SUMMARY
Colorectal cancer is a major health concern worldwide. Growing evidence for the role of the gut microbiota in the initiation of CRC has sparked interest in approaches that target these microorganisms. However, little is known about the composition and role of the microbiota associated with precancerous polyps. Here, we found distinct microbial signatures between patients with and without polyps and between polyp subtypes using sequencing and culturing techniques. We found a correlation between Bacteroides fragilis recovered and the level of inflammatory cytokines in the mucosa adjacent to the polyp. Additional analysis revealed that B. fragilis from patients with polyps are bft-negative, activate NF-κB through Toll-like receptor 4, induce a pro-inflammatory response, and are enriched in genes associated with LPS biosynthesis. This study provides fundamental insight into the microbial microenvironment of the pre-neoplastic polyp by highlighting strain-specific genomic and proteomic differences, as well as more broad compositional differences in the microbiome.
Graphical abstract
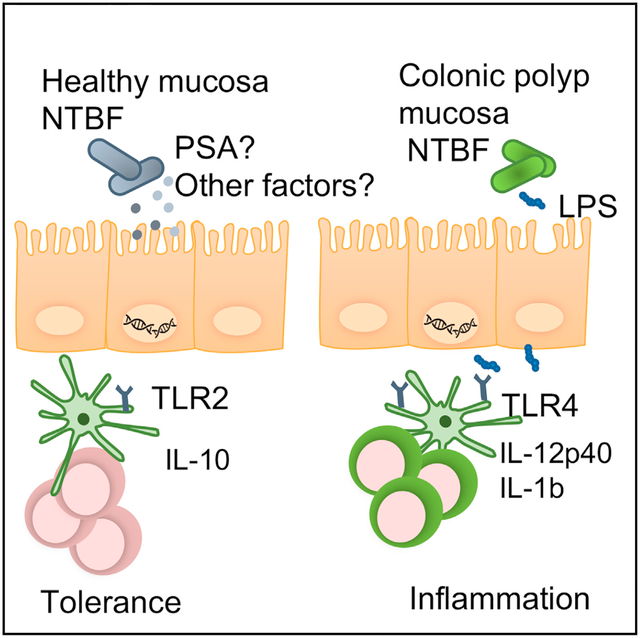
In brief
Kordahi and colleagues find that the common commensal bacteria, nontoxigenic B. fragilis (NTBF), is enriched in patients with precancerous colonic polyps. NTBF isolated from polyps is enriched in genes involved in LPS biosynthesis, which may allow for its increased ability to activate the immune system and cause inflammation.
INTRODUCTION
Colorectal cancer (CRC) is a common type of cancer that develops over the span of several years, starting with precursor lesions in the colon called polyps (Sandouk et al., 2013). Colorectal polyps result from a disruption in the normal proliferation and apoptosis cycles of the epithelial lining of the colon (Testa et al., 2018). Tubular adenomatous polyps (TAP) and sessile serrated polyps (SSP) (Lieberman et al., 2012) are two types of precancerous lesions with a relatively high malignant potential. They differ not only in their appearance, as TAP present as protruding fleshy lesions, while SSP are flattened, broad-based polyps with a saw-tooth appearance when observed histologically (Strum, 2016), but also in their molecular signatures (Mak et al., 2004).The intestinal tract is also heavily colonized by about 1014 bacteria, referred to as the microbiota, that is fundamental to human health and development (Thursby and Juge, 2017). Altered microbiomes have been associated with colorectal polyps and particular microbial species have been identified as possible drivers of oncogenesis. These include pks+ Escherichia coli and the mucosal adherent, toxin-producing strain of Bacteroides fragilis (ETBF) (Ellermann and Sartor, 2018). These bacterial strains allowed the unraveling of tumor-promoting virulence mechanisms. They further contributed to accumulating evidence that chronic infection with certain microbial species and the ensuing inflammation may contribute to tumor initiation and progression. However, their role in polyp development is unclear. Further, no study to date has characterized compositional and functional differences in the microbiota between TAP and SSP lesions and the role of putative “driver” bacteria in cancer formation. Thus, this study hypothesizes that the gut microbiome associated with TAP, SSP, and entirely polyp-free (PF) mucosal biopsies may be compositionally and functionally different and that the mucosal microbiome plays a role in the development of colon polyps.
RESULTS
Recruitment and analysis of host immune response in mucosa of patients with polyps
Forty patients undergoing routine colonoscopy for colorectal cancer screening were enrolled in the study’s cohort. All recruited patients were between the ages of 50 and 75, with 2/3 of them being women. All other characteristics (sex, age, familial history, smoking, BMI, and anemia) were not statistically significant between the polyp and PF groups (Table 1). Histological analyses of the biopsied polyps revealed that 51% of patients had TAP, 23% had SSP, 13% had hyperplastic polyps (HP), and 13% had no diagnostic alterations. The remaining patients recruited had no lesions in the colon at time of the colonoscopy and served as a healthy PF control group (Table 2). Tissue structure, morphology, and cell types were confirmed by H&E staining of sections of the actual polyp for one of each TAP, SSP, or PF biopsies (Figure S1A). Polyps with a higher malignant potential were the TAP and SSP lesions and they were mainly diagnosed in the proximal region of the colon (Figure S1B). Thus, we decided to exclude the HP samples from this study because they are more benign in nature and because they were diagnosed in the distal colon, which corresponds to a different region of the gut environment. Of the patients with polyps, two biopsies were collected from each patient, one associated near the polyp or polyp adjacent (PA) and one from macroscopically healthy mucosa located at least 10 cm away from the polyp, termed non-polyp adjacent (NPA). As a control for this study, biopsies from the proximal colon were taken from PF patients (Figure S1C).
Table 1.
Study participants’ demographics and clinical characteristics
| Polyp-free | Polyp/adenoma | p value | |
|---|---|---|---|
| Patient sample sizea | 9 (22.5) | 31 (77.5) | − |
| Age in years (mean ± SD) | 61 ± 8 | 64 ± 9 | 0.341 |
| Gender (%) | − | − | − |
| Female | 6 (67) | 19 (61) | − |
| Male | 3 (33) | 12 (39) | − |
| BMI (mean ± SD) | 24 ± 4 | 25 ± 3 | 0.307 |
| Smoker (%) | 2 (22.2) | 10 (32.3) | 0.544 |
Exclusion criteria: age < 50, BMI > 30, antibiotic use in the last 3 months, IBD, diabetes mellitus.
Table 2.
Characteristics of colon lesions, see also Figure S1
| Number (%) of patients | Lesion size in cm mean (range) | |
|---|---|---|
| Tubular adenomatous polyp (TAP) | 16 (51) | 0.55 (0.10–1.00) |
| Sessile serrated polyp (SSP) | 7 (23) | 0.59 (0.20–0.90) |
| Hyperplastic polyp (HP) | 4 (13) | 0.32 (0.20–1.00) |
| No diagnostic alterations | 4 (13) | 0.28 (0.20–0.40) |
Since the TAP and SSP tissue biopsies were sent out to the Pathology laboratory for diagnostic evaluation, we were only able to recover paraffin-embedded sections of the biopsy samples for immune and microbial characterization. Thus, we first noticed by immunohistochemistry staining that both proximal TAP and SSP polyp biopsies showed epithelial cell hyperproliferation (via Ki67 staining) and detected signs of mucosal inflammation with the pro-inflammatory markers IL-17, macrophage inflammatory protein (MIP), and myeloperoxidase (MPO) compared with the PF tissue (Figures 1A and S2). Studies have also reported that early stages of CRC present with high levels of IL-12p40 and lower levels of IL-10 (Mager et al., 2016). Using the biopsies taken adjacent to the polyp (PA), macroscopically normal-looking tissue > 10 cm away from the polyp (NPA) and from patients who were PF, we measured host cytokine levels in the tissue. We observed that proximity to the polyp (PA versus NPA) had little influence on the level of cytokines observed. In contrast, both TAP and SSP tissues had significantly more IL-12p40 than tissues from PF patients. This analysis also revealed a trend toward a higher concentration of the anti-inflammatory cytokine IL-10 in the PF tissues (Figures 1B and 1C).
Figure 1. Compositional dysbiosis and enrichment for Bacteroides fragilis in patients with colorectal polyp.
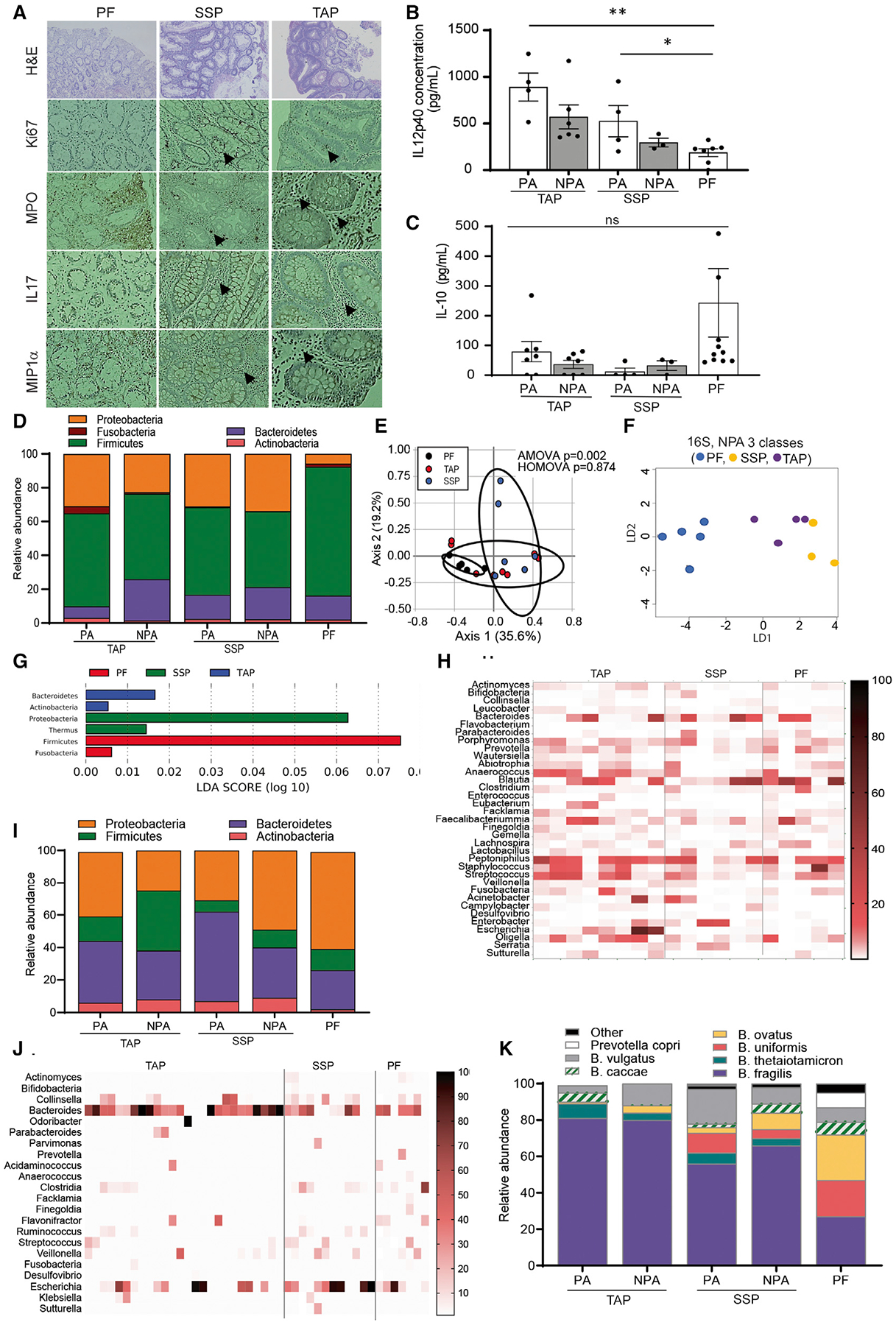
(A–C) (A) Hyperproliferation and inflammation marker expression revealed by immunohistochemistry (IHC) in polyp biopsies and PF control mucosal biopsies, no quantitative data available due to tissue restriction (B and C). Levels of cytokines in mucosal biopsy tissue samples detected by ELISA. Data are represented as the mean ± SEM of 3–4 independent experiments. (A–D), *p < 0.05. ***p < 0.001. One-way ANOVA with multiple comparisons. Levels are measured in a subset of 4 TAP patients, 2 SSP, and 3 PF patients.
