Abstract
In the male germline, the machinery to repress retrotransposons that threaten genomic integrity via the piRNA pathway is established in gonocytes. It has been reported that disruption of the piRNA pathway leads to activation of retrotransposons and arrests spermatogenesis before it enters the second meiosis; however, its effects on gonocytes have not been fully elucidated. In this study, we analyzed the effects of Asz1 deletion, which is a crucial component of the piRNA pathway, on the gonocyte transcriptome. In Asz1-null gonocytes, MIWI2, which is responsible for introducing DNA methylation to retrotransposons in a piRNA-dependent manner, disappeared from the nuclei of fetal gonocytes. Transcriptome analysis revealed that retrotransposons targeted by the piRNA pathway and non-annotated transcript variants were upregulated in gonocytes from neonatal Asz1-/- mice. These non-annotated transcript variants were chimeras generated by joining exons transcribed from retrotransposons and canonical genes. DNA methylation analysis showed that retrotransposons that induce the expression of aberrant chimeric transcripts are not fully methylated. This was consistent with the impaired nuclear localization of MIWI2 in Asz1-null gonocytes. Furthermore, heterogeneity of DNA methylation status in retrotransposons was observed in both gonocytes and their descendants. This suggests that the piRNA system in gonocytes can potentially prevent spermatogenic cell populations bearing aberrant chimeric transcripts from propagating later in spermatogenesis. In conclusion, Asz1 is required to repress retrotransposons and retrotransposon-driven aberrant chimeric transcripts in gonocytes through the piRNA pathway.
Keywords: piRNA, Retrotransposon, Spermatogenesis
Primordial germ cells (PGCs) are progenitor cells that differentiate into sperm and oocytes. Epigenetic reprogramming occurs during PGC differentiation. In mice, PGCs emerge at the base of the allantois by embryonic day 7.25 (E7.25), and subsequently, genome-wide DNA demethylation, including imprinting control regions and retrotransposons, is completed at least by E 13.5 [1]. Although germline cells must sustain genetic integrity and transmit accurate genetic information to the next generation, the germ cell genome must contend with retrotransposon activation owing to the cessation of DNA methylation during reprogramming [2]. In contrast, PGCs undergo sex differentiation directed by gonadal somatic cells between E11.5 and E12.5, after which male and female germ cells acquire sex-specific transcriptomes and epigenetic modifications [3,4,5]. To eliminate the threat of retrotransposons, PIWI-interacting RNA (piRNA) machinery is activated in the male germline, but it is still unclear how female germ cells acquire tolerance to retrotransposons [6, 7].
piRNAs are small RNAs expressed in germ cells and are indispensable to spermatogenesis, but not oogenesis [2, 8, 9]. The piRNA machinery works by the conjunction of intermitochondrial cement and pi-P-bodies, which are characteristic cytoplasmic compartments and proteins involved in piRNA production are localized on the outer mitochondrial membrane [10, 11]. In mouse gonocytes, piRNA cluster transcripts are targeted by piwi-like RNA-mediated gene silencing 2 (MILI/PIWIL2) after being sliced at the 5’ end [8, 12,13,14]. Thereafter, these piRNA precursor transcripts are subjected to 3’-end trimming and become mature primary piRNAs [15, 16]. MILI-primary piRNA complexes slice retrotransposon-related transcripts that complement the primary piRNA sequences, achieving post-transcriptional repression of retrotransposons and producing secondary piRNA sources [17]. These sliced transcripts are cut at the 5’ end again, loaded onto MILI, and trimmed at the 3′ end to produce mature secondary piRNAs. Secondary piRNAs slice complementary transcripts to enforce primary piRNA production and piRNA amplification. A subpopulation of piRNAs are loaded into piwi-like RNA-mediated gene silencing 4 (MIWI2/PIWIL4) via DEAD-box helicase 4 (DDX4/MVH) [18]. piRNA-loaded MIWI2 translocates from the cytoplasm to the nucleus and targets transcribed retrotransposon RNA using piRNA as a guide to introduce DNA methylation at these loci [17, 19, 20]. Thus, spermatogonia and spermatocytes exhibit an effective system to silence retrotransposons by piRNA in perinatal gonocytes [21].
Ankyrin repeats, SAM, and basic leucine zipper domain-containing 1 (Asz1), a component of the piRNA machinery, is known to be expressed specifically in germ cells of both sexes [22,23,24]. Since Asz1 expression depends on DNA methylation as well as Mili, its expression is induced as a result of genome-wide DNA demethylation in PGCs [25]. ASZ1 expression has been observed in mouse germ cells, from fetal gonocytes to postnatal round spermatids, with a peak in spermatocytes at the pachytene stage. ASZ1 is localized on the outer mitochondrial membrane and is indispensable for piRNA production as a molecular scaffold for intermitochondrial cement [26]. Asz1 deletion impairs piRNA production, downregulates MILI, and causes DNA demethylation of retrotransposons, such as long interspersed element 1 (LINE1) and intracisternal A-particle (IAP) in spermatogenic cells. Consequently, only male Asz1 knockout (KO) mice suffer from sterility accompanied by retrotransposon dysregulation and spermatogenesis arrest in the meiotic prophase I stage, similar to mice lacking Mili, Miwi2, Ddx4, or other piRNA machinery-related genes [23, 27]. Thus, Asz1 ensures spermatogenesis by repressing retrotransposons.
To date, the influence of impaired piRNA production on the gonocyte transcriptome has not been investigated in detail. Gonocytes are the only source of spermatogonial stem cells; therefore, it is imperative to know how retrotransposons affect the transcriptome of gonocytes. To address this question and obtain insight into ensuring robust spermatogenesis, we investigated the transcriptome of gonocytes with impaired piRNA pathway using male Asz1 KO mice. Furthermore, to understand the effects of retrotransposons on oocytes, in which the lack of piRNA machinery is a default state, we compared retrotransposon-derived transcripts of oocytes with those of gonocytes.
Materials and Methods
Animals
The mice used in this study were purchased from CLEA Japan (Tokyo, Japan). All experiments in this study were approved by the Institutional Animal Care and Use Committee of the Tokyo University of Agriculture (#300054).
Establishment of the Asz1 deleted mouse model
The Asz1 KO mouse was established based on a previous report [23]. Briefly, a targeting vector containing an Asz1-knockout construct was introduced into C57BL/6 embryonic stem (ES) cells. Next, ES cells harboring an Asz1 KO allele were injected into blastocysts from ICR mice, resulting in chimeric mice. Thereafter, Asz1+/– mice were produced by breeding chimeric and C57BL/6 mice. Finally, Asz1+/– mice were crossed with C57BL/6 mice and used to produce Asz1–/– mice.
Western blotting
Testes and livers from 3-week-old mice were lysed in 2 × sample buffer (380 mM Tris-HCl [pH 6.8], 4% SDS, and 20% glycerol) by homogenization with glass beads. Protein concentrations of each sample were measured using a Micro BCA Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA) on a SpectraMax i3 Multimode Microplate Reader (Molecular Devices, San Jose, CA, USA) and diluted with 4 × loading buffer (250 mM Tris-HCl [pH 6.8], 8% SDS, 40% glycerol, 10% β-mercaptoethanol, and 0.05% bromophenol blue). Proteins were then subjected to SDS-PAGE and transferred to polyvinylidene difluoride membranes, after which blocking and immunoreaction were performed in TBST containing 5% and 1% skim milk, respectively. The antibodies used and the dilution rates are listed in Supplementary Table 1.
Immunofluorescence of frozen sections
Testes were fixed with 4% paraformaldehyde in PBS overnight and then washed with PBS. After cryoprotection using 10% and 20% sucrose solutions, tissues were embedded in OCT Compound (Sakura Finetek Japan, Tokyo, Japan) and frozen in liquid nitrogen. Frozen sections of 10-µm thickness were sliced using a Leica CM1860 cryostat microtome (Leica Biosystems, Nussloch, Germany). Antigen retrieval was performed by microwave treatment (500 W for 15 min) in trisodium citrate buffer. Subsequently, blocking was performed in a blocking solution consisting of 10% fetal bovine serum (FBS), 3% bovine serum albumin (BSA), and 0.1% Triton-X 100 in 1 × PBS (–). Then, primary antibody treatment was performed at 4°C overnight, followed by secondary antibody treatment at room temperature for 1 h. Stained sections were mounted on VECTASHIELD Antifade Mounting Medium with 4,6-diamino-2-phenylindole (DAPI; Vector Laboratories, Burlingame, CA, USA). Finally, confocal images were obtained using an LSM710 microscope (Zeiss, Oberkochen, Germany). The antibodies used and the dilution rates are listed in Supplementary Table 1.
