Abstract
Although combination antiretroviral therapy has resulted in a considerable improvement in the treatment of human immunodeficiency virus (HIV) type 1 (HIV-1) infection, the emergence of resistant virus is a significant obstacle to the effective management of HIV infection and AIDS. We have developed a novel phenotypic drug susceptibility assay that may be useful in guiding therapy and improving long-term suppression of HIV replication. Susceptibility to protease (PR) and reverse transcriptase (RT) inhibitors is measured by using resistance test vectors (RTVs) that contain a luciferase indicator gene and PR and RT sequences derived from HIV-1 in patient plasma. Cells are transfected with RTV DNA, resulting in the production of virus particles that are used to infect target cells. Since RTVs are replication defective, luciferase activity is measured following a single round of replication. The assay has been automated to increase throughput and is completed in 8 to 10 days. Test results may be useful in facilitating the selection of optimal treatment regimens for patients who have failed prior therapy or drug-naive patients infected with drug-resistant virus. In addition, the assay can be used to evaluate candidate drugs and assist in the development of new drugs that are active against resistant strains of HIV-1.
The extent of virus replication (i.e. viral load) is the strongest single predictor of progression to AIDS and death both in antiretroviral (ARV) drug treatment-naive and -experienced human immunodeficiency virus (HIV) type 1 (HIV-1)-infected patient populations (19, 46, 48). The goal of highly active ARV therapy is to delay disease progression and prolong survival by achieving sustained suppression of viral replication (7).
Antiviral therapies that use combinations of nucleoside reverse transcriptase inhibitors (NRTIs) and protease inhibitors (PRIs) or NRTIs and nonnucleoside reverse transcriptase inhibitors (NNRTIs) produce the largest reductions in viral load and provide the greatest clinical benefit (16, 23, 24, 33, 49, 51) and are therefore the recommended treatment for HIV-1 infection in the United States (7). However, in typical clinical practice, up to 50% of patients who begin combination therapy either do not achieve or do not maintain complete suppression of virus replication (8, 18; for a review, see reference 32). Viral load rebound (i.e., virologic failure) often occurs within the first few years of treatment in patients who appear to achieve complete suppression by the existing assays during an initial course of combination therapy and is frequently accompanied by the emergence of drug-resistant viral variants. Furthermore, response to salvage therapies decreases with increasing drug experience in terms of both duration of treatment and the number of drugs with which the patient has been treated (45).
The use of routine viral load (VL) measurements to determine when to change treatment has been shown to improve treatment outcome (R. Haubrich, J. Currier, D. Forthal, G. Beall, C. Kemper, M. Dube, J. Ignosci, D. Johnson, J. Hwang, J. McCutchan, and T. C. C. T. Group, Fifth Conf. Retroviruses and Opportunistic Infections, 1998). However, VL measurements do not reveal the underlying cause(s) of treatment failure, which may include drug resistance, poor adherence, or inadequate drug absorption; nor does VL provide guidance to the physician for the selection of an effective salvage regimen. Such information may, in part, be provided by “resistance testing” performed by assays designed to measure drug susceptibility either directly (phenotyping) or indirectly by detecting mutations associated with drug resistance (genotyping) (31).
Rapid, high-throughput genotypic assays based either on the detection of specific point mutations or on complete DNA sequencing are being developed (11, 15, 63). However, the increasing number of reported drug resistance mutations and the sequence heterogeneity of HIV-1 present technical obstacles for point mutation assays. Even when complete protease (PR) and reverse transcriptase (RT) sequences are available, the large number of distinct PR and RT mutation patterns and the complex interaction of mutations have made it difficult to accurately predict drug resistance. Initially, phenotypic drug susceptibility assays used replication-competent viruses derived directly from the patient by cocultivation methods and were both labor-intensive and time-consuming (35). The development of recombinant virus assays (RVAs) that use virus stocks generated by homologous recombination between HIV-1 vectors and PR and RT sequences amplified from the patient virus have greatly simplified testing procedures and improved assay reproducibility (29, 37). However, to date RVA methods have not significantly reduced assay turnaround time (generally, 4 to 6 weeks). In the absence of rapid, reliable methods for assessment of drug susceptibility, treatment decisions regarding use of specific antiviral drugs are often empirical and are based on accepted treatment guidelines (7) and clinical experience.
This report describes a novel phenotypic assay that can be used for the rapid and accurate assessment of HIV-1 drug susceptibility. The assay, which has been automated to achieve high throughput, is used to determine the susceptibility profile of a patient's HIV-1 isolates to all currently available ARV drugs. This technology provides drug susceptibility data that physicians can use to select more effective ARV regimens when treating HIV-infected patients at the time of treatment initiation or after treatment failure.
MATERIALS AND METHODS
Antiviral drugs.
The following is a list of drugs and their sources: zidovudine (ZDV, AZT), didanosine (ddI), stavudine (d4T), and zalcitabine (ddC), Sigma Chemical (St. Louis, Mo.); lamivudine (3TC), Moravek Chemical (Brea, Calif.); nevirapine (NVP), Roxanne Laboratories (Redding, Conn.); delavirdine (DLV), Pharmacia-Upjohn (Kalamazoo, Mich.); efavirenz (EFV), DuPont Pharmaceuticals (Wilmington, Del.); saquinavir (SQV), Roche Pharmaceuticals (Nutley, N.J.); indinavir (IDV), Merck, Inc. (Blue Bell, Pa.); ritonavir (RTV), Abbott Laboratories (Abbott Park, Ill.); nelfinavir (NFV), Agouron Pharmaceuticals (San Diego, Calif.); abacavir (ABC) and amprenavir (AMP), Glaxo/Wellcome (Research Triangle Park, N.C.); and adefovir (ADV), Gilead Sciences (Foster City, Calif.).
Sample preparation and amplification.
Virus was pelleted by centrifugation at 20,400 × g for 60 min from plasma (typically, 1 ml) prepared from blood samples collected in evacuated tubes containing either EDTA, acid-citrate dextrose, or heparin as an anticoagulant. Virus particles were disrupted by resuspending the pellets in 200 μl of lysis buffer (4 M guanidine thiocyanate, 0.1 M Tris HCl [pH 8.0], 0.5% sodium lauryl sarcosine, 1% dithiothreitol). RNA was extracted from viral lysates by using oligo(dT) linked to magnetic beads (Dynal, Oslo, Norway) (47). Reverse transcription was performed with Superscript II (Gibco/BRL, Gaithersburg, Md.) with an antisense internal primer, and PR and RT sequences were amplified with the Expand High Fidelity PCR kit (Boehringer Mannheim, Indianapolis, Ind.) with a forward primer containing an ApaI site and a reverse primer containing a PinAI site (6). The 1.5-kb amplification product spans the p7-p1-p6 protease cleavage sites in the gag polyprotein, the entire PR coding region, and the RT coding region from amino acids 1 to 313.
RTVs.
A retroviral vector designed to measure antiretroviral drug susceptibility was constructed by using an infectious molecular clone of HIV-1 (1). The vector, referred to as an indicator gene viral vector (IGVV), is replication defective and contains a luciferase expression cassette inserted within a deleted region of the envelope (env) gene (see Fig. 1A). Resistance test vectors (RTVs) were constructed by incorporating amplified PR and RT regions into the IGVV by using ApaI and PinAI restriction sites and conventional cloning methods (2). RTVs were prepared as libraries (pools) in order to capture and preserve the PR and RT sequence heterogeneity of the virus in the patient. IGVVs lack PR and RT sequences, and RTVs that lack patient virus-derived inserts are excluded from RTV pools (C. J. Petropoulos et al., patent application in preparation). Amplification products were digested with ApaI and PinAI (Gibco/BRL), purified by agarose gel electrophoresis, and ligated to ApaI- and PinAI-digested IGVV DNA. Internal ApaI and PinAI recognition sites within the PR-RT segment occur infrequently (approximate frequency, 1 to 2%) in HIV-1 (Los Alamos National Laboratory sequence compendium [50] see also http://hiv-web.lanl.gov/HTML/98compendium.html). Alternative ligation strategies with additional restriction sites can be used to evaluate viruses that contain ApaI or PinAI sites within this region without changing the boundaries of the patient-derived fragment. Ligation reactions were used to transform competent Escherichia coli (Invitrogen, Carlsbad, Calif.). An aliquot of each transformation was plated onto agar, and colony counts were used to estimate the number of viral segments represented in each RTV library (generally, 500 to 5,000 clones). RTV libraries that comprised less than 100 members are not considered representative of the patient virus. RTV plasmid DNA was purified by silica column chromatography (Qiaprep; Qiagen, Valencia, Calif.).
