Abstract
Cardiac developmental disorders represent the most common of human birth defects, and anomalies in cardiomyocyte proliferation drive many of these disorders. This review highlights the molecular mechanisms of prenatal cardiac growth. Trabeculation represents the initial ventricular growth phase and is necessary for embryonic survival. Later in development, the bulk of the ventricular wall derives from the compaction process, yet the arrest of this process can still be compatible with life. Cardiomyocyte proliferation and growth form the basis of both trabeculation and compaction, and mouse models indicate that cardiomyocyte interactions with the surrounding environment are critical for these proliferative processes. The human genetics of left ventricular noncompaction cardiomyopathy suggest that cardiomyocyte cell-autonomous mechanisms contribute to the compaction process. Understanding the determinants of prenatal or early postnatal cardiomyocyte proliferation and growth provides critical information that identifies risk factors for cardiovascular disease, including heart failure and its associated complications of arrhythmias and thromboembolic events.
Keywords: cardiomyopathy, left ventricular noncompaction, myocardial development, trabeculation, noncompaction
INTRODUCTION
Cardiac Development and Specification of the Myocardium
Congenital heart defects occur in approximately 1% of all births and are the most common human birth defects (1). Cardiac structural abnormalities most frequently affect the valves, outflow tracts, and atrial and ventricular septae, often in combinations that define specific congenital cardiac syndromes. Defects in ventricular compaction, the process of ventricular growth and maturation, have recently been recognized as a cause of congenital heart disease (2). Initial reports of this cardiomyopathy, referred to as left ventricular noncompaction cardiomyopathy (LVNC), included patients with pronounced trabecular myocardium and thin compact myocardium, systolic heart failure, ventricular arrhythmia, and embolic stroke. However, increasingly sensitive imaging modalities have identified patients with LVNC who lack systolic dysfunction or rhythm abnormalities, and the likelihood of developing cardiac complications remains unclear for this subset of patients. Several genes have been associated with LVNC. Genetic mutations that alter myocardial formation during development may manifest in later life as cardiomyopathy. The process by which the ventricular myocardium forms has been molecularly dissected through gene disruption studies in mice and other model organisms, including zebrafish, a fact that emphasizes the conservation of these circuits. In this review, we briefly outline cardiac morphogenesis and describe the pathways necessary for trabeculation and compaction of the ventricular myocardium (Figure 1).
Figure 1.
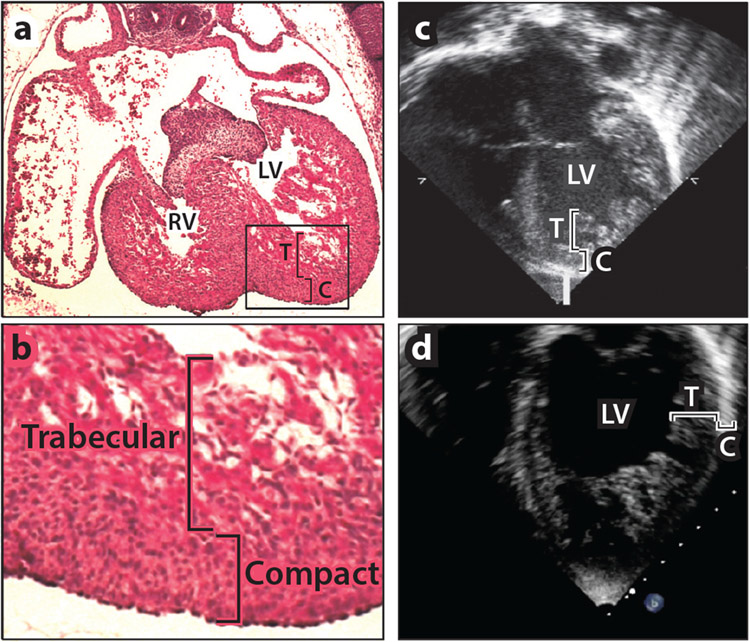
Myocardial trabeculation, compaction, and left ventricular noncompaction cardiomyopathy (LVNC). (a) Hematoxylin and eosin staining of the heart [right ventricle (RV) and left ventricle (LV)] from an embryonic day 16.5 (E16.5) mouse embryo demonstrates the presence of both trabecular (T) and compact (C) myocardium. (b) Higher-power image of the boxed area in panel a reveals that trabecular cardiomyocytes are larger and more loosely organized than the smaller, denser compact cardiomyocytes. (c,d) Echocardiographic images of a patient with LVNC, taken from the (c) parasternal short axis and (d) apical four-chamber views. These views show an abnormally thick trabecular layer and abnormally thin compact layer.
Significant progress has been made in identifying the genetics of early cardiac development, as described in many excellent reviews (3-6). Briefly, the earliest specification of cardiac progenitors begins soon after gastrulation at embryonic day (E)7 in the mouse—and, correspondingly, at E17 in humans—and requires signals from the surrounding endoderm, including those from the fibroblast growth factor (FGF) and bone morphogenetic protein (BMP) families, as well as the expression of mesodermal transcription factors such as Mesp1/2 and Tbx6. These factors coordinate gene expression programs that drive migration of cardiac mesodermal cells to the cardiac crescent, where progenitors express the transcription factors GATA 4, 5, and 6, as well as Nkx2-5 and Tbx5. The cardiac progenitors then migrate to the midline and form the linear heart tube, a two-layer structure that comprises an inner endocardial layer and outer myocardial layer. Soon thereafter, the linear heart tube loops to the right, creating an outer curvature from which the left and right ventricles form, and this looping positions the endocardial cushions (the future atrioventricular valves) near the conotruncus (the future ventricular outflow tracts). Nkx2-5, T box, and GATA family members continue to be expressed throughout this process. The transcription factor Mef2c determines myocyte fate, which is further specified by Hand1 and Hand2 for the left and right ventricles, respectively. Islet1 contributes to outflow tract development.
After cardiac looping and the establishment of left-right asymmetry, the heart undergoes significant growth that leads to septation, resulting in four chambers and enlargement of the ventricular walls, particularly in the left ventricle. During this period, which begins at about E9.5 in mice, corresponding to E26 in humans (6), considerable morphological changes occur in the ventricular walls. Initially in this heart growth phase, cardiomyocytes proliferate from the subepicardial layer as a meshwork of contractile bundles called trabecular myocardium. Trabecular cardiomyocytes are relatively large and are juxtaposed with endothelium, theoretically to provide nutrients and gas exchange before the coronary vasculature develops. The process, known as ventricular compaction, begins at approximately E11.5 in mice, corresponding to E31 in humans, and continues until birth, during which cardiomyocytes in the outer regions of the ventricles proliferate at a significantly higher rate than cardiomyocytes in the inner regions and individual cardiomyocytes adopt a smaller, denser morphology. The coronary vasculature develops in concert with compaction, and proliferation within the trabecular myocardium decreases such that the compact myocardium comprises nearly all of the left ventricular wall thickness at birth. Gene deletion studies in mice have identified at least 60 genes from diverse molecular pathways that contribute to trabeculation and compaction (Table 1), an observation supporting the notion that these are complex processes requiring integration of multiple signals at the proper place and time.
Table 1.
Genes that contribute to cardiomyocyte proliferation and growth during trabeculation and compaction
| Role | Gene | Full gene name | Stage of impact | Reference(s) |
|---|---|---|---|---|
| Endocardial-myocardial interactions | Adam17 | TACE | Compaction | (21) |
| Bmp10 | Bone morphogenetic protein 10 | Trabeculation | (27) | |
| Ejnb2 | EphrinB2 | Trabeculation | (11) | |
| Erbb2 | ErbB2 | Trabeculation | (12-15) | |
| Erbb4 | ErbB4 | Trabeculation | (12-15) | |
| Fkbp1a | FK506-binding protein 1a | Compaction | (18, 19) | |
| Hhex | Hematopoietically expressed homeobox | Compaction | (42) | |
| Jag1 | Jagged 1 | Compaction | (20) | |
| Mib1 | mindbomb homolog 1 | Compaction | (20) | |
| Myocd | Myocardin | Trabeculation | (24) | |
| Nf1 | Neurofibromatosis 1 | Compaction | (33, 34) | |
| Notch1 | Notch 1 | Trabeculation Compaction | (11, 20) | |
| Notch2 | Notch 2 | Trabeculation | (22, 23) | |
| Nrg1 | Neuregulin 1 | Trabeculation | (12-15) | |
| Numb | Numb gene homolog (Drosophila) | Compaction | (22) | |
| Numbl | Numb-like | Compaction | (22) | |
| Rbpj | Recombination signal binding protein for immunoglobulin κJ region | Trabeculation | (11) | |
| Tek | Endothelial-specific receptor tyrosine kinase | Trabeculation | (37) | |
| Tbx20 | T-box | Trabeculation | (32) | |
| Vegfa | Vascular endothelial growth factor A | Trabeculation Compaction | (39, 40, 41) | |
| Epicardial-myocardial interactions | Epo | Erythropoietin | Compaction | (53, 56) |
| Epor | Erythropoietin receptor | Compaction | (56) | |
| Fgf16 | Fibroblast growth factor 16 | Trabeculation Compaction | (58-60) | |
| Fgf20 | Fibroblast growth factor 20 | Compaction | (58, 61) | |
| Fgf9 | Fibroblast growth factor 9 | Compaction | (58) | |
| Igf2 | Insulin-like growth factor 2 | Compaction | (57) | |
| Rxrα | Retinoid X receptor α | Compaction | (48, 49, 51, 54) | |
| Vcam1 | Vascular cell adhesion molecule 1 | Compaction | (46) | |
| Extracellular matrix-myocardial interactions | Adamts1 | A disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 1 | Compaction | (67) |
| Fbln1 | Fibulin-1 | Compaction | (66) | |
| Has1 | Hyaluronic acid synthase 1 | Trabeculation | (65) | |
| Itgb1 | Integrin β1 | Compaction | (69) | |
| Ptk2 | PTK2 protein tyrosine kinase 2; also known as Focal Adhesion Kinase (FAK) | Compaction | (71) | |
| Vcan | Versican | Trabeculation | (63, 66) | |
| Intracardiomyocyte interactions | Adm | Adrenomedullin | Compaction | (87, 88) |
| Calcrl | Calcitonin receptor-like | Compaction | (87, 88) | |
| Calr | Calreticulin | Compaction | (86) | |
| Daam1 | Dishevelled-associated activator of morphogenesis 1 | Compaction | (85) | |
| Foxp1 | Forkhead box P1 | Compaction | (83) | |
| Nkx2-5 | NK2 homeobox 5 | Trabeculation | (26) | |
| Taz | Tafazzin | Compaction | (121-123) | |
| Yap1 | Yes-associated protein 1 | Trabeculation | (73) | |
| Ywhae | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, ε polypeptide; also known as 14-3-3ε | Compaction | (82) | |
| Zfpm2 | Zinc finger protein, multitype 2; also known as Friend of GATA 2 (FOG2) | Compaction | (77) | |
| Epigenetic interactions | Jarid2 | Jumonji, AT-rich interactive domain 2 | Compaction | (135-137) |
| Smarca4 | SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily a, member 4; also known as Brg1 | Trabeculation | (67) | |
| Human LVNC genes | ACTC1 | Actin, α, cardiac muscle 1 | Myofibrillogenesis | (99, 113) |
| ACTN2 | Actinin α2 | Myofibrillogenesis | (117) | |
| CASZ1 | Castor zinc finger 1 | Myofibrillogenesis | (117) | |
| DTNA | Dystrobrevin α | Costamere assembly | (121) | |
| HCN4 | Hyperpolarization-activated, cyclic nucleotide-gated K+ 4 | Conduction system | (124, 125) | |
| LDB3 | LIM domain binding 3; also known as Cypher and ZASP | Myofibrillogenesis | (119, 120) | |
| LMNA | Lamin A | Nuclear membrane | (130) | |
| MYBPC3 | Myosin-binding protein C, cardiac | Myofibrillogenesis | (100, 108-111) | |
| MYH7 | Myosin, heavy polypeptide 7, cardiac muscle, β | Myofibrillogenesis | (97-105 | |
| TNNT2 | Troponin T2, cardiac | Myofibrillogenesis | (99, 114) | |
| TPM1 | Tropomyosin 1, α | Myofibrillogenesis | (108) |
CELL-CELL INTERACTIONS IN TRABECULAR AND COMPACT MYOCARDIAL GROWTH
Endocardial-Myocardial Interactions
Failure to initiate trabecular myocardial growth results in embryonic lethality, presumably due to insufficient contractile function and poor perfusion throughout the embryo. Furthermore, trabeculation appears to be a prerequisite for compaction, as all mouse mutations that fail to trabeculate die by E14.5 with a hypoplastic left ventricular wall; indeed, no human cardiomyopathies due to a lack of trabeculation have been described. Genetic studies in mice suggest that trabeculation requires interactions between endocardium and myocardium, and that these interactions later in development contribute to ventricular compaction. These endocardial-myocardial pathways are driven in large part by signaling in the Neuregulin (NRG1)/ErbB, Notch, and BMP10 pathways.