Analysis of the synaptonemal complex
Meiotic chromosome spreads from the testes of 3-week-old mice were prepared as previously described [28]. Testes were recovered from male mice, and the tunica albuginea was removed from the testes. Some seminiferous tubules were transferred into hypotonic extraction buffer and incubated for 40 min. The seminiferous tubules were then transferred into a 50 mM sucrose solution and minced using forceps and a scalpel, followed by pipetting. Thereafter, the cell suspensions were dispersed on glass slides, and then the slides were soaked in a 1% paraformaldehyde solution containing 0.15% Triton X-100. Glass slides were subsequently placed in a humidity box at room temperature for 2 h. Finally, the glass slides were washed with Milli-Q water and air-dried. Immunostaining was performed as described for immunostaining of frozen sections.
Purifying germ cells using cell sorter
For gonocyte collection, the tunica albuginea of the testes from mice at 1 day post-partum (dpp) was removed, and seminiferous tubules were minced using forceps and a scalpel. Next, seminiferous tubules were treated with 1 mg/ml collagenase (Wako, Osaka, Japan) and 1 mg/ml DNase I (Sigma-Aldrich, St. Louis, MO, USA), followed by 0.25% trypsin-EDTA to disperse the seminiferous tubules. Next, the cell suspensions were incubated with CD16/32 antibody on ice for 15 min for blocking. Subsequently, APC/Fire750 anti-mouse EpCAM antibody was added to the cell suspensions and incubated on ice for 1 h. Gonocytes were then purified using BD FACS Aria II (BD Biosciences, Franklin Lakes, NJ, USA). The purity of gonocytes was preliminarily assessed by staining sorted neonatal gonocytes with anti-GENA antibody (Supplementary Fig. 1A–B).
Fluorescence-activated cell sorter (FACS) purification of spermatogenic cells from 2-week-old mice was performed as described for gonocyte purification. Anti-MVH antibody was used to verify purity instead of anti-GENA antibody (Supplementary Fig. 1C–D).
To purify oocytes, ovaries were removed from 1 dpp mice and subjected to enzymatic digestion, as previously reported [29]. Cells were then incubated with CD16/32 antibody on ice for 15 min and Brilliant Violet 421 anti-mouse c-kit antibody on ice for 1 h followed by FACS (Supplementary Fig. 1E). Oocyte purity was confirmed by counting the number of c-kit-positive cells among the total cells (Supplementary Fig. 1F). The dilution rates of the antibodies are listed in Supplementary Table 1.
RNA-sequencing analysis
Duplicate biological libraries of gonocytes from wild-type (WT) and Asz1–/– mice were prepared and subjected to paired-end RNA sequencing (RNA-seq) on a NextSeq 500 platform (Illumina, San Diego, CA, USA). Total RNA was extracted from gonocytes using the RNeasy Plus Micro Kit (Qiagen, Hilden, Germany). cDNA synthesis was performed using the SMART-Seq v4 Ultra Low Input RNA Kit for Sequencing (Takara, Shiga, Japan), and libraries were constructed using the Nextera XT DNA Library Prep Kit (Illumina). Sequencing data are available in the DNA Data Bank of Japan database under the accession number DRA013153.
Data analyses for RNA-seq
The UCSC mm10 mouse genome and gene annotations were downloaded from Illumina iGenomes (http://jp.support.illumina.com/sequencing/sequencing_software/igenome.html). Annotation files for retrotransposons and repeats were obtained from the repeat masker track on the UCSC genome browser (https://genome.ucsc.edu/).
RNA-seq reads were trimmed using Trimmomatic (version 0.36), and only paired reads were subjected to the following analyses (Supplementary Fig. 2) [30]. To count the reads of genes without retroelements, reads associated with retrotransposons were removed by mapping them to the linearized repeat sequence from the repeat masker track. The remaining reads were aligned with the genome as paired reads. Hisat2 (version 2.1.0) with default settings was used to align reads, and reads mapped on the mitochondrial sequence were removed using bedtools (version 2.27.1) [31, 32]. In contrast, for counting reads on retrotransposons, trimmed reads that could be mapped on exons as paired reads were removed using bedtools. Reads were mapped using Hisat2 with the following options: -k 10000 --no-spliced-alignment --score-min C,-1. Read counts were performed using the mapped data separately for retrotransposons and other genes. featureCounts (version 1.6.2) was used for read counts with the following options: -B -P -M -O --fraction [33]. The read count tables were combined and subjected to differential expression analysis and RPM calculation using the edgeR package (version 3.22.5) with RPM 5 as the cutoff.
To visualize RNA-seq read coverages, bam files of replicates were merged and converted to bigwig files using CPM normalization in deepTools (version 3.5.1) [34]. BigWig files were visualized using the IGV genome browser [35].
RNA-seq data of WT non-growing oocytes from 1 dpp female mice were downloaded from DRA005345 [36] and used for analysis.
Identification of aberrant chimeric transcripts
Reads mapped to the genome were assembled using StringTie software (version 1.3.4.d) using the gene annotation file as a guide [37]. Aberrant chimeric transcripts were defined using the following criteria: transcript variants have 5' exon(s) that have not been annotated in NCBI RefSeqGene (GRCm38); the read counts of the 5' exons of the transcript variants were significantly increased in Asz1–/– gonocytes (FDR < 0.05), and canonical transcripts accompanied with non-annotated 5' exons were significantly upregulated in Asz1–/– gonocytes (FDR < 0.01). When multiple aberrant chimeric transcript variants were identified at a locus, the variant containing the 5' exon with the highest read counts was selected for analysis. The EdgeR package was used for all the differential expression analyses.
Expression analysis of IAP-derived- Fanconi anemia, complementation group D2
Total RNA was extracted from FACS-purified gonocytes and non-growing oocytes from 1 dpp mice using an RNeasy Plus Micro Kit (Qiagen) according to the manufacturer’s protocol. Subsequently, genomic DNA removal and first-strand cDNA synthesis were performed using a PrimeScrip RT reagent Kit with gDNA Eraser (Takara). The absolute expression levels of IAP-Fanconi anemia, complementation group D2 (IAP-Fancd2), and Tbp were investigated by quantitative RT-PCR (qRT-PCR) using Power SYBR Green Master Mix (Thermo Fisher Scientific) on a QuantStudio 3 real-time PCR system (Applied Biosystems, Waltham, MA, USA), and compared with standard curves. The expression levels of IAP-Fancd2 were normalized to those of Tbp as an internal control, and statistical analysis was performed. Sequencing and bisulfite sequencing analyses were performed as described previously [38]. Details are shown in Supplementary Information and Supplementary Table 2.
Analysis of chromatin immunoprecipitation-sequencing data
Chromatin immunoprecipitation sequencing (ChIP-seq) data were downloaded from DRA006633 [39]. ChIP-seq reads were trimmed using Trimmomatic (version 0.36), and only paired reads were subjected to subsequent analyses. Reads were mapped to the genome using Bowtie2 with default settings. Reads mapped on the ENCODE blacklist regions, the Y chromosome, and the mitochondrial genome were removed using bedtools. To determine the dimethylation and trimethylation of histone H3 lysine 9 (H3K9me2 and H3K9me3) accumulation, peak calling was performed on each of the two mapping datasets using MACS2 (ver. 2.1.4) with the following options: -f BAMPE -g mm --broad -p 1e-5 [40]. Overlapping peaks in both datasets were extracted using bedtools. Visualization of read coverage was performed for RNA-seq data analysis.
Statistical analysis
Dunnett’s test was performed for gene expression analyses using the Multicomp package (ver. 1.4.17) using R software (ver. 3.6.1) [41].
