Abstract
The zygotic genome is transcriptionally silent immediately after fertilization. In mice, initial activation of the zygotic genome occurs in the middle of the one-cell stage. At the mid-to-late two-cell stage, a burst of gene activation occurs after the second round of DNA replication, and the profile of transcribed genes changes dramatically. These two phases of gene activation are called minor and major zygotic gene activation (ZGA), respectively. As they mark the beginning of the gene expression program, it is important to elucidate gene expression regulation during these stages. This article reviews the outcomes of studies that have clarified the profiles and regulatory mechanisms of ZGA.
Keywords: Chromatin structure, Epigenetic factors, Preimplantation development, Zygotic gene activation
Introduction
Immediately after fertilization, the zygotic genome is transcriptionally inert. The initiation of gene expression by the zygotic genome is called zygotic gene activation (ZGA), which occurs according to species-specific timing [1]. Mice undergo transcription initiation during the S phase of the one-cell stage. Development from a one-cell embryo to a multicellular adult organism is achieved via changes in gene expression patterns according to the gene expression program. Thus, the gene expression pattern changes dramatically after cleavage into the two-cell embryo. Therefore, clarifying the gene expression program and its management is essential for understanding the regulation of embryonic development. This article reviews gene expression patterns and their regulation during the one- and two-cell stages in mice, which represent the beginning of the gene expression program, i.e., the transcriptional cascade.
ZGA Timing
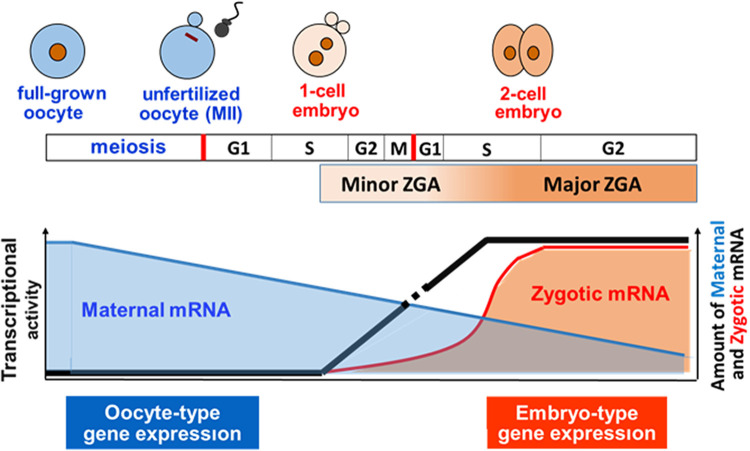
In growing oocytes, genes are actively transcribed in an oocyte-specific pattern [2]. During this period, transcribed mRNAs are highly stable and stored in oocytes as maternal mRNAs. However, gene transcription ceases when the oocytes are fully grown. After fertilization, zygotes remain transcriptionally silent, and their development is regulated by maternal mRNA [2, 3]. Early studies have suggested that zygotic genome transcription is initiated at the two-cell stage. Gel electrophoresis analyses of newly synthesized proteins detected via the incorporation of [35S]methionine showed no differences in the profiles of these proteins between unfertilized oocytes and late one-cell-stage embryos, suggesting that the proteins were synthesized from maternal mRNA, not zygotic mRNA, as a template. However, soon after cleavage into the two-cell stage, proteins were detected with a molecular weight of ~70 kDa; this was not observed in oocytes or one-cell embryos [4, 5]. Since these proteins were eliminated by treatment with α-amanitin, an RNA polymerase II inhibitor [6, 7], they were thought to have originated from zygotic transcripts. Therefore, the initiation of transcription after fertilization is thought to occur at the early two-cell stage. Gel electrophoresis analysis of the newly synthesized proteins revealed that their profiles changed dramatically from the mid-to-late two-cell stage, suggesting that gene expression patterns shifted considerably during this period [7, 8]. This global activation of zygotic genes at the mid-to-late two-cell stage was labeled major ZGA to distinguish it from minor ZGA during the early two-cell stage. However, later studies revealed that minor ZGA begins in the middle of the one-cell stage. BrUTP incorporation into nascent RNA was first detected during the S/G2 phase in one-cell embryos in an in vitro transcription assay [9] and in vivo following microinjection of BrUTP [10]. When a male transgenic mouse was crossed with a wild-type female mouse, transgene expression was observed in one-cell-stage embryos [11, 12]. The microinjection of reporter genes also showed that transcription occurred at the one-cell stage [13,14,15]. Thus, it has been established that minor ZGA occurs between the middle of the one-cell stage and the early two-cell stage and that major ZGA proceeds during the mid-to-late two-cell stage in mice (Fig. 1).
Fig. 1.
Zygotic gene activation (ZGA) in mice. The zygotic genome is transcriptionally inert immediately after fertilization and is activated at the mid-S phase of the one-cell stage. The gene expression pattern changes dramatically during the second round of DNA replication. Genome activation before and after DNA replication is called minor and major ZGA, respectively. Transcribed mRNAs that accumulate during oocyte growth (maternal RNA) remain after fertilization to support development until major ZGA occurs.
