Abstract
Streptogramins are polypeptide antibiotics inhibiting protein synthesis by the prokaryotic ribosome. Gram-positive organisms are susceptible to streptogramins, while most gram-negative bacteria are intrinsically resistant. We have found a genomic fragment from a Yersinia enterocolitica isolate with an open reading frame coding for a polypeptide similar to the virginiamycin acetyltransferases found in various plasmids from gram-positive bacteria. The susceptible Escherichia coli strain DB10 was transformed to resistance to the type A streptogramins and to mixed (A + B) streptogramins upon introduction of a plasmid containing that gene. In addition, we showed streptogramin acetylating activity in vitro dependent on the presence of the Y. enterocolitica sat gene. Southern blot hybridization experiments showed that the sat gene was present in all the Y. enterocolitica isolates examined. These data together show that the gene in the Y. enterocolitica chromosome encoded an active streptogramin acetyltransferase. The deduced sequence of the Y. enterocolitica Sat protein was close to those of sat gene products found in gram-positive bacteria and cyanobacteria, suggesting a common evolutionary origin.
Streptogramins are a group of cyclic peptide antibiotics produced by some Streptomycetes. They are divided into two classes, A and B, according to chemical structure (9). Both A and B streptogramins act to block protein synthesis by inhibition of the peptidyl transferase domain in the 50S subunit of the prokaryotic ribosome. Compounds of the A and B classes act synergistically. This property makes the A + B mixtures active against many bacterial pathogens (20). The poor solubility in water of these antibiotics has reduced their use in human medicine. However, some streptogramins (virginiamycin) have been used as animal feed additives. Recently, new streptogramins have been developed with increased water solubility and hence better pharmacological properties (7). The quinupristin-dalfopristin combination has proved useful in fighting infections produced by dangerous antibiotic-resistant strains of staphylococci or enterococci (14, 16, 22). Resistance to the class A streptogramins takes place either by active efflux mediated by the gene vga in staphylococci (3) or by inactivation via enzymatic acetylation of the antibiotic (12). This inactivation is catalyzed by streptogramin (virginiamycin) acetyltransferases, the products of sat or vat genes (1, 5, 23). These are small (24-kDa) enzymes, closely related to each other, and the sequences of their carboxyl-terminal halves are very similar to those of other acetyltransferases (5). Resistance to class B streptogramins can be obtained by efflux, modification of the drug, or rRNA methylation, which also results in resistance to macrolides and lincosamide (18). Due to the interplay of A and B streptogramins, resistance to the A compounds usually results in resistance to the mixture.
Gram-negative organisms are intrinsically resistant to the streptogramins. Since their ribosomes are sensitive to inhibition, it was thought that the mechanism of this intrinsic resistance was the exclusion of the antibiotics from the cytoplasm (15). Streptogramins are poorly hydrophilic molecules whose molecular size is greater than 500 Da. The gram-negative outer membrane constitutes a strong permeability barrier to streptogramin entrance into the periplasm. No carrier for streptogramins through the inner membrane is known. The hydrophobic pathway could be the main route for streptogramins into the bacterial cytoplasm. Organisms of the genus Neisseria and Haemophilus are more permeable to larger hydrophilic molecules, and consequently they can be susceptible to streptogramins (28). In addition to the permeability-related intrinsic resistance, resistance to class B streptogramins in Escherichia coli and other enterobacteria can also be mediated by the ermB gene, probably acquired from gram-positive organisms (18). Resistance to streptogramins of the A group by mechanisms other than the permeability barrier has never been reported, either in Yersinia spp. or in other enterobacteria.
