Abstract
The combination of Ocimum sanctum and Trigonella foenum-graecum L leaf water extract synergistically acts as a reducing and capping agent for the synthesis of narrow polydisperse silver nanoparticles (Ag NPs) with controlled sizes depending on the precursor (AgNO3) concentration in the plant extract. The toxicity of 40 nm-sized green synthesized Ag NPs is less than that of 10 nm-sized NPs. The Ag NP solution in Ocimum sanctum and Trigonella foenum-graecum L leaf water extract shows synergic antibacterial effect on Gram-negative bacteria by effecting the ester group of the lipids (hydrolysis) and also breaking the amide bonds of the bacterial chemical constituents, which leads to their rapid death.
Combining Ocimum sanctum and Trigonella foenum-graecum L leaf water extracts synergistically act as a reducing and capping agent for the synthesis of narrow polydisperse silver nanoparticles with controlled sizes depending on the precursor (AgNO3) concentration.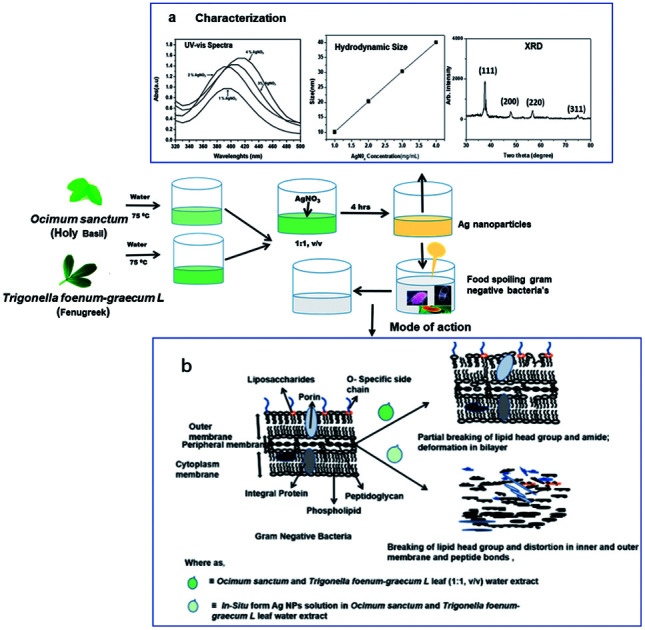
Introduction
Chemical reactions that cause offensive sensory changes in foods are mediated by a variety of microbes that use food as a carbon and energy source.1–3 These organisms include prokaryotes (bacteria) such as Escherichia coli, Lactobacillus acidophilus, Staphylococcus aureus, Bacillus cereus, Pseudomonas (P) putida, and P. fluorescens, leading to adverse effects in food preservation.4,5Pseudomonas fragi is a psychrotrophic species that is mainly responsible for the spoilage of meat stored aerobically at refrigeration temperatures. P. fragi is more widely distributed than other Pseudomonas, and has been isolated from water, soil, plant materials, and other natural media. This species grows well at temperatures ranging from 2 °C to 35 °C.6,7 The prevalence and growth conditions of P. fragi contribute towards its successful proliferation on foods, especially fresh meat, and the strict chill chain applied during fresh meat production from slaughtering to final distribution selectively favours its development.8 Moreover, P. fragi has been suggested to promote the growth and survival of several food borne pathogens, such as S. aureus and Listeria monocytogenes.9 Therefore, information about P. fragi growth and biological responses to preservation conditions during food storage is urgently needed to improve food safety.
Many Gram-negative bacteria, including Pseudomonas, secrete acyl homoserine lactones (AHLs) to regulate the expression of certain genes, such as virulence factors, as a function of cell density.10 These AHL quorum-sensing signals may regulate proteolytic enzyme production and iron chelation during the spoilage of some foods.11 Microbial spoilage is of particular concern and can be caused by either psychrotolerant spore-formers, which likely originate from raw milk, or by post pasteurization contamination.12–15
Recently, researchers have focused on nanotechnology-based techniques for food preservation.16–20 Silver nanoparticles (Ag NPs) are well known as an antibacterial agent21–23 and are employed in the textile as well as in the food/cosmetic industries as an additive to control microorganisms.24,25 In the past, researchers have successfully used Ag NPs as a strong biocide agent for various micro-organisms26–28 and fungi.29 However, recent interest on Ag NPs has particularly focused on the increasing threat of antibiotic resistance due to the excessive use/abuse of antibiotics.30
In general, the mode of antibacterial action of Ag ions or Ag NPs depends on the (a) attachment of Ag ions or Ag NPs to the cell wall, which disrupts the cell-wall permeability and respiration of the bacteria, and the (b) interaction with loan pair electron containing atoms such as the phosphorus, nitrogen or sulphur groups of DNA, as well as the proteins of bacteria to penetrate inside mammalian cells.31 Ag NPs exhibit improved antibacterial properties compared to bulk silver, mainly due to the high surface area and fraction of their surface atoms, so as a result, more of the smaller NPs are incorporated inside the bacteria, promoting their efficacy in a sustained manner.32,33 In addition, Ag NPs are relatively nontoxic to human cells.34 Considering their wide range of applications, Ag NPs have been synthesized using various methods over the years. In terms of chemical reduction methods, reducing agents such as NaBH4, citrate, or ascorbate are the most commonly used chemicals.35 However, such reducing agents and synthetic organic material templates may be associated with environmental toxicity or biological hazards.36
Recently, the synthesis of Ag NPs via a biological reduction method has been developed as a promising technique because of its special advantages, such as mild reaction conditions, nontoxic reagents, and absence of synthetic chemical additives as the stabilizing or reducing agents of the NPs.30 Because of their environmentally-friendly nature, biosynthetic processes are being researched for the synthesis of Ag NPs.37 In biosynthesis methods, the reaction time for the reduction of silver ions requires more time due to the slow rate of the reaction. For example, the synthesis of Ag NPs by bacteria requires 24–120 h to complete the reduction process, with high polydispersity and aggregation of the NPs.38
Ocimum sanctum (holy basil) (OS) is a well-known medicinal plant that is used as a folk medicine in the treatment of hepatic amoebiasis and rheumatism. It is also employed in the treatment of various disease and issues, such as infections, arthritis, chronic fever, fertility, and eye disease, due to its antimicrobial, antifungal, anticancer, hepatoprotective, antispasmodic, analgesic, antiemetic, and cardioprotective properties.39–41 This medicinal herb has also been shown to reduce blood glucose levels, making it an effective treatment of diabetes. There are many chemical constituents present in OS, such as ascorbic acid, oleanolic acid, rosmarinic acid, ursolic acid, eugenol, linalool, carvacrol, β-elemene, β-caryophyllene, germacrene, amino acids, etc.39–41 Similarly, Trigonella foenum-graecum L leaf (TFG) is also rich in ascorbic acid and many chemicals, such as sapiens, steroids, phytosterol terpenes, fatty acids, and phenolic acids, and has several medicinal properties, including antifungal and antibacterial activities.42 There have been several reports in the literature on using OS and TFGL extracts for the synthesis of a broad range of polydisperse NPs.43–46
The purpose of the current research is broadly based on adding value to agriculture products using simple techniques. For this purpose, we selected two medicinal plants, OS and TFG, to optimize their antibacterial effects, as well as use them to synthesize narrow disperse Ag NPs and measure their antibacterial properties. Herein, we used leaf extracts of OS and TFG (1 : 1, v/v) in deionized water and determined their antibacterial effects, as well as using them in the reduction and stabilizing of silver ions in Ag NPs. The reduction of Ag ions took place within 4 h, hence representing a substantial reduction in their preparation time. Synthesized Ag NPs were characterized by UV-visible and attenuated total reflectance/Fourier-transform infrared (ATR/FT-IR) spectroscopy, X-ray diffraction (XRD), dynamic light scattering, and high-resolution transmission electron microscopy (HR-TEM) (for details of the extraction, synthesis of the NPs and their characterization procedures, see the ESI†). The toxicological properties of various sized Ag NPs were studied. We chose NPs with insignificant toxicity for the studies on the synergic effect of the concentration-dependent antimicrobial activity of green Ag NPs against Pseudomonas fragi, Pseudomonas fluorescens, and Salmonella, in presence and absence of OS and TFG leaf extracts in water. Also, OS and TFG leaf extracts in water alone were tested for antibacterial activities for comparison purposes.