ZGA Gene Profiles
It is necessary to identify the genes expressed at each stage of the gene expression program to understand its regulation. Genes transcribed during major ZGA have been extensively investigated through genome-wide analysis via DNA microarrays and RNA sequencing (RNAseq) [16,17,18,19,20]. These analyses identified up to 10,000 genes expressed in late two-cell-stage embryos. The profiles of these genes differ greatly from those of oocytes. More than 3,000 genes exhibited increased expression levels at the late two-cell stage compared to MII-stage oocytes [16]. Most oocyte-specific genes are not transcribed, whereas genes that function specifically at the two-cell stage are expressed, such as ZO-1 (Tip1), which forms tight junctions [17]. The expression patterns of oocyte signaling genes are also greatly altered during the late two-cell stage. For example, Fzd2, Smad7, and Jag2 show increased expression, and Smad1 exhibits decreased expression at the late two-cell stage [17]. Gene ontology analysis has revealed that genes that are transiently expressed at the two-cell stage are enriched in transcription, metabolism, and the cell cycle [18]. Gene network analysis has suggested that c-Myc is a candidate master regulatory gene during major ZGA [21]. Genome-wide analysis of non-coding RNA has revealed that various types of long non-coding and small RNAs are also transcribed during major ZGA [22]. An analysis of transposons has shown that LINE1 is also actively expressed at the two-cell stage [23].
Genome-wide analyses have also identified several hundreds to thousands of genes transcribed during minor ZGA [17,18,19,20]. These genes have been reported to be transcribed from zygotic genes when their expression levels increase by 1.5–2 times relative to MII-stage oocytes during the one-cell or early two-cell stage. However, the accuracy of these analyses was not high because a large amount of maternal mRNA, which is much higher than that of zygotic mRNA, remains at these stages after fertilization. Furthermore, due to increased RNA adenylation after fertilization [24], RNAseq using poly(A)-selected RNA may mistake the increases caused by polyadenylation changes for those by one-cell transcription per se [19, 25]. Therefore, it is difficult to conclude that these genome-wide analyses correctly identified genes expressed during minor ZGA, except for MuERV-L, which was confirmed as a minor ZGA gene via evaluation of its high expression at the one-cell stage by reverse-transcription PCR [26].
In the precise RNAseq analysis using total RNA, rather than poly(A)-selected RNA, 23 genes likely to be transcribed at the one-cell stage were found, and their expression levels were 30-fold higher in late one-cell-stage embryos than in MII-stage oocytes and decreased following treatment with 5,6-dichlorobenzimidazole riboside, an RNA polymerase II activity inhibitor (Aoki, unpublished data). Interestingly, all of these transcripts contained intronic sequences, indicating inefficient splicing activity during the one-cell stage [27]. Subsequently, mRNA levels were analyzed using introns in RNAseq data to identify thousands of genes as minor ZGA genes [27]. Further analysis suggested that ~90% of genes were transcribed at the one-cell stage, although each gene had low expression levels [28]. Interestingly, most oocyte-specific genes were not transcribed, and intergenic regions were extensively transcribed at low levels in one-cell-stage embryos, indicating that transcription occurs globally over a large part of the genome [27]. Compared with the global, promiscuous expression found throughout the whole genome of one-cell-stage embryos, selective expression occurs in late two-cell-stage embryos [18], and the profiles of transcribed genes during one- and two-cell stages widely differed from each other, exhibiting a correlation coefficient of only 0.309 (Aoki, unpublished data) and up- and down-regulation of approximately 4,000 genes during the one- to two-cell stage transition [20].
ZGA Regulation
Triggering ZGA
The mechanism regulating transcription initiation after fertilization has not yet been clarified. However, a pioneering study proposed a hypothesis for this mechanism in Xenopuslaevis embryos, a histone titration model [29,30,31]. Unfertilized oocytes have large amounts of histones stockpiled in their cytoplasm. These histones, produced in 11 cleavage events after fertilization, are sufficient to form nucleosomes for 15,000–20,000 nuclei. The structure of nucleosomes containing histones prevents the access of transcription factors to their target DNA, causing transcriptional silencing. However, after 12 rounds of replication, these histones are no longer sufficient to constitute the complete form of nucleosomes due to the exponentially increased volume of DNA. At this point, transcription factors gain access to their DNA target sites to initiate transcription. Artificially increasing DNA content by polyspermic fertilization or microinjection of exogenous DNA induces transcription earlier.