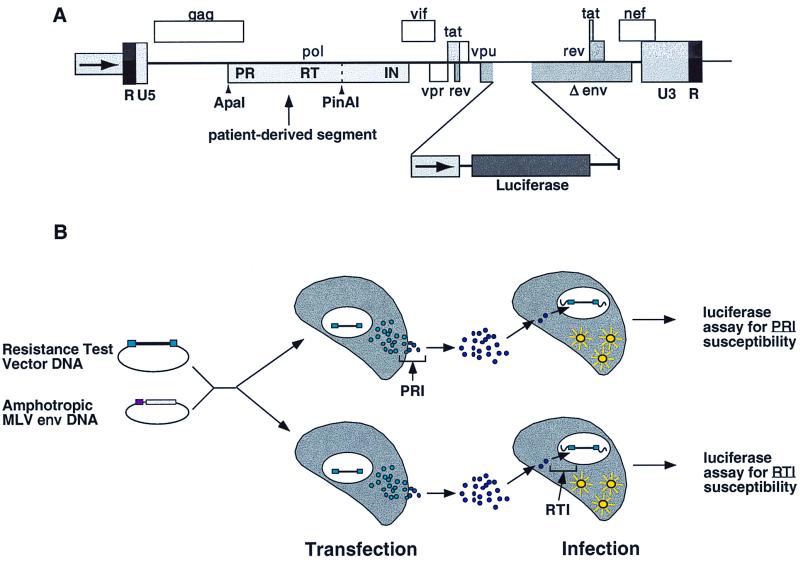
FIG. 1.
RTV structure and overview of drug susceptibility assay. (A) Amplified PR and RT gene segments from patient plasma samples are inserted into an indicator gene viral vector by using ApaI and PinAI restriction sites (vertical arrows). The ApaI site is located upstream of the gag polyprotein p7-p1-p6 cleavage sites. The PinAI site is located at amino acid 313 of RT. To monitor virus replication, a luciferase indicator gene cassette was inserted into a deleted region of the env gene, preventing HIV-1 envelope protein expression. (B) Pseudotyped virus particles are produced by cotransfecting cells with RTV DNA and a plasmid that expresses the envelope proteins of amphotropic murine leukemia virus (MLV). Following transfection, virus particles are harvested and are used to infect fresh target cells. The ability of virus particles to complete a single round of replication is assessed by measuring luciferase activity in target cells. The antiviral activities of PRIs and RTIs are measured by adding PRIs to transfected cells and RTIs to infected cells.
Drug susceptibility assay.
The virus stocks used for drug susceptibility testing were produced by cotransfecting human embryonic kidney 293 cell cultures (host cells) (AIDS Research and Reference Reagent Program, National Institutes of Health) with RTV plasmid DNA and an expression vector encoding the Env proteins of amphotropic murine leukemia virus 4070A (26, 39) (see Fig. 1B). To measure susceptibility to PRIs, the cells were trypsinized at approximately 16 h after transfection and were distributed into 96-well plates containing serial PRI dilutions spanning an empirically determined range for each drug. Viral stocks generated in the presence of PRIs were harvested at approximately 48 h after transfection and were used to infect fresh 293 cell cultures (target cells) in 96-well plates in the absence of drug. To measure susceptibility to RT inhibitors (RTIs), viral stocks generated in the absence of drug were harvested approximately 48 h after transfection and were used to infect fresh 293 cell cultures in 96-well plates containing serial RTI dilutions spanning an empirically determined range for each drug. Replication was monitored by measuring luciferase expression in infected target cells at approximately 48 h after infection. Determination of the virus titer prior to infection is not necessary and has been demonstrated by comparing the 50% inhibitory concentrations (IC50s) generated over a wide range of virus inocula (neat, 1:10, 1:100, and 1:1,000) and transfection efficiencies (data not shown). These observations are consistent with the single-replication-cycle format of this assay. RTVs containing mutations that disrupt the active site of either PR (D25G) or RT (D185G) were used to demonstrate that the ability of the virus to complete a single round of replication (i.e., produce luciferase activity) is dependent on functional PR and RT activities provided by the inserted PR-RT segment (data not shown).
Data are displayed by plotting the percent inhibition of luciferase activity versus log10 drug concentration. The percent inhibition was derived as follows: [1 −(luciferase activity in the presence of drug/luciferase activity in the absence of drug)] × 100. Mean percent inhibition for each drug concentration was determined from replicate determinations by a bootstrapping procedure (61). Inhibition curves defined by the four-parametric sigmoidal function f(x) = a −[b/(1 + (x/c)d)], were fit to the data by nonlinear least-squares and bootstrapping and were used to calculate the drug concentrations required to inhibit virus replication by 50% (IC50). The fold change in drug susceptibility is determined by comparing the IC50 for the sample virus to the IC50 for a drug-sensitive reference virus (strain CNDO) containing the PR and RT sequences of the NL4-3 strain of HIV-1 (1).
RESULTS
Assay description.
RTVs were constructed by amplifying PR and RT sequences derived from patient plasma samples and inserting the amplification products into a modified HIV-1 vector derived from the NL4-3 molecular clone (Fig. 1A; see the experimental protocol described above for details). Samples with a VL at or above the detection limit of standard VL assays (400 to 500 RNA copies/ml) can be amplified efficiently (52). Viral stocks were prepared by cotransfecting 293 cell cultures with RTV DNA and an expression vector that produces the envelope proteins from an amphotropic murine leukemia virus. Pseudotyped virus particles were harvested from the transfected cell cultures and were used to infect fresh 293 cells. RTVs contain a luciferase gene cassette within the env region and the production of luciferase in target cells is dependent on the completion of one round of virus replication. Drug susceptibility was measured by adding serial concentrations of PRIs to transfected cells or RTIs to infected cells (Fig. 1B). Drugs that inhibit virus replication reduce luciferase activity in a dose-dependent manner, providing a quantitative measure of drug susceptibility.
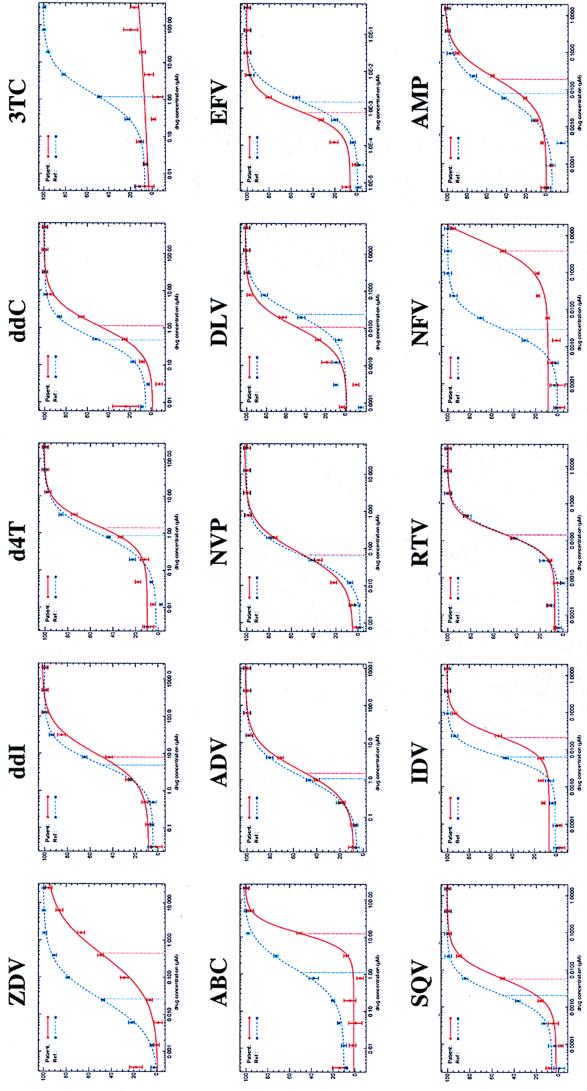
The phenotypic drug susceptibility profile for a representative patient virus is shown in Fig. 2. Inhibition of luciferase activity was plotted versus drug concentration (log10) for each drug tested. Drug susceptibility was measured by comparing the IC50 for the RTV stock derived from the patient virus with the IC50 for a drug-sensitive reference RTV stock (strain CNDO) that contains PR and RT sequences derived from the NL4-3 strain of HIV-1 (1). The assay measures the susceptibility of patient-derived HIV-1 PR and RT to all ARV drugs approved for use in the United States. In comparison to the reference virus tested in parallel, this patient's virus exhibited large reductions in susceptibility to 3TC (>150-fold), NFV (141-fold), ZDV (15.9-fold), and ABC (9.2-fold); i.e., the inhibition curves are shifted toward higher drug concentrations for the patient's virus (Fig. 2). Less pronounced reductions in susceptibility to IDV (3.3-fold), SQV (2.7-fold), and ddC (2.1-fold) were also observed.