(D) Phylum-level abundance in 16S rRNA sequencing of 6 PA and 6 NPA TAP biopsies, 3 PA and 3 NPA SSP biopsies, and 6 PF biopsies.
(E) Principal Coordinate of Analysis (PCoA) of microbiota composition by 16S at phylum level. Statistics are analysis of molecular variation (AMOVA) and homogeneity of molecular variance (HOMOVA).
(F and G) Linear discriminant analysis (LDA) of phylum-level abundance in 16S of mucosal biopsies in TAP NPA, SSP NPA, and PF biopsies.
(H) Heatmap representation of genus level abundance after 16S rRNA profiling.
(I) Phylum level abundance in culturomics on 14 PA and NPA TAP biopsies, 10 PA and NPA SSP biopsies, and 9 PF biopsies.
(J) Heatmap representation of genus level abundance in culturomics.
(K) Species level abundance of Bacteroides species in culturomics. See also Figures S2–S4.
Distinct mucosal microbial signatures can discriminate presence and type of polyp
Intestinal inflammation has been associated with changes in the composition of the gut microbiota. Therefore, we analyzed the mucosal microbiota in the proximal colon of a subset of patients by 16S rRNA sequencing to see if this could explain the phenotypic differences observed in the host tissue. As expected, we found that the composition of the microbiota associated with a polyp was distinct from the mucosa of PF patients and from macroscopically normal-looking mucosa taken from patients with both TAP and SSP lesions. These differences were mostly due to a significantly higher abundance of Firmicutes in the PF mucosa (Figures 1D and S3A). The overall mucosal microbiota composition of biopsies from patients with TAP, SSP, and PF was also significantly distinct (analysis of molecular variation [AMOVA], p value = 0.002) when analyzed using principal component analysis (PCA), suggesting a microbiota signature for each type of tissue (Figure 1E). Interestingly, when comparing the mucosa of patients with polyps, proximity to the polyp also influenced the composition. Indeed, biopsies taken from the macroscopically normal-looking tissue located >10 cm from the TAP (NPA) had more Bacteroidetes on average than the biopsy that was adjacent to the polyp (PA) (Figures 1D and S3B). The biopsy taken adjacent to the polyp (PA) showed no significant differences between SSP and TAP (Figure S4B). In contrast, when comparing the biopsies taken at a distance from the polyp (NPA), we found that patients with TAP had more Bacteroidetes on average than patients with SSP (Figure S3B). PA and NPA biopsies from patients with SSP had more Proteobacteria on average (Figure S3C). Other phyla, such as Actinobacteria (Figure S3D) and Fusobacteria (Figure S3E), did not feature any particular trend or significant alteration between the groups of study. Additionally, a linear discriminant analysis (LDA) performed on the 16S compositional data of NPA biopsies from patients with TAP and SSP as well as on the PF samples featured a discrimination between the groups (Figure 1F) with TAP NPA biopsies containing predominantly Bacteroidetes, SSP NPA biopsies being predominated by Proteobacteria such as Enterobacter species, and PF biopsies containing mostly Firmicutes dominated by Blautia species (Figures 1G and 1H). We used an LDA analysis on the 16S data from NPA and PA biopsies from patients with TAP and SSP compared with PF tissues to evaluate whether there were distinct microbial signatures associated with each type of tissue. As TAP and SSP develop through different molecular pathways, this differentiation may be linked to commensal bacteria in the local microenvironment. Thus, identification of specific microbiomes correlating with the type of polyp lesions developing in the patient may open new avenues for microbiome-based diagnostics and help pathologists determine the risk of developing a given type of lesion. Here, the PCA analysis performed on the 16S data from NPA biopsies (located 10 cm further away from the polyp) showed a better discrimination in the microbiome composition between patients with SSP, TAP, and PF tissues (Figure 1E). We did not see this discrimination when analyzing the composition of the PA tissues (biopsy taken adjacent to the polyp) from these same patients (Figure S4A). Thus, we believe that the microbiome of the NPA tissue is a better predictor of the type of polyp present in the colon as it may be less transformed than the PA tissue and more reflective of the microbiota that drove the transformation in the very early stages of development. Stool collection prior to colonoscopy was not performed on these patients, but analysis of stool will be an important aspect of future studies to understand if changes in the stool microbiome reflects local microenvironments within the colon. However, we focused on the mucosal biopsies because we wanted to understand the microbial and molecular context at the site of transformation.
Bacteroides fragilis is recovered at a higher frequency in patients with polyps
Since little work has been done with the polyp microbiome, we set out to curate a culture library of relevant species that could be used to assess biological function and could be tested in several in vitro and in vivo assays. When paired with matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI TOF MS), culturomics allowed us to assess and compare what bacteria was viable across the various groups at the species and even at the strain level. Using non-selective media and under anaerobic conditions, we were able to grow over eighty species of bacteria. In the 16S analysis, we see a significant enrichment of Firmicutes when comparing the microbial composition of PA and NPA biopsies in patients with TAP (one-way ANOVA, p value = 0.0232 and 0.0345, respectively) and SSP (one-way ANOVA, p value = 0.0234 and 0.0172, respectively) relatively to the PF biopsies with the PF tissue being more enriched in Firmicutes (Figure S3A). This finding is validated by the culturomics data that also show a significant enrichment of Firmicutes in the PF tissues compared with patients with TAP regardless of whether the biopsy was NPA or PA (Two-tailed TTEST, p value = 0.0007 and 0.0003, respectively) (Figure S3F). Other phyla do not feature a particular trend in the culturomics data (Figures S3G, S3H, and S3J). However, when we compared the ratios of the major phyla that we were able to culture to those identified by 16S sequencing, we found that our culturomics technique enriched for Bacteroidetes, which were 3-fold in our culturing approach (Figures 1I and S4B). We recovered the most Bacteroides from tissues derived from patients with TAP, followed by SSP (Figures 1I and 1J). While PF samples had the least amount of Bacteroides, they had a higher amount of E. coli recovered than samples from patients with either polyp type (Figure 1J). Analysis of the Bacteroides genera revealed that greater than 80% and 60% of the Bacteroides recovered were B. fragilis from TAP or SSP, respectively, and proximity of the sample to the polyp had little to no impact on this abundance. In contrast, there was much more heterogeneity in the species of Bacteroides recovered from PF samples where B. fragilis only accounted for 25% of Bacteroides recovered (Figure 1K). Statistically, there were no differences in B. fragilis’ abundances between PA and NPA tissues from patients with both TAP and SSP (Figures S4C and S4D). However, there was a significantly higher abundance of B. fragilis in the PA and NPA tissues from patients with TAP (one-way ANOVA p value = 0.0012 and 0.0158, respectively) (Figures S4E and S4F) and a higher average value of B. fragilis in PA and NPA tissues from patients with SSP (Figures S4G and S4H), although it did not reach significance. Overall, we found that tissue from patients with both TAP and SSP are enriched in B. fragilis compared with PF tissue. Further, these data demonstrate that there are microbial signatures that can discriminate between polyp-associated and polyp- free tissue and can even distinguish polyp type and proximity to the polyp.
B. fragilis isolates from patients with polyps lack the fragilysin gene and promote pro-inflammatory cytokines despite the presence of PSA
The enterotoxigenic strain of B. fragilis (ETBF) has been correlated with CRC in various human studies and mouse models (Sears, 2009) due to oncogenic properties (Sears, 2001) attributed to the expression of the B. fragilis toxin, or bft, while non-enterotoxigenic strains (nontoxigenic B. fragilis [NTBF]) are considered normal healthy commensal bacteria. As we recovered a predominance of B. fragilis from our patients with polyps, we hypothesized that these isolates may express bft. To assess this, we selected 39 different B. fragilis isolates recovered from 10 different patients for colony PCR with gene-specific primers for the bft gene. Out of these 10 patients, 3 had SSP lesions, 4 had TAP lesions, and 3 patients were PF. All the isolates grew in a similar fashion in blood media (Figure S4I) and PCR revealed that none of the selected patients with polyps were colonized with ETBF except for one TAP patient and 2 PF patients (Figure S4J) (bft+ isolates were removed from their respective groups and re-categorized and analyzed as ETBF+ isolates). Although only a few isolates were tested across a handful of people, our data potentially hint to the fact that a proximal colon environment with polyps does not systematically select for ETBF+ strains. This remains to be confirmed in larger cohorts.
We wanted to see if the increase in B. fragilis associated with polyp tissue correlated with host inflammatory response in the tissue. Using our cytokine data generated in Figure 1, we examined whether there was a correlation with the amount of NTBF isolates recovered. Indeed, we observed a significantly positive correlation between the abundance of NTBF recovered and IL-12p40 measured in the host’s tissue (p = 0.0002) (Figure 2A) and a non-significant negative correlation between the abundance of NTBF recovered and IL-10 measured in the same host’s tissue (p = 0.1862) (Figure 2B). We also noticed a significant correlation between the presence of NTBF isolates in TAP and the size of the polyps (p = 0.0191) (Figure 2C).
Figure 2. NTBF recovery correlates with inflammation in host tissue.