Results and discussion
Characterization of the Ag NPs
Analysis of the UV-vis, DLS, and XRD data
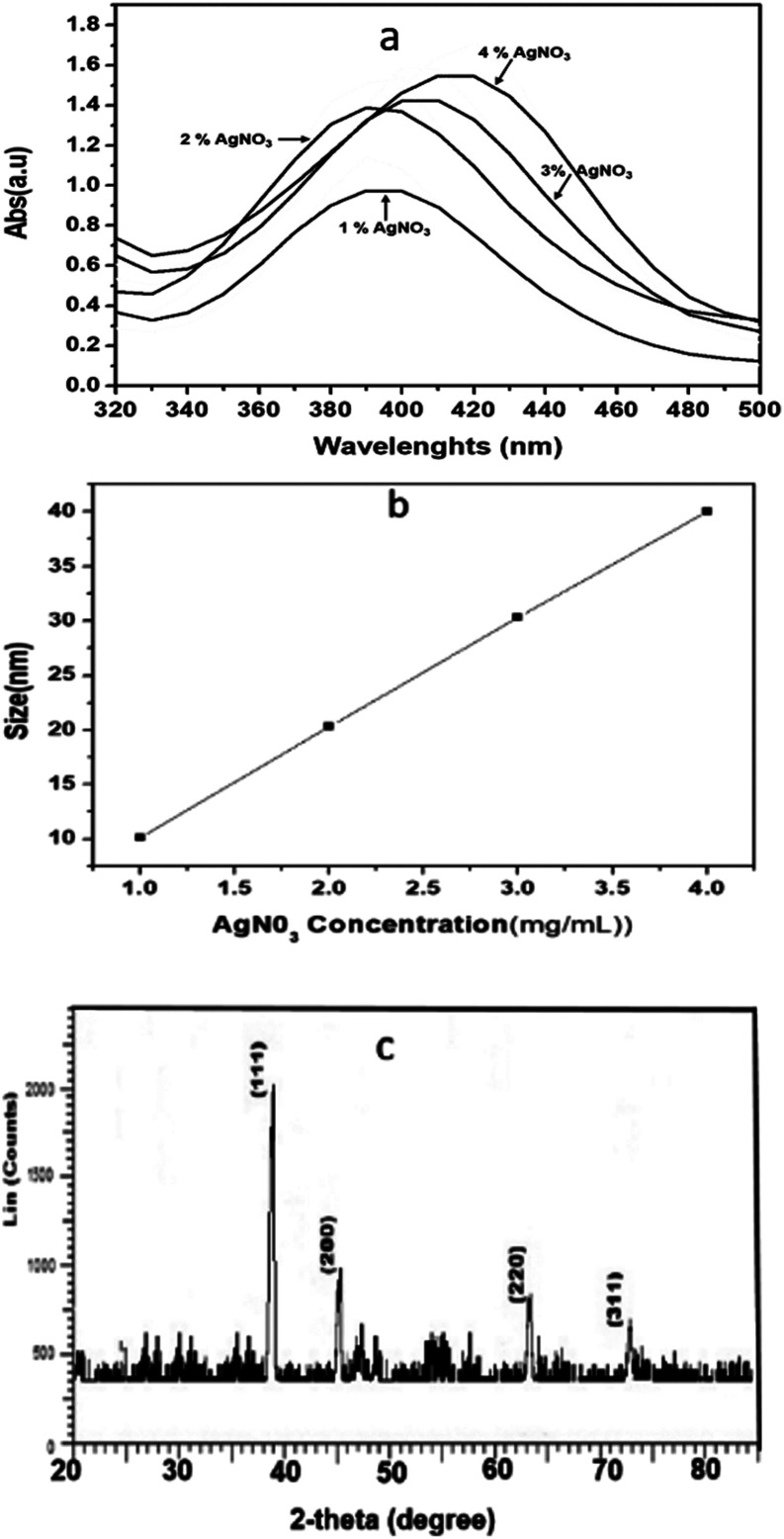
The UV-vis spectra of the biochemically synthesized green Ag NPs are shown in Fig. 1a. The spectra illustrate that with an increasing concentration of silver salt added during synthesis, the number of nuclei formed increased and bigger NPs were formed than at lower concentration, with a red shift accordingly observed in the plasma band of the Ag NPs. Absorption spectra were recorded at 397, 407, 412, and 429 nm for Ag NPs synthesized at 1, 2, 3, and 4 wt%, respectively, which reflect the effect that the precursor concentration has on the size of the Ag NPs.47,48
Fig. 1. (a) UV-vis spectra, (b) DLS data showing the number average mean hydrodynamic size of Ag NPs synthesized using a mixture of Ocimum sanctum (OS) and Trigonella foenum-graecum L (TFG) leaf water extracts (1 : 1, v/v) at precursor (AgNO3) concentrations of 1, 2, 3, and 4 wt% with respect to the leaf weight, and (c) the powder XRD pattern of a lipolyzed sample showing characteristics peak of the Ag NPs (4 weight% AgNO3 with respect to the leaf weight of OS + TFG, 40 nm).
