Abstract
Chemical risk assessments follow a long-standing paradigm that integrates hazard, dose–response, and exposure information to facilitate quantitative risk characterization. Targeted analytical measurement data directly support risk assessment activities, as well as downstream risk management and compliance monitoring efforts. Yet, targeted methods have struggled to keep pace with the demands for data regarding the vast, and growing, number of known chemicals. Many contemporary monitoring studies therefore utilize non-targeted analysis (NTA) methods to screen for known chemicals with limited risk information. Qualitative NTA data has enabled identification of previously unknown compounds and characterization of data-poor compounds in support of hazard identification and exposure assessment efforts. In spite of this, NTA data have seen limited use in risk-based decision making due to uncertainties surrounding their quantitative interpretation. Significant efforts have been made in recent years to bridge this quantitative gap. Based on these advancements, quantitative NTA data, when coupled with other high-throughput data streams and predictive models, are poised to directly support 21st-century risk-based decisions. This article highlights components of the chemical risk assessment process that are influenced by NTA data, surveys the existing literature for approaches to derive quantitative estimates of chemicals from NTA measurements, and presents a conceptual framework for incorporating NTA data into contemporary risk assessment frameworks.
Keywords: Non-targeted analysis, Risk characterization, Exposure modeling, Quantitation
1. Introduction
Advances in analytical mass spectrometry and high-throughput data processing capabilities have instigated a paradigm shift in molecular analysis over the past twenty years. At the turn of the millennium, biologically, environmentally, and pharmacologically relevant chemical assessments were based on targeted analyses of small panels of chemicals. Targeted methods are accurate and robust, but a strict focus on defined chemical panels leads only to an improved understanding of those pre-selected compounds. Modern chemical inventories have expanded drastically, with generic chemical registries like Pubchem and CAS containing hundreds of millions of registered chemical substances (CAS; Kim et al., 2020). Even a conservative chemical database, such as the EPA Comptox Chemicals Dashboard, lists over a million substances (McEachran et al., 2017). In contrast to these millions of chemical substances, only a fraction of known compounds have public monitoring and/or analysis methods in common use. In the United States, the EPA substance registry service indicates that ~ 80,000 chemical substances are tracked or regulated by the agency (USEPA, xxxx). Therefore, traditional monitoring methods are useful for, conservatively, only ~ 5% of known chemical species, with limited physicochemical, toxicity, or exposure data available for the many chemicals of emerging concern (Brack et al., 2012; Naidu et al., 2016; Weinberg et al., 2019; Egeghy et al., 2012). Recent years have ushered in new methods to fill the gap of targeted analyses. Burgeoning chemical “-omics” fields (e.g. exposomics, metabolomics) utilize holistic, non-targeted analytical approaches to interrogate compounds in complex environmental and biological systems (Rappaport and Smith, 2010; Wild, 2005; German et al., 2005).
Non-targeted analysis (NTA) methods have unique value in that they can garner informative chemical measurements from samples of interest without the need for predefined chemical targets. The workhorse techniques for NTA are nuclear magnetic resonance (NMR), which is well described in a recent review article, (Giraudeau, 2020) and high-resolution mass spectrometry (HRMS), which we discuss here. HRMS methods (often coupled with separation techniques) utilize ionization sources paired with high resolving power mass detectors to isolate and identify chemicals based on their observed accurate masses, isotopic fingerprints, and MS/MS fragments. Applications of HRMS-based NTA abound, with demonstrated capabilities in screening for disease (López-López et al., 2018; Trivedi et al., 2017) and exposure biomarkers, (Pourchet et al., 2020) detecting environmental contaminant sources, (Ruff et al., 2015; Brack et al., 2019; Focazio et al., 2008) discovering emerging environmental pollutants, (McMahen et al., 2016; Bletsou et al., 2015) and monitoring effectiveness of pollutant treatment technologies (Newton et al., 2018; McCord et al., 2020).
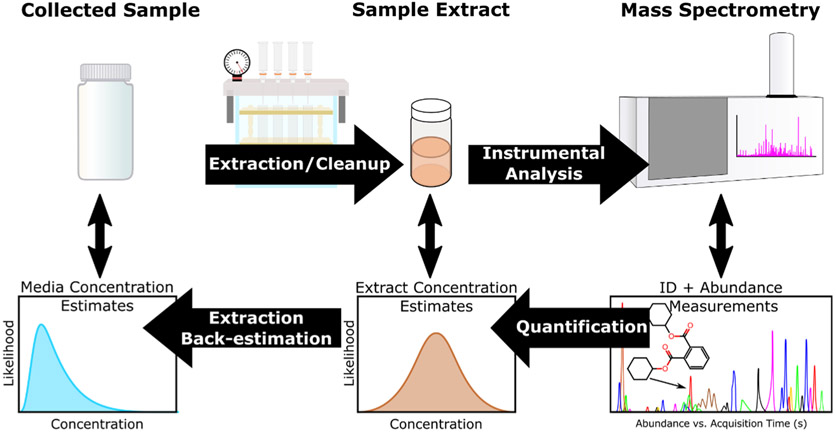
A unifying feature of NTA studies is a primarily qualitative focus on the unambiguous identification of individual species—and there are large concerted efforts to improve and formalize chemical identification processes (Schymanski and Neumann, 2013; Ulrich et al., 2019; Domingo-Almenara et al., 2018). Quantitative interpretations of NTA data to date have been largely based on relative quantitation, where measured chemical responses are compared across two or more sample groups (e.g. fold-change comparison) for prioritization of chemicals, without attempt to link them directly to fixed concentration values. Seldom has there been absolute quantitation, where direct measurements of compounds are translated to estimated sample (e.g., drinking water, blood) concentrations. Ideally, there would be approaches to convert measurements of feature response in NTA experiments to informative sample concentrations (Fig. 1).
Fig. 1.
Relationship between sample processing for MS analysis (top) and level of chemical information derived (bottom). NTA techniques primarily focus on the identification chemical species in MS analysis (bottom-right), but quantitative modeling approaches allow estimation of the likelihood of given concentrations in prepared sample extracts and parent samples (bottom-left).
The lack of well-defined concentration estimates from NTA measurements is a fundamental challenge in using NTA data to support chemical safety evaluations. As strictly a screening and prioritization tool, NTA has proven useful for directing future research activities. For example, non-targeted screening occurrence is an explicit component of the EU funded NORMAN emerging contaminant prioritization scheme. (NORMAN, xxxx) However, existing risk assessment paradigms ultimately require quantitative estimates of chemical abundance (for example, within a consumer product, in an environmental matrix, or within a living organism) to support defensible decisions by regulatory bodies. In this article, we demonstrate the importance of quantitative predictions of chemical abundance, based on NTA data, for supporting risk-based decisions in context; we further survey existing literature on approaches for deriving quantitative estimates from NTA measurements; and finally, we present a conceptual framework for incorporating quantitative NTA (qNTA) estimates into contemporary risk assessment workflows.
2. The need for quantitative estimates of chemical concentration
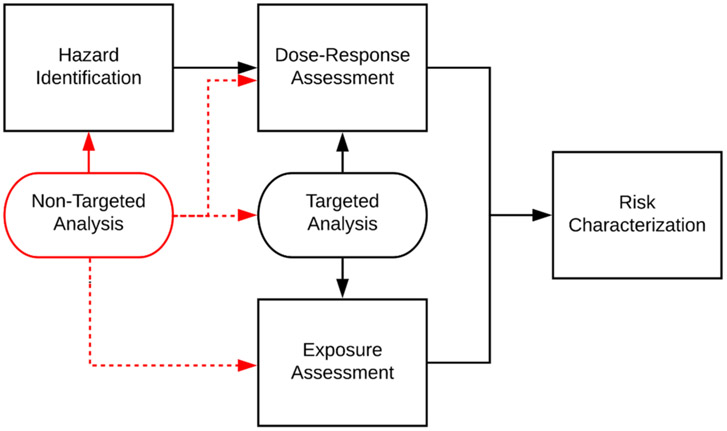
The 1983 National Research Council (NRC) risk assessment paradigm provides a historical framework for evaluating the safety of chemical species using knowledge of the chemicals’ inherent hazards and dose–response relationships, and the anticipated exposures within target populations (Fig. 2).(Council, 1983) This framework was designed around single-chemical evaluations, reflecting the available technology and views at the end of the 20th century. The early 21st century has witnessed a shift in scientific perspective and experimental capabilities, making way for an era of high-throughput (HT) testing and systems-level evaluations. New approach methodologies (NAMs) consisting of clear experimental (e.g., in vitro toxicity testing) and computational (e.g., in silico molecular modeling) advancements have the potential to improve the efficiency of chemical safety assessments; a number of agencies have developed NAM test systems and guidelines (USEPA List of Alternative Test). Nevertheless, the original NRC risk assessment paradigm is no less relevant today than in 1983; chemicals are still evaluated for safety one at a time, using information about the underlying hazard, dose–response relationship, and anticipated exposures. The fundamental advancement is in the way that hazard, dose–response, and exposure data are collected and interpreted. Moving forward, experimental data from NTA studies can enhance all aspects of the risk assessment paradigm, from hazard identification onward.
Fig. 2.
Integration of targeted and non-targeted analysis data (ovals) into the traditional risk assessment paradigm (rectangles). Current non-targeted analysis provides primarily qualitative data for hazard identification (via compound discovery) and occurrence information (presence/absence) for exposure assessment; this influences the development of targeted analysis methods for follow-up quantitative examinations. Further connection of NTA to existing dose–response, exposure assessment, and risk-characterization tools, designed for compatibility with targeted analysis approaches, requires reformulation of NTA data to adhere to workflows designed for quantitative inputs.
2.1. Hazard identification
Hazard identification is considered the initiating step for traditional risk assessment. Chemicals are identified as requiring formal evaluation and then examined for associations with environmental, ecological, or human health-related endpoints. For commercial chemicals, hazard identification is a component of review performed by regulatory bodies (for example, the U.S. EPA under the Toxic Substances Control Act [TSCA] or the Federal Insecticide, Fungicide, and Rodenticide Act [FIFRA], or the European Chemicals Agency under the legislation on the Registration, Evaluation, and Authorization of Chemicals [REACH]). However, only a subset of chemicals with human and ecological exposure routes are identified via legal disclosure mandates. There are, for example, TSCA specific exceptions for low-volume chemicals (CFR 723) and byproducts serving no “separate commercial purpose” (CFR 720). Finally, there are naturally occuring products/extracts which may pose health risks. Thus, there remains a need for hazard identification beyond existing chemical safety legislation focused on narrow lists of chemicals of commerce.
Non-targeted analysis approaches are well suited for hazard identification because compound detection is often sufficient for initial steps. In wide-scale screening investigations, NTA can identify chemicals not included on registration lists, highlight occurrence of emerging contaminants, and support downstream hazard classification (NORMAN). Such screening efforts may seek to identify “known unknown” chemicals with limited occurrence data, or implement more in-depth analyses to identify novel “unknown unknown” substances (Schymanski and Williams, 2017). For example, NTA methods have identified novel halogenated transformation products from water disinfection, (Ding et al., 2013; Tao et al., 2020) minor products of perfluorinated chemical manufacturing, (McCord et al., 2020; McCord and Strynar, 2019) and generic chemical components of unknown/variable (but registered) complex mixtures (Salvito et al., 2020).
In additional to environmental screening, non-targeted methods can support effect-directed analyses (EDA), wherein complex samples/mixtures are first fractionated, and fractions then individually screened for bioactivity (primarily using in vitro assays) (Burgess et al., 2013; Brack et al., 2019). NTA enables follow-up evaluation of risk drivers within active fractions via compound identification. As a recent example, Tian et al. used EDA and examined sequential fractions of a tire rubber extract, using NTA methods, to identify a quinone transformation product that causes lethality in coho salmon (Tian et al., 2021). Other examples include the use of EDA/NTA to identify estrogenic and antiandrogenic compounds in water (Pochiraju et al., 2020; Muschket et al., 2018) and biological matrices (Dusza et al., 2022).
