Abstract
Pseudomonas aeruginosa COL-1 was identified in a blood culture of a 39-year-old-woman treated with imipenem in Marseilles, France, in 1996. This strain was resistant to β-lactams, including ureidopenicillins, ticarcillin-clavulanic acid, cefepime, ceftazidime, imipenem, and meropenem, but remained susceptible to the monobactam aztreonam. The carbapenem-hydrolyzing β-lactamase gene of P. aeruginosa COL-1 was cloned, sequenced, and expressed in Escherichia coli DH10B. The deduced 266-amino-acid protein was an Ambler class B β-lactamase, with amino acid identities of 32% with B-II from Bacillus cereus; 31% with IMP-1 from several gram-negative rods in Japan, including P. aeruginosa; 27% with CcrA from Bacteroides fragilis; 24% with BlaB from Chryseobacterium meningosepticum; 24% with IND-1 from Chryseobacterium indologenes; 21% with CphA-1 from Aeromonas hydrophila; and 11% with L-1 from Stenotrophomonas maltophilia. It was most closely related to VIM-1 β-lactamase recently reported from Italian P. aeruginosa clinical isolates (90% amino acid identity). Purified VIM-2 β-lactamase had a pI of 5.6, a relative molecular mass of 29.7 kDa, and a broad substrate hydrolysis range, including penicillins, cephalosporins, cephamycins, oxacephamycins, and carbapenems, but not monobactams. As a metallo-β-lactamase, its activity was zinc dependent and inhibited by EDTA (50% inhibitory concentration, 50 μM). VIM-2 conferred a resistance pattern to β-lactams in E. coli DH10B that paralleled its in vitro hydrolytic properties, except for susceptibility to ureidopenicillins, carbapenems, and cefepime. blaVIM-2 was located on a ca. 45-kb plasmid that in addition conferred resistance to sulfamides and that was not self-transmissible either from P. aeruginosa to E. coli or from E. coli to E. coli. blaVIM-2 was the only gene cassette located within the variable region of a novel class 1 integron, In56, that was weakly related to the blaVIM-1-containing integron. VIM-2 is the second carbapenem-hydrolyzing metalloenzyme characterized from a P. aeruginosa isolate outside Japan.
Among the class B metalloenzymes, two carbapenem-hydrolyzing β-lactamases have been genetically characterized in Pseudomonas aeruginosa: IMP-1 and VIM-1 (7, 10, 15). Both enzymes possess the broadest substrate of hydrolysis range among P. aeruginosa β-lactamases, including penicillins, cephalosporins, cephamycins, oxacephamycins, and carbapenems, but not monobactams. Their activity is zinc dependent and is inhibited by EDTA. Since 1991, IMP-1 has spread among gram-negative rods, including P. aeruginosa, Pseudomonas putida, Pseudomonas fluorescens, Burkholderia cepacia, Alcaligenes xylosoxidans, and members of the family Enterobacteriaceae in Japan (7). According to the results of a 1996 to 1997 survey of IMP-1-producing gram-negative bacteria in Japan, 1.3% of P. aeruginosa isolates and 4.4% of Serratia marcescens isolates produced IMP-1 through acquisition of plasmids (H. Kurokawa, T. Yagi, N. Shibata, K. Shibayama, and Y. Arakawa, Letter, Lancet 354:955, 1999). Other uncharacterized carbapenem-hydrolyzing β-lactamases have been reported in P. aeruginosa and Acinetobacter baumannii isolates in Europe (5; N. M. Woodford, M.-F. I. Palepou, G. S. Babini, J. Bates, and D. M. Livermore, Letter, Lancet 352:546–547, 1998). VIM-1 has been described recently from several P. aeruginosa Italian isolates and shares 28% amino acid identity with IMP-1 (10; G. Cornaglia, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1482, 1999). blaIMP-1 is plasmid or chromosome located, while blaVIM-1 was identified as chromosome borne only (7, 10).
blaVIM-1 and blaIMP-1 are encoded within the variable region of class 1 integrons (1, 9, 10). Integrons are genetic structures capable of capturing gene cassettes. Class 1 integrons, which are most commonly isolates from antibiotic-resistant clinical isolates, possess two conserved segments (5′-CS and 3′-CS) located on either side of the integrated genes (20). Gene cassettes are discrete mobile units comprising a gene, usually an antibiotic resistance gene, and a recombination site that is recognized by the integrase (20). The cassette-associated recombination sites, known as 59-base elements, are located downstream of inserted genes and are of variable length (23). Integron-located resistance genes provide them with a wide potential for expression and dissemination.
In this work, we report analysis of the β-lactamase content and genetic support of P. aeruginosa COL-1 isolated in France in 1996, which hydrolyzed imipenem but remained susceptible to monobactams.
MATERIALS AND METHODS
Bacterial strains.