Here we describe the presence of a gene encoding streptogramin A acetyltransferase activity in the chromosome of a Yersinia enterocolitica isolate. The possible role of this gene in the evolution of bacterial resistance to streptogramins is also discussed.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The following Y. enterocolitica strains were used in this work: P1403 and P219 (biotype 1A); WA and 1354 (biotype 1B); My79b and IP97 (biotype 2); and IP4124, IP22273, IP22274, H6, H14, Y56, and Y60 (biotype 4). Most of the strains were from our laboratory collection and have already been described (11). Strains IP4124, IP22273, and IP22274 were obtained from Jeanette N. Pham. The E. coli strain DB10, a fusidic acid-susceptible derivative of E. coli PR7 (10), and DB11, another E. coli mutant susceptible to streptogramins (1), were used as recipients for recombinant plasmids. Staphylococcus aureus BM3002 and Enterococcus faecium BM4145 (obtained from P. Courvalin) were used as control streptogramin A-resistant strains known to produce streptogramin A acetyltransferases. S. aureus ATCC 28213 was used as a control susceptible strain in MIC determinations. To clone genomic DNA fragments, we used the high-copy-number plasmid vector pK18 (21), which contains a kanamycin resistance selection marker.
Antibiotics.
Virginiamycin M1 (a class A streptogramin) and polymyxin B nonapeptide were purchased from Sigma. Dalfopristin (RP54476, another class A streptogramin), quinupristin (RP57669, a class B streptogramin), and a 70:30 mixture of the latter two compounds (Synercid, also referred to as RP59500), were the generous gifts of Rhône Poulenc Rorer Laboratories.
Genetic and DNA methods.
Bacterial transformation, DNA purification, cloning, and DNA sequencing were carried out by standard methods basically as described in reference 24.
Analysis of the DNA sequence.
The deduced sequence of the Yersinia sat gene product was compared against nucleotide sequences in the nonredundant (GenBank, EMBL, DDBJ, and PDB) databases, using the program TBLASTN 2.0.9 (6) in the Blastserver at the National Center for Biotechnology Information (NCBI) web site (http://www.ncbi.nlm.nih.gov/BLAST/). Preliminary sequence data from unfinished genomes were obtained from the website of The Institute for Genomic Research (TIGR) (http://www.tigr.org/). Multiple sequence comparisons and tree construction were performed with the PILEUP program from the Genetics Computer Group (GCG), version 10, package (13), in the National node of the European Molecular Biology Network in the Centro Nacional de Biotecnología (EMBNET/CNB), Madrid, Spain, using its website (http://www.es.embnet.org/).
Southern blot hybridization of Y. enterocolitica chromosomes.
Chromosomal DNA from stationary-phase cultures of the different strains was purified after lysis with guanidine thiocyanate as described elsewhere (17), digested with the restriction endonucleases EcoRI and HindIII, separated in a 0.8% agarose gel, and transferred to positively charged nylon membranes (Boehringer Mannheim). A 624-bp EcoRV/XbaI fragment containing the sat gene from the Y56 chromosome was labeled with digoxigenin and used to probe the Yersinia chromosomes under stringent conditions (42°C in 50% formamide), including a final high-stringency wash at 68°C in 1:10-diluted SSC (0.15 M NaCl plus 0.015 M sodium citrate) containing 0.1% sodium dodecyl sulfate. Blots were developed using a chemiluminescence detection kit from Roche.
MIC determinations.
MICs were determined on Mueller-Hinton agar plates containing doubling concentrations of antibiotics between 1 and 256 μg ml−1. MICs were also determined in the presence of 3 μg of the membrane permeabilizing agent polymyxin B nonapeptide ml−1 (27). Inocula of 104 CFU were spotted onto the plate.
Determination of streptogramin A acetyltransferase activity.
In vitro streptogramin A acetyltransferase activity was determined spectrophotometrically, essentially as described previously (23, 26). Crude enzyme extracts were prepared from late-stationary-phase cells by sonic disruption in Tris (50 mM) (pH 7.8) buffer containing 50 μM β-mercaptoethanol. Crude extracts were clarified by centrifugation at 20,000 × g for 20 min at 4°C, and supernatants were directly used in enzymatic reactions. The assay mixture contained 0.1 mM streptogramin A, 0.1 mM acetyl coenzyme A, and 1 mM 5,5′-dithiobis-2-nitrobenzoic acid (DTNB). Protein concentrations in extracts were determined by the method of Bradford (8) using a commercial reagent (Bio-Rad).
Nucleotide sequence accession number.
The nucleotide sequence of the satA gene from Y. enterocolitica Y56 has been deposited in the GenBank Data Library with the accession number AF170730.
RESULTS
Nucleotide sequence of the Y. enterocolitica sat gene: sequence analysis and comparison.