Neuregulin 1 and ErbB4/ErbB2.
The Neuregulin signaling pathway was the first to be recognized as a contributor to cardiac trabeculation. NRG1, which is produced in the endocardium, contains an epidermal growth factor–like (EGF) domain that binds the cardiomyocyte receptor ErbB4, an EGF-family receptor tyrosine kinase (7, 8). Upon binding of NRG1, ErbB4 dimerizes with ErbB2, which triggers autophosphorylation and activation of MAPK and PI3K/Akt second messenger pathways to promote cardiomyocyte proliferation and differentiation (9, 10). Nrg1 expression appears to be regulated by endothelial Notch signaling pathways (described below) (11). In the absence of Nrg1 or ErbB2/ErbB4, cardiomyocytes fail to proliferate and to form the trabecular myocardium, which leads to death by E11.5 due to heart failure (12-15).
Notch1.
Notch signaling pathways comprise four transmembrane receptors (Notch1–4) and five transmembrane ligands (Jagged1, Jagged2, Delta-like1, Delta-like3, and Delta-like4) that contribute to development in virtually all organ systems, including the vasculature, heart, lung, liver, intestines, and kidney (16). In this system, the receiving cell expresses Notch, whereas the signaling cell expresses the ligand. Upon ligand binding, Notch is processed into Notch extracellular domain (NECD) and Notch intracellular domain (NICD). NICD translocates to the nucleus of the receiving cell, binds the transcriptional repressor RBPJK (also known as CSL and CBF1), and converts it into a transcriptional activator (17). By driving differential gene expression between the signal and receiver cells, Notch signaling mediates the cell fate decisions that are critical for proper vasculogenesis and organogenesis.
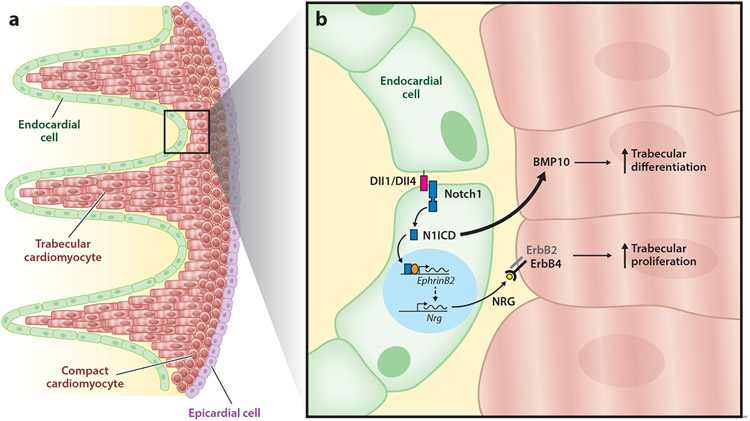
At E9.5 in the mouse heart, Notch1 is expressed throughout the ventricular endocardium with comparable expression levels from the trabecular crypts near the compact myocardium to the tips of the trabeculae (11). However, the Notch1 intracellular domain (N1ICD) protein as well as the endocardial ligands Deltal1 and Deltal4 strongly localize to endocardial nuclei at the bases of trabeculae only, which suggests that Notch signaling is concentrated in the region of trabecular myocardium that is closest to the thin compact myocardium (11). Global deletion or early-stage endothelial deletion of Notch1 and RBPJk using Tie2-Cre produces the same lethal phenotype—with a lack of trabeculation and a thick, ragged endocardium—by E11.5 (11). Expression analyses in RBPJk-null mice have demonstrated reduced expression of endocardial EphrinB2 and NRG1 as well as myocardial BMP10. It was shown that N1ICD activates the transcription of EphrinB2 directly, and EphrinB2 is known to drive expression of NRG1. However, BMP10 expression was observed to be independent of the EphrinB2-NRG1 pathway. Thus, endocardial Notch1 signaling mediates cardiomyocyte proliferation through the EphrinB2-NRG1 pathway and differentiation through the BMP10 pathway and therefore functions as a master regulator of trabecular growth and maturation (Figure 2).
Figure 2.
Notch signaling during trabeculation. (a) The heart contains three layers: the endocardium (green), myocardium (red), and epicardium (purple). The myocardium is further divided into trabecular and compact cardiomyocytes. Throughout the trabecular phase of cardiac growth, the compact myocardial layer just beneath the epicardium remains thin, and bundles of trabecular cardiomyocytes are juxtaposed with endocardium. (b) During trabecular growth, endocardial Notch1 binds endocardial Delta-like ligand 1 (Dll1) and Delta-like ligand 4 (Dll4), triggering cleavage and endocytosis of the Notch1 intracellular domain (N1ICD) and promoting activation of EphrinB2 transcription. EphrinB2 triggers Neuregulin (Nrg) transcription. NRG is released from the endocardium and binds myocardial ErbB4, which dimerizes with ErbB2 and drives trabecular cardiomyocyte proliferation. N1ICD also contributes to bone morphogenetic protein 10 (BMP10) expression in trabecular cardiomyocytes to promote differentiation.
In the developing myocardium, endocardial Notch1 signaling also contributes to the compaction process, and alterations in this pathway can disrupt compaction through at least two mechanisms. Global or endothelial-specific deletion of Fkbp1a leads to ventricular noncompaction and embryonic lethality at E14.5 (18, 19). In endothelial-specific Fkbp1a-null E12.5 embryonic hearts, N1ICD is found throughout the endocardium rather than being restricted to the trabecular bases (18). Myocardial Bmp10 expression and cardiomyocyte proliferation are significantly increased in Fkbp1a-mutant hearts, which suggests that unrestricted Notch1 signaling at E12.5 may drive inappropriate proliferation that either increases trabecular myocardium or prohibits the proper formation of compact myocardium (18).
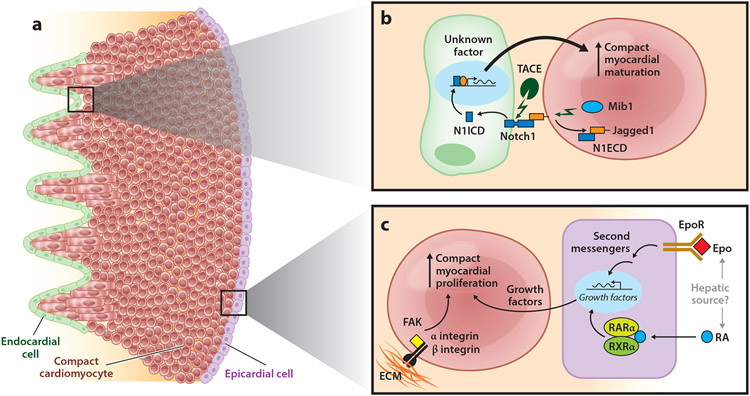
A second Notch1-mediated mechanism also contributes to ventricular compaction, as recently demonstrated in murine models and two families with LVNC (20). The Notch pathway regulator mindbomb homolog 1 (Mib1) is an E3 ubiquitin ligase expressed in myocardium that triggers endocytosis of myocardial Jagged1 after it binds the endocardial Notch1 ECD. In mice, cardiomyocyte-specific deletion of Mib1 with cTnt-Cre interferes with dissociation of ligand-bound N1ECD from the N1ICD and thereby inhibits activated Notch1 signaling in the endocardium. In contrast to mice with early endocardial Notch1 deletion, which fail to trabeculate, animals with myocardial Mib1 deletion demonstrate ventricular noncompaction and increased cardiomyocyte proliferation (20). Dominant MIB1 mutations were identified in two unrelated Spanish families, each with multiple family members with LVNC (20). In one family, a premature stop codon at position 530 in the ankyrin repeat domain region was described, and the other family was found to carry a missense variant (V943F) in the coiled-coil domain. Further studies are required to determine how Mib1 deletion, which inhibits endocardial N1ICD production, leads to the same phenotype as Fkbp1a deletion, which increases endocardial N1ICD production. However, in accordance with these results, loss of TACE (TNF-α ADAM metalloprotease-converting enzyme), a protease that cleaves ligand-bound NECD from the NICD, also promotes increased cardiomyocyte proliferation and ventricular noncompaction (21). Finally, induction of endocardial Notch1 deletion between E9 and E12 phenocopies the loss of myocardial Mib1 deletion (20). Together, these findings suggest that early endocardial Notch1 signaling drives cardiomyocyte proliferation and development of trabeculae, whereas Notch1 signaling later in development slows trabecular proliferation and contributes to the transition from the trabecular morphology to the compact morphology (Figures 2 and 3).
Figure 3.
Major proliferative signaling pathways that contribute to compaction. (a) The compact myocardium (red circles) expands dramatically via proliferative signals from the endocardium (green), epicardium (purple), and extracellular matrix (ECM) (orange). (b) During compact myocardial growth, endocardial Notch1 binds the myocardial ligand Jagged1, triggering cleavage by the protease TACE and myocardial internalization of Notch1 extracellular domain (N1ECD) and Jagged1 in a mindbomb homolog 1 (Mib1)-dependent pathway. Both the TACE and Mib1 steps are required for endocardial internalization of Notch1 intracellular domain (N1ICD). The endocardial N1ICD target gene(s) that promotes compact myocardial maturation remains unknown. (c) The epicardium responds to erythropoietin (Epo) and retinoic acid (RA), both possibly from a hepatic source, through the erythropoietin receptor (EpoR) and RA receptor dimer RXRα/RARα (retinoid X receptor α/retinoic acid receptor α). Receptor binding triggers transcription of growth factors, including insulin-like growth factor and fibroblast growth factor family members, which bind myocardial receptors to promote compact myocardial proliferation. Fibroblasts in the compact myocardium also produce ECM components, including fibronectin and collagen. β1 integrin (in concert with its α integrin heterodimer partner) binds these ECM ligands, activates focal adhesion kinase (FAK), and contributes to compact myocardial proliferation.
Notch2, Numb, and Numblike.
In contrast to Notch1, Notch2 protein is found in myocardium (22). Notch2 mRNA is expressed in both trabecular and compact myocardium, whereas Notch2 intracellular domain (N2ICD) is restricted to trabecular myocardium, suggesting that the Notch2 ligands Delta-like1, Jagged1, and Jagged2 may be specific to endocardium and/or cardiomyocytes in the trabecular region. Animals expressing a hypomorphic form of Notch2 die from multiple developmental abnormalities, including kidney glomerular defects and the lack of cardiac trabeculation (23), whereas ectopic expression of N2ICD in compact myocardium causes increased proliferation throughout the ventricular walls and leads to ventricular noncompaction, ventricular septal defect (VSD), and lethality between E12 and E15 (22). N2ICD degradation by Numb and Numblike, which are partially redundant proteins expressed throughout the myocardium, also contributes to cardiomyocyte proliferation and ventricular morphology. Cardiac-specific Numb excision in Numblike null mice phenocopies ectopic N2ICD expression, including increased cardiomyocyte proliferation throughout the ventricular wall (22). Taken together, studies of Notch signaling during heart development illustrate a complex system in which expression of the components (ligands and receptors) at the proper place (endocardial versus myocardial tissue) and proper time (early versus mid-to-late cardiac development) must be tightly coordinated to drive the cell fate decisions that allow normal cardiac trabeculation and compaction.