The bacterial strains and plasmids used in this work are listed in Table 1. The P. aeruginosa COL-1 isolate was identified with the API-20 NE system (bioMérieux, Marcy l'Etoile, France).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or phenotype | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH10B | araD139 Δ(ara, leu)7697 deoR endA1 galK1 galU nupG recA1 rpsL F′-mcrA Δ(mrr-hsdRMS-mrcBC)Φ80dlacZΔM15 ΔlacX74 | Gibco BRL, Paris, France |
| JM109 | endA1 gyrA96 hsdR17 Δ(lac proA) relA recA1 supE44 thi F′ (lacIqlacZΔM15 proAB+ traD36) | 17 |
| In vitro-obtained rifampin-resistant JM109 | Rifampin resistant | 17 |
| P. aeruginosa | ||
| COL-1 | Carbapenem-resistant clinical isolate | This study |
| MKAM 12 | Carbapenem-resistant clinical isolate producing IMP-1 | Y. Arakawa |
| In vitro-obtained ciprofloxacin-resistant P. aeruginosa PU21 | Ciprofloxacin resistant ilv leu streptromycin resistant | 17 |
| Plasmids | ||
| pNOR-2000 | 45-kb natural plasmid that encoded blaVIM-2 | This study |
| pBK-CMV | Neomycin resistant kanamycin resistant | Stratagene, Inc. (Ozyme, Saint-Quentin-en-Yvelines, France) |
| pNOR-2001 | Recombinant plasmid containing a 3,843-bp BamHI insert encoding blaVIM-2 into pBK-CMV | This study |
Antimicrobial agents and susceptibility testing.
The antimicrobial agents and the agar dilution technique for MIC determination have been described elsewhere (16). Antibiotic-containing disks were used for routine antibiograms by the disk diffusion assay (Sanofi-Diagnostics Pasteur, Marnes-La-Coquette, France).
Molecular techniques.
A search for blaVIM-1- or blaIMP-1-like genes in P. aeruginosa was performed by PCR amplification with the following sets of primers: for blaVIM-1, VIM-1A (5′-TCTACATGACCGCGTCTGTC-3′) and VIM-1B (5′-TGTGCTTTGACAACGTTCGC-3′; and for blaIMP-1, IMP-1A (5′-CTACCGCAGCAGAGTCTTTGC-3′) and IMP-1B (5′-GAACAACCAGTTTTGCCTTACC-3′) (10, 15). Whole-cell DNA of P. aeruginosa COL-1 or of P. aeruginosa MKAM 12, which produced IMP-1 (Table 1), was extracted as described previously (16) and used as a template in these PCR experiments (22). BamHI- or HindIII-restricted genomic DNA of P. aeruginosa COL-1 was ligated into either BamHI or HindIII-restricted pBK-CMV phagemid as described previously (16). Selection (amoxicillin [30 μg/ml] or imipenem (2 μg/ml) and kanamycin [30 μg/ml]) and analysis of recombinant plasmids and the electroporation technique used have been described previously (16).
Transfer of resistance genes into in vitro-obtained rifampin-resistant Escherichia coli JM109 or ciprofloxacin-resistant P. aeruginosa PU21 was attempted by liquid and solid conjugation assays at 30 and 37°C (17). Transconjugant selection was performed on Trypticase soy (TS) agar plates containing rifampin (200 μg/ml), ciprofloxacin (4 μg/ml) and amoxicillin (30 μg/ml), or imipenem (2 μg/ml). Plasmid DNA extraction of P. aeruginosa COL-1 was attempted by different methods as described previously (17). The plasmid extract from P. aeruginosa COL-1 culture was electroporated into E. coli DH10B with selection on amoxicillin- or imipenem-containing TS plates. Conjugations were repeated with an E. coli DH10B electrotransformant as the donor and rifampin-resistant E. coli strain JM109 as the recipient.
To identify the location of the β-lactamase gene, whole-cell DNA of P. aeruginosa and unrestricted and restricted plasmid DNAs of P. aeruginosa COL-1 and of a corresponding E. coli electrotransformant were run on a 0.7% agarose gel, transferred onto a Hybond N+ membrane (Amersham Pharmacia Biotech, Orsay, France), and hybridized with a PCR-obtained 801-bp internal probe for blaVIM-2 (VIM-2A; 5′-ATGTTCAAACTTTTGAGTAGTAAG-3′ and VIM-2B; CTACTCAACGACTGAGCG-3′). The nonradioactive ECL (enhanced chemiluminescence) random prime system was used (Amersham Pharmacia Biotech). Briefly, it includes a nucleic acid labeling, hybridization, and detection system based on a combination of enhanced chemiluminescence detection and random primer labeling of DNA.
DNA sequencing and protein analysis.
The cloned DNA fragment inserted into recombinant plasmid pNOR-2001 was sequenced on both strands with an Applied Biosystems sequencer (ABI 373). The nucleotide and deduced amino acid sequences were analyzed and compared to sequences available over the internet as described previously (16).
β-Lactamase extraction and purification.
β-Lactamase extraction was obtained from 6 liters of TS broth culture of E. coli DH10B(pNOR-2001) as described previously (16). Similar unpurified β-lactamase extract was obtained from a 10-ml culture of P. aeruginosa COL-1 subsequently resuspended in 0.5 ml of sodium phosphate buffer.