It is noteworthy that, when dealing with complex mixtures, analyte identification is limited to the form of the chemical substance that can be observed by mass spectrometry. This generally does not include counterion salts, adjuvants, dispersants, and other common mixture components of registered chemical substances. Therefore, it is not easy to pair observed analytes to hazard information for parent substances without a means to correlate the two. Overcoming this challenge requires that mappings exist between the chemicals identified in NTA experiments and the substances registered in public databases. The MS-Ready (McEachran et al., 2018) module of the EPA Comptox Dashboard is one such tool that provides these mapping and facilitates correlation between observable MS analytes and chemical substances with known or predicted hazard data.
Overall, compound identification in NTA experiments remains nontrivial, with annotation uncertainty propagating to downstream steps of risk evaluation (as we later discuss) (Pourchet et al., 2020). Nevertheless, the growing body of research indicates that NTA is well suited to aid the identification of emerging stressors that may pose unacceptable health risks to humans and/or other ecological species.
2.2. Dose-response assessment
Having identified a hazard, the risk-assessment paradigm envisioned the determination of a dose–response relationship via in vivo treatment studies, primarily using whole animals. In this context, targeted endpoints would be selected by a risk assessor consistent with the initial hazard identification (e.g. endocrine disruptor potential). These endpoints are measured in model systems with the goal of defining a clear point-of-departure (POD) on a single dose–response curve for that endpoint. With the application of uncertainty factors (related to intra-/inter-species variability from in vivo models, etc.) the POD becomes the basis for a chemical-specific reference dose (RfD) or reference concentration (RfC). This approach is prohibitively resource intensive for the modern chemical landscape, which is replete with chemicals of emerging concern (Council, 1984). An updated vision of hazard identification and dose–response assessment relationship was therefore set forth by the National Research Council (NRC) in their 2007 report, “Toxicity Testing in the 21st Century”.(Council, 2007) Therein, the NRC proposes a vision of toxicity testing centered around biomolecular pathways and mechanistic in vitro assays suitable for high-throughput chemical screening. Such assays are still carried out in a concentration–response format, but potential in vivo relationships are extrapolated from in vitro results using toxicokinetic modeling strategies and uncertainty factors (Kavlock and Dix, 2010; Paul Friedman et al., 2020). The work of Friedman et al. indicates that the use of the in vitro to in vivo extrapolation (IVIVE) introduces a maximum of 10-fold error compared to traditional targeted approaches, and that the estimation error is protective (i.e. overestimation of response) (Paul Friedman et al., 2020).
Non-targeted analysis has limited application in dose–response assessments when consideration is given to only known target analytes. There are, however, multiple scenarios in which NTA can augment both in vitro and in vivo dose–response studies. First, when testing chemicals which elicit adverse response only after metabolic activation, NTA is well suited to rapidly identify both anticipated and unexpected metabolites (Catron et al., 2019; Weitekamp et al., 2019) (note: an increasing number of metabolically competent in vitro test systems (DeGroot et al., 2018; Deisenroth et al., 2020)) now allow tracking of parent depletion and product formation as a function of dose and time, and with respect to the measured assay response). Second, a large portion of registered chemical substances are of Unknown or Variable Composition, Complex Reaction Products and Biological Materials (UVCB Substance) (Agency USEP,xxxx). For these compounds, toxicological screening of mixture fractions can be employed in a concentration–response format alongside NTA approaches to characterize active chemical(s). As described for EDA above, this is bottlenecked by difficulties mapping detections to parent chemical substances, and the accuracy of NTA annotations (Schymanski and Williams, 2017; McEachran et al., 2018). Further, the ability to recover and detect relevant portions of chemical mixtures is dependent on sample preparation and the applicable analysis domain (e. g. LC-MS, GC–MS, solvent solubility, etc.). Recent work by Lowe et al. has attempted to model the applicability of different analytical techniques to chemical subsets based on their structure and properties, (Lowe et al., 2021) but this is mostly useful for improving certainty in annotation, and defining the bias of your measurements. In simple cases where it is possible to assume good method applicability, quantitative NTA methods can theoretically inform dose–response curve measurements, experimental PODs, and (given appropriate uncertainty factors) eventual RfDs/RfCs. These thresholds are the critical point of comparison for quantitative measurements in exposure assessment.
2.3. Exposure assessment
Traditional exposure assessments couple quantitative measurements of chemicals in environmental media (e.g., drinking water, consumer products, blood) with activity (e.g., location, product use) and “exposure factor” information, (USEPA,xxxx) such as consumption rates, to yield final exposure estimates; these can be interpreted with respect to a RfD or RfC to characterize risk and establish “no-effect” concentrations for specific media. The task of exposure estimation has recently broadened in accordance with the rapid expansion of the known chemical universe. The NRC recommended emphasis on high-throughput exposure estimation strategies in their 2012 report “Exposure Science in the 21st Century: A Vision and A Strategy.”(Council, 2012) In accordance with NRC recommendations, efforts from organizations such as the NORMAN network have begun to incorporate non-targeted screening approaches to hazard prioritization and EPA’s ExpoCast project (Wambaugh et al., 2019) has developed HT exposure prediction models and frameworks for harmonizing exposure estimates across disparate existing models (Rager et al., 2016; Sobus et al., 2018; Wambaugh et al., 2013; Wambaugh et al., 2014; Ring et al., 2017; Ring et al., 2019). To date, exposure estimates have been generated for hundreds-of-thousands of known chemicals; for example the work of Ring et al. used machine learning and thirteen previously developed exposure models, built on the limited CDC NHANES exposure dataset, to calculate consensus predictions for the median intake rate of nearly 500,000 chemicals based on their chemical structure (Ring et al., 2019; Sobus et al., 2015; Control, 2006). Prediction uncertainty can be large for some chemicals due to a lack of available chemical measurement data for model parameterization, evaluation, and calibration. The study of Ring et al. exhibited credible intake intervals of up to eight orders of magnitude (Ring et al., 2019). A considerable increase in chemical monitoring data is therefore needed to reduce uncertainties in HT exposure predictions. Most existing environmental and biological monitoring studies focus on targeted chemical panels, yielding measurement data for only a few well-known compounds, (Egeghy et al., 2012; Sobus et al., 2015) with major data gaps for metabolites, degradants, and other unknowns. NTA studies can help fill these gaps, providing broad chemical measurements that inform presence and putative exposure sources, routes, and pathways and allowing for prioritization of chemicals of interest (Sobus et al., 2018).
2.4. Risk characterization
Risk characterization requires the full synthesis of available hazard, dose–response, and exposure information, and computational and high-throughput techniques, including NTA, are anticipated to be the backbone of future efforts in the field (Thomas et al., 2019; Academies and of Sciences, Medicine, 2017). Efficient quantitative data streams can predict the likelihood and severity of potential health risks consistent with an exposure (Academies and of Sciences, E., Medicine, , 2017). For example, quantitative structure–activity relationship (QSAR) models can be used to predict environmental fate, (Mansouri et al., 2018) determine bioactive exposure thresholds, (Pearce et al., 2017) and estimate receptor activity (Mansouri et al., 2016). These types of models are built from reference data for thousands of chemical compounds, but accept simple chemical structures as input. The constructed models leverage existing chemical data to predict outputs for novel chemicals by structural comparisons; limited application of these approaches is beginning to be seen in regulatory decision making. For example, the EPA endocrine disruptor screening program allows high-throughput computational approaches for tier 1 evaluation (USEPA Use of High Throughput).
Implementation of NTA to support high-throughput risk assessment has been seen in a limited capacity, with most applications identifying structures of emerging contaminants in need of characterization. Specifically, results of NTA experiments have been used to prioritize chemicals requiring further study (via targeted analysis) based on perceived exposure and hazard potential (NORMAN; Rager et al., 2016). The toxicological prioritization index (ToxPi) (Reif et al., 2010) is one framework used to prioritize chemical detections based on the potential for health risks. While originally conceived to visually examine chemicals using targeted toxicity data, the ToxPi framework has been extended to consider NTA experimental data (i.e., the detection frequencies and measured intensities of identified chemicals) as quantitative surrogates of exposure (Newton et al., 2018; Rager et al., 2016; Sobus et al., 2018). These efforts have proven suitable for provisional chemical evaluation and ranking. Yet, a clear delineation remains between the type of data produced in NTA studies and the type needed to support eventual quantitative risk characterizations. Simply put, targeted analytical methods remain the source of quantitative experimental data used in formal risk calculations. Since targeted quantitative methods: 1) require the procurement/synthesis of standards; 2) are time/resource intensive; and 3) are not readily available for the majority of newly discovered compounds, scientific advances are needed that will allow quantitative NTA (qNTA) data to be directly utilized for rapid risk characterization. Achieving this end will require significant advances on current qNTA techniques.
3. Current approaches for quantitative predictions in non-targeted analysis
Targeted quantitative mass spectrometry is a gold-standard analytical technique for many chemicals, allowing absolute quantification and high precision/accuracy when incorporating external and/or internal standardization procedures. With sufficient resources, quantification of chemicals can be readily explored using extensive targeted analyses in tandem with, or as a follow up to NTA. For example, the work of Moschet et al. screened nearly every water soluble pesticide in Switzerland, with follow-up targeted quantitation, (Moschet et al., 2014) and a study by Gago-Ferrero et al. screened for thousands of emerging contaminants with similar follow-up (Gago-Ferrero et al., 2015). These exhaustive studies represent the best case scenario for combined analysis, with a well characterized chemical panel, available standards, and significant resources to develop and apply quantitative methods for hundreds if not thousands of chemicals detected in an initial NTA screen. In general, this level of cost and time commitment to broad quantitative screening is infeasible. Further, screening experiments are most attractive for conditions where other methods do not currently exist, and for under-studied and/or novel compounds with limited chemical information where targeted analysis is not only difficult, but impossible. As a result, targeted quantitative approaches for chemicals of emerging concern require supplementation with NTA experiments.
While there is a strong impetus to carry out NTA screening, it lacks structured sets of reference materials, surrogate chemicals, and matched internal standards common in targeted assays, and therefore cannot match the quantitative accuracy or precision of targeted methods. At present, there is no universally accepted method for quantifying chemicals detected/identified in an NTA experiment. There are, however, several approaches for using NTA data in a quantitative context. The first approach is the common process of “relative quantitation” and involves the comparison of measured intensities or normalized response values rather than concentration values. This quantitative treatment does not yield concentration estimates but has been used to compare samples for chemical prioritization and follow-up work. True quantitative approaches yield concentration estimates either using calibration (mimicking traditional quantitative methods) or direct response modeling. Each quantitative approach has intrinsic levels of uncertainty due to the numerous factors that influence chemical response in mass spectrometry. The sections below describe important sources of quantitative uncertainties and discuss the extent to which they’ve been considered in previous qNTA applications.
