Abstract
RNA-binding motif 5 (RBM5) is a pro-death tumor suppressor gene in cancer cells. It remains to be determined if it is neurotoxic in the brain or rather if it plays a fundamentally different role in the central nervous system (CNS). Brain-specific RBM5 knockout (KO) mice were given a controlled cortical impact (CCI) traumatic brain injury (TBI). Markers of acute cellular damage and repair were measured in hippocampal homogenates 48 h post-CCI. Hippocampal CA1/CA3 cell counts were assessed 7 days post-CCI to determine if early changes in injury markers were associated with histological outcome. No genotype-dependent differences were found in the levels of apoptotic markers (caspase 3, caspase 6, and caspase 9). However, KO females had a paradoxical increase in markers of pro-death calpain activation (145/150-spectrin and breakdown products [SBDP]) and in DNA repair/survival markers. (pH2A.x and pCREB). CCI-injured male KOs had a significant increase in phosphorylated calcium/calmodulin-dependent protein kinase II (pCaMKII). Despite sex/genotype-dependent differences in KOs in the levels of acute cell signaling targets involved in cell death pathways, 7 day hippocampal neuronal survival did not differ from that of wild types (WTs). Similarly, no differences in astrogliosis were observed. Finally, gene analysis revealed increased estrogen receptor α (ERα) levels in the KO hippocampus in females and may suggest a novel mechanism to explain sex-dimorphic effects on cell signaling. In summary, RBM5 inhibition did not affect hippocampal survival after a TBI in vivo but did modify targets involved in neural signal transduction/Ca2+ signaling pathways. Findings here support the view that RBM5 may serve a purpose in the CNS that is dissimilar from its traditional pro-death role in cancer.
Keywords: gender, RBM5, RNA binding protein, sexual dimorphism, trauma
Introduction
The nuclear splicing factor RNA binding motif 5 (RBM5) regulates gene expression and exon definition of select mRNAs.1 The biological role of RBM5 has been studied primarily in cancer, where it promotes cell death and inhibits metastasis.2–6 Reports on RBM5-regulated mRNAs in non-cancerous tissues of the body identified genes not previously seen in tumors.7,8 In normal mouse spermatids, RBM5 inhibition altered the expression of genes involved in actin-binding, microtubule-cytoskeleton homeostasis, and endocytosis.7 This resulted in reproductive dysfunction but only in males, indicating for the first-time the possibility that RBM5 mediates sex-dimorphic effects in animals.7 In the normal brain cortex, we showed that RBM5 inhibition by gene knockout (KO) altered the expression of Csnka2ip, Gm756, Serpina3n, and glial fibrillary acidic protein (GFAP).8 Moreover, KO altered cassette exon splicing of regulating synaptic membrane exocytosis 2 (RIMS2), a gene involved in the regulation of voltage-gated Ca2+ channels and pre-synaptic neurotransmission.8,9 Finally, Rab4 levels (a mediator of endocytosis) was increased in RBM5 knockdown rat neurons in vitro and resulted in serotonin transporter downregulation in the membrane.10
On the other hand, central nervous system (CNS) culture studies support a traditional pro-death role of RBM5 in neuronal cell death. RBM5 knockdown in neuronal-like SH-SY5Y and PC12 cells decreased caspase activation following injury with the chemical toxin staurosporine, or hydrogen peroxide, respectively.11,12 Conversely, RBM5 overexpression in primary rat cortical neurons exacerbated damage after mechanical stretch injury.13 Thus, RBM5 likely plays multiple roles in CNS cells including pro-death functions and modulating aspects of neural signal transduction. To better understand if RBM5 inhibition alters cell death signaling and/or neuronal viability in vivo, here we subjected male/female brain-specific RBM5 wild types (WTs) versus KOs to a controlled cortical impact (CCI) traumatic brain injury (TBI) and assayed hippocampal homogenates across a variety of cell-injury markers at 48 h post-injury, and measured CA1/CA3 neuronal cell counts and GFAP staining at 7 days post-injury.
Methods
Reagents
PVDF membrane:
-
0.
2μM pore size, Cat# SVFX8301XXXX101, Lot# VA760606L (MDI Technologies; Harrisburgh, PA, USA)
Antibodies:
Cell Signaling Technology (Danvers, MA, USA): Anti-Phospho-Histone H2A.X (Ser139) , Cat# 9718S; Anti-H2A.X Total, Cat# 2595S; Anti-Phospho-CaMKII (Thr286/287), Cat# 3361S; Anti-pan CaMKII Total, Cat# 3362S; Anti-Phospho-CREB (Ser133), Cat# 9198S; Anti-CREB Total, Cat# 9197S.
Abcam (Cambridge, MA, USA): Anti-Caspase-9, Cat# ab184786; Anti-Caspase-3, Cat# ab184787; Anti-Caspase-6, Cat# ab185645; Anti-Estrogen Receptor Alpha, Cat# ab241557 (Clone CRET9D) - created and validated in ERα KO and Erβ KO mice by the vendor.
Proteintech (Chicago, IL, USA): Anti-RBM5, Cat# 19930-1-AP.
Sigma (St. Louis, MO, USA): Anti-RBM10, Cat# HPA034972.
Enzo Life Sciences (Farmingdale, NY, USA): Anti-α-II-Spectrin and breakdown products (SBDPs), Cat# BML-FG6090.
Animals
Studies were approved by the institutional animal care and use committee (IACUC) of the University of Pittsburgh. Tissue analysis at the University of South Florida was approved by a material transfer agreement. Mice were maintained on a 12 h light/dark cycle. Conditional RBM5 KOs have been previously described.8 Heterozygous male/female Rbm5tm1Ozg mice were crossed to generate homozygous Rbm5tm1Ozg mice. F1 colonies were generated by crossing homozygous Rbm5tm1Ozg mice with hemizygous B6.Cg-Tg(Nes-cre)1Kln/J mice. F1 heterozygous Rbm5tm1Ozg(-CRE) were bred with F1 heterozygous Rbm5tm1Ozg (+CRE) to generate F2 littermates for experiments. Mice were randomized to injury at the age that they became available.
Genotyping
Tail snips were harvested and processed via the Promega Wizard SV Genomic Purification System (Promega; Madison, WI, USA). Polymerase chain reactions (PCRs) were run on a Biorad T100 Thermalcycler (BioRad; Hercules, CA, USA). Samples were loaded onto 3% agarose gels with Syber Safe Stain (ThermoFisher Scientific; Waltham, MA, USA). Primer pairs were previously reported.8
CCI-TBI
Adult 12–15-week-old male/female Cre+ RBM5 WTs, heterozygous (Hets), and KOs were used for experiments. In brief, mice were anesthetized with isoflurane (4% induction/2% maintenance) in nitrous oxide (N2O)/%O2 (2:1). Shams received surgical preparations without craniotomy. Injured mice were randomized to a CCI with a pneumatic impactor device at the insult level of 6 m/sec and 1.2 mm depth. Forty ipsilateral hippocampi were homogenized for protein extracts for Western blot analysis of cell death signaling targets (n = 10 WT-Sham, n = 10 WT-CCI, n = 10 KO-Sham, and n = 10 KO-CCI). Each group contained five males and five females. A total of 14 ipsilateral cortices were used for RNA extraction to use with the 84 gene RT2 Cell Death Profiler Array (n = 7 WT-CCI and n = 7 KO-CCI). To confirm increased ESR1 at the protein level, 20 contralateral hippocampi and 20 contralateral cortices from WT/KO male and female CCI-injured mice were used for protein extracts. A separate cohort of 60 mice were used for histological analysis of hippocampal CA1/CA3 neuronal counts (n = 10 WT-Sham, n = 10 Het-Sham, n = 10 KO-Sham, n = 10 WT-CCI, n = 10 Het-CCI, and n = 10 KO-CCI). Each group contained five males and five females.