The β-lactamase extract of E. coli DH10B(pNOR-2001) was dialyzed overnight in 50 mM Bis-Tris buffer (pH 6.5). The β-lactamase extract was loaded onto a preequilibrated Q-Sepharose column (Amersham Pharmacia Biotech). The β-lactamase was eluted in 200 mM NaCl and subsequently dialyzed overnight against 50 mM phosphate buffer containing 150 mM NaCl (pH 7.0). This prepurified extract was loaded onto a 1.6- by 47-cm gel filtration column packed with Superdex 75 (Amersham Pharmacia Biotech) equilibrated with 50 mM phosphate buffer (pH 7.0) containing 150 mM NaCl. The fraction containing the β-lactamase activity was dialyzed overnight against 30 mM cacodylate buffer (pH 6.5) containing 50 μM ZnCl2 prior to a 10-fold concentration with Centrisart-C30 columns (Sartorius, Goettingen, Germany). At each purification step, the β-lactamase activity was determined qualitatively by nitrocefin hydrolysis (Oxoid, Dardilly, France) or quantitatively in a spectrophotometer with 100 μM imipenem (297 nm, −Λɛ = 9,210 M−1 cm−1) as the substrate in the dialysis buffer. One unit of enzyme activity was defined as the amount of enzyme that hydrolyzed 1 μmol of substrate per min at 30°C. The protein content was measured by using the Biorad DC protein assay (Bio-Rad), and the specific activities of the crude extract and of the purified β-lactamase from E. coli DH10B(pNOR-2001) were compared.
Analytical IEF.
The β-lactamase extract from P. aeruginosa COL-1 and the purified β-lactamase from E. coli DH10B(pNOR-2001) were subjected to analytical isoelectric focusing (IEF) as described previously (16). The focused β-lactamases were detected by overlaying the gel with 1 mM nitrocefin (Oxoid) or with an iodine starch gel containing 0.5% (wt/vol) imipenem in 100 mM phosphate buffer (pH 7.0).
Kinetic measurements and Mr determination.
Purified β-lactamase from a culture of E. coli DH10B(pNOR-2001) was used for determination of kinetic parameters (kcat, Km) performed at 30°C in 30 mM sodium cacodylate buffer (pH 6.5) supplemented with 50 μM ZnCl2 as described previously (16). Inactivation by Zn2+ removal was studied at 30°C in cacodylate buffer in the presence of different concentrations of EDTA, with 100 μM imipenem as the reporter substrate. The 50% inhibitory concentration (IC50) was determined for EDTA. Reactivation by Zn2+ (2 mM) was assayed by measuring activity after incubation with EDTA-treated (2 mM) enzyme for 15 min at 30°C.
The relative molecular mass (Mr) of the purified β-lactamase from E. coli DH10B(pNOR-2001) was determined by gel filtration with a 1.6- by 47-cm column packed with Superdex 75 (Amersham Pharmacia Biotech) equilibrated and eluted with 50 mM phosphate buffer (pH 7.0) containing 150 mM NaCl. Each elution peak was tested for β-lactamase activity by using nitrocefin as a substrate. The peak that showed the highest β-lactamase activity was linearly plotted against the logarithm of the molecular masses of standard proteins (Amersham Pharmacia Biotech) to determine the Mr of the purified β-lactamase.
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper have been assigned to the EMBL/GenBank nucleotide sequence database under accession no. AF 191564.
RESULTS
Origin of the P. aeruginosa COL-1 isolate and preliminary susceptibility testing.
P. aeruginosa COL-1 was isolated in 1996 at the Institut Paoli-Calmettes in Marseilles, France. A 39-year-old-French woman was hospitalized for chronic myelogenous leukemia and pancytopenia before the performance of an allogeneic bone marrow transplantation. She had not travelled recently to Italy or Japan, and no information is available on any patient transfer from Italian hospitals concomitant with her hospital stay. The patient had fever and received a course of imipenem and amikacin. Despite this treatment, she died of septic shock 5 days later. The day after her death, blood cultures inoculated 3 days earlier grew a carbapenem-resistant P. aeruginosa isolate, COL-1. Antibiotic susceptibility testing by disk diffusion suggested an uncommon mechanism of resistance, since the isolate was resistant to most β-lactams, including ureidopenicillins, ureidopenicillins–β-lactamase inhibitors, narrow-spectrum cephalosporins, cefepime, ceftazidime, imipenem, and meropenem, but remained fully susceptible to aztreonam (data not shown). These results were confirmed by MIC analysis (Table 2). Disk diffusion testing revealed that P. aeruginosa COL-1 was also resistant to kanamycin, tobramycin, streptomycin, spectinomycin, tetracycline, and chloramphenicol; of intermediate susceptibility to fluoroquinolones and rifampin; and susceptible to fosfomycin.
TABLE 2.
MICs of β-lactams for P. aeruginosa COL-1, E. coli DH10B harboring recombinant plasmid pNOR-2001, and E. coli reference strain DH10B
| β-Lactam(s)a | MIC (μg/ml)
|
||
|---|---|---|---|
| P. aeruginosa COL-1 | E. coli DH10B (pNOR-2001)b | E. coli DH10B | |
| Amoxicillin | >512 | >512 | 4 |
| Amoxicillin + CLA | >512 | 512 | 4 |
| Ticarcillin | >512 | >512 | 4 |
| Ticarcillin + CLA | >512 | 512 | 4 |
| Piperacillin | 64 | 4 | 1 |
| Piperacillin + TZB | 16 | 4 | 1 |
| Cephalothin | >512 | 256 | 2 |
| Cefoxitin | >512 | 128 | 1 |
| Ceftazidime | 256 | 16 | 0.5 |
| Cefotaxime | >512 | 8 | 0.06 |
| Cefepime | 64 | 0.06 | 0.03 |
| Aztreonam | 0.25 | 0.12 | 0.12 |
| Meropenem | 128 | 0.25 | 0.06 |
| Imipenem | 128 | 1 | 0.12 |
CLA, clavulanic acid at a fixed concentration of 2 μg/ml; TZB, tazobactam at a fixed concentration of 4 μg/ml.