3.1. Relative quantitation by compound response comparisons
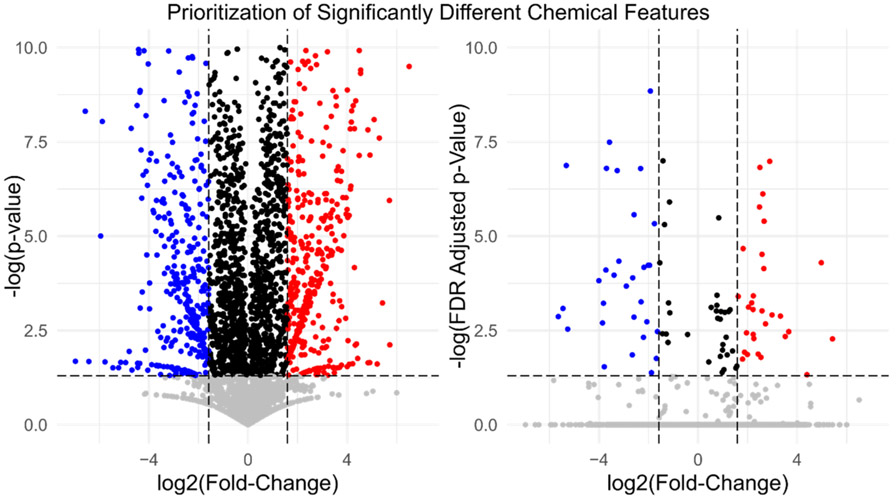
Relative quantification is the default method of comparison in NTA experiments. While not an absolute concentration measure, instrument response is assumed to be proportional to concentration, allowing comparison of analyte response across samples. The naïve assumption of linear response relationships and minimal matrix effects are seldom accurate; data normalization approaches are therefore often required to help correct for instrument variability and non-linearity (Yi et al., 2016; Di Guida et al., 2016). Simple relative comparisons are nevertheless easy and useful; an investigator could examine the fold-change in observed response between an exposed and unexposed population (i.e., Responseexposed/Responseunexposed) to identify statistically significant chemicals for prioritization (Fig. 3). Indeed, binary comparisons of two sample sets (e.g., case vs. control, upstream vs. downstream, treated vs. untreated) have been used in environmental analyses to identify geographic origins of emerging contaminants(Nakayama et al., 2010; Lindstrom et al., 2011; Strynar et al., 2015) and to associate disinfection byproducts with specific treatment methods (Tang et al., 2016; Liberatore et al., 2020). With thousands of measurements conducted in a typical NTA experiment, traditional significance thresholds of statistical comparison would yield an unacceptably high false-positive rate. Controlling the so-called false discovery rate (FDR) can involve adjusting p-values for multiple hypothesis testing, as proposed by Benjamini-Hochberg (Benjamini and Hochberg, 1995) (Fig. 3), or using other significance criteria such as Storey q-value (Storey, 2003; Storey, 2002) or local FDRs (Chong et al., 2015). Comparisons and statistical corrections are commonplace and enabled by data processing software (Xia et al., 2009; Misra, 2021).
Fig. 3.
Prioritizing chemical features by statistically significant fold change using volcano plots of individual chemical features compared between two study groups (data reprocessed from Rager et al.56). The fold-change (FC) for each feature intensity (where FC = MeanIntensityGroup1 / MeanIntensityGroup2) is plotted versus p-values for the unadjusted (left) and Benjamini–Hochberg corrected (right) group differences. Dashed lines show designated p-value (horizontal lines; p = 0.05) and FC (vertical lines; FC = 3) thresholds. Red chemical features are elevated (above designated thresholds) in study group 1 and blue chemical features are elevated in study group 2.
Time-based comparisons (e.g., time 1 vs. time 2) have further helped identify environmental metabolites in wastewater, (Gago-Ferrero et al., 2015) assess real-time pollutant levels in the surface water, (Hollender et al., 2017) and characterize emerging contaminant levels in human blood (Plassmann et al., 2016; Plassmann et al., 2018). In these examples, observed changes in the relative abundance of detected chemicals was the basis for follow-up investigation, such as contacting point sources to confirm chemical releases (Hollender et al., 2017) or performing targeted analyses to confirm chemical identities and estimate sample concentrations (Plassmann et al., 2018).
There are clear limitations to the direct use of measured HRMS intensities and response factors for relative quantitation. First, numerous factors (e.g., instrument cleanliness and tuning, matrix effects) influence HRMS response measures for any given chemical, and analyte response can be non-linear over large concentration ranges. The most straightforward normalization approach is to correct all measured intensities using reference compounds or internal standards (i.e. applying a scalar correction factor to measured intensities from a sample so that reference responses are constant across samples); (Sobus et al., 2018) although additional correction may be necessary for concentrations that fall outside the linear response range (Yu et al., 2020). Measurements can also be compared relative to a reference sample or pooled QC sample to ensure inter comparability between analytical batches and methods (Liu et al., 2020; Go et al., 2015). This has the further benefit that relative measurements can be converted to absolute measurements post hoc if concentrations are determined in the reference sample (Liu et al., 2020). Comparative measurements can thus reduce the number of necessary targeted measurements for quantification in addition to allowing retroactive concentration determination. However, without complementary quantitative measurements, normalization can only improve intra-chemical comparisons between samples, but not cross-chemical comparisons. Furthermore, they do not yield specific concentration estimates in prepared sample extracts or the original sample medium (Fig. 1). For this reason, relative quantitation methods are viable for prioritizing future work, and for retaining valuable sample information for future interrogation, but alone cannot produce the data needed for other components of risk characterization. Ultimately, methods for absolute determination of concentration are needed.
3.2. Quantification by surrogate standard
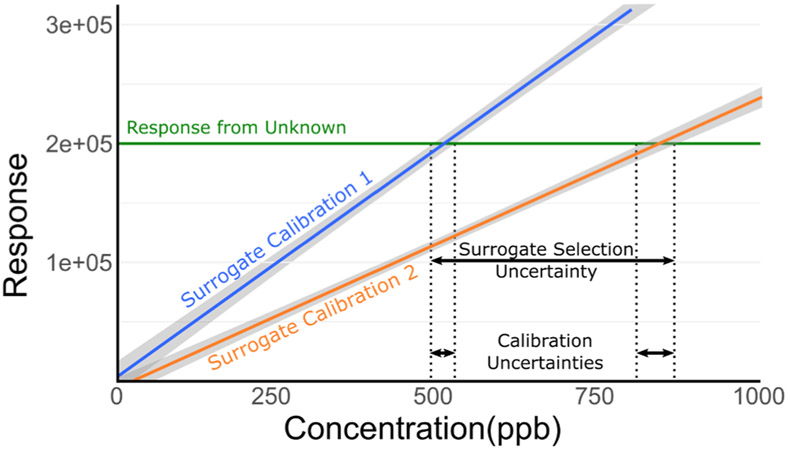
Without a direct reference standard, a straightforward alternative for compound quantitation involves using a calibration curve (i.e. sample concentration vs. instrument response/internal standard correct response) from a chemical surrogate (Fig. 4). The goal in surrogate selection would be to identify a known chemical that is likely to mimic the analytical behaviors (e.g., chromatographic separation, ionization, and HRMS detection) of the compound in question. A single surrogate can be selected in a variety of ways. For example, a parent chemical may be chosen as a surrogate for a putative metabolite, or a well-known compound from a specific chemical class (e.g., perfluorinated carboxylic acid) may act as a surrogate for an emerging chemical in that same class.
Fig. 4.
Estimation of the concentration of an unknown compound from a measured intensity (horizontal green line) and multiple surrogate calibration curves (blue and orange lines) with 95% prediction bands (grey envelopes). In this theoretical example, the uncertainty incurred from surrogate selection (i.e., calibration curve 1 vs. 2) far outweighs the calibration estimate uncertainty associated with either individual curve.
Significant challenges can exist when attempting to identify chemical surrogates for quantification. Specifically, a wide range of observed experimental inaccuracies indicate that the differences in estimated concentration when using a “closely matched” surrogate vs. a poorly matched surrogate can far outweigh the intrinsic error of any individual calibration curve (Fig. 4). Experimental examples using calibration curves of parent chemicals to quantify related metabolites have yielded inaccuracies of between four-fold for simple methyl- and hydroxylmetabolites (Dahal et al., 2011) and up to seventy-fold for more complex metabolic products (Hatsis et al., 2017). Using calibration curves of structurally similar surrogates, inaccuracies have been reported ranging from three-fold for small molecules in food, (Pieke et al., 2017) to an order-of-magnitude or more for perfluorinated contaminants in surface water, (McCord et al., 2018) and up to several hundred-fold for drugs in biological samples (Liigand et al., 2018). The measured inaccuracies derive from multiple sources, both the intrinsic variance of analytical measurement (i.e. curve preparation, instrument variability, internal standard addition etc.) and the inaccuracy introduced by applying calibration models between chemicals (i.e. deviations from predicted chemical similarity, inaccuracy of regression models etc.).
It is desirable for a closely matched surrogate to account for the intrinsic effects of the ionization, solvent composition, and matrix effects on analyte response. To date, the most accurate results from surrogate applications have been observed when considering chemicals within a defined class and with close retention times, (Pieke et al., 2017; Cech et al., 2001) or when extrapolating across bracketed homologous compounds (Kamga et al., 2014). In GC–MS experiments, observed quantitative error has been much smaller than in LC-MS experiments. In one study by Banerjee et al., an average calibration curve from a panel of surrogate compounds was able to estimate concentrations for several hundred pesticides with an average error of 10% from their true values, demonstrating significant similarity in response across the chemical space (Banerjee et al., 2012). Other work by Bu et al. observed maximum errors of only two-fold for select classes of organic contaminants when using an average curve (Bu et al., 2014). For the selected organic classes, the comparatively simpler chromatographic and ionization source interactions likely contribute to the more accurate quantitative predictions.
While the desire for closely matched surrogate is well known, methods for quantitatively describing surrogate fit are limited. A study by Aalizadeh (Aalizadeh et al., 2021) did use common substructural elements (Cao et al., 2008; Bajusz et al., 2015) to quantitatively define surrogate similarity and domain of applicability for surrogate usage with great success, as well as a mechanism for adjusting to future instrumental conditions, but only provided post hoc accuracy estimates of their fit, rather than a model for predicting estimate uncertainty. In fact, all the remaining referenced works provided only retrospective accuracy determination for tested compounds and lack a framework to bound future quantitative estimates or select appropriate surrogates beyond intuition. Crucially, the future utility of quantitative chemical estimates is intrinsically linked to the bounded accuracy of that estimate. Thus, there is a need to develop predictive models of uncertainty to accompany methods for quantitative estimation.
3.3. Response modeling from chemical structure
Since the process of selecting appropriate surrogate calibrators for small molecules contains fundamental challenges, some researchers have opted to model the ionization response of compounds based on their molecular structure and chemical properties. This approach bases quantitative predictions on the physicochemical rationale for observed responses using models containing multiple factors that can influence compound response.
Mass spectrometry relies on the ionization of chemicals for analysis and is therefore inherently confounded by signal suppression due to matrix effects, (Taylor, 2005) as well as instrument-specific parameters that can influence the response factor of any given analyte. Selection of ionization type (e.g. electrospray [ESI], chemical ionization [CI], electron ionization [EI]) affects the end results due to interactions with physicochemical properties; chemical parameters such as hydrophobicity (logP),(Null et al., 2003; Henriksen et al., 2005; Cech and Enke, 2000) polar-/non-polar surface area, (Cech and Enke, 2000; Walker et al., 2011; Golubović et al., 2016) molecular weight, (Mehta et al., 2016; Cox et al., 2003) and both liquid and gas-phase acidity(Sunner et al., 1988; Ehrmann et al., 2008; Richter and Schwarz, 1978) are known to contribute to ionization potential, along with structural parameters like carbon number and ionization cross section (Bergmann et al., 2018; Kim et al., 2014; Szulejko et al., 2013). Solvent effects from buffer pH, concentration of organic solvent, (Cech et al., 2001; Liigand et al., 2014) temperature, (Page et al., 2007) and even flow rate (Smith et al., 2004) can yield variable responses for individual chemicals from experiment-to-experiment. We direct interested readers to a recent review article by Kruve (Kruve, 2020) for an in-depth discussion of the physicochemical and instrumental influences on ionization behaviors.