Western blot
Western blot was performed using our published methodology.14,15 Brain tissues were homogenized with a Bead Mill 24 (Fisher Scientific) set to 5.2 m/sec (∼ 5500 rpm) at two cycles, with a 5 min on-ice interval, to generate equivalent homogenates. Samples (20–30 μg protein/well) were loaded onto 26-well Criterion TGX gels and electrophoresed with a Criterion Cell at 200V (Biorad). Protein was transferred to PVDF membranes with a plate-electrode Criterion Blotter (Biorad) and run 40 min/100 V/4°C. Membranes were stained with Reversible Swift Membrane Stain (Fisher Scientific) and scanned (600 dpi) for densitometry. Blots were incubated with primary/secondary antibodies and imaged on a 9.1 MP iBright™ CL1500 Imaging System (Fisher Scientific).
Histology
Histology was assessed as described by our group in the CCI model:16,17 3 mm coronal brain sections were formalin-fixed paraffin-embedded (FFPE) and cut using a microtome (5 μM sections). Sections were mounted on glass slides and stained with Hematoxylin and Eosin (H&E; Thermo Scientific – Shandon; Pittsburgh, PA, USA) or anti-GFAP antibody. Images were captured on a Nikon Eclipse 90i microscope (Nikon; Melville, NY, USA). Live cells (CA1 and CA3) in the stratum pyramidale hippocampus were counted, divided by the length of the CA1/CA3, and reported as cells/0.1 mm. Analysis of H&E and neuronal nuclear protein (NeuN) was performed using NIS Elements Software. The technician quantifying CA1/CA3 cell counts was blind to genotype/injury. Whole field analysis of GFAP staining intensity with ImageJ was performed as previously described, with minor modifications.8
RNA Isolation
Cortices were homogenized in Trizol® and incubated 15 min with chloroform (1:5 volume) at room temperature. Samples were centrifugated 15 min/12,000g. The aqueous phase was isolated and RNA precipitated with 1:1 volume of isopropanol. The RNA pellet was washed, air dried, and re-suspended in nuclease-free water. RNA concentration/purity were validated by Nanodrop (ThermoFisher Scientific). RNA integrity was confirmed by Tape Station® 4200 (Agilent; Santa Clara, CA, USA). The eRIN was >8.4 for all samples.
Reverse transcription (RT)2 cell death profiler PCR array
RT was performed on 400 ng of gDNA-free, total RNA with the RT2 easy 1st strand kit (Qiagen; Germantown, MD, USA). cDNA was diluted in 1X RT3 SYBR Green buffer + ROX to 3 pg/mL; 10 μL of mastermix + cDNA solution was added to 384-well assay plates per Format E manufacturer instructions. Each 384 well plate had equal group representation to minimize batch effects. Thermocyling was performed on an AB7900HT standard block with enzyme activation at 95°C for 10 min followed by 40 cycles of 15 sec at 95°C and 1 min at 60°C. Raw Ct values were determined using SDS 2.4 (ThermoFisher) with automatic baseline and manual threshold settings.
Statistical analysis
Densitometry of (1) total protein stain (TPS)/membrane and (2) targets of interest were measured with UN-SCAN-IT software (Silk Scientific; Orem, UT, USA). Target values were standardized by dividing each data point by the densitometry of the TPS corresponding to the target's lane.14,15 Standardized densitometic values within each blot were then normalized by setting the highest value in each blot to 100% (i.e., for across blot pooling of data). The 40 hippocampal ipsilateral extracts for cell death signaling analysis were split across two 26-well criterion gels to ensure equal representation of genotype, sex, and injury on each blot. Data were analyzed using a three-way analysis of variance (ANOVA) and Tukey's multiple comparison post-hoc test. Data were expressed as the relative difference in target expression. Cell counts were obtained and expressed as cells/0.1 mm. Data were analyzed using a two-way ANOVA and Tukey's multiple comparison test. Data were significant at p < 0.05. Histological data from H&E and NeuN were also analyzed via the Bland–Altman method for estimation of bias. Quantitative PCR (qPCR) data were analyzed in GeneGlobe Data Analysis Center (https://geneglobe.qiagen.com/us/analyze). Expression levels were normalized using the average geometric mean of housekeeping controls Atg12, Atp6v1g2, and Bcl2a1a. Fold regulation was reported based on the ΔΔCT method. Uncorrected p values were calculated based on a Student's t test of the replicate 2^(-ΔCT) values for each gene in the control versus the treatment group(s). Gene changes were considered significant if: (1) p < 0.05 and (2) fold change was <-2 or 2>.
Results