Results
Asz1–/– mice phenotype
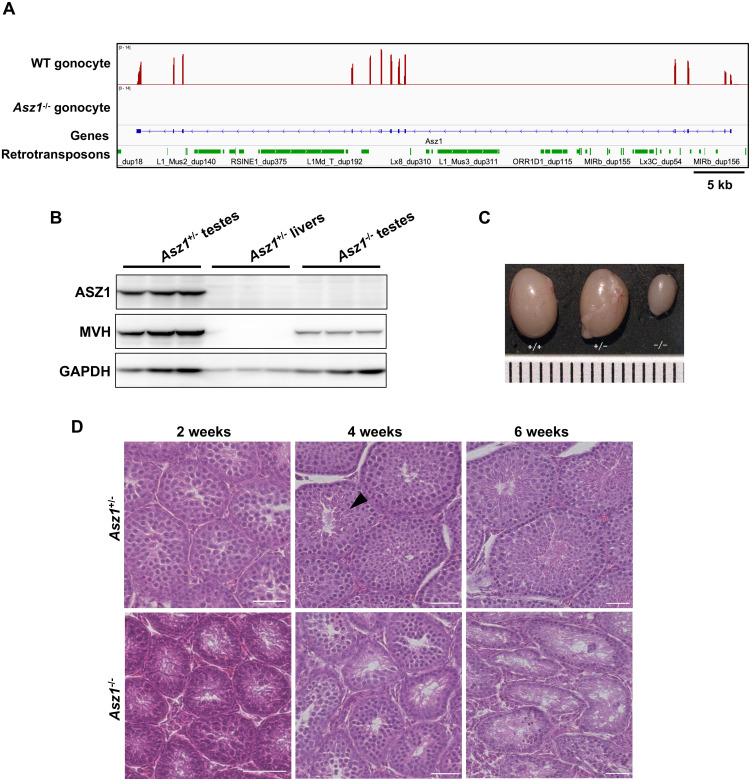
Initially, we confirmed the presence of RNA-seq reads mapped to Asz1 in gonocytes from 1dpp WT mice but not in Asz1–/– mice (Fig. 1A). Western blotting showed that ASZ1 was completely depleted in the testes of 3-week-old Asz1–/– mice, in contrast to the testes of WT mice (Fig. 1B). The testes of 6-week-old Asz1–/– mice contracted more severely than those of WT and Asz1+/– male mice (Fig. 1C). Histological analysis clearly showed that spermatocytes were observed in both Asz1+/– and Asz1–/– testes from 2-week-old mice (Fig. 1D). However, elongated spermatids appeared in the testes of 4-week-old Asz1+/– mice, but not in those of similarly aged Asz1–/– mice. Consequently, spermatogenic cells were degenerated in 6-week-old male Asz1–/– mice. This phenotype corresponds to that previously reported in Asz1-null mice [23].
Fig. 1.
Defects in spermatogenesis in Asz1–/– mice. (A) Expression of Asz1 in gonocytes of 1 dpp Asz1+/– and Asz1–/– mice. (B) Western blotting of testes from 3-week-old wild-type (left) and Asz1–/– mice (right) and livers from 3-week-old Asz1+/– mice (negative control, middle). Western blot analysis were performed using three biologically independent samples, however, three lanes for each sample are technical replicates in this gel image. (C) Testes from 6-week-old wild-type (left), Asz1+/– (middle), and Asz1–/– mice (right). The ruler scale is 1 mm wide. (D) Histological analysis of testes from 2-, 4-, and 6-week-old Asz1+/– (top) and Asz1–/– mice (bottom). An arrowhead indicates an elongated spermatid. Scale bars indicate 100 μm.
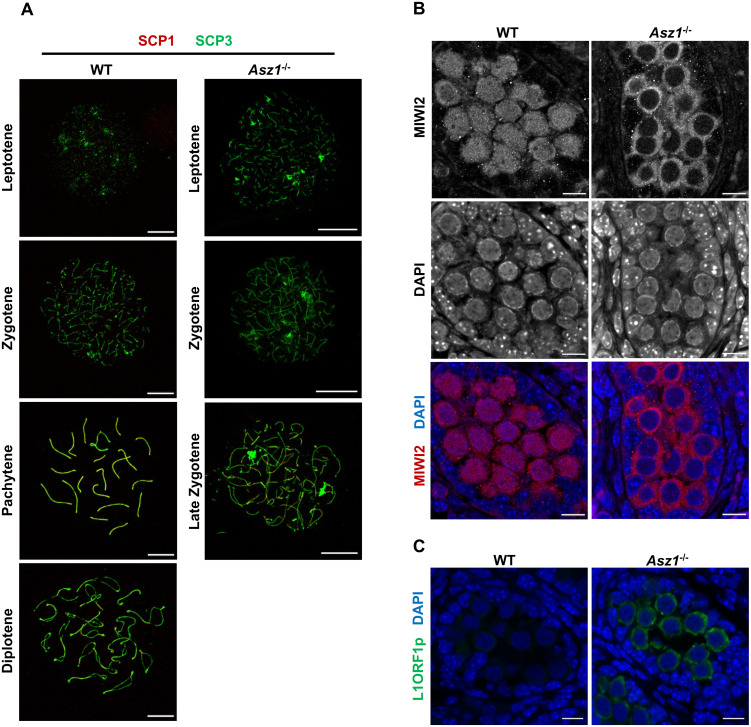
To assess when the spermatogenic defect was most prominent, immunostaining was performed against the synaptonemal complex (Fig. 2A). Spermatocytes were observed in the testes of 3-week-old WT mice at all stages of meiotic prophase I. In contrast, spermatocytes of Asz1–/– mice did not reach the pachytene stage. The most advanced spermatocytes in Asz1–/– testes were in the late zygotene stage, in which homologous chromosomes underwent partial synapsis. This finding indicates that spermatogenesis in Asz1–/– mice is arrested at the same stage not only in Mili–/– mice but also in Miwi2–/– mice [12, 21].
Fig. 2.
Failure of synapsis in spermatocytes and of nuclear localization of MIWI2 in gonocytes of Asz1–/– mice. (A) Immunofluorescence of the synaptonemal complex in spermatocytes from the testes of 3-week-old wild-type (left) and Asz1–/– (right) mice. SCP1 and SCP3 were labeled red and green, respectively. Scale bars indicate 10 μm. (B) Localization of MIWI2 (red) in gonocytes from testes at E17.5 in wild-type (left) and Asz1–/– (right) mice. Nuclei (blue) were counterstained with DAPI. Scale bars indicate 10 μm. (C) Expression of LINE1 open reading frame1 protein (L1ORF1p, green) in gonocytes from testes at E17.5 in wild-type (left) and Asz1–/– (right) fetuses. Nuclei (blue) were counterstained with DAPI. Scale bars indicate 10 μm.
Disruption of nuclear localization of MIWI2 in the gonocytes of Asz1–/– mice
Asz1 deficiency has been reported to cause downregulation of MILI, which induces post-transcriptional repression of retrotransposons in gonocytes [23]. However, it is still unclear whether Asz1-deletion affects MIWI2 expression, which introduces DNA methylation marks on retrotransposons within a narrow time frame. Therefore, we investigated MIWI2 expression in the fetal testes. Immunofluorescence analysis revealed that MIWI2 was expressed in the gonocytes of both testes from WT and Asz1–/– embryos at E17.5 (Fig. 2B). However, although MIWI2 was detected in both the cytoplasm and nuclei of WT fetal gonocytes, nuclear localization of MIWI2 was completely eliminated in the gonocytes of Asz1–/– embryos. This suggests that transcriptional repression of retrotransposon elements is disrupted in Asz1–/– gonocytes, presumably due to a lack of robust DNA methylation. Indeed, prominent upregulation of LINE1 open reading frame1 protein (L1ORF1p) in Asz1–/– gonocytes (Fig. 2C) was consistent with the fact that piRNA pathway dysfunction causes retrotransposon derepression in gonocytes [21, 23].