E. coli DH10B(pNOR-2001) expressed the carbapenem-hydrolyzing β-lactamase VIM-2.
Cloning, sequencing, and analysis of the genetic support of the β-lactamase gene.
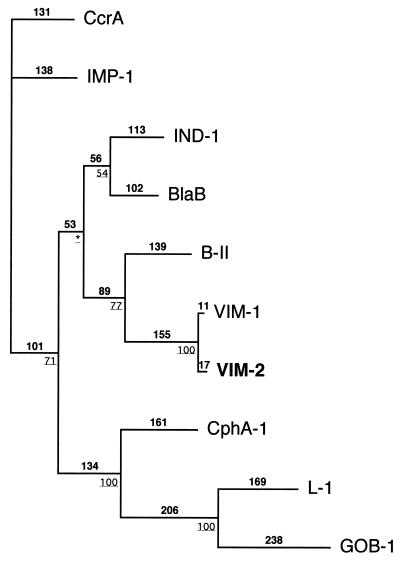
Preliminary PCR-based experiments failed to detect blaIMP-1 or a blaVIM-1-like gene in the P. aeruginosa COL-1 isolate, although no blaVIM-1-containing strain was used as a positive control. Ten recombinant E. coli clones were obtained after cloning experiments and selection on amoxicillin-containing plates. One of them, recombinant plasmid pNOR-2001 (Fig. 1), produced a β-lactamase as assessed by a positive nitrocefin test. Analysis of the nucleotide sequence from the 3,843-bp insert in pNOR-2001 revealed an 801-bp-long open reading frame (ORF) encoding a 266-amino-acid protein, named VIM-2 (Fig. 2). Amino acid sequence analysis of this protein revealed a putative cleavage site between the alanine and serine residues at positions 20 and 21, respectively (14) (Fig. 2). The G+C content of this ORF was 56%, a value that did not lie within the expected range of the G+C content of P. aeruginosa genes (ranging from 60.1 to 69.5%). The codon usage differed as well from that of P. aeruginosa genes (28). The deduced amino acid sequence of this ORF showed low amino acid identity with most of the Ambler class B carbapenem-hydrolyzing β-lactamases, ranging from 32% to 4% for B-II from Bacillus cereus to GOB-1 from Chryseobacterium meningosepticum, respectively (Table 3). It was most closely related to VIM-1 (90% amino acid identity), a recently identified metallo-β-lactamase isolated from an Italian P. aeruginosa clinical isolate (10). VIM-1 and VIM-2 clustered within a subgroup of carbapenem-hydrolyzing β-lactamases (Fig. 3). The conserved amino acids among carbapenem-hydrolyzing β-lactamases that may bind either to Zn2+ ions or a water molecule near or within their putative active site were found in VIM-2 (3, 18, 24, 27): His-86, His-88, Asp-90, His-149, His-225, Cys-168, and His-210 (Fig. 4). These amino acids were identical for VIM-2 and VIM-1 (Fig. 4). Amino acid changes in VIM-2 compared to the sequence of VIM-1 occurred mostly within the NH2- or COOH-terminal regions (Fig. 4).
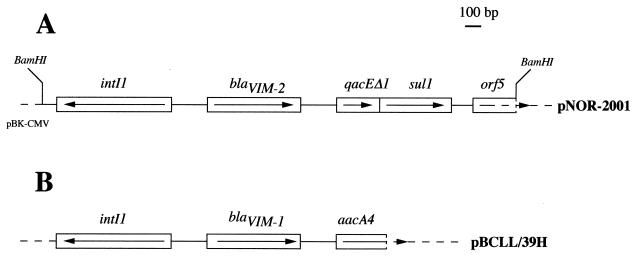
FIG. 1.
(A) Schematic map of the recombinant plasmid pNOR-2001 encoding blaVIM-2 (arrows indicate its translational orientation) and (B) comparison with the blaVIM-1-containing integron as cloned into recombinant plasmid pBCLL/39H (10). For pNOR-2001, the solid line represents the cloned insert from P. aeruginosa COL-1 with the ORFs that are boxed, and the dotted lines indicate the vector pBK-CMV.
FIG. 2.
Nucleotide sequence of the 3,843-bp fragment of the cloned BamHI-fragment of pNOR-2001 containing the blaVIM-2 coding region and its integron. The deduced amino acid sequence is designated in the single-letter code below the nucleotide sequence. The start codons of IntI1, blaVIM-2, qacEΔ1, and sul1 genes are indicated by horizontal arrows, and their stop codons are indicated by asterisks. Only the start and the end of the integrase, qacEΔ1, sul1, and orf5 genes are represented. The −35 and −10 sequences of the promoters Pc, P2, and Pint are underlined; RBS indicates the putative ribosome binding site for blaVIM-2. The conserved core and inverse core sites located at the blaVIM-2 cassette boundaries are boxed, and the composite 59-base element is italicized. The cassette boundaries are indicated by vertical arrows. The left part of the attI1 site is underlined with a dotted line.
TABLE 3.