The wide range of properties that contribute to different chemical responses in MS experiments is daunting. Indeed, sensitivity differences of several orders-of-magnitude have been observed across even fairly similar compounds (Liigand et al., 2018; Kruve, 2020). Yet, it’s encouraging that robust model-based quantitative approaches have seen widespread application in related fields (e.g., proteomics) that utilize comparable MS platforms (Bantscheff et al., 2007; Shuford et al., 2017). Models that predict compound response can incorporate disparate chemical and instrumental factors and, most importantly from a quantitative perspective, provide estimated errors that can bound the accuracy of any individual concentration estimate. Further, cross-platform compatibility can be encouraged by transferring predicted response factors between platforms using a panel of reference compounds (Panagopoulos Abrahamsson et al., 2020). A key consideration for ionization modeling approaches is the reliance chemical structure information. Any quantitative prediction model that relies on chemical structure is therefore limited in the domain of applicability by the chemicals used to construct it. Furthermore, quantitative predictions can only be as accurate as the preceding NTA identification. Assigning risk to an unknown based on uncertain identification can be fraught, but for scenarios where multiple potential structures exist, the hazard classification can be estimated for multiple structures based on worst-case scenarios of classification, or weighted based on annotation likelihood, if such values are available.
Multiple models have been constructed that use physicochemical properties or chemical descriptors to predict chemical ionization efficiency often normalized to a reference standard (Liigand et al., 2018; Leito et al., 2008; Oss et al., 2010; Kruve et al., 2014; Chalcraft et al., 2009). Some challenges have been reported with predicting chemical ionization behavior related to specific ionization sources. Work by Chalcraft et al. reported an average ratio between measured and predicted response of 1.08 ± 0.75 for chemicals that ionize via positive mode ESI (Chalcraft et al., 2009). But the same study could not establish a model for compounds that exhibit multiple ionization states, either multiple charges or adducts, or that ionize in negative mode ESI; the authors speculated that multiple models would be needed for different chemical subclasses. Another study by Leito et al. developed a regression model based on ten compounds and reported an RSD consistency of 0.16 log units (~1.5 fold),(Leito et al., 2008) but later studies using larger chemical datasets have since yielded higher prediction errors due to the larger complexity of the chemical space modeled (Oss et al., 2010; Kruve et al., 2014). Other modeling efforts have yielded errors of 26% for sugar metabolites,(Ghosh and Jones, 2015) up to 13% for lipids,(Cífková et al., 2012) 37% for steriods,(Alymatiri et al., 2015) and as little as 1% for ion-paired oligonucleotides.(Basiri et al., 2017) Built-for-purpose models therefore seem to exhibit lower prediction error than more general models due to the narrower chemical space. However, an exhaustive recent modeling study by Liigand et al. considered both positive and negative ionization mode, as well as a hundred LC conditions and several hundred chemicals in multiple chemical classes to train a random forest, machine learning model to obtain average prediction errors of approximately 2-fold (Liigand et al., 2020). This implies that a general ionization response model for an MS source is hindered by only lack of appropriate dataset(s) to capture the wide range of chemical space, rather than an incompatibility of the approach with quantitative prediction. Importantly, we note these prediction errors are considerably smaller than those observed with computational exposure modeling, and on par with those experienced by in vitro extrapolation efforts. Therefore, continued collection of ion efficiency data across large and diverse chemicals sets represents a path forward for qNTA.
4. Sample concentrations from quantitative predictions
Previously described approaches (section 3.2) for surrogate calibration can naturally incorporate experimental recovery by utilizing matrix-matched extracted calibration curves. This relies on the assumption that chemical surrogates accurately mimic preparation losses of the chemicals of interest. With limited exceptions that used matrix-matched surrogate calibration, (Aalizadeh et al., 2021; Alygizakis et al., 2021) the quantitative research efforts described above have focused on estimating in-solution concentrations for compounds in prepared standards or extracts. For risk characterization, quantitative estimates must extend back to the original sample matrix, rather than relying on extract concentrations alone (Fig. 1). Most analyses of environmental or biological matrices require some amount of sample preparation (including extraction and/or concentration steps) prior to MS analysis. While NTA experiments strive to minimize bias, sample preparation procedures can strongly affect recoveries for individual compounds. Thus, the estimated concentration in a prepared extract does not directly translate to the environmental concentration. A naive baseline assumption is that the preparation procedure is well suited for the analyte(s) of interest, and that all chemicals are transferred into the final extract (i.e., recovery = 100%). In practice, adjustment is needed to estimate concentrations in both targeted and qNTA methods.
Solid phase extraction (SPE) is a common preparatory technique for concentration of dilute environmental samples, and there are hundreds of SPE methods, such as the dispersive SPE QuEChERS approach (Perestrelo et al., 2019). Extracts can also be prepared using direct solvent extraction, for example multi-solvent extraction of soil(Fisher et al., 1997) or small molecule solvent extractions from biological samples (Vuckovic et al., 2020). Recovery of chemicals of interest is dependent on the same chemicals factors that influence MS response (e.g. pH, hydrophobicity, pKa) along with the choice of extraction solvents and SPE phases. Comparing common preparation techniques reveals that optimized methods can achieve near 100% recovery, but naïve applications of the same methods can have yields as low as 10%.(Chambers et al., 2007) This sample preparation variability adds another layer of uncertainty to qNTA predictions. Most NTA studies that have calculated recoveries did so for benchmarking purposes (Parry and Young, 2016; Crimmins et al., 2014; McCord and Strynar, 2019) with only limited examples of correction for predicted concentrations. A notable exception is the previously mentioned work of Aalizadeh et al, who applied chemical similarity measures to predict matrix effects and analyte recovery for chemicals based on surrogates (Aalizadeh et al., 2021). Importantly, robust recovery estimates are only known for chemicals with available standards. Therefore, there is significant uncertainty in chemical recovery for unknowns which compounds with other sources of error when determining quantitative estimates (note that recovery has a theoretical upper bound of 100%, but no definable lower bound [e. g., 10%, 1%, 0.1%...]). It is possible to account for the interaction of chemical structure with matrix, sample preparation, and analysis conditions using recovery surrogates. However, recovery surrogates from a lack of quantitative measures for surrogate suitability, just as was discussed for surrogate calibration (Aalizadeh et al., 2021; Alygizakis et al., 2021). Surrogate and purely computational models of recovery estimation will require expansive training sets to predict the recoveries for arbitrary chemicals and conditions. The development of these models is a final barrier to generating true concentrations needed to carry out a risk characterization from qNTA experiments.
5. Risk characterization as part of an NTA workflow
There are three anticipated scenarios when identifying chemical substances in qNTA studies, differentiated by the amount of existing guidance information available. In the simplest scenario, direct comparison can be made between estimated concentrations and an existing guidance or reference level, such as a drinking water maximum contaminant level. While straightforward, this scenario is seldom expected given: 1) the comparative paucity of chemicals with source-specific regulatory guidance/reference levels; and 2) the primary focus of NTA studies being data-poor and/or novel compounds.
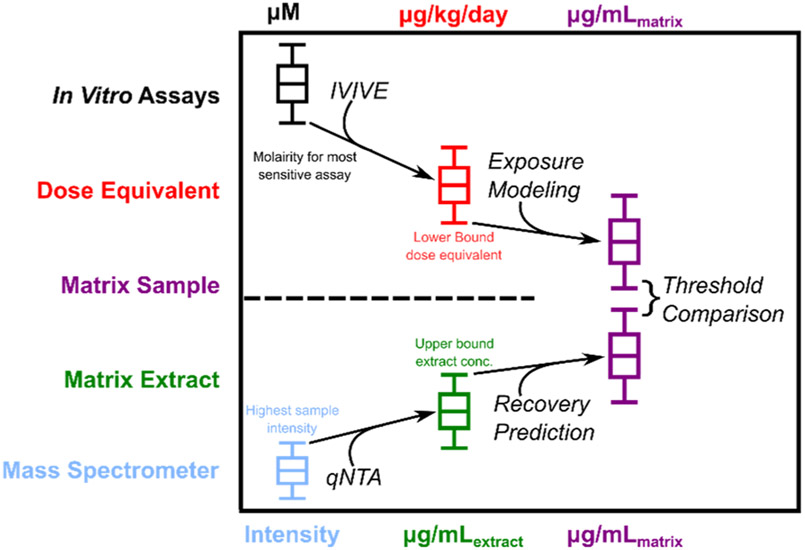
In a second scenario (Fig. 5), the identified chemical would not have fixed source-specific guidance/reference levels, but instead have only provisional bioactivity or toxicity data and provisional risk thresholds. EPA’s Toxicity Forecaster (ToxCast) project, for example, has generated in vitro bioactivity data for thousands of chemicals across hundreds of assays (Thomas et al., 2018). Using this readily available data, a lower-bound bioactivity threshold (e.g., 5th percentile) could be selected from a distribution of all existing potency measures (e.g., AC50 values, which are concentrations associated with half-maximal assay activity) (Richard et al., 2016) to represent a surrogate POD in the absence of traditional in vivo dose–response data (Paul Friedman et al., 2020). Linking these measures to qNTA estimates would require several modeling efforts. First, toxicokinetic modeling (for example, using EPA’s HTTK modeling platform) (Pearce et al., 2017) would be needed for in vitro to in vivo extrapolation, yielding a plausible distribution of dose-equivalent values that correspond to the lower-bound potency measure. This would answer the question of what dose levels (e.g., μg/kg/day) are expected to produce steady-state blood levels that are consistent with in vitro assay concentrations. Next, source-specific exposure modeling (e.g. EPA’s SHEDS-HT modeling platform (Isaacs et al., 2014), or numerous QSAR models from the NORMAN ecotox database) would be needed to determine what matrix concentrations (e.g., ng/L of drinking water) would be protective given the lower bound dose-equivalent value. Finally, the acceptable matrix concentrations and qNTA estimated concentrations could be compared to enable provisional risk evaluation for particular single source or aggregate exposure (s).
Fig. 5.
Toxicity/bioactivity assay measures (e.g. ToxCast AC50, top left) can be scaffolded to lower bound acceptable concentrations via in vitro to in vivo extrapolation (IVIVE) and exposure modeling. Chemical instrument response from NTA experiments (bottom left) can likewise be converted to protective upper-bound estimates via quantitative NTA modeling and estimation of sample recovery. Risk assessment decision making can then be based on the comparison between upper-bound estimated concentration and a lower-bound acceptable concentration. Even sizable estimation errors may be acceptable for risk-based prioritization if the degree of separation is large.
The third scenario pertains to identified chemicals for which there is no existing bioactivity or toxicity data – this is likely the most common scenario for chemicals identified in NTA experiments. Here, there are no existing hazard-based measures and, instead, researchers must rely on chemical read-across techniques (Cohen Hubal et al., 2010; Rovida et al., 2020) or in silico toxicity predictions (US-EPA, User’s Guide for TEST (version 4.2)(Toxicity Estimation Software Tool): A Program to Estimate Toxicity from Molecular Structure. Washington (USA): US-EPA, 2016; Raies and Bajic, 2016). Given hazard-based metrics derived from read-across and predictive models, the risk-based interpretation of qNTA data can use computational results as provisional estimates for exposure and/or toxicity. It is expected that most estimated upper-bound concentrations based on NTA data will be smaller than risk-limiting lower-bound concentrations based on bioactivity/toxicity data. This would suggest that environmental levels of individual chemicals are too low to elicit an adverse biological response. If this circumstance holds true, many observed compounds could be considered “low-risk” despite potentially large uncertainties in concentration estimates. For example, the credible intake ranges of Ring et al. spanned up to eight orders of magnitude, but of ~ 500,000 chemicals modeled only ~ 2,000 were expected to have exposures exceeding 0.1 mg/kg/day (Ring et al., 2019). Quantitative NTA estimates discussed in Section 3 broadly possess errors of only one or two orders of magnitude, which is similar to the uncertainty of the toxicokinetic modeling. Substantial overlap between concentration estimates and effects-based thresholds would indicate a need for more robust predictions of the anticipated exposure levels and/or toxicity thresholds due to increased risk potential. In the short term, it will still be necessary to acquire or synthesize chemical standards and to apply targeted analytical methods for chemical substances with a high degree of risk. However, this focused use of resources on higher risk chemicals is more feasible than attempting to perform rigorous quantitation on all compounds detected in an NTA experiment.