FIG. 2.
Phenotypic drug susceptibility profile. Phenotypic drug susceptibility testing was performed with a plasma sample obtained from a patient receiving combination antiretroviral therapy. The viral load was 78,620 RNA copies per ml. Susceptibility to a panel of 15 antiretroviral drugs is shown. A reference virus (strain CNDO) that exhibits wild-type levels of susceptibility to all drugs was tested in parallel. Inhibition curves shifted to the right (higher drug concentration) of the reference curve for the drug-susceptible virus indicate reduced drug susceptibility. Inhibition curves shifted to the left (lower drug concentration) of the reference curve for the drug-susceptible virus indicate increased drug susceptibility. Fold differences in drug susceptibility were determined by comparing the IC50 for the reference virus to the IC50 for the sample virus. Drug abbreviations are defined in Materials and Methods. Dashed blue line, drug-sensitive reference virus; solid red line, patient virus. The patient virus genotype was as follows: for RT, M41L, D67N, M184V, L210W, and T215Y; for PR, L10F, D30N, L63P, V77I, N88D.
Assay performance.
The ability of this assay to accurately measure alterations in drug susceptibility was demonstrated with a comprehensive panel of isogenic viruses generated by site-directed mutagenesis (56). Over 100 viruses containing one or more mutations engendering resistance to PR or RT inhibitors have been constructed and tested. The drug susceptibilities of representative viruses containing some of the more common and/or well-characterized mutations are displayed in Table 1. Measurements of altered susceptibility for all drug classes (NRTIs, NNRTIs, and PRIs) were consistent with those in the existing scientific literature in terms of both the magnitude and specificity of altered drug susceptibility (for reviews, see references 4, 31, and 57). For example, the M184V/I mutations in RT dramatically reduce susceptibility to 3TC (20, 58, 64), and the D30N mutation in PR reduces susceptibility to NFV but not to other PRIs (54). The assay correctly demonstrated incremental reductions in susceptibility to ZDV when mutations associated with ZDV resistance were incorporated sequentially into viruses (36). The assay also accurately measured resensitization to ZDV when the M184V or Y181C mutation was added to ZDV-resistant viruses (5, 42, 43). The assay precisely characterized viruses containing complex combinations of mutations known to confer multi-NRTI resistance (i.e., the Q151M combination [34, 62] and the T69SSX combination [13, 41, 66, 67]) or PRI cross-resistance (9, 65; for reviews, see references 4 and 17).
TABLE 1.
Drug susceptibilities of viruses containing mutations associated with drug resistancea
| Mutation | Fold change in susceptibility (IC50 for sample virus/IC50 for reference virus)
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ZDV | d4T | 3TC | ddC | ddI | ABC | DLV | NVP | EFV | SQV | RTV | IDV | NFV | AMP | |
| NRTI | ||||||||||||||
| M41L,T215Y | 11.2 | 1.5 | 2.0 | 1.0 | 0.9 | 2.6 | ||||||||
| M41L,D67N,K70R,T215Y,K219Q | 54.6 | 1.9 | 3.0 | 0.8 | 0.9 | 2.8 | ||||||||
| M184V | 0.4 | 0.8 | >150 | 1.4 | 0.9 | 3.8 | ||||||||
| M184I | 0.3 | 0.8 | >150 | 1.5 | 1.2 | 3.1 | ||||||||
| M41L,D67N,K70R,M184V,T215Y,K219Q | 4.8 | 1.4 | >150 | 1.8 | 1.4 | 3.4 | ||||||||
| M41L,D67N,K70R,Y181C,T215Y,K219Q | 7.3 | 1.4 | 1.1 | 0.4 | 0.5 | 1.0 | ||||||||
| K65R | 0.4 | 1.7 | 12.5 | 2.5 | 1.8 | 2.6 | ||||||||
| L74V | 1.1 | 0.9 | 2.6 | 1.2 | 1.8 | NTb | ||||||||
| V75T | 0.6 | 1.9 | 3.5 | 1.4 | 1.2 | NT | ||||||||
| M41L,A62V,T69SSA,L210W,T215Y | >1,000 | 13.3 | 21.9 | 2.1 | 3.0 | 16.4 | ||||||||
| Q151M | 7.4 | 2.9 | 1.4 | 2.2 | 6.8 | NT | ||||||||
| A62V,V75I,F77L,F116Y,Q151M,M184V | 307.4 | 11.5 | >150 | 16.1 | 18.3 | 28.9 | ||||||||
| NNRTI | ||||||||||||||
| L100I | 30.0 | 3.1 | 10.0 | |||||||||||
| K101E | 4.9 | 11.6 | 5 | |||||||||||
| K103N | 52.0 | 55.0 | 26.0 | |||||||||||
| V106A | 5.0 | 64.0 | 1.7 | |||||||||||
| V108I | 1.3 | 3 | 1.7 | |||||||||||
| Y181C | 35.0 | 161.0 | 3.0 | |||||||||||
| Y188L | 9.0 | >500 | 109.0 | |||||||||||
| G190A | 0.5 | 75.0 | 7.6 | |||||||||||
| G190S | 0.4 | 206.0 | 47.0 | |||||||||||
| K101E,G190A | 2.0 | >500 | 123.0 | |||||||||||
| K101E,G190S | 1.4 | >500 | >500 | |||||||||||
| K103N,Y181C | >250 | >500 | 31 | |||||||||||
| K103N,G190A | 37.0 | >500 | 213.0 | |||||||||||
| PRI | ||||||||||||||
| D30N,N88D | 0.5 | 0.4 | 0.5 | 7.1 | 0.4 | |||||||||
| D30N,M46I,L63P,N88D | 0.9 | 1.0 | 1.2 | 24.7 | 0.8 | |||||||||
| M46I,L63P,V82T,I84V | 1.0 | 16.1 | 2.8 | 2.2 | 3.0 | |||||||||
| L10R,M46I,L63P,V82T,I84V | 1.7 | 22.4 | 3.9 | 3.6 | 4.4 | |||||||||
| L10I,L63P,A71V,I84V,L90M | 9.0 | 12.0 | 3.6 | 6.2 | 3.6 | |||||||||
| L10I,M46I,L63P,A71V,I84V,L90M | 4.9 | 11.3 | 4.2 | 11.5 | 4.5 | |||||||||
| G48V,L90M | 12.9 | 4.4 | 3.7 | 3.0 | 2.3 | |||||||||
Resistance test vectors containing defined amino acid substitutions were generated either by site-directed mutagenesis (56) or by isolating RTV clones derived from patient viruses. PR and RT amino acid sequences were verified by nucleic acid sequence analyses.
NT, not tested.
Two approaches were taken to evaluate assay reproducibility. The ability of the cell-based portion of the assay to generate reproducible measurements of drug susceptibility was evaluated by repeatedly testing a drug-sensitive reference RTV (strain CNDO) and a multidrug-resistant reference RTV strain (strain R268). The mean IC50s, standard deviations and 95% confidence intervals derived from 13 separate determinations with 15 drugs are shown in Table 2. The reproducibility of sample preparation (RTV assembly) was evaluated by processing separate aliquots of plasma from nine subjects infected with viruses that exhibited distinct drug susceptibility profiles. The results of these paired determinations are shown in Table 3. For 99% of the determinations (106 of 107), IC50s from replicate assays differed by less than 2.5-fold (the actual value of the single determination that exceeded 2.5-fold was 2.7-fold). The variations for 94% of the determinations (101 of 107) were less than twofold. PR and RT amino acid sequences for each virus were consistent with the observed phenotypic drug susceptibility profiles (Table 3).