BMP10, Myocardin, and Tbx20.
BMP10, a member of the TGFβ superfamily, is expressed exclusively in trabecular myocytes from E9 to E13.5. Bmp10 expression is driven by endocardial Notch1 signaling, as described above, with direct transcriptional activation in cardiomyocytes by Myocardin, although the relationship between Myocardin and Notch signaling is not well described (11, 24). BMP10 is also considered a marker of trabecular myocardium that shows an expanded expression pattern in many mice exhibiting hypertrabeculation/noncompaction (25, 26). Mice lacking BMP10 die at E10–10.5 due to a hypoplastic heart with lack of trabeculation (27). BMP10 signaling appears to mediate trabecular differentiation as well as proliferation—the latter as demonstrated by hypertrabeculation in a Bmp10-overexpressing mouse—via a Smad1-Tbx20-mediated pathway (26, 28).
Tbx20 mRNA shows a complex expression pattern, with transcripts initially detected in early cardiac progenitors of the cardiac crescent and later found in endocardium and myocardium, including adult myocardium (29, 30). Mutations in human TBX20 are associated with adult congenital defects, including atrial septal defects, mitral valve abnormalities, and dilated cardiomyopathy (DCM) (31). Tbx20-null mice fail to trabeculate and die by E10.5, which suggests its major action in cardiac development occurs during mid-development rather than upon initial differentiation of cardiac progenitors; indeed, the proliferation pathway mediated by Tbx20 involves derepression of NMyc1, which promotes cell division and cardiac growth (32).
Additional endocardial-myocardial trabeculation and compaction pathways.
Neurofibromin 1 inhibits Ras signaling via its GTPase-activating function and contributes to neural and musculoskeletal development; the absence of human NF1 leads to neurofibromatosis type 1, an autosomal dominant disease of wide phenotypic variability that includes neurofibromas with the potential for malignant transformation (33). Interestingly, absence of Nf1 in mice leads to embryonic lethality due to cardiac abnormalities, including atrioventricular canal (AVC) defects and ventricular noncompaction, the latter seen only in endothelial-specific Nf1 deletion (33, 34). Loss of endothelial and endocardial Nf1 does not alter endothelial NRG or myocardial ErbB2/ErbB4, but endothelial Nfatc1 (nuclear factor of activated T cells, cytoplasmic, calcineurin-dependent 1) localizes primarily to the nucleus rather than the cytoplasm, as seen in wild-type cells (33). Humans with neurofibromatosis type 1 due to NF1 mutations have few cardiac defects, and this discrepancy between human and mouse phenotypes may reflect compensation by the remaining wild-type allele or related genes in cardiac development.
Angiopoietin 1 (Angpt1) is the major ligand for the receptor tyrosine kinase Tek, which is expressed primarily on endothelial cells. Global deletion of Angpt1 or Tek, as well as cardiac-specific deletion of Angpt1, lead to lethality between E9.5 and E12.5 owing to vascular defects and lack of cardiac trabeculation (35-37).
Nonendothelial cells produce vascular endothelial growth factor (VEGF) to promote sprouting angiogenesis at the hypoxic front of hypoperfused tissue (38). In the developing heart, trabeculation and compaction require a tight balance of myocardial VEGF. Specifically, Vegf haploinsufficiency leads to lethality at E9.5 due to impaired cardiovascular development, including an absence of trabeculation (39, 40); by contrast, stabilization of the Vegf transcript equivalent to a threefold increase in VEGF protein leads to abnormal coronary artery maturation and ventricular noncompaction with death by E14.5 (41). Similarly, deletion of the homeobox gene Hhex, which either directly or indirectly represses Vegf transcription, leads to upregulation of VEGF protein levels and ventricular noncompaction (42). These findings demonstrate that in addition to the endocardial-to-myocardial signaling systems described above, myocardial-to-endocardial pathways contribute to trabecular and compact growth. Additional studies are required to determine whether and how myocardial VEGF promotes endocardial proliferation as a prerequisite for trabeculation—as has been suggested for endocardial-to-mesenchymal transformation in the AVC (43)—and whether excessive endocardial proliferation contributes to hypertrabeculation and/or noncompaction.
Epicardial-Myocardial Interactions
The epicardium proliferates from the proepicardial organ at the venous pole of the heart starting at E9.5 in mice, and the entire myocardial surface is covered by E11 (44). In parallel fashion, ventricular compaction begins as early as E10.5, and the compact myocardium is recognizably thicker by ~E11.5–12.5. Epicardial-myocardial interactions appear to be a major driver of the compaction process. Indeed, mechanical disruption of the nascent epicardium in chick embryos prevents normal compaction, as does genetic disruption of vascular cell adhesion molecule-1, which is required for adhesion of the proliferating epicardium to the underlying myocardium (45, 46). The epicardial-myocardial pathways that support compaction include retinoic acid (RA), erythropoietin (Epo), insulin-like growth factor, and FGF signaling.
Retinoid X receptor α.
Retinoid X receptor α (RXRα) is a member of the RXR family of nuclear receptors that binds retinoids and appears to function primarily as a heterodimeric partner of retinoic acid receptor (RAR) family members (47). RXRα-deficient mice show mid-to-late gestational lethality beginning at E12.5, with living embryos detectable at E16.5 but none left by E18.5. Cardiac defects in the mutants include noncompaction and VSD, which are presumed to be the cause of embryonic lethality (48, 49). By contrast, developmental cardiac defects have not been described in RARα-, RARβ-, or RARγ-deleted mice, although ventricular noncompaction was noted in a portion of RARα;RARγ double-null mice (50). Mitotic index analyses revealed decreased proliferation in the subepithelial cardiomyocytes of both RXRα-null and RARα-null animals. In wild-type E14.5 embryos, highly organized sarcomeres and myofibrils are a common feature in trabecular myocardium but are rare in compact myocardium, where sarcomere assembly is at an early stage. The subepithelial cardiomyocytes from both RXRα-null and RARα-null mice contain well-developed sarcomeres at E14.5 and as early as E8.5 (48, 51). The observation of highly structured sarcomeres in the subepithelial myocardium with a decreased mitotic index in the absence of RXRα suggests that RA signaling contributes to ventricular compaction by delaying differentiation and promoting proliferation.
Although RXRα mutants express a clear noncompaction phenotype, the source of RA, site of receptor activity, and precise role of the epicardium in this signaling pathway remain somewhat enigmatic. Embryonic chick heart explant studies demonstrated that RA does not directly stimulate cardiomyocyte growth in the absence of epicardium but rather promotes myocardial proliferation in an epicardial-dependent fashion, which implies production of a growth factor by the epicardium after activation by RA (52). Retinaldehyde dehydrogenase 2 catalyzes production of RA during early embryonic development, but its expression appears to be limited to the splanchnic mesoderm at E9.5–10.5 and restricted from the epicardium until E12.5 (after the initiation of compaction), which suggests a nonepicardial source of RA during early compaction (53). Conditional deletion of RXRα using Gata5-Cre, which excises in epicardium, indicates an epicardial cell-autonomous role for RXRα in compaction. A caveat of this approach is that Gata5-Cre is also active in hepatic capsule epithelium, which lies adjacent to the heart before the development of the diaphragm between E10.5 and E15.5. Therefore, the hepatic capsule may contribute as a source of RXRα activity to promote ventricular compaction (54, 55). Additional studies suggest that the hepatic epithelium is a major site of RA and RXRα signaling during early compaction and that this pathway triggers the release of hepatic Epo, which diffuses into the nascent epicardium to stimulate the release of additional mitogens (53) (Figure 3).
Erythropoietin and erythropoietin receptor.
Epo is a glycoprotein that stimulates erythropoiesis, and the embryos of mice lacking either Epo or its receptor EpoR die at ~E13.5 from failed hematopoiesis, liver abnormalities, and ventricular noncompaction (56). The Epo receptor is expressed in the heart by E10.5 and localizes to epicardium and endocardium, but not myocardium, at E12.5 (56). In a signaling pathway parallel to RA, cultured embryonic chick explants respond to Epo in an epicardial-dependent fashion to stimulate cardiomyocyte proliferation, potentially via a growth factor produced by the epicardium (Figure 3) (52). Taken together, these studies suggest a model in which hepatic epithelial RA/RXRα signaling stimulates the production and release of Epo, which diffuses into the newly formed epicardium and activates epicardial production of growth factors that trigger myocardial proliferation and growth. Further studies—such as conditional excision of RXRα and EpoR using epicardial Cre drivers that do not excise in the hepatic epithelium—are needed to confirm aspects of this model.
Insulin-like growth factor 2 and fibroblast growth factors 9, 16, and 20.
Insulin-like growth factor 2 (IGF2) is expressed in epicardium and endocardium at E11.5–14.5, whereas its primary receptor, IGFR1, is expressed in myocardium at E12.5–14.5; the insulin receptor also binds IGF2 and is expressed at low levels in myocardium (57). Mice lacking functional IGF2 and cardiac-specific-Igfr2;Insr double mutants demonstrate decreased proliferation in the compact zone at E11.5 and noncompaction at E12.5. However, the proliferative rate recovers by E14.5, and compaction is largely restored by birth, which suggests that diffusion of the epicardially derived IGF trophic signal becomes rate limiting and that additional trophic signals after E14.5 are delivered by the nascent coronary vasculature (57).
Likewise, FGFs 9, 16, and 20 appear to contribute to the early compaction process. Fgf9 mRNA is expressed in right ventricular (RV) epicardium and endocardium at E10.5, but it is restricted to endocardium by E12.5, with highest expression at the RV apex near the septum. Fgf9-null mice demonstrate impaired proliferation and abnormal compaction at E12.5, but only at the apicolateral RV and septum; this type of mutant mouse is born with a normally compacted heart (58). Fgf16 transcripts are expressed in basal LV epicardium and endocardium at E10.5 and expand to basal LV, RV, and septal endocardium by E12.5. Disruption of Fgf16 exon 1 led to lack of trabeculation and embryonic lethality by E14.5, whereas disruption of Fgf16 exon 2 decreased cardiomyocyte proliferation at E14.5 but did not produce embryonic lethality (58-60). Fgf20 mRNA is expressed in the basal and apical LV epicardium and endocardium at E10.5 and is largely restricted to the RV endocardium by E12.5, and Fgf20-null mice show no cardiac defects (58, 61). Cardiomyocytes express the redundant FGF receptors Fgfr1 and Fgfr2, and Fgfr1;Fgfr2 double-knockout mice have small hearts with decreased proliferation throughout the apex and base of both LV and RV at E13.5, but compaction recovers later in development (58). Taken together, these data indicate that several FGFs in the developing heart have both overlapping and unique expression patterns, but with the exception of Fgf16, none appear to be absolutely necessary for compaction. It is possible that FGF9/FGF16/FGF20 and FGFR1/FGFR2 signaling pathways contribute to early compaction at E12.5, perhaps initiating compaction or maintaining myocytes in a proliferative or nondifferentiated state, but that other signaling networks can compensate for their loss.
Extracellular Matrix-Myocardial Interactions
The extracellular matrix (ECM) plays a significant role in both trabeculation and compaction. Shortly after heart tube formation, a relatively thick layer of ECM called the cardiac jelly lies between the endocardium and myocardium and is necessary for proper initiation and progression of trabeculation. In the compact layer, cardiac fibroblasts proliferate in concert with cardiomyocytes and deposit ECM, which appears to be required to support compaction.
Versican, hyaluronic acid, ADAMTS1, and Fibulin-1.