Percent identity between the amino acid sequences of class B carbapenem-hydrolyzing β-lactamasesa
| β-Lactamase | % Identity
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| VIM-2 | VIM-1 | B-II | IMP-1 | CcrA | BlaB | IND-1 | CphA-1 | L-1 | GOB-1 | |
| VIM-1 | 90 | |||||||||
| B-II | 32 | 35 | ||||||||
| IMP-1 | 31 | 28 | 31 | |||||||
| CcrA | 27 | 27 | 28 | 34 | ||||||
| BlaB | 24 | 24 | 32 | 27 | 25 | |||||
| IND-1 | 24 | 24 | 33 | 28 | 27 | 40 | ||||
| CphA-1 | 21 | 23 | 26 | 19 | 20 | 25 | 23 | |||
| L-1 | 11 | 13 | 12 | 9 | 11 | 9 | 11 | 12 | ||
| GOB-1 | 4 | 4 | 10 | 12 | 10 | 11 | 12 | 13 | 18 | |
The origins of the β-lactamases are as follows: VIM-2, P. aeruginosa COL-1; VIM-1, P. aeruginosa VR 143-97 (10); B-II, B. cereus (8); IMP-1, various gram-negative rods, including P. aeruginosa (15); BlaB, C. meningosepticum CCUG4310 (21); IND-1, Chryseobacterium indologenes 001 (2); CcrA, Bacteroides fragilis TAL 3636 (19); CphA-1, Aeromonas hydrophila AE036 (12); L-1, Stenotrophomonas maltophilia IID1275 (25); GOB-1, C. meningosepticum (GenBank accession no. AF90141).
FIG. 3.
Dendrogram obtained for 10 representative Ambler class B carbapenem-hydrolyzing β-lactamases by parsimony analysis (16). The alignment used for tree calculation was performed with Clustal W (16) followed by minor adjustments in order to reduce the number of gaps and to maintain the alignment of the amino acid residues identified as critical for activity of some class B carbapenem-hydrolyzing β-lactamases. Branch lengths are drawn to scale and are proportional to the number of amino acid changes. The percentage values at branching points (underlined) refer to the number of times a particular node was found in 100 bootstrap replications (the star indicates uncertainty about nodes with bootstrap values of less than 50%). The distance along the vertical axis has no significance. The origins of the β-lactamases are given in Table 3.
FIG. 4.
Comparison of the amino acid sequence of VIM-2 with that of VIM-1. Identical amino acid residues are indicated by asterisks, and functionally equivalent amino acid substitutions are indicated by colons. Boldface amino acids are those of the putative active sites of VIM-1 and of VIM-2. The numbering is according to the B-II sequence of B. cereus (18).
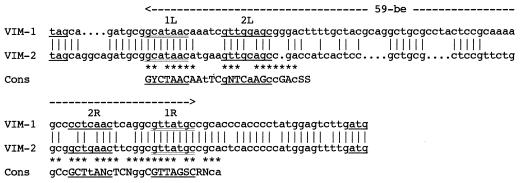
Further sequencing of the cloned fragment in pNOR-2001 revealed key signatures of the class 1 integron, such as (i) a 5′-CS containing an intI1 integrase gene with its own promoter region, (ii) an attI1 recombination site, and (iii) a 3′-CS containing qacEΔ1 and sulI1 (Fig. 2). The initiation codon (ATG) of blaVIM-2 was preceeded by two putative promoter regions named Pc (regions −35 [TGGACA] and −10 [TAAGCT]) and P2 (regions −35 [TTGTTA] and −10 [TACAGT]), which lie within the integrase structural gene (Fig. 2). The secondary promoter P2 identified in some class 1 integrons was in its active form, since the insertion of three guanosine molecules 119 bases downstream of the promoter Pc between the −35 and −10 regions of P2 brought the spacing to 17 bp (11). The blaVIM-2 gene cassette, which was inserted in the attI1 recombination site, has a core site (GTTATGC) and an inverse core site (GCATAAC) (Fig. 2 and 5). The 59-base element was 72 bp long. This class 1 integron, named In56, contained only the blaVIM-2 gene cassette (Fig. 2). The G+C content of this 59-base element was 58%. The 59-base elements for blaVIM-1 and blaVIM-2 cassettes clearly differed in size and structure (Fig. 5). Only the right and left ends of the 59-base element shared significant homology, while the center part required three gaps to be introduced in the blaVIM-2 59-base element in order to obtain an optimal alignment (Fig. 5).
FIG. 5.
Comparison of the sequences of the 72 bp of the blaVIM-2 59-base element (59-be) and the 81 bp of the blaVIM-1 59-base element present in the circular form of the β-lactamase gene cassettes to a 59-base element consensus sequence given below. The inverse core and core sequences are double underlined. L1, L2, R1, and R2 are four regions found to be highly conserved within 59-base elements of class 1 integrons (20, 23). Consensus bases (Cons) in uppercase letters are present in two-thirds or more of the 59-base elements, and bases in lowercase letters are present in half or more of the 59-base elements. Stars indicate bases of the 59-base element of blaVIM-2 that fit the consensus. R, purine; Y, pyrimidine; S, C or G; N, undetermined base.