TABLE 2.
Reproducibility of drug susceptibility determinationa
| Virus and statistic | NRTI IC50 (μM)
|
NNRTI IC50 (nM)
|
PRI IC50 (nM)
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ZDV | ddI | d4T | ddC | 3TC | ABC | ADV | NVP | DLV | EFV | SQV | IDV | RTV | NFV | AMP | |
| CNDO | |||||||||||||||
| Mean IC50 | 0.030 | 4.57 | 0.66 | 0.68 | 1.77 | 1.74 | 1.38 | 78.0 | 32.0 | 2.0 | 3.0 | 6.0 | 14.0 | 3.0 | 12.0 |
| SD | 0.005 | 1.04 | 0.13 | 0.23 | 0.48 | 0.50 | 0.28 | 11.0 | 6.6 | 0.3 | 0.7 | 0.9 | 2.1 | 0.7 | 2.4 |
| 95% CI | 0.003 | 0.57 | 0.07 | 0.12 | 0.26 | 0.27 | 0.15 | 6.0 | 3.6 | 0.2 | 0.4 | 0.5 | 1.1 | 0.4 | 1.3 |
| R268 | |||||||||||||||
| Mean IC50 | 2.81 | 9.96 | 1.96 | 1.60 | >300 | 22.17 | 3.26 | 3,817.9 | 807.3 | 32.2 | 470.3 | 338.0 | >3.0 | 163.8 | 52.3 |
| SD | 1.18 | 2.66 | 0.57 | 0.52 | NAb | 10.37 | 0.85 | 1,379.0 | 248.0 | 8.0 | 134.7 | 64.4 | NA | 42.3 | 12.3 |
| 95% CI | 0.64 | 1.45 | 0.31 | 0.28 | NA | 5.64 | 0.46 | 749.4 | 134.9 | 4.5 | 73.2 | 35.0 | NA | 23.0 | 6.7 |
The mean IC50 of each drug, standard deviation (SD; derived from 13 independent determinations), and 95% confidence interval (CI; derived from 13 independent determinations) are listed for drug-sensitive (strain CNDO) and multidrug-resistant (strain R268) reference viruses. The multidrug-resistant reference virus contains RT mutations M41L, D67N, K103N, M184V, L210W, T215Y, and K219Q and PR mutations L10I, M36I, L63P, A71V, and V82T.
NA, not applicable.
TABLE 3.
Reproducibility of drug susceptibility assay starting with plasmaa
| Sampleb | Fold change in susceptibility (IC50 for sample virus/IC50 for reference virus)
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NRTI
|
NNRTI
|
PRI
|
||||||||||
| ZDV | ddI | d4T | ddC | 3TC | NVP | DLV | SQV | IDV | RTV | NFV | AMP | |
| 178A | 39.8 | 1.40 | 1.57 | 1.77 | —c | 0.38 | 0.27 | 3.12 | 6.42 | 12.05 | 3.97 | 3.24 |
| 178B | 51.1 | 1.71 | 2.02 | 1.76 | — | 0.52 | 0.42 | 3.09 | 6.09 | 25.40 | 7.04 | 3.67 |
| 268A | 37.1 | 2.50 | 2.12 | 2.14 | — | 1.13 | 23.9 | 91.0 | 20.3 | — | 34.7 | 5.23 |
| 268B | 30.0 | 2.86 | 2.25 | 1.88 | — | 1.45 | 28.4 | 121.4 | 21.9 | — | 36.5 | 3.69 |
| 372A | 2.79 | 1.22 | 1.46 | 2.20 | — | 194.5 | — | 0.76 | 4.41 | 5.89 | 3.36 | 2.25 |
| 372B | 2.67 | 1.86 | 1.13 | 1.17 | — | 236.1 | 145.6 | 2.08 | 9.66 | 5.84 | 5.06 | 2.15 |
| 644A | 1.04 | 1.24 | 0.84 | NDd | 1.56 | 581.2 | 2.78 | 1.00 | 3.56 | 4.40 | 5.05 | 0.90 |
| 644B | 2.17 | 0.98 | 0.64 | 1.09 | 0.82 | 622.3 | 1.78 | 0.47 | 2.17 | 4.57 | 2.42 | 0.68 |
| 648A | 1.51 | 0.56 | 1.15 | 0.64 | 0.83 | 1.00 | 1.59 | 0.37 | 0.93 | 0.90 | 11.14 | 0.43 |
| 648B | 1.36 | 0.70 | 0.80 | 1.14 | 0.48 | 0.88 | 1.99 | 0.32 | 1.36 | 0.69 | 15.48 | 0.31 |
| 649A | 3.33 | 0.99 | 0.72 | 0.71 | 0.92 | 0.37 | 0.62 | 1.17 | 1.71 | 2.09 | 2.01 | 2.30 |
| 649B | 2.81 | 0.83 | 0.71 | 0.82 | 0.93 | 0.38 | 0.59 | 1.62 | 2.03 | 2.00 | 3.42 | 2.87 |
| 675A | 5.09 | 1.75 | 1.08 | 1.96 | — | 4.70 | 1.45 | 0.92 | 0.86 | 1.81 | 2.29 | 0.36 |
| 675B | 4.03 | 2.31 | 1.15 | 2.15 | — | 7.33 | 2.73 | 0.92 | 0.53 | 2.22 | 2.10 | 0.51 |
| 682A | 21.4 | 1.51 | 2.83 | 0.66 | — | 0.17 | 0.21 | 29.1 | 32.16 | 203.2 | 121.7 | 10.8 |
| 682B | 38.9 | 1.83 | 3.86 | 1.15 | — | 0.26 | 0.10 | 37.4 | 42.31 | 136.8 | 167.3 | 8.87 |
| 686A | 22.1 | 0.87 | 2.02 | 0.37 | 3.50 | 0.15 | 0.18 | 37.2 | 33.18 | 203.8 | 97.2 | 13.9 |
| 686B | 15.5 | 0.74 | 0.92 | 0.72 | 2.01 | 0.20 | 0.14 | 53.9 | 37.60 | 135.6 | 73.7 | 19.4 |
Duplicate aliquots of nine patient plasma samples were processed in parallel.
Patient virus genotypes were as follows: sample 178, RT, M41L, D67N, K70R, M184V/I, L210W, and T215Y; PR, L10V, M46I/L, L63P, A71T/V, V82T, and I84V; sample 268, RT, M41L, D67N, K103K/N, M184V, L210W, T215Y, K219Q, and P236P/L; PR, L10I, K20R, M36I, I54V, L63P, A71V, V82T, I84V, and L90M; sample 372, RT, D67N, T69D, K70R, K103N, M184V, and K219Q; PR, M46I and A71V; sample 644, RT, K101E and G190S; PR, L10I, L63P, A71T/I, V82T, and L90L/M; sample 648, RT, K70K/R and T215T/Y; PR, K20R, D30N, M36I, L63H, and A71V; sample 649, RT, D67N, and K219Q; PR, L10I, and L63A; sample 675, RT, M41L, L74V, A98G, M184V, and T215Y; PR, L10I, M36I, L63S, and L90M; sample 682, RT, M41L, D67N, M184V, L210W, and T215Y; PR, L10I, K20K/R, M36I, M46L, I54V, L63P, A71V, V82A, and L90M; sample 686, RT, M41L, L210W, and T215F; PR, L10I, M46M/I, I54V, L63P, A71V, V82A, and L90M.
—, no inhibition of luciferase activity was observed at the highest concentration of drug tested.
ND, not determined.
