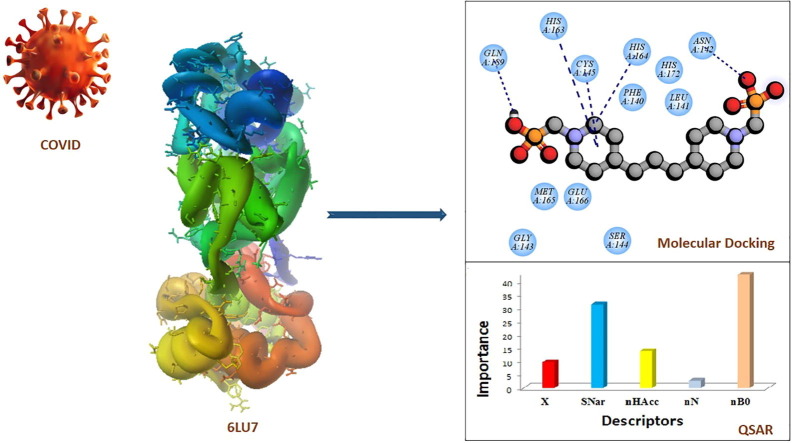
Graphical abstract
This paper consists of new mono, bis, and tetraphosphonate compounds that were selected against COVID-19 Mpro. Computational studies based on molecular docking, the quantitative structure–activity relationship (QSAR), and the pharmacophore studies were conducted to detailed study the inhibitory potency of compounds against COVID-19 Mpro and the factors influencing inhibitory potency.
Keywords: Phosphonate derivatives, Coronavirus inhibitors, Docking, DFT calculation, QSAR
Abstract
The global coronavirus (COVID-19) outbreak has prompted scientists to discover a cure for the disease. So far, phosphorus-based drugs have been proposed. These drugs have good inhibitory activity against the main protease (Mpro). Hence, in order to introduce a group of inhibitors the coronavirus, 51 compounds containing different mono, bis, and tetra phosphonates as Remdesivir derivatives, 32 of which are new, were synthesized and characterized by 31P, 13C, and 1H NMR and IR spectroscopy. Their biological activities were also investigated by Molecular Docking, QSAR, and Pharmacophore. Van der Waals, hydrogen bonding, and hydrophobic interactions were studied for all compounds as well as binding energy (△G, Kcal/mole) and the inhibitory constant Ki (μM) obtained by Molecular Docking. The results showed that the topology of the ligands and the change of the different groups attached to them can be effective in the placement position in the active site of the enzyme (Glu 166 and Gln 189). And bisphosphonates have a high interaction tendency with Mpro COVID-19. Compound L24 was identified as the best inhibitor with the −6.38 kcal/mol binding energy. The quantitative structure–activity relationship (QSAR) findings demonstrated that the polarity and topology of molecules in all phosphonate derivatives were important parameters affecting the effecting on the binding energy and inhibitory ability of compounds. The DFT and pharmacophore results are in good accordance with those of QSAR and molecular docking. This study can be helpful to gain a better understanding of the interactions between the Mpro of virus and its inhibitors in order to attain drugs with more effect on coronavirus (COVID-19).
1. Introduction
Research based on chemistry and biology greatly contributes to the production of bio-inhibitors. According to the electronic and structural properties of phosphonates such as the phosphorus-carbon stable covalent bond and their high potential in biology, Phosphonate chemistry can be introduced as a suitable candidate for the production of biomaterials [1], [2], [3]. Phosphonates have a wide range of application in radiopharmaceuticals [4], enzyme inactivators [5], HIV protease inhibitor [6], cardiovascular drugs [7], anti-parasitic drugs [8], and antiviral drug [9]. On 11th March 2020, the World Health Organization (WHO) categorized COVID-19, as a pandemic disease [10], [11]. To date, there is no definitive treatment for COVID-19; however, scientists are working hard to prevent and find its safe treatment albeit, some useful drugs were introduced by them [12]. One group of these proposed treatments was antiviral drugs based on phosphorus amide and phosphonates such as tenofovir, Adefovir, Sofosbuvir, Foscarnet, Uprifosbuvir, and Remdesivir, but any country didn't approve them as a reliable treatment for coronavirus [12], [13], [14], [15]. Thus, produced intensifying drugs based on phosphorus compounds, HIV and malaria drugs were tested for COVID-19 [16], [17].
Quantitative Structure-Activity relationship (QSAR) is a tool to discover the relationship between the structure and activity of chemical compounds. In the face of unknown disease particularly COVID-19, this technique and molecular docking have brought benefits to design and produce new drugs [18], [19], [20], [21].
In this research, due to the extensive biological properties of phosphonates as well as their ability to inhibit the main protease, 51 phosphonate derivatives including mono, bis, and tetra (Scheme 1, Scheme 2, Scheme 3 ) have been used to study interactions with COVID-19 (PDB ID 6LU7) (https://doi.org/10.2210/pdb6LU7/pdb). The parameters affecting the inhibitory activity of the compounds were obtained from QSAR studies [22]. Binding energies for all ligands were calculated by molecular docking [23], [24]. Also, it was shown that the presence of different groups in the chemical structure of phosphonates could change the binding energy. We hope that these results can be effective in drug designing to definitive treatment of coronavirus.
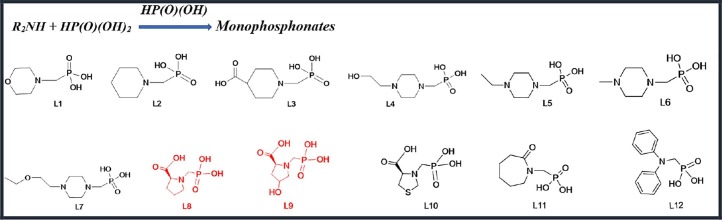
Scheme 1.
Monophosphonate derivatives have been used.
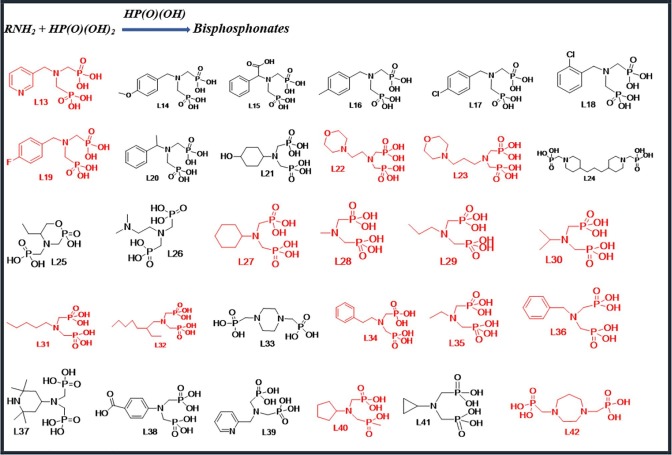
Scheme 2.
Bisphosphonate derivatives have been used.
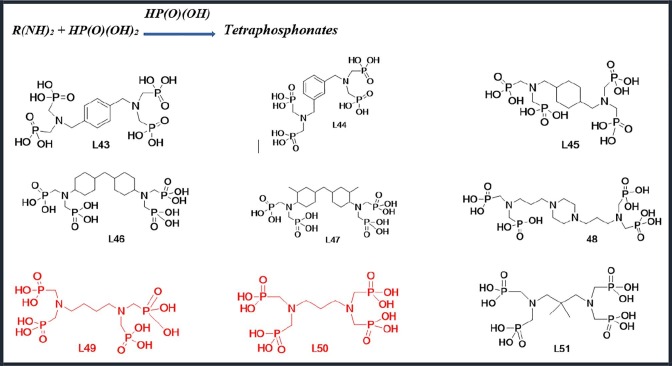
Scheme 3.
Tetraphosphonate derivatives have been used.
2. Experimental
2.1. Materials and methods
The compounds used to synthesize the ligands were commercial products of Merck. 1H, 13C and 31P NMR spectra were recorded at room temperature on a Bruker Avance DRS 500 MHz spectrometer. The chemical shifts of 1H, 13C and 31P were obtained in D2O solvent, with reference TMS as an internal standard. The IR spectra of the synthesized ligands were recorded on a Shimadzu IR-60 model spectrometer using KBr pellets in the range 400–4000 cm−1.
2.2. Synthesis
2.2.1. Synthesis of monophosphonates
All of the ligands were synthesized based on a Mannich reaction [25]. 1 mmol of Secondary (2°) amine was blended in 5 mL 37% hydrochloric acid, 5 mL deionised water, and 1.5 mmol phosphorous acid. This mixture was refluxed at100–120 °C for 1.5 h, then 2 mmol of paraformaldehyde was added dropwise it over a period of 1 h and the solution was refluxed again for one hour. The solution was dried at room temperature, and after washing with ethanol, was obtained a pure product. The Ligands L8 and L9 have been synthesized in the previous work [26], but other ligands synthesized and were characterized with 1H, 13C, 31P NMR and IR spectroscopy (see Supplementary information).
2.2.1.1. N-morpholinemethylenephosphonicacid (L1)
31PNMR (202.46 MHz, D2O): δ = 6.69 ppm. 1H NMR (500.13MHz,D2O): 47 δ = 3.27 ppm (–CH2-N, d, 2H, 2 JP,H = 12.7 Hz), 3.48–4.01 ppm(CH2,S,), 13CNMR (125.77 MHz,D2O): δ = 50.42 ppm(1 JP,C = 135.9 Hz), 50.95 ppm, 60.92 ppm. IR(KBr,ν,cm−1): ν = 3425(W,NH+), 3065(m,CH), 2850(m,CH), 2735(m,P-OH), 2335(m,C-N), 1084–1235 (s, ν asPO3), 956–1045(s, ν sPO3), 749(s,P-C), 540(s,p = o), 465(m), 434(m). m.p., 210 °C.
2.2.1.2. N-piperidinemethylenephosphonicacid (L2)
31PNMR (202.46 MHz, D2O): δ = 6.99 ppm. 1H NMR(500.13 MHz,D2O): δ = 3.37 ppm (–CH2-N, d, 2H, 2JP,H = 12.4 Hz), 3.48–4.01 ppm (CH2,S,), 13CNMR(125.77 MHz,D2O): δ = 47.42 ppm, 50.42 ppm (1 JP,C = 134.5 Hz), 51.05 ppm, 60.92 ppm. IR(KBr,ν,cm−1): ν = 3450(m,NH+), 3040(m,CH), 2755(m,P-OH), 2585(m), 1278(m,C-N), 1107–1221 (s, ν asPO3), 972–1038(s, ν sPO3), 709(m,P-C), 587(m,p = o), 527(w), 416(m). m.p., 205 °C.
2.2.1.3. N-piperidinecarboxylicacidmethylenephosphonicacid (L3)
31PNMR (202.46 MHz, D2O): δ = 7.50 ppm. 1H NMR(500.13 MHz,D2O): δ = 1.08 (CH,t, H), 1.91–3.48(CH2,m), 3.66(CH2,d,2H 2JP,H = 12.3 Hz). 13CNMR (125.77 MHz,D2O): δ = 24.09 (s), 37.84(s), 52.5(d, 1 JP,C = 136.800 Hz,CH2), 53.84(s), 176.84(s).IR(KBr,ν,cm−1): ν = 3415(w,NH+), 3005(m,CH), 2885(m,CH), 2765(m,P-OH), 2640(m), 1283(m,C-N), 1145–1230(s, ν asPO3), 945–1014(s, ν sPO3), 771 (w,P-C), 583(s,P=O), 506(m), 433(m). m.p., 183 °C.
