Abstract
Evaluation of drug interactions is an essential step in the new drug development process. Regulatory agencies, including U.S. Food and Drug Administrations and European Medicines Agency, have been published documents containing guidelines to evaluate potential drug interactions. Here, we have streamlined in vitro experiments to assess metabolizing enzyme-mediated drug interactions and provided an overview of the overall process to evaluate potential clinical drug interactions using in vitro data. An experimental approach is presented when an investigational drug (ID) is either a victim or a perpetrator, respectively, and the general procedure to obtain in vitro drug interaction parameters is also described. With the in vitro inhibitory and/or inductive parameters of the ID, basic, static, and/or dynamic models were used to evaluate potential clinical drug interactions. In addition to basic and static models which assume the most conservative conditions, such as the concentration of perpetrators as Cmax, dynamic models including physiologically-based pharmacokinetic models take into account changes in in vivo concentrations and metabolizing enzyme levels over time.
Keywords: Drug Interactions, Metabolism, Cytochrome P-450 Enzyme Inhibitors, Cytochrome P-450 Enzyme Inducers
INTRODUCTION
Drug-drug interactions (DDIs) are crucial concerns in drug development process and clinical practice. The in vitro techniques for drug interaction assessment continue to be developed, and various approaches have been established to translate in vitro data into in vivo prediction of potential drug interactions. Regulatory agencies such as Food and Drug Administration in the U.S. (USFDA) and European Medicines Agency (EMA) detailed these methods through the documented guidelines [1,2]. DDI referred to herein stands for pharmacokinetic (PK) DDI and more specifically implies metabolizing enzyme- and transporter-mediated DDI.
In order to predict potential DDI using in vitro data, both cases, where an investigational drug (ID) acts as a perpetrator or as a victim (substrate), should be considered. If an ID acts as a perpetrator, it can increase or decrease exposures of a substrate, inducing potential toxicity or loss of efficacy. In this case, a basic model and static mechanistic or dynamic model are sequentially used to determine the magnitude of clinical DDI, in which in vitro absorption, distribution, excretion, and metabolization (ADME) data and an inhibitory constant or induction parameters are used. On the other hand, if an ID acts as a victim, which means that the drug is metabolized ≥ 25% by a specific enzyme, clinical DDI studies with strong inhibitors and/or inducers should be conducted [1,2].
A general framework for evaluating clinical DDI using in vitro data is well documented in the guidance issued by USFDA [1], and there have been many publications on an in vitro-in vivo extrapolation (IVIVE) approach for predicting DDI. However, this tutorial focuses more on the following contents: 1) specific methods and information about in vitro experiments, 2) an overview of the overall process to evaluate clinical DDI potentials using in vitro data, and 3) an introductory and comprehensive description of a translational DDI approach for scientists who are not familiar with drug metabolism and pharmacokinetics. This tutorial only covers metabolizing enzyme-mediated DDI, but it does not discuss the interactions with therapeutic proteins, gastric pH change-dependent DDI, transporter-mediated DDI, and pharmacodynamic DDI.
EXPERIMENTAL APPROACHES FOR OBTAINING IN VITRO PARAMETERS IN NEED FOR DDI PREDICTION
ID as a substrate: metabolism parameters
The approach to be introduced here is a general and frequently used method. However, it is not standardized and can be flexibly adjusted depending on characteristics of an ID.
Prior to conducting a reaction phenotyping study for metabolizing enzyme identification, metabolic stability screening in various experimental conditions is generally performed. Various in vitro systems, including human hepatocytes (HHs), human liver microsomes (HLMs), S9, cytosol, and/or recombinant human CYP (rhCYP) isozymes, are used for experiments, depending on which enzymes are investigated. From the experiments, a metabolic pathway and metabolic clearance (CLint) of the ID can be identified. The numerous kinds of in vitro systems are summarized in Table 1 [3], and the examples of metabolic stability tests using HLM are shown in Table 2.
Table 1. In vitro drug-metabolizing experimental systems.
| In vitro system | CYP | MAO | UGT | ST | GST |
|---|---|---|---|---|---|
| Microsomes | O | X | O | X | O* |
| S9 | O | X | O | O | O |
| Cytosol | X | X | X | O | O† |
| Hepatocytes | O | O | O | O | O |
CYP, cytochrome P450; MAO, monoamine oxidase; UGT, UDP-glucuronosyltransferase; ST, sulfonyltransferase; GST, glutathione-S-transferase.
*Membrane-bound GST; †Soluble GST.
Table 2. The examples of metabolic stability screening using HLM.
| Metabolic Pathways (enzymes) | None | ES, CES | ES, CES, CYP, FMO | ES, CES, UGT | ES, CES, CYP | ES, CES, FMO |
|---|---|---|---|---|---|---|
| Buffer* | O | O | O | O | O | O |
| HLM | X | O | O | O | O | O |
| NADPH† | X | X | O | X | O | O |
| UDPGA‡ | X | X | X | O | X | X |
| SKF525A§ | X | X | X | X | X | O |
| Pre-incubation | 37°C, 5 min | 37°C, 5 min | 37°C, 5 min | Ice, 30 min | 45°C, 30 min∥ | 37°C, 5 min |
ES, esterase; CES, carboxylesterase; CYP, cytochrome P450; FMO, flavin-containing monooxygenase; HLM, human liver microsome; UGT, UDP-glucuronosyltransferase; GST, glutathione-S-transferase.