The chondroitin sulfate proteoglycan versican and the mucopolysaccharide hyaluronic acid (HA) are ECM factors that make up a significant portion of the cardiac jelly. These components are produced by the endocardium and myocardium and are well described for their contribution to atrioventricular cushion remodeling during valve morphogenesis (62-65). In addition to their atrioventricular cushion defects, mice lacking Has-1, the enzyme that produces HA, and Vcan, encoding versican, fail to trabeculate (63, 65, 66). These observations demonstrate the necessity of cardiac jelly for early heart development and proliferation of the trabecular myocardium, possibly via modulation of the diffusion of myocardial VEGF and endocardial Nrg in the cardiac jelly (67). Alternatively, HA and versican may directly or indirectly stimulate cardiomyocytes. HA is known to contribute to Ras activation during endothelial cell invasion in cultured atrioventricular explants (65), and such a signaling pathway may be at play within cardiomyocytes during trabeculation.
As trabeculation proceeds, the cardiac jelly progressively disappears until the endocardium is in contact with the myocardium, and this ECM breakdown process plays a critically important role in cardiac development. ADAMTS1 (a disintegrin and metalloproteinase with thrombospondin motif) and its cofactor Fibulin-1 cleave versican beginning around E12.5, after which the cardiac jelly is degraded and trabecular proliferation eventually declines; inappropriately early ADAMTS1 activity leads to premature breakdown of cardiac jelly and lack of trabecular cardiomyocyte proliferation (67). Conversely, attenuation of ADAMTS1 activity in mice lacking Fibulin-1 leads to trabecular hyperproliferation and ventricular noncompaction (66).
β1 integrin.
The integrins are transmembrane proteins encoded by α and β subtypes that form functional heterodimers, bind components of the ECM, and transmit intracellular signals upon ligand binding. Ligand specificity is determined by the α and β subtypes within the heterodimer; the β1 subunit can pair with one of multiple α subunits to form heterodimers that bind collagen, laminin, fibronectin, and vitronectin (68). The developing heart is enriched in ECM components—including fibronectin and collagen—produced by embryonic cardiac fibroblasts, and these fibroblasts proliferate in parallel with cardiomyocytes in the compact region (69). Cardiomyocyte-specific deletion of β1 integrin, which contributes to mechanical anchoring of cardiomyocytes to the ECM via costameres in adult cardiomyocytes, has significant effects on embryonic cardiac growth. Lethality occurs in half of the mutant mice between E16.5 and P7, with significantly smaller heart size from E14.5 onward and ventricular noncompaction in all mutants (69). The rate of myocardial proliferation significantly decreases at the midpoint of compaction (E16.5) in the absence of cardiomyocyte β1 integrin. Taken together, these results point toward a fibroblast-ECM-cardiomyocyte signaling pathway that contributes to normal proliferation, compaction, and overall cardiac growth during development (Figure 3).
Focal adhesion kinase and focal adhesion kinase–related nonkinase.
Focal adhesion kinase (FAK) is a nonreceptor tyrosine kinase encoded by the Ptk2 gene that relays signals from activated integrins, whereas focal adhesion kinase–related nonkinase (FRNK), the C-terminal portion of FAK, is transcribed from an intronic promoter within Ptk2 perinatally and acts to inhibit FAK signaling (70). Cardiomyocyte-specific deletion of Ptk2 using the Mlc2a-Cre driver results in decreased cardiomyocyte proliferation at E14.5, VSD, and ventricular noncompaction (71). Likewise, overexpression of FRNK early in embryonic heart development leads to decreased cardiomyocyte proliferation and ventricular noncompaction (70). These observations are in keeping with a role for β1 integrin/FAK signaling in cardiomyocyte proliferation and compaction in the developing heart.
Other Cardiomyocyte Factors
As described above, cardiac growth and development require interactions between cardiomyocytes and the surrounding environment of endocardium, epicardium, and ECM. In addition, some gene products contribute to proliferation and growth in a cell-autonomous manner, in parallel with or independently of the interactions involving surrounding cells and matrix. Other genes play a role in cardiomyocyte proliferation, but whether or how their protein products facilitate interactions with surrounding cells remains unclear.
Yap1.
During organogenesis, the Hippo signaling pathway mediates organ size via phosphorylation of Yap1 (Yes-associated protein 1), which prevents nuclear localization and activation of TEAD-family transcription factors. Growth factor stimulation inhibits the Hippo pathway and permits Yap1- and TEAD-mediated transcription of cell cycle genes, leading to proliferation and organ growth (72). In the heart, Yap1 is highly expressed in cardiomyocytes at E17.5 and in the early postnatal period, but it is significantly downregulated between 8 and 12 weeks of age (73). Yap1-null mice do not survive beyond E8.5, and cardiomyocyte-specific deletion of Yap1 is embryonically lethal by E12.5, with lack of trabeculation noted in hearts from mutant animals (73). In contrast, cardiomyocyte-specific overexpression of Yap1 is lethal by E16.5; hearts from these animals display a moderate increase in compact myocardium and significantly increased trabecular myocardium, which together cause left ventricular cavity obliteration (73). Cardiomyocyte proliferation is enhanced in both the compact and trabecular myocardium of Yap1-overexpressing mutants, likely via increased expression of cell cycle genes, but the magnitude of increase is markedly higher in the trabecular myocardium (73). These observations suggest that Yap1 activity is downregulated in trabecular myocardium and either sustained or upregulated in compact myocardium during the process of ventricular compaction.
Nkx2-5.
Nkx2-5 is required early in development for cardiac looping, and this homeobox gene continues to function in conduction system differentiation as well as trabecular cardiomyocyte maturation. Specifically, excision of Nkx2-5 within ventricular cardiomyocytes leads to hypoplasia of the atrioventricular node and hyperproliferation specifically within the trabecular myocardium and not the compact myocardium (26). The compact layer remains relatively normal in these mutant mice, and hypertrabeculation does not appear to impact survival. Bmp10 expression remains high in the expanded trabecular myocardium, even in adult cardiomyocyte-Nkx2-5-mutant mice (26). Over 40 mutations in human NKX2-5 have been associated with congenital heart disease, many encompassing atrial septation defects and associated cardiac conduction disease, but noncompaction has not been described (26, 74-76). These results suggest that the ventricular cardiomyocyte-specific activity of Nkx2-5 drives trabecular myocardium toward differentiation and away from proliferation (26, 74-76).
Friend of GATA 2.
Members of the friend of GATA (FOG) transcription factor family physically interact with GATA factors to repress gene transcription activity. FOG-2 is highly expressed in the heart (77). In the absence of Zfpm2, the gene that encodes FOG-2, the epicardium expresses low levels of intercellular adhesion molecule (ICAM) and VEGFR2, and the coronary vasculature fails to develop despite the presence of the normal epicardium; furthermore, hearts display ventricular noncompaction (77). Cardiomyocyte-specific overexpression of Zfpm2 in Zfmp2-null mice rescues coronary vascularization as well as compaction (77). Early deletion of FOG-2 using an NKX2.5-driven Cre recapitulates the complex congenital defects seen in global FOG-2 deletion (77-79). Cardiomyocyte-restricted deletion of FOG-2 after cardiac looping reproduces the phenotype of thinned ventricular walls and deficient vasculature, an observation that links defective cardiomyocyte proliferation to deficient coronary artery development. FOG-2 interacts with the nucleosome remodeling and deacetylase (NuRD) complex, and a point mutation that disrupts the FOG-2-NuRD complex interaction induces a proliferation defect similar to that induced by cardiac-restricted FOG-2 deletion (80).
Other genes that regulate cardiomyocyte trabeculation and compaction.
The 14-3-3 protein family in mammals comprises seven members that bind phosphoserine or phosphothreonine proteins and thereby modify multiple cell processes, including cell cycle progression (81). 14-3-3ε is highly expressed in the brain and heart during development, and mice that lack 14-3-3ε die in the perinatal period (82). Hearts from 14-3-3ε-null mice are noncompacted at E18.5, and proliferation is significantly decreased in the compact myocardium owing in part to upregulation of the cell cycle progression–inhibiting protein p27Kip1 (82). Although 14-3-3ε is expressed in cardiomyocytes, cell autonomy still needs to be confirmed through a cardiomyocyte-specific 14-3-3ε-·καΆ model.
Foxp1 belongs to the Fox family of DNA-binding proteins with a forkhead or winged-helix domain (83). Foxp1 is expressed in endocardium and myocardium, and animals with global deletion of Foxp1 die at E14.5 with atrioventricular cushion defects and ventricular noncompaction due to increased proliferation in trabecular myocardium as well as decreased proliferation in compact myocardium (83). As with 14-3-3ε, endocardial versus myocardial specificity for Foxp1 activity remains to be determined.
Actin-polymerizing proteins modify the sarcomere thin filaments as well as the nonsarcomere cytoskeletal components in cardiomyocytes; one such protein, Dishevelled-associated activator of morphogenesis 1 (Daam1), is a formin protein localized to the cell membrane in endocardium, epicardium, and myocardium that mediates F-actin elongation at the barbed end (84). Mice homozygous for a Daam1 genetrap mutation show a wide range of cardiac structural abnormalities, including outflow tract defects, VSD, and possible ventricular noncompaction (85). Interestingly, Daam1-mutant mice display no differences from controls in cardiac-specific gene expression (Nkx2-5, Tbx20, BMP, Mlc2a, and Mlc2v) at E10.5 and no differences in proliferation or apoptosis at E12.5, but these measures may change later in development (85). Sarcomere and intercalated disk abnormalities have been seen via transmission electron microscopy in Daam1-deficient cardiomyocytes, and these cells display poor adherence to collagen, but more studies are required to determine the mechanism underlying these cardiac defects (85).
Calreticulin is a calcium-binding protein localized to the endoplasmic reticulum (86). crt-null mice die during embryonic development, between E15.5 and E18.5, with ventricular noncompaction that is possibly due to decreased nuclear translocation of Nfatc4, a calcineurin-dependent factor (86).
Adrenomedullin is a potent vasodilator that is expressed in the adrenal medulla as well as in cardiomyocytes, vascular smooth muscle, and aortic endothelium during mouse development. The calcitonin receptor-like receptor (Calcrl) binds adrenomedullin. Genetic deletion of either Adm or Calcrl phenocopies deletion of the other, with embryonic lethality at E14.5, noncompaction, and hydrops fetalis that is greater in magnitude than would be expected from heart failure alone (87, 88). Indeed, subsequent studies revealed that the major defect in these mice lies in lymphatic vasculature development. Whether the cardiac effects are primary or secondary to abnormal lymphatic formation remains to be determined (89).
HUMAN GENETICS OF LEFT VENTRICULAR NONCOMPACTION
Human cardiomyopathy is classified as dilated, hypertrophic, arrhythmogenic, or noncompaction (2). There is an increasing appreciation for the clinical overlap of these forms of cardiomyopathy, and this overlap is partially explained by shared genetic predisposition. Noncompaction cardiomyopathy is seen in both children and adults as an increase in the ratio of trabeculated to compacted myocardium, typically in multiple segments of the left ventricle (Figure 1). LVNC may be seen in the setting of normal or impaired systolic function. The clinical consequences of LVNC range from no symptoms to heart failure, arrhythmias, and thromboembolic events. With the growing use of cardiac magnetic resonance imaging and its ability to delineate detailed features of the myocardium better than other imaging modalities like echocardiography, hypertrabeculated segments can be seen in nearly all hearts, and the ratio of trabeculated to compact myocardium changes with age (90). In a study of normal adults without cardiac disease, 18% of subjects were found to have ≥1 of the traditional criteria for LVNC (91). Notably, those with hypertrabeculation had significantly higher serum pro-BNP levels compared to the general population, consistent with functional impairment, albeit mild, of the heart. LVNC is known to be heritable and is most commonly autosomal dominant, although X-linked recessive and autosomal recessive forms also make important contributions to inheritance (92). Family members of those with LVNC have a higher incidence of LVNC, and a higher incidence of premature death, than what is seen in the general population (93).