However, this process does not occur in mice. An increase in DNA content by polyspermic fertilization did not affect the transcriptional activity in one-cell-stage embryos. Furthermore, neither a decrease in DNA content by parthenogenesis nor DNA synthesis inhibition by aphidicolin prevents transcription initiation in one-cell-stage embryos [9]. A maternal transcript may be involved in transcription induction in mouse embryos. Some maternal mRNAs are not translated before fertilization but are translated concurrently with the elongation of their poly-A tails [32,33,34]. A transcription factor translated from this type of maternal mRNA may have a specific function to activate the zygotic genome. Indeed, the inhibition of poly-A tail elongation was found to prevent transcriptional activation after fertilization [35]. Alternatively, there may be a protein that inhibits transcription in oocytes, and its inhibitory activity may decrease after fertilization. In rabbits, when the nucleus of an embryo at the 32-cell stage, at which time embryonic genes are actively transcribed, is transplanted into an enucleated unfertilized oocyte, transcriptional activity decreases rapidly and then resumes when the reconstituted embryo reaches the stage at which normal embryos begin zygotic gene expression [36]. To date, no gene has been identified as a ZGA trigger, although dozens of genes have been shown to be involved in ZGA.
Mechanism regulating minor ZGA
Global, promiscuous gene expression during minor ZGA may be caused by the loosened chromatin structure at the one-cell and early two-cell stages before the second round of DNA replication. Pioneering studies have demonstrated that transcription does not depend on enhancers during minor ZGA. Although reporter genes were shown to require enhancers for expression following microinjection in late two-cell-stage embryos, they were expressed without an enhancer in one-cell-stage embryos. This change to enhancer-dependent transcription involved a second round of DNA replication [37,38,39,40]. Since one function of enhancers is to loosen the chromatin structure to allow transcription factors to access gene promoters [41, 42], chromatin structure appears loose in one-cell-stage embryos. Consistent with this finding, when late two-cell-stage embryos were treated with butyrate, a histone deacetylase inhibitor expected to loosen the chromatin structure, active transcription occurred from the reporter gene without an enhancer as seen in one-cell-stage embryos [37, 38]. Fluorescence recovery after photobleaching analysis also suggested that the chromatin structure was greatly loosened in one-cell-stage embryos [43]. Although these experimental results strongly imply that chromatin structure is loosened in one-cell-stage embryos, a global analysis of DNase I-sensitive sites (DHSs) showed that DHSs were least frequent at the one-cell stage during preimplantation development, suggesting that the chromatin structure was tightest in one-cell-stage embryos [44]. However, this experiment used stringent DHS selection criteria at > 1 read per million. As low-level transcription occurs from numerous sites in one-cell-stage embryos, one would expect low numbers of reads per million at each site, resulting in the exclusion of these sites from the DHS count. Consistent with this interpretation, high-throughput chromosome conformation capture analysis revealed that the topologically associated domain where DHSs are enriched is obscure in one-cell-stage embryos [45].
The mechanism by which the chromatin structure is loosened in one-cell-stage embryos remains to be clarified. However, some epigenetic factors have been identified as candidates involved in this process. The nucleosomes of one-cell-stage embryos are composed of a unique set of histone variants. In nearly all cell types, all three non-centromeric histone H3 variants (H3.1, H3.2, and H3.3) are incorporated within the nucleosome [46]. However, these mainly comprise of H3.3 in one-cell-stage embryos [47], which should loosen the chromatin structure because H3.3 and H3.1/2 are involved in the formation of loose and tight chromatin structures, respectively (reviewed in [48]). H2A variant composition is unique to the nucleosomes of one-cell-stage embryos. H2A.X and TH2A, suggested to be involved in chromatin loosening [49, 50], are abundantly incorporated within the nucleosomes, and macroH2A, which forms condensed heterochromatin, is absent from the chromatin of one-cell-stage embryos [50, 51].
Thus, pioneering studies using reporter genes have suggested that transcription does not depend on enhancers in one-cell-stage embryos. Further studies have shown that short motifs without core promoter elements maintain transcriptional activity. A reporter gene assay using the Tktl1 promoter, expressed at the one-cell stage, showed that only the 56-bp sequence upstream of the TSS, containing GC and TATA boxes, had transcriptional activity in one-cell-stage embryos. GC box mutation completely abolished this activity, whereas 40% of the activity remained following mutation of the TATA box [52]. We extended these experiments by constructing a short sequence containing only the GC box and core promoter elements (BRE and Inr) in a reporter gene assay. This short sequence showed transcriptional activity in one-cell embryos but not in late two-cell embryos (Aoki, unpublished data). When the second round of DNA replication was inhibited, this one-cell specific transcriptional activity remained at the late two-cell stage, suggesting changes in gene expression regulation during the transition from minor to major ZGA. Interestingly, replacing the core promoter elements with a nonsense sequence did not decrease transcriptional activity, implying that gene expression is activated only by the GC box in one-cell-stage embryos. Transcriptional activity was also not altered by replacing C with A in the GC box (GGGCGG) and remained at 50% following C-to-T replacement (Aoki, unpublished data). These results suggest that GC box-like motifs without enhancer or promoter motifs induce transcription and that a number of these types of sequences are present throughout the genome, causing global, promiscuous gene expression during minor ZGA (Fig. 2).