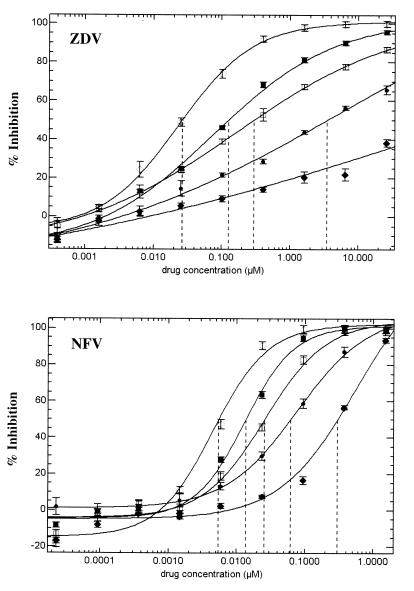
Assay sensitivity was assessed with virus preparations containing incremental mixtures of drug-susceptible and drug-resistant viruses or DNA preparations containing incremental mixtures of drug-susceptible and drug-resistant RTV DNA. The results of representative experiments that evaluated the sensitivity of the assay to mixtures of wild-type and NFV-resistant or wild-type and ZDV-resistant virus are displayed in Fig. 3. These data demonstrate that the assay readily distinguished mixtures that comprised 25, 50, or 75% resistant virus from the samples with 100% drug-sensitive or 100% drug-resistant virus.
FIG. 3.
Detection of drug-resistant virus subpopulations. RTV DNA representing genetically defined wild-type or drug-resistant mutant viruses were mixed at various ratios and were cotransfected into 293 cells to generate virus stocks. The drug susceptibility of each mixture was determined by the phenotypic assay. (A) Mixtures of wild-type virus and a patient virus clone containing RT mutations M41L, A62V, T69SSA, L74V, M184V, L210W, and T215Y tested against ZDV. (B) Mixtures of wild-type virus and a patient virus clone containing PR mutations K20R, L24I, G48V, I54V, V77I, and V82A tested against NFV. ◊, 0% mutant and 100% wild type; ■, 25% mutant and 75% wild type; □, 50% mutant and 50% wild type; ●, 75% mutant and 25% wild type; ⧫, 100% mutant and 0% wild type. Vertical dashed lines denote the IC50.
DISCUSSION
The phenotypic assay presented here can rapidly, accurately, and reproducibly measure the susceptibility of HIV-1 to all currently available ARV drugs. The assay is intended to aid physicians in the selection of more efficacious treatment regimens after treatment failure or at the initiation of therapy in drug-naive patients at risk of infection with resistant virus.
This assay has several advantages over the existing phenotypic drug susceptibility assays (29, 35, 37). First and foremost, the assay can be completed in 8 to 10 days, whereas most phenotypic assays typically require 4 to 8 weeks for completion. RTV plasmid DNAs are assembled by DNA ligation and are propagated in bacteria. This approach is efficient and reproducible and does not rely on the relatively inefficient and random process of homologous recombination to incorporate patient-derived PR and RT amplification products into HIV-1 vectors containing PR and RT deletions. Transfection of 293 cells with a standard amount of RTV DNA is a rapid method of producing consistent high-titer virus stocks and does not require virus outgrowth from patient samples or recombinant virus outgrowth following homologous recombination. Since RTVs are restricted to a single round of replication, there is little opportunity for selection of virus subpopulations that may not accurately reflect the susceptibility of the initial virus population in the patient. Viruses with drug resistance mutations that exhibit different in vitro replication kinetics compared to those of wild-type strains have been reported (10, 21, 22, 38). The single-replication-cycle format of the assay and the large dynamic range of the luciferase reaction also eliminate the need to determine virus titers prior to the infection of target cells. In addition to reducing the turnaround time, configuration of the assay in a single-cycle format with HIV-1 vectors containing an indicator gene (RTVs) provides increased sensitivity and reproducibility over those of existing assays that require multiple replication cycles and that are performed with indicator cell lines. Such improvements in sensitivity and reproducibility enable this assay to distinguish viruses that exhibit subtle (2.5- to 4-fold) differences in susceptibility that cannot be reliably measured by existing assays (3, 44). In general, this assay can detect subpopulations of drug-resistant viruses when they are present at a frequency of 20 to 25%, but this is dependent on several factors, including the differential drug susceptibilities of the variants in the mixture, the relative fitness of the variants, the biochemistry of the mutant RT or PR, and the mechanism of drug inhibition. As with any phenotypic assay, it is important to appreciate that the lower limit of detection of mixtures may vary (from 10 to 40%) depending on the mutant virus.
Treatment failure (i.e., the loss of suppression of virus replication) now affects a significant number of infected individuals receiving highly active ARV therapy (8, 18, 24, 30; for a review, see reference 32). The emergence of multidrug-resistant virus represents a significant obstacle in maintaining long-term suppression of virus replication in both treatment-naive and -experienced patients. Regaining sustained viral suppression in patients who have failed one or more previous treatment regimens is difficult with currently available salvage treatment regimens (J. Gallant, C. Hall, and S. Barnett, Fifth Conf. Retroviruses and Opportunistic Infections, 1998; S. Hammer, K. Squires, V. Degruttola, M. Fischl, R. Bassett, L. Demeter, K. Hertogs, and B. Larder, Sixth Conf. Retroviruses and Opportunistic Infections, 1999). Furthermore, several recent studies have described poor responses to initial treatment regimens in patients newly infected with drug-resistant HIV-1 (3, 28, 44; R. Grant, F. Hecht, N. Bandrapalli, C. Petropoulos, T. Gittens, M. Warmerdam, N. Hellmann, M. Chesney, M. Busch, and J. Kahn, submitted for publication).
Several small retrospective studies have examined the potential clinical utility of drug resistance testing for HIV-1 (12, 25, 40, 53, 55, 68). These analyses demonstrated that the baseline phenotype and genotype were predictive of drug treatment outcome. More recently, two prospective trials investigated the use of genotyping to assist in the selection of salvage treatment regimens (14; J. Baxter, D. Mayers, D. Wentworth, J. Neaton, and T. Merigan, Sixth Conf. Retroviruses and Opportunistic Infections, 1999). In both studies, both the proportion of patients who obtained complete suppression (defined as <500 copies/ml [Baxter et al., Sixth Conf. Retroviruses and Opportunistic Infections] or <200 copies/ml [14]) and the magnitude of viral load suppression were greater in the treatment arms that included resistance testing. Although the results of these studies are encouraging, only 29 and 32% of the patients in the genotyping arms obtained complete suppression of virus replication at 12 weeks and 6 months, respectively (14; Baxter et al., Sixth Conf. Retroviruses and Opportunistic Infections). This suggests that there is an insufficient understanding, at both the descriptive and the mechanistic levels, of the correlation between genotype and changes in phenotypic drug susceptibility. Of course, other factors in addition to the emergence of drug-resistant viral variants may contribute to treatment failure. The results of recent studies also revealed significant variability in the genotypic test results obtained from different laboratories, most notably, when mixtures of sensitive and resistant viruses were present in the same specimen (59, 60). Consequently, a direct measurement of drug susceptibility provided by a simple and rapid phenotypic assay may provide more consistent and relevant information for ARV drug treatment decisions. As with all resistance assays (whether they are phenotypic or genotypic assays), results are weighted in favor of the majority species present at the time of sampling. This can obscure the detection of minority species, especially in cases in which clinically important reductions in drug susceptibility are small or are associated with impaired replication fitness. Consequently, the interpretation of test results should always be performed with these points in mind and in the context of other relevant clinical data, especially drug treatment history. Modifications intended to enhance the detection of minor populations of drug-resistant viruses or viruses with reduced fitness are in progress.
Ultimately, the clinical utility of this and other resistance assays can be assessed only in carefully designed prospective clinical trials; these studies are in progress. The assay can also be used to monitor the frequency of drug-resistant virus transmission in newly infected individuals (3, 28, 44; Grant et al., submitted for publication), to characterize baseline variation in susceptibilities to various drugs in treatment-naive patients (3, 44; Grant et al., submitted for publication), and to gain a better understanding of how and why current treatments fail (27, 52). Finally, the assay can be used to screen for or assist in the development of new drugs that are active against resistant strains of HIV-1.