6. The future of quantitative NTA
Our review has highlighted the motivating need for higher-throughput chemical safety assessments in the modern chemical landscape and explored the existing data/methodology gaps between current NTA implementation and its incorporation into a rapid risk characterization workflow as a quantitative data source. Moving forward, we can foresee qNTA experiments that contribute to every step of the risk assessment process, providing chemical identities to support components of existing frameworks (e.g., hazard identification) and sample concentration estimates to facilitate next generation dose, exposure, and risk assessments. The goal is to enable seamless linkage between outputs of qNTA experiments and computational toxicology studies that will yield rapid risk categorizations (e.g., high, medium, low) for emerging chemicals.
Current NTA approaches rely on data processing pipelines to yield large chemical feature lists and putative structures. Future developments should retain and improve the automated nature of data analysis to allow high-throughput processing while improving the accuracy of compound identifications and subsequent characterization. Further, the adoption of one or more general qNTA techniques should provide for bounded estimates of concentrations, with continuous improvements to prediction accuracy and domain of applicability. Existing methods function well for specific chemical sub-classes but may have limited utility outside of their defined chemical domain. Ideal quantitative methods should therefore strive to extend to any arbitrary chemical structure or possess the ability to flag chemicals (or chemicals sub-classes) that are likely to be not well estimated. Finally, while there is clearly a desire to maximize the accuracy of any predictive model, it must be universally recognized that quantitative estimates will possess intrinsic error and future methods must bound this error to facilitate protective decision making.
Integration of qNTA methods with other HT chemical evaluation tools offers a means to radically advance the utility of NTA data. In addition to enabling detection of chemicals, qNTA will allow the immediate prioritization of emerging contaminants based on anticipated risk; importantly, this utility extends to unexamined chemicals not covered by traditional chemical registration efforts. Quantitative NTA will further provide a pathway for efficient hazard screening in rapid-response scenarios, such as chemical spills or natural disasters, where chemical exposures may be heterogenous in scale and complexity. Finally, qNTA will provide a means to rapidly communicate chemical-related risks to potentially affected populations in all manner of environmental and biological monitoring studies. Provided that existing challenges can be addressed, it is likely that qNTA will ssupport rapid and defensible risk-based decisions for hundreds to thousands of understudied chemical stressors.
Acknowledgements
This document has been reviewed by the U.S. Environmental Protection Agency, Office of Research and Development, and approved for publication. The views expressed in this article are those of the authors and do not necessarily represent the views or policies of the U.S. Environmental Protection Agency. Special thanks to Barbara Wetmore, Brett Blackwell, and Rusty Thomas for comments, suggestions, and technical review on the manuscript text.
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- CAS Content: Substances. https://www.cas.org/about/cas-content. [Google Scholar]
- Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, Li Q, Shoemaker BA, Thiessen PA, Yu B, Zaslavsky L, Zhang J, Bolton EE, 2020. PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res 49 (D1), D1388–D1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachran AD, Sobus JR, Williams AJ, 2017. Identifying known unknowns using the US EPA’s CompTox Chemistry Dashboard. Anal. Bioanal. Chem 409 (7), 1729–1735. [DOI] [PubMed] [Google Scholar]
- USEPA Substance Registry Services (SRS). https://sor.epa.gov/sor_internet/registry/substreg/home/overview/home.do. [Google Scholar]
- Brack W, Dulio V, Slobodnik J, 2012. The NORMAN Network and its activities on emerging environmental substances with a focus on effect-directed analysis of complex environmental contamination. Environmental Sciences Europe 24, (1), 29. [Google Scholar]
- Naidu R, Arias Espana VA, Liu Y, Jit J, 2016. Emerging contaminants in the environment: Risk-based analysis for better management. Chemosphere 154, 350–357. [DOI] [PubMed] [Google Scholar]
- Weinberg N, Nelson D, Sellers K, Byrd J, 2019. Insights from TSCA Reform: a Case for Identifying New Emerging Contaminants. Current Pollution Reports 5 (4), 215–227. [Google Scholar]
- Egeghy PP, Judson R, Gangwal S, Mosher S, Smith D, Vail J, Cohen Hubal EA, 2012. The exposure data landscape for manufactured chemicals. Sci. Total Environ 414, 159–166. [DOI] [PubMed] [Google Scholar]
- Rappaport SM, Smith MT, 2010. Epidemiology. Environment and disease risks. Science 330 (6003), 460–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild CP, 2005. Complementing the genome with an “exposome”: the outstanding challenge of environmental exposure measurement in molecular epidemiology. In AACR 14 (8), 1847–1850. [DOI] [PubMed] [Google Scholar]
- German JB, Hammock BD, Watkins SM, 2005. Metabolomics: building on a century of biochemistry to guide human health. Metabolomics 1 (1), 3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudeau P, 2020. NMR-based metabolomics and fluxomics: developments and future prospects. Analyst 145 (7), 2457–2472. [DOI] [PubMed] [Google Scholar]
- López-López Á, López-Gonzálvez Á, Barker-Tejeda TC, Barbas C, 2018. A review of validated biomarkers obtained through metabolomics. Expert Rev. Molecular Diagnostics 18 (6), 557–575. [DOI] [PubMed] [Google Scholar]
- Trivedi DK, Hollywood KA, Goodacre R, 2017. Metabolomics for the masses: The future of metabolomics in a personalized world. New Horiz. Transl. Med 3 (6), 294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourchet M, Debrauwer L, Klanova J, Price EJ, Covaci A, Caballero-Casero N, Oberacher H, Lamoree M, Damont A, Fenaille F, Vlaanderen J, Meijer J, Krauss M, Sarigiannis D, Barouki R, Le Bizec B, Antignac J-P, 2020. Suspect and non-targeted screening of chemicals of emerging concern for human biomonitoring, environmental health studies and support to risk assessment: From promises to challenges and harmonisation issues. Environ. Int 139, 105545. 10.1016/j.envint.2020.105545. [DOI] [PubMed] [Google Scholar]
- Ruff M, Mueller MS, Loos M, Singer HP, 2015. Quantitative target and systematic non-target analysis of polar organic micro-pollutants along the river Rhine using high-resolution mass-spectrometry–identification of unknown sources and compounds. Water Res 87, 145–154. [DOI] [PubMed] [Google Scholar]
- Brack W, Hollender J, de Alda ML, Müller C, Schulze T, Schymanski E, Slobodnik J, Krauss M, 2019. High-resolution mass spectrometry to complement monitoring and track emerging chemicals and pollution trends in European water resources. Environ. Sci. Europe 31 (1), 62. [Google Scholar]
- Focazio MJ, Kolpin DW, Barnes KK, Furlong ET, Meyer MT, Zaugg SD, Barber LB, Thurman ME, 2008. A national reconnaissance for pharmaceuticals and other organic wastewater contaminants in the United States — II) Untreated drinking water sources. Sci. Total Environ 402 (2-3), 201–216. [DOI] [PubMed] [Google Scholar]
- McMahen RL, Strynar MJ, McMillan L, DeRose E, Lindstrom AB, 2016. Comparison of fipronil sources in North Carolina surface water and identification of a novel fipronil transformation product in recycled wastewater. Sci. Total Environ 569-570, 880–887. [DOI] [PubMed] [Google Scholar]
- Bletsou AA, Jeon J, Hollender J, Archontaki E, Thomaidis NS, 2015. Targeted and non-targeted liquid chromatography-mass spectrometric workflows for identification of transformation products of emerging pollutants in the aquatic environment. TrAC, Trends Anal. Chem 66, 32–44. [Google Scholar]
- Newton SR, McMahen RL, Sobus JR, Mansouri K, Williams AJ, McEachran AD, Strynar MJ, 2018. Suspect screening and non-targeted analysis of drinking water using point-of-use filters. Environ. Pollut 234, 297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCord JP, Strynar MJ, Washington JW, Bergman EL, Goodrow SM, 2020. Emerging Chlorinated Polyfluorinated Polyether Compounds Impacting the Waters of Southwestern New Jersey Identified by Use of Nontargeted Analysis. Environ. Sci. Technol. Lett 7 (12), 903–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schymanski EL, Neumann S, 2013. The critical assessment of small molecule identification (CASMI): challenges and solutions. Metabolites 3 (3), 517–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich EM, Sobus JR, Grulke CM, Richard AM, Newton SR, Strynar MJ, Mansouri K, Williams AJ, 2019. EPA’s non-targeted analysis collaborative trial (ENTACT): genesis, design, and initial findings. Anal. Bioanal. Chem 411 (4), 853–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo-Almenara X, Montenegro-Burke JR, Benton HP, Siuzdak G, 2018. Annotation: A Computational Solution for Streamlining Metabolomics Analysis. Anal. Chem 90 (1), 480–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NORMAN-Network NORMAN Prioritisation framework for emerging substances: critical review. https://norman-data.eu/NORMAN%20Documents/Discussion_updated%20prioritisation%20scheme_WG-1.pdf. [Google Scholar]
- Council NR, Risk Assessment in the Federal Government: Managing the Process. The National Academies Press: Washington, DC, 1983; p 205. [PubMed] [Google Scholar]
- USEPA List of Alternative Test Methods and Strategies (or New Approach Methodologies [NAMs]). https://www.epa.gov/assessing-and-managing-chemicals-under-tsca/strategic-plan-reduce-use-vertebrate-animals-chemical. [Google Scholar]
- 40 CFR § 723.50. In 40.
- 40 CFR § 720.30(h). In.