Conjugation experiments failed to transfer β-lactam resistance from P. aeruginosa COL-1 to rifampin-resistant E. coli DH10B or ciprofloxacin-resistant P. aeruginosa strains. However, plasmid extraction from P. aeruginosa COL-1 followed by electroporation into E. coli gave a ca. 45-kb natural plasmid, pNOR-2000 (data not shown). This plasmid was not self-transferable from E. coli to E. coli. This plasmid conferred a similar resistance profile to β-lactams, as was found for recombinant plasmid pNOR-2001, as well as resistance to sulfamides. Hybridization experiments confirmed the presence of a plasmid of similar size in P. aeruginosa COL-1 as in the E. coli DH10B electroporant (data not shown).
Biochemical properties of VIM-2 and resistance pattern conferred by VIM-2.
IEF analysis of the β-lactamase preparation revealed that E. coli DH10B(pNOR-2001) produced only a single β-lactamase with a pI of 5.6. For P. aeruginosa COL-1, an additional band of β-lactamase activity with a pI of 9.0 was found that likely corresponded to the chromosomal P. aeruginosa AmpC cephalosporinase (26), the pI of 5.6 being revealed only after imipenem hydrolysis detection.
VIM-2 was purified 400-fold from a culture of E. coli DH10B(pNOR-2001) with a specific activity of 17.8 U · mg of protein−1 with imipenem as the substrate. The Mr of the mature β-lactamase was 29.7 kDa.
Kinetic parameters revealed that VIM-2 has a broad hydrolysis profile, including most β-lactams, except monobactams (aztreonam), cefsulodin, cefepime, and cefpirome (Table 4). VIM-2 activity was higher against imipenem than against meropenem. Its activity was inhibited by EDTA (IC50, 50 μM) and was restored in the presence of 2 mM ZnCl2. Thus, VIM-2 could be included in the functional group 3a of the Bush β-lactamase classification that includes most metalloenzymes, except those from Aeromonas sp., Myroides odoratus, and Legionella gormanii, which show a restricted hydrolysis spectrum to carbapenems (3, 18).
TABLE 4.
Kinetic parameters of the purified β-lactamase VIM-2
| Antimicrobial agent | kcat (s−1) | Km (μM) | kcat/Km (μM−1 · s−1) |
|---|---|---|---|
| Benzylpenicillin | 55.8 | 49 | 1.14 |
| Amoxicillin | 29.7 | 54 | 0.55 |
| Ticarcillin | 31.7 | 46 | 0.69 |
| Piperacillin | 32.7 | 72 | 0.45 |
| Cephalothin | 56.2 | 44 | 1.28 |
| Cefoxitin | 2.8 | 24 | 0.12 |
| Cefuroxime | 12.1 | 22 | 0.55 |
| Cefoperazone | 29.8 | 49 | 0.61 |
| Cefsulodin | 26.0 | 521 | 0.05 |
| Cefotaxime | 27.5 | 32 | 0.86 |
| Cefpirome | 9.2 | 123 | 0.07 |
| Cefepime | 4.7 | 184 | 0.03 |
| Ceftazidime | 88.7 | 98 | 0.90 |
| Aztreonam | <0.5 | NDa | —b |
| Moxalactam | 14.8 | 80 | 0.18 |
| Meropenem | 1.4 | 5 | 0.28 |
| Imipenem | 9.9 | 10 | 0.99 |
ND, not determinable.
—, in this case, the hydrolysis parameters could not be calculated.
The natural plasmid pNOR-2000 (data not shown) or the recombinant plasmid pNOR-2001 conferred resistance to aminopenicillins and narrow- and extended-spectrum cephalosporins and a reduced susceptibility to piperacillin, cefepime, and carbapenems in E. coli DH10B. However, the MIC of aztreonam for E. coli DH10B(pNOR-2001) remained unchanged compared to those for the parental E. coli DH10B strain (Table 2).
DISCUSSION
The carbapenem-hydrolyzing β-lactamase VIM-2 shared 90% amino acid identity with VIM-1. It has been obtained from an isolate from the French Riviera region (Marseilles) that is only 300 km from Verona, where VIM-1 had been isolated (10). Moreover, patient transfers were common between Italian hospitals and Marseilles hospitals until 1994, thus underlining a possible regional outbreak of organisms producing related enzymes.
Both VIM-1 and VIM-2 can be classified in the protein sequence-based subclass B1 of metallo-β-lactamases (3). The amino acids that may be involved in the catalytic site of these enzymes were identical (18, 24, 27). Once cloned onto a plasmid vector and expressed in E. coli, both enzymes provided a similar pattern of decreased susceptibility to β-lactams, except aztreonam. However, their level of resistance to carbapenems remained low. As suggested from results of experiments performed with another carbapenem-hydrolyzing β-lactamase (an IMP-1-like enzyme), the permeability coefficient of each β-lactam may play a major role in explaining the level of resistance to each β-lactam in gram-negative bacteria that produce metalloenzymes (13), thus explaining the low level of resistance to ureidopenicillins in P. aeruginosa COL-1 (Table 1). VIM-2 did not significantly hydrolyze either cefsulodin, cefepime, or cefpirome. The high MIC of cefepime for P. aeruginosa COL-1 could be due to its low permeability coefficient (13). Interestingly, P. aeruginosa isolates that expressed either VIM-1 or VIM-2 β-lactamases were fully susceptible to aztreonam only and resistant to most aminoglycosides, thus limiting the choice of active drugs in clinical use.