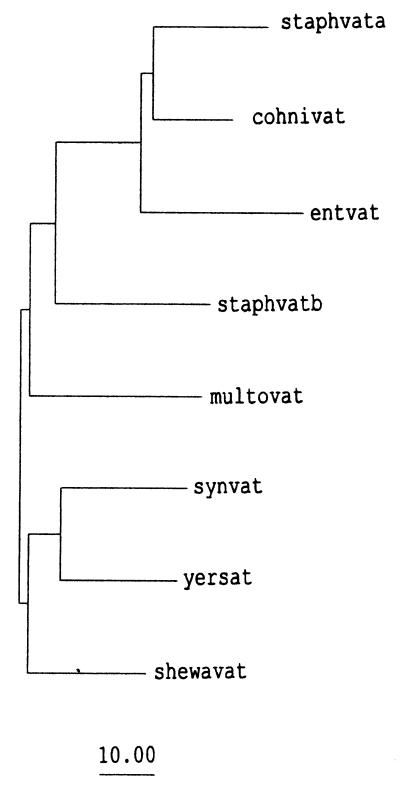
While characterizing the genome region of Y. enterocolitica Y56 around the blaA gene (25), we obtained a 1.5-kb EcoRI fragment which was cloned in the E. coli kanamycin resistance vector pK18. This plasmid was called pAS3. The 1.5-kb EcoRI DNA fragment was further subcloned using XbaI and EcoRV restriction sites, and its complete nucleotide sequence was determined on the two DNA strands, using universal and specific DNA primers. The G+C content of the sequence was 49.4%. This value is similar to the overall G+C content of Y. enterocolitica DNA (48.5% ± 1.5%). A 663-bp open reading frame (ORF) was found in the sequence. The translated product from this ORF (221 amino acids; 24.4 kDa) was used to investigate the presence of homologous genes in the nonredundant nucleotide sequence database using the TBLASTN program. We found that the gene was homologous with streptogramin A acetyltransferases from Synechocystis spp., S. aureus, and E. faecium. Comparison was also carried out with the nucleotide sequence of unfinished genomes kindly provided by TIGR. Two additional homologous genes were found in the genomes of Shewanella putrefaciens and Pasteurella multocida. The deduced amino acid sequence of the Y. enterocolitica ORF was aligned with those of the virginiamycin acetyltransferases from the other bacteria (Fig. 1). The evolutionary distances among the aligned sequences were calculated and used to construct the phylogenetic tree shown in Fig. 2. Based on the high degree of similarity among the proteins, we assumed that the Y. enterocolitica gene encoded a streptogramin A acetyltransferase and called it sat.
FIG. 1.
Multiple alignment of Vat proteins from different microorganisms. Amino acids are represented by the one-letter standard code. Gaps are represented by dots. A dash indicates that the residue in that position is the same as that in the Y. enterocolitica sequence shown in the top line. Residues conserved in all the sequences are boldfaced. Abbreviations: yersat, Y. enterocolitica Y56 Sat (GenBank accession number AF170730); synvat, Vat of Synechocystis spp. (D13960); staphvata, S. aureus VatA (L07778); staphvatb, S. aureus VatB (U19459); cohnivat, S. cohnii VatC (AF015628); shewavat, S. putrefaciens putative Sat; multovat, P. multocida PM70 putative Sat; entvat, E. faecium SatA (L12033). Preliminary sequence data for S. putrefaciens and P. multocida PM70 were obtained from the website of The Institute for Genomic Research (http://www.tigr.org/).
FIG. 2.
Phylogenetic tree showing the evolutionary relationship of the Y. enterocolitica sat gene product with other streptogramin acetyltransferases. The tree was elaborated with the program GROWTHTREE from the GCG package (13) from a distance matrix calculated from the alignment shown in Fig. 1. The length of each branch is proportional to the number of substitutions per 100 residues. Bar, 10 substitutions per 100 amino acids. Abbreviations are as explained in the legend to Fig. 1.
Contribution of the sat gene to streptogramin resistance in E. coli and Y. enterocolitica.