*Phosphate buffer or Tris buffer; †MgCl2 can be added; ‡Alamethicin is added before pre-incubation; §SKF525A is a nonspecific CYP inhibitor; ∥Heat (45°C) inactivates FMO activity.
Regarding the identification of specific metabolizing enzymes of an ID, two methods are widely used and recommended by USFDA guidance for the industry as well [1]. The first one is to use chemicals, drugs, and antibodies as selective inhibitors of enzymes, and the other is to use recombinant human enzymes. Both methods of measuring parent depletion and metabolite formation can be utilized. However, when examining the contribution of individual metabolizing enzymes to metabolite formation, the formation rate of specific metabolites should be measured. If metabolite information is available, the maximum reaction velocity (Vmax) and the substrate concentration producing a reaction velocity of 50% of Vmax (Km) can be determined with enzyme kinetic analysis.
After identifying the specific enzymes that are responsible for ID metabolism and obtaining CLint from the experiment, the fraction of drug metabolized by a specific enzyme, fm, should be calculated. In general, the higher the fm of a specific enzyme is, the greater the potential of the specific enzyme-mediated DDI can occur. The process of calculating fm is well-described in the previous tutorial [4].
ID as an inhibitor: inhibition parameters
The potential of an ID to inhibit the metabolism of other drugs should be examined. The regulatory agencies recommend in vitro inhibition studies on 6 CYP isoforms, including CYP1A2, CYP2B6, CYP2C9, CYP2C19, CYP2D6, and CYP3A in both reversible and irreversible (time-dependent inhibition, TDI) manner [1,2], and UGT isoforms including UGT1A1 and UGT2B7 as well [2]. The list of in vitro selective marker reactions of probe substrates, inhibitors, and inducers recommended by EMA and/or USFDA are summarized in Table 3.
Table 3. Examples of in vitro marker reactions, inhibitors, and inducers for specific enzymes.
| Enzymes | Marker reaction | Inhibitor | Inducer |
|---|---|---|---|
| CYP1A2 | Phenacetin O-deethylation, 7-ethoxyresorufin-O-deethylation | α-Naphthoflavone, furafylline* | Omeprazole, lansoprazole |
| CYP2B6 | Efavirenz hydroxylation, bupropion hydroxylation | Sertraline, phencyclidine*, thiotepa*, ticlopidine* | Phenobarbital |
| CYP2C8 | Paclitaxel 6α-hydroxylation, amodiaquine N-deethylation | Montelukast, quercetin, phenelzine* | Rifampicin |
| CYP2C9 | S-Warfarin 7-hydroxylation, diclofenac 4'-hydroxylation | Sulfaphenazole, tienilic acid* | Rifampicin |
| CYP2C19 | S-Mephenytoin 4'-hydroxylation | S-(+)-N-3-benzyl-nirvanol, nootkatone, ticlopidine*, loratadine | Rifampicin |
| CYP2D6 | Bufuralol 1'-hydroxylation, dextromethorphan O-demethylation | Quinidine, paroxetine* | - |
| CYP3A | Midazolam 1'-hydroxylation, testosterone 6β-hydroxylation | Itraconazole, ketoconazole, azamulin*, troleandomycin*, verapamil* | Rifampicin |
*Time-dependent inhibitors; †No selective inhibitor is available in vitro for CYP2C19- and CYP2B6-mediated metabolisms.
IC50 represents the concentration of an inhibitor to cause 50% inhibition at one selected substrate concentration and is commonly used for reversible inhibition screening. Determination of IC50 is a simple assay, and multiple inhibitor concentrations are tested for inhibition at a single substrate concentration. The most important concern here is that the substrate concentration should be close to Km for achieving high precision results. If IC50 of the ID is low at a specific enzyme, Ki (the enzyme inhibition constant) assay can proceed. At this time, 3 or more substrate concentrations including Km should be selected. It provides information about the type of reversible inhibition of the ID (competitive, non-competitive, uncompetitive, or mixed inhibition) and the potency of the inhibition. Ki is used for quantitative assessment of DDI potential. In the absence of assayed Ki values, IC50 values can be used to calculate Ki (Ki = IC50/2 if the substrate concentration used for IC50 assay is similar to Km) [5]. However, it is recommended to use Ki values obtained from the experiment as IC50 is not a constant and tends to have high interlaboratory variability. A representative example of reversible inhibition is shown in Fig. 1.
Figure 1. A representative example of IC50 and Ki determination assay. Dextromethorphan was used as a probe substrate of CYP2D6 activity. (A) IC50 determination assay was conducted in the presence of 5 μM dextromethorphan whose concentration is known as Km of dextromethorphan of CYP2D6. (B) Ki determination assay of quinidine (a CYP2D6 inhibitor) in the presence of 3 different concentrations of dextromethorphan was performed, and the results were a good fit for the competitive inhibition type when linearized on a Dixon plot. Figures are adapted from Cho et al. [6] with permission.