Protein markers of cell death, cell survival, and Ca2+-signaling mediators in CCI-Injured RBM5 WT versus KOs
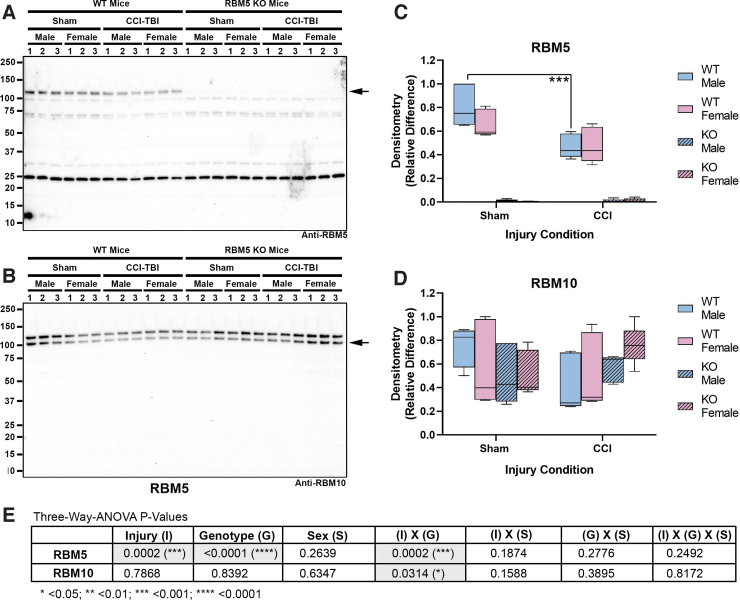
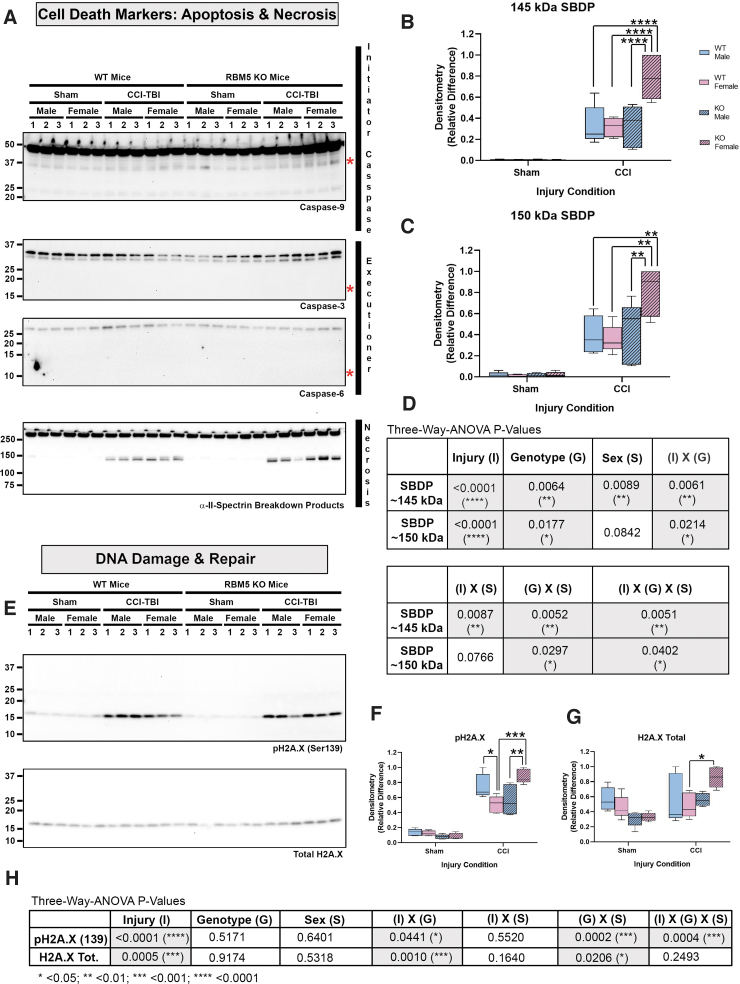
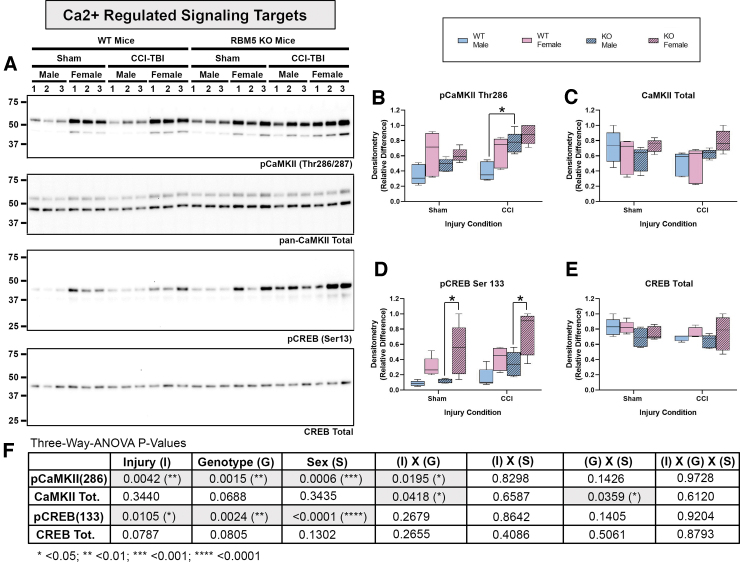
RBM5 KO was confirmed by Western blot analysis. Hippocampal RBM5 migrated at 120 kDa on sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE) and decreased in male WT mice 48 h post-injury (Fig. 1A and C and Supplementary Fig. S1C). No genotype differences in the gene paralogue RBM10 were observed (Fig. 1B and D and Fig. S1D). Next, markers of apoptosis and DNA damage were measured in hippocampal extracts (Fig. 2). Cleaved caspase 3 and caspase 6 were below the level of detection and did not increase after a CCI (Fig. 2A and Supplementary Fig. S2). Cleaved caspase 9 (∼ 35 kDa) was detectable at baseline but did not increase further after a CCI (Fig. 2A and Fig. S2). Antibody validation studies in staurosporine-treated mouse neurons supported the fidelity of anti-caspase antibodies (Supplementary Fig. S3). In contrast, necrosis-associated SBDPs were significantly increased 48 h after CCI in WTs and KOs. Moreover, injured female KOs had significantly higher SBDP levels (Fig. 2A–D and Fig. S2G). Changes in the levels of the DNA damage and repair marker pH2A.X was also sex and genotype dependent. In WTs, pH2A.X levels were significantly higher in injured males (Fig. 2E, F and Fig. S2H). In KOs, pH2A.X levels were significantly higher in injured females (Fig. 2E, F, and H and Fig. S2H). Further, injured KO females had a significant increase in total H2A.X levels compared with injured WT females (Fig. 2E, G, and H and Fig. S2I). Finally, phosphorylation (activation) levels of key Ca2+-regulated signaling effectors that influence neuronal survival were explored. Phosphorylated calcium/calmodulin-dependent protein kinase II (pCaMKII) levels were significantly altered by injury, genotype, and sex (Fig. 3A, B, and F and Supplementary Fig. S4B). Also, injured male KOs had significantly increased pCaMKII levels compared with injured WT males (Fig. 3B and Fig. S4B). Phosphorylated CREB levels were also significantly affected by injury, genotype, and sex (Fig. 3A, D, and F and Fig. S4F). Female sham or injured KOs had significantly increased pCREB levels compared with male sham or injured KOs, respectively (Fig. 3A and D and Fig. S4F).
FIG. 1.
Hippocampal RNA-binding motif 5 (RBM5) and RBM10 levels in sham versus controlled cortical impact (CCI)-injured mice. (A) Representative blot of hippocampal RBM5 levels in sham versus CCI male/female wild type (WT) and knockout (KO) (n = 3/group). (B) Representative blot of hippocampal RBM10 levels in sham versus CCI male/female WT and KO (n = 3/group). (C and D) Densitometry values (n = 5/group), normalized to total protein loading, were analyzed by three-way analysis of variance (ANOVA) and Tukey's Multiple Comparison Test. Data were significant at p < 0.05. Box plots show minimum, maximum, interquartile range (IQR), and median. Color image is available online.
FIG. 2.
Hippocampal levels of apoptosis, necrosis, and DNA damage markers in sham vs. controlled cortical impact (CCI)-injured mice. (A) Representative blots of hippocampal caspase-9, caspase-3, caspase-6, and total and cleaved α-II-spectrin breakdown products (SBDPs) in sham versus CCI male/female wild type (WT) and knockout (KO) (n = 3/group). Red asterisks indicate the predicted kDa of caspase cleavage products for each antibody (B and C) Densitometric analysis (n = 5/group) of calpain-cleaved 145 kDa and 150 kda SBDPs, normalized to total protein loading, and analyzed by three-way analysis of variance (ANOVA) and Tukey's Multiple Comparison Test. Data were significant at p < 0.05. Box plots show minimum, maximum, interquartile range (IQR), and median. (D) Table shows p values for the main effects and interactions on SBDP levels. (E) Representative blot of hippocampal total and phosphorylated H2A.X (pH2A.X Ser139) in sham versus CCI male/female WT and KO (n = 3/group). (F and G) Densitometric analysis (n = 5/group) of pH2A.X and total H2A.X, normalized to total protein loading, and analyzed by three-way ANOVA and Tukey's Multiple Comparison Test. Data were significant at p < 0.05. Box plots show minimum, maximum, IQR, and median. (H) Table shows p values for the main effects and interactions on pH2A.X and H2A.X total levels. Asterisks in the graphs indicate post-hoc significance (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001). Color image is available online.
FIG. 3.