Taking into account the structural similarity between VIM-1 and VIM-2 and the similar MIC data, it is likely that the biochemical properties of VIM-1 are close to those of VIM-2. Although related to B-II from B. cereus, VIM-2 did not share its peculiar property of better hydrolyzing meropenem than imipenem (3, 6, 18). The extended hydrolysis profile of VIM-2 was different from the restricted hydrolysis profile found for CphA-1 and ImiS from Aeromonas species (3, 18). Therefore, VIM-2 could be included in biochemical group 3a (3).
While the G+C content of blaVIM-1 is not typical of P. aeruginosa genes, it could correspond to that of genes found in members of the family Enterobacteriaceae. Upstream of blaVIM-2, two putative promoters, Pc and P2, were found (Fig. 2). Compared to other Pc sequences, the Pc promoter for blaVIM-2 is a weak promoter (4, 11). P2 expression may be responsible for up to 90% of blaVIM-2 transcription, as described for other integron-located genes (4, 11). blaVIM-1 and blaVIM-2 are located on different class 1 integrons not related to blaIMP-1 integrons, and the corresponding 59-base elements were different in size and structure (Fig. 5). A similar situation was observed for dfrA1, dfrA5, and dfrA7 genes, which share 70% amino acid identity, but have unrelated 59-base elements, with the first and last 20 bp of these 59-base elements showing similarity to the consensus (20, 23). The catB2 and catB3 cassettes also contain quite different 59-base elements (20). The fact that closely related genes such as the VIM-1 and VIM-2 genes have different 59-base elements supports the hypothesis of a separate origin of the genes and of the 59-base element in each cassette. The class 1 integron for blaVIM-2 contained only this gene cassette, as opposed to the class 1 integron that contained blaVIM-1 together with at least another gene cassette (Fig. 1) (10).
The dendrogram analysis revealed that VIM-2 and VIM-1 clustered in the same carbapenem-hydrolyzing β-lactamase subgroup and that neither of them is related to the chromosome-borne class B carbapenem-hydrolyzing β-lactamases (Fig. 3). It may now be time to detect gram-negative rods that produce these novel expanded-spectrum β-lactamases to prevent their spread. Their detection should be performed with a PCR technique using, for example, the following consensus primer sequences: VIMB, 5′-ATGGTGTTTGGTCGCATATC-3′; and VIMF, 5′-TGGGCCATTCAGCCAGATC-3′.
ACKNOWLEDGMENTS
This work was funded by the Ministère de l'Education Nationale et de la Recherche, Université Paris XI, Faculté de Médecine Paris Sud (UPRES, JE-2227); and the French network on β-lactamase research “Les β-lactamases: de l'observation clinique à la structure,” France.
We thank Y. Arakawa for the gift of the IMP-1-containing P. aeruginosa isolate MKAM 12.
REFERENCES
- 1.Arakawa Y, Murakami M, Suzuki K, Ito H, Wacharotayankun R, Ohsuka S, Kato N, Ohta M. A novel integron-like element carrying the metallo-β-lactamase gene blaIMP. Antimicrob Agents Chemother. 1995;39:1612–1615. doi: 10.1128/aac.39.7.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bellais S, Léotard S, Poirel L, Naas T, Nordmann P. Molecular characterization of a carbapenem-hydrolyzing β-lactamase from Chryseobacterium (Flavobacterium) indologenes. FEMS Microbiol Lett. 1999;171:127–132. doi: 10.1111/j.1574-6968.1999.tb13422.x. [DOI] [PubMed] [Google Scholar]
- 3.Bush K. Metallo-enzymes: a class apart. Clin Infect Dis. 1998;27(Suppl. 1):S48–S53. doi: 10.1086/514922. [DOI] [PubMed] [Google Scholar]
- 4.Collis C M, Hall R M. Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob Agents Chemother. 1995;39:155–162. doi: 10.1128/aac.39.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Da Silva G J, Leitão R, Peixe L. Emergence of carbapenem-hydrolyzing enzymes in Acinetobacter baumannii clinical isolates. J Clin Microbiol. 1999;37:2109–2110. doi: 10.1128/jcm.37.6.2109-2110.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Felici A, Amicosante G, Oratore A, Strom R, Ledent P, Joris B, Fanuel L, Frère J-M. An overview of the kinetic parameters of class B β-lactamases. Biochem J. 1993;291:151–155. doi: 10.1042/bj2910151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirakata Y, Izumikawa K, Yamaguchi T, Takemura H, Tanaka H, Yoshida R, Matsuda J, Nakano M, Tomono K, Maesaki S, Kaku M, Yamada Y, Kamihira S, Kohno S. Rapid detection and evaluation of clinical characteristics of emerging multiple-drug-resistant gram-negative rods carrying the metallo-β-lactamase gene blaIMP. Antimicrob Agents Chemother. 1998;42:2006–2011. doi: 10.1128/aac.42.8.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hussain M, Carlino A, Madonna M J, Lampen J O. Cloning and sequencing of the metallothioprotein β-lactamase II gene of Bacillus cereus 569/H in Escherichia coli. J Bacteriol. 1985;164:223–229. doi: 10.1128/jb.164.1.223-229.