ACKNOWLEDGMENTS
We are grateful to Stephen Goff, David Ho, Stephen Hughes, Clifford Lane, Douglas Richman, and Robert Schooley for guidance; to Diane Havlir, Martin Markowitz, Daniel Kuritzkes, and James Kahn for providing patient samples; to Nicholas Hellmann, Rainer Ziermann, Ellen Paxinos, Gabrielle Heilek-Snyder, and Michelle Sartoris for discussion and critical review of the manuscript; to Linda Logan and Holly D'Souza for technical assistance with amplification sensitivity studies; to Sebastian Bonhoeffer for assistance with data analysis; to Merck, Glaxo/Wellcome, Bristol Myers-Squibb, Roxanne, Pharmacia-Upjohn, DuPont, Roche, Abbott, Agouron, and Gilead for providing antiretroviral drugs; and to Lisa Paborsky and Jennifer Fischer for assistance in preparing the manuscript. Finally, we acknowledge the enthusiastic support and constant encouragement of Martin Goldstein.
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F, editor. Current protocols in molecular biology. 1 and 2. New York, N.Y: John Wiley & Sons, Inc.; 1999. [Google Scholar]
- 3.Boden D, Hurley A, Zhang L, Cao Y, Guo Y, Duran M, Tsay J, Ip J, Farthing C, Limoli K, Parkin N, Markowitz M. HIV-1 drug resistance in newly infected individuals. JAMA. 1999;282:1135–1141. doi: 10.1001/jama.282.12.1135. [DOI] [PubMed] [Google Scholar]
- 4.Boden D, Markowitz M. Resistance to human immunodeficiency virus type 1 protease inhibitors. Antimicrob Agents Chemother. 1998;42:2775–2783. doi: 10.1128/aac.42.11.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Byrnes V W, Emini E A, Schleif W A, Condra J H, Schneider C L, Long W J, Wolfgang J A, Graham D J, Gotlib L, Schlabach A J, Wolanski B S, Blahy O M, Quintero J C, Rhodes A, Rothy E, Titus D L, Sardana V V. Susceptibilities of human immunodeficiency virus type 1 enzyme and viral variants expressing multiple resistance-engendering amino acid substitutions to reverse transcriptase inhibitors. Antimicrob Agents Chemother. 1994;38:1404–1407. doi: 10.1128/aac.38.6.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Capon D. Compositions and methods for determining anti-viral drug susceptibility and resistance and anti-viral drug screening. U.S. patent 5,837,464. Nov. 1998. [Google Scholar]
- 7.Carpenter C C J, Cooper D A, Fischl M A, Gatell J M, Gazzard B G, Hammer S M, Hirsch M S, Jacobsen D M, Katzenstein D A, Montaner J S, Richman D D, Saag M S, Schechter M, Schooley R T, Thompson M A, Vella S, Yeni P G, Volberding P A. Antiretroviral therapy in adults: updated recommendations of the International AIDS Society-USA panel. JAMA. 2000;283:381–390. doi: 10.1001/jama.283.3.381. [DOI] [PubMed] [Google Scholar]
- 8.Casado J, Perez-Elias M, Antela A, Sabido R, Marti-Belda P, Dronda F, Blazquez J, Quereda C. Predictors of long-term response to protease inhibitor therapy in a cohort of HIV-infected patients. AIDS. 1998;12:F131–F135. doi: 10.1097/00002030-199811000-00005. [DOI] [PubMed] [Google Scholar]
- 9.Condra J, Schleif W, Blahy O, Gabryelski L, Graham D, Quintero J, Rhodes A, Robbins H, Roth E, Shivaprakash M, Titus D, Yan T, Teppler H, Squires K, Deutsch P, Emini E. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature. 1995;374:569–571. doi: 10.1038/374569a0. [DOI] [PubMed] [Google Scholar]
- 10.Croteau G, Doyon L, Thibeault D, McKercher G, Pilote L, Lamarre D. Impaired fitness of human immunodeficiency virus type 1 variants with high-level resistance to protease inhibitors. J Virol. 1997;71:1089–1096. doi: 10.1128/jvi.71.2.1089-1096.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deeks S G, Abrams D I. Genotypic-resistance assays and antiretroviral therapy. Lancet. 1997;349:1489–1490. doi: 10.1016/S0140-6736(05)62092-2. [DOI] [PubMed] [Google Scholar]
- 12.Deeks S G, Hellmann N S, Grant R M, Parkin N T, Petropoulos C J, Becker M, Symonds W, Chesney M, Volberding P A. Novel four-drug salvage treatment regimens after failure of a human immunodeficiency virus type 1 protease inhibitor-containing regimen: antiviral activity and correlation of baseline phenotypic drug susceptibility with virologic outcome. J Infect Dis. 1999;179:1375–1381. doi: 10.1086/314775. [DOI] [PubMed] [Google Scholar]
- 13.de Jong J, Goudsmit J, Lukashov V, Hillebrand M, Baan E, Huismans R, Danner S, ten Veen J, de Wolf F, Jurriaans S. Insertion of two amino acids combined with changes in reverse transcriptase containing tyrosine-215 of HIV-1 resistant to multiple nucleoside analogs. AIDS. 1999;13:75–80. doi: 10.1097/00002030-199901140-00010. [DOI] [PubMed] [Google Scholar]
- 14.Durant J, Clevenbergh P, Halfon P, Delgiudice P, Porsin S, Simonet P, Montagne N, Boucher C A, Schapiro J M, Dellamonica P. Drug-resistance genotyping in HIV-1 therapy: the VIRADAPT randomised controlled trial. Lancet. 1999;353:2195–2199. doi: 10.1016/s0140-6736(98)12291-2. [DOI] [PubMed] [Google Scholar]
- 15.Eastman P S, Boyer E, Mole L, Kolberg J, Urdea M, Holodniy M. Nonisotopic hybridization assay for determination of relative amounts of genotypic human immunodeficiency virus type 1 zidovudine resistance. J Clin Microbiol. 1995;33:2777–2780. doi: 10.1128/jcm.33.10.2777-2780.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Egger M, Hirschel B, Francioli P, Sudre P, Wirz M, Rickenbach M, Malinverni R, Vernazza P, Battegay M. Impact of new antiretroviral combination therapies in HIV infected patients in Switzerland: prospective multicentre study. Br Med J. 1997;315:1194–1199. doi: 10.1136/bmj.315.7117.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Erickson J, Gulnik S, Markowitz M. Protease inhibitors: resistance, cross-resistance, fitness and the choice of initial and salvage therapies. AIDS. 1999;13(Suppl. A):S189–S204. [PubMed] [Google Scholar]
- 18.Fatkenheuer G, Theisen A, Rockstroh J, Grabow T, Wicke C, Becker K, Wieland U, Pfister H, Reiser M, Hegener P, Franzen C, Schwenk A, Salzberger B. Virological treatment failure of protease inhibitor therapy in an unselected cohort of HIV-infected patients. AIDS. 1997;11(14):F113–F116. doi: 10.1097/00002030-199714000-00001. [DOI] [PubMed] [Google Scholar]
- 19.Fiscus S A, Hughes M D, Lathey J L, Pi T, Jackson B, Rasheed S, Elbeik T, Reichman R, Japour A, Byington R, Scott W, Griffith B P, Katzenstein D A, Hammer S M. Changes in virologic markers as predictors of CD4 cell decline and progression of disease in human immunodeficiency virus type 1-infected adults treated with nucleosides. J Infect Dis. 1998;177:625–633. doi: 10.1086/514248. [DOI] [PubMed] [Google Scholar]
- 20.Gao Q, Gu Z, Parniak M A, Cameron J, Cammack N, Boucher C, Wainberg M A. The same mutation that encodes low-level human immunodeficiency virus type 1 resistance to 2′,3′-dideoxyinosine and 2′,3′-didoxycytidine confers high-level resistance to the (−) enantiomer of 2′,3′-dideoxy-3′-thiacytidine. Antimicrob Agents Chemother. 1993;37:1390–1392. doi: 10.1128/aac.37.6.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goudsmit J, de Ronde A, de Rooij E, de Boer R. Broad spectrum of in vivo fitness of human immunodeficiency virus type 1 subpopulations differing at reverse transcriptase codons 41 and 215. J Virol. 1997;71:4479–4484. doi: 10.1128/jvi.71.6.4479-4484.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goudsmit J, De Ronde A, Ho D, Perelson A. Human immunodeficiency virus fitness in vivo: calculations based on a single zidovudine resistance mutation at codon 215 of reverse transcriptase. J Virol. 1996;70:5662–5664. doi: 10.1128/jvi.70.8.5662-5664.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gulick R M, Mellors J W, Havlir D, Eron J J, Gonzalez C, McMahon D, Richman D D, Valentine F T, Jonas L, Meibohm A, Emini E A, Chodakewitz J A. Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. N Engl J Med. 1997;337:734–739. doi: 10.1056/NEJM199709113371102. [DOI] [PubMed] [Google Scholar]