The effects of piRNA pathway dysfunction on the gonocyte transcriptome
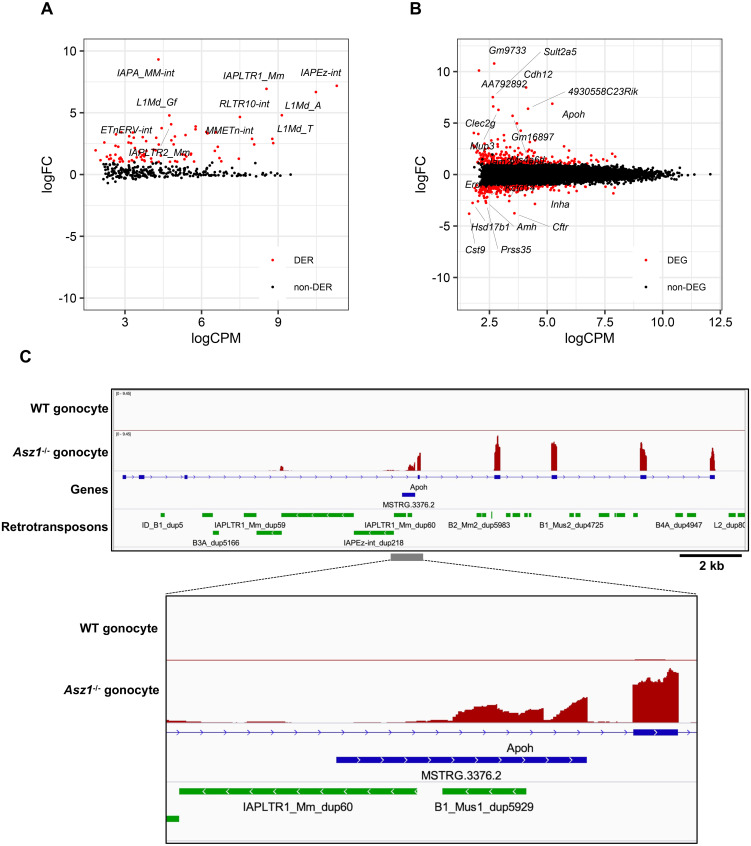
Next, the influence of retrotransposon derepression on the gonocyte transcriptome was investigated. Our RNA-seq data also showed that LINE and long terminal repeat (LTR)-type retrotransposons were upregulated in 1 dpp Asz1–/– mouse gonocytes compared to those of WT mice (Fig. 3A). Furthermore, differential gene expression analysis revealed that 275 and 216 genes, excluding retrotransposons, were significantly upregulated and downregulated, respectively, in Asz1–/– 1 dpp mouse gonocytes relative to those of WT mice (Fig. 3B). Since gene ontology analysis of these 491 differentially expressed genes (DEGs) revealed enrichment of only a few terms (data not shown), we focused on DEGs individually. Features that can explain abnormal spermatogenesis or the association with retrotransposon activity were not found in downregulated DEGs but in upregulated DEGs (Fig. 3B and Supplementary Table 3). The expression levels of apolipoprotein H (Apoh), which is expressed in adult testes and is suggested to be involved in apoptosis [42], were approximately 100-fold higher in Asz1–/– mouse gonocytes than in WT mice (Fig. 3B and 3C). Interestingly, the Apoh transcription start site was newly formed in intron 3, which was absent in canonical exons 1 to 3 (Fig. 3C). A manual view of the alignment reads revealed that some reads were mapped across the new exon and the original Apoh exon 4. Thus, the transcript variant of Apoh was upregulated in Asz1–/– mouse gonocytes. To understand the cause of the increased expression of the transcript variant at the Apoh loci, we investigated the reference sequence of Apoh intron 3. As shown in Fig. 3C, IAPLTR1_Mm_dup60, which is an LTR retrotransposon, and B1_Mus1_dup5929, a short interspersed element (SINE), is located in Apoh intron 3. To date, there has been no evidence that SINE expression is regulated by piRNAs. Indeed, our data did not show upregulation of any SINE-type retrotransposons (Supplementary Fig. 3). Taken together, we hypothesized that transcription from IAPLTR1_Mm_dup60 was activated in Asz1–/– gonocytes (Fig. 3A), and IAPLTR1_Mm_dup60 may trigger the generation of the ectopic transcription start site of Apoh (hereafter referred to as IAP-Apoh). Specifically, an aberrant chimeric transcript variant is activated by the promoter activity of a derepressed retrotransposon.
Fig. 3.
RNA-seq analyses in gonocytes of Asz1–/– mice. (A, B) MA plots depicting changes in expression levels of retrotransposons (A) and genes (B) in gonocytes of Asz1–/– mice relative to those of wild-type mice. Differentially expressed retrotransposons (DERs) and genes (DEGs) in gonocytes of Asz1–/– and wild-type mice were defined by statistical significance (FDR < 0.01) and a greater than 2-fold change. DERs and DEGs are shown as red plots. Non-DERs and non-DEGs are shown as black plots. (C) IGV genome browser tracks showing Apoh in gonocytes of wild-type and Asz1–/– mice. MSTRG.3376.2 is the aberrant exon of the Apoh transcript variant identified by StringTie software.
Detection of aberrant chimeric transcript variants in gonocytes
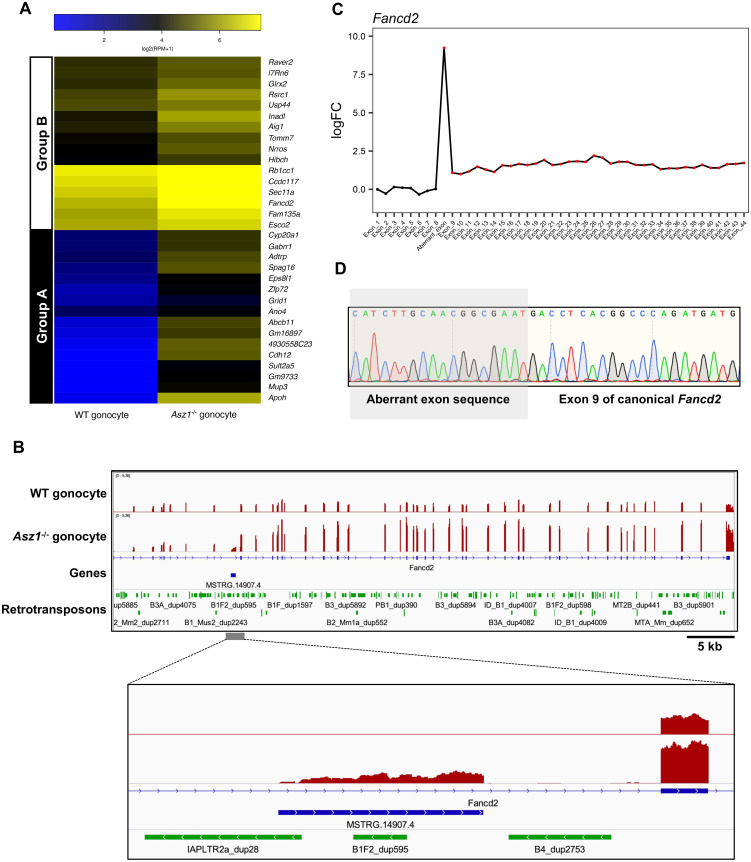
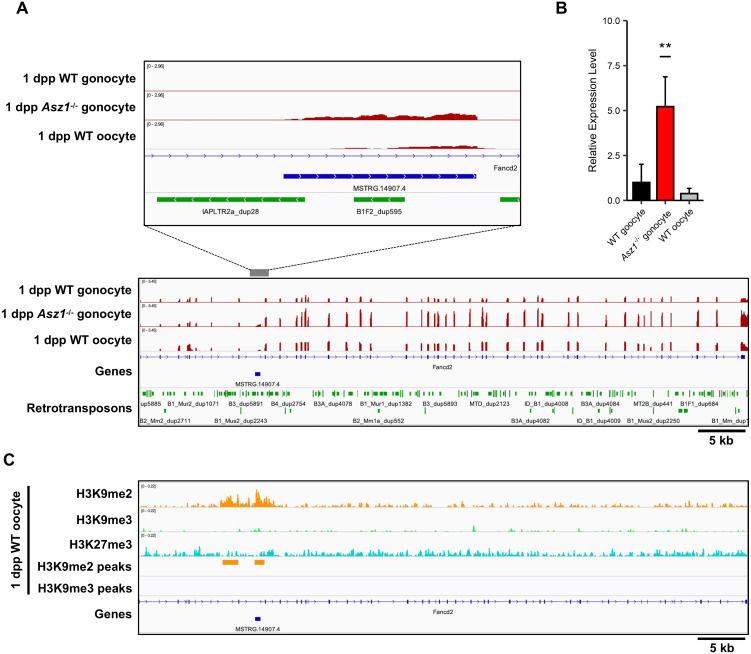
As described above, it has been suggested that derepressed retrotransposons drive the transcription of neighboring genes, rendering the gonocyte genome susceptible to generating transcript variants. Therefore, we attempted to detect retrotransposon-driven transcript variants among DEGs in Asz1–/– gonocytes. As a result, 32 transcripts were identified as aberrant chimeric transcript variants (Supplementary Table 4). Most of them lacked one or more exons of canonical transcripts because retrotransposons are located in intronic regions of the genes, and retrotransposon-driven transcript variants are shared with the 3' exons of the canonical transcripts. Therefore, the expression levels of the aberrant chimeric transcript variants were comprehensively investigated (Fig. 4A). These transcripts were divided into two groups based on hierarchical clustering of their expression patterns between WT and Asz1–/– mouse gonocytes. Group A contained transcripts that were not constitutively expressed in WT mouse gonocytes but were expressed by upstream retrotransposons in Asz1–/– mouse gonocytes, such as Apoh. In contrast, transcripts classified into group B exhibited higher expression levels in Asz1–/– mice gonocytes than in those of WT mice due to retrotransposon-driven aberrant transcripts in Asz1–/– mouse gonocytes. We focused on the expression of Fancd2 among the transcripts in group B (Fig. 4B). An integrative genomic viewer revealed that canonical Fancd2 is expressed in gonocytes of both WT and Asz1–/– mice. However, the aberrant chimeric transcript variant of Fancd2 (hereafter referred to as IAP-Fancd2) was expressed upstream of canonical Fancd2 exon 8 only in Asz1–/– mouse gonocytes. This led to increased read counts of canonical Fancd2 exon 8 and its 3' exons, resulting in the detection of Fancd2 upregulation (Fig. 4C). Sequencing analysis of IAP-Fancd2 demonstrated that the splice site predicted from a genomic context and the exon 8 of canonical Fancd2 were joined (Fig. 4D). In addition to IAP-Fancd2, one or more LTR-or LINE-type retrotransposons were located within 1 kb upstream of 28 out of 32 aberrant chimeric transcript variants (Supplementary Table 5).