2.2.1.4. 4-(2-methylenephosphonicacidpiperazine) ethanol (L4)
31PNMR (202.46 MHz, D2O): δ = 7.23 ppm. 1H NMR(500.13 MHz,D2O): δ = 1.21 ppm (CH2,d), 2.28 ppm(–CH2, m), 3.74 ppm(–CH2-N, d, 2H, 2 JP,H = 12.8 Hz). 13CNMR (125.77 MHz,D2O): δ = 13.05 ppm (s), 23.05 ppm (s), 43.05 ppm (s),45.62 ppm(d, 1 JP,C = 135.1 Hz,CH2), 56.63 (s). IR(KBr,ν,cm−1): ν = 3420(w,NH+), 3010(m,CH), 2730(m,P-OH), 2605(m), 1258(m,C-N), 1120–1220(s, ν asPO3), 948–1004(s, ν s PO3), 781(w,P-C),555(m,p = o), 504(m), 4531 (w). m.p., 235 °C.
2.2.1.5. 4-ethylpiperazinemethylenephosphonicacid (L5)
31PNMR (202.46 MHz, D2O): δ = 7.33 ppm. 1H NMR(500.13 MHz,D2O): δ = 0.706 (CH3, t,3H), 1.17 (CH2,m,2H), 1.60(CH2,m,2H), 3.31(CH2,t,2H), 3.41 (CH2, d,4H, 2JP, H = 12.1 Hz). 13CNMR(125.77 MHz,D2O): δ = 10.26 (s), 20.02(s), 24.86(s), 48.3 (d 1 JP,C = 135.1 Hz,CH2), 54.40(s).IR(KBr,ν,cm−1): ν = 3420(w,NH+), 2950(m,CH), 2750 (m,P-OH), 2640(m), 2500(m), 1271(m,C-N), 1150–1212(s, ν asPO3), 940–1014(s, ν sPO3), 775 (w,P-C), 583(s,P=O), 517(m), 484(m). m.p., 186 °C.
2.2.1.6. 4-methylpiperazinemethylphosphonicacid (L6)
31PNMR (202.46 MHz, D2O): δ = 7.33 ppm. 1HNMR(500.13 MHz,D2O): δ = 0.706 (CH3,t, 3H), 1.17 (CH2, m, 2H), 1.60(CH2,m,2H), 3.31(CH2,t,2H), 3.41(CH2,d,4H, 2JP, H = 12.1 Hz).13CNMR(125.77 MHz,D2O): δ = 10.26 (s),20.02(s),24.86(s), 48.3(d 1 JP,C = 135.1 Hz,CH2), 54.40(s). IR(KBr,ν,cm−1): ν = 3420(w,NH+), 2950(m,CH), 2750 (m,P-OH), 2640(m), 2500(m), 1271(m,C-N), 1150–1212(s, ν asPO3), 940–1014(s, νsPO3), 775 (w,P-C), 583(s,P=O), 517(m), 484(m). m.p., 186 °C.
2.2.1.7. 4-(2- ethoxyethyl)piperazinemethylenephosphonicacid (L7)
31PNMR (202.46 MHz, D2O): δ = 7.25 ppm (t, 2JP,H = 12.2 Hz),1H NMR (500.13 MHz,D2O): δ = 0.706 (CH3,t, 3H), 1.17 (CH2, m,), 1.60(CH2,m,2H), 3.36(CH2,t,2H), 3.44(CH2,d,4H,2JP,H = 12.2 Hz).13CNMR(125.77 MHz,D2O): δ = 10.26 (s),20.02(s),24.86(s), 48.3(d 1 JP,C = 135.1 Hz,CH2), 52.10(s). 54.40(s).IR(KBr,ν,cm−1): ν = 3405(w,NH+), 3000 (m,CH), 2750(m,P-OH), 2520(m), 1282(m,C-N), 1134–1236(s, ν asPO3), 934–1034(s, ν sPO3), 759(w,P-C), 557(s,p = o), 521(m), 451(m). m.p., 215 °C.
2.2.1.8. (R)-3-thiazolidine-4-carboxylicacidmethylenephosphonicacid (L10)
31PNMR (202.46 MHz, D2O):δ = 7.29 ppm 1H NMR(500.13 MHz,D2O): δ = 3.26–3.66 (CH2,m,), 4.38(CH2,d,2H, 2JP,H = 12.1 Hz), 4.54(CH,m) 13CNMR(125.77 MHz,D2O): δ = 27.2 (s), 53.75(d, 1JP,C = 133.59 Hz,CH2), 58.19(s),69.32(s),172.63(s) IR(KBr,ν,cm−1): ν = 3435(w,NH+), 2980 (m,CH), 2755(m,P-OH), 2295(m), 1274(m,C-N), 1078–1192(s, ν asPO3), 913–1015(s, ν sPO3), 759(w,P-C), 521(s,p = o), 461(m), 411(m). m.p., 218 °C.
2.2.1.9. Caprolactammethylenephosphonicacid (L11)
31PNMR (202.46 MHz, D2O): δ = 7.87 ppm. 1H NMR (500.13 MHz, D2O): δ = 1.17 2.27(CH2, s), 3.43(CH2, d, 2H, 2JP, H = 12.1 Hz), 3.63(CH2, s). 13CNMR (125.77 MHz, D2O): δ = 22.36 (s), 23.18(s), 24.48(ss), 32.94(s), 50.07(d, 1 JP, C = 137.1 Hz, CH2), 56.23, 178.17(s). IR (KBr, ν, cm−1): ν = 3420(s, NH+), 2985(s, CH), 2875(s, CH), 2750(s, P-OH), 2625(s), 1461(m), 1235(m, C-N), 1167(s, ν asPO3), 938–1067(s, ν sPO3), 780 (m, PC), 537(m), 451. m.p., 245 °C.
2.2.1.10. Diphenylamine methylphosphonicacid (L12)
31P NMR (202.46 MHz, D2O): 7.41(s) ppm. 1H NMR (500.13 MHz, D2O): δ = 2.75 (d, 2H, CH2), 4.17 (s, 4H, 2CH2), 7.28 (s, 10H, Ph). 13C NMR (125.77 MHz, D2O): δ = 50.61 (d, 1 JP, C = 132.5 Hz, CH2), 127.93 (s), 128.25 (s), 130.06 (s), 135.18 (s). IR data (KBr, cm−1): ν = 3415(w,NH+), 2374 (m), 1171(s, ν asPO3), 937 (s, ν sPO3), 450 (m).
2.2.2. Synthesis of bisphosphonates
The method mentioned above is also used to synthesize this group of compounds. 1 mmol amount of Primary (1°) amine was mixed with 5 mL 37% hydrochloric acid, 5 mL deionised water, 3 mmol phosphorous acid and refluxed at100–120 °C for 1.5 h. In the last step, 4 mmol of paraformaldehyde was added to the mixture like the previous method. Finally, a powder was obtained that was characterized with 1H, 13C, 31P NMR, and IR spectroscopy (see Supplementary Information). Ligands L13, L19, L22, L23, L27, L28, L29, L30, L31, L32, L34, L35, L36, L40, and L42 have been synthesized in the previous works [26], [27].
2.2.2.1. 4-methoxybenzylimino-bis(methylenephosphonicacid) (L14)
31PNMR (202.46 MHz, DMSO): δ = 7.99 ppm (m). 1H NMR(500.13 MHz,D2O): δ = 3.32 ppm (CH2,d, 4H, 2JP,H = 10.90 Hz), 3.85 (–CH3,s,3H),4.55 (–CH2,s,2H), 6.85–7.01 (m, 4H). IR(KBr,ν,cm−1): ν = 3400(w,NH+), 3025 (m,CH), 2730(m,P-OH), 2330(m), 1294(m,C-N), 1184–1254(m, ν asPO3), 937 (s, ν sPO3), 769(w,P-C), 531(s,p = o), 493(m), 447(m). m.p., 251 °C.
2.2.2.2. 2-phenylaglycineimino-bis(methylenephosphonicacid) (L15)
31PNMR (202.46 MHz, D2O): δ = 7.48 ppm. 1H NMR(500.13 MHz,D2O): δ = 3.07 (CH2,t, 2H),3.52,3.68 (CH2,d,4H,2JP,H = 12.1 Hz), 7.29–7.33 (C6H5,m,5H). 13CNMR (125.77 MHz,D2O): δ = 26.7(s),48.7(d,1 JP,C = 134.5 Hz,CH2),55.46(s),124.62(s),126.25(s),126.33(s),133.15(s).IR(KBr,ν,cm−1):ν = 3415(w,NH+),3040(m,CH),2870(m,CH), 2775(m,P-OH), 2590(m), 1450(w), 1237(m,C-N), 1161 1200(s, ν asPO3), 940–1004(s, νsPO3), 744 (w,P-C), 575(m), 493(m), 405(m). m.p., 237 °C.
2.2.2.3. 4-methylbenzylimino-bis(methylenephosphonicacid) (L16)
31PNMR (202.46 MHz, D2O): δ = 7.32 ppm. 1H NMR(500.13 MHz,D2O): δ = 3.07 (CH2,t, 2H), 3.52, 3.68(CH2,d,4H,2 JP,H = 12.7 Hz), 7.29 7.33(C6H5,m,5H). 13C NMR (125.77 MHz, D2O): δ = 26.7 (s), 48.7(d, 1 JP, C = 132.5 Hz, CH2), 55.46(s), 124.62(s), 126.25(s), 126.33(s), 133.15(s). IR(KBr,ν,cm−1): ν = 3415(w,NH+), 3040(m,CH), 2870(m,CH), 2775(m,P-OH), 2590(m), 1450(w), 1237(m,C-N), 1161–1200(s, ν asPO3), 940–1004(s, ν sPO3), 744 (w,P-C), 575(m), 493(m), 405(m). m.p., 217 °C.
2.2.2.4. 4-chlorobenzylimino-bis(methylenephosphonicacid) (L17)
31P NMR (101.25 MHz, D2O): δ (ppm) = 4.77. 1H NMR (250.13 MHz, D2O): δ (ppm) = 3.36 (d, 2JPH = 12.51 Hz, 4H; 2CH2), 4.60 (s, 2H; CH2), 7.42 (s, 2H; Ph), 7.43 (s, 2H; Ph). IR (KBr, cm−1): ν = 2990 (m), 2758 (m, br, OH), 1584 (m), 1493 (m), 1162 (vs, νas PO3), 936 (vs, νs PO3), 830 (s), 795 (s), 706 (m), 578 (vs), 523 (m), 492 (m), 413 (m).m.p., 233 °C.
2.2.2.5. 2-chlorobenzylimino-bis(methylenephosphonicacid) (L18)
31P NMR (101.25 MHz, D2O): δ (ppm) = 7.41. 1H NMR (250.13 MHz, D2O): δ (ppm) = 3.46 (d, 2JPH = 12.3 Hz, 4H; 2CH2), 4.450 (s, 2H; CH2), 7.22 (s, 2H; Ph), 7.57 (s, 2H; Ph). IR (KBr, cm−1): ν = 2990 (m), 2758 (m, br, OH), 1584 (m), 1493 (m), 1162 (vs, νas PO3), 936 (vs, νs PO3), 830 (s), 795 (s), 706 (m), 578 (vs), 523 (m), 492 (m), 413 (m).m.p., 237 °C.
2.2.2.6. S-(-)-1-phenylethylimino-bis(methylenephosphonicacid) (L20)
31PNMR (202.46 MHz, DMSO): δ = 9.50 ppm (m). 1H NMR(500.13 MHz, DMSO): δ = 1.68 (s,3H, CH3), 3.40(m, 2 JH,H = 15.09, 2 JP,H = 12.1 Hz,4H,CH2),5.11(s,1H, CH), 7.45–7.52(m, 5H, Ar-H), 13CNMR(125.77 MHz, DMSO): δ = 12.09 (s), 48.10 (d, 1JP,C = 134.3 Hz, CH2),65.17(s), 120.6(s), 129.01(s), 129.98(s), 132.75(s), IR(KBr,ν,cm−1): ν = 3400(w,NH+), 3025 (m,CH), 2730(m,P-OH), 2330(m), 1294(m,C-N), 1184–1254(m, ν asPO3), 937 (s, ν sPO3), 769(w,P-C), 531(s,p = o), 493(m), 447(m). m.p., 241 °C.