The hydrodynamic diameters (d) of the Ag NPs were calculated using the Stokes–Einstein equation d = kBT/3πηD, where kB is the Boltzmann constant, T is the absolute temperature, η is the solvent viscosity, and D is the diffusion coefficient. CONTIN algorithms were used in the Laplace inversion of the autocorrelation function to obtain the size distribution. The number average hydrodynamic sizes of the Ag NPs at 1, 2, 3, and 4 wt% with respect to leaf weight, were 10.13, 20.21, 30.21, and 40 nm, respectively with a narrow polydispersity index (0.22–0.29) (Fig. 1b). Fig. 1c shows the characteristic two theta peaks of the green Ag NPs. The characteristics peaks of (111), (200), (220), and (311) can be attributed to the reflection of the face centred cubic (fcc) lattice of the Ag NPs (compared with JCPDS file no: 89-3722). As the concentration of the precursor increased, the plasma band of the Ag NPs red shifted, which indicates the increase in the size of Ag (Fig. 1a) was confirmed by measuring the hydrodynamic size via DLS (Fig. 1b). The narrow polydispersity index (0.22 to 0.29) measured by DLS is supported by the sharp XRD peaks (Fig. 1c, low bandwidth of the peaks and ESI, Fig. S1†).47 However, for the synthesis of Ag NPs in OS or TFG leaf water extracts, a polydisperse hydrodynamic size of Ag NPs was observed (ESI, Fig. S2†).
Morphology of the Ag nanoparticles
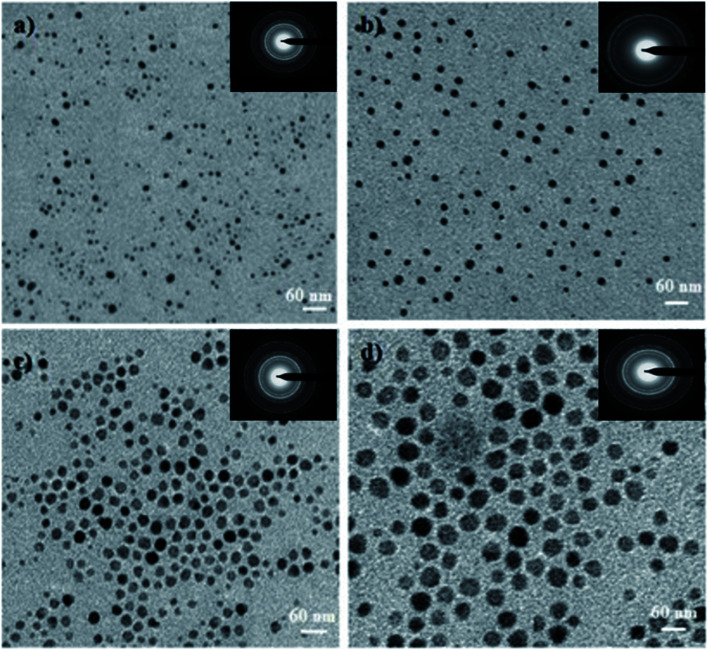
The average size of the Ag NPs synthesized via a green synthesis route was calculated from the TEM images to be 10, 20, 30, and 40 nm at weight 1, 2, 3, and 4 wt%, respectively, in a mixed aqueous solution of OS and TFG extracts (Fig. 2a–d). All of the NPs were found to be spherical in shape, in correlation with data from both DLS and UV-vis spectroscopy measurements (Fig. 2).48,49 TEM micrographs (ESI, Fig. S3†) and XRD spectra (ESI, Fig. S4†) of the Ag NPs synthesized in OS and TFG leaf water extracts are shown in the ESI.† TEM and XRD data of the citrate-capped synthesized Ag NPs are also shown in the ESI (Fig. S5†).
Fig. 2. TEM micrographs of the Ag NPs synthesized using a mixture of Ocimum sanctum and Trigonella foenum-graecum L leaf at concentrations of (a) 1, (b) 2, (c) 3, and (d) 4 wt% of AgNO3 with respect to the plant leaf weight. The inset images show the corresponding selected area electron diffraction (SAED) patterns.
Toxicity analysis
MTT assay
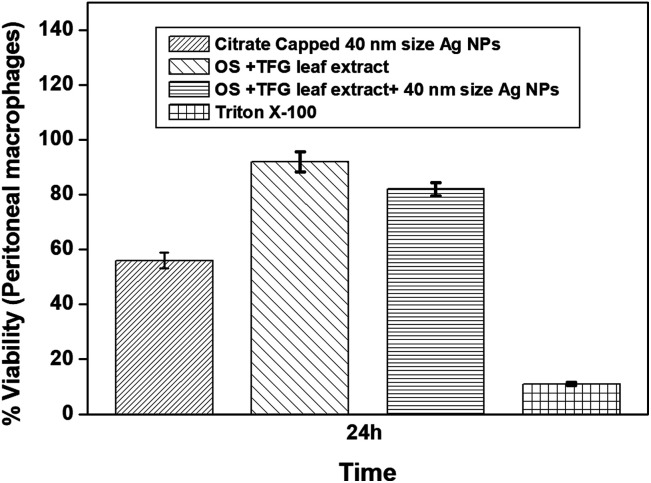
A dose dependent response was observed in MTT assay for calculating the LD50 values and cytotoxicity data of the Ag NPs, citrate-capped Ag NPs (40 nm size), and AgNO3, as presented in Fig. 3 (the method for the MTT assay is shown in the ESI). The LD50 values for the 10 nm Ag NPs was the most sensitive amongst all the NPs (ESI, Fig. S3a†) in both macrophages, while the 40 nm spherical shaped Ag NPs (Fig. 3) were found to be the least sensitive in macrophages, where the cell viability was found to decrease with a decrease in the size of the Ag NPs. Also, the toxicity of the Ag NPs was found to be less than those of AgNO3 in water (ESI, Fig. S6†) and the 40 nm sized citrate-capped Ag NPs (Fig. 3). The LD50 values of the 10 nm Ag NPs are much lower than those of the 40 nm spherical Ag NPs after 24 h of exposure time.