Hippocampal levels of Ca2+ pathway signaling markers in sham versus controlled cortical impact (CCI)-injured mice. (A) Representative blots of hippocampal total and phosphorylated calcium/calmodulin-dependent protein kinase II (CaMKII), and total and phosphorylated cAMP-response element binding protein (CREB) in sham versus CCI male/female wild type (WT) and knockout (KO) (n = 3/group). (B–E) Densitometric analysis (n = 5/group) of pCaMKII Thr286, total CaMKII, CREB Ser133, and total CREB, normalized to total protein loading, and analyzed by three-way analysis of variance (ANOVA) and Tukey's Multiple Comparison Test. Data were significant at p < 0.05. Box plots show minimum, maximum, interquartile range (IQR), and median. (F) Table shows p values for the main effects and interactions on the levels of Ca2+ pathway targets. Asterisks in the graphs indicate post-hoc significance (*p < 0.05). Color image is available online.
Histological analysis of neuropathology in CCI-injured RBM5 WT versus KOs
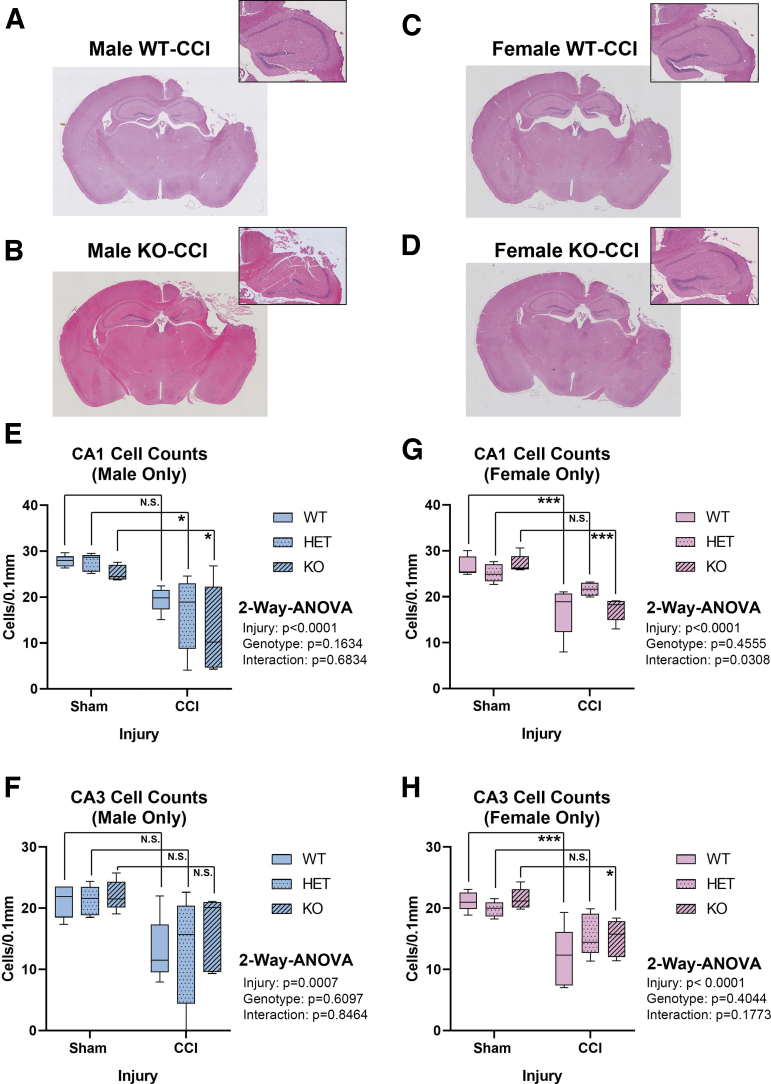
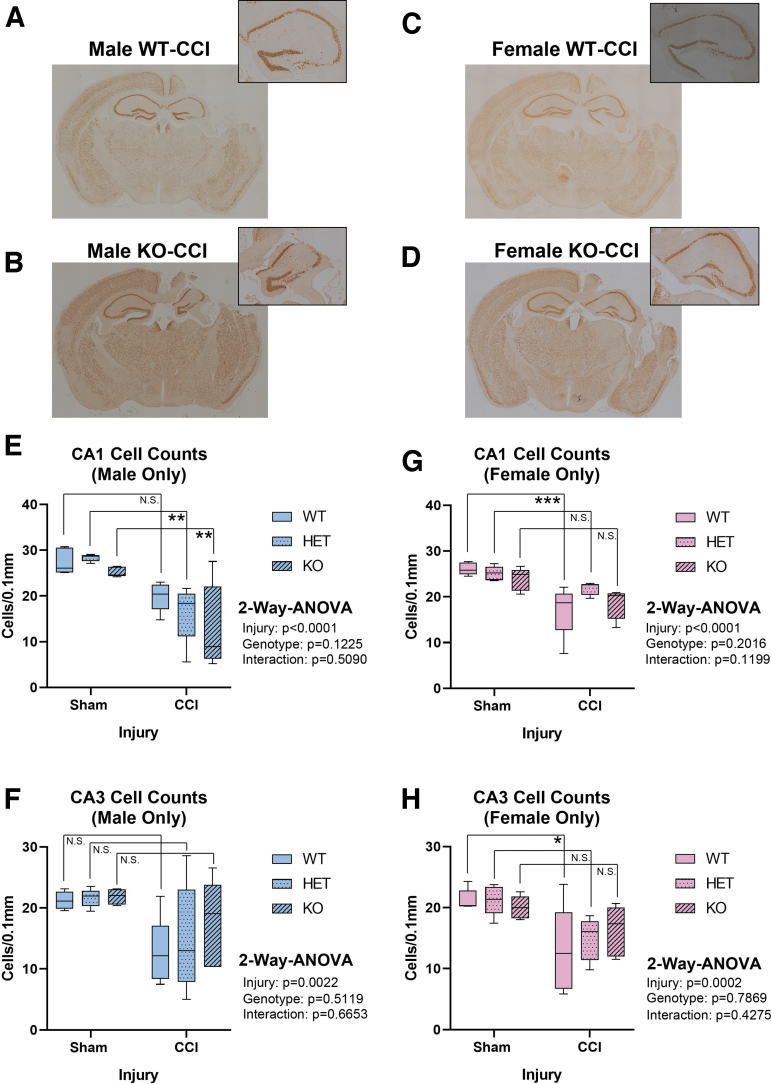
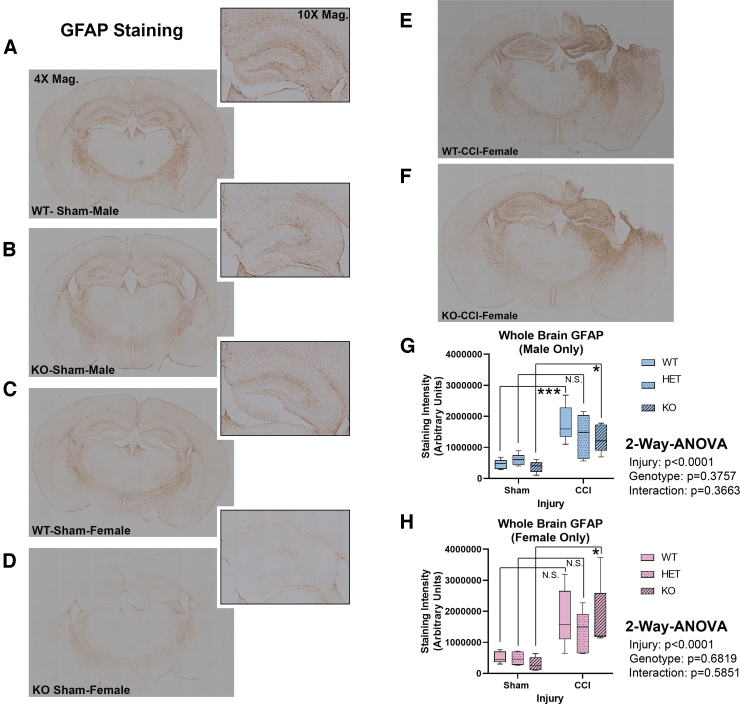
Hippocampal CA1/CA3 total cell counts were quantified at 7 days post-injury using H&E stain (Fig. 4A–D). Extensive damage was seen in the ipsilateral hemisphere – consistent with TBI phenotypes produced by the CCI model. Hemizygous or complete RBM5 KO did not increase CA1/CA3 total cell counts 7 days post-CCI in males (Fig 4E and F) or in females (Fig 4G and H). Next, we verified H&E results by NeuN-positive counts, and specifically to confirm the effect of KO on hippocampal CA1/CA3 neurons. NeuN counts supported H&E findings (Fig. 5). Both staining approaches showed low bias and high agreement (Supplementary Fig. S5). Finally, GFAP staining was performed to determine the extent of astrogliosis in CCI-injured WT compared with KOs. GFAP staining increased with CCI injury but was not significantly affected by genotype (Fig. 6).