IC50, the concentration of an inhibitor to cause 50% inhibition; Ki, the enzyme inhibition constant, Km, Michaelis constant, the substrate concentration producing a reaction velocity of 50% of Vmax.
For TDI, a single point assay and IC50 shift assay are generally performed to screen whether an ID is a potential irreversible inhibitor or not. In the single-point assay, the percent of inhibition of an ID is calculated using metabolite formation of a probe substrate in the presence or absence of NADPH in the pre-incubation step. On the other hand, in the IC50 shift assay, IC50 values are compared in the presence or absence of NADPH in the pre-incubation step. If the IC50 value in the presence of NADPH shift to the left more than 1.5-fold, it is considered as a potential irreversible inhibitor. In that case, further approaches for determining inactivation kinetic parameters, including KI (concentration of inhibitor at which the inactivation rate is at half-maximum) and kinact (the maximal rate of inactivation), are warranted. In this experiment, metabolite formations of the probe substrate are measured with various concentrations of the inhibitor at various pre-incubation times (0 to 40 minutes), and the inactivation rate constant (kobs) can be obtained by regression using the following Eq. 1.
| (Eq. 1) |
Imax,u represents the maximum unbound concentration of the interacting (perpetrator) drug at steady state. The examples of TDI results were shown Figs. 2 and 3.
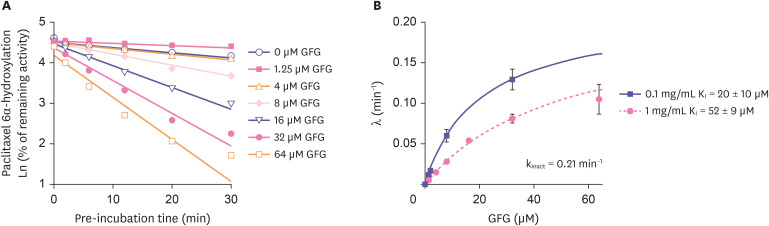
Figure 2. Examples of IC50 shift assay. Paclitaxel 6α-hydroxylation is a marker reaction of CYP2C8. (A) Gemfibrozil inhibited CYP2C8 with IC50 values of 120 μM (without NADPH preincubation) and 150 μM (with NADPH preincubation), indicating that gemfibrozil competitively inhibited CYP2C8. (B) On the other hand, GFG inhibited CYP2C8 with IC50 values of 24 μM (without NADPH preincubation) and 1.8 μM (with NADPH preincubation), indicating that GFG irreversibly inhibited CYP2C8. Figures are adapted from Ogilvie et al. [7] with permission.
IC50, the concentration of an inhibitor to cause 50% inhibition; GFG, gemfibrozil glucuronide.
Figure 3. An example of determination of inactivation kinetic parameters. (A) Based on the result from IC50 shift assay (Fig. 2), KI and kinact of GFG against CYP2C8 activity were determined, and (B) kobs (λ) was also calculated using KI and kinact. The kinact value was 0.21 1/min which meant that 21% of CYP2C8 was inactivated every minute by GFG. Figures are adapted from Ogilvie et al. [7] with permission.
GFG, gemfibrozil glucuronide; kobs, the observed inactivation rate constant; KI, the enzyme inactivation constant; kinact, the maximal rate of inactivation.
ID as an inducer: induction parameters
To investigate the induction potential of an ID in vitro, HH is preferred. HHs from at least 3 donors are recommended, and both positive and negative control groups should be added to validate the experimental system. Incubation of HHs with an ID generally lasts for 48–72 hours to allow complete induction. The mRNA levels and/or enzyme activity should be assessed. In other words, because enzyme activity can be masked if concomitant inhibitors are present, the measurement of mRNA level is preferred. The USFDA guidance recommends conducting an induction study for CYPs, including CYP1A2, CYP2B6, and CYP3A, induced via different nuclear receptors. If CYP3A4 is induced by the ID, additional induction studies of CYP2Cs should be conducted because CYP2C is induced by the pregnane X receptor (PXR), the same nuclear receptor for CYP3A induction. If in vitro induction results are positive, further induction studies to determine Emax (the maximum induction effect determined in vitro) and EC50 (the concentration causing half-maximal effect determined in vitro) are warranted. Here, Emax typically represents the highest fold-induction observed at experimental concentrations. Therefore, at least 4 different concentrations of inducer are required to determine Emax and EC50 values. If the maximal fold-induction has not been determined in the experiment, or if Emax exhibits high variability between donors, the maximal fold-induction obtained from the positive control can be used for calibration.