Fig. 3. Plots of cell viability (MTT assay) studied for citrate-capped Ag NPs (40 nm), leaf extracts and Ag NPs synthesized using a mixture of Ocimum sanctum and Trigonella foenum-graecum L leaf extract in water compared to the negative control (Triton X-100) after 24 h of incubation at the same concentration of 0.8 mg mL−1 (confidence interval ± 1.09). This shows that the green synthesized Ag NPs are less cytotoxic in comparison to the citrate-capped Ag NPs.
Hemolysis assay
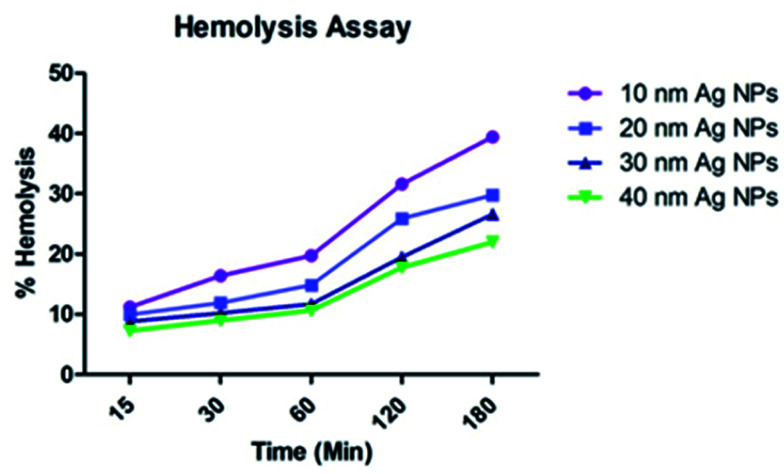
The results of hemolytic experiments indicate that the 10, 20, 30, and 40 nm NPs (Fig. 4) show slightly decreased blood compatibility, but are non-toxic up to 1 mg mL−1 towards erythrocytes, which places significant emphasis on the usefulness of these nano-systems in the treatment of many diseases.48
Fig. 4. Size dependent toxicity of the green-synthesized Ag NPs towards erythrocytes.
Antimicrobial activity
The antibacterial activity of the green-synthesized Ag NPs was investigated against the Gram-negative strains P. fragi, P. fluorescens, and Salmonella, which are responsible for food spoilage and cause food poisoning, typhoid fever, and paratyphoid fever. This is the first report, as per our knowledge, demonstrating the synergic effect of Ag NPs and OS and TFG leaf extracts on the antibiotic resistant microbes P. fragi, P. fluorescens, and Salmonella. Thus, this information will be useful for making food preservatives as well as drug delivery formulations for the treatment of diseases related to microbial infection. The minimum inhibition concentrations (MICs) against 105 CFU mL−1 of P. fragi, P. fluorescens, and Salmonella were calculated to be 2.10, 2.18, and 2.20 μg mL−1, respectively, whereas the minimum bactericidal concentration (MBC) values for the respective bacteria were 3.80, 4.50, and 4.30 μg mL−1 (Table 1). However, the MIC value of the citrate-capped Ag NPs was in the range of 7.20 to 8.21 μg mL−1 for the above-mentioned Gram-negative bacteria and for the combined solution of OS and TFG leaf extracts in water (1 : 1, v/v), the MIC values were 62 ± 5.3 μg mL−1, respectively (Table S1†), and the aqueous OS and TFG leaf extracts alone exhibit roughly 4 and 3.7 times higher MIC values, respectively, than when the two are combined. The MBC value for each sample is roughly double their respective MIC value, which is in line with reported studies.46 The data from the antibacterial experiments carried out using a disc diffusion method are shown in the ESI (Fig. S7†).
Antimicrobial action, minimum inhibition concentration (MIC), and minimum bactericidal concentration (MBC) of plant leaf extract (with and without 40 nm sized Ag NPs) against selected Gram-negative bacteriaa.
| Systems | Concentration | |||||||
|---|---|---|---|---|---|---|---|---|
| 0.1 mg mL−1 | 0.2 mg mL−1 | 0.4 mg mL−1 | 0.6 mg mL−1 | 0.8 mg mL−1 | Control | MIC (μg mL−1) | MBC (μg mL−1) | |
| Pseudomonas fragi treated with | (Zone of inhibition in mm) | |||||||
| Sample A | 11 | 14 | 17 | 19 | 21 | 0 | 2.1 | 3.8 |
| Sample B | 5 | 8 | 10 | 13 | 15 | 0 | 7.2 | 15 |
| Sample C | 0 | 0 | 2 | 5 | 7 | 0 | 58 | 132 |
| Sample D | 0 | 0 | 0 | 4 | 6 | 0 | 98 | 101 |
| Sample E | 0 | 0 | 0 | 3 | 5 | 0 | 110 | 232 |
| Pseudomonas fluorescens treated with | ||||||||
| Sample A | 12 | 16 | 19 | 22 | 25 | 0 | 2.18 | 4.5 |
| Sample B | 5 | 9 | 11 | 14 | 16 | 0 | 7.5 | 14.2 |
| Sample C | 0 | 0 | 3 | 6 | 8 | 0 | 65 | 142 |
| Sample D | 0 | 0 | 0 | 5 | 7 | 0 | 104 | 195 |
| Sample E | 0 | 0 | 0 | 4 | 6 | 0 | 121 | 240 |
| Salmonella treated with | ||||||||
| Sample A | 12 | 14 | 15 | 17 | 21 | 0 | 2.2 | 4.3 |
| Sample B | 6 | 9 | 10 | 12 | 15 | 0 | 8.1 | 15.2 |
| Sample C | 0 | 0 | 2 | 4 | 6 | 0 | 55.3 | 104 |
| Sample D | 0 | 0 | 0 | 3 | 6 | 0 | 88 | 170 |
| Sample E | 0 | 0 | 0 | 3 | 5 | 0 | 101 | 194 |
Sample A: in situ 40 nm-sized green-synthesized silver nanoparticles in a combination of aqueous extracts of Ocimum sanctum and Trigonella foenum-graecum L; sample B: 40 nm-sized citrate-capped Ag NPs; sample C: combination of aqueous extracts of Ocimum sanctum and Trigonella foenum-graecum L; sample D: aqueous extract of Trigonella foenum-graecum L; sample E: aqueous extract of Ocimum sanctum leaf.