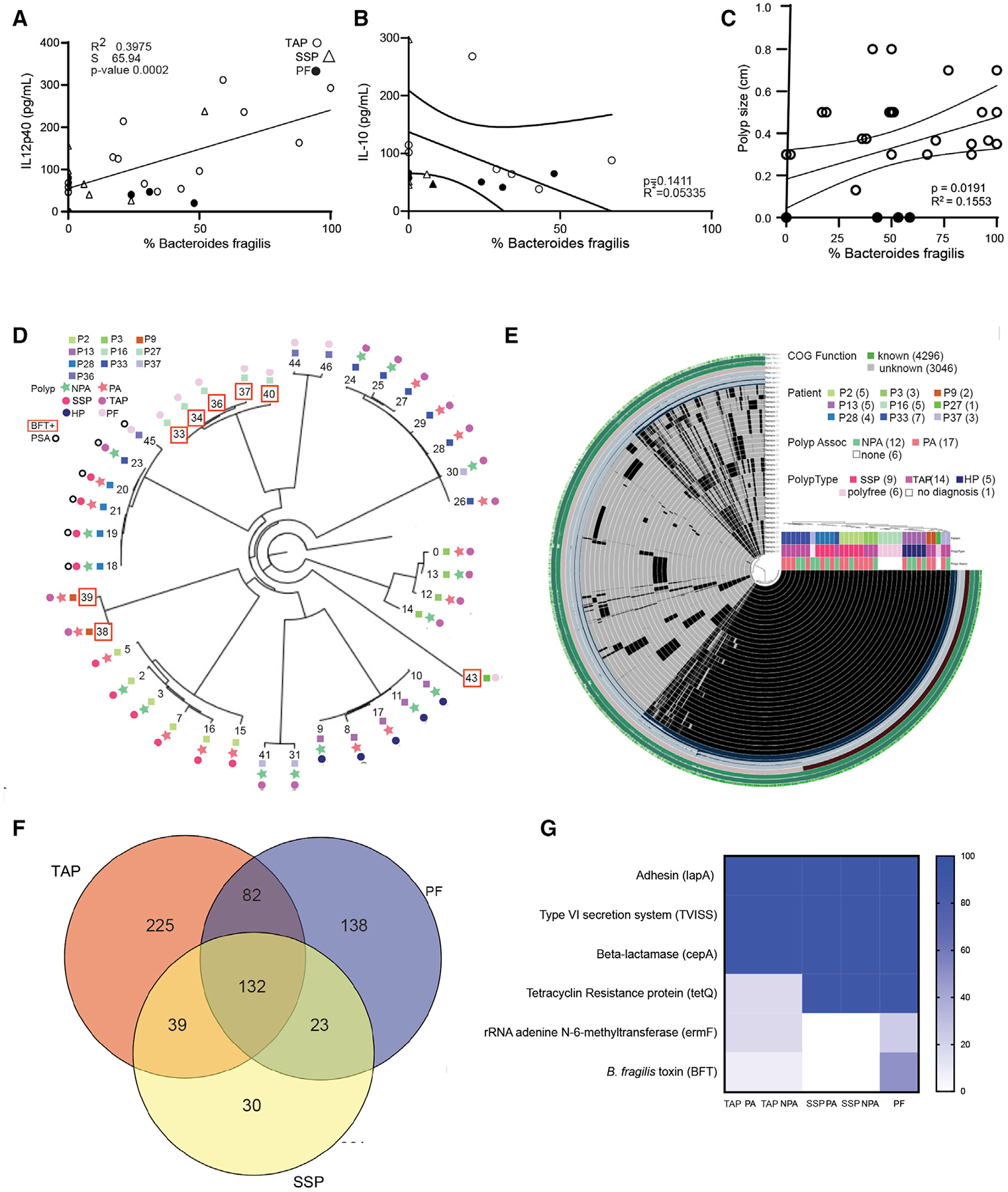
(A and B) Correlation between mucosal IL-12p40 and IL-10 concentration (pg/ml) and abundance of B. fragilis isolated in vitro. Statistics are Pearson correlation coefficient of all data points with each data point representing a biological replicate of NTBF isolate recovered from seven PA and seven NPA TAP biopsies (o), 4 PA and 3 NPA SSP biopsies (Δ), and 7 PF biopsies (•).
(C) Correlation between polyp size and abundance of NTBF isolated. Statistics are Pearson correlation coefficient of all data points, each data point representing a biological replicate of NTBF isolate recovered from TAP (o) or PF (•) biopsy tissue.
(D) Whole genome assembly and reconstitution of 39 B. fragilis clinical isolates from, 9 PA and 76 NPA biopsies from 4 TAP patients (Patient 3, 9, 33, and 37), 6 PA and 3 NPA isolated from 2 SSP patients (Patient 2 and 28), 9 PF isolates from 3 different patients (Patient 16, 27, and 36), and 2 PA and 3 NPA isolates from an HP (Patient 13). Patients positive for PSA gene (o) and bft (peach open box). Each patient involved in the analysis is labeled with a specific color on the tree, the tree also features the 15 PA (dark green star) and 10 NPA (Light green star) isolates recovered from 4 TAP (blue dot), 2 SSP (peach dot), 1 HP (black dot), and 9 PF (grey dot).
(E) Genome reconstitution of the B. fragilis clinical isolates featuring PA (green box) versus NPA (peach box) from TAP (blue box), SSP (pink box), HP (black box), and PF (grey box).
(F) Venn diagram of single nucleotide variants (SNVs) in the genome of the PA and NPA from TAP, SSP, and PF patients.
(G) Heatmap representing the proportion of virulence genes present in the genome of PA and NPA B. fragilis isolates from TAP, SSP, and PF patients.
The NTBF strain NCTC9343 has been shown to induce IL-10 through the recognition of capsular polysaccharide A (PSA) genes by TLR2 (Mazmanian et al., 2008). Based on this premise, we hypothesized that the NTBF strains isolated from PF patients were enriched in PSA compared with the NTBF isolated from patients with polyps. We performed whole genome sequencing (WGS) and a pangenome analysis of the same subsets of isolates that we had selected for colony PCR. Our analysis confirmed the lack of bft in our isolates, but it also revealed that most isolates had an incomplete PSA gene, making it impossible to express PSA (Figure 2D). However, the NTBF isolates from one of the 2 SSA patients were PSA-positive but were also able to produce robust quantities of pro-inflammatory IL-12p40 and IL-1β. Thus, the absence of PSA could not explain the positive correlation between NTBF and inflammatory IL-12p40.
Whole genome sequencing of isolates reveals presence of genes associated with virulence in NTBF isolates from patients with polyps
Pangenome analysis detected a total of 664 single nucleotide variant (SNV) positions with both major and minor alleles and identified a core genome that was 99.9% homologous between all the isolates (Figure 2E). Among the subjects within TAP, SSP, and PF groups, we identified one or more isolates with SNVs unique to their respective groups (Figure 2D). Figure 2F shows a Venn diagram of the respective overlapping and unique SNV counts. We found variants unique to the PF (n = 138 SNVs), SSP only (n = 30 SNVs), TAP only (n = 225 SNVs), and SSP TAP only (n = 39 SNVs) groups as well as B. fragilis gene annotations and allele counts among the isolates analyzed (Figure 2F). The pangenome analysis further showed that there was no statistical difference between many of the genes associated with virulence (Wexler, 2007) among the NTBF isolates (Figure 2G). For instance, it has been reported that B. fragilis’ PSA induces the anti-inflammatory function of Tregs through TLR2 signaling, which could be a mechanism that allows the persistence of B. fragilis on mucosal surfaces of the gut (Round et al., 2011). Livanis et al. also showed that B. fragilis expresses a Type 6 secretion system (T6SS) that deploys toxins able to antagonize other species in the gut (Chatzidaki-Livanis et al., 2016). All the B. fragilis isolates in this study carried genes encoding PSA and T6SS in their genomes. They also carried adhesins that improve attachment to the gut mucosa (Wexler, 2007; Figure 2G), and the presence of various antibiotic resistance genes (cepA, tetQ, and ermF) that can enable B. fragilis isolates to resist clearance by antibiotics (Figure 2G).
NTBF isolates from patients with polyps induce distinct cytokine signatures when co-cultured with monocytes
NTBF has been shown to induce IL-10, which prevents colonization by enterotoxigenic B. fragilis strains (Casterline et al., 2017) and ameliorates disease in animal models of IBD and CRC (Casterline et al., 2017). Thus, we wanted to assess the immunogenic and inflammatory potential of the NTBF isolates in vitro by co-culturing cell-free supernatants of saturated NTBF cultures with a monocytic cell line (THP-1) and measure a panel of cytokines involved in cancer pathways (Grivennikov et al., 2012; Ning et al., 2011; Masucci et al., 2019; Andersen et al., 2013). We found that NTBF isolates from patients with polyps induced significantly more pro-inflammatory responses than NTBF isolates from PF patients regardless of polyp type (Figures 3A–3C). Specifically, NTBF derived from TAP (only PA) and SSP (PA and NPA) tissues induced significantly more IL-12p40 than isolates from PF patients, whereas NTBF derived from TAP (both PA and NPA) and SSP (PA and NPA) tissues induced significantly more IL-8 and IL-1β compared with PF isolates (Figure 3A). Despite their very low level of production of IL-1β, IL8, and IL-12p40, NTBF from PF patients were able to stimulate IL-10 secretion from THP-1 cells, albeit significantly less than TAP (PA & NPA) and SSP (NPA). Interestingly, NTBF from SSP (PA) tissue induced a significantly weaker IL-10 response than the other isolates obtained from polyp tissues (Figure 3D). The ratio of IL-10 to IL-12p40 may be an important factor to consider as some studies have reported that early stages of CRC tendto present with highlevelsofIL-12p40 and lower levels of IL-10 (Mager et al., 2016). We calculated the ratio of IL-10 to IL-12p40 and found that NTBF from PF patients induced a clear IL-10-dominant tolerogenic profile, while NTBF isolates from all other groups promoted an IL-12p40 dominant response (Figure 3E). Interestingly, we observed that the ratio of IL-10 to IL-12p40 in vitro when co-culturing TAP or SSP NTBF supernatants with THP1 cell lines mirrored the same cytokine ratio measured in host tissues (Figure 3F). The ratio of IL-12p40 to IL-10 was also noticeably skewed toward IL-12p40 in both the patient tissue and the isolates from SSP compared with TAP (Figures 3E and 3F). Taken together, we found that NTBF isolated from patients with polyp induced a pro-inflammatory response, while NTBF from PF patients had a more tolerogenic profile and a higher ratio of IL-10. Differences in the cytokine profile and magnitude of response also seems to differentiate isolates from SSP and TAP.
Figure 3. NTBF isolates from patients with polyps induce inflammation in vitro and signal through TLR4.