DATA INTERPRETATION
Based on collected in vitro DDI data, DDI potential can be predicted in three ways: a basic, static, or dynamic model. Despite the fact that all of these models require in vitro DDI parameters, their complicatedness and sensitivity vary according to how they are used. According to the USFDA, basic models are the first-line DDI evaluation tools to determine whether further evaluation is necessary or not. If the basic model does not rule out the DDI potential, a static or dynamic model is recommended for further evaluation to assess DDI risks and proactively plan the DDI study in clinical development [1]. The static model approach is relatively simple and direct but tends to overpredict DDI risks because it does not consider time-varying changes and assumes that the concentration of perpetrator stays at maximum. On the other hand, the dynamic model approach is more demanding but most informative, because it allows time-dependent changes of concentration and enzyme abundance. Thus, between static and dynamic models, it is recommended to choose a static model as startup approaches and pursue a dynamic model subsequently depending on the predicted DDI potential, potential concomitant drugs and sponsor’s financial/human resource capacity.
Basic model
The basic model does not incorporate drug disposition, and it only determines whether an ID is a potential perpetrator or not based on its in vitro DDI parameters (i.e., Ki, KI, kinact, EC50) and Imax,u. It calculates the ratios of intrinsic clearance values (R1, R2, R3) of a probe substrate for an enzyme pathway in the absence and presence of the interacting drug by each mechanism using corresponding in vitro DDI parameters mentioned above.
| For reversible inhibition | (Eq. 2) |
| For TDI | (Eq. 3) |
| For induction | (Eq. 4) |
In the equations, kdeg represents the apparent first-order degradation rate constant of the affected enzyme and d refers to the scaling factor and is usually assumed to be 1 [1]. If R1 ≥ 1.02, R1,gut ≥ 11, R2 ≥ 1.25, or R3 ≤ 0.8, the potential DDI risks should be further evaluated according to USFDA guidance [1]. For accessing the TDI and induction potentials in the basic model, 50 and 10 times of Imax,u are used respectively for the conservative results. Remember that the unbound fractions of Imax and Ki (or KI) are different. The former is the fraction of unbound in plasma (fu), whereas the latter is the fraction of an unbound drug in in vitro incubation system (fu,incubation). For the assessment of CYP induction, fold-change methods and/or correlation methods can also be used instead of the R3 value. These methods are summarized in Table 4. If induction kinetic parameters including Emax and EC50 are not available, fold-change methods can be used. In addition, RIS in correlation methods is suitable for CYP3A induction since sufficient data are only available for PXR activation [8].
Table 4. Basic methods for the assessment of CYP induction.
| Methods | Fold-change methods | Correlation methods |
|---|---|---|
| Descriptions | 1) mRNA levels increase in a concentration-dependent manner | 1) RIS = (Emax × Imax,u)/(EC50 + Imax,u) |
| 2) The fold-change of mRNA levels relative to the vehicle control is ≥ 2-fold at the expected hepatic concentrations of the drug | 2) Calculate Imax,u/EC50 values | |
| 3) The increase is more than 20% of the response of the positive control: | ||
| % of positive control = (mRNA fold increase of test drug treated cells − 1) × 100/(mRNA fold increase of positive control − 1) |
CYP, cytochrome P450; RIS, relative induction score; Emax, the maximum induction effect determined in vitro; Imax,u, the maximal unbound plasma concentration of the interacting (perpetrator) drug at steady-state; EC50, the concentration causing half-maximal effect determined in vitro.
By evaluating the R1, R2, R3 values of each enzyme, the potential DDI risk of the ID as a perpetrator is determined. This method is the most conservative because it assumes that the victim drug is 100% eliminated by the enzyme (fm, enzyme = 1), and exposure of the perpetrator drug stays at maximum. Thus, it is the first and most important model to use for DDI evaluation which can rule out further evaluations in mechanistic models and determine the necessity of clinical DDI study using index drugs [1].