- Schymanski EL, Williams AJ, 2017. Open Science for Identifying “Known Unknown” Chemicals. Environ. Sci. Technol 51 (10), 5357–5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding G, Zhang X, Yang M, Pan Y, 2013. Formation of new brominated disinfection byproducts during chlorination of saline sewage effluents. Water Res 47 (8), 2710–2718. [DOI] [PubMed] [Google Scholar]
- Tao D, Wang R, Shi S, Yun L, Tong R, Peng Y, Guo W, Liu Y, Hu S, 2020. The identification of halogenated disinfection by-products in tap water using liquid chromatography–high resolution mass spectrometry. Sci. Total Environ 740, 139888. 10.1016/j.scitotenv.2020.139888. [DOI] [PubMed] [Google Scholar]
- McCord J, Strynar M, 2019. Identification of Per- and Polyfluoroalkyl Substances in the Cape Fear River by High Resolution Mass Spectrometry and Nontargeted Screening. Environ. Sci. Technol 53 (9), 4717–4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvito D, Fernandez M, Jenner K, Lyon DY, Knecht J, Mayer P, MacLeod M, Eisenreich K, Leonards P, Cesnaitis R, León-Paumen M, Embry M, Déglin SE, 2020. Improving the Environmental Risk Assessment of Substances of Unknown or Variable Composition, Complex Reaction Products, or Biological Materials. Environ. Toxicol. Chem 39 (11), 2097–2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess RM, Ho KT, Brack W, Lamoree M, 2013. Effects-directed analysis (EDA) and toxicity identification evaluation (TIE): Complementary but different approaches for diagnosing causes of environmental toxicity. Environ. Toxicol. Chem 32 (9), 1935–1945. [DOI] [PubMed] [Google Scholar]
- Brack W, Aissa SA, Backhaus T, Dulio V, Escher BI, Faust M, Hilscherova K, Hollender J, Hollert H, Müller C, Munthe J, Posthuma L, Seiler T-B, Slobodnik J, Teodorovic I, Tindall AJ, de Aragão Umbuzeiro G, Zhang X, Altenburger R, 2019. Effect-based methods are key. The European Collaborative Project SOLUTIONS recommends integrating effect-based methods for diagnosis and monitoring of water quality. Environ. Sci. Europe 31 (1), 10. [Google Scholar]
- Tian Z, Zhao H, Peter KT, Gonzalez M, Wetzel J, Wu C, Hu X, Prat J, Mudrock E, Hettinger R, Cortina AE, Biswas RG, Kock FVC, Soong R, Jenne A, Du B, Hou F, He H, Lundeen R, Gilbreath A, Sutton R, Scholz NL, Davis JW, Dodd MC, Simpson A, McIntyre JK, Kolodziej EP, 2021. A ubiquitous tire rubber–derived chemical induces acute mortality in coho salmon. Science 371 (6525), 185–189. [DOI] [PubMed] [Google Scholar]
- Pochiraju SS, Linden K, Gu AZ, Rosenblum J, 2020. Development of a separation framework for effects-based targeted and non-targeted toxicological screening of water and wastewater. Water Res 170, 115289. 10.1016/j.watres.2019.115289. [DOI] [PubMed] [Google Scholar]
- Muschket M, Di Paolo C, Tindall AJ, Touak G, Phan A, Krauss M, Kirchner K, Seiler T-B, Hollert H, Brack W, 2018. Identification of Unknown Antiandrogenic Compounds in Surface Waters by Effect-Directed Analysis (EDA) Using a Parallel Fractionation Approach. Environ. Sci. Technol 52 (1), 288–297. [DOI] [PubMed] [Google Scholar]
- Dusza HM, Manz KE, Pennell KD, Kanda R, Legler J, 2022. Identification of known and novel nonpolar endocrine disruptors in human amniotic fluid. Environ. Int 158, 106904. 10.1016/j.envint.2021.106904. [DOI] [PubMed] [Google Scholar]
- McEachran AD, Mansouri K, Grulke C, Schymanski EL, Ruttkies C, Williams AJ, 2018. “MS-Ready” structures for non-targeted high-resolution mass spectrometry screening studies. J. Cheminformatics 10 (1), 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Council NR, 1984. Toxicity testing: strategies to determine needs and priorities. National Academies Press. [PubMed] [Google Scholar]
- Council NR, Toxicity Testing in the 21st Century: A Vision and a Strategy. The National Academies Press: Washington, DC, 2007; p 216. [Google Scholar]
- Kavlock R, Dix D, 2010. Computational Toxicology as Implemented by the U.S. EPA: Providing High Throughput Decision Support Tools for Screening and Assessing Chemical Exposure, Hazard and Risk. J. Toxicology Environ. Health, Part B 13 (2-4), 197–217. [DOI] [PubMed] [Google Scholar]
- Paul Friedman K, Gagne M, Loo LH, Karamertzanis P, Netzeva T, Sobanski T, Franzosa JA, Richard AM, Lougee RR, Gissi A, Lee JJ, Angrish M, Dorne JL, Foster S, Raffaele K, Bahadori T, Gwinn MR, Lambert J, Whelan M, Rasenberg M, Barton-Maclaren T, Thomas RS, 2020. Utility of In Vitro Bioactivity as a Lower Bound Estimate of In Vivo Adverse Effect Levels and in Risk-Based Prioritization. Toxicol. Sci 173 (1), 202–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catron TR, Swank A, Wehmas LC, Phelps D, Keely SP, Brinkman NE, McCord J, Singh R, Sobus J, Wood CE, Strynar M, Wheaton E, Tal T, 2019. Microbiota alter metabolism and mediate neurodevelopmental toxicity of 17β-estradiol. Sci. Rep 9 (1) 10.1038/s41598-019-43346-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitekamp CA; Phelps D; Swank A; McCord JP; Sobus JR; Catron T; Keely S; Brinkman N; Zurlinden T; Wheaton E, Triclosan-selected host-associated microbiota perform xenobiotic biotransformations in larval zebrafish. Toxicological Sciences 2019, 172, (1), 109–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGroot DE, Swank A, Thomas RS, Strynar M, Lee MY, Carmichael PL, Simmons SO, 2018. mRNA transfection retrofits cell-based assays with xenobiotic metabolism. J. Pharmacol. Toxicol. Methods 92, 77–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisenroth C, DeGroot DE, Zurlinden T, Eicher A, McCord J, Lee M-Y, Carmichael P, Thomas RS, 2020. The Alginate Immobilization of Metabolic Enzymes (AIME) Platform Retrofits an Estrogen Receptor Transactivation Assay with Metabolic Competence. Toxicol. Sci [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agency, U. S. E. P. Chemical Substances of Unknown or Variable Composition, Complex Reaction Products and Biological Materials (UVCB Substance) on the TSCA Inventory. https://www.epa.gov/sites/production/files/2015-05/documents/uvcb.pdf. [Google Scholar]
- Lowe CN, Isaacs KK, McEachran A, Grulke CM, Sobus JR, Ulrich EM, Richard A, Chao A, Wambaugh J, Williams AJ, 2021. Predicting compound amenability with liquid chromatography-mass spectrometry to improve non-targeted analysis. Anal. Bioanal. Chem 413 (30), 7495–7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agency, U. S. E. P. EPA’s Exposure Factors Handbook (EFH). https://www.epa.gov/expobox/about-exposure-factors-handbook. [Google Scholar]
- Council NR, Exposure Science in the 21st Century: A Vision and a Strategy. The National Academies Press: Washington, DC, 2012; p 210. [PubMed] [Google Scholar]
- Wambaugh JF, Bare JC, Carignan CC, Dionisio KL, Dodson RE, Jolliet O, Liu X, Meyer DE, Newton SR, Phillips KA, Price PS, Ring CL, Shin H-M, Sobus JR, Tal T, Ulrich EM, Vallero DA, Wetmore BA, Isaacs KK, 2019. New approach methodologies for exposure science. Current Opinion Toxicology 15, 76–92. [Google Scholar]
- Rager JE, Strynar MJ, Liang S, McMahen RL, Richard AM, Grulke CM, Wambaugh JF, Isaacs KK, Judson R, Williams AJ, Sobus JR, 2016. Linking high resolution mass spectrometry data with exposure and toxicity forecasts to advance high-throughput environmental monitoring. Environ. Int 88, 269–280. [DOI] [PubMed] [Google Scholar]
- Sobus JR, Wambaugh JF, Isaacs KK, Williams AJ, McEachran AD, Richard AM, Grulke CM, Ulrich EM, Rager JE, Strynar MJ, Newton SR, 2018. Integrating tools for non-targeted analysis research and chemical safety evaluations at the US EPA. J. Eposure Sci. Environ. Epidemiol 28 (5), 411–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wambaugh JF, Setzer RW, Reif DM, Gangwal S, Mitchell-Blackwood J, Arnot JA, Joliet O, Frame A, Rabinowitz J, Knudsen TB, Judson RS, Egeghy P, Vallero D, Cohen Hubal EA, 2013. High-Throughput Models for Exposure-Based Chemical Prioritization in the ExpoCast Project. Environ. Sci. Technol 47 (15), 8479–8488. [DOI] [PubMed] [Google Scholar]
- Wambaugh JF, Wang A, Dionisio KL, Frame A, Egeghy P, Judson R, Setzer RW, 2014. High Throughput Heuristics for Prioritizing Human Exposure to Environmental Chemicals. Environ. Sci. Technol 48 (21), 12760–12767. [DOI] [PubMed] [Google Scholar]
- Ring CL, Pearce RG, Setzer RW, Wetmore BA, Wambaugh JF, 2017. Identifying populations sensitive to environmental chemicals by simulating toxicokinetic variability. Environ. Int 106, 105–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring CL, Arnot JA, Bennett DH, Egeghy PP, Fantke P, Huang L, Isaacs KK, Jolliet O, Phillips KA, Price PS, Shin H-M, Westgate JN, Setzer RW, Wambaugh JF, 2019. Consensus Modeling of Median Chemical Intake for the U.S. Population Based on Predictions of Exposure Pathways. Environ. Sci. Technol 53 (2), 719–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobus JR, DeWoskin RS, Tan Y-M, Pleil JD, Phillips MB, George BJ, Christensen K, Schreinemachers DM, Williams MA, Hubal EAC, Edwards SW, 2015. Uses of NHANES Biomarker Data for Chemical Risk Assessment: Trends, Challenges, and Opportunities. Environ. Health Perspect 123 (10), 919–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Control, C. f. D.; Prevention, National Center for Health Statistics (NCHS). National Health and Nutrition Examination Survey Questionnaire (or Examination Protocol, or Laboratory Protocol). http://www.cdc.gov/nchs/nhanes.htm 2006. [Google Scholar]
- Sobus JR, Wambaugh JF, Isaacs KK, Williams AJ, McEachran AD, Richard AM, Grulke CM, Ulrich EM, Rager JE, Strynar MJ, Newton SR, 2018. Integrating tools for non-targeted analysis research and chemical safety evaluations at the US EPA. J. Exposure Sci. Environ. Epidemiol 28 (5), 411–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas RS; Bahadori T; Buckley TJ; Cowden J; Deisenroth C; Dionisio KL; Frithsen JB; Grulke CM; Gwinn MR; Harrill JA; Higuchi M; Houck KA; Hughes MF; Hunter ES III; Isaacs KK; Judson RS; Knudsen TB; Lambert JC; Linnenbrink M; Martin TM; Newton SR; Padilla S; Patlewicz G; Paul-Friedman K; Phillips KA; Richard AM; Sams R; Shafer TJ; Setzer RW; Shah I; Simmons JE; Simmons SO; Singh A; Sobus JR; Strynar M; Swank A; Tornero-Valez R; Ulrich EM; Villeneuve DL; Wambaugh JF; Wetmore BA; Williams AJ, The Next Generation Blueprint of Computational Toxicology at the U.S. Environmental Protection Agency. Toxicological Sciences 2019, 169, (2), 317–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Academies of Sciences, E.; Medicine, Using 21st Century Science to improve Risk-Related Evaluations. The National Academies Press: Washington, DC, 2017; p 200. [PubMed] [Google Scholar]