Genetic testing for inherited cardiomyopathy now relies on the use of gene panels in which 10–100 genes are sequenced simultaneously (94). As whole-exome and whole-genome analyses become more routine, it is likely that gene panels will be replaced by exome/genome analysis (95, 96). Many genes implicated in the development of LVNC are also implicated in the genesis of DCM and hypertrophic cardiomyopathy (HCM), and this observation has promoted the use of large gene panels covering sarcomere, cytoskeletal, metabolic, mitochondrial, and other genes for human genetic testing.
Sarcomere Gene Mutations that Correlate with Human LVNC
The sarcomere is made up of thin and thick filaments, and mutations in sarcomeric genes are among the most frequent causes of inherited cardiomyopathies. Although these mutations are often considered in the context of HCM and DCM, mutations in genes encoding sarcomere proteins have also been linked to LVNC.
β myosin heavy chain.
An individual myosin subunit contains two heavy chains and two each of the regulatory and essential light chains. In the human adult myocardium, β myosin heavy chain contributes to the majority of the thick filament protein component of the sarcomere. Mutations in MYH7, the gene encoding β myosin heavy chain, have been identified in families with LVNC (97-105). MYH7 mutations are more commonly associated with HCM and DCM, where the majority of mutations are missense variants that fall into the subfragment 1 “head” or motor domain region of myosin. Most subfragment 1 mutations map within the functionally significant portions of the protein, including the regions that regulate actin binding and ATPase activity, although more rare, missense mutations in the rod region can lead to cardiomyopathy. The first report of LVNC-associated MYH7 mutations identified splice site variants within the head domain, which in principle could act through either a dominant negative or a haploinsufficient mechanism (99). However, since that time, additional missense variants, including those that extend into the rod region, have been described in LVNC. Missense variants within the head region may act by reducing force production by the sarcomere. Missense variants that perturb thick filament formation may reduce force transmission within the sarcomere.
In general, an attractive hypothesis has been that HCM-associated variants in MYH7 are activating mutations linked to increased contractility and the hypercontractile state seen in HCM, whereas DCM- or LVNC-associated variants are linked with reduced function. However, this may be an oversimplification, and the effect of any given variant on the myocardium may actually produce a range of phenotypic outcomes throughout the lifetime of the myocardium, depending on heart morphology and the presence of alternative sarcomere proteins generated through expression or splicing events. For example, the MYH7 mutation Arg234His has been associated with both LVNC and HCM, which supports the notion that sarcomere gene mutations manifest a clinical spectrum of phenotypes (99, 106). How the same mutation can cause different cardiomyopathies requires further study, but this clinical spectrum likely involves additional polymorphisms and epigenetic and/or environmental interactions that sensitize the sarcomere and/or cardiomyocyte to a particular remodeling pathway.
Myosin-binding protein C3.
During systolic cardiac contraction, proper force generation depends on the rate and extent of crossbridge cycling between actin and myosin filaments, and myosin-binding protein C3 (MYBPC3) constrains this cycling to appropriately limit the systolic phase (107). As with MYH7, many mutations in MYBPC3 have been associated with HCM. MYBPC3 mutations make up the second-most common cause of LVNC after MYH7 mutations (100, 108-110). Notably, compared with MYH7 mutations, a greater proportion of HCM-associated MYBPC3 mutations are frameshifting or truncating. Early-onset cardiomyopathy associated with features of hypertrabeculation and noncompaction can be seen with compound heterozygous mutations in MYBPC3 (100, 110, 111). Mouse models support the idea that MYBPC3 causes disease, as Mybpc3-null animals lack M lines; a patient with a compound heterozygous MYBPC3 mutation was also found to have disrupted and in some areas absent M lines (100, 112). The observation that mutations in both MYH7 and MYBPC3 lead to noncompaction supports the idea that normal sarcomere function is required for the transition from trabeculated to compacted myocardium in humans, as it is in mice.
Other sarcomere genes in LVNC.
Mutations within the thin filament genes ACTC1, TNNT2, and TPM1, as well as the Z-disk gene ACTN2, have all been associated with LVNC (99, 108, 113, 114, 115). Of note, the cardiac α-actin mutation Glu202Lys and TNNT2 mutation Arg131Trp have been described in HCM and DCM as well (99, 106, 113, 116). CasZ1 is a recently identified zinc finger protein that falls within the human congenital heart disease–associated deletion region of chromosome 1p36, and the gene product either directly or indirectly stimulates transcription of many sarcomere components—including TNNI2, TNNT1, and ACTA1 (117).
LDB3 protein, also known as Cypher/ZASP, contributes to the sarcomere Z disk as one of several proteins that mediate physical interactions between α-actinin and actin (118). Mutations within LBD3 are associated with DCM, arrhythmia, and LVNC (119). Among mice, the global-Lbd3-null animals die perinatally owing to muscle weakness and heart failure. In the diaphragm muscle, Z lines are seen via transmission electron microscopy during embryonic development, but the electron-dense Z-line region disappears after birth; in the heart, Z lines are abnormally wide during development and progressively widen until death (120). These observations suggest that Lbd3 is not required for Z-line or sarcomere assembly, but rather is necessary for Z-line and/or sarcomere maintenance during the mechanical process of contraction.
Alpha-dystrobrevin, encoded by DTNA, is a member of the dystrophin complex, which associates with costameres to link sarcomeres at the myocyte periphery with the cardiomyocyte membrane and ECM. Mutations in DTNA were identified early in the study of genetic causes of LVNC (121). Whereas mutations in other components of the dystrophin complex are known to cause skeletal and cardiac myopathy, the mechanism of α-dystrobrevin-mediated LVNC remains unknown.
X-Linked LVNC
Tafazzin (TAZ), also known as G4.5, was one of the first genes connected with human LVNC (121, 122). This X-linked gene is associated with Barth syndrome, is highly expressed in cardiac and skeletal muscle, and encodes a phospholipid-lysophospholipid transacylase that catalyzes the final step in cardiolipin synthesis; the unbalanced cardiolipin profile in Taz mutants adversely affects mitochondrial function (123). Taz knockdown in mice leads to ventricular noncompaction due to decreased cardiomyocyte proliferation at E13.5. These observations suggest a link between mitochondrial bioenergetics and cardiomyocyte proliferation that deserves further investigation.
Arrhythmias and LVNC
The relationship between arrhythmia risk and LVNC is not well understood and requires further study. The risk of ventricular arrhythmias is increased in LVNC, and in first-degree relatives the risk of premature death is heightened (93). This overlap between arrhythmias and LVNC may reflect the concomitant development of the cardiac conduction system and the maturing myocardium in the same developmental windows, such that perturbation of the myocardium may have a secondary adverse effect on the developing conduction system. However, in some cases, the responsible gene may be leading to independent pathologies in the cardiac conduction system and the myocardium. Recently, the HCN4 gene, which encodes the potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4, was linked to LVNC in multiple families. HCN4 is known to be expressed highly in the sinus node and to contribute to pacemaker activity. Mutations in HCN4 have now been associated with LVNC and concomitant bradycardia (124, 125). Whether noncompaction is a primary effect of HCN4 mutation or a secondary effect of decreased cardiac output/force generation from bradycardia requires further investigation.
The nuclear envelope proteins lamin A and lamin C are produced from a single gene, LMNA. Lamins A and C are found in most postmitotic cell types, including heart, muscle, and brain. Mutations in LMNA lead to a constellation of phenotypes. Among LMNA-associated diseases, cardiomyopathy—with or without associated skeletal muscle disease—is a frequent consequence (126, 127). The mechanism by which LMNA mutations cause disease is not fully known and likely employs multiple pathways, including the stability of chromatin structure (128). An early study identified a link between LVNC and LMNA mutations (129). In LMNA-associated cardiomyopathy, cardiac conduction system disease is common and can include atrial- and ventricular-associated abnormal rhythms (130).
Genetic Assessment of LVNC: Lessons from Mouse Models
In mouse models, many of the genes linked to cardiomyocyte proliferation—and therefore noncompaction-like phenotypes—yield broad effects necessitating tissue-specific genetic analysis. Cell type–specific deletion of many genes studied in mice was required to dissect the genes’ roles in proliferation, trabeculation, and compaction. Because the products of many of these genes perform functions in other cells, the absence of the genes would be expected to cause embryonic lethality. By contrast, many of the mutations linked to human LVNC alter the resultant protein rather than eliminating the protein’s function. Many of the genes implicated through genetic deletion in mice should likely be included in gene panel testing for cardiomyopathy. Mutations in two genes implicated by mouse studies, Mib1 and Nkx2-5, have been associated with human LVNC as disease-associated variants, and Nxk2-5 is a cardiomyocyte-specific gene (20, 76). In fact, the majority of human LVNC cases with an established genetic diagnosis are due to mutations in cardiomyocyte-specific genes, particularly those that encode components of the sarcomere, the basic myocyte contractile unit (108).
Large databases of human genetic variation now emerging in the form of the 1000 Genomes Project, the NHLBI Exome Variant server, and the Exome Aggregation Consortium have yielded information that supports a higher-than-expected prevalence of pathogenic mutations, including those that are anticipated to cause cardiomyopathy and LVNC (131, 132) (http://exac.broadinstitute.org). The observation that hypertrabeculation and LVNC can be found in seemingly normal individuals is consistent with the genetic landscape of the overall population.
EPIGENETICS OF LEFT VENTRICULAR NONCOMPACTION
Epigenetics refers to features of DNA other than the sequence itself that affect gene expression. These features generally relate to chromatin-level regulation—particularly DNA methylation, ATP-dependent chromatin remodeling that changes nucleosome structure, and covalent histone modifications that alter DNA-histone interactions (reviewed in 133). Chromatin-level regulation contributes to gene expression in cardiac development, including the expression of genes involved in cardiomyocyte proliferation, trabeculation, and compaction.
Jarid2
Jarid2 (Jumonji, AT-rich interactive domain 2) is a nuclear factor found in both endocardium and myocardium; whereas most members of the Jumonji family function as histone demethylases, Jarid2 appears to promote histone H3-K9 methylation via recruitment of G9a and GLP methyl-transferases (134). Global Jarid2 deletion leads to ventricular noncompaction, VSD, double outlet right ventricle, and perinatal lethality (135). Reporter gene studies demonstrate that Jarid2 expression begins in the trabecular cardiomyocytes at E10.5, as the mitotic index in this region declines, and that expression expands to the compact myocardium at low levels beginning at E12.5 and then dramatically increases in this region at postnatal day 7. These findings suggest that Jarid2 promotes cardiomyocyte cell cycle exit, an idea supported by the observation of increased proliferation in Jarid2-null hearts (136). Indeed, Jarid2 appears to act as a transcriptional repressor of the cell cycle progression gene cyclin D1 via both cardiomyocyte cell-autonomous and non-cell-autonomous mechanisms: Jarid2 directly binds the cyclin D1 promoter to repress expression in cardiomyocytes, and it also binds an intronic Notch1 repressor element in endocardium to repress Nrg and ErbB2/ErbB4 signaling, as previously described (136, 137).
Brg1
Brg1 is a member of the SWI/SNF (BAF) chromatin-remodeling family that is expressed in endocardium and myocardium and mediates associations between DNA and histones/nucleosomes as well as interacts with transcription factors to affect promoter and repressor functions (reviewed in 133, 138). In endocardium, Brg1 acts as a transcriptional repressor of ADAMTS1, a matrix metalloprotease that cleaves versican in trabecular ECM, as described above; the absence of endocardial Brg1 leads to lack of trabeculation due to inappropriately early derepression of Adamts1 expression with resultant premature degradation of the cardiac jelly (64).