FIG. 4.
Seven day Hematoxylin and Eosin (H&E) post-injury hippocampal CA1 and CA3 sub-field total cell counts in sham versus controlled cortical impact (CCI)-injured mice. (A–D) Representative H&E-stained whole brain sections (4 × magnification) showing insult severity in male/female wild type (WT) and knockout (KO). Magnified (10 × ) images of the hippocampus are shown at the top right. Quantification of CA1 and CA3 cell counts in males (E and F) and (G and H) females across all genotypes (n = 5/group) and analyzed by two-way analysis of variance (ANOVA) and Tukey's Multiple Comparison Test. Data were significant at p < 0.05. Box plots show minimum, maximum, interquartile range (IQR), and median. Asterisks indicate post-hoc significance (*p < 0.05, ***p < 0.001, N.S., not significant). Color image is available online.
FIG. 5.
Seven day neuronal nuclear protein (NeuN) post-injury hippocampal CA1 and CA3 sub-field neuronal counts in sham versus controlled cortical impact (CCI)-injured mice. (A–D) Representative NeuN-stained whole brain sections (4 × magnification) showing insult severity in male/female wild type (WT) and knockout (KO). Magnified (10 × ) images of the hippocampus are shown at the top right. Quantification of CA1 and CA3 neuronal counts in males (E and F) and females (G and H) across all genotypes (n = 5/group) and analyzed by two-way analysis of variance (ANOVA) and Tukey's Multiple Comparison Test. Data were significant at p < 0.05. Box plots show minimum, maximum, interquartile range (IQR), and median. Asterisks indicate post-hoc significance (*p < 0.05, **p < 0.01, ***p < 0.001, N.S., not significant). Color image is available online.
FIG. 6.
Seven day glial fibrillary acidic protein (GFAP) levels in sham versus controlled cortical impact (CCI)-injured mice. (A–D) Representative whole brain sections (4 × magnification) showing 7 day GFAP staining in male/female wild type (WT) and knockout (KO). Magnified (10 × ) images of the hippocampus are shown at the top right of each representative image (E and F) Representative 7 day GFAP staining in female WT versus KO brain sections. (G and H) ImageJ quantification of GFAP staining in whole brain sections from male/female sham versus CCI-injured mice across all genotypes (n = 5/group). Staining intensity was analyzed by two-way analysis of variance (ANOVA) and Tukey's Multiple Comparison Test. Data were significant at p < 0.05. Box plots show minimum, maximum, interquartile range (IQR), and median. Asterisks in the graphs indicate post-hoc significance (*p < 0.05, ***p < 0.001, N.S., not significant). Color image is available online.
Increased estrogen receptor levels in female RBM5 KOs
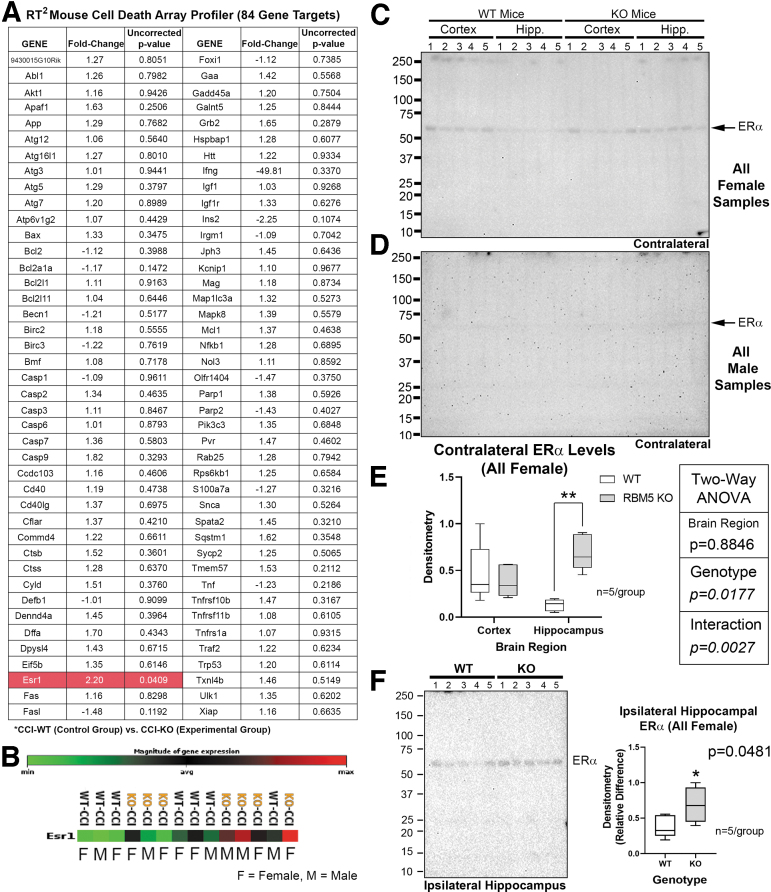
We utilized the quantitative 84 gene RT2 Cell Death Profiler Array to further characterize cell death genes/pathways potentially altered by RBM5 KO. Only the ESR1 (ERα) gene was significantly increased in ipsilateral cortex in injured KOs vs. WTs (Fig. 7A and B). Western blots of the contralateral cortex and hippocampus in injured KOs versus WTs was performed to validate increased ESR1 at the protein level. ERα was only detectable in females and significantly increased in the hippocampus of KOs (Fig. 7C–E, and Supplementary Fig. S6). We further confirmed that ERα levels in the injured hippocampus were significantly increased in female KOs compared with female WTs (Fig. 7F). Finally, we confirmed increased ERα levels in the ipsilateral hippocampus of injured KO males, but levels in KO females remained much higher (Supplementary Fig. S7).
FIG. 7.