2.2.2.7. 4-hydroxycyclohexylimino-bis(methylenephosphonicacid) (L21)
31PNMR (202.46 MHz, D2O): δ = 8.50 ppm. 1H NMR(500.13 MHz,D2O): δ = 3.07 (CH2,t, 2H), 3.52 (CH2, d, 4H, 2JP,H = 13.25 Hz), 3.68(CH2,d,2H, 2JP,H = 13.0 Hz), 7.29–7.33(C6H5,m,5H). 13CNMR (125.77 MHz, D2O): δ = 26.7 (s), 36.1 (s), 48.7(d, 1 JP,C = 137.4 Hz,CH2),55.46(s),68.62(s). IR(KBr,ν,cm−1): ν = 3415(w,NH+), 3040(m,CH), 2870(m,CH), 2775(m,P-OH), 2590(m), 1450(w), 1237(m,C-N), 1161–1200(s, ν asPO3), 940–1004(s, ν sPO3), 744 (w,P-C), 575(m), 493(m), 405(m). m.p., 217 °C.
2.2.2.8. 1,3-bis(4- piperidyl- methylenephosphonicacid)propane (L24)
31PNMR (202.46 MHz, D2O): δ = 7.33 ppm. 1H NMR (500.13 MHz, D2O): δ = 1.14–1.544 (CH2, m 2H), 1.82(CH2, m 2H), 2.92(CH2, m 2H), 3.11 (CH2, d, 4H, 2JP,H = 11.0 Hz), 3.23(CH2, 2H,), 3.57(CH2,m,2H). 13CNMR (125.77 MHz, D2O): δ = 21.89 (s), 28.89 (s), 30.52 (s), 34.27 (s), 51.7(d, 1 JP, C = 136.5 Hz, CH2), 54.83(s). IR(KBr,ν,cm−1): ν = 3415 (w,NH+), 3040(m,CH), 2870(m,CH), 2775(m,POH), 2590(m), 1450(w), 1237(m,C-N), 1161–1200(s, ν asPO3), 940–1004(s, ν sPO3), 744 (w,P-C), 575(m), 493(m), 405(m). m.p., 257 °C.
2.2.2.9. 4,4′-(1,3-propanediol)pipyridine-bis(methylenephosphonicacid) (L25)
31PNMR (202.46 MHz, D2O): δ = 4.1 ppm, 7.3 ppm 1HNMR(500.13 MHz,D2O): δ = 0.94 (CH3,t, 3 JH,H = 7.3 Hz 3H), 1.74(CH2,t, 3JH,H = 7.1 Hz, 2H), 3.4 (CH2, d, 4H,2 JP,H = 13.1 Hz), 3.44(CH,m, 3 JH,H = 8.2 Hz.1H), 3.56(br, 1H), 3.62 (t, 2 JH,H = 14.75 Hz 2JP,H = 14.2 Hz, 1H), 4.23 (dt, 2 JH,H = 13.75 Hz 3 JP,H = 9.4 Hz, 3 JH,H = 4.3 Hz, 1H), 4.35 (t, 2 JH,H = 13.6 Hz 3JP,H = 12.4 Hz, 1H). 13CNMR(125.77 MHz,D2O): δ = 8.7 (s), 17.2, 47.5(d, 1 JP,C = 129.3 Hz,CH2), 49.4(d, 1 JP,C = 139.1 Hz,CH2),62.2(s),63.5(s). IR(KBr,ν,cm−1): ν = 3415(w,NH+), 3040(m,CH), 2870(m,CH), 2775(m,P-OH), 2590(m), 1450(w), 1237(m,C-N), 1161–1200(s, ν asPO3), 940–1004(s, ν sPO3), 744 (w,P-C), 575(m), 493(m), 405(m). m.p., 211 °C.
2.2.2.10. N,N-dimethylethylendiimino-bis(methylenephosphonicacid) (L26)
31PNMR (202.46 MHz, D2O): δ = 4.1 ppm, 7.3 ppm 1HNMR(500.13 MHz,D2O): δ = 0.94 (CH3,t, 3 JH,H = 7.3 Hz 3H), 1.74(CH2,t, 3 JH,H = 7.1 Hz, 2H), 3.4 (CH2, d, 4H, 2JP,H = 13.1 Hz), 3.44(CH,m, 3 JH,H = 8.2 Hz.1H), 3.56(br, 1H), 3.62 (t, 2 JH,H = 14.75 Hz 2JP,H = 14.2 Hz, 1H), 4.23 (dt, 2 JH,H = 13.75 Hz 3JP,H = 9.4 Hz, 3 JH,H = 4.3 Hz, 1H), 4.35 (t, 2 JH,H = 13.6 Hz 3JP,H = 12.4 Hz, 1H). 13CNMR (125.77 MHz, D2O): δ = 8.7 (s), 17.2, 47.5(d, 1 JP, C = 129.3 Hz, CH2), 49.4(d, 1 JP, C = 139.1 Hz,CH2),62.2(s),63.5(s). IR(KBr,ν,cm−1): ν = 3415(w,NH+), 3040(m,CH), 2870(m,CH), 2775(m,P-OH), 2590(m), 1450(w), 1237(m,C-N), 1161–1200(s, ν asPO3), 940–1004(s, ν sPO3), 744 (w,P-C), 575(m), 493(m), 405(m). m.p., 211 °C.
2.2.2.11. Pipirazine1,4-bis(methylenephosphonicacid) (L33)
31PNMR (202.46 MHz, D2O): δ = 6.18 ppm. 1H NMR (500.13 MHz, D2O): δ = 3.42 (CH2, d, 4H, 2JP, H = 12.8 Hz), 3.81(CH2,s,8H). IR(KBr,ν,cm−1): ν = 3400 (w,NH+), 3030 (m,CH), 2775(m,P-OH), 2405(m), 1254(m,C-N), 1071–1217(s, ν asPO3), 916–1025(s, ν sPO3), 763(w,P-C), 520(m,δ P=O), 430(m). Calcd for C6H16N2O6P2: C, 26.29; H, 5.88; N, 10.22. Found: C, 26.25; H, 5.86; N, 10.19.
2.2.2.12. [(Benzhydryl-phosphonomethyl-amino)-methyl]-phosphonicacid (L37)
1H NMR (D2O, ppm): δ = 1.39 (s, 6H, 2CH3), 1.41 (s, 6H, 2CH3), 1.86(t, 2H, 2CH2(α)), 2.32(d, 2H, 2CH2(β)), 3.51 (d, 4H, 2CH2), 4.43 (m, 1H, CH). 31P NMR (D2O, ppm): δ = 7.38 (t). 13C NMR (D2O): 23.60 (d), 29.20 (t), 34.41 (d), 48.14 (t), 49.21 (t), 56.97(d, 1JP,C = 138.2 Hz,CH2). IR data (KBr, cm−1): ν = 3415 (w, NH+), 2978 (s), 2724 s, 1643 m, 1475 m, 1196 (s, νs PO3), 1051 (s, νs PO3), 509 w.
2.2.2.13. 4-benzoicacidimino-bis(methylenephosphonicacid) (L38)
31PNMR (202.46 MHz, D2O): δ = 19.6 ppm. (IR (KBr, ν, cm−1): ν = 3429 (w, NH+), 2925 (m, CH), 2823 (m, CH), 2765 (m, P-OH), 2610 (m), 1660 (w, COOH), 1501 (m), 1426 (m), 1275 (m, C-N), 1167 (s, νas PO3), 1075 (s, νs PO3), 773 (w, P-C), 529 (m), 460 (m), 409 (m). m.p., 395 °C.
2.2.2.14. Pyridine (2-ilmethyl)azanedile-bis(methylenephosphonicacid) (L39)
31PNMR (121 MHz, DMSO): δ = 17.4 ppm. 1HNMR (300 MHz, DMSO): δ = 4.00 ppm (CH2, d, 2 JP,H = 10.75 Hz, 4H), 4.28 ppm (CH2, s, 2H), 5.20–5.90 ppm (OH, m, 4H), 7.36–8.03 ppm (Ar, m). IR (KBr, ν, cm−1): ν = 3414 (w, NH + ), 3140 (s), 3067 (m, CH), 2938 (m, CH), 2849 (m, CH), 2753 (m, P-OH), 1628 (m, C-C), 1578 (m, C-C), 1453 (m, C-C), 1373 (m), 1214 (s, νas PO3), 928–1107 (s,νs PO3), 812 (w, P C), 766 (m), 638 (m), 546 (m). m.p., 215 °C.
2.2.2.15. Cyclopropylimino-bis(methylenephosphonicacid) (L41)
31PNMR (202.46 MHz, D2O): δ = 8.27 ppm (t, 2JP,H = 11.7 Hz), 1H NMR (500.13 MHz,D2O):δ = 0.83 (m, 4H,CH2), 3.30 (m,1H,CH), 3.48 (d, 2JP,H = 11.7 Hz,4H, CH2). 13CNMR (125.77 MHz,D2O):δ = 13.96 (s), 50.32(d, 1JP,C = 138.2 Hz,CH2), 54.58(s).IR (KBr,ν,cm−1): ν = 3405(w,NH+), 3000 (m,CH), 2750(m,P-OH), 2520(m), 1282(m,C-N), 1134–1236(s, ν asPO3), 934–1034(s, ν sPO3), 759(w,P-C), 557(s,δp = o), 521(m),451(m). Calcd For C5H13NO6P2:C, 24.50; H, 5.35; N, 5.71. Found: C, 24.47; H, 5.32; N, 5.68.
2.2.3. Synthesis of tetraphosphonates
Ligands L43 to L51 were synthesized by Mannich reaction. This synthesis method is exactly the same as the previous two batches, but 1millimol of diamine and 6 mmol phosphorous acid was used, here. The results were characterized with 1H, 13C, 31P NMR, and IR spectroscopy (see Supplementary Information). The data of L43, L49, and L50 is brought in another paper [26].
2.2.3.1. 1,3-xylenediimino-tetra(methylenephosphonicacid) (L44)
31PNMR (202.46 MHz, D2O): δ = 7.69 ppm. 1H NMR(500.13 MHz,D2O): δ = 3.09 ppm (CH2,d, 8H, 2JP,H = 10.6 Hz), 4.57 (–CH2,s,4H), 7.31–7.62 (Ar-H, m, 4H). 13CNMR (125.77 MHz,D2O): δ = 52.91 ppm (d, 1JP,C = 125.3 Hz,CH2), 58.89 (s), 131.03(s), 131.62(s). IR(KBr,ν,cm−1): ν = 3405(w,NH+), 2965(s,CH), 2845(s,CH), 2720(s,P-OH), 2545(s), 1284(m,C-N), 1166–1229(s, ν asPO3), 935–1008(s, ν s PO3), 746(S,P-C), 573(s,p = o), 485(m), 421 (M). m.p., 245 °C.
2.2.3.2. 1,4-cyclohexanediyldimethanediimino-tetra(methylenephosphonicacid) (L45)
31PNMR (202.46 MHz, D2O): δ = 7.78 ppm. 1H NMR(500.13 MHz,D2O): δ = 0.97 1.97(CH2,t,8H), 2.83(CH,s,2H), 3.23 (CH2, d, 8H,2JP,H = 12.2 Hz), 3.43(CH2,m,4H).IR(KBr,ν,cm−1): ν = 3415(w,NH+), 3040(m,CH), 2870(m,CH), 2775(m,P-OH), 2590(m), 1450(w), 1237(m,C-N), 1161–1200(s, ν asPO3), 940–1004(s, ν sPO3), 744 (w,P-C), 575(m), 493(m), 405(m). m.p., 221 °C.