To determine the contribution of the sat gene to the resistance to streptogramins in E. coli, we used the susceptible strains DB10 and DB11. The MICs of a class A streptogramin (dalfopristin), a class B streptogramin (quinupristin), and the 70:30 dalfopristin-quinupristin mixture, were determined for the E. coli strains with and without plasmids and for a collection of Y. enterocolitica strains representative of the different biotypes (data not shown). Upon introduction of the sat gene-containing plasmid pAS3 in the E. coli strains, the MIC of the class A streptogramin increased from 2 to 8 μg ml−1 for E. coli DB10 and from 4 to 32 μg ml−1 for E. coli DB11. The MIC for the Y. enterocolitica strain Y56 was 64 μg ml−1. Detailed results of these experiments are summarized in Table 1.
TABLE 1.
MICs of streptogramins for E. coli strains with and without plasmid pAS-3 and for Y. enterocolitica Y56
| Strain | MIC (μg ml−1) ofa:
|
||
|---|---|---|---|
| A | B | A + B | |
| E. coli | |||
| DB10 | 2 | 64 | 2 |
| DB10 (pK18) | 2 | 64 | 2 |
| DB10 (pAS3) | 8 | 256 | 8 |
| DB11 | 4 | 256 | 4 |
| DB11 (pK18) | 4 | 256 | 4 |
| DB11 (pAS3) | 32 | 256 | 32 |
| E. faecium BM4145 | 64 | 256 | 32 |
| Y. enterocolitica Y56 | 64 | >256 | 32 |
A, the class A streptogramin dalfopristin; B, the class B streptogramin quinupristin; A + B, a 70:30 dalfopristin-quinupristin mixture.
In vitro activity of streptogramin acetyltransferase.
The activity of the Y. enterocolitica sat gene was further investigated in vitro by determination of the acetylating activity in crude protein extracts from Y. enterocolitica and E. coli with and without the sat plasmid pAS3 on class A streptogramins. We used two different substrates for this reaction, namely, virginiamycin M1 and dalfopristin. Extracts from Y. enterocolitica Y56 and E. coli DB10 containing plasmid pAS3 showed acetylating activity on the two class A streptogramins. The activity of the extracts from E. coli DB10 containing only the vector pK18 was negligible (Table 2).
TABLE 2.
In vitro acetylating activities of crude extractsa on the class A streptogramins virginiamycin M1 and dalfopristin
| Extract | Activity (nmol of antibiotic acetylated/min/mg of protein) on:
|
|
|---|---|---|
| Virginiamycin M1 | Dalfopristin | |
| E. coli DB10 pK18 | 0.65 ± 0.15 | 0.46 ± 0.11 |
| E. coli DB10 pAS3 | 42.4 ± 7 | 31.27 ± 3 |
| S. aureus BM3002 | 129.7 ± 8.2 | 63.49 ± 6.4 |
| E. faecium BM4145 | 161.83 ± 12.3 | 82.68 ± 8.5 |
| Y. enterocolitica Y56 | 16.2 ± 2 | 11.06 ± 1.4 |
Extracts were prepared as described in the text.
Investigation of the presence of sat genes in chromosomes from other Y. enterocolitica isolates.
To determine whether the presence of the sat gene was a peculiarity of the Y. enterocolitica strain Y56 or, on the contrary, was commonplace in the chromosome of Y. enterocolitica, we probed the chromosomes of a collection of Y. enterocolitica strains with the sat gene probe. All the strains tested hybridized with the probe under high-stringency conditions, indicating that they contained at least one copy of the gene (Fig. 3). We observed basically three different sizes of hybridizing bands among the different strains, which correlated with their biotype. Biotype 1A strains showed a band of about 4 kb, strains of biotypes 1B and 2 showed a band of 3.4 kb, and biotype 4 strains showed a band of 1.5 kb. A similar result had been obtained previously when the same strains were probed for the presence of bla genes (11); these results probably reflect differences in genomic organization in the different Y. enterocolitica biotypes. The sat-containing bands in the biotype 4 strains showed small but noticeable size differences. The band from Y56 (which is the same 1.5-kb EcoRI fragment cloned in plasmid pAS3) was about 100 bp larger than the bands from strains IP22274 and H14. The reason for this size polymorphism is not understood at the moment. A fainter, smaller hybridization band was also observed in most strains. The nature of this second hybridization band is also unknown, but it could be due to the presence of an additional acetyltransferase gene in the Y. enterocolitica chromosome.