- Mansouri K, Grulke CM, Judson RS, Williams AJ, 2018. OPERA models for predicting physicochemical properties and environmental fate endpoints. J. Cheminformatics 10 (1), 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce RG, Setzer RW, Strope CL, Wambaugh JF, Sipes NS, 2017. httk: R Package for High-Throughput Toxicokinetics. J. Stat. Softw 79 (4), 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansouri Kamel, Abdelaziz Ahmed, Rybacka Aleksandra, Roncaglioni Alessandra, Tropsha Alexander, Varnek Alexandre, Zakharov Alexey, Worth Andrew, Richard Ann M., Grulke Christopher M., Trisciuzzi Daniela, Fourches Denis, Horvath Dragos, Benfenati Emilio, Muratov Eugene, Wedebye Eva Bay, Grisoni Francesca, Mangiatordi Giuseppe F., Incisivo Giuseppina M., Hong Huixiao, Ng Hui W., Tetko Igor V., Balabin Ilya, Kancherla Jayaram, Shen Jie, Burton Julien, Nicklaus Marc, Cassotti Matteo, Nikolov Nikolai G., Nicolotti Orazio, Andersson Patrik L., Zang Qingda, Politi Regina, Beger Richard D., Todeschini Roberto, Huang Ruili, Farag Sherif, Rosenberg Sine A., Slavov Svetoslav, Hu Xin, Judson Richard S., 2016. CERAPP: Collaborative Estrogen Receptor Activity Prediction Project. Environ. Health Perspect 124 (7), 1023–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- USEPA Use of High Throughput Assays and Computational Tools in the Endocrine Disruptor Screening Program, https://www.epa.gov/endocrine-disruption/use-high-throughput-assays-and-computational-tools-endocrine-disruptor. [Google Scholar]
- Reif David M., Martin Matthew T., Tan Shirlee W., Houck Keith A., Judson Richard S., Richard Ann M., Knudsen Thomas B., Dix David J., Kavlock Robert J., 2010. Endocrine profiling and prioritization of environmental chemicals using ToxCast data. Environ. Health Perspect 118 (12), 1714–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moschet Christoph, Wittmer Irene, Simovic Jelena, Junghans Marion, Piazzoli Alessandro, Singer Heinz, Stamm Christian, Leu Christian, Hollender Juliane, 2014. How a Complete Pesticide Screening Changes the Assessment of Surface Water Quality. Environ. Sci. Technol 48 (10), 5423–5432. [DOI] [PubMed] [Google Scholar]
- Gago-Ferrero Pablo, Schymanski Emma L., Bletsou Anna A., Aalizadeh Reza, Hollender Juliane, Thomaidis Nikolaos S., 2015. Extended Suspect and Non-Target Strategies to Characterize Emerging Polar Organic Contaminants in Raw Wastewater with LC-HRMS/MS. Environ. Sci. Technol 49 (20), 12333–12341. [DOI] [PubMed] [Google Scholar]
- Yi L, Dong N, Yun Y, Deng B, Ren D, Liu S, Liang Y, 2016. Chemometric methods in data processing of mass spectrometry-based metabolomics: A review. Anal. Chim. Acta 914, 17–34. [DOI] [PubMed] [Google Scholar]
- Di Guida R, Engel J, Allwood JW, Weber RJM, Jones MR, Sommer U, Viant MR, Dunn WB, 2016. Non-targeted UHPLC-MS metabolomic data processing methods: a comparative investigation of normalisation, missing value imputation, transformation and scaling. Metabolomics 12 (5), 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama SF, Strynar MJ, Reiner JL, Delinsky AD, Lindstrom AB, 2010. Determination of perfluorinated compounds in the Upper Mississippi River Basin. Environ. Sci. Technol 44 (11), 4103. [DOI] [PubMed] [Google Scholar]
- Lindstrom AB; Strynar MJ; Delinsky AD; Nakayama SF; McMillan L; Libelo EL; Neill M; Thomas L, Application of WWTP biosolids and resulting perfluorinated compound contamination of surface and well water in Decatur, Alabama, USA. Environ. Sci. Technol 2011, 45, (19), 8015. [DOI] [PubMed] [Google Scholar]
- Strynar M, Dagnino S, McMahen R, Liang S, Lindstrom A, Andersen E, McMillan L, Thurman M, Ferrer I, Ball C, 2015. Identification of novel perfluoroalkyl ether carboxylic acids (PFECAs) and sulfonic acids (PFESAs) in natural waters using accurate mass time-of-flight mass spectrometry (TOFMS). Environ. Sci. Technol 49, (19), 11622. [DOI] [PubMed] [Google Scholar]
- Tang YN, Xu Y, Li F, Jmaiff L, Hrudey SE, Li XF, 2016. Nontargeted identification of peptides and disinfection byproducts in water. J. Environ. Sci 42, 259–266. [DOI] [PubMed] [Google Scholar]
- Liberatore Hannah K., Westerman Danielle C., Allen Joshua M., Plewa Michael J., Wagner Elizabeth D., McKenna Amy M., Weisbrod Chad R., McCord James P., Liberatore Richard J., Burnett David B., Cizmas Leslie H., Richardson Susan D., 2020. High-Resolution Mass Spectrometry Identification of Novel Surfactant-Derived Sulfur-Containing Disinfection Byproducts from Gas Extraction Wastewater. Environ. Sci. Technol 54 (15), 9374–9386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Yoav, Hochberg Yosef, 1995. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. Roy. Stat. Soc.: Ser. B (Methodol.) 57 (1), 289–300. [Google Scholar]
- Storey JD, 2003. The positive false discovery rate: a Bayesian interpretation and the q-value. Ann. Stat 31, 2013–2035. [Google Scholar]
- Storey JD, 2002. A direct approach to false discovery rates. J. Royal Statistical Soc.: Series B (Statistical Methodology) 64 (3), 479–498. [Google Scholar]
- Chong EY, Huang Y, Wu H, Ghasemzadeh N, Uppal K, Quyyumi AA, Jones DP, Yu T, 2015. Local false discovery rate estimation using feature reliability in LC/MS metabolomics data. Sci. Rep 5 (1), 17221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J; Psychogios N; Young N; Wishart DS, MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic acids research 2009, 37, (Web Server issue), W652–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra BB, 2021. New software tools, databases, and resources in metabolomics: updates from 2020. Metabolomics 17 (5), 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollender Juliane, Schymanski Emma L., Singer Heinz P., Ferguson P. Lee, 2017. Nontarget Screening with High Resolution Mass Spectrometry in the Environment: Ready to Go? Environ. Sci. Technol 51 (20), 11505–11512. [DOI] [PubMed] [Google Scholar]
- Plassmann Merle M., Tengstrand Erik, Åberg K. Magnus, Benskin Jonathan P., 2016. Non-target time trend screening: a data reduction strategy for detecting emerging contaminants in biological samples. Anal. Bioanal. Chem 408 (16), 4203–4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plassmann Merle M., Fischer Stellan, Benskin Jonathan P., 2018. Nontarget Time Trend Screening in Human Blood. Environ. Sci. Technol. Lett 5 (6), 335–340. [Google Scholar]
- Yu Huaxu, Xing Shipei, Nierves Lorenz, Lange Philipp F., Huan Tao, 2020. Fold-Change Compression: An Unexplored But Correctable Quantitative Bias Caused by Nonlinear Electrospray Ionization Responses in Untargeted Metabolomics. Anal. Chem 92 (10), 7011–7019. [DOI] [PubMed] [Google Scholar]
- Liu Ken H., Nellis Mary, Uppal Karan, Ma Chunyu, Tran ViLinh, Liang Yongliang, Walker Douglas I., Jones Dean P., 2020. Reference Standardization for Quantification and Harmonization of Large-Scale Metabolomics. Anal. Chem 92 (13), 8836–8844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go Young-Mi, Walker Douglas I., Liang Yongliang, Uppal Karan, Soltow Quinlyn A., Tran ViLinh, Strobel Frederick, Quyyumi Arshed A., Ziegler Thomas R., Pennell Kurt D., Miller Gary W., Jones Dean P., 2015. Reference Standardization for Mass Spectrometry and High-resolution Metabolomics Applications to Exposome Research. Toxicological Sci. Official J. Soc. Toxicology 148 (2), 531–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahal Upendra P., Jones Jeffrey P., Davis John A., Rock Dan A., 2011. Small molecule quantification by liquid chromatography-mass spectrometry for metabolites of drugs and drug candidates. Drug Metab. Dispos 39 (12), 2355–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatsis Panos, Waters Nigel J., Argikar Upendra A., 2017. Implications for Metabolite Quantification by Mass Spectrometry in the Absence of Authentic Standards. Drug metabolism and disposition: the biological fate of chemicals 45 (5), 492–496. [DOI] [PubMed] [Google Scholar]
- Pieke EN, Granby K, Trier X, Smedsgaard J, 2017. A framework to estimate concentrations of potentially unknown substances by semi-quantification in liquid chromatography electrospray ionization mass spectrometry. Anal. Chim. Acta 975, 30–41. [DOI] [PubMed] [Google Scholar]
- McCord JP, Newton S, Strynar M, 2018. Validation of quantitative measurements and semi-quantitative estimates of emerging perfluoroethercarboxylic acids (PFECAs) and hexfluoroprolyene oxide acids (HFPOAs). J. Chromatogr. A 1551, 52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liigand P, Liigand J, Cuyckens F, Vreeken RJ, Kruve A, 2018. Ionisation efficiencies can be predicted in complicated biological matrices: A proof of concept. Anal. Chim. Acta 1032, 68–74. [DOI] [PubMed] [Google Scholar]
- Cech Nadja B., Krone Jennifer R., Enke Christie G., 2001. Predicting Electrospray Response from Chromatographic Retention Time. Anal. Chem 73 (2), 208–213. [DOI] [PubMed] [Google Scholar]
- Kamga Albert W., Behar Fancoise, Hatcher Patrick G., 2014. Quantitative Analysis of Long Chain Fatty Acids Present in a Type I Kerogen Using Electrospray Ionization Fourier Transform Ion Cyclotron Resonance Mass Spectrometry: Compared with BF3/MeOH Methylation/GC-FID. J. Am. Soc. Mass Spectrom 25 (5), 880–890. [DOI] [PubMed] [Google Scholar]
- Banerjee K, Utture S, Dasgupta S, Kandaswamy C, Pradhan S, Kulkarni S, Adsule P, 2012. Multiresidue determination of 375 organic contaminants including pesticides, polychlorinated biphenyls and polyaromatic hydrocarbons in fruits and vegetables by gas chromatography–triple quadrupole mass spectrometry with introduction of semi-quantification approach. J. Chromatogr. A 1270, 283–295. [DOI] [PubMed] [Google Scholar]
- Bu Q, Wang D, Liu X, Wang Z, 2014. A high throughout semi-quantification method for screening organic contaminants in river sediments. J. Environ. Manage 143, 135–139. [DOI] [PubMed] [Google Scholar]
- Aalizadeh Reza, Panara Anthi, Thomaidis Nikolaos S., 2021. Development and Application of a Novel Semi-quantification Approach in LC-QToF-MS Analysis of Natural Products. J. Am. Soc. Mass Spectrom 32 (6), 1412–1423. [DOI] [PubMed] [Google Scholar]
- Cao Y, Jiang T, Girke T, 2008. A maximum common substructure-based algorithm for searching and predicting drug-like compounds. Bioinformatics (Oxford, England) 24 (13), i366–i374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajusz D, Rácz A, Héberger K, 2015. Why is Tanimoto index an appropriate choice for fingerprint-based similarity calculations? J. Cheminformatics 7 (1), 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor Paul J., 2005. Matrix effects: the Achilles heel of quantitative high-performance liquid chromatography–electrospray–tandem mass spectrometry. Clin. Biochem 38 (4), 328–334. [DOI] [PubMed] [Google Scholar]
- Null Allison P., Nepomuceno Angelito I., Muddiman David C., 2003. Implications of Hydrophobicity and Free Energy of Solvation for Characterization of Nucleic Acids by Electrospray Ionization Mass Spectrometry. Anal. Chem 75 (6), 1331–1339. [DOI] [PubMed] [Google Scholar]
- Henriksen Trine, Juhler René K., Svensmark Bo, Cech Nadja B., 2005. The relative influences of acidity and polarity on responsiveness of small organic molecules to analysis with negative ion electrospray ionization mass spectrometry (ESI-MS). J. Am. Soc. Mass Spectrom 16 (4), 446–455. [DOI] [PubMed] [Google Scholar]
- Cech Nadja B., Enke Christie G., 2000. Relating Electrospray Ionization Response to Nonpolar Character of Small Peptides. Anal. Chem 72 (13), 2717–2723. [DOI] [PubMed] [Google Scholar]