2.2.3.3. 4,4′-(diimino-tetra(methylenephosphonicacid))dicyclohexylmethane (L46)
31PNMR (202.46 MHz, DMSO): δ = 7.97 ppm (m). 1H NMR(500.13 MHz, DMSO): δ = 1.02 (m, 2H, CH2), 1.07–2.13 (m, CH2),2.21(s, 2H, CH), 3.40(d, 2 JP,H = 10.5 Hz,8H,CH2),4.63(q, 3 JH,H = 6.8 Hz,1H, CH), 7.34(m, 3H, Ar-H), 7.53 (d, 2H, Ar-H), 10.83(s, 4H, OH). 13C NMR(125.77 MHz, DMSO): δ = 16.35 (s), 25.45 (s) 30.35 (s), 32.45 (s) 48.50 (d, 1JP,C = 128.7 Hz, CH2),57.7(s). IR(KBr,ν,cm−1): ν = 3400(w,NH+), 3025 (m,CH), 2730(m,P-OH), 2330(m), 1294(m,C-N), 1184–1254(m, ν asPO3), 937 (s, ν sPO3), 769(w,P-C), 531(s,p = o), 493(m), 447(m). m.p., 257 °C.
2.2.3.4. 4,4′-methylenebis(2-methylcyclohexanediimino-tetra(methylenephosphonicacid)) (L47)
31PNMR (202.46 MHz, DMSO): δ = 8.21 ppm (m). 1H NMR(500.13 MHz, DMSO): δ = 1.02 (m, 6H, CH3), 1.22 (m, 2H, CH), 1.47–2.13 (m, CH2),2.21(s, 2H, CH), 3.40(d, 2 JP,H = 10.5 Hz,8H,CH2),4.63(q, 3 JH,H = 6.8 Hz,1H, CH), 7.34(m, 3H, Ar-H), 7.53 (d, 2H, Ar-H), 10.83(s, 4H, OH). 13CNMR(125.77 MHz, DMSO): δ = 16.35(s),17.21(s), 25.45 (s) 30.35 (s), 32.45 (s) 48.50 (d, 1JP,C = 128.7 Hz, CH2),57.7(s). IR(KBr,ν,cm−1): ν = 3400(w,NH+), 3025 (m,CH), 2730(m,POH),2330(m), 1294(m,C-N), 1184–1254(m, ν asPO3), 937 (s, νsPO3),769(w,P-C), 531(s,p = o), 493(m), 447(m). m.p., 214 °C.
2.2.3.5. Pipirazine-1,4-bis(3-iminopropyl)tetra(methylenephosphonicacid) (L48)
31PNMR (202.46 MHz, D2O): δ = 7.15 ppm. 1H NMR(500.13 MHz,D2O): δ = 2.23 (CH2,s, 4H), 3.29 (CH2,s,4H),3.47(CH2, d,8H, 2JP,H = 12.6 Hz),3.64(CH2,s,8H). 13CNMR (125.77 MHz,D2O): δ = 18.22 (s),48.36(s),51.5(d, 1 JP,C = 136.00 Hz,CH2), 53.16(s), 56.99(s).IR(KBr,ν,cm−1): ν = 3415(w,NH+), 3040(m,CH), 2870(m,CH), 2775(m,P-OH), 2590(m),1450(w), 1237(m,C-N), 1161–1200(s, ν asPO3), 940–1004(s, ν sPO3), 744(w,P-C), 575(m), 493(m), 405(m). m.p., 236 °C.
2.2.3.6. 2,2′-dimethylpropane-1,3diiminotetra(methylenephosphonicacid) (51)
31PNMR (202.46 MHz, DMSO): δ = 7.35 ppm (m). 1H NMR(500.13 MHz, DMSO): δ = 0.67 (m, 6H, CH3), 3.23(m, 2 JP,H = 12.6 Hz,12H,CH2). 13CNMR (125.77 MHz, DMSO):δ = 15.34 (s), 17.18 (s),52.09 (d, 1JP,C = 136.5 Hz, CH2),56.3(s). IR (KBr,ν,cm1): ν = 3415(w,NH+), 3040(m,CH), 2870(m,CH), 2775(m,P-OH), 2590(m), 1450(w), 1237(m,C-N), 1161–1200(s, ν asPO3), 940–1004(s, ν sPO3), 744 (w,P-C), 575(m), 493(m),405(m). m.p., 233 °C.
2.3. Preparation of protein and ligand for docking study
The X-ray crystallographic structures of COVID-19 main protease (PDB ID 6LU7) are available on the Protein Data Bank (PDB (http: //https://www.pdb.org.) database, and this was saved by the PDB format. The file of receptor protein contains A and B chains that chain A was used for the preparation receptor protein. Water molecules and Crystallized ligand in crystal structures of 6LU7 were removed using BIOVIA Discovery Studio 2019 (DS) [28]. To create pdbqt file, polar hydrogen atoms and Kollman united atom charges were added to the receptor protein by Auto Dock Tools-1.5.6 (ADT) program [29], [30]. The molecular structures of the ligands (L1-L51) were drawn in GaussView 6.0.16 software and optimized in the Gaussian 09 W using basis set b3lyp/6–311 + g (d,p) [31] and the log file of each ligand saved with pdb format in GaussView. For all ligands, Gasteiger charge was assigned, and non-polar hydrogens were merged by Auto Dock Tools (ADT); then pdbqt files of the ligands were generated for docking simulation.
2.4. Docking study using Auto Dock Tools (ADT)
Molecular docking simulations were carried out in Auto Dock Tools and from ligplot+ 1.4.5 [32] used to analyze of docking results and interactions between ligand and protein. The automatic grid was determined grid points 72 × 78 × 82 A° and grid center (X, Y, Z) of −12.789 14.313 71.074 in grid box. Therefore, the binding of the ligands with receptor protein was carried out via 100 numbers of GA Run of the genetic algorithm.
2.5. QSAR Calculations.
2.5.1. Data set
In order to study the inhibition of COVID-19 by phosphonates, a series of mono, bis, and tetra phosphonates were selected to investigate their binding affinity in the SARS-CoV-2 Mpro active site (S4). These compounds were previously synthesized by our group [26], [27]. In this study, binding energies (△G) values of the compounds from Docking in Table 1 were used as dependent variables.
Table 1.
Docking parameters of 6LU7 activities of monophosphonates.
| No | ΔG bindig (kcal/mol) | Electrostatic Energy (kcal/mol) | vdW + Hbond + desolv Energy (kcal/mol) | Final Intermolecular Energy (kcal/mol) | Est. Inhibition Constant, Ki (µM) |
|---|---|---|---|---|---|
| L1 | −4.68 | −1.31 | −4.56 | −5.87 | 370.17 |
| L2 | −4.86 | −1.39 | −4.66 | −6.06 | 271.71 |
| L3 | −4.69 | −1.40 | −5.07 | −6.48 | 367.09 |
| L4 | −4.74 | −2.05 | −4.77 | −6.82 | 337.50 |
| L5 | −4.49 | −1.43 | −4.56 | −5.98 | 511.73 |
| L6 | −4.48 | −2.34 | −3.34 | −5.67 | 519.94 |
| L7 | −4.92 | −0.45 | −6.86 | −7.31 | 246.47 |
| L8 | −5.22 | −1.75 | −5.26 | −7.01 | 149.55 |
| L9 | −4.34 | −1.12 | −5.31 | −6.43 | 659.71 |
| L10 | −4.55 | −1.35 | −4.99 | −6.34 | 460.40 |
| L11 | −4.73 | −1.28 | −4.64 | −5.92 | 343.16 |
| L12 | −5.91 | +0.05 | −7.75 | −7.70 | 46.92 |
2.5.2. Molecular optimization and calculation of descriptors
It is essential to build numerical descriptors of a set of inhibitors in order to build QSAR models. Descriptors can represent quantitative properties that depend on the structure of the molecules. Advantage of use of theoretical descriptors is that they are free of the uncertainty of experimental measurements and can be calculated for compounds not synthesized [33]. Molecular descriptors were computed using the DRAGON software for a set of 51 phosphonates as coronavirus inhibitors [34]. A total of 1478 molecular descriptors of different kinds (0, 1, 2, and 3D) were used to describe the chemical diversity of the compounds. The descriptor typology is: a) constitutional (atom and group fragments), b) functional groups, c) atom centered fragments, d) empirical, e) topological, f) walk count, g) various autocorrelations from the molecular graph, h) Randic molecular profiles from the geometry matrix, i) geometrical, j) WHIMs, k) GETAWAYs descriptors and various indicator descriptors. The meaning of these molecular descriptors and the calculation procedure is summarized in the manual to the DRAGON software and are explained in detail, with related literature references, in the Handbook of Molecular Descriptors by Todeschini and Consonni [34].
In addition, the chemical quantum descriptors were calculated by the DFT B3LYP/6-31G(d, p) method. These descriptors include energies of highest occupied molecular orbital (HOMO), lowest unoccupied molecular orbital (LUMO), the energy difference between LUMO, HOMO (gap), dipole moment (μ), electrophilicity (ω), density (ρ), molecular weight (Mw), molecular volume (Mv), Sanderson electronegativity (χ), chemical hardness (ὴ) and chemical power (Cπ) (see Table S1). Therefore, the set of descriptors contains D = 1496 features [35], [36].
2.5.3. Chemometric methods
The correlation between biological activity and structural properties was obtained using the variable selection Genetic Algorithm-A stepwise multiple linear regression (GA-SW-MLR) methods. We applied the Genetic Algorithm to select from all the calculated descriptors only the best combinations of those and the most relevant for obtaining models with the highest predictive power for the binding energy of compounds with COVID-19 Mpro. Genetic Algorithms have been employed in recent years as a powerful tool to optimize many problems in drug design [33], [37], [38]. This approach allows the selection of the models with the following characteristics: high correlation coefficient R, low standard deviation S, and the least number of descriptors involved [39]. In order to evaluate the generated models, leave-one-out cross-validation is used. In this algorithm, one compound is left in each step as a prediction set, and the model is developed using the remaining molecules as the training set [40]. For further exhaustive testing of the prediction power of the model, in addition to LOO-CV, the leave-multiple-out cross-validation (LMO-CV) algorithm was also carried out. Then the ΔG of this group was predicted by the model developed by using the remaining observations as the training set [40].
3. Results and discussion
3.1. Molecular docking study
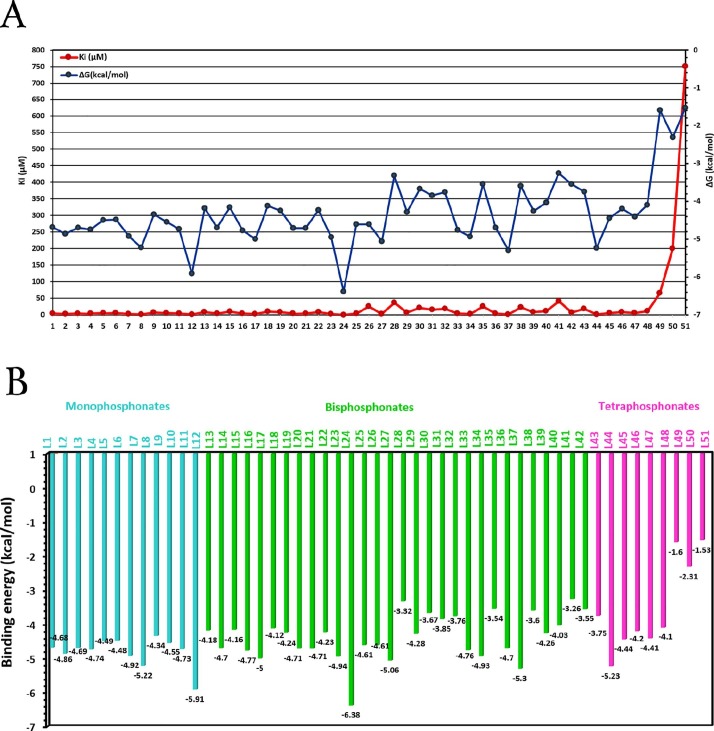
Computer techniques such as molecular docking have been very effective in drug designing against new diseases. This method provides better insight into the binding of ligands to protein, bond lengths, binding energies, and types of interactions. Furthermore, the molecular docking considers the effect of various factors on the ligand ranging from ring size to aliphatic chain length to the presence of different atoms, and to electronegative and electropositive groups when interacting with the protein. In this work, the interactions between the different phosphonate compounds and the X-ray crystal structure of COVID-19 (PDB ID: 6LU7) were studied by molecular docking. The aim of this study is to investigate the inhibitory potency of a group of phosphonate derivatives against the coronavirus and to evaluate the most effective electronic and structural factors in this process. Three series of compounds were classified as mono, bis, and tetra phosphonates, respectively. The binding energies were measured for the mono and bisphosphonates in the range of −3.26 to −6.38 kcal/mol, Well-known antiviral drugs (Table 4) were compared to the synthesized compounds, in terms of Docking parameters, Remdesivir (-6.75 kcal/mol), Adefovir (-4.18 kcal/mol), Tenofovir (-4.97 kcal/mol), and Foscarnet (-3.13 kcal/mol). Therefore, it can be anticipated that mono and bisphosphonates could be effective against COVID-19 as possible medicinal agents.