Static model
When the basic model does not rule out the DDI potential, mechanistic modeling can be used for further evaluation, and a static model is one of them. The static model approach is relatively simple and useful for preliminary mechanistic evaluation [9,10]. It was suggested by Fahmi et al. [11] and also called as “Modified Rowland-Matin model” [12,13,14]. Unlike the basic model, which only determines DDI potential without considering PKs of a victim drug, the static model predicts the overall exposure changes of a victim drug in terms of area under the concentration-time curve (AUC) ratio by incorporating elimination pathway (fm, FG) of the victim drug and in vitro DDI parameters of the perpetrator drug. Also, it takes into account intestinal metabolism for oral administration. Basically, when assuming that absorption rate is unaffected, the AUC ratio in the presence and absence of a perpetrator drug can be expressed using the ratio of intrinsic clearance in intestine and liver as below:
| (Eq. 5) |
F’G and FG represent the fraction available after intestinal metabolism of the victim drug with or without the perpetrator drug, respectively. CL’int,H and CLint,H represent the total intrinsic hepatic clearance of the victim drug with or without the perpetrator drug, respectively. Like the basic model, the ratio of intrinsic clearance can be expressed using in vitro DDI parameters and the maximum concentration of a perpetrator drug. However, the static model simultaneously assesses competitive inhibition, TDI, and induction effect [11]. Moreover, when a victim drug is eliminated by multiple pathways, DDIs only affect the related intrinsic clearances by factoring the contribution fraction to the total clearance (fm). The expected net effects of intrinsic clearance changes can be expressed as Eq. 6 and Eq. 7, and the AUC ratio (Eq. 8) can be illustrated by substituting Eq. 6 and Eq. 7 into Eq. 5:
| (Eq. 6) |
| (Eq. 7) |
| (Eq. 8) |
| (Eq. 9) |
| (Eq. 10) |
| (Eq. 11) |
where A and B is the reversible and TDI term, respectively, and C is the induction term to describe the DDI effect on the intrinsic clearance of the enzyme. Most parameters and structures in A, B, and C terms are comparable to those in R1, R2, and R3 terms of basic models, and they are applied to either gut or liver in the same way. However, instead of using a maximum unbound concentration in plasma, the static model uses maximum unbound concentration in the liver or intestine (Ig(H)). For hepatic intrinsic clearance, A, B, and C terms should be applied to all metabolic pathways of a victim drug. Only CYP3A enzyme-mediated pathway is included for intestinal intrinsic clearance, and the fraction metabolized in the intestine (1 − Fg) is considered fraction metabolized by CYP3A enzyme (fm,CYP3A) unless there is evidence of other major mechanisms in intestinal cells. However, predicting a change of intestinal clearance is not as direct as predicting that of hepatic clearance due to uncertainty of (1) intestinal concentration, (2) metabolizing pathway, and (3) other influencing factors (i.e., permeability, residence time, enterohepatic recirculation) [15]. Moreover, even though the model can simultaneously account for influences of competitive inhibition, TDI, and induction of enzymes in both liver and intestine, the USFDA recommends evaluating inhibition potential and induction potential separately in case of the combination of inhibition and induction to prevent false negative prediction. The detailed descriptions of the parameter values are provided in the USFDA guidance, and PIPET provides the application that can assess DDI risks using basic and static models (pipetapps.com).
Dynamic model
Although both static and basic models are based on the assumption that the DDI effect is independent of change of time, concentrations and enzyme levels of both victim and perpetrator change with time, and the DDI effect in vivo between them is a dynamic process. These models assume that the concentration of a perpetrator is maintained at maximum (e.g., Imax,u, Ig(H)), which can lead to the overprediction of DDI effects [15]. Thus, the dynamic model has its strength for accurate DDI prediction as it considers the changes of in vivo concentrations and the enzyme level with time. Physiologically based pharmacokinetic (PBPK) modeling is one of the best methods minutely expressing concentration over time in each organ, incorporating those parameters and time-varying concentration of drugs and predicting DDI effects dynamically.
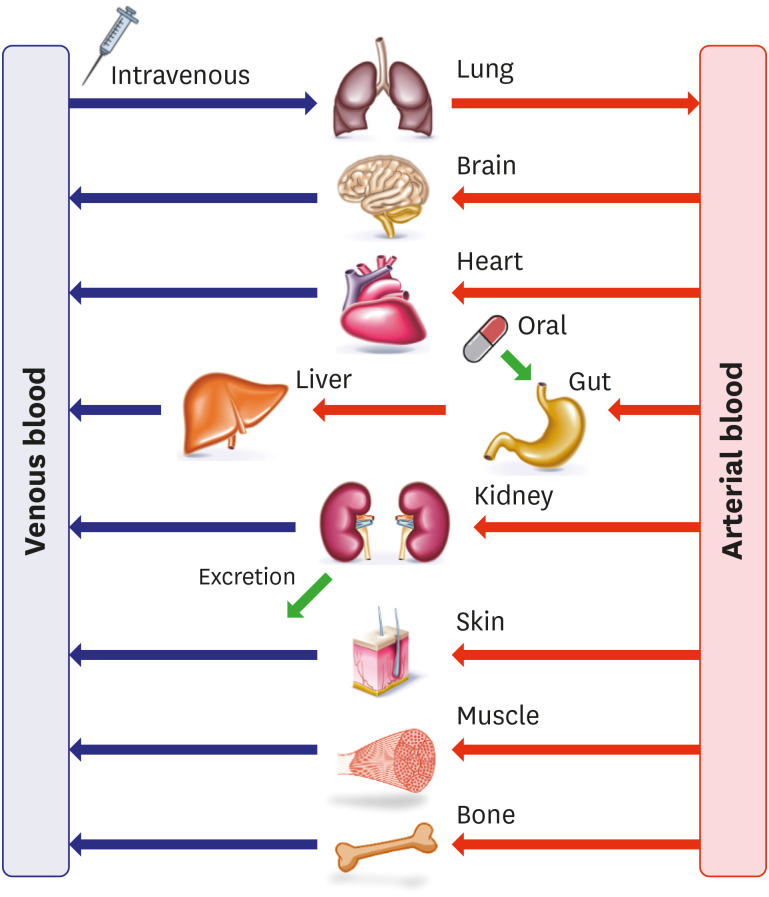
The PBPK model is mathematically expressed by dividing a human body into multiple compartments according to each organ, as shown in Fig. 4 [16]. The compartments are linked by tissue blood flow and expressed using differential equations (Eq. 12, Eq. 13). In the equations, VT, QT, CT, CV, and CA represent the tissue volume, the tissue blood flow, the drug concentrations in tissue, venous, and arterial compartments, respectively. The model integrates physiology data such as blood-flow rates and tissue volumes and PK properties of an ID such as tissue to plasma partition coefficient (Kp) and permeability. Compartments in a PBPK model generally include organs associated with ADME, but these may vary depending on the drug properties or DDI mechanism of interest. The PBPK model can be used in various cases, and DDI evaluation is one of its major applications. According to a recently published survey of the PBPK-based submissions to the USFDA, about 56–67% cases of the submissions utilized PBPK modeling to evaluate the DDI potential [17,18].