Fig. 2.
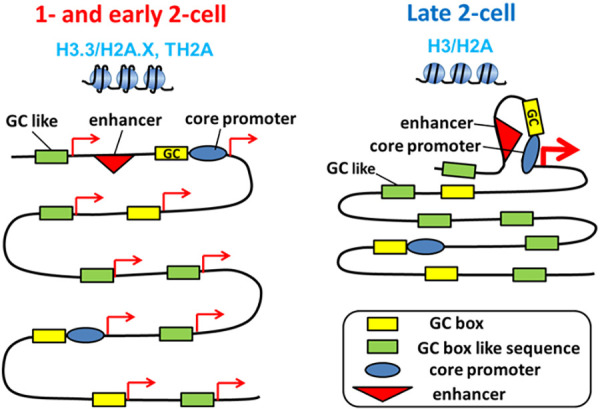
Change in the regulatory mechanism of gene expression during minor and major ZGA. In one-cell- and early two-cell-stage embryos (during minor ZGA), chromatin structure is loosened because the nucleosome contains a unique combination of histone variants (H3.3/H2A.X, TH2A). This type of chromatin structure appears to allow transcription with GC box-like motifs as promoters, causing global, promiscuous expression from the genome, including intergenic regions. However, at the late two-cell stage (during major ZGA), a core promoter and enhancers are required for active transcription, as occurs in most cell types, due to the tight chromatin structure, which contains all the histone variants.
Mechanism regulating the transition from minor to major ZGA
Gene expression patterns change dramatically during the transition from minor to major ZGA, from promiscuous, enhancer-independent expression to regulated, enhancer-dependent expression. Changes in chromatin structure appear to play an important role in this alteration. Chromatin structure essentially represses transcription to prevent transcription factors from gaining access to their DNA target motifs, which explains why enhancers are required for active transcription. However, as described above, the chromatin structure is very loose in one-cell-stage embryos, creating a permissive state for transcription [53]. This transition from a permissive to a repressive state involves the second round of DNA replication. Indeed, fluorescence recovery after photobleaching analysis showed that inhibition of this transition by aphidicolin, a DNA polymerase inhibitor, led to the persistence of a loose chromatin structure at the late two-cell stage [43].
Epigenetic factors appear to be involved in the change in chromatin structure during the transition from minor to major ZGA. As described above, nucleosomes lack H3.1/2, which is involved in forming a loose chromatin structure (i.e., permissive chromatin) in one-cell-stage embryos. This state continues after cleavage into the two-cell stage. However, the nucleosome incorporates these proteins following the second round of DNA replication [47, 54], which appears to be involved in establishing a tight chromatin structure (i.e., repressive chromatin) at the late two-cell stage. Global changes in H3K4me3 localization within the genome may also be involved in altering chromatin structure. Although H3K4me3 has been detected in broad signals covering vast genome regions in one-cell and early two-cell embryos, it is later localized in regions adjacent to the TSSs of active genes in the late two-cell stages, as observed in most other cell types [55, 56]. H1FOO, a variant of linker histone H1 involved in the loosening of the chromatin structure, is abundant in the chromatin of one-cell embryos, whereas it is eliminated in late two-cell embryos [57].
After the transition from minor to major ZGA, gene expression changes from a promiscuous global pattern to a regulated specific pattern. A recent report demonstrated that minor ZGA is required for major ZGA to occur. After transient inhibition of minor ZGA by treatment with 5,6-dichlorobenzimidazole riboside, a reversible Pol II-mediated transcription inhibitor, the pattern of minor ZGA persisted during the time of major ZGA [58]. Recent studies showed that Dux, which is transiently expressed during minor ZGA, regulates the expression of certain genes during major ZGA ([59,60,61,62]) to address how minor ZGA induces the activation of specific genes involved in major ZGA, despite inefficient transcript splicing for the production of dysfunctional proteins during minor ZGA [27]. Dux is an intronless gene. DUX4, a human ortholog of mouse Dux, is present at the ends of tandem repeats with its paralogs. These tandem repeats are heterochromatinized to prevent their expression in most cell types. However, DUX4 expression has been found in patients with facioscapulohumeral muscular dystrophy (FSHD), with short tandem repeats that appear to allow partial loosening of the chromatin structure [63,64,65,66]. In mice, Dux and its paralogs are also found within tandem repeats, which are expected to be heterochromatinized [67]. However, Dux may be expressed during minor ZGA since the chromatin structure is loosened [62]. Interestingly, more than a dozen paralogs are expressed in addition to Dux during minor ZGA in mice, unlike in human FSHD [62], which causes efficient expression during the relatively short period of minor ZGA (Fig. 3).
Fig. 3.
