2 that induces a thymidine kinase with altered substrate specificity. Antimicrob Agents Chemother. 1987;31:1117–1125. doi: 10.1128/aac.31.7.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fillet A M, Dumont B, Caumes E, Visse B, Agut H, Bricaire F, Huraux J M. Acyclovir-resistant varicella-zoster virus—phenotypic and genetic characterization. J Med Virol. 1998;55:250–254. [PubMed] [Google Scholar]
- 14.Fillet A M, Visse B, Caumes E, Dumont B, Gentilini M, Huraux J M. Foscarnet-resistant multidermatomal zoster in a patient with AIDS. Clin Infect Dis. 1995;21:1348–1349. doi: 10.1093/clinids/21.5.1348. [DOI] [PubMed] [Google Scholar]
- 15.Gaudreau A, Hill E, Balfour H H, Erice A, Boivin G. Phenotypic and genotypic characterization of acyclovir-resistant herpes simplex viruses from immunocompromised patients. J Infect Dis. 1998;178:297–303. doi: 10.1086/515626. [DOI] [PubMed] [Google Scholar]
- 16.Gershon A A, Forghani B. Varicella-zoster virus. In: Lennette E H, Lennette D A, Lennette E T, editors. Diagnostic procedures for viral, rickettsial, and chlamydial infections. Washington, D.C.: American Public Health Association; 1995. pp. 601–613. [Google Scholar]
- 17.Gershon A A, Silverstein S J. Varicella-zoster virus. In: Richman D D, Whitley R J, Hayden F G, editors. Clinical virology. New York, N.Y: Churchill Livingstone Inc.; 1997. pp. 421–444. [Google Scholar]
- 18.Hammer S M, Inouye R T. Antiviral agents. In: Richman D D, Whitley R J, Hayden F G, editors. Clinical virology. New York, N.Y: Churchill Livingstone Inc.; 1997. pp. 185–250. [Google Scholar]
- 19.Hill E L, Hunter G A, Ellis M N. In vitro and in vivo characterization of herpes simplex virus clinical isolates recovered from patients infected with human immunodeficiency virus. Antimicrob Agents Chemother. 1991;35:2322–2328. doi: 10.1128/aac.35.11.2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoppenjans W B, Bibler M R, Orme R L, Solinger A M. Prolonged cutaneous herpes zoster in acquired immunodeficiency syndrome. Arch Dermatol. 1990;126:1048–1050. [PubMed] [Google Scholar]
- 21.Jacobson M A, Berger T G, Fikrig S, Becherer P, Moohr J W, Stanat S C, Biron K K. Acyclovir-resistant varicella zoster virus infection after chronic oral acyclovir therapy in patients with the acquired immunodeficiency syndrome (AIDS) Ann Intern Med. 1990;112:187–191. doi: 10.7326/0003-4819-112-3-187. [DOI] [PubMed] [Google Scholar]
- 22.Lacey S F, Suzutani T, Powell K L, Purifoy D J, Honess R W. Analysis of mutations in the thymidine kinase genes of drug-resistant varicella-zoster virus populations using the polymerase chain reaction. J Gen Virol. 1991;72:623–630. doi: 10.1099/0022-1317-72-3-623. [DOI] [PubMed] [Google Scholar]
- 23.Linnemann C C J, Biron K K, Hoppenjans W G, Solinger A M. Emergence of acyclovir-resistant varicella zoster virus in an AIDS patient on prolonged acyclovir therapy. AIDS. 1990;4:577–579. doi: 10.1097/00002030-199006000-00014. [DOI] [PubMed] [Google Scholar]
- 24.Lokke J, Weismann K, Mathiesen L, Klem T. Atypical varicella-zoster infection in AIDS. Acta Dermato-Venereol. 1993;73:123–125. doi: 10.2340/0001555573123125. [DOI] [PubMed] [Google Scholar]
- 25.Meylan P R, Miklossy J, Iten A, Petignat C, Meuli R, Zufferey J, Sahli R. Myelitis due to varicella-zoster virus in an immunocompromised patient without a cutaneous rash. Clin Infect Dis. 1995;20:206–208. doi: 10.1093/clinids/20.1.206. [DOI] [PubMed] [Google Scholar]
- 26.Morfin F, Thouvenot D, Turenne-Tessier M, Lina B, Aymard M, Ooka T. Phenotypic and genetic characterization of thymidine kinase from clinical strains of varicella-zoster virus resistant to acyclovir. Antimicrob Agents Chemother. 1999;43:2412–2416. doi: 10.1128/aac.43.10.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nugier F, Colin J N, Aymard M, Langlois M. Occurrence and characterization of acyclovir-resistant herpes simplex virus isolates: report on a two-year sensitivity screening survey. J Med Virol. 1992;36:1–12. doi: 10.1002/jmv.1890360102. [DOI] [PubMed] [Google Scholar]
- 28.Pahwa S, Biron K, Lim W, Swenson P, Kaplan M H, Sadick N, Pahwa R. Continuous varicella-zoster infection associated with acyclovir resistance in a child with AIDS. JAMA. 1988;260:2879–2882. [PubMed] [Google Scholar]
- 29.Parker A C, Craig J I, Collins P, Oliver N, Smith I. Acyclovir-resistant herpes simplex virus infection due to altered DNA polymerase. Lancet. 1987;ii:1461. doi: 10.1016/s0140-6736(87)91157-3. [DOI] [PubMed] [Google Scholar]