Fig. 4.
Expression of aberrant chimeric transcripts in gonocytes of Asz1–/– mice. (A) Heat map showing expression levels of canonical genes in gonocytes of wild-type mice and canonical genes and their variants in gonocytes of Asz1–/– mice. Group A contains genes that are not constitutively expressed in gonocytes of wild-type mice but are expressed from their upstream or intronic retrotransposon in gonocytes of Asz1–/– mice. Group B contains genes that are expressed in gonocytes of wild-type mice but are significantly upregulated, accompanied by retrotransposon-driven non-annotated exon(s) in gonocytes of Asz1–/– mice. (B) IGV genome browser tracks showing Fancd2 in gonocytes of wild-type and Asz1–/– mice. MSTRG.14907.4 is the aberrant exon of the Fancd2 transcript variant identified by StringTie software. (C) Expression levels (log2-fold change) of each exon in gonocytes of Asz1–/– mice relative to those in wild-type mice. Annotated Fancd2 contains 44 exons. The aberrant chimeric variant (MSTRG.14907.4 in Fig. 4B) is transcribed from IAP between Fancd2 exon 8 and 9 in gonocytes of Asz1–/– mice. Namely, the aberrant exon indicates the first exon of IAP-Fancd2. A red dot indicates significantly upregulated expression of an exon (FDR < 0.05). (D) Sequencing of the PCR product confirmed the splice junction site of IAP-Fancd2. Aberrant exons were transcribed from IAPLTR2a_dup28, resulting in the expression of an aberrant chimeric transcript variant, MSSTRG.14907.4.
Taken together, our results indicate that aberrant chimeric transcript variants disrupt the normal transcriptome profile of gonocytes by derepressing retrotransposons in Asz1–/– mice.
DNA methylation in Asz1–/– mice gonocytes
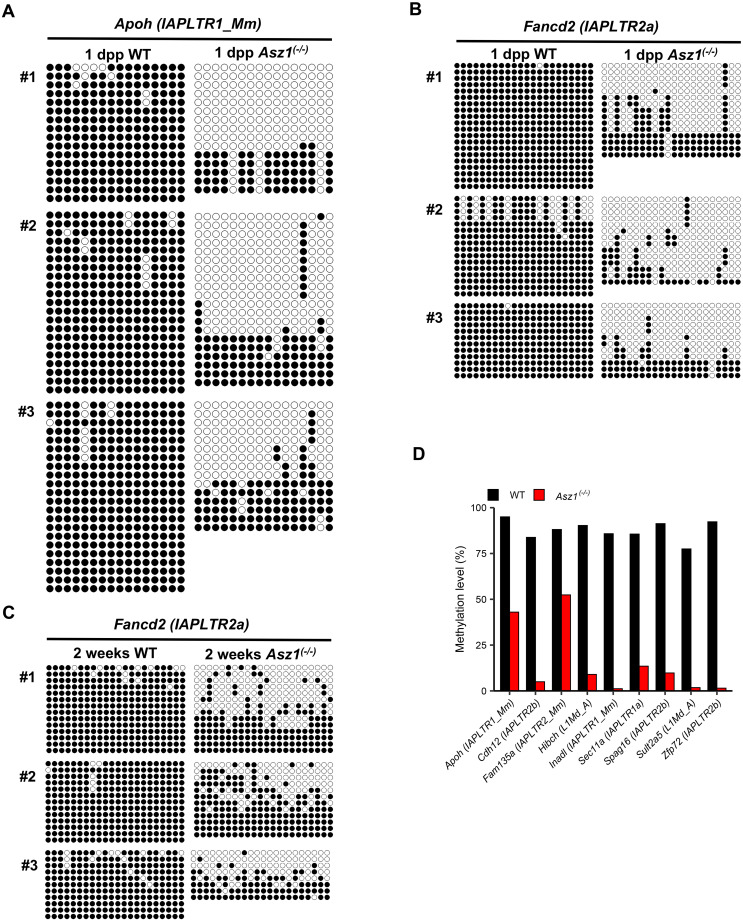
Next, we examined the DNA methylation status of the retrotransposons that cause transcript variants in gonocytes from 1 dpp mice. At the IAPLTR1_Mm_dup60 locus, which drove IAP-Apoh transcription, both hyper- and hypomethylated DNA strands were detected in the gonocytes of Asz1–/– mice, although they were almost completely hypermethylated in the gonocytes of WT mice (Fig. 5A). DNA methylation analyses of IAPLTR2a_dup28, which drove IAP-Fancd2 transcription, also showed similar tendencies (Fig. 5B). To understand whether gonocytes harboring hypermethylated alleles could survive in the testes of Asz1–/– mice, we performed DNA methylation analysis in testicular germ cells, that is, gonocyte descendants. Bimodal methylation was also observed in testicular germ cells from 2-week-old Asz1–/– mice (Fig. 5C). Although the average DNA methylation levels of retrotransposons associated with nine another retrotransposon-driven transcript variants were lower in gonocytes of Asz1–/– mice than in WT mice at 2 weeks of age (Fig. 5D), hypermethylated alleles were also detected at each locus. Thus, loss of DNA methylation of retrotransposons in Asz1–/– mouse gonocytes may be the underlying cause for derepression of retrotransposons and expression of aberrant chimeric transcript variants.
Fig. 5.
Bisulfite sequencing analyses of retrotransposons located within 1 kb upstream of aberrant chimeric transcript variants in spermatogenic cells. (A–C) DNA methylation status of IAPLTR_Mm associated with IAP-Apoh (A) and IAPLTR2a associated with IAP-Fancd2 (B) in gonocytes of 1 dpp wild-type and Asz1–/– mice, and IAPLTR2a associated with IAP-Fancd2 in spermatogenic cells from 2-week-old wild-type and Asz1–/– mice (C). Each line indicates the analyzed DNA strand. Black and white circles indicate methylated and unmethylated cytosine residues of cytosine-guanine dinucleotides. #1–#3 indicate the biologically independent samples. (D) DNA methylation status of retrotransposons associated with aberrant chimeric transcript variants in spermatogenic cells from 2-week-old wild-type (black bars) and Asz1–/– mice (red bars). DNA methylation analysis was performed on the two biological samples.