ATR/FT-IR spectroscopy analysis
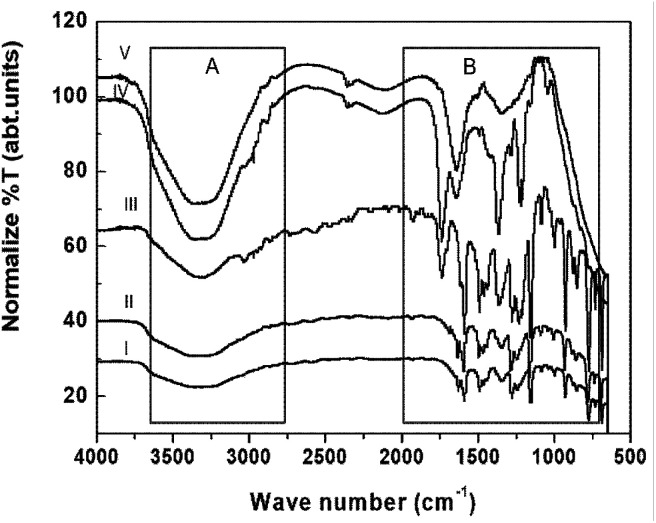
ATR/FT-IR spectroscopy analysis was carried out to determine the interactions between or among the functional groups of the constituents of the systems and the experimental data are shown in Fig. 5. The possible functional groups of the chemicals in the aqueous OS and TFG leaf extracts involved in the NP synthesis were identified by ATR/FT-IR spectroscopy analysis. The broad bands in the range of 3336 to 3100 cm−1 can be attributed to H–O–H, –O–H stretching of phenols, hydrogen-bonded groups, amines, and amino acids.50 The band at 3000–3020 cm−1 indicates the presence of aromatic compound –C–H stretching and in the range of 2990 to 2700 cm−1 aliphatic –C–H peaks were observed for all of the samples (Fig. 5, zone A). The peak at 2724 cm−1 indicates the presence of the –C–H stretching of an aldehyde. After the reduction of AgNO3, a shift in the band of amide II (1590 to 1572 cm−1) and 1640 cm−1 was observed and a peak near to 1687 cm−1, along with a –C–O stretching peak at 1232 cm−1, appeared that can be attributed to a carboxyl group (Fig. 5, spectra II, zone B). The peaks observed at 848, 731, and 680 cm−1 correspond to the C–H stretching of alkenes. The hydrogen-bonded C–N band at 996 cm−1 shifts to 1005 cm−1, which indicates partial positive charge on nitrogen atoms in presence of Ag NPs (Fig. 5, spectra I and II, zone B).50–54
Fig. 5. FT-IR/ATR spectra of (I) aqueous extracts of Ocimum sanctum (OS) and Trigonella foenum-graecum L (TFG) leaf, (II) green-synthesized Ag NPs in a solution of aqueous extracts of OS and TFG leaf, (III) P. fragi Gram-negative bacteria in aqueous solution, (IV) P. fragi Gram-negative bacteria treated with an aqueous solution of extracts of OS and TFG leaf, and (V) P. fragi Gram-negative bacteria treated with green-synthesized Ag NPs in an aqueous solution of extracts of OS and TFG leaf.
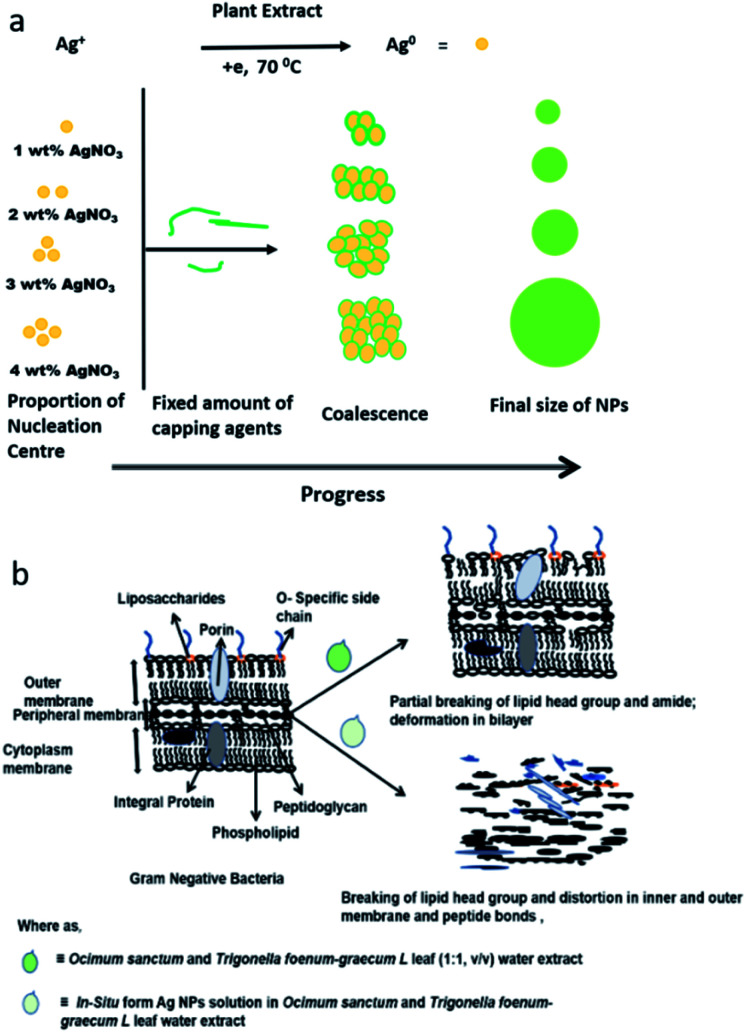
The FT-IR/ATR data suggests that several loan pair containing and polar functional groups present in the plant extract are capable of covering and stabilizing the Ag NP surface (which has positive charge on the surface of the NPs due to plasmon resonance) during nucleation, with concentration dependent narrow polydisperse NPs formed in aqueous medium as a result (Fig. 1b and 2). During the synthesis of the NPs, atoms and ions are reduced one by one and act as individual centres for nucleation and further coalescence due to high binding energy between two atoms or atoms and unreduced ions in comparison to the binding energy of atom-solvent and atom-capping ligand.55 Consequently, the atoms dimerize or become associated with unreduced metal ions. Also, due to their large surface area, NPs possess high surface energy and as a result thermodynamically unstable colloids can associate with each other via van der Waals forces of attraction. However, control of the final size of the NPs depends on the restraints applied to the coalescence by balancing the attractive forces with repulsive forces through electrostatic/static stabilization (Fig. 6a).56 The capping agent that binds the precursor (AgNO3) and as well as Ag nanoparticles on their surface and shows strong effect on final size of the synthesised metal NPs. As the concentration of the precursor increased at a fixed concentration of the capping agent, covering of the NP surfaces by ligand reduced as van der Waals forces of attraction became more dominant, which led to larger sized NPs being formed. ATR data suggest that the plant extract contains amine, carbonyl, and phenolic-groups, which have a high affinity for metal atoms (Fig. 5, spectrum I). Concentration dependent size control of the Ag NPs in the aqueous OS and TFG leaf extract also indicates that the intermolecular interactions of plant extract constituents with Ag nuclei are stronger than their interactions with solvent.