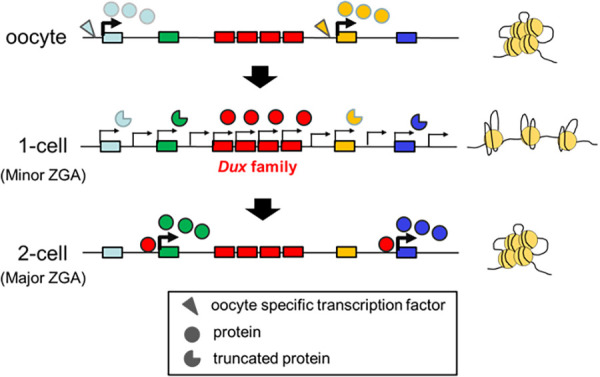
Involvement of Dux in the transition from minor to major ZGA. In oocytes, tandem repeats of Dux and its paralogs (Dux family genes) form transcriptionally silent heterochromatin. However, their chromatin structures become loosened in one-cell stage embryos, inducing transcription of numerous Dux family genes. In these embryos, the transcripts of most genes do not produce functional proteins due to inefficient splicing. However, functional proteins are translated from Dux transcripts because Dux is an intronless gene. The DUX protein activates major ZGA genes until its elimination at the late two-cell stage when the chromatin structure tightens.
Perspectives
Understanding the profile and regulatory mechanism of ZGA has progressed extensively within this decade via the development of analytical methods, such as next-generation sequencing technology. However, numerous issues remain to be addressed before a full understanding of ZGA can be obtained. Although the composition of a unique set of histone variants in the nucleosome appears to be involved in loosening chromatin structure during minor ZGA, the mechanism by which only particular variants are incorporated into the nucleosome remains to be elucidated. The means by which dynamic histone replacement occurs soon after fertilization, which is important for erasing epigenetic information in oocytes to reprogram gene expression, also requires clarification. Although recent studies have shown that Dux activates major ZGA genes, it regulates only hundreds of genes, and the mechanisms that activate thousands of other genes remain unknown. Minor ZGA gene inhibition involves the second round of DNA replication. However, this mechanism does not seem to be responsible for repressing all the minor ZGA genes. For example, Dux expression is inhibited at the late two-cell stage, independent of the second round of DNA replication [62]. LINE1 is reportedly responsible for Dux repression [68]. Considering the recent rapid progress of research in this field, it is expected that these issues will be clarified in the near future.
Conflict of interests
The author declare no conflict of interests.
Acknowledgments
I would like to express my gratitude to Dr. Richard M. Schultz for his help with the ZGA research. I also thank my students for their involvement in this study. This study was supported in part by grants-in-aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (nos. 17K19318, 18H03970, 19H05752, and 21H04752).
References
- 1.Schultz RM. The molecular foundations of the maternal to zygotic transition in the preimplantation embryo. Hum Reprod Update 2002; 8: 323–331. [DOI] [PubMed] [Google Scholar]
- 2.Schultz RM. Regulation of zygotic gene activation in the mouse. BioEssays 1993; 15: 531–538. [DOI] [PubMed] [Google Scholar]
- 3.Telford NA, Watson AJ, Schultz GA. Transition from maternal to embryonic control in early mammalian development: a comparison of several species. Mol Reprod Dev 1990; 26: 90–100. [DOI] [PubMed] [Google Scholar]
- 4.Van Blerkom J, Brockway GO. Qualitative patterns of protein synthesis in the preimplantation mouse embryo. II. During release from facultative delayed implantation. Dev Biol 1975; 46: 446–451. [DOI] [PubMed] [Google Scholar]
- 5.Conover JC, Temeles GL, Zimmermann JW, Burke B, Schultz RM. Stage-specific expression of a family of proteins that are major products of zygotic gene activation in the mouse embryo. Dev Biol 1991; 144: 392–404. [DOI] [PubMed] [Google Scholar]
- 6.Levey IL, Brinster RL. Effects of alpha-amanitin on RNA synthesis by mouse embryos in culture. J Exp Zool 1978; 203: 351–360. [DOI] [PubMed] [Google Scholar]
- 7.Flach G, Johnson MH, Braude PR, Taylor RA, Bolton VN. The transition from maternal to embryonic control in the 2-cell mouse embryo. EMBO J 1982; 1: 681–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bolton VN, Oades PJ, Johnson MH. The relationship between cleavage, DNA replication, and gene expression in the mouse 2-cell embryo. J Embryol Exp Morphol 1984; 79: 139–163. [PubMed] [Google Scholar]
- 9.Aoki F, Worrad DM, Schultz RM. Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev Biol 1997; 181: 296–307. [DOI] [PubMed] [Google Scholar]
- 10.Bouniol C, Nguyen E, Debey P. Endogenous transcription occurs at the 1-cell stage in the mouse embryo. Exp Cell Res 1995; 218: 57–62. [DOI] [PubMed] [Google Scholar]