Figure 4. General scheme of physiologically based pharmacokinetic model structure.
| For non-eliminating tissue | (Eq. 12) |
| For eliminating tissue | (Eq. 13) |
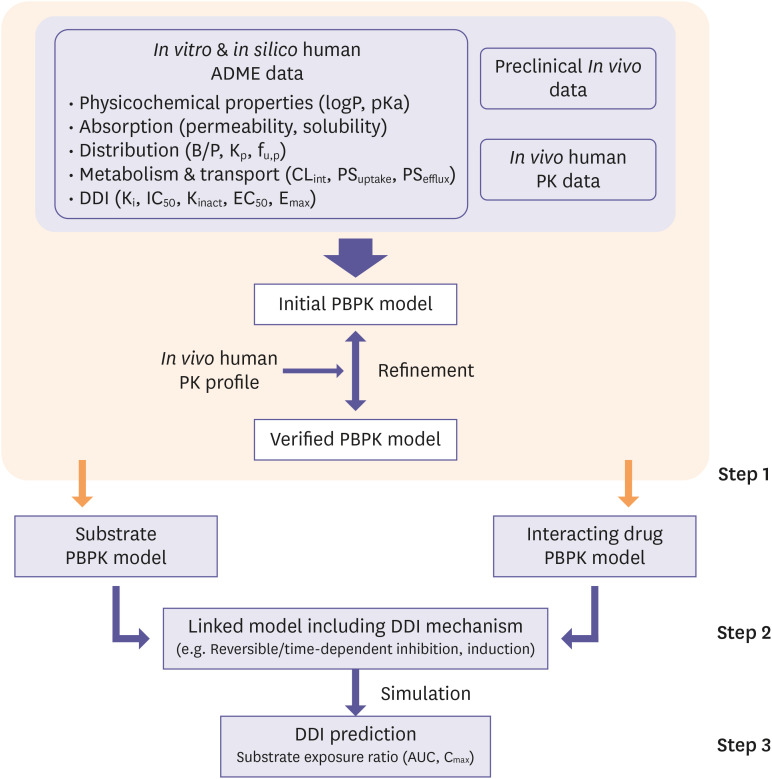
PBPK modeling and DDI assessment should be performed in a step-by-step manner (Fig. 5) [1]. First, PBPK models of both victim and perpetrator drugs should be developed with a mechanistic understanding of drugs and verified by clinical data. If necessary, the model can be refined to describe PK profiles during the verification process, but applicable rationale should be provided. Second, the developed PBPK models should be linked to each other, incorporating all of the DDI mechanisms based on in vitro DDI parameters by following equations of CLint and active enzyme abundance (Eact,enzyme) (Eq. 14, Eq. 15) [19] where the [I] refers to time-varying unbound concentration of the perpetrator drug in the tissue compartment. The change in CLint by the perpetrator, which is a reversible inhibitor, an irreversible inhibitor, or an inducer is included in Eq. 14 and Eq. 15.
Figure 5. Process of DDI assessment using PBPK model adapted from USFDA guidance. Step 1. Building verified PBPK models of a substrate and interacting drugs which can describe clinical PK profiles appropriately using in vitro/in silico/in vivo human ADME data. Step 2. Linking the models by including all DDI mechanisms based on in vitro DDI parameters. Step 3. Simulating the linked model with the proper dosing regimen and predicting DDI potential based on the anticipated exposure change of substrate (e.g., AUC ratio, Cmax ratio).
DDI, drug-drug interaction; PBPK, physiologically based pharmacokinetic; USFDA, Food and Drug Administration in the U.S.; PK, pharmacokinetic; ADME, absorption, distribution, metabolism, and excretion; AUC, area under the concentration-time curve.
| (Eq. 14) |
| (Eq. 15) |
Finally, simulation should be performed using the linked model to evaluate DDI potential by predicting plasma or tissue exposure ratio of a substrate (e.g., AUC, Cmax). When using PBPK modeling for DDI assessment, in-depth justification and credible sources should be provided on any model assumptions. Moreover, the physiological and biochemical plausibility of the model, parameter and variablity, sensitivity analysis, and uncertainty measures should be clarified [1]. These processes are demanding that a validated PBPK model from published literature or commercial software such as Simcyp, PK-Sim, and GastroPlusTM can be utilized [20,21,22].