FIG. 3.
Hybridization of Y. enterocolitica chromosomal DNA digested with EcoRI/HindIII with a probe containing the sat gene from Y. enterocolitica Y56. Lanes: 1, P1403; 2, P219; 3, WA; 4, 13514; 5, My79b; 6, IP97; 7, IP4124; 8, IP22273; 9, IP22274; 10, H-6; 11, H-14; 12, Y-56; 13, Y-60; 14, E. coli chromosomal DNA; 15, molecular mass markers. The sizes of the markers in kilobases are shown on the right.
DISCUSSION
Resistance to class A streptogramins by inactivation catalyzed by streptogramin acetyltransferase is common among gram-positive organisms, and sequences of sat genes from S. aureus (1, 5), Staphylococcus cohnii (2), E. faecium (23), etc., have been reported. In all these cases, sat genes were located in plasmids. Here we report the isolation of a sat gene from the chromosome of a Y. enterocolitica isolate. The gene product was first identified by sequence comparison by its high similarity with other streptogramin acetyltransferases. The sequence comparison search also revealed high similarities with the carboxyl termini of other acetyltransferases such as the chloramphenicol acetyltransferase of Tn2424, indicating that streptogramin acetyltransferases belong to a broad family of enzymes with different substrates, as had been previously described (5, 19).
The functionality of the Y. enterocolitica sat gene was demonstrated by an increase in the dalfopristin and quinupristin-dalfopristin MICs upon transfer of the sat gene to susceptible E. coli strains, and by determination of in vitro acetylating activity on the class A streptogramins dalfopristin and virginiamycin M1. The fact that the presence of this gene in a gram-negative organism has remained unnoticed until now may be due to the high level of intrinsic resistance to streptogramins in gram-negative organisms (15, 28). For most of the Y. enterocolitica and E. coli laboratory strains, the MICs of class A streptogramins were higher than 64 μg ml−1. Since these values are much higher than the regular therapeutic levels, susceptibility testing of gram-negative organisms is considered useless and is not performed. On the other hand, the elevated intrinsic resistance to streptogramins in gram-negative bacteria devoid of sat genes, such as E. coli, raises questions about the contribution of the sat gene to resistance in these bacteria. How much of the resistance to class A streptogramins in Yersinia isolates is due to the activity of the sat gene? We cannot give an exact answer to this question, since we have not found any Y. enterocolitica strain devoid of the sat gene. However, we can obtain an approximate answer by use of the membrane-permeabilizing agent polymyxin B nonapeptide. The MICs of class A streptogramins for the Yersinia strains were reduced to 32 μg ml−1 in the presence of this agent (data not shown). This resistance level was similar to that obtained with E. coli DB11 upon acquisition of the satA-containing plasmid pAS3. From these observations we may conclude that the elevated level of resistance observed in Y. enterocolitica isolates, and in most E. coli strains, resulted from the simultaneous action of several mechanisms, some of which still remain unknown.
We have demonstrated that the sat gene was present in all Y. enterocolitica isolates examined. This suggested that the gene is part of the bacterial genome, rather than an acquisition of a particular strain where the gene was found. Furthermore, the analysis of unfinished bacterial genomic sequences revealed that sat genes are often present in bacterial chromosomes of the gamma division of the purple bacteria. They were present in P. multocida, a close relative of Yersinia, as well as in S. putrefaciens. A sat gene is also present in the chromosome of the cyanobacterium Synechocystis. The presence of sat genes in all these genera, and the similarity of the product of this gene to a broad family of acetylases with many different substrates, may indicate that it plays a physiological role consisting in the acetylation of some unidentified substrate and that streptogramin acetylation is an undesired collateral effect. Furthermore, the chromosomal location of this gene contrasted with the plasmid location of sat genes in gram-positive bacteria, suggesting that plasmid genes could have originated from their chromosomal counterparts (or vice versa) after a process of gene mobilization and transfer, well documented in many of these organisms. This hypothesis could also be supported by the level of identity found between the deduced sequences of Sat proteins from gram-positive and gram-negative bacteria.