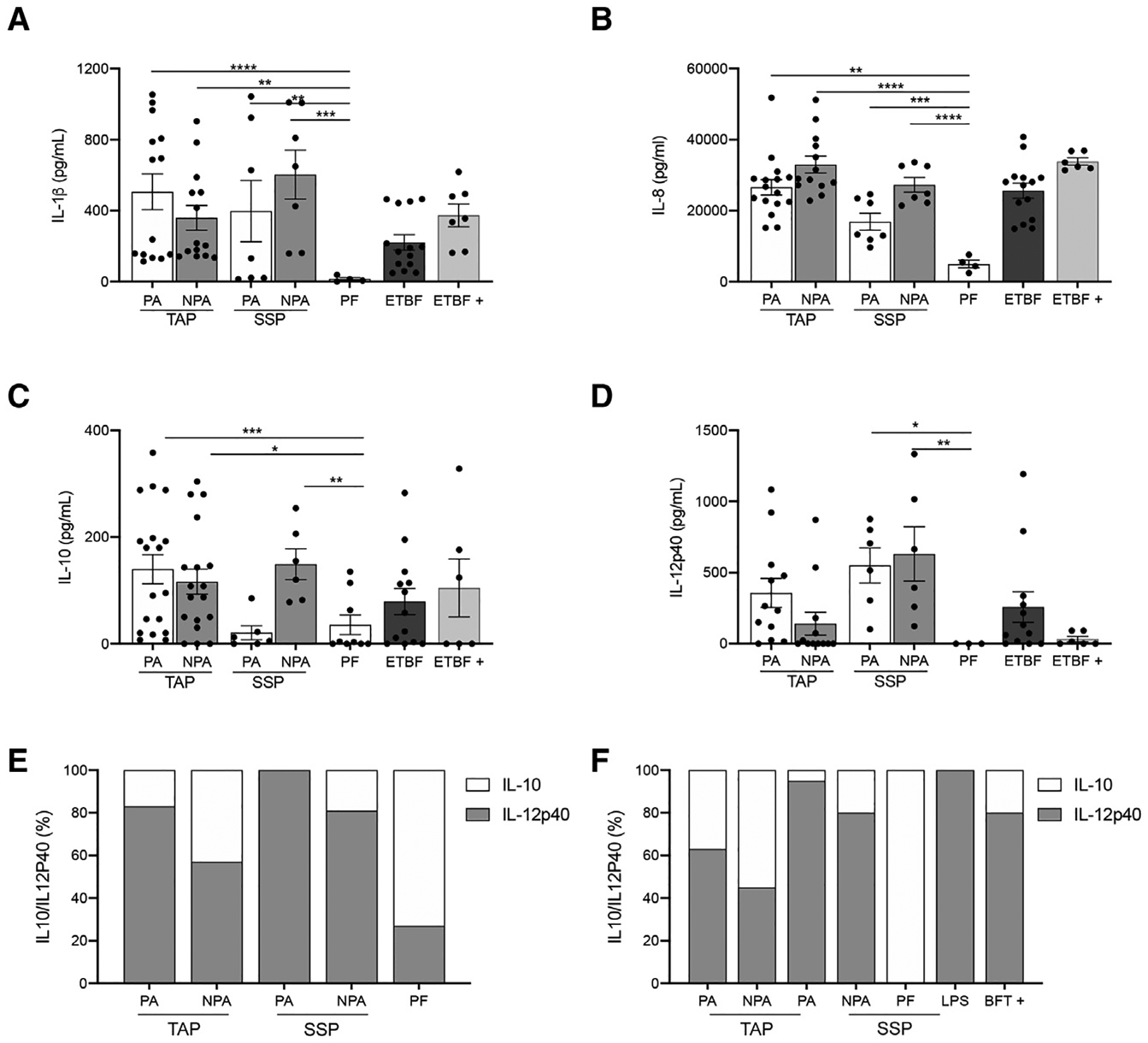
(A–D) Levels of cytokines detected from co-culturing Bacteroides fragilis supernatants with monocytic cell line by ELISA. Data are represented as the mean ± SEM of 3–4 independent experiments including 16 PA and 16 NPA isolates from 4 different TAP patients, 8 PA and 8 NPA clinical isolates from 2 different SSP patients, and 4 clinical isolates from 1 PF patient. ETBF+ represents the bft positive study isolates that include 4 PA and 4 NPA isolates from 1 TAP patient and 8 isolates from 2 PF patients. ETBF represents a control bft positive strain from ATCC. (A–D), *p < 0.05. ***p < 0.001. One-way ANOVA with multiple comparisons.
(E) Ratio of IL10/IL-12p40 expression detected from co-culturing B. fragilis supernatants with monocytic cell line by ELISA.
(F) Ratio of IL10/IL-12p40 expression in biopsy tissue samples detected by ELISA. See also Figure S4.
NTBF isolates from patients with polyps activate TLR4 and express genes associated with LPS biosynthesis
As transformed tissue grows, changes in the production of tissue factors and/or secreted cellular metabolites may alter the local microenvironment. This changing landscape may make it difficult for some commensal bacteria to survive. However, other microbes may turn on new genetic and metabolic programs that allow for their survival, while being detrimental to the host. In many cases these changes impact the cell surface of the bacteria as they try and evade the immune system. Traditionally, innate responses against B. fragilis have been shown to involve TLR2 sensing of polysaccharide capsid and other lipoproteins12. Therefore, we hypothesized that we would see differences in TLR2 activation between NTBF isolated from patients with and without polyps. Using aliquots of cell-free supernatants generated for cytokine studies, we assessed TLR2 activation using HEK cells transfected with an NF-κB-dependent firefly luciferase reporter. Despite the differences in cytokine production observed between isolates from patients with and without polyps, we observed that all the NTBF isolates were able to activate TLR2 in a similar fashion (Figure 4A). In addition to polysaccharides and lipoproteins that can be detected via TLR2, B. fragilis also makes a unique LPS that has been shown to bind weakly to TLR4 (Alhawi et al., 2009). Using cells transfected with TLR4/MD2- and NF-κB-dependent firefly luciferase reporter, the NTBF isolates from patients with polyps were able to significantly activate TLR4 to a much greater extent than NTBF isolates from PF patients (Figure 4B).
Figure 4. Persistent NTBF isolates from patients with polyps are proteomically distinct and enriched with LPS biosynthesis genes.
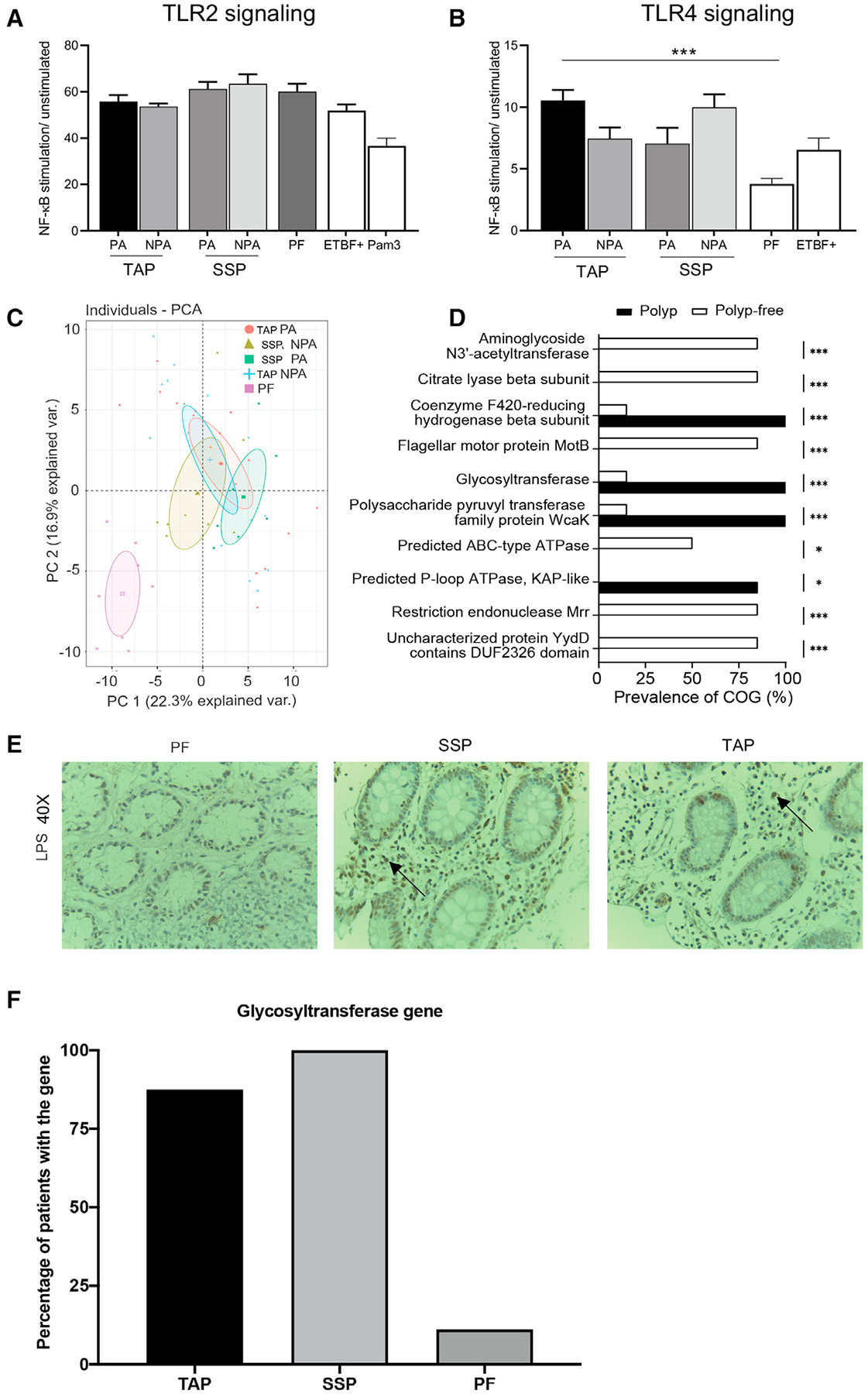
(A) Fold change NF-kB stimulation of HEK293 cells transfected with TLR2 and infected with supernatants from B. fragilis isolates relative to unstimulated control in relative light unit (RLU). Results are means ± SEM from 3 independent experiments including 16 PA and 16 NPA isolates from 4 different TAP patients, 8 PA and 8 NPA clinical isolates from 2 different SSP patients, and 4 clinical isolates from 1-PF patient. ETBF+ represents the bft positive study isolates that include 4 PA and 4 NPA isolates from 1 TAP patient and 8 isolates from 2 PF patients (by one-way ANOVA with multiple comparisons, *p < 0.05).
(B) Fold NF-kB stimulation of HEK293 cells transfected with TLR4 and infected with B. fragilis isolates supernatants relative to unstimulated control. Results are means ± SEM from 3 independent experiments (by one-way ANOVA with multiple comparisons, *p < 0.05).
(C) Principal component analysis based on peptide mass fingerprint profile of various NTBF B. fragilis isolates. Data are mean ± SEM of ± of 2–3 independent experiments with n = 4–6. Each dot represents the average of technical replicates. Differences are maintained after growing isolates multiple times.