The USFDA suggests utilizing a PBPK model to predict the effect of enzyme modulators if the model can describe clinical data on DDIs for a strong enzyme inhibitor or inducer. However, according to a recent update from USFDA, even though PBPK modeling showed good predictive performance for CYP-mediated inhibition, it had a limitation on predicting CYP-mediated induction and transporter-mediated DDI [17]. PBPK modeling is still in progress to bridge knowledge gaps and improve predictive performance. Nevertheless, the capability of PBPK modeling on DDI prediction is growing rapidly with accumulating knowledge on PBPK models and the development of in vitro experiments, and it is expected to broaden its role in the process of new drug development.
CONCLUSION
This tutorial has streamlined in vitro methods to evaluate metabolizing enzyme-mediated DDI and briefly introduced data interpretation methods for clinical DDI prediction recommended by regulatory agencies. The importance of DDI prediction is getting more attention, and so is the ability to understand and interpret in vitro data for DDI prediction. Therefore, from the early stage of new drug development, it is recommended that in vitro ADME data should be generated with consideration in mind that these data will be utilized to translate IVIVE and predict clinical potential of DDI.
Glossary
- CA
drug concentration in arterial compartment
- CLint
intrinsic clearance, metabolic clearance
- CLint, H
the total intrinsic hepatic clearance
- CLint,G
the total intrinsic gut clearance
- CLint,met
metabolic intrinsic clearance
- CT
drug concentration in tissue compartment
- CV
drug concentration in venous compartment
- CYP
cytochrome P450
- DDI
drug-drug interaction
- Eact
abundance of active enzyme
- EC50
the concentration causing half-maximal effect determined in vitro
- Emax
the maximum induction effect determined in vitro
- fm
the fraction of drug metabolized by an enzyme
- fu
the fraction of unbound drug in plasma
- fu,incubation
the fraction of unbound drug in in vitro incubation system
- HH
human hepatocyte
- HLM
human liver microsome
- IC50
the concentration of an inhibitor to cause 50% inhibition
- ID
investigational drug
- Ig(H)
the maximal unbound concentration of the interacting (perpetrator) drug in gut (liver)
- Imax,u
the maximal unbound plasma concentration of the interacting (perpetrator) drug at steady-state
- ISEF
intersystem extrapolation factor
- IVIVE
in vitro-in vivo extrapolation
- kdeg
the apparent first-order degradation rate constant of the affected enzyme
- Ki
the enzyme inhibition constant
- KI
the enzyme inactivation constant
- kinact
the maximal rate of inactivation
- Km
Michaelis constant, the substrate concentration producing a reaction velocity of 50% of Vmax
- kobs
the observed inactivation rate constant
- Kp
tissue to plasma partition coefficient
- QT
tissue blood flow
- RIS
relative induction score
- rhCYP
recombinant human CYP
- TDI
time-dependent inhibition
- UGT
uridine 5′-diphospho-glucuronosyltransferase
- Vmax
the maximum rate of metabolite formation
- VT
tissue volume
Footnotes
Conflict of Interest: - Authors: Nothing to declare
- Reviewers: Nothing to declare
- Editors: Nothing to declare
- Conceptualization: Choi S, Yim DS.
- Supervision: Bae SH.
- Writing - original draft: Choi S.
- Writing - review & editing: Yim DS.
References
- 1.U.S. Food and Drug Administration. In vitro drug interaction studies – Cytochrome P450 enzyme- and transporter-mediated drug interactions. Guidance for industry [Internet] [Accessed March 15, 2022]. https://www.fda.gov/media/134582/download .
- 2.European Medicines Agency. Guideline on the investigation of drug interactions [Internet] [Accessed March 15, 2022]. https://www.ema.europa.eu/en/documents/scientific-guideline/guideline-investigation-drug-interactions-revision-1_en.pdf .