IAP-Fancd2 is repressed in neonatal oocytes independent of the piRNA pathway
MIWI2, which is critical for DNA methylation of retrotransposons, is not expressed in oocytes during oogenesis [7]. Furthermore, our previous study showed that DNA methyltransferase 3a2 (DNMT3A2) and 3L (DNMT3L) are not expressed in non-growing oocytes from 1 dpp female mice [43]. These results prompted us to investigate the expression of a retrotransposon-driven transcript variant IAP-Fancd2 in neonatal oocytes. RNA-seq data of non-growing oocytes from 1 dpp mice exhibited low expression of IAP-Fancd2 (Fig. 6A). qRT-PCR analysis also revealed much lower expression levels of IAP-Fancd2 in 1 dpp oocytes than in Asz1–/– mouse gonocytes (Fig. 6B). To understand the mechanism of IAP-Fancd2 repression in non-growing oocytes lacking MIWI2, DNMT3A2, and DNMT3L expression, we investigated repressive histone modifications. ChIP-seq data showed that H3K9me2 was enriched at the transcriptional start site of IAP-Fancd2 in non-growing oocytes of 1 dpp mice (Fig. 6C). Similar to IAP-Fancd2, H3K9me2 and H3K9me3 were enriched in IAP-Apoh in non-growing oocytes (Supplementary Fig. 4). Repressive histone marks on these regions were also observed in male and female germ cells of E13.5 fetuses, and gonocytes of the 1 dpp mice (Supplementary Fig. 5 and Supplementary Fig. 6). These data suggest that repression of retrotransposon-driven aberrant transcripts may be achieved through histone modification in a piRNA-independent manner in early spermatogenesis and oogenesis.
Fig. 6.
Expression of IAP-Fancd2 in non-growing oocytes of 1dpp mice. (A) IGV genome browser tracks showing comparative expression levels of IAP-Fancd2 among gonocytes of 1 dpp wild-type and Asz1–/– mice and non-growing oocytes of 1 dpp wild-type mice. (B) qRT-PCR analyses of IAP-Fancd2 in gonocytes of 1 dpp wild-type and Asz1–/– mice and non-growing oocytes of 1dpp wild-type mice. Despite the absence of DNA methylation, the expression level of IAP-Fancd2 in non-growing oocytes of wild-type mice was much lower than that in gonocytes of Asz1–/– mice. Data are mean ± SD, and multiple comparisons were performed using Dunnett’s test. ** P < 0.01. (C) IGV genome browser tracks showing ChIP-seq profiles of genomic regions of Fancd2 in non-growing oocytes of 1 dpp wild-type mice. Significant enrichment of H3K9me2 was detected by MACS2 in non-growing oocytes of 1 dpp wild-type mice. Previously published data sets (DRA005345 and DRA006633) of RNA-seq and ChIP-seq in non-growing oocytes were used.
Discussion
The effects of the absence of the fetal piRNA pathway on gonocytes are not fully understood. In the present study, we investigated the transcriptomic effects of fetal piRNA dysfunction in gonocytes using Asz1 KO mice. We revealed that gonocytes before mitotic resumption give rise to aberrant chimeric transcripts due to a lack of piRNA machinery.
It has been shown that the phenotype of Asz1–/– mice is similar to that of Mili-null mice. ASZ1 colocalizes with MILI on the mitochondrial outer membrane and is essential for piRNA production, presumably by providing a scaffold with MILI [12, 21, 23, 26]. It is also known that loss of Asz1 depletes MILI in gonocytes [23]. MILI regulates post-transcriptional repression of retrotransposons, whereas MIWI2 binds to piRNA produced by MILI and recruits DNA methyltransferases to a retrotransposon sequence to repress its transcription [6, 19]. First, we demonstrated that Asz1–/– male mice exhibited the same phenotypes as Mili-null mice. As reported previously, meiotic spermatocytes of Asz1–/– mice did not progress beyond the zygotene stage (Fig. 2A), which is consistent with previous reports on Mili- or Miwi2-null mice [12, 21]. Furthermore, we found that MIWI2 was localized in both the nuclei and cytoplasm of WT mice gonocytes but was excluded from the nuclei of Asz1–/– mice, presumably due to the depletion of MILI in Asz1–/– mice (Fig. 2B). Loss of DNA methylation at retrotransposon elements and loss of retrotransposon expression was also observed in Asz1–/– mouse gonocytes (Fig. 5 and Fig. 3A). Thus, we emphasize that Asz1 deletion results in MILI and MIWI2 dysfunction in gonocytes.
Next, the effects of Asz1-deletion on the transcriptome profiles were investigated in the gonocytes of 1 dpp mice. Gonocytes at the center of the testicular cord are the origin of spermatogonial stem cells and the 1st wave of spermatogenic cells. However, there have been few reports on transcriptome analysis of gonocytes lacking piRNA machinery [44]. Here, we showed that only a small number of DEGs were identified in Asz1–/– mouse gonocytes compared to those in WT mice. This is consistent with the fact that fetal piRNAs are unlikely to target exons of protein-coding genes, and gonocytes lacking piRNA machinery do not exhibit prominent abnormal phenotypes [10, 45].
Currently, DEGs in gonocytes do not delineate the pathways. It remains unknown whether up- or downregulation of these genes is a secondary effect of certain genes. Among the DEGs, Apoh, one of the highly upregulated genes in Asz1–/– mice, consisted of non-annotated transcript variants that were transcribed by the promoter of the derepressed IAP retrotransposon, which is located in the Apoh intron (Fig. 3C). It has been reported that Apoh is upregulated in the testes of Mili–/– mice at 10 days old and is upregulated by IAP retrotransposon promoter activity in spermatogonia of Dnmt3l- or Miwi2-knockout mice [20, 46]. Therefore, we revealed that the expression of aberrant chimeric transcript variants is initiated in gonocytes before the differentiation of spermatogenic cells in mice lacking piRNAs. In addition, upregulation of aberrant chimeric transcripts in Asz1–/– mouse gonocytes was induced by a lack of DNA methylation rather than post-transcriptional repression mechanisms. Indeed, we found that, in most cases, the retrotransposon element that is governed by piRNA is located within 1 kb upstream of aberrant chimeric transcript variants (Supplementary Table 5) and is not fully methylated in Asz1–/– mouse gonocytes (Fig. 5A–B). Interestingly, only a few aberrant chimeric transcript variants had retrotransposon elements, such as SINE. To the best of our knowledge, there are no studies reporting that SINE is regulated by piRNAs. Therefore, it remains unknown why these aberrant transcripts were expressed when Asz1 was deleted.
We demonstrated that Apoh, Sult2a5, Cdh12, etc., were expressed in gonocytes of in Asz1–/– mice, but not those of WT mice. Therefore, these retrotransposon-dependent transcript variants were easily detected as DEGs (Fig. 3B). On the other hand, we found that transcript variants of constitutively expressed genes generated by retrotransposons in WT mouse gonocytes which are essential for normal spermatogenesis, such as IAP-Fancd2, were significantly upregulated in Asz1–/– mice. These aberrant chimeric transcript variants may have been difficult to detect because they are hidden by the expression of canonical transcripts in mice lacking piRNA machinery. Aberrant chimeric transcript variants, such as IAP-Fancd2, have not been observed in spermatogenic cells, including gonocytes, in Miwi2-, Pld6-, or Dnmt3l-null mice [47]. Our results suggest that the piRNA pathway plays a role in repressing aberrant chimeric mRNA via DNA methylation in gonocytes earlier than in spermatocytes. In addition to DNA damage, the accumulation of aberrant chimeric transcripts during differentiation of gonocytes to spermatocytes by derepression of retrotransposons may be one of causes for inhibiting normal spermatogenesis [21]. Further studies are required to understand whether aberrant chimeric transcripts affect spermatogenesis.
In gonocytes and spermatogenic cells of Asz1–/– mice, the DNA methylation status of retrotransposons located in a region upstream of aberrant chimeric transcript variants exhibited hyper- and hypomethylation (Fig. 5). Hypomethylation of retrotransposons may be due to MIWI2 dysfunction in Asz1–/– mice, whereas hypermethylated alleles of retrotransposons in Asz1–/– mice could be a result of their resistance to genome-wide DNA demethylation during PGC differentiation or de novo DNA methylation independent of the piRNA pathway in gonocytes [4, 48]. Furthermore, such heterogeneous DNA methylation status might induce heterogeneous expression of aberrant chimeric transcript variants among spermatogenic cells in Asz1–/– mice. Gonocytes differentiated from PGCs in fetal testes resume mitotic proliferation and acquire spermatogonial stem cell capability between 2 and 4 dpp in mice [49, 50]. Therefore, the piRNA pathway, including ASZ1, has the potential to prevent heterogeneity in spermatogonial stem cells.