(D) Prevalence of clusters of orthologous proteins based on comparative genome analysis between isolates from patients with polyps and polyp-free patients.
(E) LPS expression revealed by immunohistochemistry (IHC) in CRC patients’ biopsies (40×). Comparison of PF human normal colon tissue section and NTBF positive SSP and TAP tumor sections. Representative picture for 3–4 sections from one biopsy from each type of tissue (PF, SSP, or TAP), no quantitative data available due to tissue restriction.
(F) Percentage of glycosyltransferase gene present in the host gut tissue based on PCR performed on 7 patients with TAP, 6 patients with SSP, and 9-PF patients. See also Figure S4.
Proteomic differences may account for the differential stimulation of TLR4 and lead to the pro- inflammatory phenotype of the TAP and SSP NTBF isolates. Using a MALDI TOF-based approach that highlights strain-level phenotypic differences in bacteria recently published by our group (Chac et al., 2020, 2021), we observed that the NTBF isolates from patients with polyps had significantly different proteomic profiles compared with the NTBF isolates from PF patients (Figure 4C). To determine which genes encoded for these phenotypic differences, we performed an untargeted comparative genome analysis of all the NTBF isolates. We observed 4 top hits that were significantly enriched in the isolates from patients with polyps compared with the PF isolates: a gene encoding for a coenzyme F420-reducing hydrogenase, a gene encoding for a polysaccharide pyruvyl-transferase family protein, a gene encoding for a predicted P-loop ATPase, and lastly a gene encoding for a glycosyltransferase involved in LPS biosynthesis (FDR-corrected p < 0.001 from a Rao score test) (Figure 4D). Knowing that TLR4 recognizes LPS as a microbial ligand (Park and Lee, 2013; Reife et al., 2006) and that LPS can signal through TLR4 to induce both IL-12p40 and IL-1β (Bodnar, 2002), a difference in LPS biosynthesis could very possibly explain the phenotypic differences observed between the NTBF isolates. Moreover, a qualitative assessment of LPS expression in the host tissue showed stronger staining in the TAP and SSP tissue sections compared with the PF mucosal tissue (Figures 4E, S5A, and S5B). This observation supports the findings of Nejman et al. who reported that the human tumor microbiome is enriched in LPS and composed of tumor type-specific intracellular bacteria (Nejman et al., 2020). To confirm that B. fragilis’ LPS was indeed responsible for TLR4 signaling and pro-inflammation, we isolated LPS from B. fragilis isolates recovered from patients with polyps and saw that they signaled through TLR4 reporter cells in a concentration-dependent fashion (Figure S5C). Lastly, using the data from whole genome sequencing, we designed conventional primers to test for the presence of the glycosyltransferase gene in the colon biopsies of patients with polyps. We found that the glycosyltransferase gene was present in 87.5% of the patients with TAP (8 out 9 patients) and 100% of the patients with SSP (6 out of 6 patients) versus only 11.1% of the PF patients (3 out of 9) (Figure S5D).
DISCUSSION
These data suggest that the precancerous colon polyp microenvironment is enriched with NTBF that stimulate TLR4, rather than TLR2, leading to the induction of pro-inflammatory cytokines. Using a genomic approach, we identified 4 genes in NTBF isolates from persons with polyps. Further, the gene corresponding to a glycosyltransferase involved in LPS biosynthesis was significantly enriched in the polyp tissue but not in the tissue from PF patients. Thus, it is possible that during the early stages of polyp formation, a dysbiotic and inflamed gut microenvironment allows for the selection or colonization by NTBF enriched in LPS biosynthesis genes. These isolates can activate NF-kB via TLR4 and induce inflammation, which may contribute directly to the growth of colon tumors or may modify the microenvironment making it more hospitable for colonization by oncogenic species, such as enterotoxigenic B. fragilis. While the enterotoxigenic strain of B. fragilis has already been correlated to colon cancer development in various studies (DeJea et al., 2018; Chung et al., 2018; Tomkovich et al., 2019), our study distinguishes itself from this literature as it has discovered a potential role played by the non-toxigenic strain of B. fragilis in the early stages of colon cancer progression.
In conclusion, this study is the first, to our knowledge, to characterize the composition of the mucosa associated with precancerous colon polyps by sub-type using 16S sequencing and a culture-based approach to examine the role of human mucosal NTBF isolates in the early stages of colorectal carcinogenesis. We found that there is a distinct microbial composition in the mucosa adjacent (PA) to and near but non-adjacent (NPA) to pre-neoplastic colonic polyps compared with the mucosa of patients who are PF. While we were able to recover NTBF from PF biopsies, we recovered a much higher number from patients with polyps. Despite over 99% genetic homology constituting the core genome of B. fragilis, the NTBF strains from patients with polyps were significantly enriched in LPS biosynthesis genes. They were also proteomically distinct, activated TLR4 and induced a pro-inflammatory cytokine response compared with PF NTBF isolates. Taken together, these data suggest a role for commensal NTBF in the early stages of neoplasia in which either NTBF with LPS biosynthesis genes colonizes individuals and predisposes them to polyps or whereby adaptation to the polyp microenvironment selects NTBF strains that can increase or modify its LPS, activate TLR4, and promote local inflammation that can enhance polyp growth or potentially make the tissue more permissive to oncogenic members of the microbiome such as ETBF, pks+ E. Coli, or Fusobacterium nucleatum. We believe that the findings of this study provide fundamental insight into the mechanisms underlying the microbiota’s capacity to induce CRC and are a step forward in the discovery of potential microbiome-based diagnostic and therapeutic targets against CRC.
Limitations of the study
The authors acknowledge that the complexity of the human biopsy sample could lead to false negatives for the gr25 gene. Future studies will utilize real-time PCR and a reference gene to achieve more quantitative results.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, William DePaolo (wdepaolo@medicine.washington.edu).
Materials availability
Bacterial strains used in this study are available upon request.
Data and code availability
Whole generation sequencing data and 16S data have been deposited at zenodo (https://zenodo.org/record/5090347) and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Ki67 | Thermo | Cat# RM9106, RRID:AB_2341197 |
| Anti-IL-17 | Santa Cruz | Cat# SC-7927, RRID:AB_2124997 |
| Anti-LPS core | Hyacult Biotech | Cat# WN1 222–5, RRID:AB_2750644 |
| Anti-MPO | DAKO | Cat# A0398 |
| Anti-CCL3 | Pierce | Cat# OA1653721, RRID: AB_1956667 |
| Bacterial and virus strains | ||
| B fragilis NCTC9343 | ATCC | Cat# 25285 |
| B. thetaiotamicron | ATCC | Cat# 29148 |
| Biological samples | ||
| B fragilis clinical isolates | This study | N/A |
| Proximal colon biopsies | This study | N/A |
| Critical commercial assays | ||
| Human IL-8 ELISA | Becton Dickinson | Cat # 555244 |
| Human IL-10 ELISA | Becton Dickinson | Cat # 555157 |
| Human IL-12p40 ELISA | Becton Dickinson | Cat # 555171 |
| Human IL-1b ELISA | Invitrogen | Cat # KHC001 |
| Nextera DNA Flex Library Prep kits | Illumina | Cat# 20018705 |
| bacterial DNA extraction kit | Qiagen | Cat#158567 |
| dual luciferase assay reporter system | Promega | Cat # E2920 |
| Deposited data | ||
| 16S Phylum level LDA analysis of biopsies | This study | https://doi.org/10.5281/zenodo.5090347 |
| 16S DNA fastQ files | This study | https://doi.org/10.5281/zenodo.5090347 |
| Whole genome sequencing of B fragilis isolates | This study | https://doi.org/10.5281/zenodo.5090347 |
| Whole genome sequencing fastQ files | This study | https://doi.org/10.5281/zenodo.5090347 |
| Experimental models: Cell lines | ||
| Human: HEK293 cells | ATCC | CRL-1573 |
| Human: THP-1 cells | ATCC | TIB-202 |
| Experimental models: Organisms/strains | ||
| Human colon biopsies | This study | N/A |
| B. fragilis clinical isolates | This study | N/A |
| Oligonucleotides | ||
| Primer bft Forward, GCGAACTCGGTTTAPTGCAGT Reverse, GTTGTAPGACATCCCACTGGC26 | DeJea et al., 2018 | N/A |
| Primer glycosyltransferase Forward, ACCTAAGAGCGCACGGAA and Reverse, AGGTCGTCCGAATAPGCCA. | This paper | N/A |
| Software and algorithms | ||
| Comparative genomics | Galperin et al 2015. https://github.com/StoreyLab/qvalue | N/A |
| ASNP identification | Seemann, 2020 | N/A |
| CLC genomics suite | Qiagen | N/A |
| LDA | https://huttenhower.sph.harvard.edu/galaxy/] | N/A |
| Anvio | Eren et al., 2015. http://merenlab.org/software/anvio/ | N/A |
All original code has been deposited at zenodo and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human subjects
Between May 2017 and September 2018, 40 patients (mean age 64 ± 9 years old) were enrolled after obtaining informed consent through the Gastroenterology department of the University of Washington, Seattle Washington, (IRB # 34095A). Exclusion criteria included age <50, body mass index (BMI) > 30 kg/m2, any use of antibiotics within the past 3 months, any active inflammation, and inability to sign informed consent. All colon mucosa samples were collected by designated gastroenterologist Dr. Cynthia Ko and reviewed by a pathologist with expertise in GI pathology. Polyp associated mucosa (PA) and healthy non-polyp associated (NPA) tissue located about 10 cm away from the lesion were obtained from the same subject collected by endoscopy biopsy forceps, generating a total of 240 samples.
Cell lines
HEK293 cells used for transfectant experiments were purchased through ATCC (CRL-1573) and are derived from female embryonic tissue. HEK cells were grown in DMEM media containing 10% heat inactivated FBS, and Penicillin/Streptomycin at 37C. We have not authenticated our stock of HEK cells however, we routinely check the ATCC web pages as this resource houses the most up to date information on cell line misidentification and works closely with cell line repositories.
THP-1 cells are a human monocytic cell line derived from a male donor and purchased through ATCC (TIB-202). THP-1 cells were grown in RPMI containing 10% heat inactivated FBS, 0.05mM 2-mercaptoethanol and Penicillin/Streptomycin at 37C. THP-1 cells have not been authenticated, however the presence of MD2. CD14, MyD88, TLR2 and TLR4 cells were confirmed by PCR.
Bacterial strains
B. fragilis NCTC9343 was purchased through ATCC (25285); B. thetaiotaomicron strain VPI5482 was purchased through ATCC (29148). ETBF strain 86-5443-2-2 was donated by Dr. Cynthia Sears. This strain was originally isolated from a piglet. Δbft2 ETBF was derived from this strain by the Sears’ group and kindly donated for this study. Commercial strains were derived from frozen stocks and grown anaerobically at 37°C in pre-reduced chopped meat media (ATCC medium 1490). Authentication was performed by MALDI-TOF and presence of bft was confirmed by PCR.