Increased hippocampal estrogen receptor α (ERα/ESR1) levels in female knockouts (KOs.) (A) Quantitative reverse transcription polymerase chain reaction (RT-PCR) analysis of 84 cell death gene targets in the contused ipsilateral cortex of male/female wild type (WT) versus KO controlled cortical impact (CCI)-injured mice. Only ERα (highlighted red in table) was significantly increased in KOs versus WTs. GeneGlobe generated p values are uncorrected. (B) Heat map of ERα expression levels in each of the 14 samples suggesting increased levels in both male and female KOs at the mRNA level. (C and E) Images of the blot and densitometric analysis (n = 5/group) of ERα levels in the contralateral cortex and hippocampus in CCI-injured females (WT-CCI vs. KO-CCI). (D) Images of the blot of ERα levels in the contralateral cortex and hippocampus in CCI-injured males (WT-sham vs. KO-sham). No signals were detected in males. (E) Densitometric analysis (n = 5/group) of ERα levels in the contralateral cortex and hippocampus in CCI-injured females (WT vs. KO). Signals were normalized to total protein loading, and analyzed by two-way analysis of variance (ANOVA) and Tukey's Multiple Comparison Test. (F) Blot and densitometric analysis (n = 5/group) of ERα levels in the ipsilateral hippocampus in CCI-injured females (WT vs. KO). Signals were normalized to total protein loading, and analyzed by an unpaired students t test. Data were significant at p < 0.05. Box plots show minimum, maximum, interquartile range (IQR), and median. Asterisks in the graphs indicate post-hoc significance (*p < 0.05, **p < 0.01). Color image is available online.
Discussion
Neuronal survival after TBI in RBM5 KOs
The hippocampus is a key investigational target in CCI.18,19 We hypothesized that RBM5 in KO would protect the hippocampus.11–13 However, 7 day neuronal death and astrogliosis was no different in WTs than in KOs. Increased data dispersion with an overall lower median for hippocampal cell counts in KO versus WT males suggested worse outcome, which agrees with the only other study that assessed RBM5 inhibition on viability in normal (non-cancerous) tissue in vivo and found increased death of primary spermatids in males.7 Also, CCI injured female KOs (but not male KOs) had increased calpain cleavage products (145/150 SBDP), a classic marker of necrosis. It is possible that increased necrosis in female KOs resulted from a shift in cell death pathways. However, apoptotic markers, including cleaved/activated caspase-3, caspase-6, and caspase-9, did not differ in KOs and WTs. In addition, although tumor necrosis factor (TNF)α plays a role in mediating RBM5-induced cell death in cancer cells, we did not observe altered levels of TNFα mRNA among the 84 cell death genes analyzed in injured KOs (Fig. 7).20 There are more than a dozen cell death mechanisms and it is possible that other processes not investigated here were altered in Kos.21 An alternative hypothesis of altered Ca2+ homeostasis might explain increased SBDP levels in female KOs, which will be discussed next.
Increased levels of calcium-activated targets in KOs
In hippocampal neurons, the post-excitotoxic increase in 145/150 SBDP levels is triggered by the initial Ca2+ surge and μ-calpain activation.22–24 Therefore, another construct is that the initial Ca2+ surge in injured female KOs was greater either in magnitude or in duration, and led to increased SBDP levels. Relatedly, we reported that RBM5 knockdown in rat cortical neurons in vitro, and from KO in brain cortices ex vivo, produced a limited number of gene changes enriched for targets involved in neurotransmission.8,10 Here we analyzed several Ca2+-regulated targets known to modulate cell death. Ca2+/CaMKII is a prototypical Ca2+-activated effector of neural signaling but it also promotes neuroprotection or neurotoxicity.25,26 Hippocampal pCaMKII (Thr286) levels were significantly increased in injured male KOs versus injured male WTs. Female KOs had a similar increase but this did not reach significance, possibly because of a ceiling effect. We also measured the neuroprotectant cAMP response element-binding protein (pCREB), which is activated by synaptic Ca2+ in neurons.27,28 Female KOs (but not male KOs) had augmented pCREB levels in shams and in CCI-injured mice. Future studies are needed to measure real-time calcium influx in glutamate-treated KO neurons. Cell signaling changes in the brain in KOs likely result from direct RBM5 inhibition in CNS cells; however, Nestin-Cre mice have scattered expression of Cre in a number of peripheral organs. Therefore, we cannot rule out the possibility that peripheral effects play a role.29
RBM5 is a novel gene target in the study of sexual dimorphism in mammals
O'Bryan and colleagues first reported on the sex-dimorphic effects of RBM5 gene disruption.7 We observed robust sex-dependent differences in hippocampal levels of SBDPs, pH2A.X, H2A.X total, pCaMKII, and pCREB. In light of these findings, it is essential that sex as a variable be considered in future RBM5 studies.
We also found that female KOs had a modest increase in the levels of hippocampal ERα. Given that females were not ovexed, it is possible that a novel RBM5/ERα/estrogen signaling axis mediated some of the sex differences seen on cell signaling targets analyzed here. Consistent with that possibility, estrogens potently increase hippocampal pCREB levels in mice.30,31 Whether increased estrogen signaling in the brain could have provided transgenic females a neuroprotective advantage underpowered to detect here remains to be determined, but merits exploration.32,33
Increased pCREB in female KOs may also have implications for neuro-oncology. Constitutive CREB activation strongly correlates with tumor grade in glioblastoma.34 A recent Genome-Wide Association Study (GWAS) on genetic risk factors for gliomas identified a novel female-specific risk locus in the 3p21.31 region.35 The ∼200 kb risk locus (49400kb–49600kb) is close to the RBM5 gene locus (50088.919–50119.021kb), which is also found in the 3p21.3 region.36,37 Future studies are warranted to explore if women have a higher incidence of certain cancers in which RBM5 is downregulated.
Limitations
The small group size for histological assessment was the result of three factors. (1) Our breeding scheme ensured one copy of the CRE transgene in mice, but resulted in culling approximately half of F2 litters for the absence of CRE expression.38 (2) Male/female CRE+KOs were born at a lower-than-expected Mendelian ratio (∼ 7% based on data from ∼119 F2 litters across a ∼3 year period). The combined consequence was ∼35 KOs per 1000 genotyped pups. (3) Finally, the robust sex-dependent differences on molecular findings precluded combining male/female histology data to increase statistical power. Two alternative mouse strains to study RBM5 inhibition in vivo also have drawbacks. In one strain males are sterile.7 The second strain produced post-natal death of homozygotes.39
We chose the 48 h time point for our cell signaling analysis because, in a different mouse model of TBI with a second insult, hippocampal RBM5 levels and caspase-cleavage products increased at the 48 h time point.11 And other reports have demonstrated an exacerbation of neuronal death by apoptosis by the addition of second insults after CCI.40 Nevertheless, additional studies in CCI without a second insult merit investigation, as caspase activation may be more robust at earlier time points and provide a better environment to test if RBM5 KO inhibits apoptotic pathways, which was not addressed here. RBM5 gene KO also needs to be explored in other models of brain injury in which the threshold for neuroprotection may differ. Finally, future studies are warranted to characterize RBM5 KO in alternative CRE strains. In summary, despite challenges using genetic approaches to study RBM5 inhibition in vivo, this is the first study to successfully investigate the effect of homozygous RBM5 deletion on neuronal survival in a model of brain injury in males and in females.
Supplementary Material
Acknowledgments
We are grateful to the University of Pittsburgh Genomics Core for assistance in RNA studies.