- 11.Matsumoto K, Anzai M, Nakagata N, Takahashi A, Takahashi Y, Miyata K. Onset of paternal gene activation in early mouse embryos fertilized with transgenic mouse sperm. Mol Reprod Dev 1994; 39: 136–140. [DOI] [PubMed] [Google Scholar]
- 12.Christians E, Campion E, Thompson EM, Renard JP. Expression of the HSP 70.1 gene, a landmark of early zygotic activity in the mouse embryo, is restricted to the first burst of transcription. Development 1995; 121: 113–122. [DOI] [PubMed] [Google Scholar]
- 13.Latham KE, Solter D, Schultz RM. Acquisition of a transcriptionally permissive state during the 1-cell stage of mouse embryogenesis. Dev Biol 1992; 149: 457–462. [DOI] [PubMed] [Google Scholar]
- 14.Vernet M, Bonnerot C, Briand P, Nicolas JF. Changes in permissiveness for the expression of microinjected DNA during the first cleavages of mouse embryos. Mech Dev 1992; 36: 129–139. [DOI] [PubMed] [Google Scholar]
- 15.Ram PT, Schultz RM. Reporter gene expression in G2 of the 1-cell mouse embryo. Dev Biol 1993; 156: 552–556. [DOI] [PubMed] [Google Scholar]
- 16.Hamatani T, Carter MG, Sharov AA, Ko MS. Dynamics of global gene expression changes during mouse preimplantation development. Dev Cell 2004; 6: 117–131. [DOI] [PubMed] [Google Scholar]
- 17.Wang QT, Piotrowska K, Ciemerych MA, Milenkovic L, Scott MP, Davis RW, Zernicka-Goetz M. A genome-wide study of gene activity reveals developmental signaling pathways in the preimplantation mouse embryo. Dev Cell 2004; 6: 133–144. [DOI] [PubMed] [Google Scholar]
- 18.Zeng F, Baldwin DA, Schultz RM. Transcript profiling during preimplantation mouse development. Dev Biol 2004; 272: 483–496. [DOI] [PubMed] [Google Scholar]
- 19.Xue Z, Huang K, Cai C, Cai L, Jiang CY, Feng Y, Liu Z, Zeng Q, Cheng L, Sun YE, Liu JY, Horvath S, Fan G. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature 2013; 500: 593–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park SJ, Komata M, Inoue F, Yamada K, Nakai K, Ohsugi M, Shirahige K. Inferring the choreography of parental genomes during fertilization from ultralarge-scale whole-transcriptome analysis. Genes Dev 2013; 27: 2736–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeng F, Schultz RM. RNA transcript profiling during zygotic gene activation in the preimplantation mouse embryo. Dev Biol 2005; 283: 40–57. [DOI] [PubMed] [Google Scholar]
- 22.Svoboda P. Long and small noncoding RNAs during oocyte-to-embryo transition in mammals. Biochem Soc Trans 2017; 45: 1117–1124. [DOI] [PubMed] [Google Scholar]
- 23.Jachowicz JW, Bing X, Pontabry J, Bošković A, Rando OJ, Torres-Padilla ME. LINE-1 activation after fertilization regulates global chromatin accessibility in the early mouse embryo. Nat Genet 2017; 49: 1502–1510. [DOI] [PubMed] [Google Scholar]
- 24.Pikó L, Clegg KB. Quantitative changes in total RNA, total poly(A), and ribosomes in early mouse embryos. Dev Biol 1982; 89: 362–378. [DOI] [PubMed] [Google Scholar]
- 25.Deng Q, Ramsköld D, Reinius B, Sandberg R. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science 2014; 343: 193–196. [DOI] [PubMed] [Google Scholar]
- 26.Kigami D, Minami N, Takayama H, Imai H. MuERV-L is one of the earliest transcribed genes in mouse one-cell embryos. Biol Reprod 2003; 68: 651–654. [DOI] [PubMed] [Google Scholar]
- 27.Abe K, Yamamoto R, Franke V, Cao M, Suzuki Y, Suzuki MG, Vlahovicek K, Svoboda P, Schultz RM, Aoki F. The first murine zygotic transcription is promiscuous and uncoupled from splicing and 3′ processing. EMBO J 2015; 34: 1523–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamamoto R, Aoki F. A unique mechanism regulating gene expression in 1-cell embryos. J Reprod Dev 2017; 63: 9–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: II. Control of the onset of transcription. Cell 1982; 30: 687–696. [DOI] [PubMed] [Google Scholar]
- 30.Prioleau MN, Huet J, Sentenac A, Méchali M. Competition between chromatin and transcription complex assembly regulates gene expression during early development. Cell 1994; 77: 439–449. [DOI] [PubMed] [Google Scholar]
- 31.Prioleau MN, Buckle RS, Méchali M. Programming of a repressed but committed chromatin structure during early development. EMBO J 1995; 14: 5073–5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Temeles GL, Schultz RM. Transient polyadenylation of a maternal mRNA following fertilization of mouse eggs. J Reprod Fertil 1997; 109: 223–228. [DOI] [PubMed] [Google Scholar]