Although MIWI2 is not expressed during oogenesis, IAP-Fancd2 was repressed to the basal level in neonatal oocytes, suggesting that H3K9me2 is involved in repressing retrotransposons. Repressive histone marks on retrotransposon have already been enriched in both male and female germ cells by E13.5. In male germlines, however, demethylation of H3K9 and H3K27 on retrotransposons and H3K4 trimethylation occurs, leading to transient upregulation of retrotransposons [51]. This process occurs concomitantly with the transcription of piRNA clusters, including antisense retrotransposon sequences and piRNA production [52]. To establish DNA methylation and repress retrotransposons, lysine (K)-specific demethylase (KDM) 1A and 5 B interacting with MIWI2 are thought to catalyze the demethylation of H3K4 on retrotransposons in fetal gonocytes [53]. The present study demonstrated that di- and trimethylation of H3K9 was enriched in the IAP-Fancd2 and IAP-Apoh loci in male germ cells of E13.5 fetuses and gonocytes of 1 dpp mice. It is unknown whether these repressive histone marks are locally maintained from E13.5, or reestablished after DNA methylation in the perinatal stage. In contrast, in the female germline, oocytes might depend on H3K9me2 to repress retrotransposons instead of DNA methylation via the piRNA pathway. Indeed, it has been reported that chimeric transcripts mainly with IAP are generated in female germ cells in SET domain bifurcated 1 (Setdb1, encoding a histone lysine methyltransferase) conditional knockout mouse fetuses at E13.5, produced by crossing tissue-nonspecific alkaline phosphatase-Cre recombinase (TNAP-Cre) mice and Setdb1 flox mice [54]. These conditional knockout mice had reduced ovary size, and the maternal Setdb1-deleted allele was never transmitted to their descendants. However, the effect of SETDB1 deletion on oocytes has not been investigated because of the drastic loss of oocytes. In this regard, an oocyte-specific gene knockdown system that exhibits a milder phenotype than that a gene KO system which can avoid germ cell loss may be able to elucidate how retrotransposon derepression affects oocyte development during oogenesis [55].
In conclusion, we found that Asz1 repressed retrotransposons and retrotransposon-driven aberrant chimeric transcripts in gonocytes through DNA methylation via the piRNA pathway. The piRNA pathway has a potential role in the homogenous transcriptome of gametes to ensure the integrity of spermatogenesis. In contrast, a piRNA-independent repressive mechanism of retrotransposon-driven aberrant chimeric transcripts was observed in oocytes. Further studies are required to understand the impact of retrotransposons on oocyte development.
Conflict of interests
The authors declare that there are no conflicts of interests.
Supplementary
Acknowledgments
We thank Mr. Tomoya HASUNUMA and the members of the Animal Life Science Research Center (Tokyo University of Agriculture) for their technical support. The authors also thank the NODAI Genome Research Center, Tokyo University of Agriculture for the sequencing. This work was supported in part by KAKENHI (18H05547 to Y.O.).
References
- 1.Saitou M, Kagiwada S, Kurimoto K. Epigenetic reprogramming in mouse pre-implantation development and primordial germ cells. Development 2012; 139: 15–31. [DOI] [PubMed] [Google Scholar]
- 2.Crichton JH, Dunican DS, Maclennan M, Meehan RR, Adams IR. Defending the genome from the enemy within: mechanisms of retrotransposon suppression in the mouse germline. Cell Mol Life Sci 2014; 71: 1581–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adams IR, McLaren A. Sexually dimorphic development of mouse primordial germ cells: switching from oogenesis to spermatogenesis. Development 2002; 129: 1155–1164. [DOI] [PubMed] [Google Scholar]
- 4.Kobayashi H, Sakurai T, Miura F, Imai M, Mochiduki K, Yanagisawa E, Sakashita A, Wakai T, Suzuki Y, Ito T, Matsui Y, Kono T. High-resolution DNA methylome analysis of primordial germ cells identifies gender-specific reprogramming in mice. Genome Res 2013; 23: 616–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sakashita A, Kawabata Y, Jincho Y, Tajima S, Kumamoto S, Kobayashi H, Matsui Y, Kono T. Sex specification and heterogeneity of primordial germ cells in mice. PLoS One 2015; 10: e0144836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aravin AA, Sachidanandam R, Bourc’his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell 2008; 31: 785–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kabayama Y, Toh H, Katanaya A, Sakurai T, Chuma S, Kuramochi-Miyagawa S, Saga Y, Nakano T, Sasaki H. Roles of MIWI, MILI and PLD6 in small RNA regulation in mouse growing oocytes. Nucleic Acids Res 2017; 45: 5387–5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 2007; 316: 744–747. [DOI] [PubMed] [Google Scholar]
- 9.Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, Surani MA, Sakaki Y, Sasaki H. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 2008; 453: 539–543. [DOI] [PubMed] [Google Scholar]
- 10.Aravin AA, van der Heijden GW, Castañeda J, Vagin VV, Hannon GJ, Bortvin A. Cytoplasmic compartmentalization of the fetal piRNA pathway in mice. PLoS Genet 2009; 5: e1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watanabe T, Chuma S, Yamamoto Y, Kuramochi-Miyagawa S, Totoki Y, Toyoda A, Hoki Y, Fujiyama A, Shibata T, Sado T, Noce T, Nakano T, Nakatsuji N, Lin H, Sasaki H. MITOPLD is a mitochondrial protein essential for nuage formation and piRNA biogenesis in the mouse germline. Dev Cell 2011; 20: 364–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, Lin H, Matsuda Y, Nakano T. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development 2004; 131: 839–849. [DOI] [PubMed] [Google Scholar]
- 13.Nishimasu H, Ishizu H, Saito K, Fukuhara S, Kamatani MK, Bonnefond L, Matsumoto N, Nishizawa T, Nakanaga K, Aoki J, Ishitani R, Siomi H, Siomi MC, Nureki O. Structure and function of Zucchini endoribonuclease in piRNA biogenesis. Nature 2012; 491: 284–287. [DOI] [PubMed] [Google Scholar]
- 14.Vourekas A, Zheng K, Fu Q, Maragkakis M, Alexiou P, Ma J, Pillai RS, Mourelatos Z, Wang PJ. The RNA helicase MOV10L1 binds piRNA precursors to initiate piRNA processing. Genes Dev 2015; 29: 617–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ernst C, Odom DT, Kutter C. The emergence of piRNAs against transposon invasion to preserve mammalian genome integrity. Nat Commun 2017; 8: 1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishimura T, Nagamori I, Nakatani T, Izumi N, Tomari Y, Kuramochi-Miyagawa S, Nakano T. PNLDC1, mouse pre-piRNA Trimmer, is required for meiotic and post-meiotic male germ cell development. EMBO Rep 2018; 19: e44957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoshimura T, Watanabe T, Kuramochi-Miyagawa S, Takemoto N, Shiromoto Y, Kudo A, Kanai-Azuma M, Tashiro F, Miyazaki S, Katanaya A, Chuma S, Miyazaki JI. Mouse GTSF1 is an essential factor for secondary piRNA biogenesis. EMBO Rep 2018; 19: e42054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Takamatsu K, Chuma S, Kojima-Kita K, Shiromoto Y, Asada N, Toyoda A, Fujiyama A, Totoki Y, Shibata T, Kimura T, Nakatsuji N, Noce T, Sasaki H, Nakano T. MVH in piRNA processing and gene silencing of retrotransposons. Genes Dev 2010; 24: 887–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kojima-Kita K, Kuramochi-Miyagawa S, Nagamori I, Ogonuki N, Ogura A, Hasuwa H, Akazawa T, Inoue N, Nakano T. MIWI2 as an effector of DNA methylation and gene silencing in embryonic male germ cells. Cell Reports 2016; 16: 2819–2828. [DOI] [PubMed] [Google Scholar]