Table 4.
Results of the docking of antiviral agents against COVID-19.
| Est. Inhibition Constant, Ki (µM) | Final Intermolecular Energy (kcal/mol) | vdW + Hbond + desolv Energy (kcal/mol) | Electrostatic Energy (kcal/mol) | ΔGbinding (kcal/mol) | Drug name |
|---|---|---|---|---|---|
| 859.90 | −6.57 | −6.27 | −0.30 | −4.18 | Adefovir |
| 6.47 | −10.96 | −10.81 | −0.14 | −7.08 | Foscarnet |
| 229.18 | −10.33 | −10.24 | −0.10 | −4.97 | Tenofovir |
| 11.23 | −11.82 | −11.46 | −0.37 | −6.75 | Remdesivir |
3.2. Visualization of docking results
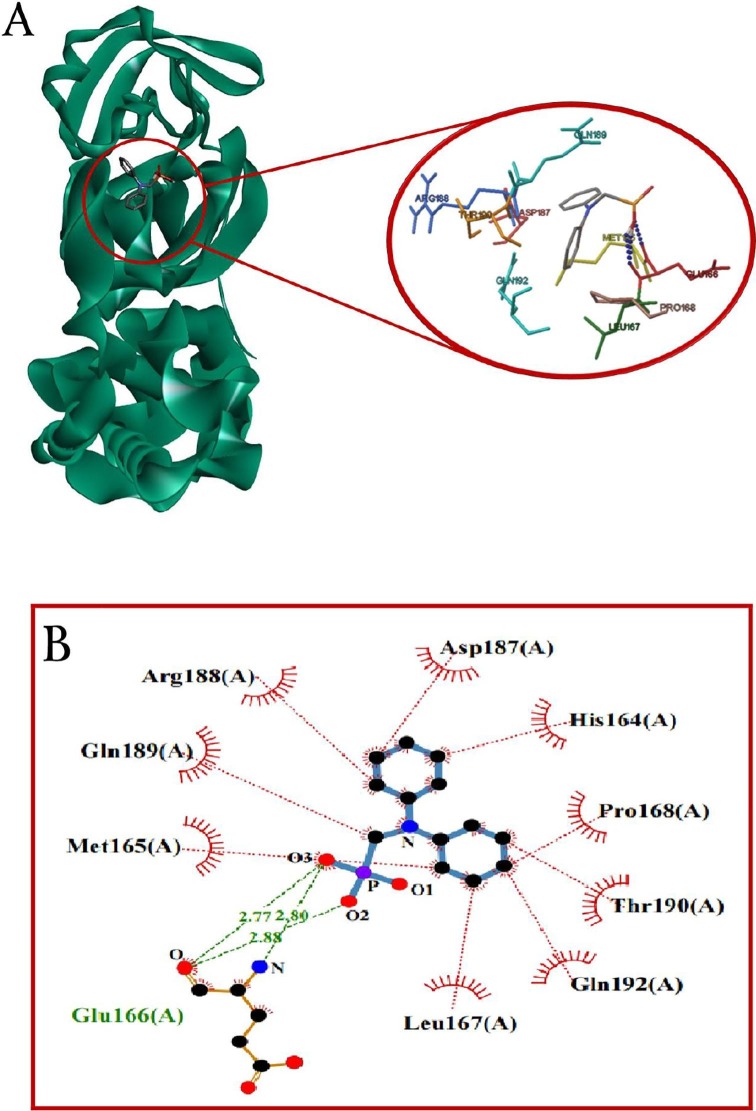
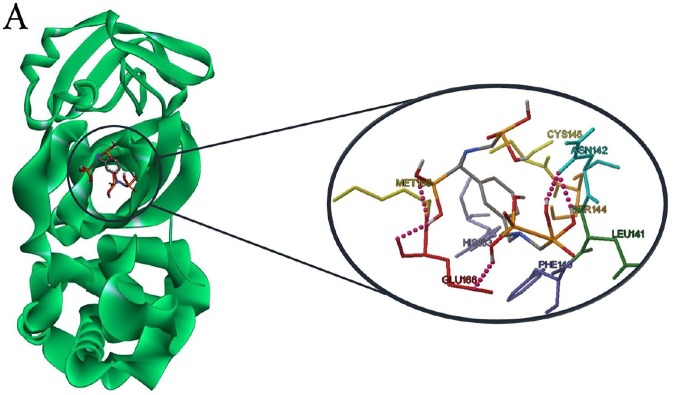
Results of docking studies of monophosphonates and 6LU7 are reported in Table 1. In this series of phosphonate derivatives, there are twelve cyclic monophosphate ligands, the new ones shown in black (Scheme 1). Further exact investigation shows that H- bond is mostly formed through the amino acid residues Ser 144, Leu 141, Cys 145, and His 163 with monophosphonates. Furthermore, amino acid residues Met 165, Leu 167, Thr 190, and Arg 188 have the most presence in hydrophobic areas. Binding energy has changed in the △G = -5.91 to −4.34 Kcal/mole range for monophosphonates (Table 1). L12 compound containing two nitrogen-attached phenyl groups is found as an aromatic one in the monophosphonate series. This compound has the best binding energy and inhibitory constant among monophosphonates. Docking simulation of L12 with the protein 6LU7 revealed binding energy △G = − 5.91 Kcal/mole and the inhibitory constant (Ki) 46.92 μM. 3D interaction model of L12 with the main protease of COVID-19 is shown in Fig. 1 A. Also, hydrogen bonds were observed between the C=O functional group of Glu 166 and O-H groups of L12 at a distance of 2.77 and 2.88 A°. The hydrophobic interactions were shown between residues and different sections of the ligand L12 with red color (Fig. 1B). The π-Sulfur and π-alkyl interactions were observed between the phenyl ring of L12 with the sulfur atom of Met 165 and the alkyl group of Pro 168 at a distance 5.40 A° and 5.47 A°, respectively (Fig. 1C).
Fig. 1.
(A) 3D model molecular interactions of L12 at the binding site of 6LU7. The hydrogen bonds are shown by blue dots. (B) and (C) 2D interaction diagram of L12 with the target protein. π-Alkyl interactions are indicated. Hydrogen bonds are shown in by green dots, and hydrophobic interactions are marked with red dotted lines.
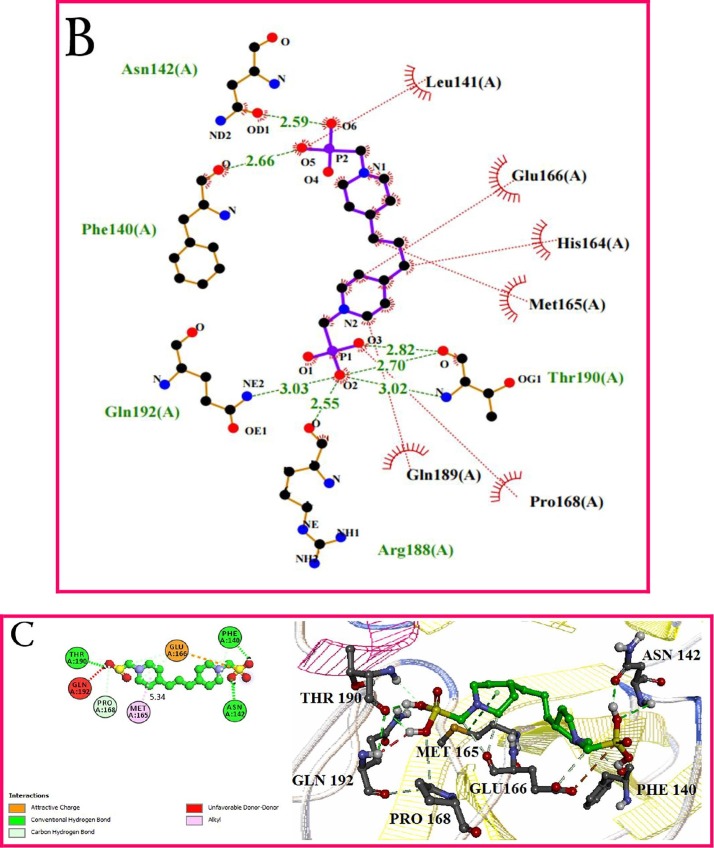
In this series, cyclic and linear derivatives of bisphosphonates (L13-L42) have been studied some of which are aromatic (Scheme 2). Newly synthesized bisphosphonates are shown in black in Scheme 2. Molecular docking data of the interactions among the bisphosphonates and 6LU7 are gathered in Table 2. The most effective amino acids were identified in hydrogen bonds and hydrophobic interactions between Glu 166, Ser 144, Gln 189, His 163 and Met 169, His 164, Phe 140, Pro 168, respectively. According to the obtained numerical values from the molecular docking (Table 2 ), binding energy and Inhibition Constant of cyclic bisphosphonates are better than those of aliphatics. L24 has a symmetrical structure containing two piperidine rings and has the lowest binding energy (△G) and Inhibition Constant (Ki) among all subject series. The interaction results of L24 and protein 6LU7 showed binding energy (△G) −6.38 Kcal/mole and the inhibitory constant (Ki) 21.02 μM (Table. 2). The plausible binding mode of L24 and the active site of 6LU7 is displayed in Fig. 2 A. As can be seen, acidic hydrogens were involved in the formation of hydrogen bonds with Asn142, Phe140, Gln192, Arg188, and Thr190 residues from the active site of the Mpro, respectively. The hydrophobic interactions were shown in red color between residues and different sections of the compound L12 (Fig. 2B). Furthermore, the phenyl ring was connected to Met165 via π-Alkyl bonds with distance 5.34 A°. These interactions have led to more stability of the target complex (Fig. 2C).
Table 2.