- 33.Oh B, Hwang SY, Solter D, Knowles BB. Spindlin, a major maternal transcript expressed in the mouse during the transition from oocyte to embryo. Development 1997; 124: 493–503. [DOI] [PubMed] [Google Scholar]
- 34.Oh B, Hwang S, McLaughlin J, Solter D, Knowles BB. Timely translation during the mouse oocyte-to-embryo transition. Development 2000; 127: 3795–3803. [DOI] [PubMed] [Google Scholar]
- 35.Aoki F, Hara KT, Schultz RM. Acquisition of transcriptional competence in the 1-cell mouse embryo: requirement for recruitment of maternal mRNAs. Mol Reprod Dev 2003; 64: 270–274. [DOI] [PubMed] [Google Scholar]
- 36.Kanka J, Hozák P, Heyman Y, Chesné P, Degrolard J, Renard JP, Fléchon JE. Transcriptional activity and nucleolar ultrastructure of embryonic rabbit nuclei after transplantation to enucleated oocytes. Mol Reprod Dev 1996; 43: 135–144. [DOI] [PubMed] [Google Scholar]
- 37.Wiekowski M, Miranda M, DePamphilis ML. Regulation of gene expression in preimplantation mouse embryos: effects of the zygotic clock and the first mitosis on promoter and enhancer activities. Dev Biol 1991; 147: 403–414. [DOI] [PubMed] [Google Scholar]
- 38.Wiekowski M, Miranda M, DePamphilis ML. Requirements for promoter activity in mouse oocytes and embryos distinguish paternal pronuclei from maternal and zygotic nuclei. Dev Biol 1993; 159: 366–378. [DOI] [PubMed] [Google Scholar]
- 39.Henery CC, Miranda M, Wiekowski M, Wilmut I, DePamphilis ML. Repression of gene expression at the beginning of mouse development. Dev Biol 1995; 169: 448–460. [DOI] [PubMed] [Google Scholar]
- 40.Latham KE, Schultz RM. Embryonic genome activation. Front Biosci 2001; 6: D748–D759. [DOI] [PubMed] [Google Scholar]
- 41.Majumder S, Miranda M, DePamphilis ML. Analysis of gene expression in mouse preimplantation embryos demonstrates that the primary role of enhancers is to relieve repression of promoters. EMBO J 1993; 12: 1131–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smale ST, Kadonaga JT. The RNA polymerase II core promoter. Annu Rev Biochem 2003; 72: 449–479. [DOI] [PubMed] [Google Scholar]
- 43.Ooga M, Fulka H, Hashimoto S, Suzuki MG, Aoki F. Analysis of chromatin structure in mouse preimplantation embryos by fluorescent recovery after photobleaching. Epigenetics 2016; 11: 85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu F, Liu Y, Inoue A, Suzuki T, Zhao K, Zhang Y. Establishing chromatin regulatory landscape during mouse preimplantation development. Cell 2016; 165: 1375–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ke Y, Xu Y, Chen X, Feng S, Liu Z, Sun Y, Yao X, Li F, Zhu W, Gao L, Chen H, Du Z, Xie W, Xu X, Huang X, Liu J. 3D chromatin structures of mature gametes and structural reprogramming during mammalian embryogenesis. Cell 2017; 170: 367–381.e20. [DOI] [PubMed] [Google Scholar]
- 46.Hake SB, Garcia BA, Duncan EM, Kauer M, Dellaire G, Shabanowitz J, Bazett-Jones DP, Allis CD, Hunt DF. Expression patterns and post-translational modifications associated with mammalian histone H3 variants. J Biol Chem 2006; 281: 559–568. [DOI] [PubMed] [Google Scholar]
- 47.Kawamura M, Funaya S, Sugie K, Suzuki MG, Aoki F. Asymmetrical deposition and modification of histone H3 variants are essential for zygote development. Life Sci Alliance2021; 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Funaya S, Aoki F. Regulation of zygotic gene activation by chromatin structure and epigenetic factors. J Reprod Dev 2017; 63: 359–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li A, Yu Y, Lee SC, Ishibashi T, Lees-Miller SP, Ausió J. Phosphorylation of histone H2A.X by DNA-dependent protein kinase is not affected by core histone acetylation, but it alters nucleosome stability and histone H1 binding. J Biol Chem 2010; 285: 17778–17788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shinagawa T, Takagi T, Tsukamoto D, Tomaru C, Huynh LM, Sivaraman P, Kumarevel T, Inoue K, Nakato R, Katou Y, Sado T, Takahashi S, Ogura A, Shirahige K, Ishii S. Histone variants enriched in oocytes enhance reprogramming to induced pluripotent stem cells. Cell Stem Cell 2014; 14: 217–227. [DOI] [PubMed] [Google Scholar]
- 51.Nashun B, Yukawa M, Liu H, Akiyama T, Aoki F. Changes in the nuclear deposition of histone H2A variants during pre-implantation development in mice. Development 2010; 137: 3785–3794. [DOI] [PubMed] [Google Scholar]