In addition to their use in human medicine, some streptogramins (virginiamycin) are used as growth promoters in farm animals. It has been reported that this practice resulted in the selection of staphylococci and enterococci resistant to virginiamycin and to other streptogramins in poultry (29). Y. enterocolitica is a pathogen closely associated with pigs, one of the animal species fed virginiamycin. This association may have selected for an acetylase gene with a higher affinity for streptogramins, capable of conferring resistance to these drugs. While the finding of sat genes in Y. enterocolitica can be of little relevance to the susceptibility of this species to streptogramins, due to its high level of intrinsic resistance, it can be very important from an epidemiological point of view, since we may have identified a reservoir of sat genes ready to be mobilized to other human pathogens whose resistance to streptogramins may represent a very serious clinical problem.
ACKNOWLEDGMENTS
We are indebted to Rhône Poulenc Rorer for the generous gift of the streptogramins quinupristin, dalfopristin, and Synercid. We are also grateful to Jeannette N. Pham for providing some of the Yersinia strains and to E. Carniel and D. Postid of the Pasteur Institute, Paris, France, for the typing of Y. enterocolitica.
This work was financed by a grant from the Spanish Fondo de Investigación Sanitaria.
REFERENCES
- 1.Allignet J, el Solh N. Diversity among the gram-positive acetyltransferases inactivating streptogramin A and structurally related compounds and characterization of a new staphylococcal determinant, vatB. Antimicrob Agents Chemother. 1995;39:2027–2036. doi: 10.1128/aac.39.9.2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allignet J, Liassine N, el Solh N. Characterization of a staphylococcal plasmid related to pUB110 and carrying two novel genes, vatC and vgbB, encoding resistance to streptogramins A and B and similar antibiotics. Antimicrob Agents Chemother. 1998;42:1794–1798. doi: 10.1128/aac.42.7.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allignet J, Loncle V, el Solh N. Sequence of a staphylococcal plasmid gene, vga, encoding a putative ATP-binding protein involved in resistance to virginiamycin A-like antibiotics. Gene. 1992;117:45–51. doi: 10.1016/0378-1119(92)90488-b. [DOI] [PubMed] [Google Scholar]
- 4.Allignet J, Loncle V, Mazodier P, el Solh N. Nucleotide sequence of a staphylococcal plasmid gene, vgb, encoding a hydrolase inactivating the B components of virginiamycin-like antibiotics. Plasmid. 1988;20:271–275. doi: 10.1016/0147-619x(88)90034-0. [DOI] [PubMed] [Google Scholar]
- 5.Allignet J, Loncle V, Simenel C, Delepierre M, el Solh N. Sequence of a staphylococcal gene, vat, encoding an acetyltransferase inactivating the A-type compounds of virginiamycin-like antibiotics. Gene. 1993;130:91–98. doi: 10.1016/0378-1119(93)90350-c. [DOI] [PubMed] [Google Scholar]
- 6.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barriere J C, Bouanchaud D H, Paris J M, Rolin O, Harris N V, Smith C. Antimicrobial activity against Staphylococcus aureus of semisynthetic injectable streptogramins: RP 59500 and related compounds. J Antimicrob Chemother. 1992;30(Suppl. A):1–8. doi: 10.1093/jac/30.suppl_a.1. [DOI] [PubMed] [Google Scholar]
- 8.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 9.Cocito C. Antibiotics of the virginiamycin family, inhibitors which contain synergistic components. Microbiol Rev. 1979;43:145–198. doi: 10.1128/mr.43.2.145-192.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Datta N, Hedges R W, Becker D, Davies J. Plasmid-determined fusidic acid resistance in Enterobacteriaceae. J Gen Microbiol. 1974;83:191–196. doi: 10.1099/00221287-83-1-191. [DOI] [PubMed] [Google Scholar]