Docking parameters of 6LU7 activities of bisphosphonates.
| No | ΔG bindig (kcal/mol) | Electrostatic Energy (kcal/mol) | vdW + Hbond + desolv Energy (kcal/mol) | Final Intermolecular Energy (kcal/mol) | Est. Inhibition Constant, Ki (µM) |
|---|---|---|---|---|---|
| L13 | −4.18 | −0.34 | −6.82 | −7.16 | 860.78 |
| L14 | −4.70 | −0.33 | −7.65 | −7.98 | 359.03 |
| L15 | −4.16 | −0.32 | −7.41 | −7.73 | 899.73 |
| L16 | −4.77 | +0.03 | −7.78 | −7.76 | 316.21 |
| L17 | −5.00 | −0.47 | −7.52 | −7.99 | 215.21 |
| L18 | −4.12 | −0.01 | −7.09 | −7.10 | 955.06 |
| L19 | −4.24 | −0.05 | −7.17 | −7.23 | 776.92 |
| L20 | −4.71 | −0.38 | −7.31 | −7.69 | 355.07 |
| L21 | −4.71 | −0.51 | −7.18 | −7.69 | 355.50 |
| L22 | −4.23 | +0.03 | −7.54 | −7.51 | 797.46 |
| L23 | −4.94 | −0.39 | −8.13 | −8.52 | 240.12 |
| L24 | −6.38 | −1.24 | −8.72 | −9.96 | 21.02 |
| L25 | −4.61 | −0.21 | −6.19 | −6.40 | 418.43 |
| L26 | −3.55 | −1.34 | −5.49 | −6.83 | 2510 |
| L27 | −5.06 | −0.39 | −7.35 | −7.74 | 195.80 |
| L28 | −3.32 | −1.07 | −4.63 | −5.71 | 3680 |
| L29 | −4.28 | −1.29 | −5.97 | −7.26 | 731.61 |
| L30 | −3.67 | −0.19 | −6.16 | −6.35 | 2060 |
| L31 | −3.85 | −0.10 | −7.33 | −7.43 | 1510 |
| L32 | −3.76 | +0.05 | −7.98 | −7.94 | 1750 |
| L33 | −4.76 | −2.16 | −4.99 | −7.15 | 321.60 |
| L34 | −4.93 | −0.17 | −8.04 | −8.21 | 245.42 |
| L35 | −3.54 | −0.52 | −5.70 | −6.22 | 2550 |
| L36 | −4.70 | −0.17 | −7.52 | −7.68 | 358.28 |
| L37 | −5.30 | −0.40 | −7.58 | −7.98 | 130.81 |
| L38 | −3.60 | +0.05 | −6.93 | −6.88 | 2290 |
| L39 | −4.26 | −0.14 | −7.10 | −7.25 | 751.14 |
| L40 | −4.03 | −0.52 | −6.19 | −6.71 | 1110 |
| L41 | −3.26 | −0.01 | −5.94 | −5.94 | 4080 |
| L42 | −4.27 | −0.45 | −6.21 | −6.66 | 738.48 |
Fig. 2.
(A) 3D model interactions between L24 and binding site of 6LU7. (B) and (C) 2D representation of docking of L24 with binding site of the main protease of COVID-19. Hydrogen bonds and hydrophobic interactions are shown in the form of green and red dotted lines.
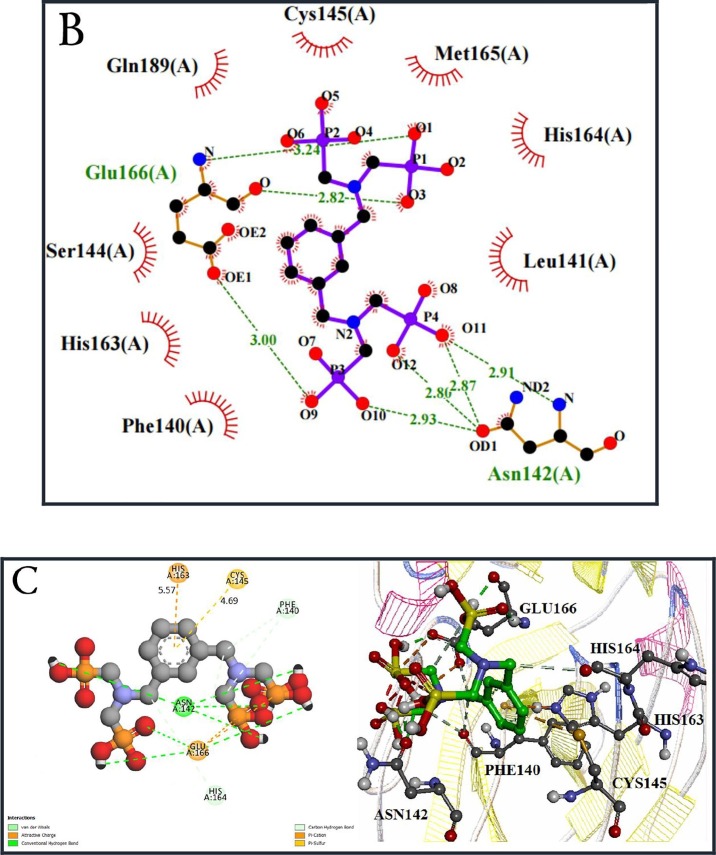
Tetraphosphonates include two classes of cyclic and linear compounds that new derivatives are shown in black (Scheme 3). Docking studies showed that the amino acid residues Glu 166, Ser144, His 163, Cys 143 and Met 165, His 164, Gln 189, Phe 140 had the most interaction in the hudrogen and hydrophobic bonding with tetraphosphonates. It was observed that the binding energy is reduced in aliphatics. Docking simulation of L44 with 6LU7 protein revealed binding energy −5.23 Kcal/mole and the inhibitory constant (Ki) 145.73 μM. This aromatic compound has the best interactions with protein 6LU7 in tetraphosphonates (Fig. 3 A). The hydrophobic residues interactions of 6LU7 with L44 were shown by the red dashed line (Fig. 3B, C). Several hydrogen bonds are seen between the hydrogen of O-H group in L44 with the oxygen of C=O group in Asn142, Glu166, the nitrogen of N-H in Glu166, and Asn142 (Fig. 3B). Also, π–sulfur and π–cation stacking interactions have been respectively observed between the phenyl ring of L44 with the sulfur atom of Cys 145 and the NH3 + of residue His 163 at a distance 4.69 A° and 5.57 A° (Fig. 3C).
Fig. 3.
(A) 3D interaction diagram of L44 with the main protease of COVID-19. (B) and (C) 2D interaction diagram diagram of L44 with the protease. π–sulfur, and π–cation are indicated. Hydrogen-bonding and hydrophobic interactions between L44 and 6LU7 in the binding site are marked with green and red dotted lines, respectively.
As observed, investigation of the relationship between the binding energy (ΔG) and inhibition constant (Ki) showed that the bisphosphonates had a higher inhibitory effect and lower binding energy than those of other derivatives (Fig. 4 A), And L24 can be considered as the most active compound.
Fig. 4.
(A) 2D graph of Ki vs. binding (ΔG) for 51 Ligands. (B) Binding energy of mono, bis, and tetra phosphonates against 6LU7 protease.
The increasing number of phosphonate functional groups could not be considered as an effective factor for inhibition and interactions in active sites of the protein (Fig. 4B). Also, the connection desire of the phosphonates to Mpro depends on the type of their topology, inhibitors' substituents, and their ability to form interactions with active sites of the protease.
Docking results for the effect of some phosphonate-derived drugs on the crystal structure of COVID-19 (PDB ID: 6LU7) are presented with binding energy values (ΔG) −4.18 to −7.08 (kcal/mol) (Table 4). The binding energy of mono, bis, and tetraphosphonates are in the mentioned range (except linear tetraphosphonates). Moreover, the amino acid residues involved in hydrogen bonding for these drugs are similar to the amino acid residues for L12, L24, and L44 that the maximum of repetition and similarity is observed for residues Glu 166 and Gln 189. Binding map for L49-L51 is provided in S1-S3. it can be said that ΔG values are related to the type of amino acid that reacts with the ligands. So the mentioned drugs, in Table 4, can be used as a reference to confirm the inhibition of 6 LU7 by synthesized compounds (See Table 3 ).
Table 3.
Docking parameters of 6LU7 activities of tetraphosphonates.
| No | ΔG bindig (kcal/mol) | Electrostatic Energy (kcal/mol) | vdW + Hbond + desolv Energy (kcal/mol) | Final Intermolecular Energy (kcal/mol) | Est. Inhibition Constant, Ki (µM) |
|---|---|---|---|---|---|
| L43 | −3.72 | −0.57 | −9.12 | −9.69 | 1860 |
| L44 | −5.23 | −1.64 | −8.36 | −10.01 | 145.73 |
| L45 | −4.44 | −0.15 | −10.26 | −10.41 | 551.97 |
| L46 | −4.20 | +0.16 | −10.32 | −10.17 | 834.95 |
| L47 | −4.41 | −0.14 | −10.24 | −10.38 | 584.15 |
| L48 | −4.01 | −2.43 | −8.74 | −11.17 | 1150 |
| L49 | −1.60 | −1.34 | −6.53 | −7.87 | 6530 |
| L50 | −2.31 | −0.58 | −7.69 | −8.28 | 20,000 |
| L51 | −1.53 | −1.21 | −6.29 | −7.50 | 75,000 |
According to this research and to obtain a logical relationship between the structure and activity of compounds, the QSAR study was performed.
3.3. QSAR analysis
QSAR calculations in the Genetic Algorithm -stepwise multiple linear regression (GA–SW–MLR) method were conducted to specify the parameters affecting the binding energy and inhibitory ability of each category mono (model 1), bis(model 2), tetra(model 3), and all phosphonates derivatives (model 4) to the main protease of COVID-19.
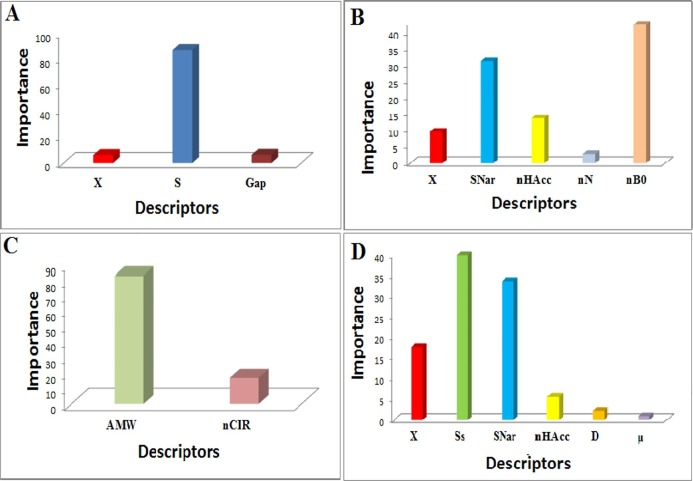
It can be seen from Fig. 5 and Table 5 , which the energy difference between LUMO and HOMO (Gap), Chemical softness (S), and electronic chemical potential (χ) as the main descriptors were chosen for mono phosphonates. Also, χ, Narumi simple topological index (SNar), number of Nitrogen atoms (nN), number of non-H bonds (nB0), and number of hydrogen bond acceptors (nHAcc) descriptors were chosen for bisphosphonates. Average molecular weight (AMW) and number of circuits (nCIR) were chosen for tetra phosphonates. On the other hand, six descriptors were selected as the most influencing descriptors for all phosphonates derivatives, which are the χ, nHAcc, Narumi simple topological index (SNar), Sum of Kier–Hall electro-topological states (Ss), density (D), and dipole moment (μ). A detailed description of these descriptors was given (Todeschini and Consonni, 2008) [38]. After selecting the most appropriate descriptors, the next step calculating was the linear relationship between each category descriptors and the biological activity (binding energy) of COVID-19 inhibitors (Table 5).
Fig. 5.
The Absolute relative means effects of selected descriptors in GA-SW-MLR models. A) model 1, B) model 2, C) model 3, and D) model 4.
Table 5.
QSAR equations and obtained statistical parameters from QSAR models.
| No | QSAR equations | Training |
Validation |
||||
|---|---|---|---|---|---|---|---|
| Rt2 | RMSEt | Q2LOO | RMSELOO | R2LMO | RMSELMO | ||
| Model 1 | (△G) = 40.7407 + 98.3112(Gap)-2.4502(S)-10.7921(X) | 0.935 | 0.101 | 0.858 | 0.264 | 0.807 | 0.189 |
| Model 2 | (△G) = -5.445 + 7.772(X) + 0.059(Ss)-0.234(nN)-0.266 (nB0) −0.199(nHACC) | 0.700 | 0.351 | 0.664 | 0.427 | 0.615 | 0.422 |
| Model 3 | (△G) = 6.904–0.975(AMW) −2.003(nCIR) | 0.861 | 0.526 | 0.829 | 0.5468 | 0.753 | 0.587 |
| Model 4 | (△G) = -2.732–0.089 nHAcc −0.412 SNar −17.240 X - 0.085 Ss + 0.165 D + 0. 102 μ | 0.843 | 0.341 | 0.788 | 0.397 | 0.732 | 0.387 |
R2t is a correlation coefficient of the training set; RMSEt is a root mean square error of the training set; Q2LOO is a correlation coefficient of leave-one-out cross-validation; RMSELOO is a root mean square error LOO–CV.