- 20.Manakov SA, Pezic D, Marinov GK, Pastor WA, Sachidanandam R, Aravin AA. MIWI2 and MILI have differential effects on piRNA biogenesis and DNA methylation. Cell Reports 2015; 12: 1234–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bao J, Zhang Y, Schuster AS, Ortogero N, Nilsson EE, Skinner MK, Yan W. Conditional inactivation of Miwi2 reveals that MIWI2 is only essential for prospermatogonial development in mice. Cell Death Differ 2014; 21: 783–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan W, Rajkovic A, Viveiros MM, Burns KH, Eppig JJ, Matzuk MM. Identification of Gasz, an evolutionarily conserved gene expressed exclusively in germ cells and encoding a protein with four ankyrin repeats, a sterile-α motif, and a basic leucine zipper. Mol Endocrinol 2002; 16: 1168–1184. [DOI] [PubMed] [Google Scholar]
- 23.Ma L, Buchold GM, Greenbaum MP, Roy A, Burns KH, Zhu H, Han DY, Harris RA, Coarfa C, Gunaratne PH, Yan W, Matzuk MM. GASZ is essential for male meiosis and suppression of retrotransposon expression in the male germline. PLoS Genet 2009; 5: e1000635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lim AK, Lorthongpanich C, Chew TG, Tan CWG, Shue YT, Balu S, Gounko N, Kuramochi-Miyagawa S, Matzuk MM, Chuma S, Messerschmidt DM, Solter D, Knowles BB. The nuage mediates retrotransposon silencing in mouse primordial ovarian follicles. Development 2013; 140: 3819–3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hackett JA, Reddington JP, Nestor CE, Dunican DS, Branco MR, Reichmann J, Reik W, Surani MA, Adams IR, Meehan RR. Promoter DNA methylation couples genome-defence mechanisms to epigenetic reprogramming in the mouse germline. Development 2012; 139: 3623–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J, Wang Q, Wang M, Jiang M, Wang Y, Sun Y, Wang J, Xie T, Tang C, Tang N, Song H, Cui D, Chao R, Ding S, Ni B, Chen X, Wang Y. GASZ and mitofusin-mediated mitochondrial functions are crucial for spermatogenesis. EMBO Rep 2016; 17: 220–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang H, Gao Q, Peng X, Choi S-Y, Sarma K, Ren H, Morris AJ, Frohman MA. piRNA-associated germline nuage formation and spermatogenesis require MitoPLD profusogenic mitochondrial-surface lipid signaling. Dev Cell 2011; 20: 376–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peters AHFM, Plug AW, van Vugt MJ, de Boer P. A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res 1997; 5: 66–68. [DOI] [PubMed] [Google Scholar]
- 29.Shimamoto S, Nishimura Y, Nagamatsu G, Hamada N, Kita H, Hikabe O, Hamazaki N, Hayashi K. Hypoxia induces the dormant state in oocytes through expression of Foxo3. Proc Natl Acad Sci USA 2019; 116: 12321–12326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014; 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 2015; 12: 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 2010; 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014; 30: 923–930. [DOI] [PubMed] [Google Scholar]
- 34.Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, Manke T. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 2016; 44(W1): W160-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol 2011; 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sakashita A, Wakai T, Kawabata Y, Nishimura C, Sotomaru Y, Alavattam KG, Namekawa SH, Kono T. XY oocytes of sex-reversed females with a Sry mutation deviate from the normal developmental process beyond the mitotic stage†. Biol Reprod 2019; 100: 697–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 2015; 33: 290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumaki Y, Oda M, Okano M. QUMA: quantification tool for methylation analysis. Nucleic Acids Res 2008; 36: W170-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kawabata Y, Kamio A, Jincho Y, Sakashita A, Takashima T, Kobayashi H, Matsui Y, Kono T. Sex-specific histone modifications in mouse fetal and neonatal germ cells. Epigenomics 2019; 11: 543–561. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS). Genome Biol 2008; 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hothorn T, Bretz F, Westfall P. Simultaneous inference in general parametric models. Biom J 2008; 50: 346–363. [DOI] [PubMed] [Google Scholar]
- 42.Giuffrida V, Pezzino FM, Romano F, Litrico L, Garofalo MR, Nicotra G, Libra M, D’Amico F, Castrogiovanni P, Imbesi R, Averna M, Sanfilippo S, D’Agata R, Vicari E, Calogero AE, Travali S. Gene expression in mouse spermatogenesis during ontogenesis. Int J Mol Med 2006; 17: 523–528. [PubMed] [Google Scholar]
- 43.Hara S, Takano T, Fujikawa T, Yamada M, Wakai T, Kono T, Obata Y. Forced expression of DNA methyltransferases during oocyte growth accelerates the establishment of methylation imprints but not functional genomic imprinting. Hum Mol Genet 2014; 23: 3853–3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schöpp T, Zoch A, Berrens RV, Auchynnikava T, Kabayama Y, Vasiliauskaitė L, Rappsilber J, Allshire RC, O’Carroll D. TEX15 is an essential executor of MIWI2-directed transposon DNA methylation and silencing. Nat Commun 2020; 11: 3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Unhavaithaya Y, Hao Y, Beyret E, Yin H, Kuramochi-Miyagawa S, Nakano T, Lin H. MILI, a PIWI-interacting RNA-binding protein, is required for germ line stem cell self-renewal and appears to positively regulate translation. J Biol Chem 2009; 284: 6507–6519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vasiliauskaitė L, Berrens RV, Ivanova I, Carrieri C, Reik W, Enright AJ, O’Carroll D. Defective germline reprogramming rewires the spermatogonial transcriptome. Nat Struct Mol Biol 2018; 25: 394–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Inoue K, Ichiyanagi K, Fukuda K, Glinka M, Sasaki H. Switching of dominant retrotransposon silencing strategies from posttranscriptional to transcriptional mechanisms during male germ-cell development in mice. PLoS Genet 2017; 13: e1006926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Molaro A, Falciatori I, Hodges E, Aravin AA, Marran K, Rafii S, McCombie WR, Smith AD, Hannon GJ. Two waves of de novo methylation during mouse germ cell development. Genes Dev 2014; 28: 1544–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McLean DJ, Friel PJ, Johnston DS, Griswold MD. Characterization of spermatogonial stem cell maturation and differentiation in neonatal mice. Biol Reprod 2003; 69: 2085–2091. [DOI] [PubMed] [Google Scholar]
- 50.Yoshida S, Sukeno M, Nakagawa T, Ohbo K, Nagamatsu G, Suda T, Nabeshima Y. The first round of mouse spermatogenesis is a distinctive program that lacks the self-renewing spermatogonia stage. Development 2006; 133: 1495–1505. [DOI] [PubMed] [Google Scholar]
- 51.Yamanaka S, Nishihara H, Toh H, Eijy Nagai LA, Hashimoto K, Park S-J, Shibuya A, Suzuki AM, Tanaka Y, Nakai K, Carninci P, Sasaki H, Siomi H. Broad heterochromatic domains open in gonocyte development prior to de novo DNA methylation. Dev Cell 2019; 51: 21–34.e5. [DOI] [PubMed] [Google Scholar]
- 52.Watanabe T, Cui X, Yuan Z, Qi H, Lin H. MIWI2 targets RNAs transcribed from piRNA-dependent regions to drive DNA methylation in mouse prospermatogonia. EMBO J 2018; 37: e95329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nagamori I, Kobayashi H, Nishimura T, Yamagishi R, Katahira J, Kuramochi-Miyagawa S, Kono T, Nakano T. Relationship between PIWIL4-Mediated H3K4me2 Demethylation and piRNA-Dependent DNA Methylation. Cell Reports 2018; 25: 350–356. [DOI] [PubMed] [Google Scholar]
- 54.Liu S, Brind’Amour J, Karimi MM, Shirane K, Bogutz A, Lefebvre L, Sasaki H, Shinkai Y, Lorincz MC. Setdb1 is required for germline development and silencing of H3K9me3-marked endogenous retroviruses in primordial germ cells. Genes Dev 2014; 28: 2041–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sasaki K, Takaoka S, Obata Y. Oocyte-specific gene knockdown by intronic artificial microRNAs driven by Zp3 transcription in mice. J Reprod Dev 2021; 67: 229–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.