Authors' Contributions
T.C.J. conceived the study. T.C.J. and P.M.K. contributed to the study design. J.F., K.J.F., V.A.V., and K.S. contributed to experiments and data acquisition. T.C.J., P.M.K., and J.F. contributed to data analysis. T.C.J. and J.F. drafted the manuscript. P.M.K., K.F.J., K.S., and V.A.V. edited the draft. All authors approved the final submitted version of the manuscript.
Funding Information
This work was supported by National Institutes of Health (NIH)/National Institute of Neurological Disorders and Stroke (NINDS) grants R01NS105721 to T.C.J., by the University of South Florida Morsani College of Medicine start-up funds to T.C.J., and by the Ake N. Grenvik Chair in Critical Care Medicine to P.M.K.
Author Disclosure Statement
T.C.J. and P.M.K. are inventors of a USPTO patent (No. 9610266) titled: Small molecule inhibitors of RNA binding motif (RBM) proteins for the treatment of acute cellular injury. The other authors have nothing to disclose
Supplementary Material
References
- 1. Jackson, T.C., and Kochanek, P.M. (2020). RNA binding motif 5 (RBM5) in the CNS-moving beyond cancer to harness RNA splicing to mitigate the consequences of brain injury. Front. Mol. Neurosci. 13, 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Oh, J.J., Razfar, A., Delgado, I., Reed, R.A., Malkina, A., Boctor, B., and Slamon, D.J. (2006). 3p21.3 tumor suppressor gene H37/Luca15/RBM5 inhibits growth of human lung cancer cells through cell cycle arrest and apoptosis. Cancer Res. 66, 3419–3427. [DOI] [PubMed] [Google Scholar]
- 3. Fushimi, K., Ray, P., Kar, A., Wang, L., Sutherland, L.C., and Wu, J.Y. (2008). Up-regulation of the proapoptotic caspase 2 splicing isoform by a candidate tumor suppressor, RBM5. Proc. Natl. Acad. Sci. U. S. A. 105, 15,708–15,713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kobayashi, T., Ishida, J., Musashi, M., Ota, S., Yoshida, T., Shimizu, Y., Chuma, M., Kawakami, H., Asaka, M., Tanaka, J., Imamura, M., Kobayashi, M., Itoh, H., Edamatsu, H., Sutherland, L.C., and Brachmann, R.K. (2011). p53 transactivation is involved in the antiproliferative activity of the putative tumor suppressor RBM5. Int. J. Cancer 128, 304–318. [DOI] [PubMed] [Google Scholar]
- 5. Rintala-Maki, N.D., and Sutherland, L.C. (2004). LUCA-15/RBM5, a putative tumour suppressor, enhances multiple receptor-initiated death signals. Apoptosis 9, 475–484. [DOI] [PubMed] [Google Scholar]
- 6. Mourtada-Maarabouni, M., Sutherland, L.C., and Williams, G.T. (2002). Candidate tumour suppressor LUCA-15 can regulate multiple apoptotic pathways. Apoptosis 7, 421–432. [DOI] [PubMed] [Google Scholar]
- 7. O'Bryan, M.K., Clark, B.J., McLaughlin, E.A., D'Sylva, R.J., O'Donnell, L., Wilce, J.A., Sutherland, J., O'Connor, A.E., Whittle, B., Goodnow, C.C., Ormandy, C.J., and Jamsai, D. (2013). RBM5 is a male germ cell splicing factor and is required for spermatid differentiation and male fertility. PLoS Genet. 9, e1003628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jackson, T.C., Janesko-Feldman, K., Gorse, K., Vagni, V.A., Jackson, E.K., and Kochanek, P.M. (2020). Identification of novel targets of RBM5 in the healthy and injured brain. Neuroscience 440, 299–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kaeser, P.S., Deng, L., Fan, M., and Sudhof, T.C. (2012). RIM genes differentially contribute to organizing presynaptic release sites. Proc. Natl. Acad. Sci. U. S. A. 109, 11,830–11,835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jackson, T.C., Kotermanski, S.E., and Kochanek, P.M. (2017). Whole-transcriptome microarray analysis reveals regulation of Rab4 by RBM5 in neurons. Neuroscience 361, 93–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jackson, T.C., Du, L., Janesko-Feldman, K., Vagni, V.A., Dezfulian, C., Poloyac, S.M., Jackson, E.K., Clark, R.S., and Kochanek, P.M. (2015). The nuclear splicing factor RNA binding motif 5 promotes caspase activation in human neuronal cells, and increases after traumatic brain injury in mice. J. Cereb. Blood Flow Metab. 35, 655–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang, J., Cui, Z., Feng, G., Bao, G., Xu, G., Sun, Y., Wang, L., Chen, J., Jin, H., Liu, J., Yang, L., and Li, W. (2015). RBM5 and p53 expression after rat spinal cord injury: implications for neuronal apoptosis. Int. J. Biochem. Cell Biol. 60, 43–52. [DOI] [PubMed] [Google Scholar]
- 13. Jackson, T.C., Kotermanski, S.E., Jackson, E.K., and Kochanek, P.M. (2018). BrainPhys(R) increases neurofilament levels in CNS cultures, and facilitates investigation of axonal damage after a mechanical stretch-injury in vitro. Exp. Neurol. 300, 232–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jackson, T.C., Kotermanski, S.E., and Kochanek, P.M. (2018). Infants uniquely express high levels of RBM3 and other cold-adaptive neuroprotectant proteins in the human brain. Dev. Neurosci. 40, 325–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jackson, T.C., Gorse, K., Herrmann, J.R., and Kochanek, P.M. (2021). Hippocampal and prefrontal cortical brain tissue levels of irisin and GDF15 receptor subunits in children. Mol. Neurobiol. 58, 2145–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hemerka, J.N., Wu, X., Dixon, C.E., Garman, R.H., Exo, J.L., Shellington, D.K., Blasiole, B., Vagni, V.A., Janesko-Feldman, K., Xu, M., Wisniewski, S.R., Bayir, H., Jenkins, L.W., Clark, R.S., Tisherman, S.A., and Kochanek, P.M. (2012). Severe brief pressure-controlled hemorrhagic shock after traumatic brain injury exacerbates functional deficits and long-term neuropathological damage in mice. J. Neurotrauma 29, 2192–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jackson, T.C., Dixon, C.E., Janesko-Feldman, K., Vagni, V., Kotermanski, S.E., Jackson, E.K., and Kochanek, P.M. (2018). Acute physiology and neurologic outcomes after brain injury in SCOP/PHLPP1 KO mice. Sci. Rep. 8, 7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Frankowski, J.C., Kim, Y.J., and Hunt, R.F. (2019). Selective vulnerability of hippocampal interneurons to graded traumatic brain injury. Neurobiol. Dis. 129, 208–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Patel, A.D., Gerzanich, V., Geng, Z., and Simard, J.M. (2010). Glibenclamide reduces hippocampal injury and preserves rapid spatial learning in a model of traumatic brain injury. J. Neuropathol. Exp. Neurol. 69, 1177–1190. [DOI] [PubMed] [Google Scholar]