- 24.Hammer S M, Squires K E, Hughes M D, Grimes J M, Demeter L M, Currier J S, Eron J J, Jr, Feinberg J E, Balfour H H, Jr, Deyton L R, Chodakewitz J A, Fischl M A. A controlled trial of two nucleoside analogues plus indinavir in persons with human immunodeficiency virus infection and CD4 cell counts of 200 per cubic millimeter or less. AIDS Clinical Trials Group Study Team. N Engl J Med. 1997;337:725–733. doi: 10.1056/NEJM199709113371101. [DOI] [PubMed] [Google Scholar]
- 25.Harrigan P, Hertogs K, Verbiest W, Pauwels R, Larder B, Kemp S, Bloor S, Yip B, Hogg R, Alexander C, Montaner J. Baseline HIV drug resistance profile predicts response to ritonavir-saquinavir protease inhibitor therapy in a community setting. AIDS. 1999;13(14):1863–1871. doi: 10.1097/00002030-199910010-00008. [DOI] [PubMed] [Google Scholar]
- 26.Hartley J W, Rowe W P. Naturally occuring murine leukemia viruses in wild mice: characterization of a new amphotropic class. J Virol. 1976;19:19–25. doi: 10.1128/jvi.19.1.19-25.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Havlir D, Whitcomb J, Hellmann N, Petropoulos C, Collier A, Hirsch M, Tebas P, Richman D. Drug susceptibility in HIV isolates obtained after viral rebound from patients receiving an indinavir containing regimen. JAMA. 2000;283:229–234. doi: 10.1001/jama.283.2.229. [DOI] [PubMed] [Google Scholar]
- 28.Hecht F M, Grant R M, Petropoulos C J, Dillon B, Chesney M A, Tian H, Hellmann N S, Bandrapalli N I, Digilio L, Branson B, Kahn J O. Sexual transmission of an HIV-1 variant resistant to multiple reverse-transcriptase and protease inhibitors. N Engl J Med. 1998;339:307–311. doi: 10.1056/NEJM199807303390504. [DOI] [PubMed] [Google Scholar]
- 29.Hertogs K, De Béthune M-P, Miller V, Ivens T, Schel P, Cauwenberge A V, Van Den Eynde C, Van Gerwen V, Azijn H, Van Houtte M, Peeters F, Staszewski S, Conant M, Bloor S, Kemp S, Larder B, Pauwels R. A rapid method for simultaneous detection of phenotypic resistance to inhibitors of protease and reverse transcriptase in recombinant human immunodeficiency virus type 1 isolates from patients treated with antiretroviral drugs. Antimicrob Agents Chemother. 1998;42:269–276. doi: 10.1128/aac.42.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirsch M, Steigbigel R, Staszewski S, Mellors J, Scerpella E, Hirschel B, Lange J, Rawlins S K, S, Meiboh A, Leavitt R. A randomized, controlled trial of indinavir, zidovudine, and lamivudine in adults with advanced human immunodeficiency virus type 1 infection and prior antiretroviral therapy. J Infect Dis. 1999;180:659–665. doi: 10.1086/314948. [DOI] [PubMed] [Google Scholar]
- 31.Hirsch M S, Conway B, D'Aquila R T, Johnson V A, Brun-Vezient F, Clotet B, Demeter L M, Hammer S M, Jacobsen D M, Kuritzkes D R, Loveday C, Mellors J W, Vella S, Richman D D. Antiretroviral drug resistance testing in adults with HIV infection: implications for clinical management. JAMA. 1998;279:1984–1991. doi: 10.1001/jama.279.24.1984. [DOI] [PubMed] [Google Scholar]
- 32.Hirschel B, Opravil M. The year in review: antiretroviral treatment. AIDS. 1999;13(Suppl. A):S177–S187. [PubMed] [Google Scholar]
- 33.Hogg R, Yip B, Kully C, Craib K, O'Shaunghnessy M, Schechter M, Montaner J. Improved survival among HIV-infected patients after initiation of triple-drug antiretroviral regimens. Can Med Assoc J. 1999;160:659–665. [PMC free article] [PubMed] [Google Scholar]
- 34.Iversen A, Shafer R, Wehrly K, Winters M, Mullins J, Cheseboro B, Merigan T. Multidrug-resistant human immunodeficiency virus type 1 strains resulting from combination antiretroviral therapy. J Virol. 1996;70:1086–1090. doi: 10.1128/jvi.70.2.1086-1090.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Japour A J, Mayers D L, Johnson V A, Kuritzkes D R, Beckett L A, Arduino J-M, Lane J, Black R J, Reichelderfer P S, D'Aquila R T, Crumpacker C S The RV-43 Study Group and the AIDS Clinical Trials Group Virology Committee Resistance Working Group. Standardized peripheral blood mononuclear cell culture assay for determination of drug susceptibilities of clinical human immunodeficiency virus type 1 isolates. Antimicrob Agents Chemother. 1993;37:1095–1101. doi: 10.1128/aac.37.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kellam P, Boucher C, Tijnagel J, Larder B. Zidovudine treatment results in the selection of human immunodeficiency virus type 1 variants whose genotypes confer increasing levels of drug resistance. J Gen Virol. 1994;75:341–351. doi: 10.1099/0022-1317-75-2-341. [DOI] [PubMed] [Google Scholar]
- 37.Kellam P, Larder B A. Recombinant virus assay: a rapid, phenotypic assay for assessment of drug susceptibility of human immunodeficiency virus type 1 isolates. Antimicrob Agents Chemother. 1994;38:23–30. doi: 10.1128/aac.38.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keulen W, Back N K, van Wijk A, Boucher C A, Berkhout B. Initial appearance of the 184Ile variant in lamivudine-treated patients is caused by the mutational bias of human immunodeficiency virus type 1 reverse transcriptase. J Virol. 1997;71:3346–3350. doi: 10.1128/jvi.71.4.3346-3350.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Landau N R, Page K A, Littman D R. Pseudotyping with human T-cell leukemia virus type 1 broadens the human immunodeficiency virus host range. J Virol. 1991;65:162–169. doi: 10.1128/jvi.65.1.162-169.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lanier R, Danehower S, Daluge S, Cutrell A, Tisdale M, Pearce G, Spreen B, Lafon S, Kemp S, Bloor S, Larder B. Genotypic and phenotypic correlates of response to abacavir (ABC, 1592) Antivir Ther. 1998;3(Suppl. 1):36. [Google Scholar]
- 41.Larder B, Bloor S, Kemp S, Hertogs K, Desmet R, Miller V, Sturmer M, Staszewski S, Ren J, Stammers D, Stuart D, Pauwels R. A family of insertion mutations between codons 67 and 70 of human immunodeficiency virus type 1 reverse transcriptase confer multinucleoside analog resistance. Antimicrob Agents Chemother. 1999;43:1961–1967. doi: 10.1128/aac.43.8.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Larder B, Kellam P, Kemp S. Convergent combination therapy can select viable multidrug-resistant HIV-1 in vitro. Nature. 1993;365:451–453. doi: 10.1038/365451a0. [DOI] [PubMed] [Google Scholar]
- 43.Larder B A, Kemp S D, Harrigan P R. Potential mechanism for sustained antiretroviral efficacy of AZT-3TC combination therapy. Science. 1995;269:696–699. doi: 10.1126/science.7542804. [DOI] [PubMed] [Google Scholar]
- 44.Little S, Daar E, D'Aquila R, Keiser P, Connick E, Whitcomb J, Hellmann N, Petropoulos C, Pitt J, Rosenberg E, Koup R, Richman D. Reduced antiretroviral drug susceptibility among patients with primary HIV infection. JAMA. 1999;282:1142–1149. doi: 10.1001/jama.282.12.1142. [DOI] [PubMed] [Google Scholar]