- 30.Roberts G B, Fyfe J A, Gaillard R K, Short S A. Mutant varicella-zoster virus thymidine kinase: correlation of clinical resistance and enzyme impairment. J Virol. 1991;65:6407–6413. doi: 10.1128/jvi.65.12.6407-6413.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sacks S L, Wanklin R J, Reece D E, Hicks K A, Tyler K L, Coen D M. Progressive esophagitis from acyclovir-resistant herpes simplex. Clinical roles for DNA polymerase mutants and viral heterogeneity? Ann Intern Med. 1989;111:893–899. doi: 10.7326/0003-4819-111-11-893. [DOI] [PubMed] [Google Scholar]
- 32.Safrin S. Nucleosides and foscarnet—clinical aspects. In: Richman D D, editor. Antiviral drug resistance. Chichester, England: John Wiley & Sons Ltd.; 1996. pp. 103–122. [Google Scholar]
- 33.Safrin S, Berger T G, Gilson I, Wolfe P R, Wofsy C B, Mills J, Biron K K. Foscarnet therapy in five patients with AIDS and acyclovir-resistant varicella-zoster virus infection. Ann Intern Med. 1991;115:19–21. doi: 10.7326/0003-4819-115-1-19. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Preparation and transformation of competent E. coli. In: Sambrook J, Fritsch E F, Maniatis T, editors. Molecular cloning: a laboratory manual. 2nd ed. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. pp. 1.82–1.84. [Google Scholar]
- 35.Sasadeusz J J, Tufaro F, Safrin S, Schubert K, Hubinette M M, Cheung P K, Sacks S L. Homopolymer mutational hot spots mediate herpes simplex virus resistance to acyclovir. J Virol. 1997;71:3872–3878. doi: 10.1128/jvi.71.5.3872-3878.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sawyer M H, Inchauspe G, Biron K K, Waters D J, Straus S E, Ostrove J M. Molecular analysis of the pyrimidine deoxyribonucleoside kinase gene of wild-type and acyclovir-resistant strains of varicella-zoster virus. J Gen Virol. 1988;69:2585–2593. doi: 10.1099/0022-1317-69-10-2585. [DOI] [PubMed] [Google Scholar]
- 37.Snoeck R, Gerard M, Sadzot-Delvaux C, Andrei G, Balzarini J, Reymen D, Ahadi N, De Bruyn J M, Piette J, Rentier B. Meningoradiculoneuritis due to acyclovir-resistant varicella zoster virus in an acquired immune deficiency syndrome patient. J Med Virol. 1994;42:338–347. doi: 10.1002/jmv.1890420404. [DOI] [PubMed] [Google Scholar]
- 38.Standring-Cox R, Bacon T H, Howard B A. Comparison of a DNA probe assay with the plaque reduction assay for measuring the sensitivity of herpes simplex virus and varicella-zoster virus to penciclovir and acyclovir. J Virol Methods. 1996;56:3–11. doi: 10.1016/0166-0934(95)01889-1. [DOI] [PubMed] [Google Scholar]
- 39.Summers W C, Raksin P. A method for selection of mutations at the tdk locus in Escherichia coli. J Bacteriol. 1993;175:6049–6051. doi: 10.1128/jb.175.18.6049-6051.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzutani T, Lacey S F, Powell K L, Purifoy D J, Honess R W. Random mutagenesis of the thymidine kinase gene of varicella-zoster virus. J Virol. 1992;66:2118–2124. doi: 10.1128/jvi.66.4.2118-2124.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swierkosz E M, Scholl D R, Brown J L, Jollick J D, Gleaves C A. Improved DNA hybridization method for detection of acyclovir-resistant herpes simplex virus. Antimicrob Agents Chemother. 1987;31:1465–1469. doi: 10.1128/aac.31.10.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Talarico C L, Phelps W C, Biron K K. Analysis of the thymidine kinase genes from acyclovir-resistant mutants of varicella-zoster virus isolated from patients with AIDS. J Virol. 1993;67:1024–1033. doi: 10.1128/jvi.67.2.1024-1033.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tappero J W, Perkins B A, Wenger J D, Berger T G. Cutaneous manifestations of opportunistic infections in patients infected with human immunodeficiency virus. Clin Microbiol Rev. 1995;8:440–450. doi: 10.1128/cmr.8.3.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tebas P, Scholl D, Jollick J, McHarg K, Arens M, Olivo P D. A rapid assay to screen for drug-resistant herpes simplex virus. J Infect Dis. 1998;177:217–220. doi: 10.1086/517357. [DOI] [PubMed] [Google Scholar]
- 45.Whitley R J, Roizman B. Herpes simplex viruses. In: Richman D D, Whitley R J, Hayden F G, editors. Clinical virology. New York, N.Y: Churchill Livingstone Inc.; 1997. pp. 375–410. [Google Scholar]
- 46.Wunderli W, Miner R, Wintsch J, Vongunten S, Hirsch H H, Hirschel B. Outer retinal necrosis due to a strain of varicella-zoster virus resistant to acyclovir, ganciclovir, and sorivudine. Clin Infect Dis. 1996;22:864–865. doi: 10.1093/clinids/22.5.864. [DOI] [PubMed] [Google Scholar]