CONCLUSIONS
The orchestration of myocardial development is under tight control and heavily influenced by the concomitant development of vascular supply of the myocardium. In early development of the myocardium, the trabeculated myocardium is thought to derive nutrients from blood within the pulsating ventricular cavity. With development of the coronary arteries, the compacted myocardium extracts oxygen from this vascular source and becomes less reliant on the trabeculated myocardium. Disruptions in the genetic pathways that specify trabeculation by and large significantly impair survival. In contrast, disruption of the compaction process, either completely or in part, may be compatible with postnatal life. However, in many cases abnormal compaction manifests later in life with impaired cardiac function, which may reflect continued molecular dysfunction. For example, in the case of MYH7 or TAZ mutations, defective sarcomeres or mitochondrial function, respectively, can cause cardiomyopathy. However, the suboptimal morphology of the myocardium itself, with a reduced compacted layer, may also be a continued source of heart dysfunction.
The excessive left ventricular trabeculation observed in a measurable fraction of the normal population may be linked to an increased risk of heart failure, arrhythmias, or thromboembolic events later in life. Broader genetic and image profiling of the human population will help clarify the risks of hypertrabeculated myocardium.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Hoffman JI, Kaplan S. 2002. The incidence of congenital heart disease. J. Am. Coll. Cardiol 39:1890–900 [DOI] [PubMed] [Google Scholar]
- 2.Maron BJ, Towbin JA, Thiene G, Antzelevitch C, Corrado D, et al. 2006. Contemporary definitions and classification of the cardiomyopathies: an American Heart Association Scientific Statement from the Council on Clinical Cardiology, Heart Failure and Transplantation Committee; Quality of Care and Outcomes Research and Functional Genomics and Translational Biology Interdisciplinary Working Groups; and Council on Epidemiology and Prevention. Circulation 113:1807–16 [DOI] [PubMed] [Google Scholar]
- 3.Srivastava D 2006. Genetic regulation of cardiogenesis and congenital heart disease. Annu. Rev. Pathol 1:199–213 [DOI] [PubMed] [Google Scholar]
- 4.Harvey RP. 2002. Patterning the vertebrate heart. Nat. Rev. Genet 3:544–56 [DOI] [PubMed] [Google Scholar]
- 5.Van Vliet P, Wu SM, Zaffran S, Puceat M. 2012. Early cardiac development: a view from stem cells to embryos. Cardiovasc. Res 96:352–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sylva M, van den Hoff MJ, Moorman AF. 2014. Development of the human heart. Am. J. Med. Genet. A 164A:1347–71 [DOI] [PubMed] [Google Scholar]
- 7.Citri A, Yarden Y. 2006. EGF-ERBB signalling: Towards the systems level. Nat. Rev. Mol. Cell Biol 7:505–16 [DOI] [PubMed] [Google Scholar]
- 8.Zhao YY, Feron O, Dessy C, Han X, Marchionni MA, Kelly RA. 1999. Neuregulin signaling in the heart. Dynamic targeting of erbB4 to caveolar microdomains in cardiac myocytes. Circ. Res 84:1380–87 [DOI] [PubMed] [Google Scholar]
- 9.Kang J-O, Sucov HM. 2005. Convergent proliferative response and divergent morphogenic pathways induced by epicardial and endocardial signaling in fetal heart development. Mech. Dev 122:57–65 [DOI] [PubMed] [Google Scholar]
- 10.Hertig CM, Kubalak SW, Wang Y, Chien KR. 1999. Synergistic roles of neuregulin-1 and insulin-like growth factor-I in activation of the phosphatidylinositol 3-kinase pathway and cardiac chamber morphogenesis. J. Biol. Chem 274:37362–69 [DOI] [PubMed] [Google Scholar]
- 11.Grego-Bessa J, Luna-Zurita L, del Monte G, Bolós V, Melgar P, et al. 2007. Notch signaling is essential for ventricular chamber development. Dev. Cell 12:415–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gassmann M, Casagranda F, Orioli D, Simon H, Lai C, et al. 1995. Aberrant neural and cardiac development in mice lacking the ErbB4 neuregulin receptor. Nature 378:390–94 [DOI] [PubMed] [Google Scholar]
- 13.Lee KF, Simon H, Chen H, Bates B, Hung MC, Hauser C. 1995. Requirement for neuregulin receptor erbB2 in neural and cardiac development. Nature 378:394–98 [DOI] [PubMed] [Google Scholar]
- 14.Meyer D, Birchmeier C. 1995. Multiple essential functions of neuregulin in development. Nature 378:386–90 [DOI] [PubMed] [Google Scholar]
- 15.Kramer R, Bucay N, Kane DJ, Martin LE, Tarpley JE, Theill LE. 1996. Neuregulins with an Ig-like domain are essential for mouse myocardial and neuronal development. PNAS 93:4833–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shawber CJ, Kitajewski J. 2004. Notch function in the vasculature: insights from zebrafish, mouse and man. Bioessays 26:225–34 [DOI] [PubMed] [Google Scholar]
- 17.Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. 1995. Signalling downstream of activated mammalian Notch. Nature 377:355–58 [DOI] [PubMed] [Google Scholar]
- 18.Chen H, Zhang W, Sun X, Yoshimoto M, Chen Z,et al. 2013. Fkbp1a controls ventricular myocardium trabeculation and compaction by regulating endocardial Notch1 activity. Development 140:1946–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shou W, Aghdasi B, Armstrong DL, Guo Q, Bao S, et al. 1998. Cardiac defects and altered ryanodine receptor function in mice lacking FKBP12. Nature 391:489–92 [DOI] [PubMed] [Google Scholar]
- 20.Luxán G, Casanova JC,Martínez-Poveda B, Prados B, D’Amato G,et al. 2013.Mutationsin the NOTCH pathway regulator MIB1 cause left ventricular noncompaction cardiomyopathy. Nat. Med, 19:193–201 [DOI] [PubMed] [Google Scholar]
- 21.Shi W, Chen H, Sun J, Buckley S, Zhao J, et al. 2003. TACE is required for fetal murine cardiac development and modeling. Dev. Biol 261:371–80 [DOI] [PubMed] [Google Scholar]
- 22.Yang J, Bücker S, Jungblut B, Bottger T, Cinnamon Y, et al. 2012. Inhibition of Notch2 by Numb/Numblike controls myocardial compaction in the heart. Cardiovasc. Res 96:276–85 [DOI] [PubMed] [Google Scholar]
- 23.McCright B, Gao X, Shen L, Lozier J, Lan Y, et al. 2001. Defects in development of the kidney, heart and eye vasculature in mice homozygous for a hypomorphic Notch2 mutation. Development 128:491–502 [DOI] [PubMed] [Google Scholar]
- 24.Huang J, Elicker J, Bowens N, Liu X, Cheng L, et al. 2012. Myocardin regulates BMP10 expression and is required for heart development. J. Clin. Investig 122:3678–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galfré E, Pitt SJ, Venturi E, Sitsapesan M, Zaccai NR, et al. 2012. FKBP12 activates the cardiac ryanodine receptor Ca2+-release channel and is antagonised by FKBP12.6. PLOS ONE 7:e31956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pashmforoush M, Lu JT, Chen H, Amand TS, Kondo R, et al. 2004. Nkx2-5 pathways and congenital heart disease; loss of ventricular myocyte lineage specification leads to progressive cardiomyopathy and complete heart block. Cell 117:373–86 [DOI] [PubMed] [Google Scholar]
- 27.Chen H, Shi S, Acosta L, Li W, Lu J, et al. 2004. BMP10 is essential for maintaining cardiac growth during murine cardiogenesis. Development 131:2219–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang W, Chen H, Wang Y, Yong W, Zhu W, et al. 2011. Tbx20 transcription factor is a downstream mediator for bone morphogenetic protein-10 in regulating cardiac ventricular wall development and function. J. Biol. Chem 286:36820–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen T, Aneas I, Sakabe N, Dirschinger RJ, Wang G, et al. 2011. Tbx20 regulates a genetic program essential to adult mouse cardiomyocyte function. J. Clin. Investig 121:4640–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kraus F, Haenig B, Kispert A. 2001. Cloning and expression analysis of the mouse T-box gene Tbx20. Mech. Dev 100:87–91 [DOI] [PubMed] [Google Scholar]
- 31.Kirk EP, Sunde M, Costa MW, Rankin SA, Wolstein O, et al. 2007. Mutations in cardiac T-box factor gene TBX20 are associated with diverse cardiac pathologies, including defects of septation and valvulogenesis and cardiomyopathy. Am.J. Hum. Genet 81:280–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cai CL, Zhou W, Yang L, Bu L, Qyang Y, et al. 2005. T-box genes coordinate regional rates of proliferation and regional specification during cardiogenesis. Development 132:2475–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gitler AD, Zhu Y, Ismat FA, Lu MM, Yamauchi Y, et al. 2003. Nf1 has an essential role in endothelial cells. Nat. Genet 33:75–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brannan CI, Perkins AS, Vogel KS, Ratner N, Nordlund ML, et al. 1994. Targeted disruption of the neurofibromatosis type-1 gene leads to developmental abnormalities in heart and various neural crestderived tissues. Genes Dev. 8:1019–29 [DOI] [PubMed] [Google Scholar]
- 35.Suri C, Jones PF, Patan S, Bartunkova S, Maisonpierre PC, et al. 1996. Requisite role of angiopoietin-1, a ligand for the TIE2 receptor, during embryonic angiogenesis. Cell 87:1171–80 [DOI] [PubMed] [Google Scholar]
- 36.Jeansson M, Gawlik A, Anderson G, Li C, Kerjaschki D, et al. 2011. Angiopoietin-1 is essential in mouse vasculature during development and in response to injury. J. Clin. Investig, 121:2278–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dumont DJ, Gradwohl G, Fong GH, Puri MC, Gertsenstein M, et al. 1994. Dominant-negative and targeted null mutations in the endothelial receptor tyrosine kinase, tek, reveal a critical role in vasculogenesis of the embryo. GenesDev. 8:1897–909 [DOI] [PubMed] [Google Scholar]
- 38.Ferrara N, Gerber HP, LeCouter J. 2003. The biology of VEGF and its receptors. Nat. Med 9:669–76 [DOI] [PubMed] [Google Scholar]
- 39.Ferrara N, Carver-Moore K, Chen H, Dowd M, Lu L, O’Shea KS, et al. 1996. Heterozygous embryonic lethality induced by targeted inactivation of the VEGF gene. Nature 380:439–42 [DOI] [PubMed] [Google Scholar]
- 40.Carmeliet P, Ferreira V, Breier G, Pollefeyt S, Kieckens L, et al. 1996. Abnormal blood vessel development and lethality in embryos lacking a single VEGF allele. Nature 380:435–39 [DOI] [PubMed] [Google Scholar]
- 41.Miquerol L, Langille BL, Nagy A. 2000. Embryonic development is disrupted by modest increases in vascular endothelial growth factor gene expression. Development 127:3941–46 [DOI] [PubMed] [Google Scholar]
- 42.Hallaq H, Pinter E, Enciso J, McGrath J, Zeiss C,et al. 2004.A null mutation of Hhex results in abnormal cardiac development, defective vasculogenesis and elevated Vegfa levels. Development 131:5197–209 [DOI] [PubMed] [Google Scholar]
- 43.Mass E, Wachten D, Aschenbrenner AC, Voelzmann A, Hoch M. 2014. Murine Creld1 controls cardiac development through activation of calcineurin/NFATc1 signaling. Dev. Cell 28:711–26 [DOI] [PubMed] [Google Scholar]
- 44.Jenkins SJ, Hutson DR, Kubalak SW. 2005. Analysis of the proepicardium-epicardium transition during the malformation of the RXRα−/− epicardium. Dev. Dyn 233:1091–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pennisi DJ, Ballard VLT, Mikawa T. 2003. Epicardium is required for the full rate of myocyte proliferation and levels of expression of myocyte mitogenic factors FGF2 and its receptor, FGFR-1, but not for transmural myocardial patterning in the embryonic chick heart. Dev. Dyn 228:161–72 [DOI] [PubMed] [Google Scholar]