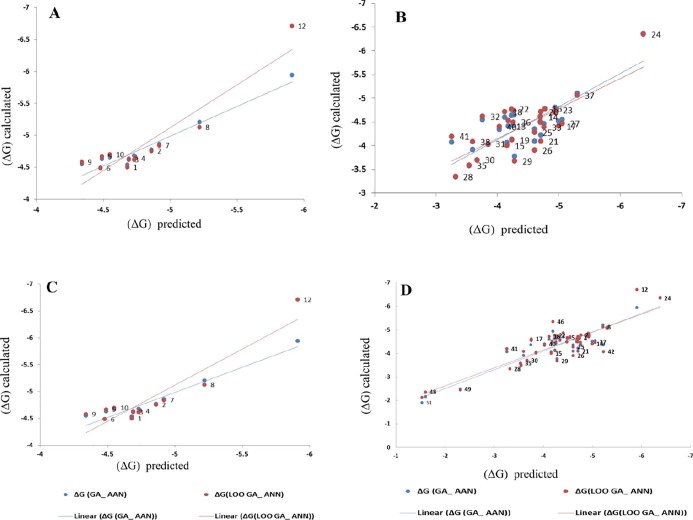
The predicted values of the binding energies value for the compounds in each model are plotted versus the experimental values of COVID-19 inhibitors in Fig. 6 . The cross-validation results of GA–SW–MLR models are shown in Table 5. The consistency and reliability of these models were validated using the leave-one-out (LOO) and leave-multi-out (LMO) cross-validation techniques. The consistency of the Q2 values for LOO and LMO data sets indicated that the proposed models were reliable. The observed GA–SW–MLR predicted values of the △G for all COVID-19 inhibitors studied in this work are shown in S2-S5.
Fig. 6.
The diagram of (ΔG) values from docking versus the predicted (ΔG) values from the GA-SW-MLR models for phosphosphonat compounds. A) model 1, B) model 2, C) model 3, and D) model 4.
In the final step, to interpret the inhibition mechanism, the most significant predictors appearing in the GA–SW–MLR models were examined. In GA–SW–MLR models, the mean effect of each descriptor could be considered as the relative mean effects [41]. Fig. 5 shows the importance of each variable for the QSAR models.
The appearance of χ and S, and Gap molecular descriptors in the QSAR model 1 shows the critical role of the electronegativity, chemical hardness, and gap energy of the molecules in mono phosphonates inhibition behavior against the main protease of COVID-19. The selection of these descriptors in this model implies that the inhibition mechanism was an electrical-based procedure. The behavior of bisphosphonates in the QSAR model 2 shows that structural descriptors (nB0, SNar, nHAcc) are more momentous than electronic descriptors (X). The nB0 and SNar were two of the effective factors of this model, which displays the importance of number non-hydrogen bonds (π − π stacking, hydrophobic interactions) and molecule topology in the inhibition mechanism. Indeed, SNar is a Narumi simple topological index related to molecular branching proposed [42]. These descriptor coefficients are negative, meaning that increasing non-hydrogen bonds and the molecular branching can lead to the reduction of the value of the binding energy of compounds with COVID-19 Mpro and better interact with the connection sites.
Fig. 5 data show that the average molecular weight (AMW) and the number of circuits (nCIR) 9are negative correlated with the binding energy of tetra phosphonates with COVID-19 Mpro. The nCIR is a structural descriptor, which is related to molecular flexibility. Indeed it includes both rings and circuits (a circuit is a larger loop around two or more rings). And inhibitory ability increases with the increase in average molecular weight, the number of rings, and circuits.
In the QSAR study of model 4 with all phosphonate derivatives mentioned in this work, it was shown that structural (SNar, nHAcc) and electronic parameters (χ Ss, μ, D) affect the biological behavior of these derivatives.
Descriptor χ was one of the parameters, affecting the inhibitory behavior of mono and bis and all phosphonates against COVID-19 Mpro. The electronic chemical potential [χ = (EHOMO + ELUMO)/2] is the escaping tendency of electrons from a species in its ground state and is the negative of absolute electronegativity [41]. The compound has greater interaction with the active site of COVID-19 Mpro when greater χ, the less bonding energy. The Ss is calculated by dividing the sum of Kier − Hall electro-topological states by the number of non-hydrogen atoms [43]. This predictor is related to the electronic essence of the molecules, namely the number of valence electrons and the principal quantum number of atoms in molecules [39]. With increasing Ss, the polarity of these compounds increases, and the electronic interactions between the inhibitors and the active site COVID-19 Mpro increase.
The nHAcc is the number of hydrogen bond acceptors (number of OH and NH atoms). Examination of this factor showed that as the number of OH and NH atoms in the structure increased, the bonding energy decreased, and the inhibitory power increased.
The μ and ρ descriptors are the dipole moment and density of molecules. Fig. 5 and Table 5 show that the dipole moment and density are correlated with the binding energy. Indeed, the binding energy decreases with their increase.
Comparing the QSAR models showed that the difference between descriptors and Inhibition mechanism in models QSAR could explain the reasons for the diverse behavior of inhibitors in these three groups phosphonate. The electronic descriptor in the QSAR model mono phosphonate, structural descriptors in the QSAR model tetra phosphonate, and both structural and electron parameters in the QSAR model bisphosphonate were effective on binding energy. Molecule topology, chemical potential, and the number of hydrogen bond acceptors were common descriptors between QSAR models 1, 2, and 4.
3.4. Frontier molecular orbitals
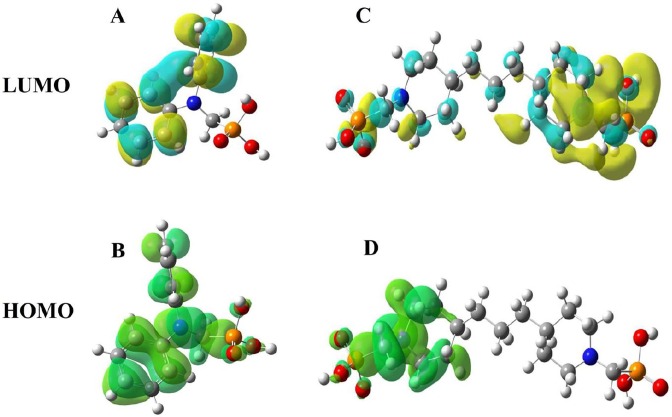
The analysis of Frontier molecular orbitals of ligands is one of the methods used to predict reactive electrophilic or nucleophilic sites [44]. The lowest unoccupied molecular orbital (LUMO) and highest occupied molecular orbital (HOMO) are called frontier molecular orbitals. HOMO-LUMO energies were calculated using DFT at B3LYP method and 6–311 + G** basis set for all the ligands. Our study revealed that, in general, the bisphosphonates had a stronger inhibitory activity compared to monophosphonates. HOMO-LUMO plots of the L12 and L24 are gathered in Fig. 7 . HOMO-LUMO gap energies of ligands L12 and L24 calculated 4.68 eV and 5.44 eV respectively; these values are comparable with the bandgap energy values of the bioactive molecules. The desirability of these values for these compounds indicates its tendency to interact with the COVID-19 acceptor as a bioactive molecule. In the case of compound L12, the HOMO plot was scattered on oxygens (phosphonate), nitrogen amine, and methyl phenyl benzenamine moieties, and LUMO was composed of the benzen rings. The ligand L24, HOMO was scattered over the nitrogen atom (1), oxygen atoms (1, 2, 3), and LUMO was spread overall structure. Frontier orbital energies can affect the inhibitory activity through charge transfer and π − π stacking between inhibitors and amino acid residues in the binding site of COVID-19 Mpro. The results of the study of molecular docking on phosphonate derivatives showed that these regions of this compound participated in important interactions with the main residues of the COVID-19 Mpro [41].
Fig. 7.
HOMO-LUMO orbitals composition analysis of ligands L12 and L24.
3.5. Pharmacophore of compounds
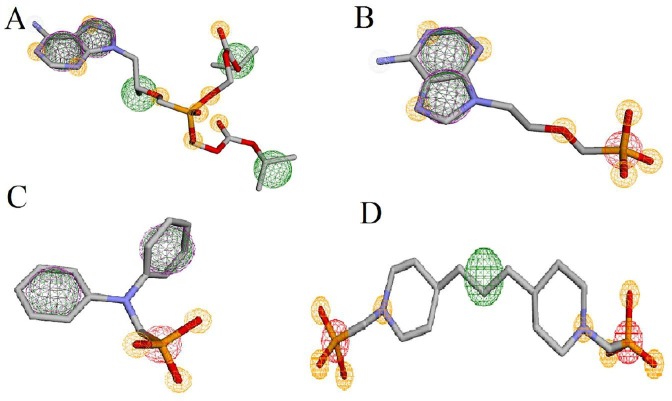
To determine the pharmacophore features which are significant for the use in drug discovery, we carried out ligand-based pharmacophore modeling. For this purpose, the strongest inhibitors (L12, L24) and reference molecules were subjected. Pharmacophore features such as aromatic ring and hydrophobic group, hydrogen-bond acceptor, hydrogen-bond donor, positive and negative ion, etc., may be useful for understanding the specific activity of molecules. The common pharmacophores from the reference molecule (Tenofovir) showed nine hydrogen bond acceptors (Yellow), five hydrophobic groups (Green), and two aromatic rings (Purple) (Fig. 8 A). The second reference molecule (Adefovir) showed seven hydrogen bond acceptors, two hydrophobic groups, and two aromatic rings (Fig. 8B), and L12 showed three hydrogen-bond acceptors, two hydrophobic groups, two aromatic rings, and one negative ion (Red) (Fig. 8C), as well as L24, displayed eight hydrogen bond acceptor, one hydrophobic group, and two negative ions (Fig. 8d). Hydrogen bond acceptors, hydrophobic groups, and negative ions were common pharmacophores in all structures of phosphonates that are helpful for binding with receptors. These results were in good agreement with those of QSAR and molecular docking.
Fig. 8.
Common pharmacophore for reference molecule, Tenofovir (A), Adefovir (B), L12(C), and L24.
4. Conclusion
The biggest crisis in recent years has been the appearance of COVID-19 disease and public health challenges. So far, many antiviral agents have been tested, but they have not been completely effective due to the side effects on other organs. Therefore, there is an attempt to find new ways to improve or control the prevalence of this disease. Phosphonate derivatives have been used here. By changing different functional groups in each category, the amount of binding energy is determined using molecular docking. The topology of the ligands and the change of the different groups attached to them can be effective in the placement position in the active site of the enzyme (Glu 166 and Gln 189).Compound L24 was identified as the best inhibitor with the −6.38 kcal/mol binding energy. In general, bisphosphonate had a higher inhibitory effect and lower binding energy than other phosphonates. QSAR calculation results using the Genetic Algorithm -Stepwise multiple linear regression (GA–SW-MLR) method for mono, bis, tetra phosphonate reveal that the difference between descriptors and Inhibition mechanism in QSAR models can explain the reasons for the diverse behavior of inhibitors. The electronic descriptor in the QSAR model of the mono phosphonate, structural descriptors in the QSAR model of the tetra phosphonate, and both structural and electron parameters in the QSAR model of bisphosphonate were effective on binding energy. Molecule topology, chemical potential, and the number of hydrogen bond acceptors were common descriptors between QSAR models mono, bis, and all phosphonate derivatives. The DFT and pharmacophore were used to verify docking and QSAR.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
The financial support of Tarbiat Modares University’s Research Council is gratefully acknowledged.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.poly.2022.115824.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Al Quntar, A. A. A., Dweik, H., Jabareen, A., Gloriozova, T. A., & Dembitsky, V. M. (2020). An Aminopyrrolidinyl Phosphonates—A New Class of Antibiotics: Facile Synthesis and Predicted Biological Activity.