- 45.Lorenzi P, Opravil M, Hirschel B, Chave J P, Furrer H J, Sax H, Perneger T V, Perrin L, Kaiser L, Yerly S. Impact of drug resistance mutations on virologic response to salvage therapy. AIDS. 1999;13(2):F17–F21. doi: 10.1097/00002030-199902040-00001. [DOI] [PubMed] [Google Scholar]
- 46.Marschner I C, Collier A C, Coombs R W, D'Aquila R T, DeGruttola V, Fischl M A, Hammer S M, Hughes M D, Johnson V A, Katzenstein D A, Richman D D, Smeaton L M, Spector S A, Saag M S. Use of changes in plasma levels of human immunodeficiency virus type 1 RNA to assess the clinical benefit of antiretroviral therapy. J Infect Dis. 1998;177:40–47. doi: 10.1086/513823. [DOI] [PubMed] [Google Scholar]
- 47.Meijer A, van Loon A, Borleffs J C, Roozendaal G. Development of molecular methods for detection and epidemiological investigation of HIV-1, HIV-2, and HTLV I/II infections 118504001. Bilthoven, The Netherlands: National Institute of Public Health and Environmental Protection; 1995. [Google Scholar]
- 48.Mellors J W, Munoz A, Giorgi J V, Margolick J B, Tassoni C J, Gupta P, Kingsley L A, Todd J A, Saah A J, Detels R, Phair J P, Rinaldo C R., Jr Plasma viral load and CD4+ lymphocytes as prognostic markers of HIV-1 infection. Ann Intern Med. 1997;126:946–954. doi: 10.7326/0003-4819-126-12-199706150-00003. [DOI] [PubMed] [Google Scholar]
- 49.Mouton Y, Alfandari S, Valette M, Cartier F, Dellamonica P, Humbert G, Lang J, Massip P, Michali D, Leclercq P, Modai J, Portier H. Impact of protease inhibitors on AIDS-defining events and hospitalizations in 10 French AIDS reference centres. AIDS. 1997;11(12):F101–F105. doi: 10.1097/00002030-199712000-00003. [DOI] [PubMed] [Google Scholar]
- 50.Myers G, Wain-Hobson S, Korber B, Smith R F, Pavlakis G N. Human retroviruses and AIDS. Los Alamos, N.M: Los Alamos National Laboratory; 1996. [Google Scholar]
- 51.Palella F J, Jr, Delaney K M, Moorman A C, Loveless M O, Fuhrer J, Satten G A, Aschman D J, Holmberg S D. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV Outpatient Study Investigators. N Engl J Med. 1998;338:853–860. doi: 10.1056/NEJM199803263381301. [DOI] [PubMed] [Google Scholar]
- 52.Parkin N, Lie Y, Hellmann N, Markowitz M, Bonhoeffer S, Ho D, Petropoulos C. Phenotypic changes in drug susceptibility associated with failure of human immunodeficiency virus type 1 (HIV-1) triple combination therapy. J Infect Dis. 1999;180:865–870. doi: 10.1086/314928. [DOI] [PubMed] [Google Scholar]
- 53.Patick A, Zhang M, Hertogs K, Griffiths L, Mazabel E, Pauwels R, Becker M. Correlation of virological response with genotype and phenotype of plasma HIV-1 variants in patients treated with nelfinavir in the US expanded access program. Antivir Ther. 1998;3(Suppl. 1):39. [Google Scholar]
- 54.Patick A K, Duran M, Cao Y, Shugarts D, Keller M R, Mazabel E, Knowles M, Chapman S, Kuritzkes D R, Markowitz M. Genotypic and phenotypic characterization of human immunodeficiency virus type 1 variants isolated from patients treated with the protease inhibitor nelfinavir. Antimicrob Agents Chemother. 1998;42:2637–2644. doi: 10.1128/aac.42.10.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Piketty C, Race E, Castiel P, Belec L, Peytavin G, si-Mohamed A, Gonzalez-Canali G, Weiss L, Clavel F, Kazatchkine M. Efficacy of a five-drug combination including ritonavir, saquinavir and efavirenz in patients who failed on a conventional triple-drug regimen: phenotypic resistance to protease inhibitors predicts outcome of therapy. AIDS. 1999;13(11):F71–F77. doi: 10.1097/00002030-199907300-00001. [DOI] [PubMed] [Google Scholar]
- 56.Sarkar G, Sommer S S. The “megaprimer” method of site-directed mutagenesis. BioTechniques. 1990;8:404–407. [PubMed] [Google Scholar]
- 57.Schinazi R F, Larder B A, Mellors J W. Mutations in retroviral genes associated with drug resistance. Intl Antivir News. 1999;7:46–69. [Google Scholar]
- 58.Schinazi R F, Lloyd R M, Jr, Nguyen M-H, Cannon D L, McMillan A, Ilksoy N, Chu C K, Liotta D C, Bazmi H Z, Mellors J W. Characterization of human immunodeficiency viruses resistant to oxathiolane-cytosine nucleosides. Antimicrob Agents Chemother. 1993;37:875–881. doi: 10.1128/aac.37.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schuurman R, Brambilla D, deGroot T, Boucher C. Second worldwide evaluation of HIV-1 drug resistance genotyping quality using the ENVA 2 panel. Antivir Ther. 1999;4(Suppl. 1):41. [Google Scholar]
- 60.Schuurman R, Demeter L, Reichelderfer P, Tijnagel J, de Groot T, Boucher C. Worldwide evaluation of DNA sequencing approaches for identification of drug resistance mutations in the human immunodeficiency virus type 1 reverse transcriptase. J Clin Microbiol. 1999;37:2291–2296. doi: 10.1128/jcm.37.7.2291-2296.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shargel L, Yu A B C. Applied biopharmaceutics and pharmacokinetics. 3rd ed. Norwalk, Conn: Appleton & Lange; 1993. [Google Scholar]
- 62.Shirasaka T, Kavlick M, Ueno T, Gao W, Kojima E, Alcaide M, Chokekijchai S, Roy B, Arnold E, Yarchoan R, Mitsuya H. Emergence of human immunodeficiency virus type 1 variants with resistance to multiple dideoxynucleosides in patients receiving therapy with dideoxynucleosides. Proc Natl Acad Sci USA. 1995;92:2398–2402. doi: 10.1073/pnas.92.6.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stuyver L, Wyseur A, Rombout A, Louwagie J, Scarcez T, Verhofstede C, Rimland D, Schinazi R, Rossau R. Line probe assay for rapid detection of drug-selected mutations in the human immunodeficiency virus type 1 reverse transcriptase gene. Antimicrob Agents Chemother. 1997;41:284–291. doi: 10.1128/aac.41.2.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tisdale M, Kemp S D, Parry N R, Larder B A. Rapid in vitro selection of human immunodeficiency virus type 1 resistant to 3′-thiacytidine inhibitors due to a mutation in the YMDD region of reverse transcriptase. Proc Natl Acad Sci USA. 1993;90:5653–5656. doi: 10.1073/pnas.90.12.5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tisdale M, Myers R E, Maschera B, Parry N R, Oliver N M, Vlair E D. Cross-resistance analysis of human immunodeficiency virus type 1 variants individually selected for resistance to five different protease inhibitors. Antimicrob Agents Chemother. 1995;39:1704–1710. doi: 10.1128/aac.39.8.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Whitcomb J, Limoli K, Wrin T, Smith D, Tian H, Parkin N, Lie Y, Petropoulos C. Phenotypic and genotypic analysis of stavudine-resistant isolates of HIV-1. Antivir Ther. 1998;3(Suppl. 1):14. [Google Scholar]
- 67.Winters M, Coolley K, Girard Y, Levee D, Hamdan H, Shafer R, Katzenstein D, Merigan T. A 6-basepair insert in the reverse transcriptase gene of human immunodeficiency virus type 1 confers to multiple nucleoside inhibitors. J Clin Invest. 1998;102:1769–1775. doi: 10.1172/JCI4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zolopa A, Shafer R, Warford A, Montoya J, Ilsu P, Katzenstein D, Merigan T, Efron B. HIV-1 genotypic resistance patterns predict response to saquinavir-ritonavir therapy in patients in whom previous protease inhibitor therapy had failed. Ann Intern Med. 1999;131:813–821. doi: 10.7326/0003-4819-131-11-199912070-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]