- 46.Kwee L, Baldwin HS, Shen HM, Stewart CL, Buck C, et al. 1995. Defective development of the embryonic and extraembryonic circulatory systems in vascular cell adhesion molecule (VCAM-1) deficient mice. Development 121:489–503 [DOI] [PubMed] [Google Scholar]
- 47.Germain P, Chambon P, Eichele G, Evans RM, Lazar MA, et al. 2006. International Union of Pharmacology. LXIII. Retinoid X receptors. Pharmacol. Rev 58:760–72 [DOI] [PubMed] [Google Scholar]
- 48.Kastner P, Grondona JM, Mark M, Gansmuller A, LeMeur M, et al. 1994. Genetic analysis of RXRα developmental function: convergence of RXR and RAR signaling pathways in heart and eye morphogenesis. Cell 78:987–1003 [DOI] [PubMed] [Google Scholar]
- 49.Sucov HM, Dyson E, Gumeringer CL, Price J, Chien KR, Evans RM. 1994. RXRα mutant mice establish a genetic basis for vitamin A signaling in heart morphogenesis. Genes Dev. 8:1007–18 [DOI] [PubMed] [Google Scholar]
- 50.Mendelsohn C, Lohnes D, Decimo D, Lufkin T, LeMeur M, et al. 1994. Function of the retinoic acid receptors (RARs) during development (II). Multiple abnormalities at various stages of organogenesis in RAR double mutants. Development 120:2749–71 [DOI] [PubMed] [Google Scholar]
- 51.Kastner P, Messaddeq N, Mark M, Wendling O, Grondona JM, et al. 1997. Vitamin A deficiency and mutations of RXRα, RXRβ and RARα lead to early differentiation of embryonic ventricular cardiomyocytes. Development 124:4749–58 [DOI] [PubMed] [Google Scholar]
- 52.Stuckmann I, Evans S, Lassar AB. 2003. Erythropoietin and retinoic acid, secreted from the epicardium, are required for cardiac myocyte proliferation. Dev. Biol 255:334–49 [DOI] [PubMed] [Google Scholar]
- 53.Brade T, Kumar S, Cunningham TJ, Chatzi C, Zhao X, et al. 2011. Retinoic acid stimulates myocardial expansion by induction of hepatic erythropoietin which activates epicardial Igf2. Development 138:139–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Merki E, Zamora M, Raya A, Kawakami Y, Wang J, et al. 2005. Epicardial retinoid X receptor α is required for myocardial growth and coronary artery formation. PNAS 102:18455–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ackerman KG, Greer JJ. 2007. Development of the diaphragm and genetic mouse models of diaphragmatic defects. Am. J. Med. Genet. C Semin. Med. Genet 145C:109–16 [DOI] [PubMed] [Google Scholar]
- 56.Wu H, Lee SH, Gao J, Liu X, Iruela-Arispe ML. 1999. Inactivation of erythropoietin leads to defects in cardiac morphogenesis. Development 126:3597–605 [DOI] [PubMed] [Google Scholar]
- 57.Li P, Cavallero S, Gu Y, Chen THP, Hughes J, et al. 2011. IGF signaling directs ventricular cardiomyocyte proliferation during embryonic heart development. Development 138:1795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lavine KJ, Yu K, White AC, Zhang X, Smith C, et al. 2005. Endocardial and epicardial derived FGF signals regulate myocardial proliferation and differentiation in vivo. Dev. Cell 8:85–95 [DOI] [PubMed] [Google Scholar]
- 59.Lu SY, Sheikh F, Sheppard PC, Fresnoza A, Duckworth ML, et al. 2008. FGF-16 is required for embryonic heart development. Biochem. Biophys. Res. Commun 373:270–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hotta Y, Sasaki S, Konishi M, Kinoshita H, Kuwahara K, et al. 2008. Fgf16 is required for cardiomyocyte proliferation in the mouse embryonic heart. Dev. Dyn 237:2947–54 [DOI] [PubMed] [Google Scholar]
- 61.Hayashi T, Ray CA, Bermingham-McDonogh O. 2008. Fgf20 is required for sensory epithelial specification in the developing cochlea. J. Neurosci 28:5991–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kern CB, Twal WO, Mjaatvedt CH, Fairey SE, Toole BP, et al. 2006. Proteolytic cleavage of versican during cardiac cushion morphogenesis. Dev. Dyn 235:2238–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yamamura H, Zhang M, Markwald RR, Mjaatvedt CH. 1997. A heart segmental defect in the anterior-posterior axis of a transgenic mutant mouse. Dev. Biol 186:58–72 [DOI] [PubMed] [Google Scholar]
- 64.Mjaatvedt CH, Yamamura H, Capehart AA, Turner D, Markwald RR. 1998. The Cspg2 gene, disrupted in the hdf mutant, is required for right cardiac chamber and endocardial cushion formation. Dev. Biol 202:56–66 [DOI] [PubMed] [Google Scholar]
- 65.Camenisch TD, Spicer AP, Brehm-Gibson T, Biesterfeldt J, Augustine ML, et al. 2000. Disruption of hyaluronan synthase-2 abrogates normal cardiac morphogenesis and hyaluronan-mediated transformation of epithelium to mesenchyme. J. Clin. Investig 106:349–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cooley MA, Fresco VM, Dorlon ME, Twal WO, Lee NV, et al. 2012. Fibulin-1 is required during cardiac ventricular morphogenesis for versican cleavage, suppression of ErbB2 and Erk1/2 activation, and to attenuate trabecular cardiomyocyte proliferation. Dev. Dyn 241:303–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stankunas K, Hang CT,Tsun ZY, Chen H, Lee NV, et al. 2008. Endocardial Brg1 represses ADAMTS1 to maintain the microenvironment for myocardial morphogenesis. Dev. Cell 14:298–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Takada Y, Ye X, Simon S. 2007. The integrins. Genome Biol. 8:215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ieda M,Tsuchihashi T, Ivey KN, Ross RS, Hong T-T, et al. 2009. Cardiac fibroblasts regulate myocardial proliferation through β1 integrin signaling. Dev. Cell 16:233–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.DiMichele LA, Hakim ZS, Sayers RL, Rojas M, Schwartz RJ, et al. 2009. Transient expression of FRNK reveals stage-specific requirement for focal adhesion kinase activity in cardiac growth. Circ. Res 104:1201–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Peng X, Wu X, Druso JE, Wei H, Park AY-J, et al. 2008. Cardiac developmental defects and eccentric right ventricular hypertrophy in cardiomyocyte focal adhesion kinase (FAK) conditional knockout mice. PNAS 105:6638–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhao B, Li L, Lei Q, Guan KL. 2010. The Hippo-YAP pathway in organ size control and tumorigenesis: An updated version. Genes Dev. 24:862–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.von Gise A, Lin Z, Schlegelmilch K, Honor LB, Pan GM, et al. 2012. YAP1, the nuclear target of Hippo signaling, stimulates heart growth through cardiomyocyte proliferation but not hypertrophy. PNAS 109:2394–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reamon-Buettner SM, Borlak J. 2010. NKX2-5: an update on this hypermutable homeodomain protein and its role in human congenital heart disease (CHD). Hum. Mutat 31:1185–94 [DOI] [PubMed] [Google Scholar]
- 75.Ashraf H, Pradhan L, Chang EI, Terada R, Ryan NJ, et al. 2014. A mouse model of human congenital heart disease: high incidence of diverse cardiac anomalies and ventricular noncompaction produced by heterozygous Nkx2-5 homeodomain missense mutation. Circ. Cardiovasc. Genet 7:423–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ouyang P, Saarel E, Bai Y, Luo C,Lv Q, et al. 2011. A de novo mutation in NKX2.5 associated with atrial septal defects, ventricular noncompaction, syncope and sudden death. Clin. Chim. Acta 412:170–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tevosian SG, Deconinck AE, Tanaka M, Schinke M, Litovsky SH, et al. 2000. FOG-2, a cofactor for GATA transcription factors, is essential for heart morphogenesis and development of coronary vessels from epicardium. Cell 101:729–39 [DOI] [PubMed] [Google Scholar]
- 78.Svensson EC, Huggins GS, Lin H, Clendenin C, Jiang F, et al. 2000. A syndrome of tricuspid atresia in mice with a targeted mutation of the gene encoding Fog-2. Nat. Genet 25:353–56 [DOI] [PubMed] [Google Scholar]
- 79.Zhou B, Ma Q, Kong SW, Hu Y, Campbell PH, et al. 2009. Fog2 is critical for cardiac function and maintenance of coronary vasculature in the adult mouse heart. J. Clin. Investig 119:1462–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Garnatz AS, Gao Z, Broman M, Martens S, Earley JU, Svensson EC. 2014. FOG-2 mediated recruitment of the NuRD complex regulates cardiomyocyte proliferation during heart development. Dev. Biol 395:50–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Healy S, Khan DH, Davie JR. 2011. Gene expression regulation through 14-3-3 interactions with histones and HDACs. Discov. Med 11:349–58 [PubMed] [Google Scholar]
- 82.Kosaka Y, Cieslik KA, Li L, Lezin G,Maguire CT, et al. 2012. 14-3-3ε plays a role in cardiac ventricular compaction by regulating the cardiomyocyte cell cycle. Mol. Cell. Biol, 32:5089–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang B, Weidenfeld J, Lu MM, Maika S, Kuziel WA, et al. 2004. Foxp1 regulates cardiac outflow tract, endocardial cushion morphogenesis and myocyte proliferation and maturation. Development 131:4477–87 [DOI] [PubMed] [Google Scholar]
- 84.Habas R, Kato Y, He X. 2001. Wnt/Frizzled activation of Rho regulates vertebrate gastrulation and requires a novel Formin homology protein Daam1. Cell 107:843–54 [DOI] [PubMed] [Google Scholar]
- 85.Li D, Hallett MA, Zhu W, Rubart M, Liu Y, et al. 2010. Dishevelled-associated activator of morphogenesis 1 (Daam1) is required for heart morphogenesis. Development 138:303–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mesaeli N, Nakamura K, Zvaritch E, Dickie P, Dziak E, et al. 1999. Calreticulin is essential for cardiac development. J. Cell Biol 144:857–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Caron KM, Smithies O. 2001. Extreme hydrops fetalis and cardiovascular abnormalities in mice lacking a functional Adrenomedullin gene. PNAS 98:615–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dackor RT, Fritz-Six K, Dunworth WP, Gibbons CL, Smithies O, Caron KM. 2006. Hydrops fetalis, cardiovascular defects, and embryonic lethality in mice lacking the calcitonin receptor-like receptor gene. Mol. Cell. Biol 26:2511–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Klein KR, Karpinich NO, Espenschied ST,Willcockson HH, Dunworth WP, et al. 2014. Decoy receptor CXCR7 modulates adrenomedullin-mediated cardiac and lymphatic vascular development. Dev. Cell 30:528–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dawson DK, Maceira AM, Raj VJ, Graham C, Pennell DJ, Kilner PJ. 2011. Regional thicknesses and thickening of compacted and trabeculated myocardial layers of the normal left ventricle studied by cardiovascular magnetic resonance. Circ. Cardiovasc. Imaging 4:139–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tizon-Marcos H, de la Paz Ricapito M, Pibarot P, Bertrand O, Bibeau K, et al. 2014. Characteristics of trabeculated myocardium burden in young and apparently healthy adults. Am. J. Cardiol, 114:1094–99 [DOI] [PubMed] [Google Scholar]