- 3.Wang B. Drug-drug interactions in pharmaceutical development. New York (NY): John Wiley & Sons; 2008. [Google Scholar]
- 4.Yim DS, Bae SH, Choi S. Predicting human pharmacokinetics from preclinical data: clearance. Transl Clin Pharmacol. 2021;29:78–87. doi: 10.12793/tcp.2021.29.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cer RZ, Mudunuri U, Stephens R, Lebeda FJ. IC50-to-Ki: a web-based tool for converting IC50 to Ki values for inhibitors of enzyme activity and ligand binding. Nucleic Acids Res. 2009;37:W441–W445. doi: 10.1093/nar/gkp253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho DY, Bae SH, Lee JK, Kim YW, Kim BT, Bae SK. Selective inhibition of cytochrome P450 2D6 by sarpogrelate and its active metabolite, M-1, in human liver microsomes. Drug Metab Dispos. 2014;42:33–39. doi: 10.1124/dmd.113.054296. [DOI] [PubMed] [Google Scholar]
- 7.Ogilvie BW, Zhang D, Li W, Rodrigues AD, Gipson AE, Holsapple J, et al. Glucuronidation converts gemfibrozil to a potent, metabolism-dependent inhibitor of CYP2C8: implications for drug-drug interactions. Drug Metab Dispos. 2006;34:191–197. doi: 10.1124/dmd.105.007633. [DOI] [PubMed] [Google Scholar]
- 8.Fahmi OA, Boldt S, Kish M, Obach RS, Tremaine LM. Prediction of drug-drug interactions from in vitro induction data: application of the relative induction score approach using cryopreserved human hepatocytes. Drug Metab Dispos. 2008;36:1971–1974. doi: 10.1124/dmd.108.021907. [DOI] [PubMed] [Google Scholar]
- 9.Lu C, Berg C, Prakash SR, Lee FW, Balani SK. Prediction of pharmacokinetic drug-drug interactions using human hepatocyte suspension in plasma and cytochrome P450 phenotypic data. III. In vitro-in vivo correlation with fluconazole. Drug Metab Dispos. 2008;36:1261–1266. doi: 10.1124/dmd.107.019000. [DOI] [PubMed] [Google Scholar]
- 10.Kosugi Y, Hirabayashi H, Igari T, Fujioka Y, Okuda T, Moriwaki T. Risk assessment of drug-drug interactions using hepatocytes suspended in serum during the drug discovery process. Xenobiotica. 2014;44:336–344. doi: 10.3109/00498254.2013.837988. [DOI] [PubMed] [Google Scholar]
- 11.Fahmi OA, Hurst S, Plowchalk D, Cook J, Guo F, Youdim K, et al. Comparison of different algorithms for predicting clinical drug-drug interactions, based on the use of CYP3A4 in vitro data: predictions of compounds as precipitants of interaction. Drug Metab Dispos. 2009;37:1658–1666. doi: 10.1124/dmd.108.026252. [DOI] [PubMed] [Google Scholar]
- 12.Ohno Y, Hisaka A, Ueno M, Suzuki H. General framework for the prediction of oral drug interactions caused by CYP3A4 induction from in vivo information. Clin Pharmacokinet. 2008;47:669–680. doi: 10.2165/00003088-200847100-00004. [DOI] [PubMed] [Google Scholar]
- 13.Shou M, Hayashi M, Pan Y, Xu Y, Morrissey K, Xu L, et al. Modeling, prediction, and in vitro in vivo correlation of CYP3A4 induction. Drug Metab Dispos. 2008;36:2355–2370. doi: 10.1124/dmd.108.020602. [DOI] [PubMed] [Google Scholar]
- 14.Bohnert T, Patel A, Templeton I, Chen Y, Lu C, Lai G, et al. Evaluation of a new molecular entity as a victim of metabolic drug-drug interactions—an industry perspective. Drug Metab Dispos. 2016;44:1399–1423. doi: 10.1124/dmd.115.069096. [DOI] [PubMed] [Google Scholar]
- 15.Lu C, Di L. In vitro and in vivo methods to assess pharmacokinetic drug- drug interactions in drug discovery and development. Biopharm Drug Dispos. 2020;41:3–31. doi: 10.1002/bdd.2212. [DOI] [PubMed] [Google Scholar]
- 16.Shin HK, Kang YM, No KT. In: Handbook of computational chemistry. Leszczynski J, Kaczmarek-Kedziera A, Puzyn T, Papadopoulos MG, Reis H, Shukla MK, editors. Cham: Springer Nature; 2017. Predicting ADME properties of chemicals; pp. 2265–2301. [Google Scholar]
- 17.Zhang X, Yang Y, Grimstein M, Fan J, Grillo JA, Huang SM, et al. Application of PBPK modeling and simulation for regulatory decision making and its impact on US prescribing information: an update on the 2018-2019 submissions to the US FDA’s Office of Clinical Pharmacology. J Clin Pharmacol. 2020;60(Suppl 1):S160–S178. doi: 10.1002/jcph.1767. [DOI] [PubMed] [Google Scholar]
- 18.Grimstein M, Yang Y, Zhang X, Grillo J, Huang SM, Zineh I, et al. Physiologically based pharmacokinetic modeling in regulatory science: an update from the US Food and Drug Administration’s Office of Clinical Pharmacology. J Pharm Sci. 2019;108:21–25. doi: 10.1016/j.xphs.2018.10.033. [DOI] [PubMed] [Google Scholar]
- 19.Almond LM, Mukadam S, Gardner I, Okialda K, Wong S, Hatley O, et al. Prediction of drug-drug interactions arising from CYP3A induction using a physiologically based dynamic model. Drug Metab Dispos. 2016;44:821–832. doi: 10.1124/dmd.115.066845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simcyp, Ltd. The population-based simulator [Internet] [Accessed March 15, 2022]. https://www.certara.com/software/simcyp-pbpk/
- 21.Bayer, Inc. Computational systems biology suite [Internet] [Accessed March 15, 2022]. https://www.open-systems-pharmacology.org .
- 22.Simulations Plus, Inc. GastroPlus [Internet] [Accessed March 15, 2022]. https://www.simulations-plus.com .