Fig. 6. Model for (a) synthesis and (b) mode of action of Ag NPs in presence of Ocimum sanctum and Trigonella foenum-graecum L leaf water extract.
The ATR spectrum of the untreated bacteria shows a strong ester carbonyl band at 1742 cm−1 (Fig. 5, zone B, spectrum III) along with additional peaks at 1390, 1166, and 1088 cm−1. Phospholipid group peaks at 1224 and 1087 cm−1 corresponding to –OPO– and –P–OCH2, respectively, were also observed (Fig. 5, spectrum III, zone B).53 These peaks can be attributed to Gram-negative bacteria, wherein the bacteria cell membrane is composed of 25% phospholipids and more than 60% protein. The presence of C–O–C and C–O vibrations in the sugar rings in the various polysaccharide components can be identified from the peaks at 1167 and 932 cm−1 (Fig. 5, spectrum III, zone B). Also, FT-IR/ATR spectral peaks observed at 2973, 2941, 2856, (zone A) and 1464 cm−1 (zone B) confirm the occurrence of –C–H stretching (–CH3, –CH2) of alkyl chains (Fig. 5, spectrum III).50 The peaks at 1445 cm−1 and in the range of 980–835 cm−1 indicate the presence of C C bonding in the fatty acid chain and were observed for untreated Gram-negative bacteria (Fig. 5, spectrum III, zone B). Besides this, amide I (1628 cm−1) and amide II (1588 cm−1) peaks of the protein in the structure of the bacteria were also identified. After treatment with an aqueous extract of OS and TFG leaf, partial breaking of ester bonds and the formation of a new broad peak near to 1654 cm−1 were observed along with a shift in the alkyl peaks at 2973, 2941, and 2856, cm−1 to 2965, 2920, and 2819 cm−1, respectively (Fig. 5, zone A). Also, the amide II peak shifts from 1588 to 1574 cm−1 (Fig. 5, spectrum IV, zone B) and the phospholipid group peaks of –OPO– and –P–OCH2 slightly shift from 1224 and 1087 cm−1 to 1218 and 1057 cm−1, respectively (Fig. 5, zone B, spectrum IV). However, for Ag NPs in an aqueous extract of OS and TFG leaf, an ester peak was not observed and a very broad peak from 1780 to 1450 cm−1 appeared (Fig. 5, spectrum V, zone B) and noticeable peak separation can be seen in the range of 3400 to 3200 cm−1 for bacteria treated with aqueous extracts of OS and TFG leaf in the presence or absence of Ag NPs (Fig. 3, spectra IV and V, zone A). Minor peaks near to 1563 and 1541 cm−1 can be seen in the spectrum of the bacteria treated with Ag NPs containing aqueous extracts of OS and TFG.50–54
Analysis of FT-IR/ATR data suggests that the chemical constituents of the aqueous extracts of OS and TFG leaf partially break the ester bonds of the phospholipids and disturb the hydrogen bonding, responsible for crosslinking and stabilizing by effecting the pore size of the Gram-negative bacteria, which is a prominent reason for their antibiotic resistance.57,58 However, in the presence of Ag NPs in an aqueous solution of OS and TFG leaf extracts, the peak for the ester bonding of the lipids that are chemical constituents of the bacteria (Fig. 6b) disappeared in the ATR spectra, also the amide band appeared to fragment. The Ag NPs are more efficient for antibacterial application in presence of OS and TFG leaf extract due to presence of several interacting groups such as phenolic, carboxylic, carbonyl, alcoholic groups, etc. in plant extract which synergistically interfering in more efficiently way with bacterial cell wall and shows more effect on cellular interaction leading to cell death.30,57–62 These results show that Ag NPs exhibit potential antimicrobial activity and food preservation activity, even at low concentration.
Conclusions
In conclusion, well-defined monodispersed Ag NPs were synthesized via a biochemical method using a combined solution of Ocimum sanctum and Trigonella foenum-graecum L leaf extracts in aqueous media (1 : 1, v/v) as a reducing and stabilizing agents for NPs with sizes of 10, 20, 30, and 40 nm, respectively. Ag NPs of 40 nm in size exhibit excellent antibacterial activities against food spoiling Gram-negative bacteria. The effect of the size of the Ag NPs on murine peritoneal macrophages showed significant cell viability at various concentrations of the NPs after 24 h of treatment, thus indicating their non-toxic nature. The green-synthesized Ag NPs can thus be used as a cost-effective and nontoxic material for use in food preservation.
Abbreviations
- TEM
Transmission electron microscopy
- MTT
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- Ag NPs
Silver nanoparticles
- LD50
Median lethal dose
- DLS
Dynamic light scattering
- EDTA
Ethylenediaminetetraacetic acid
- DPPH
2-Diphenyl-1-picrylhydrazyl
- XRD
X-ray diffraction measurements
- MIC
Minimum inhibitory concentration
- MBC
Minimum bactericidal concentration
Funding sources
The research leading to these results received funding from the Ministry of Higher Education, Research and Innovation (MoHERI) of the Sultanate of Oman under the bulk funding program with agreement number MoHERI/BFP/ASU/01/2020 and TRC block funding agreement no. TRC/BFP/ASU/01/2018.
Author contributions
M. F. C. and M. C. contributed equally in designing the experiments and writing the manuscript; S. G. provided logistic support and R. A. B. was involved in supporting M. C. in the analysis of the results. M. F. C. prepared samples and carried out UV-vis spectroscopy, FT-IR and XRD; M. F. C. also carried out toxicological and antibacterial experiments with support from S. K. M. F. A. prepared samples for TEM analysis along with S. K. and A. C.; M. C. F., S.-G., and R. A. B. were also involved in text editing.
Conflicts of interest
There are no conflicts of interest.
Supplementary Material
Acknowledgments
M. C. A. thankful to the Director, All India Institute of Medical Science, New Delhi for providing with the laboratory facilities to carry out various experiments. Authors are also grateful to the Department of Science and Technology, Government of India. M. C. thanks to The Research Council (TRC) and Ministry of Higher Education, Research and Innovation (MoHERI), Sultanate of Oman for Block Funding.
Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra08098a
Notes and references
- Oladapo A. S. Akinyosoye F. A. Abiodun O. A. J. Microbiol. Res. 2014;8:1510–1515. [Google Scholar]
- Oms-Oliu G. Soliva-Fortuny R. Martin-Belloso O. Eur. Food Res. Technol. 2007;225:301–311. doi: 10.1007/s00217-006-0415-9. [DOI] [PubMed] [Google Scholar]
- Chesson L. A. Podlesak D. W. Thompson A. H. Cerling T. E. Ehleringer J. R. J. Agric. Food Chem. 2008;56:4084–4091. doi: 10.1021/jf0733618. [DOI] [PubMed] [Google Scholar]
- Nath S. Chowdhury S. Dora K. C. Int. J. Eng. Res. Ind. Appl. 2015;5:85–95. [Google Scholar]
- Stevens K. A. Sheldon B. W. Klapes N. A. Klaenhammer T. R. Appl. Environ. Microbiol. 1991;57:3613–3615. doi: 10.1128/aem.57.12.3613-3615.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casaburi A. Piombino P. Nychas G. J. Villani F. Ercolini D. Food Microbiol. 2015;45:83–102. doi: 10.1016/j.fm.2014.02.002. [DOI] [PubMed] [Google Scholar]
- Selvakumar G. Joshi P. Nazim S. Mishra P. Bisht J. Gupta H. Biologia. 2009;64:239–245. doi: 10.2478/s11756-009-0041-7. [DOI] [Google Scholar]
- George-John E. N. Skandamis P. N. Tassou C. C. Koutsoumanis K. P. Meat Sci. 2008;78:77–89. doi: 10.1016/j.meatsci.2007.06.020. [DOI] [PubMed] [Google Scholar]
- Zhao F. Zhou G. Ye K. Wang S. Xu X. Li C. Meat Sci. 2010;100:145–149. doi: 10.1016/j.meatsci.2014.10.004. [DOI] [PubMed] [Google Scholar]
- Lade H. Paul D. Kweon J. H. BioMed Res. Int. 2014:1–25. doi: 10.1155/2014/162584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammor M. S. Michaelids C. George-John E. N. J. Food Prot. 2008;71:1510–1525. doi: 10.4315/0362-028X-71.7.1510. [DOI] [PubMed] [Google Scholar]
- Gopal N. Hill C. Ross P. R. Beresford T. P. Fenelon M. A. Cotter P. D. Front. Microbiol. 2015;6:1–18. doi: 10.3389/fmicb.2015.01418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier F. Raymond Y. Champagne C. P. Morin A. J. Agric. Food Chem. 1991;39:159–161. doi: 10.1021/jf00001a031. [DOI] [Google Scholar]
- Eneroth A. Svensson B. Molin G. Christiansson A. J. Dairy Res. 2001;68:189–196. doi: 10.1017/S002202990100485X. [DOI] [PubMed] [Google Scholar]
- Swaisgood H. E. Bozoglu F. J. Agric. Food Chem. 1984;32:7–10. doi: 10.1021/jf00121a002. [DOI] [Google Scholar]
- William D. Ana V. Stéphane P. Céline M. Marie-Hélène R. Nathalie G. Bernard C. Innovative Food Sci. Emerging Technol. 2018;46:107–121. doi: 10.1016/j.ifset.2017.09.007. [DOI] [Google Scholar]
- Parandhaman T. Dey M. D. Das S. K. Green Chem. 2019;21:5469–5500. doi: 10.1039/C9GC02291K. [DOI] [Google Scholar]
- Imran K. Deog-Hwan O. Innovative Food Sci. Emerging Technol. 2016;34:376–384. doi: 10.1016/j.ifset.2015.12.013. [DOI] [Google Scholar]
- Chen H. Seiber J. N. Hotze M. J. Agric. Food Chem. 2014;62(6):1209–1212. doi: 10.1021/jf5002588. [DOI] [PubMed] [Google Scholar]
- Wang H. Qian J. Ding F. J. Agric. Food Chem. 2018;66:395–413. doi: 10.1021/acs.jafc.7b04528. [DOI] [PubMed] [Google Scholar]
- Antony J. J. Nivedheetha M. Siva D. Pradeepha G. Kokilavani P. Kalaiselvi S. Sankarganesh A. Balasundaram A. Masilamani V. Achiraman S. Colloids Surf., B. 2013;109:20–24. doi: 10.1016/j.colsurfb.2013.03.020. [DOI] [PubMed] [Google Scholar]
- Jiang H. Manolache S. Wong A. C. L. Denes F. S. J. Appl. Polym. Sci. 2004;93:1411–1422. doi: 10.1002/app.20561. [DOI] [Google Scholar]
- Irene G. S. Miguel M. Beatriz M. Gloria A. Carolina C. Julian C. de L Jose M. L. Olmos M. E. Begoña B. de L Dolores G. Arribas M. V. M. Innovative Food Sci. Emerging Technol. 2019;51:64–72. doi: 10.1016/j.ifset.2018.04.017. [DOI] [Google Scholar]
- Iravani S. Varma R. S. Green Chem. 2019;21:4583–4603. doi: 10.1039/C9GC01759C. [DOI] [Google Scholar]
- Setua P. Chakraborty A. Seth D. Bhatta M. U. Satyam P. V. Sarkar N. J. Phys. Chem. C. 2007;111:3901–3907. doi: 10.1021/jp067475i. [DOI] [Google Scholar]
- Ye Z. Zhu H. Zhang S. Li J. Wang J. Wang E. J. Mater. Chem. B. 2021;9:307–313. doi: 10.1039/D0TB02267E. [DOI] [PubMed] [Google Scholar]
- Shafrina A. H. Malco C. C. R. Tandra G. Michael A. M. Enda C. Joseph P. K. Innovative Food Sci. Emerging Technol. 2015;27:136–143. doi: 10.1016/j.ifset.2014.10.012. [DOI] [Google Scholar]
- Fayaz A. M. Balaji K. Girilal M. Kalaichelvan P. T. Venkatesan R. J. Agric. Food Chem. 2009;57:6246–6252. doi: 10.1021/jf900337h. [DOI] [PubMed] [Google Scholar]
- Mohammadinejad R. Karimi S. Irani S. Varma R. S. Green Chem. 2016;18:20–52. doi: 10.1039/C5GC01403D. [DOI] [Google Scholar]
- Chen Z.-Y. Gao S. Zhang Y.-W. Zhou R.-B. Zhou F. J. Mater. Chem. B. 2021;9:2594–2612. doi: 10.1039/D0TB02983A. [DOI] [PubMed] [Google Scholar]