- 92.Sasse-Klaassen S, Gerull B, Oechslin E, Jenni R, Thierfelder L. 2003. Isolated noncompaction of the left ventricular myocardium in the adult is an autosomal dominant disorder in the majority of patients. Am. J. Med. Genet. A 119A:162–67 [DOI] [PubMed] [Google Scholar]
- 93.Caliskan K, Michels M, Geleijnse ML, van Domburg RT, van der Boon R, et al. 2012. Frequency of asymptomatic disease among family members with noncompaction cardiomyopathy. Am. J. Cardiol 110:1512–17 [DOI] [PubMed] [Google Scholar]
- 94.Puckelwartz MJ, McNally EM. 2014. Genetic profiling for risk reduction in human cardiovascular disease. Genes (Basel) 5:214–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Golbus JR, Puckelwartz MJ, Dellefave-Castillo L, Fahrenbach JP, Nelakuditi V, et al. 2014. Targeted analysis of whole genome sequence data to diagnose genetic cardiomyopathy. Circ. Cardiovasc. Genet 7:751–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zaidi S, Choi M, Wakimoto H, Ma L, Jiang J, et al. 2013. De novo mutations in histone-modifying genes in congenital heart disease. Nature 498:220–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hoedemaekers YM, Caliskan K, Majoor-Krakauer D, van de Laar I, Michels M, et al. 2007. Cardiac β-myosin heavy chain defects in two families with non-compaction cardiomyopathy: linking non-compaction to hypertrophic, restrictive, and dilated cardiomyopathies. Eur. Heart J 28:2732–37 [DOI] [PubMed] [Google Scholar]
- 98.Budde BS, Binner P, Waldmuller S, Hohne W, Blankenfeldt W, et al. 2007. Noncompaction of the ventricular myocardium is associated with a de novo mutation in the β-myosin heavy chain gene. PLOS ONE 2:e1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Klaassen S, Probst S, Oechslin E, Gerull B, Krings G, et al. 2008. Mutations in sarcomere protein genes in left ventricular noncompaction. Circulation 117:2893–901 [DOI] [PubMed] [Google Scholar]
- 100.Dellefave LM, Pytel P, Mewborn S, Mora B, Guris DL, et al. 2009. Sarcomere mutations in cardiomyopathy with left ventricular hypertrabeculation. Circ. Cardiovasc. Genet 2:442–49 [DOI] [PubMed] [Google Scholar]
- 101.Basu R, Hazra S, Shanks M, Paterson DI, Oudit GY. 2014. Novel mutation in exon 14 of the sarcomere gene MYH7 in familial left ventricular noncompaction with bicuspid aortic valve. Circ. Heart Fail 7:1059–62 [DOI] [PubMed] [Google Scholar]
- 102.Finsterer J, Stollberger C, Brandau O, Laccone F, Bichler K, Laing NG. 2014. Novel MYH7 mutation associated with mild myopathy but life-threatening ventricular arrhythmias and noncompaction. Int. J. Cardiol, 173:532–35 [DOI] [PubMed] [Google Scholar]
- 103.Hirono K, Hata Y, Ibuki K, Yoshimura N. 2014. Familial Ebstein’s anomaly, left ventricular noncompaction, and ventricular septal defect associated with an MYH7 mutation. J. Thorac. Cardiovasc. Surg 148:e223–26 [DOI] [PubMed] [Google Scholar]
- 104.Nomura Y, Momoi N, Hirono K, Hata Y, Takasaki A, et al. 2015. A novel MYH7 gene mutation in a fetus with left ventricular noncompaction. Can. J. Cardiol 31:103 e1–3 [DOI] [PubMed] [Google Scholar]
- 105.Yang J, Zhu M, Wang Y, Hou X, Wu H, et al. 2015. Whole-exome sequencing identify a new mutation of MYH7 in a Chinese family with left ventricular noncompaction. Gene 558:138–42 [DOI] [PubMed] [Google Scholar]
- 106.Arad M, Penas-Lado M, Monserrat L, Maron BJ, Sherrid M, et al. 2005. Gene mutations in apical hypertrophic cardiomyopathy. Circulation 112:2805–11 [DOI] [PubMed] [Google Scholar]
- 107.Stelzer JE, Dunning SB, Moss RL. 2006. Ablation of cardiac myosin-binding protein-C accelerates stretch activation in murine skinned myocardium. Circ. Res 98:1212–18 [DOI] [PubMed] [Google Scholar]
- 108.Probst S, Oechslin E, Schuler P, Greutmann M, Boye P, et al. 2011. Sarcomere gene mutations in isolated left ventricular noncompaction cardiomyopathy do not predict clinical phenotype. Circ. Cardiovasc. Genet. 4:367–74 [DOI] [PubMed] [Google Scholar]
- 109.Schaefer E, Helms P, Marcellin L, Desprez P, Billaud P, et al. 2014. Next-generation sequencing (NGS) as a fast molecular diagnosis tool for left ventricular noncompaction in an infant with compound mutations in the MYBPC3 gene. Eur. J. Med. Genet 57:129–32 [DOI] [PubMed] [Google Scholar]
- 110.Wessels MW, Herkert JC, Frohn-Mulder IM, Dalinghaus M, van den Wijngaard A, et al. 2014. Compound heterozygous or homozygous truncating MYBPC3 mutations cause lethal cardiomyopathy with features of noncompaction and septal defects. Eur. J. Hum. Genet, 23:922–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zahka K, Kalidas K, Simpson MA, Cross H, Keller BB, et al. 2008. Homozygous mutation of MYBPC3 associated with severe infantile hypertrophic cardiomyopathy at high frequency among the Amish. Heart 94:1326–30 [DOI] [PubMed] [Google Scholar]
- 112.McConnell BK, Jones KA, Fatkin D, Arroyo LH, Lee RT, et al. 1999. Dilated cardiomyopathy in homozygous myosin-binding protein-C mutant mice. J. Clin. Investig 104:1235–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Monserrat L, Hermida-Prieto M, Fernandez X, Rodriguez I, Dumont C, et al. 2007. Mutation in the alpha-cardiac actin gene associated with apical hypertrophic cardiomyopathy, left ventricular non-compaction, and septal defects. Eur. Heart J 28:1953–61 [DOI] [PubMed] [Google Scholar]
- 114.Luedde M, Ehlermann P, Weichenhan D, Will R, Zeller R, et al. 2010. Severe familial left ventricular non-compaction cardiomyopathy due to a novel troponin T (TNNT2) mutation. Cardiovasc. Res 86:452–60 [DOI] [PubMed] [Google Scholar]
- 115.Bagnall RD, Molloy LK, Kalman JM, Semsarian C. 2014. Exome sequencing identifies a mutation in the ACTN2 gene in a family with idiopathic ventricular fibrillation, left ventricular noncompaction, and sudden death. BMC Med. Genet 15:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mogensen J, Murphy RT, Shaw T, Bahl A, Redwood C, et al. 2004. Severe disease expression of cardiac troponin C and T mutations in patients with idiopathic dilated cardiomyopathy. J. Am. Coll. Cardiol, 44:2033–40 [DOI] [PubMed] [Google Scholar]
- 117.Liu Z, Li W, Ma X, Ding N, Spallotta F, et al. 2014. Essential role of the zinc finger transcription factor Casz1 for mammalian cardiac morphogenesis and development. J. Biol. Chem 289:29801–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Xi Y,Ai T, De Lange E, Li Z, Wu G,et al. 2012. Loss of function of hNav1.5 by a ZASP1 mutation associated with intraventricular conduction disturbances in left ventricular noncompaction. Circ. Arrhythm. Electrophysiol, 5:1017–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Vatta M, Mohapatra B, Jimenez S, Sanchez X, Faulkner G, et al. 2003. Mutations in Cypher/ZASP in patients with dilated cardiomyopathy and left ventricular non-compaction. J. Am. Coll. Cardiol 42:2014–27 [DOI] [PubMed] [Google Scholar]
- 120.Zhou Q, Chu PH, Huang C, Cheng CF, Martone ME, et al. 2001. Ablation of Cypher, a PDZ-LIM domain Z-line protein, causes a severe form of congenital myopathy. J. Cell Biol 155:605–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ichida F, Tsubata S, Bowles KR, Haneda N, Uese K, et al. 2001. Novel gene mutations in patients with left ventricular noncompaction or Barth syndrome. Circulation 103:1256–63 [DOI] [PubMed] [Google Scholar]
- 122.Kenton AB, Sanchez X, Coveler KJ, Makar KA, Jimenez S, et al. 2004. Isolated left ventricular non-compaction is rarely caused by mutations in G4.5, alpha-dystrobrevin and FK Binding Protein-12. Mol. Genet. Metab. 82:162–66 [DOI] [PubMed] [Google Scholar]
- 123.Phoon CK, Acehan D, Schlame M, Stokes DL, Edelman-Novemsky I, et al. 2012. Tafazzin knockdown in mice leads to a developmental cardiomyopathy with early diastolic dysfunction preceding myocardial noncompaction. J. Am. Heart Assoc 1:jah3–e000455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Milano A, Vermeer AM, Lodder EM, Barc J, Verkerk AO, et al. 2014. HCN4 mutations in multiple families with bradycardia and left ventricular noncompaction cardiomyopathy. J. Am. Coll. Cardiol 64:745–56 [DOI] [PubMed] [Google Scholar]
- 125.Schweizer PA, Schroter J, Greiner S, Haas J, Yampolsky P, et al. 2014. The symptom complex of familial sinus node dysfunction and myocardial noncompaction is associated with mutations in the HCN4 channel. J. Am. Coll. Cardiol 64:757–67 [DOI] [PubMed] [Google Scholar]
- 126.Carboni N, Politano L, Floris M, Mateddu A, Solla E, et al. 2013. Overlapping syndromes in laminopathies: a meta-analysis of the reported literature. Acta Myol. 32:7–17 [PMC free article] [PubMed] [Google Scholar]
- 127.Bonne G, Quijano-Roy S. 2013. Emery-Dreifuss muscular dystrophy, laminopathies, and other nuclear envelopathies. Handb. Clin. Neurol 113:1367–76 [DOI] [PubMed] [Google Scholar]
- 128.Mewborn SK, Puckelwartz MJ, Abuisneineh F, Fahrenbach JP, Zhang Y, et al. 2010. Altered chromosomal positioning, compaction, and gene expression with a lamin A/C gene mutation. PLOS ONE 5:e14342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hermida-Prieto M, Monserrat L, Castro-Beiras A, Laredo R, Soler R, et al. 2004. Familial dilated cardiomyopathy and isolated left ventricular noncompaction associated with lamin A/C gene mutations. Am. J. Cardiol 94:50–54 [DOI] [PubMed] [Google Scholar]
- 130.van Berlo JH, de Voogt WG, van der Kooi AJ, van Tintelen JP, Bonne G, et al. 2005. Meta-analysis of clinical characteristics of 299 carriers of LMNA gene mutations: Do lamin A/C mutations portend a high risk of sudden death? J. Mol. Med. (Berl.) 83:79–83 [DOI] [PubMed] [Google Scholar]
- 131.Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, et al. 2010. A map of human genome variation from population-scale sequencing. Nature 467:1061–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tennessen JA, Bigham AW, O’Connor TD, Fu W, Kenny EE, et al. 2012. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science 337:64–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chang CP, Bruneau BG. 2012. Epigenetics and cardiovascular development. Annu. Rev. Physiol 74:41–68 [DOI] [PubMed] [Google Scholar]
- 134.Shirato H, Ogawa S, Nakajima K, Inagawa M, Kojima M, et al. 2009. A jumonji (Jarid2) protein complex represses cyclin D1 expression by methylation of histone H3-K9. J. Biol. Chem 284:733–39 [DOI] [PubMed] [Google Scholar]
- 135.Lee Y, Song AJ, Baker R, Micales B, Conway SJ, Lyons GE. 2000. Jumonji, a nuclear protein that is necessary for normal heart development. Circ. Res 86:932–38 [DOI] [PubMed] [Google Scholar]
- 136.Toyoda M, Shirato H, Nakajima K, Kojima M, Takahashi M, et al. 2003. jumonji downregulates cardiac cell proliferation by repressing cyclin D1 expression. Dev. Cell 5:85–97 [DOI] [PubMed] [Google Scholar]
- 137.Mysliwiec MR, Bresnick EH, Lee Y. 2011. Endothelial Jarid2/Jumonji is required for normal cardiac development and proper Notch1 expression. J. Biol. Chem 286:17193–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wang X, Haswell JR, Roberts CW. 2014. Molecular pathways: SWI/SNF (BAF) complexes are frequently mutated in cancer—mechanisms and potential therapeutic insights. Clin. Cancer Res 20:21–27 [DOI] [PMC free article] [PubMed] [Google Scholar]