- 2.Harsági N., Keglevich G. The hydrolysis of phosphinates and phosphonates: a review. Molecules. 2021;26(10):2840. doi: 10.3390/molecules26102840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shevchuk M., Wang Q., Pajkert R., Xu J., Mei H., Röschenthaler G.V., Han J. Recent advances in synthesis of difluoromethylene phosphonates for biological applications. Adv. Synth. Catal. 2021 [Google Scholar]
- 4.Tsebrikova G., Baulin V., Kalashnikova I., Ragulin V., Zavel’skii V., Maruk A.Y., Lunev A., Klement’eva O., Kodina G., Tsivadze A.Y. Russ. J. Gen. Chem. 2015;85:2071–2079. [Google Scholar]
- 5.Pallitsch K., Kalina T., Stanković T. Synthetic phosphonic acids as potent tools to study phosphonate enzymology. Synlett. 2019;30(07):770–776. [Google Scholar]
- 6.Lášek T., Dobiáš J., Buděšínský M., Kozák J., Lapuníková B., Rosenberg I., Birkuš G., Páv O. Synthesis of phosphonate derivatives of 2'-deoxy-2'-fluorotetradialdose d-nucleosides and tetradialdose d-nucleosides. Tetrahedron. 2021 doi: 10.1016/j.tet.2021.132159. Apr 16:132159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu H., Yang H., Shi E., Tang W. Med. Drug Discov. 2020 doi: 10.1016/j.medidd.2020.100063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lountos G.T., Tropea J.E., Waugh D.S. Mol. Biochem. Parasitol. 2013;187:1–8. doi: 10.1016/j.molbiopara.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rusconi S, Innocenti A, Vullo D, Mastrolorenzo A, Scozzafava A, Supuran CT. Carbonic anhydrase inhibitors. Interaction of isozymes I, II, IV, V, and IX with phosphates, carbamoyl phosphate, and the phosphonate antiviral drug foscarnet. Bioorganic & medicinal chemistry letters. 2004 Dec 6;14(23):5763-7. [DOI] [PubMed]
- 10.Keretsu S., Bhujbal S.P., Cho S.J. Rational approach toward COVID-19 main protease inhibitors via molecular docking, molecular dynamics simulation and free energy calculation. Sci. Rep. 2020;10(1):1–14. doi: 10.1038/s41598-020-74468-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abosheasha, M. A., & El‐Gowily, A. H. (2020). Superiority of cilostazol among antiplatelet FDA‐approved drugs against COVID 19 Mpro and spike protein: Drug repurposing approach. Drug development research. [DOI] [PMC free article] [PubMed]
- 12.Stasi C., Fallani S., Voller F., Silvestri C. Treatment for COVID-19: an overview. Eur. J. Pharmacol. 2020;173644 doi: 10.1016/j.ejphar.2020.173644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abdelrheem D.A., Ahmed S.A., Abd El-Mageed H., Mohamed H.S., Rahman A.A., Elsayed K.N., Ahmed S.A. The inhibitory effect of some natural bioactive compounds against SARS-CoV-2 main protease: insights from molecular docking analysis and molecular dynamic simulation. J. Environ. Sci. Health Part A. 2020;55(11):1373–1386. doi: 10.1080/10934529.2020.1826192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silva Arouche, T. d., Reis, A. F., Martins, A. Y., S Costa, J. F., Carvalho Junior, R. N., & JC Neto, A. M. (2020). Interactions Between Remdesivir, Ribavirin, Favipiravir, Galidesivir, Hydroxychloroquine and Chloroquine with Fragment Molecular of the COVID-19 Main Protease with Inhibitor N3 Complex (PDB ID: 6LU7) Using Molecular Docking. Journal of Nanoscience and Nanotechnology, 20(12), 7311-7323. [DOI] [PubMed]
- 15.Shah B., Modi P., Sagar S.R. In silico studies on therapeutic agents for COVID-19: Drug repurposing approach. Life Sci. 2020;117652 doi: 10.1016/j.lfs.2020.117652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chhetri A., Chettri S., Rai P., Mishra D.K., Sinha B., Brahman D. Synthesis, characterization and computational study on potential inhibitory action of novel azo imidazole derivatives against COVID-19 main protease (Mpro: 6LU7) J. Mol. Struct. 2020;1225 doi: 10.1016/j.molstruc.2020.129230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Belhassan A., En-Nahli F., Zaki H., Lakhlifi T., Bouachrine M. Assessment of effective imidazole derivatives against SARS-CoV-2 main protease through computational approach. Life Sci. 2020;262 doi: 10.1016/j.lfs.2020.118469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toropov A.A., Toropova A.P., Veselinović A.M., Leszczynska D., Leszczynski J. SARS CoV Mpro inhibitory activity of aromatic disulfide compounds: QSAR model. J. Biomol. Struct. Dyn. 2020:1–7. doi: 10.1080/07391102.2020.1818627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amin S.A., Ghosh K., Gayen S., Jha T. Chemical-informatics approach to COVID-19 drug discovery: Monte Carlo based QSAR, virtual screening and molecular docking study of some in-house molecules as papain-like protease (PLpro) inhibitors. J. Biomol. Struct. Dyn. 2020:1–10. doi: 10.1080/07391102.2020.1780946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Płonka W., Paneth A., Paneth P. Docking and QSAR of aminothioureas at the SARS-CoV-2 S-protein–human ACE2 receptor interface. Molecules. 2020;25(20):4645. doi: 10.3390/molecules25204645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rafi M.O., Bhattacharje G., Al-Khafaji K., Taskin-Tok T., Alfasane M.A., Das A.K., Parvez M.A.K., Rahman M.S. Combination of QSAR, molecular docking, molecular dynamic simulation and MM-PBSA: analogues of lopinavir and favipiravir as potential drug candidates against COVID-19. J. Biomol. Struct. Dyn. 2020:1–20. doi: 10.1080/07391102.2020.1850355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshida F., Topliss J.G. QSAR model for drug human oral bioavailability. J. Med. Chem. 2000;43(13):2575–2585. doi: 10.1021/jm0000564. [DOI] [PubMed] [Google Scholar]
- 23.Pagadala N.S., Syed K., Tuszynski J. Software for molecular docking: a review. Biophys. Rev. 2017;9(2):91–102. doi: 10.1007/s12551-016-0247-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomsen R., Christensen M.H. MolDock: a new technique for high-accuracy molecular docking. J. Med. Chem. 2006;49(11):3315–3321. doi: 10.1021/jm051197e. [DOI] [PubMed] [Google Scholar]
- 25.Noble A., Anderson J.C. Nitro-mannich reaction. Chem. Rev. 2013;113(5):2887–2939. doi: 10.1021/cr300272t. [DOI] [PubMed] [Google Scholar]
- 26.Gholivand K., Mohammadpanah F., Yaghoubi R., Rahimzadeh Dashtaki M., Pooyan M., Rahmani H., Roshanian Z., Sharifi M., EbrahimiValmoozi A.A., Roohzadeh R. Synthesis, crystal structure, cholinesterase inhibitory activity, evaluation of insecticidal activities, and computational studies of new phosphonic acids. Mol. Diversity. 2021:1–12. doi: 10.1007/s11030-021-10283-7. [DOI] [PubMed] [Google Scholar]
- 27.Gholivand K., Ghaziani F., Yaghoubi R., Hosseini Z., Shariatinia Z. Design, synthesis and anticholinesterase activity of some new α-aminobisphosphonates. J. Enzyme Inhib. Med. Chem. 2010;25(6):827–835. doi: 10.3109/14756361003691860. [DOI] [PubMed] [Google Scholar]
- 28.Studio, D. (2008). Discovery Studio. Accelrys [2.1].
- 29.Ferreira L.G., Dos Santos R.N., Oliva G., Andricopulo A.D. Molecular docking and structure-based drug design strategies. Molecules. 2015;20(7):13384–13421. doi: 10.3390/molecules200713384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meng X.-Y., Zhang H.-X., Mezei M., Cui M. Molecular docking: a powerful approach for structure-based drug discovery. Curr. Comput. Aided Drug Des. 2011;7(2):146–157. doi: 10.2174/157340911795677602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wodrich M.D., Corminboeuf C., Schleyer P.V.R. Systematic errors in computed alkane energies using B3LYP and other popular DFT functionals. Org. Lett. 2006;8(17):3631–3634. doi: 10.1021/ol061016i. [DOI] [PubMed] [Google Scholar]
- 32.Laskowski, A. B., & Swindells, M. B. (2011). LigPlot+: Multiple ligand–protein interaction diagrams for drug discovery. Journal of chemical information and modeling 2011 51 (10), 2778 2786. [DOI] [PubMed]
- 33.Rasulev B.F., Abdullaev N.D., Syrov V.N., Leszczynski J. A quantitative structure-activity relationship (QSAR) study of the antioxidant activity of flavonoids. QSAR Comb. Sci. 2005;24(9):1056–1065. [Google Scholar]
- 34.R. Todeschini, V. Consonni, DRAGON software for the Calculation of Molecular Descriptors, web version 3.0 for Windows 2003.
- 35.Petrescu A.-M., Putz M.V., Ilia G. Quantitative structure–activity/ecotoxicity relationships (QSAR/QEcoSAR) of a series of phosphonates. Environ. Toxicol. Pharmacol. 2015;40(3):800–824. doi: 10.1016/j.etap.2015.08.032. [DOI] [PubMed] [Google Scholar]
- 36.Mebi C.A. DFT study on structure, electronic properties, and reactivity of cis-isomers of [(NC 5 H 4-S) 2 Fe (CO) 2] J. Chem. Sci. 2011;123(5):727–731. [Google Scholar]
- 37.L. Davis N.Y. Van Nostrand Reinhold Handbook of Genetic Algorithms 1991 (USA).
- 38.Devillers J. Academic Press Ltd; London: 1996. Genetic Algorithms in Molecular Modeling. [Google Scholar]
- 39.Asadpour-Zeynali K., Jalili-Jahani N. Modeling GC-ECD retention times of pentafluorobenzyl derivatives of phenol by using artificial neural networks. J. Sep. Sci. 2008;31(21):3788–3795. doi: 10.1002/jssc.200800418. [DOI] [PubMed] [Google Scholar]
- 40.Jalali-Heravi M., Mani-Varnosfaderani A. QSAR modeling of 1-(3, 3-diphenylpropyl)-piperidinyl amides as CCR5 modulators using multivariate adaptive regression spline and bayesian regularized genetic neural networks. QSAR Comb. Sci. 2009;28(9):946–958. [Google Scholar]
- 41.Gholivand K., Mohammadpanah F., Pooyan M., Valmoozi A.A.E., Sharifi M., Mani-Varnosfaderani A., Hosseini Z. Synthesis, crystal structure, insecticidal activities, molecular docking and QSAR studies of some new phospho guanidines and phospho pyrazines as cholinesterase inhibitors. Pestic. Biochem. Physiol. 2019;157:122–137. doi: 10.1016/j.pestbp.2019.03.010. [DOI] [PubMed] [Google Scholar]
- 42.Todeschini, R., & Consonni, V. (2009). Molecular descriptors for chemoinformatics: volume I: alphabetical listing/volume II: appendices, references (Vol. 41). John Wiley & Sons.
- 43.Ghorbanzadeh M., Fatemi M.H., Karimpour M. Modeling the cellular uptake of magnetofluorescent nanoparticles in pancreatic cancer cells: a quantitative structure activity relationship study. Ind. Eng. Chem. Res. 2012;51(32):10712–10718. [Google Scholar]
- 44.Gholivand K., Mohammadpanah F., Pooyan M., Roohzadeh R. Evaluating anti-coronavirus activity of some phosphoramides and their influencing inhibitory factors using molecular docking, DFT, QSAR, and NCI-RDG studies. J. Mol. Struct. 2022;1248 doi: 10.1016/j.molstruc.2021.131481. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.