- Feng G. L. Wu J. Chen G. Q. Cui F. Z. Kim T. N. Kim J. O. J. Biomed. Mater. Res. 2000;52:662–668. doi: 10.1002/1097-4636(20001215)52:4<662::AID-JBM10>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Pal S. Yoon E. J. Tak Y. K. Eung C. Choi E. C. Song J. M. J. Am. Chem. Soc. 2009;131:16147–16155. doi: 10.1021/ja9051125. [DOI] [PubMed] [Google Scholar]
- Rather L. J. Islam S. Azam M. Shabbir M. Bukhari M. N. Shahid M. Khan M. A. Haque Q. M. Mohammad F. RSC Adv. 2016;6:39080–39094. doi: 10.1039/C6RA02717B. [DOI] [Google Scholar]
- Markova Z. Siskova K. Filip J. Panacek A. Safarova K. Zboril R. Green Chem. 2012;14:2550–2558. doi: 10.1039/C2GC35545K. [DOI] [Google Scholar]
- Islam S. Batola B. S. Mohammad F. RSC Adv. 2016;6:44232–44247. doi: 10.1039/C6RA05799C. [DOI] [Google Scholar]
- Kramarenko G. G. Wilke W. W. Dayal D. Buettner G. R. Schafer F. Q. Free Radical Biol. Med. 2006;40:1615–1627. doi: 10.1016/j.freeradbiomed.2005.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srećković N. Z. Nedić Z. P. Liberti D. Monti D. M. Mihailović N. R. Stanković J. S. K. Dimitrijević S. Mihailović V. B. RSC Adv. 2021;11:35585–35599. doi: 10.1039/D1RA05570D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair B. Pradeep T. Cryst. Growth Des. 2002;2:293–298. doi: 10.1021/cg0255164. [DOI] [Google Scholar]
- Sood S. Narang D. Dinda A. K. Maulik S. K. J. Pharm. Pharmacol. 2005;57:127–133. doi: 10.1211/0022357055146. [DOI] [PubMed] [Google Scholar]
- Wink M. Molecules. 2012;17:12771–12791. doi: 10.3390/molecules171112771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmukh A. S. Deshmukh B. G. Shirole P. D. Int. J. Pharmacogn. 2015;2:550–559. [Google Scholar]
- Thomas J. E. Bandara M. Lee E. L. Driedger D. Acharya S. New Biotechnol. 2011;28:110–117. doi: 10.1016/j.nbt.2010.09.001. [DOI] [PubMed] [Google Scholar]
- Sharma V. Tiwari P. Mobin S. M. J. Mater. Chem. B. 2017;5:8904–8924. doi: 10.1039/C7TB02484C. [DOI] [PubMed] [Google Scholar]
- Chen Z.-Y. Gao S. Zhang Y.-W. Zhou R.-B. Zhou F. J. Mater. Chem. B. 2021;9:2594–2612. doi: 10.1039/D0TB02983A. [DOI] [PubMed] [Google Scholar]
- Amitabh A. P. Pal P. K. NanoImpact. 2020;19:1–14. [Google Scholar]
- Kiessoun K. Kassi Y. Oksana S. Marian B. Asian Pac. J. Trop. Biomed. 2015;5:242–248. [Google Scholar]
- Anwar M. F. Yadav D. Kapoor S. Chander J. Samim M. Drug Dev. Ind. Pharm. 2013;41:43–50. doi: 10.3109/03639045.2013.845840. [DOI] [PubMed] [Google Scholar]
- Kuang-Che L. Su-Jien L. Chih-Hong L. Chih-Song T. Yu-Jen L. Surf. Coat. Technol. 2008;202:5339–5342. doi: 10.1016/j.surfcoat.2008.06.080. [DOI] [Google Scholar]
- Chahrdoli A. Qalekhani F. Ghowsi M. Nemati H. Shokoohina Y. Fattahi A. Mater. Today Commun. 2020;25:1–9. [Google Scholar]
- Fatemeh F. Nelson M. A. Laura A. S. Vinka O.-C. Nanotechnol. Environ. Eng. 2016;1:1. doi: 10.1007/s41204-016-0001-8. [DOI] [Google Scholar]
- Yu C. Irudayaraj J. Biopolymers. 2005;77:368–377. doi: 10.1002/bip.20247. [DOI] [PubMed] [Google Scholar]
- Riding M. J. Martin F. L. Trevisani J. Llabjani V. Patel I. M. Jones K. C. Semple K. T. Environ. Pollut. 2012;163:226–234. doi: 10.1016/j.envpol.2011.12.027. [DOI] [PubMed] [Google Scholar]
- Niancao C. Yuanwei C. Lijian W. Xianglin L. Juan L. Bin W. J. Mater. Sci. 2009;44:6317–6324. doi: 10.1007/s10853-009-3870-4. [DOI] [Google Scholar]
- Gurbanov R. Simsek O. N. Gozen A. G. Severcan F. Anal. Chem. 2015;87:9653–9661. doi: 10.1021/acs.analchem.5b01659. [DOI] [PubMed] [Google Scholar]
- Abedini A. Daud A. R. Hamid M. A. A. Saion E. Nanoscale Res. Lett. 2013;8:474. doi: 10.1186/1556-276X-8-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozaffari S. Li W. Thompson C. Ivanov S. Seifert S. Lee B. Kovarik L. Karim A. M. Nanoscale. 2017;9:13772–13785. doi: 10.1039/C7NR04101B. [DOI] [PubMed] [Google Scholar]
- Fanoro O. T. Oluwafemi O. S. Pharmaceutics. 2020;12:1044. doi: 10.3390/pharmaceutics12111044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan L. Chen S. Jensen G. J. Proc. Natl. Acad. Sci. U. S. A. 2008;105:18953–18957. doi: 10.1073/pnas.0808035105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodisetti P. K. Reddy M. Proc. Natl. Acad. Sci. U. S. A. 2019;116:7825–7830. doi: 10.1073/pnas.1816893116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashin N.-Z. Edgar H. H. W. Cyrille B. ACS Infect. Dis. 2021;7:215–253. doi: 10.1021/acsinfecdis.0c00635. [DOI] [PubMed] [Google Scholar]
- Fan H. Yang G. Yumin Z. Tangjian C. Hanlin O. Lijun Y. Jinjiang L. Linqi S. Jianfeng L. ACS Appl. Mater. Interfaces. 2017;9:16880–16889. doi: 10.1021/acsami.7b03347. [DOI] [PubMed] [Google Scholar]
- Hamouda H. I. Abdel-Ghafar H. M. H Mahmoud M. H. J. Environ. Chem. Eng. 2021;9:105034. doi: 10.1016/j.jece.2021.105034. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.