- 20. Wang, K., Bacon, M.L., Tessier, J.J., Rintala-Maki, N.D., Tang, V., and Sutherland, L.C. (2012). RBM10 modulates apoptosis and influences TNF-alpha gene expression. J. Cell Death 5, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fricker, M., Tolkovsky, A.M., Borutaite, V., Coleman, M., and Brown, G.C. (2018). Neuronal cell death. Physiol. Rev. 98, 813–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brustovetsky, T., Bolshakov, A., and Brustovetsky, N. (2010). Calpain activation and Na+/Ca2+ exchanger degradation occur downstream of calcium deregulation in hippocampal neurons exposed to excitotoxic glutamate. J. Neurosci. Res. 88, 1317–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pike, B.R., Flint, J., Dave, J.R., Lu, X.C., Wang, K.K., Tortella, F.C., and Hayes, R.L. (2004). Accumulation of calpain and caspase-3 proteolytic fragments of brain-derived alphaII-spectrin in cerebral spinal fluid after middle cerebral artery occlusion in rats. J. Cereb. Blood Flow Metab. 24, 98–106. [DOI] [PubMed] [Google Scholar]
- 24. Brophy, G.M., Pineda, J.A., Papa, L., Lewis, S.B., Valadka, A.B., Hannay, H.J., Heaton, S.C., Demery, J.A., Liu, M.C., Tepas, J.J., 3rd, Gabrielli, A., Robicsek, S., Wang, K.K., Robertson, C.S., and Hayes, R.L. (2009). alphaII-Spectrin breakdown product cerebrospinal fluid exposure metrics suggest differences in cellular injury mechanisms after severe traumatic brain injury. J. Neurotrauma 26, 471–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ashpole, N.M., and Hudmon, A. (2011). Excitotoxic neuroprotection and vulnerability with CaMKII inhibition. Mol. Cell Neurosci. 46, 720–730. [DOI] [PubMed] [Google Scholar]
- 26. Deng, G., Orfila, J.E., Dietz, R.M., Moreno-Garcia, M., Rodgers, K.M., Coultrap, S.J., Quillinan, N., Traystman, R.J., Bayer, K.U., and Herson, P.S. (2017). Autonomous CaMKII activity as a drug target for histological and functional neuroprotection after resuscitation from cardiac arrest. Cell Rep. 18, 1109–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kornhauser, J.M., Cowan, C.W., Shaywitz, A.J., Dolmetsch, R.E., Griffith, E.C., Hu, L.S., Haddad, C., Xia, Z., and Greenberg, M.E. (2002). CREB transcriptional activity in neurons is regulated by multiple, calcium-specific phosphorylation events. Neuron 34, 221–233. [DOI] [PubMed] [Google Scholar]
- 28. Yan, X., Liu, J., Ye, Z., Huang, J., He, F., Xiao, W., Hu, X., and Luo, Z. (2016). CaMKII-mediated CREB phosphorylation is involved in Ca2+-induced BDNF mRNA transcription and neurite outgrowth promoted by electrical stimulation. PLoS One 11, e0162784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Heffner, C.S., Herbert Pratt, C., Babiuk, R.P., Sharma, Y., Rockwood, S.F., Donahue, L.R., Eppig, J.T., and Murray, S.A. (2012). Supporting conditional mouse mutagenesis with a comprehensive cre characterization resource. Nat. Commun. 3, 1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Koss, W.A., and Frick, K.M. (2017). Sex differences in hippocampal function. J. Neurosci. Res. 95, 539–562. [DOI] [PubMed] [Google Scholar]
- 31. Abraham, I.M., and Herbison, A.E. (2005). Major sex differences in non-genomic estrogen actions on intracellular signaling in mouse brain in vivo. Neuroscience 131, 945–951. [DOI] [PubMed] [Google Scholar]
- 32. Day, N.L., Floyd, C.L., D'Alessandro, T.L., Hubbard, W.J., and Chaudry, I.H. (2013). 17beta-estradiol confers protection after traumatic brain injury in the rat and involves activation of G protein-coupled estrogen receptor 1. J. Neurotrauma 30, 1531–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Biegon, A. (2021). Considering biological sex in traumatic brain injury. Front. Neurol. 12, 576,366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Daniel, P., Filiz, G., Brown, D.V., Hollande, F., Gonzales, M., D'Abaco, G., Papalexis, N., Phillips, W.A., Malaterre, J., Ramsay, R.G., and Mantamadiotis, T. (2014). Selective CREB-dependent cyclin expression mediated by the PI3K and MAPK pathways supports glioma cell proliferation. Oncogenesis 3, e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ostrom, Q.T., Kinnersley, B., Wrensch, M.R., Eckel-Passow, J.E., Armstrong, G., Rice, T., Chen, Y., Wiencke, J.K., McCoy, L.S., Hansen, H.M., Amos, C.I., Bernstein, J.L., Claus, E.B., Il'yasova, D., Johansen, C., Lachance, D.H., Lai, R.K., Merrell, R.T., Olson, S.H., Sadetzki, S., Schildkraut, J.M., Shete, S., Rubin, J.B., Lathia, J.D., Berens, M.E., Andersson, U., Rajaraman, P., Chanock, S.J., Linet, M.S., Wang, Z., Yeager, M., GliomaScan, c., Houlston, R.S., Jenkins, R.B., Melin, B., Bondy, M.L., and Barnholtz-Sloan, J.S. (2018). Sex-specific glioma genome-wide association study identifies new risk locus at 3p21.31 in females, and finds sex-differences in risk at 8q24.21. Sci. Rep. 8, 7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wei, M.H., Latif, F., Bader, S., Kashuba, V., Chen, J.Y., Duh, F.M., Sekido, Y., Lee, C.C., Geil, L., Kuzmin, I., Zabarovsky, E., Klein, G., Zbar, B., Minna, J.D., and Lerman, M.I. (1996). Construction of a 600-kilobase cosmid clone contig and generation of a transcriptional map surrounding the lung cancer tumor suppressor gene (TSG) locus on human chromosome 3p21.3: progress toward the isolation of a lung cancer TSG. Cancer Res. 56, 1487–1492. [PubMed] [Google Scholar]
- 37. Timmer, T., Terpstra, P., van den Berg, A., Veldhuis, P.M., Ter Elst, A., Voutsinas, G., Hulsbeek, M.M., Draaijers, T.G., Looman, M.W., Kok, K., Naylor, S.L., and Buys, C.H. (1999). A comparison of genomic structures and expression patterns of two closely related flanking genes in a critical lung cancer region at 3p21.3. Eur. J. Hum. Genet. 7, 478–486. [DOI] [PubMed] [Google Scholar]
- 38. Forni, P.E., Scuoppo, C., Imayoshi, I., Taulli, R., Dastru, W., Sala, V., Betz, U.A., Muzzi, P., Martinuzzi, D., Vercelli, A.E., Kageyama, R., and Ponzetto, C. (2006). High levels of Cre expression in neuronal progenitors cause defects in brain development leading to microencephaly and hydrocephaly. J. Neurosci. 26, 9593–9602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jamsai, D., Watkins, D.N., O'Connor, A.E., Merriner, D.J., Gursoy, S., Bird, A.D., Kumar, B., Miller, A., Cole, T.J., Jenkins, B.J., and O'Bryan, M.K. (2017). In vivo evidence that RBM5 is a tumour suppressor in the lung. Sci. Rep. 7, 16323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Clark, R.S., Kochanek, P.M., Dixon, C.E., Chen, M., Marion, D.W., Heineman, S., DeKosky, S.T., and Graham, S.H. (1997). Early neuropathologic effects of mild or moderate hypoxemia after controlled cortical impact injury in rats. J. Neurotrauma 14, 179–189. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.