Frozen stocks of B. fragilis isolates identified from patient biopsies were made in skim milk media and stored at −80C. Upon thawing, the isolates were grown anaerobically in pre-reduced modified chop meat media (ATCC medium 1490) and incubated at 37°C for 24 hours. Authentication of isolates was performed using MALDI-TOF and presence of bft was confirmed using PCR.
METHOD DETAILS
Microbiome culture
All collected biopsies were placed in cryovials (Thermo Fisher Scientific Inc. cat# 368632, USA) filled with anaerobic media (Anaerobic systems, Cat# AS-916, USA) and processed fresh in the 30 minutes following collection. Following immediate and thorough homogenization with a homogenizing pestle (Thermo Fisher Scientific Inc. Cat# K749521-1590, USA) in an anaerobic chamber and under sterile conditions, samples were plated on non-selective Tryptic Soy Agar containing 5% sheep blood (Anaerobic systems, Cat# AS-542, USA) for bacterial growth. The plates were kept under anaerobic conditions (90% N2, 5% CO2, 5% H2) at 37°C for 2 days. Colonies cultured for 48 hours were expanded in Tryptic Soy Broth + 5% defibrinated sheep blood after identification, then transferred to 50% skim milk stock (BD Difco Skim Milk, Cat# 232100, USA), and preserved at −80°C until analyzed.
Identification of bacterial cultures by MALDI-TOF
For identification, bacteria were processed according to manufacturer’s indirect protein extraction method. Briefly, Bacterial Test Standard (BTS) (Bruker Daltonics, Cat# 8255343, Germany) was used for external calibration of MALDI-TOF MS. For sample analysis, individual colonies of clinical isolates were individually transferred from agar plate cultures to a MALDI MSP 48 target polished steel BC target plate (Bruker Daltonics, Cat# 8281817, Germany), using a sterile wooden transfer device (Puritan, Cat# 25-28107, USA) then air dried. Samples were then treated with 1 μL of a 70% formic acid solution (Sigma, Cat# F0507, Germany) for protein extraction, then air dried again. Samples were also treated with 1 μL of a-cyano-4-hydroxycinnamic acid matrix solution (α-CHCA, Bruker Daltonics, Cat# 8255344, Germany), air dried, and Peptide Mass Fingerprints (PMF) were analyzed in Microflex LT MALDI-TOF mass spectrometer (Bruker Daltonics, Germany). Reads were analyzed with the following settings: linear positive ion mode, N 2 laser, λ = 355 nm, pulse duration: 150 ns, laser frequency: 200 Hz. All spectra were recorded over the range m/z 2000–20,000.
For protein extraction method of B. fragilis isolates, single B. fragilis colonies grown on blood agar (Anaerobe systems, Cat # AS-542, USA) were selected and added to 300 ml of HPLC- grade water in a 1.5 ml Eppendorf tube and then mixed thoroughly with 900 μl of 100% Ethanol. After centrifugation at 13000 rpm for 2 min twice, pellets were dried at room temperature for 5 minutes then they were directly mixed with equal volumes of 70% formic acid and acetonitrile (20–40 ul, depending on pellet size) for protein extraction. After centrifugation at 13000 rpm for 2 minutes, 1 μl of protein extract was spotted on a MALDI MSP 96 target polished steel BC plate (Bruker Daltonics, Cat# 8280800, Germany) in four technical replicates, air-dried, and overlaid with 1 μl of matrix solution. Target plate was placed in the MALDI TOF Biotyper.
MALDI TOF data analysis
Raw spectra text files were analyzed using the R package, MALDIquant(Gibb and Strimmer, 2012). The raw data were trimmed to a spectra range of 3,000 to 15,000 m/z. The spectra intensities were then square-root transformed and smoothed using the Savitzky-Golay algorithm(Gorry, 1990). Baseline noise was removed using the statistics-sensitive non-linear iterative peak clipping (or SNIP) algorithm with 100 iterations. The data were then normalized using total ion current (or TIC) calibration, which sets the total intensity to 1. Multiple spectra within the same analysis were aligned to the same x-axis using the Lowess warping method, a signal-to-noise ratio of 3, and a tolerance of 0.001. Peaks were detected from the average of at least 4 technical replicates using median absolute deviation. Principal components analyses and hierarchical clustering were also performed in R using the base stats package. Hierarchical clustering was performed on a calculated Euclidean distance matrix using Ward’s method.
16S rRNA Sequencing on selected colon mucosal biopsies
Library preparation and sequencing 16S sequencing libraries prepared by QIAseq 16S/ITS screening panel (QIAGEN Cat# 333822, USA). Extracted DNA was diluted into 1ng/uL. Seven 16S regions (V1V2, V2V3, V3V4, V4V5, V5V7, V7V9 and ITS) were captured and amplified in three different primer panel pools for each sample, according to manufacturer’s protocol. For each of the primer panel pool, 2uL of the diluted DNA was used as input and incubated at 95°C for 2 minutes, then amplified for 20 PCR cycles of 95°C for 30s, 50°C for 30s and 72°C for 2 minutes, with a final extension at 72°C for 7 minutes using an UCP Multiplex PCR kit (QIAGEN Cat# 206742, USA). Reactions from the same samples were pooled and the pooled intermediate products was cleaned up twice using 1.1X QIAseq beads (QIAGEN, Cat# 1107149, USA) to remove unused PCR primers. A final PCR reaction was done using UCP Multiplex PCR kit and QIAseq 16S/ITS indices (QIAGEN Cat# 333822, USA) to incorporate sample indices and sequencing adapters. The reaction mix was incubated at 95°C for 2 minutes and 14 cycles of 95°C for 30s, 50°C for 30s and 72°C for 2 minutes, with a final extension at 72°C for 7 minutes. The final PCR product was cleaned up using 0.9X QIAseq beads (QIAGEN, Cat# 1107149), and the cleaned-up libraries were inspected on an Agilent Tapestation (Agilent Cat# G2991AA, USA).
The 16S libraries were sequenced on an Illumina MiSeq for 1.5 M 2x276bp reads per sample.
Data analysis 16S
16S screening panel sequencing data analysis was carried out using the QIAGEN CLC Genomics Workbench (version 11.0) and the Data QC and OTU Clustering workflow from the Microbial Genomics Pro Suite Module (version 4.0). Briefly, raw reads were demultiplexed and grouped into different 16S/ITS regions (e.g., V1V2, V3V4, and ITS) using the QIAseq 16S/ITS Demultiplexer tool which classified reads into different regions using the phased 16S primer sequences. Operational taxonomic clustering for each 16S/ITS region was performed using the OTU Clustering tool, where the demultiplexed reads are aligned against the SILVA 16S database using a similarity percentage parameter at 97%, to create an alignment score for each OTU.
16S LDA Analysis
For each sequenced sample, the proportion of 16S reads was calculated for Actinobacteria, Bacteroidetes, Firmicutes, Fusobacteria, Proteobacteria and Thermus. The centered log ratio (clr) of these proportions was calculated using the ‘compositions’ R package [https://cran.r-project.org/web/packages/compositions/index.html]. LDA analyses were performed with one sequenced sample from each subject using the ‘MASS’ R package [https://cran.r-project.org/web/packages/MASS/]. The clr proportions for Actinobacteria, Bacteroidetes, Fusobacteria, Proteobacteria and Thermus were included in analysis after analysis for collinearity. The class labels for TAP, SSP, and PF were assigned to each sample. PA and NPA samples were analyzed separately with the common PF control set. The default R plot() function was used on LDA output to generate LD1 and LD2 plots. To generate Lefse plots, the sample phylum proportions with polyp category labels were written to a csv file using R. These csv files were then uploaded to the Lefse Galaxy tool [https://huttenhower.sph.harvard.edu/galaxy/] supported by the Huttenhower Lab.
bft PCR
A total of 3–5 biological replicates from each B. fragilis isolate were selected for colony PCR detection of bft using the following bft Forward and Reverse primers (281 bp product): bft-F
GCGAACTCGGTTTAPTGCAGT and bft-R GTTGTAPGACATCCCACTGGC(Dejea et al., 2018).
Glycosyltransferase PCR
DNA was extracted from colonic biopsies from 9 patients with TAP, 6 patients with SSP, and 9 PF patients and selected for PCR detection of glycosyltransferase (GR) using the following GR Forward and Reverse primers (512 bp product): GR-F ACCTAAGAGCGCACGGAA and GR-R AGGTCGTCCGAATAPGCCA.
Next Generation Sequencing
5 mL Tryptic soy broth (TSB) (BD BactoTM Cat # 6357914) + 5% Defibrinated Sheep blood (Remel, Cat # R54016, USA) liquid cultures from stock of clinical isolates were prepared and incubated under anaerobic conditions (90% N2, 5% CO2, 5% H2) at 37°C for 48 hrs. Then 1 mL of each isolate liquid culture was transferred to an Eppendorf tube and centrifuged at 13000 rpm for 60 seconds to collect bacteria. Total bacterial genome DNA of Bacteroides fragilis isolates was extracted, referring to steps in the instructions of the bacterial DNA extraction kit (Puregene yeast/Bact kit B, Qiagen, Cat#158567, USA). Extracted DNA samples were then quantified using a Qubit fluorometer (Thermo Fisher Scientific Inc., Cat# Q33238, USA) and library prepared using Illumina’s Nextera DNA Flex Library Prep kits (Cat# 20018705, USA) and following the manufacturer recommendation for the protocol. Only samples with a minimum of 100 ng DNA inputs were included. Pooled libraries were then quantified using the Qubit and diluted with Resuspension Buffer (Illumina, Cat# 20018705, USA) to 2 nM, then loaded on a NextSeq for sequencing with 50X sequencing depth.
Preprocessing and assembly of isolate sequencing data
Preprocessing and assembly were performed using existing Anvi’o pipelines (http://merenlab.org/software/anvio/) (Li et al., 2015), (Eren et al., 2015), (Eren et al., 2013). In brief, the raw FASTQ files received from sequencing minimum contig size of 1250 bps (Li et al., 2015). For each sample, the filtered reads were then realigned to the assembled contigs to yield coverage profiles which were used together with the assembled contigs to create the Anvi’o contig and profile databases. Gene-calling was performed by identifying open reading frames with Prodigal(Hyatt et al., 2010) and basic functional annotation was performed by using the hidden Markov models included with Anvi’o’s “run-hmms” function. Additionally, taxonomy was assigned to individual genes using Centrifuge(Kim et al., 2016). All the processed data was then used to manually refine the assembled contigs in the Anvi’o interactive interface (“anvi-interactive”) by selecting contigs with homogeneous coverage and taxonomy. We removed contigs displaying bovine or ovine origin as those likely came from contaminants like growth media. Completeness and redundancy for all assemblies was quantified by the presence of taxon-specific single copy core genes (SCGs). All manual assemblies showed larger than 99% completeness and less than 5% of redundancy/contamination. The resulting curated genomes and plasmids were stored in the Anvi’o contig databases.