- 52.Hamamoto G, Suzuki T, Suzuki MG, Aoki F. Regulation of transketolase like 1 gene expression in the murine one-cell stage embryos. PLoS One 2014; 9: e82087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schultz RM, Stein P, Svoboda P. The oocyte-to-embryo transition in mouse: past, present, and future. Biol Reprod 2018; 99: 160–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ishiuchi T, Abe S, Inoue K, Yeung WKA, Miki Y, Ogura A, Sasaki H. Reprogramming of the histone H3.3 landscape in the early mouse embryo. Nat Struct Mol Biol 2021; 28: 38–49. [DOI] [PubMed] [Google Scholar]
- 55.Zhang B, Zheng H, Huang B, Li W, Xiang Y, Peng X, Ming J, Wu X, Zhang Y, Xu Q, Liu W, Kou X, Zhao Y, He W, Li C, Chen B, Li Y, Wang Q, Ma J, Yin Q, Kee K, Meng A, Gao S, Xu F, Na J, Xie W. Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature 2016; 537: 553–557. [DOI] [PubMed] [Google Scholar]
- 56.Sha QQ, Zhang J, Fan HY. Function and regulation of histone H3 lysine-4 methylation during oocyte meiosis and maternal-to-zygotic transition. Front Cell Dev Biol 2020; 8: 597498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Funaya S, Ooga M, Suzuki MG, Aoki F. Linker histone H1FOO regulates the chromatin structure in mouse zygotes. FEBS Lett 2018; 592: 2414–2424. [DOI] [PubMed] [Google Scholar]
- 58.Abe KI, Funaya S, Tsukioka D, Kawamura M, Suzuki Y, Suzuki MG, Schultz RM, Aoki F. Minor zygotic gene activation is essential for mouse preimplantation development. Proc Natl Acad Sci USA 2018; 115: E6780–E6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hendrickson PG, Doráis JA, Grow EJ, Whiddon JL, Lim JW, Wike CL, Weaver BD, Pflueger C, Emery BR, Wilcox AL, Nix DA, Peterson CM, Tapscott SJ, Carrell DT, Cairns BR. Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat Genet 2017; 49: 925–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Whiddon JL, Langford AT, Wong CJ, Zhong JW, Tapscott SJ. Conservation and innovation in the DUX4-family gene network. Nat Genet 2017; 49: 935–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.De Iaco A, Planet E, Coluccio A, Verp S, Duc J, Trono D. DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat Genet 2017; 49: 941–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sugie K, Funaya S, Kawamura M, Nakamura T, Suzuki MG, Aoki F. Expression of Dux family genes in early preimplantation embryos. Sci Rep 2020; 10: 19396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hewitt JE, Lyle R, Clark LN, Valleley EM, Wright TJ, Wijmenga C, van Deutekom JC, Francis F, Sharpe PT, Hofker M, et al. Analysis of the tandem repeat locus D4Z4 associated with facioscapulohumeral muscular dystrophy. Hum Mol Genet 1994; 3: 1287–1295. [DOI] [PubMed] [Google Scholar]
- 64.Winokur ST, Bengtsson U, Feddersen J, Mathews KD, Weiffenbach B, Bailey H, Markovich RP, Murray JC, Wasmuth JJ, Altherr MR, et al. The DNA rearrangement associated with facioscapulohumeral muscular dystrophy involves a heterochromatin-associated repetitive element: implications for a role of chromatin structure in the pathogenesis of the disease. Chromosome Res 1994; 2: 225–234. [DOI] [PubMed] [Google Scholar]
- 65.Gabriëls J, Beckers MC, Ding H, De Vriese A, Plaisance S, van der Maarel SM, Padberg GW, Frants RR, Hewitt JE, Collen D, Belayew A. Nucleotide sequence of the partially deleted D4Z4 locus in a patient with FSHD identifies a putative gene within each 3.3 kb element. Gene 1999; 236: 25–32. [DOI] [PubMed] [Google Scholar]
- 66.Dixit M, Ansseau E, Tassin A, Winokur S, Shi R, Qian H, Sauvage S, Mattéotti C, van Acker AM, Leo O, Figlewicz D, Barro M, Laoudj-Chenivesse D, Belayew A, Coppée F, Chen YW. DUX4, a candidate gene of facioscapulohumeral muscular dystrophy, encodes a transcriptional activator of PITX1. Proc Natl Acad Sci USA 2007; 104: 18157–18162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Clapp J, Mitchell LM, Bolland DJ, Fantes J, Corcoran AE, Scotting PJ, Armour JA, Hewitt JE. Evolutionary conservation of a coding function for D4Z4, the tandem DNA repeat mutated in facioscapulohumeral muscular dystrophy. Am J Hum Genet 2007; 81: 264–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Percharde M, Lin CJ, Yin Y, Guan J, Peixoto GA, Bulut-Karslioglu A, Biechele S, Huang B, Shen X, Ramalho-Santos M. A LINE1-nucleolin partnership regulates early development and ESC identity. Cell 2018; 174: 391–405.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]