- Walker S. Hunter, Lilley Laura M., Enamorado Monica F., Comins Daniel L., Muddiman David C., 2011. Hydrophobic derivatization of N-linked glycans for increased ion abundance in electrospray ionization mass spectrometry. J. Am. Soc. Mass Spectrom 22 (8), 1309–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golubović J, Birkemeyer C, Protić A, Otašević B, Zečević M, 2016. Structure–response relationship in electrospray ionization-mass spectrometry of sartans by artificial neural networks. J. Chromatogr. A 1438, 123–132. [DOI] [PubMed] [Google Scholar]
- Mehta Nickita, Porterfield Mindy, Struwe Weston B., Heiss Christian, Azadi Parastoo, Rudd Pauline M., Tiemeyer Michael, Aoki Kazuhiro, 2016. Mass Spectrometric Quantification of N-Linked Glycans by Reference to Exogenous Standards. J. Proteome Res 15 (9), 2969–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox Frederick J., Johnston Murray V., Dasgupta Arnab, 2003. Characterization and relative ionization efficiencies of end-functionalized polystyrenes by matrix-assisted laser desorption/ionization mass spectrometry. J. Am. Soc. Mass Spectrom 14 (6), 648–657. [DOI] [PubMed] [Google Scholar]
- Sunner Jan., Nicol Gordon., Kebarle Paul., 1988. Factors determining relative sensitivity of analytes in positive mode atmospheric pressure ionization mass spectrometry. Anal. Chem 60 (13), 1300–1307. [Google Scholar]
- Ehrmann Brandie M., Henriksen Trine, Cech Nadja B., 2008. Relative importance of basicity in the gas phase and in solution for determining selectivity in electrospray ionization mass spectrometry. J. Am. Soc. Mass Spectrom 19 (5), 719–728. [DOI] [PubMed] [Google Scholar]
- Richter Wilhelm J., Schwarz Helmut, 1978. Chemical Ionization—A Mass-Spectrometric Analytical Procedure of Rapidly Increasing Importance. Angew. Chem., Int. Ed. Engl 17 (6), 424–439. [Google Scholar]
- Bergmann Alan J., Points Gary L., Scott Richard P., Wilson Glenn, Anderson Kim A., 2018. Development of quantitative screen for 1550 chemicals with GC-MS. Anal. Bioanal. Chem 410 (13), 3101–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y-H, Kim K-H, Szulejko JE, Bae M-S, Brown RJC, 2014. Experimental validation of an effective carbon number-based approach for the gas chromatography–mass spectrometry quantification of ‘compounds lacking authentic standards or surrogates’. Anal. Chim. Acta 830, 32–41. [DOI] [PubMed] [Google Scholar]
- Szulejko Jan E., Kim Yong-Hyun, Kim Ki-Hyun, 2013. Method to predict gas chromatographic response factors for the trace-level analysis of volatile organic compounds based on the effective carbon number concept. J. Sep. Sci 36 (20), 3356–3365. [DOI] [PubMed] [Google Scholar]
- Liigand Jaanus, Kruve Anneli, Leito Ivo, Girod Marion, Antoine Rodolphe, 2014. Effect of mobile phase on electrospray ionization efficiency. J. Am. Soc. Mass Spectrom 25 (11), 1853–1861. [DOI] [PubMed] [Google Scholar]
- Page Jason S., Kelly Ryan T., Tang Keqi, Smith Richard D., 2007. Ionization and Transmission Efficiency in an Electrospray Ionization-Mass Spectrometry Interface. J. Am. Soc. Mass Spectrom 18 (9), 1582–1590. [DOI] [PubMed] [Google Scholar]
- Smith Richard D., Shen Yufeng, Tang Keqi, 2004. Ultrasensitive and Quantitative Analyses from Combined Separations–Mass Spectrometry for the Characterization of Proteomes. Acc. Chem. Res 37 (4), 269–278. [DOI] [PubMed] [Google Scholar]
- Kruve Anneli, 2020. Strategies for Drawing Quantitative Conclusions from Nontargeted Liquid Chromatography–High-Resolution Mass Spectrometry Analysis. Anal. Chem 92 (7), 4691–4699. [DOI] [PubMed] [Google Scholar]
- Bantscheff Marcus, Schirle Markus, Sweetman Gavain, Rick Jens, Kuster Bernhard, 2007. Quantitative mass spectrometry in proteomics: a critical review. Anal. Bioanal. Chem 389 (4), 1017–1031. [DOI] [PubMed] [Google Scholar]
- Shuford Christopher M., Walters James J., Holland Patricia M., Sreenivasan Uma, Askari Nadav, Ray Kevin, Grant Russell P., 2017. Absolute Protein Quantification by Mass Spectrometry: Not as Simple as Advertised. Anal. Chem 89 (14), 7406–7415. [DOI] [PubMed] [Google Scholar]
- Abrahamsson Panagopoulos, Dimitri Park, June-Soo Singh, Randolph R, Sirota Marina, Woodruff Tracey J., 2020. Applications of Machine Learning to In Silico Quantification of Chemicals without Analytical Standards. J. Chem. Inf. Model 60 (6), 2718–2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leito Ivo, Herodes Koit, Huopolainen Merit, Virro Kristina, Künnapas Allan, Kruve Anneli, Tanner Risto, 2008. Towards the electrospray ionization mass spectrometry ionization efficiency scale of organic compounds. Rapid Commun. Mass Spectrom 22 (3), 379–384. [DOI] [PubMed] [Google Scholar]
- Oss Merit, Kruve Anneli, Herodes Koit, Leito Ivo, 2010. Electrospray Ionization Efficiency Scale of Organic Compounds. Anal. Chem 82 (7), 2865–2872. [DOI] [PubMed] [Google Scholar]
- Kruve Anneli, Kaupmees Karl, Liigand Jaanus, Leito Ivo, 2014. Negative Electrospray Ionization via Deprotonation: Predicting the Ionization Efficiency. Anal. Chem 86 (10), 4822–4830. [DOI] [PubMed] [Google Scholar]
- Chalcraft Kenneth R., Lee Richard, Mills Casandra, Britz-McKibbin Philip, 2009. Virtual Quantification of Metabolites by Capillary Electrophoresis-Electrospray Ionization-Mass Spectrometry: Predicting Ionization Efficiency Without Chemical Standards. Anal. Chem 81 (7), 2506–2515. [DOI] [PubMed] [Google Scholar]
- Ghosh Banibrata, Jones A. Daniel, 2015. Dependence of negative-mode electrospray ionization response factors on mobile phase composition and molecular structure for newly-authenticated neutral acylsucrose metabolites. Analyst 140 (19), 6522–6531. [DOI] [PubMed] [Google Scholar]
- Cífková Eva, Holčapek Michal, Lísa Miroslav, Ovčačíková Magdaléna, Lyčka Antonín, Lynen Frédéric, Sandra Pat, 2012. Nontargeted Quantitation of Lipid Classes Using Hydrophilic Interaction Liquid Chromatography-Electrospray Ionization Mass Spectrometry with Single Internal Standard and Response Factor Approach. Anal. Chem 84 (22), 10064–10070. [DOI] [PubMed] [Google Scholar]
- Alymatiri Christina M., Kouskoura Maria G., Markopoulou Catherine K., 2015. Decoding the signal response of steroids in electrospray ionization mode (ESI-MS). Anal. Methods 7 (24), 10433–10444. [Google Scholar]
- Basiri Babak, Murph Mandi M., Bartlett Michael G., 2017. Assessing the Interplay between the Physicochemical Parameters of Ion-Pairing Reagents and the Analyte Sequence on the Electrospray Desorption Process for Oligonucleotides. J. Am. Soc. Mass Spectrom 28 (8), 1647–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liigand J, Wang T, Kellogg J, Smedsgaard J, Cech N, Kruve A, 2020. Quantification for non-targeted LC/MS screening without standard substances. Sci. Rep 10 (1), 5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alygizakis Nikiforos, Galani Aikaterini, Rousis Nikolaos I., Aalizadeh Reza, Dimopoulos Meletios-Athanasios, Thomaidis Nikolaos S., 2021. Change in the chemical content of untreated wastewater of Athens, Greece under COVID-19 pandemic. Sci. Total Environ 799, 149230. 10.1016/j.scitotenv.2021.149230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perestrelo R, Silva P, Porto-Figueira P, Pereira JAM, Silva C, Medina S, Câmara JS, 2019. QuEChERS - Fundamentals, relevant improvements, applications and future trends. Anal. Chim. Acta 1070, 1–28. [DOI] [PubMed] [Google Scholar]
- Fisher JA, Scarlett MJ, Stott AD, 1997. Accelerated Solvent Extraction: An Evaluation for Screening of Soils for Selected U.S. EPA Semivolatile Organic Priority Pollutants. Environ. Sci. Technol 31 (4), 1120–1127. [Google Scholar]
- Vuckovic D, 2020. Chapter 4 - Sample preparation in global metabolomics of biological fluids and tissues. In: Issaq HJ, Veenstra TD (Eds.), Proteomic and Metabolomic Approaches to Biomarker Discovery (Second Edition). Academic Press, Boston, pp. 53–83. [Google Scholar]
- Chambers Erin, Wagrowski-Diehl Diane M., Lu Ziling, Mazzeo Jeffrey R., 2007. Systematic and comprehensive strategy for reducing matrix effects in LC/MS/MS analyses. J. Chromatogr. B 852 (1-2), 22–34. [DOI] [PubMed] [Google Scholar]
- Parry Emily, Young Thomas M., 2016. Comparing targeted and non-targeted high-resolution mass spectrometric approaches for assessing advanced oxidation reactor performance. Water Res 104, 72–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crimmins Bernard S., Xia Xiaoyan, Hopke Philip K., Holsen Thomas M., 2014. A targeted/non-targeted screening method for perfluoroalkyl carboxylic acids and sulfonates in whole fish using quadrupole time-of-flight mass spectrometry and MSe. Anal. Bioanal. Chem 406 (5), 1471–1480. [DOI] [PubMed] [Google Scholar]
- McCord JP, Strynar M, 2019. Identifying Per- and Polyfluorinated Chemical Species with a Combined Targeted and Non-Targeted-Screening High-Resolution Mass Spectrometry Workflow. JoVE 146, e59142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas RS, Paules RS, Simeonov A, Fitzpatrick SC, Crofton KM, Casey WM, Mendrick DL, 2018. The US Federal Tox21 Program: A strategic and operational plan for continued leadership. Altex 35 (2), 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard Ann M., Judson Richard S., Houck Keith A., Grulke Christopher M., Volarath Patra, Thillainadarajah Inthirany, Yang Chihae, Rathman James, Martin Matthew T., Wambaugh John F., Knudsen Thomas B., Kancherla Jayaram, Mansouri Kamel, Patlewicz Grace, Williams Antony J., Little Stephen B., Crofton Kevin M., Thomas Russell S., 2016. ToxCast Chemical Landscape: Paving the Road to 21st Century Toxicology. Chem. Res. Toxicol 29 (8), 1225–1251. [DOI] [PubMed] [Google Scholar]
- Isaacs KK; Glen WG; Egeghy P; Goldsmith M-R; Smith L; Vallero D; Brooks R; Grulke CM; Özkaynak H. k., SHEDS-HT: an integrated probabilistic exposure model for prioritizing exposures to chemicals with near-field and dietary sources. Environmental science & technology 2014, 48, (21), 12750–12759. [DOI] [PubMed] [Google Scholar]
- Hubal Cohen, Elaine A Richard, Ann M Shah, Imran Gallagher, Jane Kavlock, Robert Blancato, Jerry Edwards, Stephen W, 2010. Exposure science and the U.S. EPA National Center for Computational Toxicology. J. Exposure Sci. Environmental Epidemiology 20 (3), 231–236. [DOI] [PubMed] [Google Scholar]
- Rovida C, Barton-Maclaren T, Benfenati E, Caloni F, Chandrasekera PC, Chesné C, Cronin MTD, De Knecht J, Dietrich DR, Escher SE, Fitzpatrick S, Flannery B, Herzler M, Hougaard Bennekou S, Hubesch B, Kamp H, Kisitu J, Kleinstreuer N, Kovarich S, Leist M, Maertens A, Nugent K, Pallocca G, Pastor M, Patlewicz G, Pavan M, Presgrave O, Smirnova L, Schwarz M, Yamada T, Hartung T, 2020. Internationalization of read-across as a validated new approach method (NAM) for regulatory toxicology. Altex 37 (4), 579–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- US-EPA, User’s Guide for TEST (version 4.2)(Toxicity Estimation Software Tool): A Program to Estimate Toxicity from Molecular Structure. Washington (USA): US-EPA; 2016. [Google Scholar]
- Raies Arwa B., Bajic Vladimir B., 2016. In silico toxicology: computational methods for the prediction of chemical toxicity. WIREs Comput. Mol. Sci 6 (2), 147–172. [DOI] [PMC free article] [PubMed] [Google Scholar]