Comparative genomics
To improve functional annotations identified genes in each curated contig were first annotated by searching for clusters of orthologous proteins (COGs) in the NCBI COG database using the DIAMOND aligner(Galperin et al., 2015). Curated contigs along with annotations were then used to assemble an Anvi’o genome storage and pangenome database(Delmont and Eren, 2018). For comparative purposes, the reference genome for Bacteroides fragilis (Refseq NC_003228.3) was also added to the database. Functional enrichment analysis was performed by using the “anvi-get-enriched-functions-perpan-group” script in Anvi’o. In brief, for each annotated COG a logistic regression was fitted with a COG presence indicator as the dependent variable and the tracked phenotype as the independent variable. The significance of the logistic regression was judged by Rao’s score test and false discovery rate was controlled by using the “qvalue” R package. (https://github.com/StoreyLab/qvalue)
Immunohistochemistry staining
Formalin-fixed, paraffin-embedded human tissue of colonoscopically resected polyps or healthy mucosa from patients recruited in the study were obtained and affixed to slides by the Pathology/Histology Department, University of Washington. Sections were dewaxed in xylol and rehydrated. The sections were then incubated in 1.0% H2O2, 0.1% NaN3 in TBS for 10 min to block endogenous peroxidase activity, then washed in three changes of PBS for 5 min each. Nonspecific antibody binding was inhibited by incubating the sections in 4% skim milk powder in TBS for 15 min, followed by a brief wash in TBS. The sections were then incubated with 10% normal (nonimmune) goat serum for 20 min. Isotype controls and primary antibodies mAbs were applied overnight (MPO Dako, Cat# A0398, 1:100 dilution), (Ki67, Thermo, Cat# RM9106, 1:200 dilution), (IL-17, Santa Cruz, Cat# SC-7927, 1:50 dilution), (CCL3, Pierce, Cat # OA1653721), and (Lipopolysaccharide Core, mAb WN1 222–5, HycultBiotech #HM6011, 1:300 dilution). Sections were washed with PBS for 5min and incubated with biotinylated secondary antibodies for 30 min. Slides were then incubated with 1 drop of High Sensitivity-HRP conjugates (Thermo Scientific™, Pierce™, Cat #11806824) for 30 min then washed in three changes of PBS for 5 min. Color was developed with DAB Solution before being washed and mounted for visualization under a microscope using a bright-field illumination. The section observed at 5x and 10x magnification and staining was estimated by comparing intensity with the unstained adjacent mucosa of the same specimen. Haemotoxylin and Eosin stains (H&E) staining was also used to evaluate tissue morphology.
Preparation of biopsy samples for cytokines analysis
Extraction of proteins from mechanically homogenized biopsy samples was achieved by adding RIPA lysis buffer (Thermo Scientific™, CAT # 8900, USA) and protease-inhibitors (Thermo Scientific, CAT #A32963). The samples were then incubated on ice for 30 min with occasional shaking. The insoluble components were removed via centrifugation at 15000 g for 30 min at 4C. The protein concentration was measured using the Bradford assay (BIO RAD CAT# 5000006, USA). Samples were normalized accordingly before cytokine quantification by ELISA.
Co-culture cell assays
5mL of chopped meat cultures (Anaerobe systems, Cat # AS-811, USA) from skim milk frozen stocks of clinical isolates were prepared and incubated under anaerobic conditions (90% N2, 5% CO2, 5% H2) at 37°C for 48 hrs. Then,1 mL of each isolate liquid culture was centrifuged at 13000 rpm for 30 min to pellet bacteria and collect secreted factors in bacterial supernatant. In parallel, 100,000 cells from THP1 human cell lines (ATCC®, TIB-202™) were seeded in a 96 well plate then co-cultured with the recovered bacterial supernatant at a concentration of 1:5, at 37°C in the CO2 incubator for 24 hours. When removed from the incubator, the plates were centrifuged and supernatants of co-stimulated THP1 were transferred to a new sterile 96 wells flat bottom plate at −20°C until used for ELISA assays.
Cytokine quantification by ELISA
50 μL ELISA Diluent was added to each well followed by 100 μL of standard or supernatant sample and incubated 2 hours at room temperature. The content of the wells was aspirated and washed 5 times. 100 μL of prepared Working Detector was added to each well and the wells were incubated 1 hour at room temperature. The content of the wells was aspirated and washed 7 times. Then 100 μL TMB One-Step Substrate Reagent was added to each well and incubated for 30 min at room temperature. 50 μL Stop Solution was finally added to each well and the plate was read at OD 450 nm. Cytokine level are reported per TAP and SSP lesions in Figure 3 and compared to the healthy PF tissue, and to both ETBF strains (black bar correspond to our B. fragilis isolate, grey bar to the ETBF+ positive control). (IL-12p40, IL-8, and IL-10 Human ELISA kits BD OptEIA™ CAT# 555171, 555244, and 555157, USA) (IL-1beta Human ELISA kit, Invitrogen™, CAT# KHC001).
TLR2 AND TLR4 Assays
HEK293 cells were plated in 96-well plates and transfected the following day with plasmids encoding human TLRs, NF-kB-dependent firefly luciferase reporter, and B-actin promoter- dependent Renilla luciferase reporter. In the case of human TLR4, 0.002 μg plasmid encoding human TLR4 was co-transfected with 0.0025 μg plasmid encoding human MD-2. For the human TLR2, 0.001 μg plasmid encoding TLR2 was co-transfected with 0.002 μg plasmid encoding human mCD14. At 18 to 20 h post-transfection, test wells were stimulated in duplicates for 4 h at 37°C with B. fragilis supernatants, which were suspended in Dulbecco’s modified Eagle medium (DMEM) containing 10% human serum. Luciferase activity was assayed using a dual luciferase assay reporter system (Promega, Madison, WI). NF-kB activity was measured as the ratio of NF-kB-dependent firefly luciferase activity to B-actin promoter-dependent Renilla luciferase activity, which served as an internal standard. The data were plotted as the fold difference between the NF- kB activity of the sample and that of the unstimulated control.
SNPS analysis
Fastq DNA sequence files were first trimmed and cleaned using trimmomatic (v0.39) (Bolger et al., 2014). Cleaned and trimmed fastq files were then aligned to the reference genome fasta file (NC_006347.1_Bacteroides_fragilis_YCH46.fasta, Genome_Size 5352665bp) and variant calling was performed using the Snippy pipeline (v4.0.2) to identify bacterial single nucleotide variants (SNVs) (Seemann, 2020). The identified SNVs were then analyzed for overlapping and private categories using the R package VennDiagram (v1.6.20) calculate.overlap() function and plotted using venn.diagram() function(Chen and Boutros, 2011). Nine (n=9) subjects with 34 total collective B. fragilis isolates were analyzed. The reported SNVs were detected in one or more isolates from the respective groups (TAP (n=4 subjects among 16 isolates), SSP (n=2 subjects among 9 isolates), PF (n=3 subjects among 9 isolates)). SNVs with more than one minor allele variant at a genome position (i.e., tri- allelic) were reported in the Venn analysis for each minor allele. SNV genome positions that are monomorphic within the selected isolates and different from the reference alignment genome are not reported. SNV genome positions were annotated using the NCBI Bacteroides fragilis YCH46 gene annotation file (NCBI, n.d.).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical details of experiments are found in the figures and figure legends. Data are expressed either as the mean value ± standard error of the mean (SEM) or as individual values. For cytokine and luciferase assays, one-way ANOVAs with multiple comparisons were used to analyze the data. To analyze the correlation between % B. fragilis and cytokines measured in the host tissue, we performed the Pearson Correlation Coefficient. 16S sequencing data was evaluated for statistical significance using analysis of molecular variance (AMOVA) and homogeneity of molecular variance (HOMOVA) tests of PCoAs on mothur v.1.36.130 (https://github.com/mothur/mothur/issues/639). Graphpad Prism V8 (San Diego, CA) was used for graphical and statistical analysis. LDA analysis was performed using QIIME2. p < 0.05 was considered statistically significant. Data analysis included all bacterial isolates and all patients that were recruited for the study.
Supplementary Material
Highlights.
Differences in microbiota found between patients with polyps and between polyp subtypes
NTBF is enriched in patients with polyps
NTBF from polyps is enriched for LPS biosynthesis genes, activates TLR4, and induces IL-12
Presence of LPS genes and elevated expression of LPS are found in polyp tissues
ACKNOWLEDGMENTS
We are thankful for the clinical efforts and assistance from Dr. Cynthia Ko and GICares (Department of Medicine, University of Washington) to enroll participants and are thankful to all the patients for their participation. We also thank the Department of Histology (University of Washington) for their assistance with the IHC staining. This study was supported by the UW start-up funds (R.W.D.), Funds from the Lynn M. & Michael D. Garvey Endowed Faculty Fund (R.W.D.), and Funds from the Rodger C. Haggitt Endowed Chair Fund (W.M.G.) (R01CA220004 and U01CA152756), and S.M.G. and C.D. were supported by a Washington Research Foundation Distinguished Investigator Award and by startup funds from the Institute for Systems Biology.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.chom.2021.08.013.
DECLARATION OF INTERESTS
The authors declare the following competing interests. R.W.D. is on the advisory board of MicrobiomX. W.M.G. is on the advisory board of Guardant Health, Freenome, SEngine, and consults for Diacarta and receives research support from Jannsen and Tempus. C.W.K. receives research support from Freenome.
REFERENCES
- Alhawi M, Stewart J, Erridge C, Patrick S, and Poxton IR (2009). Bacteroides fragilis signals through toll-like receptor (TLR) 2 and not through TLR4. J. Med. Microbiol 58, 1015–1022. 10.1099/jmm.0.009936-0. [DOI] [PubMed] [Google Scholar]
- Andersen V, Holst R, Kopp TI, Tjønneland A, and Vogel U (2013). Interactions between diet, lifestyle and IL10, IL1B, and PTGS2/COX-2 gene polymorphisms in relation to risk of colorectal cancer in a prospective Danish case-cohort study. PLoS One 8, e78366. 10.1371/journal.pone.0078366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodnar KA (2002). The interaction between murine dendritic cell and Mycobacterium tuberculosis. http://d-scholarship.pitt.edu/6320/.