- 11.de la Prieta M C, Seoane A, Diaz J, Navas J, García-Lobo J M. Beta-lactamase genes and beta-lactamic susceptibility in Yersinia enterocolitica. Contrib Microbiol Immunol. 1995;13:184–187. [PubMed] [Google Scholar]
- 12.De Meester C, Rondelet J. Microbial acetylation of M factor of virginiamycin. J Antibiot. 1976;29:1297–1305. doi: 10.7164/antibiotics.29.1297. [DOI] [PubMed] [Google Scholar]
- 13.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fass R J. In vitro activity of RP 59500, a semisynthetic injectable pristinamycin, against staphylococci, streptococci, and enterococci. Antimicrob Agents Chemother. 1991;35:553–559. doi: 10.1128/aac.35.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leclercq R, Courvalin P. Intrinsic and unusual resistance to macrolide, lincosamide, and streptogramin antibiotics in bacteria. Antimicrob Agents Chemother. 1991;35:1273–1276. doi: 10.1128/aac.35.7.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leclercq R, Nantas L, Soussy C J, Duval J. Activity of RP 59500, a new parenteral semisynthetic streptogramin, against staphylococci with various mechanisms of resistance to macrolide-lincosamide-streptogramin antibiotics. J Antimicrob Chemother. 1992;30(Suppl. A):67–75. doi: 10.1093/jac/30.suppl_a.67. [DOI] [PubMed] [Google Scholar]
- 17.Lippke J A, Strzempko M N, Raia F F, Simon S L, French C K. Isolation of intact high-molecular-weight DNA by using guanidine isothiocyanate. Appl Environ Microbiol. 1987;53:2588–2589. doi: 10.1128/aem.53.10.2588-2589.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mabilat C, Courvalin P. Gene heterogeneity for resistance to macrolides, lincosamides and streptogramins in Enterobacteriaceae. Ann Inst Pasteur Microbiol. 1988;139:677–681. doi: 10.1016/0769-2609(88)90072-5. [DOI] [PubMed] [Google Scholar]
- 19.Parent R, Roy P H. The chloramphenicol acetyltransferase gene of Tn2424: a new breed of cat. J Bacteriol. 1992;174:2891–2897. doi: 10.1128/jb.174.9.2891-2897.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pechère J C. Streptogramins. A unique class of antibiotics. Drugs. 1996;51(Suppl. 1):13–19. doi: 10.2165/00003495-199600511-00005. [DOI] [PubMed] [Google Scholar]
- 21.Pridmore R D. New and versatile cloning vectors with kanamycin-resistance marker. Gene. 1987;56:309–312. doi: 10.1016/0378-1119(87)90149-1. [DOI] [PubMed] [Google Scholar]
- 22.Qadri S M, Ueno Y, Abu Mostafa F M, Halim M. In vitro activity of quinupristin/dalfopristin, RP 59500, against gram-positive clinical isolates. Chemotherapy. 1997;43:94–99. doi: 10.1159/000239542. [DOI] [PubMed] [Google Scholar]
- 23.Rende-Fournier R, Leclercq R, Galimand M, Duval J, Courvalin P. Identification of the satA gene encoding a streptogramin A acetyltransferase in Enterococcus faecium BM4145. Antimicrob Agents Chemother. 1993;37:2119–2125. doi: 10.1128/aac.37.10.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 25.Seoane A, Garcia Lobo J M. Nucleotide sequence of a new class A beta-lactamase gene from the chromosome of Yersinia enterocolitica: implications for the evolution of class A beta-lactamases. Mol Gen Genet. 1991;228:215–220. doi: 10.1007/BF00282468. [DOI] [PubMed] [Google Scholar]
- 26.Shaw W. Chloramphenicol acetyltransferase from chloramphenicol resistant bacteria. Methods Enzymol. 1975;43:737–755. doi: 10.1016/0076-6879(75)43141-x. [DOI] [PubMed] [Google Scholar]
- 27.Vaara M. Agents that increase the permeability of the outer membrane. Microbiol Rev. 1992;56:395–411. doi: 10.1128/mr.56.3.395-411.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verbist L, Verhaegen J. Comparative in-vitro activity of RP 59500. J Antimicrob Chemother. 1992;30(Suppl. A):39–44. doi: 10.1093/jac/30.suppl_a.39. [DOI] [PubMed] [Google Scholar]
- 29.Witte W. Impact of antibiotic use in animal feeding on resistance of bacterial pathogens in humans. Ciba Found Symp. 1997;207:61–71. doi: 10.1002/9780470515358.ch5. [DOI] [PubMed] [Google Scholar]