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laraki N, Galleni M, Thamm I, Riccio M L, Amicosante G, Frère J-M, Rossolini G M. Structure of In31, a blaIMP-containing Pseudomonas aeruginosa integron phyletically related to In5, which carries an unusual array of gene cassettes. Antimicrob Agents Chemother. 1999;43:890–901. doi: 10.1128/aac.43.4.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lauretti L, Riccio M L, Mazzariol A, Cornaglia G, Amicosante G, Fontana R, Rossolini G M. Cloning and characterization of blaVIM, a new integron-borne metallo-β-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob Agents Chemother. 1999;43:1584–1590. doi: 10.1128/aac.43.7.1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lévesque C, Brassard S, Lapointe J, Roy P H. Diversity and relative strength of tandem promoters for the antibiotic-resistance genes of several integrons. Gene. 1994;142:49–54. doi: 10.1016/0378-1119(94)90353-0. [DOI] [PubMed] [Google Scholar]
- 12.Massidda O, Rossolini G M, Satta G. The Aeromonas hydrophila cphA gene: molecular heterogeneity among class B metallo-β-lactamases. J Bacteriol. 1991;173:4611–4617. doi: 10.1128/jb.173.15.4611-4617.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsumura N, Minami S, Watanabe Y, Iyobe S, Mitsuhashi S. Role of permeability in the activities of β-lactams against gram-negative bacteria which produce a group 3 β-lactamase. Antimicrob Agents Chemother. 1999;43:2084–2086. doi: 10.1128/aac.43.8.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nielsen H, Engelbrecht J, Brunak S, Von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 15.Osano E, Arakawa Y, Wacharotayankun R, Ohta M, Horii T, Ito H, Yoshimura F, Kato N. Molecular characterization of an enterobacterial metallo β-lactamase found in a clinical isolate of Serratia marcescens that shows imipenem resistance. Antimicrob Agents Chemother. 1994;38:71–78. doi: 10.1128/aac.38.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poirel L, Guibert M, Girlich D, Naas T, Nordmann P. Cloning, sequence analyses, expression and distribution of ampC-ampR from Morganella morganii clinical isolates. Antimicrob Agents Chemother. 1999;43:769–776. doi: 10.1128/aac.43.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poirel L, Naas T, Guibert M, Chaibi E B, Labia R, Nordmann P. Molecular and biochemical characterization of VEB-1, a novel class A extended-spectrum β-lactamase encoded by an Escherichia coli integron gene. Antimicrob Agents Chemother. 1999;43:573–581. doi: 10.1128/aac.43.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rasmussen B A, Bush K. Carbapenem-hydrolyzing β-lactamases. Antimicrob Agents Chemother. 1997;41:223–232. doi: 10.1128/aac.41.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rasmussen B A, Gluzman Y, Tally F P. Cloning and sequencing of the class B β-lactamase gene (ccrA) from Bacteroides fragilis TAL3636. Antimicrob Agents Chemother. 1990;34:1590–1592. doi: 10.1128/aac.34.8.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Recchia G D, Hall R M. Gene cassettes: a new class of mobile elements. Microbiology. 1995;141:3015–3027. doi: 10.1099/13500872-141-12-3015. [DOI] [PubMed] [Google Scholar]
- 21.Rossolini G M, Franceschini N, Riccio M L, Mercuri P S, Perilli M, Galleni M, Frère J-M, Amicosante G. Characterization and sequence of the Chryseobacterium (Flavobacterium) meningosepticum carbapenemase: a new molecular class B β-lactamase showing a broad substrate profile. Biochem J. 1998;332:145–152. doi: 10.1042/bj3320145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 23.Stokes H W, O'Gorman D B, Recchia G D, Parsekhian M, Hall R M. Structure and function of 59-base element recombination sites associated with mobile gene cassettes. Mol Microbiol. 1997;26:731–745. doi: 10.1046/j.1365-2958.1997.6091980.x. [DOI] [PubMed] [Google Scholar]
- 24.Ullah J H, Walsh T R, Taylor I A, Emery D C, Vermas C S, Gamblin S J, Spencer J. The crystal structure of the L-1 metallo-β-lactamase from Stenotrophomonas maltophilia at 12.7 A resolution. J Mol Biol. 1998;284:125–136. doi: 10.1006/jmbi.1998.2148. [DOI] [PubMed] [Google Scholar]
- 25.Walsh T R, Hall L, Assinder S J, Nichols W W, Cartwright S J, MacGowan A P, Bennett P M. Sequence analysis of the L-1 metallo β-lactamase from Xanthomonas maltophilia. Biochim Biophys Acta. 1994;1218:199–201. doi: 10.1016/0167-4781(94)90011-6. [DOI] [PubMed] [Google Scholar]
- 26.Walther-Rasmussen J, Johnsen A H, Hoiby N. Terminal truncations in AmpC β-lactamase from a clinical isolate of Pseudomonas aeruginosa. Eur J Biochem. 1998;263:478–485. doi: 10.1046/j.1432-1327.1999.00529.x. [DOI] [PubMed] [Google Scholar]
- 27.Wang Z, Fast W, Benkovic S J. On the mechanism of the metallo-β-lactamase from Bacteroides fragilis. Biochemistry. 1999;38:10013–10023. doi: 10.1021/bi990356r. [DOI] [PubMed] [Google Scholar]
- 28.West S E H, Iglewski B H. Codon usage in Pseudomonas aeruginosa. Nucleic Acids Res. 1998;16:9323–9335. doi: 10.1093/nar/16.19.9323. [DOI] [PMC free article] [PubMed] [Google Scholar]