- Bolger AM, Lohse M, and Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casterline BW, Hecht AL, Choi VM, and Bubeck Wardenburg J (2017). The Bacteroides fragilis pathogenicity island links virulence and strain competition. Gut Microbes 8, 374–383. 10.1080/19490976.2017.1290758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chac D, Kordahi M, Brettner L, and DePaolo RW (2021). Sample preparation for detection of different bacterial strains by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry. Curr. Protoc 1, e212. 10.1002/cpz1.212. [DOI] [PubMed] [Google Scholar]
- Chac D, Kordahi M, Brettner L, Verma A, McCleary P, Crebs K, Yee C, and DePaolo RW (2020). Proteomic changes in bacteria caused by exposure to environmental conditions can be detected by matrix-assisted laser desorption/ionization – time of flight (MALDI-ToF) mass spectrometry. bioRxiv 10.1101/2020.01.24.918938. [DOI] [Google Scholar]
- Chatzidaki-Livanis M, Geva-Zatorsky N, and Comstock LE (2016). Bacteroides fragilis type VI secretion systems use novel effector and immunity proteins to antagonize human gut Bacteroidales species. Proc. Natl. Acad. Sci. USA 113, 3627–3632. 10.1073/pnas.1522510113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung L, Thiele Orberg E, Geis AL, Chan JL, Fu K, DeStefano Shields CE, Dejea CM, Fathi P, Chen J, Finard BB, et al. (2018). Bacteroides fragilis toxin coordinates a pro-carcinogenic inflammatory cascade via targeting of colonic epithelial cells. Cell Host Microbe 23, 203–214.e5. 10.1016/j.chom.2018.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejea CM, Fathi P, Craig JM, Boleij A, Taddese R, Geis AL, Wu X, DeStefano Shields CED, Hechenbleikner EM, Huso DL, et al. (2018). Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science 359, 592–597. 10.1126/science.aah3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmont TO, and Eren AM (2018). Linking pangenomes and metagenomes: the Prochlorococcus metapangenome. PeerJ 6, e4320. 10.7717/peerj.4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellermann M, and Sartor RB (2018). Intestinal bacterial biofilms modulate mucosal immune responses. J. Immunol. Sci 2, 13–18. [PMC free article] [PubMed] [Google Scholar]
- Eren AM, Esen ÖC, Quince C, Vineis JH, Morrison HG, Sogin ML, and Delmont TO (2015). Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ 3, e1319. 10.7717/peerj.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren AM, Vineis JH, Morrison HG, and Sogin ML (2013). A filtering method to generate high quality short reads using Illumina paired-end technology. PLoS One 8, e66643. 10.1371/journal.pone.0066643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY, Makarova KS, Wolf YI, and Koonin EV (2015). Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res 43, D261–D269. 10.1093/nar/gku1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibb S, and Strimmer K (2012). MALDIquant: a versatile R package for the analysis of mass spectrometry data. Bioinformatics 28, 2270–2271. 10.1093/bioinformatics/bts447. [DOI] [PubMed] [Google Scholar]
- Gorry PA (1990). General least-squares smoothing and differentiation by the convolution (Savitzky-Golay) method. Anal. Chem 62, 570–573. 10.1021/ac00205a007. [DOI] [Google Scholar]
- Grivennikov SI, Wang K, Mucida D, Stewart CA, Schnabl B, Jauch D, Taniguchi K, Yu GY,Österreicher CH, Hung KE, et al. (2012). Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature 491, 254–258. 10.1038/nature11465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt D, Chen GL, LoCascio PF, Land ML, Larimer FW, and Hauser LJ (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11, 119. 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Song L, Breitwieser FP, and Salzberg SL (2016). Centrifuge: rapid and sensitive classification of metagenomic sequences. Genome Res 26, 1721–1729. 10.1101/gr.210641.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Liu CM, Luo R, Sadakane K, and Lam TW (2015). MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676. 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- Lieberman DA, Rex DK, Winawer SJ, Giardiello FM, Johnson DA, and Levin TR (2012). Guidelines for colonoscopy surveillance after screening and polypectomy: a consensus update by the US multi-society task force on colorectal cancer. Gastroenterology 143, 844–857. 10.1053/j.gastro.2012.06.001. [DOI] [PubMed] [Google Scholar]
- Mager LF, Wasmer MH, Rau TT, and Krebs P (2016). Cytokine-induced modulation of colorectal cancer. Front. Oncol 6, 96. 10.3389/fonc.2016.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak T, Lalloo F, Evans DGR, and Hill J (2004). Molecular stool screening for colorectal cancer. Br. J. Surg 91, 790–800. 10.1002/bjs.4576. [DOI] [PubMed] [Google Scholar]
- Masucci MT, Minopoli M, and Carriero MV (2019). Tumor associated neutrophils. Their role in tumorigenesis, metastasis, prognosis and therapy. Front. Oncol 9, 1146. 10.3389/fonc.2019.01146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Round JL, and Kasper DL (2008). A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453, 620–625. 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, Rotter-Maskowitz A, Weiser R, Mallel G, Gigi E, et al. (2020). The human tumor microbiome is composed of tumor type–specific intracellular bacteria. Science 368, 973–980. 10.1126/science.aay9189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI. Genome list - genome https://www.ncbi.nlm.nih.gov/genome/browse/#!/proteins/414/300113%7CBacteroides%20fragilis%20YCH46/.
- Ning Y, Manegold PC, Hong YK, Zhang W, Pohl A, Lurje G, Winder T, Yang D, LaBonte MJ, Wilson PM, et al. (2011). Interleukin-8 is associated with proliferation, migration, angiogenesis and chemosensitivity in vitro and in vivo in colon cancer cell line models. Int. J. Cancer 128, 2038–2049. 10.1002/ijc.25562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BS, and Lee JO (2013). Recognition of lipopolysaccharide pattern by TLR4 complexes. Exp. Mol. Med 45, e66. 10.1038/emm.2013.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reife RA, Coats SR, Al-Qutub M, Dixon DM, Braham PA, Billharz RJ, Howald WN, and Darveau RP (2006). Porphyromonas gingivalis lipopolysaccharide lipid A heterogeneity: differential activities of tetra- and pentaacylated lipid A structures on E-selectin expression and TLR4 recognition. Cell. Microbiol 8, 857–868. 10.1111/j.1462-5822.2005.00672.x. [DOI] [PubMed] [Google Scholar]
- Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, and Mazmanian SK (2011). The toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science 332, 974–977. 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandouk F, Al Jerf F, and Al-Halabi MHDB (2013). Precancerous lesions in colorectal cancer. Gastroenterol. Res. Pract 2013, 457901. 10.1155/2013/457901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears CL (2001). The toxins of Bacteroides fragilis. Toxicon 39, 1737–1746. 10.1016/S0041-0101(01)00160-X. [DOI] [PubMed] [Google Scholar]
- Sears CL (2009). Enterotoxigenic Bacteroides fragilis: a rogue among Symbiotes. Clin. Microbiol. Rev 22, 349–369. Table of Contents. 10.1128/CMR.00053-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T (2020). https://github.com/tseemann/snippy
- Chen H, Boutros PC 2011. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics 12,35 (2011). doi: 10.1186/1471-2105-12-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strum WB (2016). Colorectal adenomas. N. Engl. J. Med 374, 1065–1075. 10.1056/NEJMra1513581. [DOI] [PubMed] [Google Scholar]
- Testa U, Pelosi E, and Castelli G (2018). Colorectal cancer: genetic abnormalities, tumor progression, tumor heterogeneity, clonal evolution and tumorinitiating cells. Med. Sci. (Basel) 6. 10.3390/medsci6020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thursby E, and Juge N (2017). Introduction to the human gut microbiota. Biochem. J 474, 1823–1836. 10.1042/BCJ20160510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomkovich S, Dejea CM, Winglee K, Drewes JL, Chung L, Housseau F, Pope JL, Gauthier J, Sun X, Mühlbauer M, et al. (2019). Human colon mucosal biofilms from healthy or colon cancer hosts are carcinogenic. J. Clin. Invest 129, 1699–1712. 10.1172/JCI124196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wexler HM (2007). Bacteroides: the good, the bad, and the nitty-gritty. Clin. Microbiol. Rev 20, 593–621. 10.1128/CMR.00008-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Whole generation sequencing data and 16S data have been deposited at zenodo (https://zenodo.org/record/5090347) and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Ki67 | Thermo | Cat# RM9106, RRID:AB_2341197 |
| Anti-IL-17 | Santa Cruz | Cat# SC-7927, RRID:AB_2124997 |
| Anti-LPS core | Hyacult Biotech | Cat# WN1 222–5, RRID:AB_2750644 |
| Anti-MPO | DAKO | Cat# A0398 |
| Anti-CCL3 | Pierce | Cat# OA1653721, RRID: AB_1956667 |
| Bacterial and virus strains | ||
| B fragilis NCTC9343 | ATCC | Cat# 25285 |
| B. thetaiotamicron | ATCC | Cat# 29148 |
| Biological samples | ||
| B fragilis clinical isolates | This study | N/A |
| Proximal colon biopsies | This study | N/A |
| Critical commercial assays | ||
| Human IL-8 ELISA | Becton Dickinson | Cat # 555244 |
| Human IL-10 ELISA | Becton Dickinson | Cat # 555157 |
| Human IL-12p40 ELISA | Becton Dickinson | Cat # 555171 |
| Human IL-1b ELISA | Invitrogen | Cat # KHC001 |
| Nextera DNA Flex Library Prep kits | Illumina | Cat# 20018705 |
| bacterial DNA extraction kit | Qiagen | Cat#158567 |
| dual luciferase assay reporter system | Promega | Cat # E2920 |
| Deposited data | ||
| 16S Phylum level LDA analysis of biopsies | This study | https://doi.org/10.5281/zenodo.5090347 |
| 16S DNA fastQ files | This study | https://doi.org/10.5281/zenodo.5090347 |
| Whole genome sequencing of B fragilis isolates | This study | https://doi.org/10.5281/zenodo.5090347 |
| Whole genome sequencing fastQ files | This study | https://doi.org/10.5281/zenodo.5090347 |
| Experimental models: Cell lines | ||
| Human: HEK293 cells | ATCC | CRL-1573 |
| Human: THP-1 cells | ATCC | TIB-202 |
| Experimental models: Organisms/strains | ||
| Human colon biopsies | This study | N/A |
| B. fragilis clinical isolates | This study | N/A |
| Oligonucleotides | ||
| Primer bft Forward, GCGAACTCGGTTTAPTGCAGT Reverse, GTTGTAPGACATCCCACTGGC26 | DeJea et al., 2018 | N/A |
| Primer glycosyltransferase Forward, ACCTAAGAGCGCACGGAA and Reverse, AGGTCGTCCGAATAPGCCA. | This paper | N/A |
| Software and algorithms | ||
| Comparative genomics | Galperin et al 2015. https://github.com/StoreyLab/qvalue | N/A |
| ASNP identification | Seemann, 2020 | N/A |
| CLC genomics suite | Qiagen | N/A |
| LDA | https://huttenhower.sph.harvard.edu/galaxy/] | N/A |
| Anvio | Eren et al., 2015. http://merenlab.org/software/anvio/ | N/A |
All original code has been deposited at zenodo and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


